Abstract
Biallelic expression of Igf2 is frequently seen in cancers because Igf2 functions as a survival factor. In many tumors the activation of Igf2 expression has been correlated with de novo methylation of the imprinted region. We have compared the intrinsic susceptibilities of the imprinted region of Igf2 and H19, other imprinted genes, bulk genomic DNA, and repetitive retroviral sequences to Dnmt1 overexpression. At low Dnmt1 methyltransferase levels repetitive retroviral elements were methylated and silenced. The nonmethylated imprinted region of Igf2 and H19 was resistant to methylation at low Dnmt1 levels but became fully methylated when Dnmt1 was overexpressed from a bacterial artificial chromosome transgene. Methylation caused the activation of the silent Igf2 allele in wild-type and Dnmt1 knockout cells, leading to biallelic Igf2 expression. In contrast, the imprinted genes Igf2r, Peg3, Snrpn, and Grf1 were completely resistant to de novo methylation, even when Dnmt1 was overexpressed. Therefore, the intrinsic difference between the imprinted region of Igf2 and H19 and of other imprinted genes to postzygotic de novo methylation may be the molecular basis for the frequently observed de novo methylation and upregulation of Igf2 in neoplastic cells and tumors. Injection of Dnmt1-overexpressing embryonic stem cells in diploid or tetraploid blastocysts resulted in lethality of the embryo, which resembled embryonic lethality caused by Dnmt1 deficiency.
The two most common mechanisms to induce tumor growth are the activation of oncogenes and the inactivation of tumor suppressor genes. Both genetic alterations or epigenetic events can result in activation of oncogenes (8, 9) or in the silencing of tumor suppressor genes (17, 40). Imprinted genes that affect cellular growth are particularly vulnerable targets in tumorigenesis because only one allele of an imprinted gene is expressed. Indeed, activation of the silent Igf2 allele leading to biallelic Igf2 expression is a frequent event in neoplasia, which provides the cell with a strong growth signal. Loss of imprinting (LOI) and biallelic Igf2 expression have been described to occur in over 20 different tumor types, including cancers of the liver, breast, pancreas, and colon (16, 27, 30, 35, 40). In addition, it has been shown that experimental overexpression of Igf2 in mice leads to increased cell proliferation, overgrowth, and increased probability of malignant transformation (2, 34, 38).
It has been suggested that deregulation of Igf2 expression is caused by de novo methylation of the differentially methylated domain (DMD), which controls expression of Igf2 and H19 (43, 52). In normal cells the DMD region is methylated on the paternal chromosome and unmethylated on the maternal chromosome, leading to paternal expression of Igf2 and maternal expression of H19. During tumorigenesis, the unmethylated DMD region is frequently methylated, and this has been correlated with biallelic expression of Igf2 and silencing of H19 (27, 40). In contrast, the active allele of imprinted tumor suppressor genes such as Igf2r is inactivated during tumorigenesis through genetic mechanisms and not by epigenetic changes (16). Although the imprinted status of the human Igf2r gene remains unclear, during liver tumorigenesis in rats Igf2r function is always deleted by mutations and loss of heterozygosity, which is in contrast to the LOI of Igf2 and H19 (16). These observations raise the question of whether Igf2 and H19 have an intrinsic susceptibility to de novo methylation that is different from other imprinted genes.
Inheritable DNA methylation patterns are established through a two-step process that converts two unmethylated cytosine residues within palindromic CpG dinucleotides into two methylated CpG dinucleotides with hemi-methylated DNA as an intermediate. During early development the establishment of genomic methylation patterns is accomplished by the concerted action of Dnmt3 and Dnmt1 DNA methyltransferases (MTases). Dnmt3a and Dnmt3b, both expressed in early embryos, have been shown to “de novo methylate” unmodified DNA after implantation of blastocysts and in transgenic flies, resulting in hemi-methylated DNA (26, 31). However, the activity of the hemi-MTase Dnmt1 is crucial to achieving a normal inheritable methylation level of the postgastrulation embryo, because this enzyme acts as a maintenance methylase that recognizes the hemi-methylated CpG sites established by Dnmt3a and -b, methylates the complementary CpG and, thus, converts the respective CpG site to a fully methylated state. Consistent with this notion is the observation that the deficiency of Dnmt1 leads to genomic hypomethylation and embryonic lethality after gastrulation (23).
It has been shown that the deletion of Dnmt1 in mice leads, in addition to genome-wide hypomethylation and embryonic lethality, to loss of monoallelic expression of imprinted genes (23). Dnmt1 mutant mice show biallelic expression of H19 and silencing of the active Igf2 and Igf2r alleles. In addition, deregulation of several other imprinted genes, including p57Kip2 and Kvlqt1, has been observed in Dnmt1 mutant mouse embryos, further demonstrating the involvement of DNA methylation in the maintenance of genomic imprinting (4). In contrast, Mash2 was unaffected by the lack of Dnmt1 MTase, suggesting either that not every imprinted gene is controlled by DNA methylation or that the Dnmt1 mutant embryos died prior to complete demethylation of this locus. Reexpression of the Dnmt1 cDNA in Dnmt1 homozygous mutant (Dnmt1−/−) embryonic stem (ES) cells at low levels did not result in remethylation of parental imprints despite restoration of the overall genomic methylation level (45), implying that methylation imprints can only be imposed on imprinted genes during gametogenesis and that postzygotic cells lack the ability to de novo methylate imprinting boxes.
Increased MTase activity and changes of DNA methylation patterns are commonly seen in neoplastic cells and tumors of humans and mice (3, 6, 19, 49), and this increase in MTase activity has been linked to the deregulation of tumor suppressor genes, oncogenes, and imprinted genes (47). Because de novo methylation and biallelic expression of Igf2 is a frequent alteration in neoplastic cells and tumors, we tested whether the imprinted region of Igf2 and H19 is particularly susceptible to elevated levels of Dnmt1 expression, distinguishing Igf2 and H19 from other imprinted genes. In this study, we generated ES cells with a wide range of Dnmt1 expression levels in wild-type and in Dnmt1−/− ES cells to determine whether the imprinted region of Igf2 and H19 has a different intrinsic sensitivity to postzygotic de novo methylation in comparison to other imprinted genes.
MATERIALS AND METHODS
Dnmt1 BACs.
PCR screening with two different sets of primers of a 129Sv/J mouse bacterial artificial chromosome (BAC) library from the Whitehead Institute Genome Center identified a BAC clone containing the Dnmt1 gene. The BAC clone Dnmt1-BAC has a length of 150 kb as determined by pulsed-field gel electrophoresis. The BAC clone was shown to contain the complete Dnmt1 gene by a combination of PCR and Southern hybridization techniques.
ES cell culture and transfection.
Wild-type ES cells (J1 ES cells) and Dnmt1c/c homozygous mutant ES cells (22) were cultivated on γ-irradiated murine embryonic fibroblasts (mEF) as described previously (24) or without mEF by using a high concentration of leukemia inhibitory factor (LIF; 1,000 U/ml). All transfections were done using the cationic liposome reagent DOTAP from Boehringer Mannheim. To prepare BAC DNA for transfection, bacteria containing the artificial chromosomes were grown on Luria-Bertani medium with 12.5 μg of chloramphenicol/ml. BAC DNA was prepared using standard Qiagen columns. The quality of the BAC DNA was controlled by pulsed-field gel electrophoresis. Uncut BAC DNA was used for lipofection, due to the lack of an unique restriction site for linearization.
DNA preparation and methylation analysis.
Cells were digested in lysis buffer (100 mM Tris-HCl [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl, 100 to 300 μg of proteinase K/ml) for several hours at 55°C, phenol-chloroform extracted, and precipitated with an equal volume of isopropanol. Ten micrograms of DNA was digested with the stated restriction endonuclease for 12 to 16 h. The products were resolved on an agarose gel, transferred to nylon membranes, and hybridized in Church buffer (0.5 M NaPO4 [pH 7.5], 7% SDS, 2 mM EDTA) with a radioactively labeled probe for 8 to 16 h. Radioactively labeled probes were synthesized with random labeling. Final wash was with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS at 65°C. All methylation-sensitive assays were performed two to four times with independently derived genomic DNAs. Methylation analysis of repetitive repeats was performed by comparison of the intensity of individual low-molecular-weight bands to the intensity of all bands. Details for the methylation-sensitive digests were as follows [gene name, enzyme(s), probe (reference)]: Igf2r, PvuII and MluI, region 2 probe, (41); Peg3, KpnI and SacII (25); H19, SacI and HhaI, DMD probe (nucleotides [nt] 1440 to 3332; GenBank accession no. U19619); Grf1, EcoRV and HhaI (EcoRV, NotI) (33); Snrpn, EcoRI and MluI (10); intracisternal type A particle sequence (IAP), HpaII, probe homologous to gag coding region (47a).
Northern blotting.
Poly(A)-RNA was purified using the OligoTex system (Qiagen), separated on formaldehyde-agarose gels, and transferred to nylon membranes (Hybond). Hybridization was carried out at 63°C for 8 to 16 h in Church buffer. All details for the probes are described in reference 42. The IAP probe is homologous to the gag coding region (nt 1570 to 1899; GenBank accession no. M17551). RNA loading was normalized to hybridization with Gapdh.
In vitro differentiation.
In vitro differentiation of ES cells was induced with retinoic acid in the absence of a fibroblast feeder layer and with LIF, as described in references 12 and 45. After 9 days of differentiation, a monolayer of cells was harvested and mRNA transcripts were purified. To ensure complete differentiation of the cell lines, we measured the onset of a differentiation marker (Fgf5) and the transcriptional downregulation of an ES cell marker (Oct3/4). Indeed, we found that Oct3/4 mRNA was undetectable in all differentiated cells. In addition, Fgf5 mRNA levels were increased to similar levels in all differentiated cell lines.
Western blotting.
Protein extracts were harvested from ES cells, which were grown for at least two passages without mEF and with 1,000 U of LIF/ml. ES cells were harvested in 10 volumes of sample buffer (2% SDS, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 0.01% bromophenol blue), boiled, sonicated, and separated on an SDS-7% polyacrylamide gel. All protein extracts were controlled by Coomassie-stained gel to ensure equal loading. Antibodies against the C terminus and N terminus of Dnmt1 are described in references 11 and 46. mEF were derived from tetraploid embryos as described in references 24 and 12. Cells were transformed with large SV40 antigen, harvested in 10 volumes of sample buffer, and analyzed using antibodies against the N terminus of Dnmt1 (46). Protein extracts were controlled by Coomassie-stained gel.
In vitro methylation assay.
ES cells were grown without mEF to avoid any Dnmt1 contamination from the mEF. ES cell nuclear lysates were prepared and the protein concentrations were normalized. Methyltransferase activity of the lysates was measured by their ability to incorporate the methyl group from [3H]S-adenosyl-l-methionine into a synthetic poly(dIdC) substrate or on a double-stranded oligonucleotide (30-mer) containing one unmethylated CpG site (20). The in vitro assays were performed with three independently derived sets of protein extracts and their protein concentration was normalized. All results within a set were normalized to wild-type activity, which was defined as 100%.
Bisulfite modification.
One to 2 μg of DNA was digested for 6 h with 10 U of restriction endonuclease, precipitated in ethanol, and resuspended. The DNA fragments were modified by bisulfite treatment using a final concentration of 2.0 M sodium metabisulfite (Sigma) in 474 μl for 16 h at 50°C (32, 48). The deaminated restriction fragments were desalted using a DNA clean-up column (Promega). Bisulfite-converted DNA was used for PCR amplification using primers directed at the deaminated sequence. After 40 cycles of amplification (94°C denaturation for 1 min, 55°C annealing for 1 min, 72°C extension for 1 min), PCR products were size fractionated on a 1% agarose gel. The PCR products were recovered, cloned into the pGEM T-Easy vector (Promega), and sequenced (ABI Automated Sequencer).
Reverse-phase HPLC.
Thirty micrograms of DNA was incubated with RNase A (250 μg/ml) and RNase T1 (5,000 U/ml) and recovered by ethanol precipitation. The DNA was then digested to completion with DNase and nuclease P1 and filtered with 0.2-μm spin columns. The resulting nucleotides were separated by isocratic reverse-phase high-pressure liquid chromatography (HPLC). The mobile phase was 50 mM ammonium orthophosphate (pH 4.1; flow rate, 1 ml/min) and the solid phase was a 25- by 0.4-cm, 5-μm APEX ODS column (Jones Chromatography Limited, Wales, United Kingdom). The nucleotides were detected by UV absorption (280 nm) and the area under each peak was converted to a molar equivalent by dividing by the respective extinction coefficient of each nucleotide. These have been previously determined as 11.5 × 10−3 and 10.1 × 10−3 at pH 4.3 and 280 nm for dCMP and mdCMP, respectively (39). The percent cytosine methylation was then found by the equation [mdCMP/(mdCMP + dCMP)] × 100.
RNA fluorescence in situ hybridization (RNA FISH).
ES cells were differentiated for 5 days with retinoic acid, trypsinized, and fixed onto poly-l-lysine-coated slides with 4% formaldehyde-5% acetic acid in phosphate-buffered saline (PBS) for 18 min at room temperature (RT). Subsequently, the slides were washed with PBS (three times for 5 min) and stored in 70% ethanol at −20°C. Pretreatment of slides included washing for 5 min with 70% ethanol and 0.1 M Tris, 0.15 M NaCl. Next, slides were subjected to pepsin digestion (4 min at 37°C; 0.01% pepsin in 0.01 M HCl), rinsed in water, and postfixed in 4% formaldehyde-PBS (5 min). Slides were washed with PBS (10 min), dehydrated in 70, 90, and 100% ethanol, and air dried. The hybridization mixture (1 ng of probe/μl, 50% formamide, 2× SSC, 200 ng of salmon sperm DNA/μl, 5× Denhardt solution, 50 mM phosphate buffer, 1 mM EDTA) was applied (12 ml per 24- by 24-mm coverslip), and slides were incubated at 37°C in a moisturized chamber for 12 h. Igf2 and H19 probes were nick translated (Boehringer) with digoxigenin and biotin-conjugated nucleotides, respectively. The Igf2 probe contains an 8.6-kb Igf2 fragment (accession no. U71085; nt 8017 to 16706), and the H19 probe is a 582-bp fragment (accession no. AF049091; nt 7550 to 8132). After hybridization, slides were washed in 2× SSC (four times for 10 min; 37°C) and in 0.1 M Tris, 0.15 M NaCl, 0.05% Tween 20 (twice for 5 min; RT) and then incubated in 2 mg of bovine serum albumin/ml in 0.1 M Tris, 0.15 M NaCl in a humidified chamber (30 min; RT). Detection was with subsequent incubation steps with antidigoxigenin (Boehringer), -sheep (fluorescein isothiocyanate [FITC]-conjugated; Jackson Laboratory), -rabbit (FITC-conjugated; Jackson Laboratory), -biotin (Boehringer), -mouse (rhodamine red-conjugated; Jackson Laboratory), and -horse (rhodamine red-conjugated; Jackson Laboratory) antibodies in 0.1 M Tris, 0.15 M NaCl (30 min; RT). Slides were washed twice in between each detection step with 0.1 M Tris, 0.15 M NaCl, 0.05% Tween 20, mounted with Vectashield (Vector Laboratories), and stored at 4°C. Fluorescence was detected by epifluorescence-charge-coupled device. Between 100 and 300 cells were counted for each cell line. Between 29 and 32% of ES cells in each line were informative and Igf2 expression was detected. Igf2 expression was detected in 15% of the fetal liver cells.
Tetraploid blastocyst injection.
Two-cell embryos for tetraploid electrofusion were produced by superovulation of B6D2F1 females with intraperitoneal injection of 7.5 IU of pregnant mare serum gonadotropin (Calbiochem) and 7.5 IU of human chorionic gonadotropin (HCG; Calbiochem) (46 to 50 h later). After injection of HCG, females were mated with B6D2F1 males and fertilized zygotes were isolated from the oviduct 24 h later. Zygotes were washed in HEPES-buffered Chatot-Ziomek-Bavister (CZB) with 0.1% bovine testicular hyaluronidase (2 to 10 min; RT) to remove remaining cumulus cells. Zygotes were cultured overnight at 37° to obtain two-cell embryos. Forty hours after HCG injection, the blastomeres of two-cell embryos were electrofused to produce one-cell tetraploid embryos. Electrofusion was carried out on an inverted microscope. Platinum wires were used as electrodes. Two-cell embryos were placed on the stage in a 200-μl drop of M2 medium (Sigma). Embryos were aligned with the interface between their two blastomeres perpendicular to the electrical field, and each embryo was fused with a single electrical pulse of 100 V for 100 μs. Embryos that had not undergone membrane fusion within 1 h in CZB medium at 37°C were discarded. ES cells were injected into tetraploid blastocysts with a flat-tip microinjection pipette (internal diameter, 12 to 15 μm) using a Piezo Micromanipulator (Primetech Pmm, Ibaraki, Japan). The injection pipette containing the ES cells was pressed against the zona opposite the inner cell mass. A brief pulse of the Piezo Micromanipulator was applied, and the injection needle was pushed through the zona and trophectoderm layer into the blastocoel cavity. About 10 ES cells were then expelled from the injection pipette and placed against the inner cell mass. After injection, blastocysts were placed in CZB medium at 37°C until transfer to recipient females. Ten injected blastocysts were transferred to each uterine horn of 2.5-days postcoitum (dpc) pseudopregnant Swiss females that had mated with vasectomized males. Recipient mothers were sacrificed at the indicated time points.
Regular blastocyst injection to obtain chimeric mice was performed as described in references 24 and 23. Recipient blastocysts were BALB/c derived, while injected ES cells originated from 129/SvJae mice. Teratomas were generated by the injection of 107 ES cells into the flank of isogenic 129/SvJae mice and excision 3 weeks after injection (11).
RESULTS
Genomic methylation is dependent on the Dnmt1 expression level.
A 150-kb BAC containing the complete Dnmt1 gene (Fig. 1A) was introduced into wild-type and Dnmt1−/− ES cells (22). Wild-type (Dnmt1+/+;BAC) and knockout (Dnmt1−/−;BAC) ES cells with insertion of the Dnmt1 BAC were analyzed for Dnmt1 expression (Fig. 1B). Additionally, we analyzed expression of Dnmt1−/− ES cells with random integration of a Dnmt1 minigene (Dnmt1−/−;cDNA ES cells) (46) and of Dnmt1−/− ES cells with homologous insertion of a partial Dnmt1 cDNA into the cognate Dnmt1 locus (Dnmt1chip/− ES cells) (45).
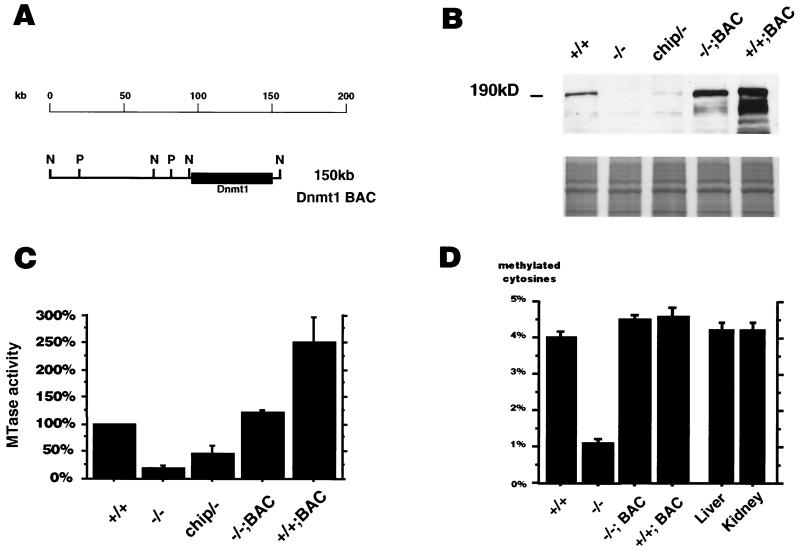
FIG. 1.
(A) Schematic diagram and restriction map of the BAC containing the Dnmt1 gene. Restriction sites are abbreviated as follows: N, NotI; P, PstI. PCR screening of a mouse 129/Sv BAC library identified a BAC clone containing the Dnmt1 gene. The 150-kb BAC clone was shown to contain the complete Dnmt1 gene by a combination of PCR and Southern hybridization techniques (data not shown). (B) Western blot analysis of ES cell extracts using an antibody which was generated against the catalytic domain of Dnmt1. Similar results were obtained with a polyclonal antibody against the N-terminal region of Dnmt1 and are therefore not shown. All protein extracts were controlled by Coomassie-stained gel to ensure equal loading. (C) In vitro MTase activity assay measuring incorporation of the methyl group from [3H]S-adenosyl-l-methionine into a synthetic poly(dIdC) substrate. The in vitro assay was performed with three independently derived sets of protein extracts and their protein concentration was normalized. All results within a set were normalized to wild-type activity, which was defined as 100%. Each value represents the mean ± SEM. (D) 5-Methyl-cytosine content of genomic DNA from different ES cells and from differentiated tissues (liver, kidney), as measured by HPLC. Each value represents the mean ± SEM.
To determine the Dnmt1 level in the different ES cells, protein extracts were analyzed by Western blotting using two different Dnmt1-specific antibodies (11, 46). A Dnmt1-specific band of 190 kDa was detected in Dnmt1+/+ ES cells, while no Dnmt1 band was seen in Dnmt1−/− ES cells (Fig. 1B). Dnmt1chip/− ES cells displayed approximately 10% of wild-type Dnmt1 protein levels. Insertion of the Dnmt1 minigene in Dnmt1−/− ES cells resulted in low Dnmt1 expression in 24 independent clones (data not shown). In contrast, the level of Dnmt1 protein was elevated to approximately 200% of wild-type levels in Dnmt1−/−;BAC ES cells and to approximately 400% in Dnmt1+/+;BAC ES cells.
To confirm that an increase in Dnmt1 expression led to elevation of MTase activity, we performed a standard in vitro methylation assay with synthetic poly(dIdC) as the substrate (Fig. 1C) (20). The results were normalized to wild-type activity, which was defined as 100%. Dnmt1−/− ES cells had residual activity of 20%, which probably reflects Dnmt3a/b activity (31, 37, 51). Dnmt1chip/− ES cells showed an activity level of 47%, while Dnmt1−/−;BAC ES cells had a MTase activity elevated to about 130%. Dnmt1+/+;BAC ES cells further increased MTase activity to 251%. We also measured de novo methylation activity on an unmethylated oligonucleotide (30-mer) and detected no increased MTase activity in Dnmt1+/+;BAC wild-type ES cells when compared to Dnmt1+/+ and Dnmt1−/− ES cells (data not shown).
Next, we determined whether the observed Dnmt1 overexpression caused changes in the genomic DNA methylation level. The 5-methyl-cytosine content of genomic DNA was measured by reverse-phase HPLC. A total of 4.0% of all cytosine residues were methylated in the genome of Dnmt1+/+ ES cells, while the 5-methyl-cytosine content in Dnmt1−/− ES cells was only 1.1% (Fig. 1D). Dnmt1−/−;BAC ES cells displayed an increase in 5-methyl-cytosine content to about 4.5%, while Dnmt1+/+;BAC ES cells contained about 4.6%. The biological significance of the 0.6% increase in 5-methyl-cytosine content in Dnmt1+/+;BAC ES cells was demonstrated by nearest neighbor analysis, which showed that the level of methylation at CpG dinucleotides increased from 62% in Dnmt1+/+ ES cells to 72% in Dnmt1+/+;BAC ES cells (36). Therefore, insertion of the Dnmt1 BAC into ES cells resulted in the generation of two cell lines with elevated Dnmt1 expression, increased MTase activity, and hypermethylated genomic DNA. We note that wild-type ES cells are slightly less methylated than somatic tissues, as observed previously (36).
Methylation assays with methyl-sensitive restriction enzymes were used to compare methylation of specific genomic sequences such as retroviral elements, centromeric repeats, CpG islands from nonimprinted genes, and differentially methylated regions of imprinted genes. The minor satellite centromeric repeat and the IAP (47a), both repetitive sequences, showed a high level of DNA methylation in wild-type ES cells but were severely hypomethylated in Dnmt1−/− ES cells, as indicated by the increased density of the lower-molecular-weight bands (Fig. 2A). DNA methylation of bulk repetitive DNA was almost restored to normal in Dnmt1chip/− ES cells but not in Dnmt1−/−;cDNA ES cells. Dnmt1−/−;BAC and Dnmt1+/+;BAC ES cells de novo methylated both repetitive elements to a level higher than that observed in wild-type ES cells. It has been shown that expression of IAP elements is controlled by methylation (47a). Northern analysis confirmed that the level of IAP expression was inversely correlated to the level of DNA methylation detected at the IAP retroviral elements (Fig. 2B). All three major classes of IAP elements were highly expressed in Dnmt1−/− cells (50-fold induction). IAP expression was reduced in Dnmt1−/−;cDNA cells compared to Dnmt1−/− cells and was only expressed at barely detectable levels in Dnmt1chip/−, Dnmt1−/−;BAC, Dnmt1+/+;BAC, and Dnmt1+/+ ES cells. We conclude that Dnmt1 overexpression in wild-type or Dnmt1−/− ES cells caused hypermethylation of repetitive genomic DNA, including centromeric repeats and retroviral IAP elements, and that IAP expression was highly sensitive to genomic demethylation.
FIG. 2.
(A) Methylation-sensitive restriction enzyme assays with HpaII using two repetitive probes, minor satellite centromeric repeat and an IAP. In these assays, low-molecular-weight bands indicate demethylated restriction sites, while high-molecular-weight bands indicate methylated restriction sites. (B) Northern blot analysis of IAP expression in differentiated cells. RNA loading was controlled for by hybridization with Gapdh.
DNA methylation of imprinted genes.
The methylation status of several imprinted genes, including Igf2r (41), Peg3 (25), Grf1 (33), and Snrpn (21; data not shown), was determined at their differentially methylated regions (Fig. 3). Dnmt1+/+ cells displayed one unmethylated allele and one methylated allele in each of the four imprinted genes, while both alleles were unmethylated in the Dnmt1−/− ES cells (Fig. 3 and data not shown). Reexpression of Dnmt1 by the CHIP construct, cDNA construct, or BAC in Dnmt1−/− ES cells did not result in remethylation of the unmethylated alleles, even when Dnmt1 was expressed at high levels, as observed in Dnmt1−/−;BAC ES cells. Similarly, when high levels of Dnmt1 were expressed in wild-type ES cells (Dnmt1+/+;BAC), no de novo methylation of the unmethylated allele was detected. In addition, we measured the methylation level of CpG islands of several nonimprinted genes, such as p21 (nt 3900 to 4500; accession no. MMU24171), p16 (promoter and exon1), and telomerase (promoter and exon1), by bisulfite sequencing and detected no significant increase in DNA methylation in the Dnmt1+/+;BAC ES cells compared to levels in Dnmt1+/+ ES cells. Therefore, Dnmt1 overexpression in ES cells did not lead to changes in DNA methylation of the imprinted region of Igf2r, Peg3, Snrpn, and Grf1 or of CpG islands of nonimprinted genes.
FIG. 3.
The imprinted region of the genes Igf2r (A), Peg3 (B), and Grf1 (C) are resistant to de novo methylation by overexpression of Dnmt1 in ES cells. For the Igf2r gene, we assayed an MluI site in region 2, which resulted in a maternally methylated band and a paternally unmethylated band in wild-type ES cells (42). For the Peg3 gene, a SacII site was assayed which is normally methylated on the maternal allele (25). A differentially methylated HhaI site was assayed for the Grf1 gene, which is paternally methylated in wild-type cells (34).
A strikingly different result was obtained when the methylation status of several imprinted HhaI sites within the DMD of the imprinted genes Igf2 and H19 was assayed (Fig. 4A). Dnmt1+/+ ES cells contained one fully methylated paternal band and several weaker undermethylated lower bands from the maternal allele (Fig. 4B) (1, 44, 48). Dnmt1−/− ES cells were completely unmethylated and displayed several lower-molecular-weight bands. Minimal remethylation of the imprinted region was observed in Dnmt1chip/− ES cells and Dnmt1−/−;cDNA ES cells, which express Dnmt1 at a low level. In contrast, Dnmt1−/−;BAC ES cells showed significant remethylation of the imprinted H19/Igf2 region. Remarkably, the Dnmt1+/+;BAC ES cells also showed an increase in methylation of the undermethylated maternal allele, as indicated by the disappearance of low-molecular-weight bands. To confirm these results, we measured the DNA methylation status of 10 CpG dinucleotides within the DMD (nt 1089 to 1481; accession no. U19619) by using genomic bisulfite sequencing (data not shown). Previously, this region was shown to be differentially methylated in Dnmt1+/+ ES cells, with one allele completely methylated and the other undermethylated (48). The overall methylation level in Dnmt1+/+ ES cells in this region was between 56 and 78% (48). Dnmt1−/− ES cells displayed a DNA methylation level of 3.3%, which was slightly lower than that in Dnmt1chip/− ES cells (8.8%) (48). In contrast, Dnmt1−/−;BAC ES cells remethylated this region to approximately 70%. Dnmt1+/+;BAC ES cells increased DNA methylation in this region to approximately 88%. Therefore, we conclude that the unmethylated allele(s) of imprinted region of H19 and Igf2 is susceptible to de novo methylation in ES cells, which overexpress Dnmt1. This is in contrast to the imprinted regions of Igf2r, Snrpn, Peg3, Grf1, and Snrpn, which were completely resistant to de novo methylation.
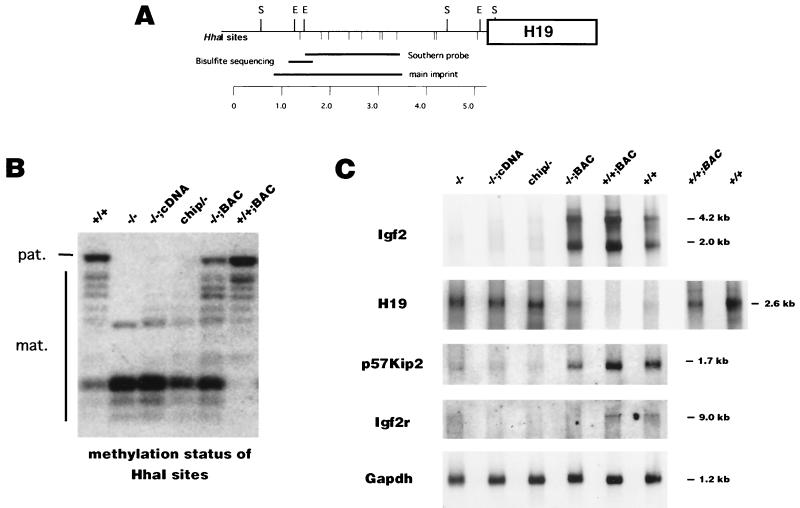
FIG. 4.
(A) Map of the upstream region of H19, which includes the DMD that regulates expression of Igf2 and H19. Location of the main imprinted region and the Southern probe (nt 1440 to 3332; GenBank accession no. U19619) are shown. Restriction sites are abbreviated as follows: E, EcoRI; S, SacI. HhaI sites are indicated beneath the line. The region (nt 1089 to 1481; accession no. U19619) which was assayed by bisulfite modification is also indicated. (B) Methylation analysis of the DMD Igf2/H19 imprinted region, performed using the methylation-sensitive restriction enzyme HhaI and the methylation-insensitive enzyme SacI. ES cells show one paternal methylated band and there are several undermethylated lower bands from the maternal allele. (C) Northern blot analysis of Igf2, H19, p57Kip2, and Igf2r expression in differentiated cells. RNA loading was controlled for by hybridization with Gapdh. The last two lanes on the right show a longer exposure of H19 expression only for the wild-type and Dnmt1+/+;BAC cells.
Expression of imprinted genes.
To determine if de novo methylation of the DMD region resulted in altered expression of H19 and Igf2, we measured Igf2 and H19 expression in the different ES cell lines. Because undifferentiated ES cells do not express H19 and Igf2 (45), expression of both genes was induced upon differentiation in vitro by culturing the cells in the presence of retinoic acid and without LIF and embryonic fibroblasts (12, 45). Differentiation was monitored by the appearance of the differentiation marker Fgf5 and the repression of the ES cell marker Oct3/4 9 days after retinoic acid exposure (data not shown). As expected, several Igf2 RNA transcripts of different sizes were detected in differentiated Dnmt1+/+ ES cells (Fig. 4C) (7). Little or no Igf2 expression was observed in Dnmt1−/− cells, Dnmt1chip/− cells, or Dnmt1−/−;cDNA cells, which is consistent with the DMD region being unmethylated. In contrast, Igf2 was highly expressed in Dnmt1−/−;BAC cells, where the DMD region was de novo methylated on both alleles. Furthermore, Igf2 expression level in Dnmt1+/+;BAC cells was increased to approximately twice the level in wild-type cells (mean ± standard error of the mean [SEM], 1.86 ± 0.084; n = 3), suggesting biallelic expression. Conversely, the H19 gene was expressed at a higher level than in wild-type cells when the DMD region was unmethylated, such as in Dnmt1−/−, Dnmt1chip/−, and Dnmt1−/−;cDNA cells (Fig. 4C). Expression of H19 was significantly downregulated in Dnmt1−/−;BAC cells to a level still higher than in wild-type cells. H19 expression was reduced below the wild-type expression level in Dnmt1+/+;BAC cells, which showed increased methylation at the DMD imprinted box. These results indicate that DMD methylation controls expression of Igf2 and H19. Igf2 was only active when the DMD was methylated, and Igf2 expression was completely silenced when the DMD was unmethylated. H19 expression was inhibited by methylation of DMD but with lesser stringency.
We also investigated the expression level of Igf2r in differentiated ES cells (Fig. 4C). Igf2r was expressed at comparable levels in wild-type and Dnmt1+/+;BAC cells, but it was not detected in Dnmt1−/−, Dnmt1chip/−, Dnmt1−/−;cDNA, and Dnmt1−/−;BAC cells. This is consistent with the lack of remethylation of the Igf2r imprinted region in these cells, which is only expressed when the gene is methylated. Importantly, the unmethylated and inactive copies of Igf2r, carried in Dnmt1−/− and Dnmt1+/+ ES cells, cannot be reactivated through an epigenetic mechanism because the Igf2r imprinted region 2 is resistant to de novo methylation even when the Dnmt1 MTase is overexpressed.
Similar to Igf2 expression, expression of p57Kip2 was also deregulated in differentiated ES cells overexpressing Dnmt1 (Fig. 4C). While no or only residual p57Kip2 expression was detected in Dnmt1−/−, Dnmt1chip/−, or Dnmt1−/−;cDNA cells, reactivation of p57Kip2 was observed in Dnmt1−/−;BAC cells by Northern blotting. p57Kip2 expression was also increased in Dnmt1+/+;BAC cells compared to that in wild-type cells, suggesting that p57Kip2 is activated by DNA methylation. Because of the absence of restriction site polymorphisms in our ES cell lines, we cannot determine if the increased p57Kip2 expression is the result of biallelic expression or upregulation of expression of the active maternal allele. Our results are in contrast with a former report that suggests that the maternal allele of p57Kip2 is activated in Dnmt1−/− embryos as measured by nonquantitative reverse transcription-PCR (RT-PCR) (4). Our Northern analysis suggests that the detected expression of the maternal p57Kip2 allele in dying Dnmt1−/− embryos by nonquantitative RT-PCR is likely the result of increased nonspecific expression at the p57Kip2 locus by activated endogenous retroviral elements. Due to the lack of a defined imprinted region, we were unable to analyze the imprinted p57Kip2 gene in the Dnmt1-overexpressing cells for methylation differences.
Biallelic expression of Igf2.
The increased Igf2 expression in Dnmt1−/−;BAC or Dnmt1+/+;BAC cells could be due either to activation of the inactive maternal Igf2 allele, resulting in biallelic Igf2 expression, or to overexpression of the active paternal Igf2 allele. To distinguish between these two possibilities, we performed Igf2 RNA FISH. As expected, wild-type ES cells as well as control wild-type fetal liver cells showed monoallelic Igf2 expression (Fig. 5A). A small percentage of fetal liver cells (8%) and ES cells (7%) showed biallelic Igf2 expression, which is consistent with previously reported results (18). In contrast, Igf2 expression was biallelic in 68% of the Dnmt1+/+;BAC cells, demonstrating that Dnmt1 overexpression causes activation of the silent maternal allele (Fig. 5A).
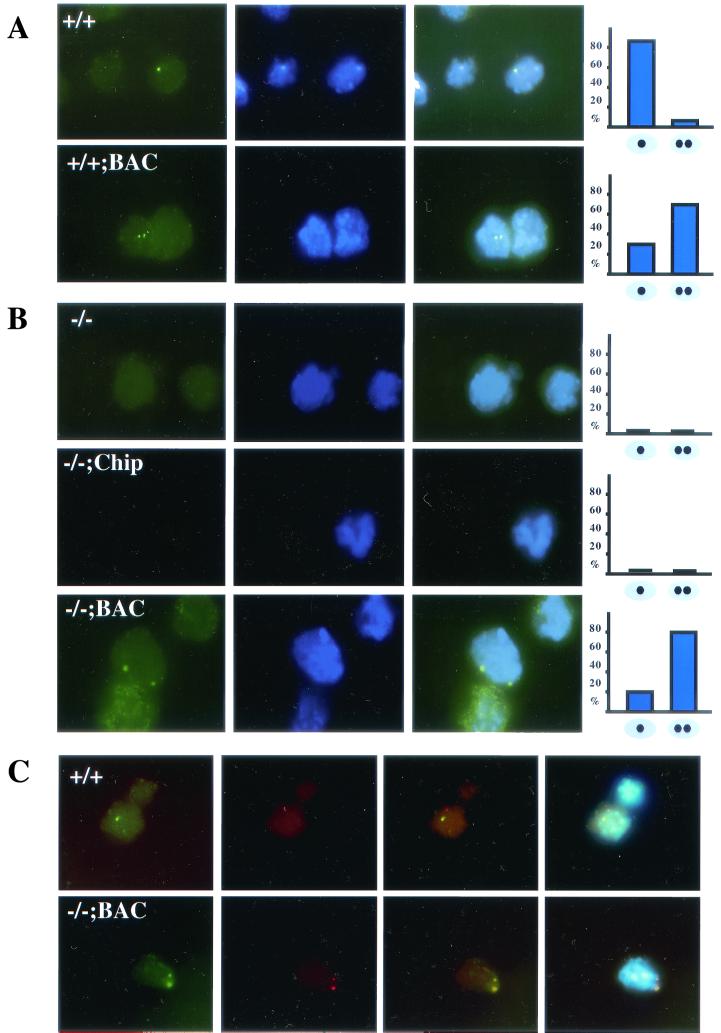
FIG.5.
Igf2 RNA FISH in wild-type (A), Dnmt1+/+;BAC (A), Dnmt1−/− (B), Dnmt1chip/− (B), and Dnmt1−/−;BAC (B) cells. Content of images for each cell line is as follows, from left to right: Igf2 expression (green), nuclear 4′,6′-diamidino-2-phenylindole (DAPI) staining (blue), and merged image of Igf2 and DAPI. The graph to the right indicates the monoallelic versus biallelic Igf2 expression observed in each cell line. (C) Igf2 and H19 RNA FISH of wild-type and Dnmt1−/−;BAC cells. Images, from right to left, are Igf2 expression (green), H19 expression (red), a merged imaged of Igf2 and H19 expression, and a merged image of Igf2 and H19 expression with nuclear DAPI staining.
To independently confirm that Dnmt1 overexpression causes activation of silenced Igf2 alleles, we also analyzed Igf2 expression in Dnmt1−/− cells, Dnmt1chip/− cells, and Dnmt1−/−;BAC cells by RNA FISH. No expression of Igf2 was detected in Dnmt1−/− or Dnmt1chip/− cells, as measured by FISH analysis (Fig. 5B). In contrast, a high level of Dnmt1 expression from the Dnmt1-BAC transgene in Dnmt1−/−;BAC cells caused activation of both silenced Igf2 alleles, as indicated by 81% biallelic expression in these cells (Fig. 5B). When globin expression was analyzed as a control for biallelic expression, 91% of fetal liver cells expressed both alleles (data not shown). We conclude that overexpression of Dnmt1 activates inactive Igf2 alleles.
We also analyzed H19 expression in wild-type and Dnmt1−/−;BAC cells by RNA FISH analysis. While wild-type ES cells showed monoallelic expression of the paternal Igf2 allele and of the maternal H19 allele (Fig. 5C), Dnmt1−/−;BAC cells demonstrated biallelic expression of both Igf2 and H19 (Fig. 5C). This is consistent with the high level of H19 expression in Dnmt1−/−;BAC cells as measured by Northern analysis, indicating that partial methylation might not be sufficient for silencing of the H19 gene.
Overexpression of Dnmt1 causes embryonic lethality.
To determine whether overexpression of the Dnmt1 MTase would impair the developmental potential of totipotent ES cells, we injected Dnmt1+/+;BAC and Dnmt1−/−;BAC ES cells into wild-type blastocysts. Injection of each of the 129Sv/Jae-derived Dnmt1+/+;BAC and Dnmt1−/−;BAC ES cells into over 300 BALB/c blastocysts resulted in few newborns but no chimeric pups (Fig. 6A). In contrast, injection of Dnmt1chip/− ES cells into wild-type blastocysts generated chimeric mice, with the germ line contribution of the injected cells indicating that the rescued Dnmt1−/− ES cells can regain developmental totipotency (45).
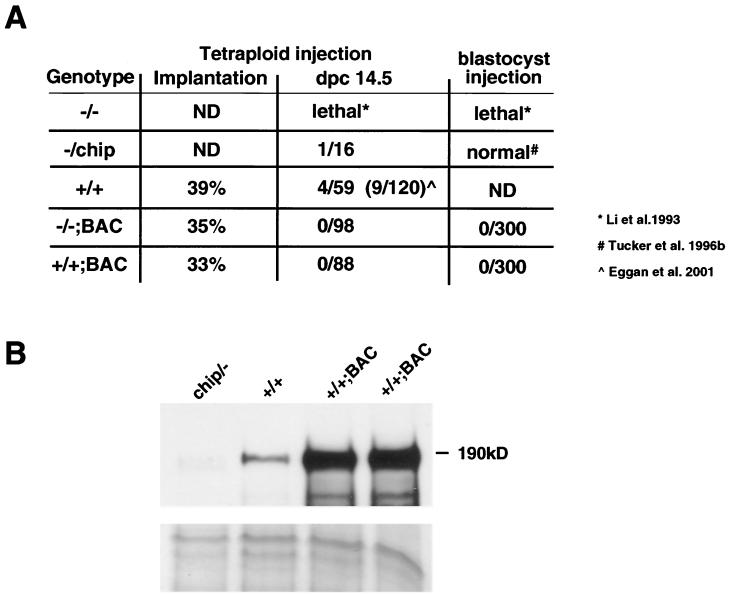
FIG. 6.
(A) Summary of the development (implantation) and survival of ES cell tetraploid blastocyst-derived mice or ES cell blastocyst-derived mice. Implantation is indicated in percent, while survial at dpc 14.5 is indicated in total numbers. ND, not determined. (B) Western blot analysis of mEF cell extracts using an antibody against the N-terminal domain of Dnmt1. All protein extracts were controlled by Coomassie-stained gel to ensure equal loading.
To further investigate the developmental potency of these ES cells, we injected Dnmt1+/+;BAC cells and Dnmt1−/−;BAC cells into tetraploid host blastocysts (Fig. 6A). Because tetraploid blastocysts, generated by electrofusion of the blastomeres in a two-cell embryo, cannot contribute to embryonic lineages, the composite embryos give rise to mice that are entirely derived from the descendants of ES cells injected into the blastocyst (28, 29). Wild-type control cells (Dnmt1+/+) and targeted control cells (Dnmt1chip/−) implanted efficiently into the uterus of pseudopregnant females and showed normal development at dpc 14.5, similar to previously reported results (5). Although Dnmt1+/+;BAC cells and Dnmt1−/−;BAC ES cells injected into tetraploid blastocysts showed a similar percentage of implantation, 33% (29 of 88) and 35% (34 of 98), respectively, embryonic development of these ES cells was severely impaired after implantation (Fig. 6A). None of the embryos derived from Dnmt1+/+;BAC cells survived past dpc 14.5 (0 of 88), with three malformed and developmentally delayed embryos recovered between dpc 10.5 and 12.5. Similarly, embryos derived from Dnmt1−/−;BAC ES cells were unable to develop beyond dpc 12.5 (0 of 98), with one malformed and developmentally delayed embryo recovered at dpc 10.5. Even though Dnmt1-overexpressing ES cells exhibit developmental deficiencies, Dnmt1−/−;BAC ES cells (7 of 8) and Dnmt1+/+;BAC ES cells (8 of 8) were able to form teratomas at a similar rate as wild-type ES cells (8 of 8) when injected into the flank of 129Sv/Jae mice. In contrast, Dnmt1−/− ES cells were unable to form teratomas (0 of 8).
Protein extracts of Dnmt1+/+;BAC, Dnmt1chip/−, and wild-type mEF, which were cultured from the recovered tetraploid embryos, were analyzed by Western blotting to determine the Dnmt1 expression level (Fig. 6B). All analyzed fibroblast cell lines were transformed with SV40 large T antigen, as we were unable to long-term culture the nontransformed Dnmt1+/+;BAC mEF. Dnmt1+/+;BAC fibroblasts from two independent embryos displayed significant overexpression when compared to the Dnmt1 protein level of wild-type fibroblasts. Dnmt1chip/− fibroblasts displayed approximately 10% of wild-type Dnmt1 protein levels. Thus, Dnmt1 expression levels in differentiated fibroblasts recapitulate the expression profile of the corresponding ES cells. We conclude that Dnmt1 overexpression in Dnmt1+/+;BAC and Dnmt1−/−;BAC ES cells results in embryonic lethality resembling the embryonic fatality observed in Dnmt1-deficient ES cells.
DISCUSSION
The establishment of genomic methylation patterns requires the concerted activity of two enzymes, the de novo MTase Dnmt3 and the hemi-MTase Dnmt1 (26, 31). Our study defines three classes of sequences which are subject to gain of methylation at different levels of Dnmt1 expression, as summarized in Fig. 7. (i) Retroviral elements like IAP and other repetitive sequences such as centromeric repeats are highly susceptible to gain of methylation even under conditions of low Dnmt1 expression (Dnmt1chip/− cells). (ii) Unmethylated alleles of most imprinted genes, including Igf2r, Peg3, Snrpn, and Grf1 and the CpG islands of several nonimprinted genes, are completely resistant to de novo methylation. (iii) The DMD imprinted region, which has been shown to control expression of Igf2 and H19, is resistant to de novo methylation at low Dnmt1 expression levels but becomes fully methylated at higher levels of Dnmt1 expression.
FIG. 7.
Summary of methylation and expression status of repetitive sequences such as IAP and of the imprinted genes Igf2r and Igf2 in cells with different levels of Dnmt1 expression, as measured by Western blot analysis (shown in second column). Three classes of genes (IAP, Igf2r, and Igf2) are subject to postzygotic de novo methylation at different levels of Dnmt1 expression, as illustrated. Repetitive IAP sequences are highly susceptible to de novo methylation. Igf2r and other imprinted genes, including Peg3, Snrpn, and Grf1 and the CpG islands of several nonimprinted genes, are completely resistant to de novo methylation. The imprinted region of Igf2 and H19 was resistant to de novo methylation at low Dnmt1 levels but became fully methylated at higher levels of Dnmt1 expression. The maternal and paternal alleles of Igf2 and Igf2r are indicated. The expression levels of IAP, Igf2r, and Igf2 are indicated by the number of + signs, while a − sign indicates no expression. Monoallelic or biallelic Igf2 expression in differentiated cells is indicated as the percentage of total Igf2-expressing cells. Embryonic survival of the ES cell tetraploid blastocyst-derived mice is summarized in the last column. Symbols: □, unmethylated; ▧, partially methylated (low); ▨, partially methylated (high); ▪, methylated; oval with one dot, monoallelic Igf2 expression; oval with two dots, biallelic Igf2 expression.
Several lines of evidence indicate that the Dnmt1 MTase catalyzes methylation of hemi-methylated DNA and therefore is likely not responsible per se for the initiation of de novo methylation of unmethylated sequences (26, 31). However, ES cells and early embryos express Dnmt3a/b MTases (37, 51) which are able to de novo methylate unmethylated DNA in vitro (30a) and in vivo (31). Because the methylation activity of the Dnmt3a/b MTases is low, the final level of genomic DNA methylation in ES cells depends on the Dnmt1 expression level in the cells. Our results are consistent with the notion that the establishment of a normal methylation pattern depends on cooperation between Dnmt3 and Dnmt1 (26, 31). Dnmt1 “fixes” the Dnmt3-initiated de novo methylation of CpG dinucleotides, which results in an increase of methylation to reach a final level of DNA methylation (36).
Methylation imprints are imposed upon imprinted genes during gametogenesis when the two alleles are in different compartments. The regions of parent-specific methylation, denoted as the “imprinting box,” are resistant to the wave of global demethylation, which occurs during cleavage and removes methylation from nonimprinted genes. Equally important, the nonmethylated imprinted allele cannot be postzygotically de novo methylated and is resistant to the wave of global de novo methylation that methylates all nonimprinted genes after implantation (15). Thus, the resistance to demethylation and the resistance to de novo methylation of the respective imprinted alleles assure that the parent-specific methylation mark imposed during gametogenesis remains unaltered and distinguishes the two alleles of imprinted genes in the adult. A previous study using Dnmt1chip/− ES cells supported the idea that ES cells and postzygotic embryos lack the enzymatic machinery to de novo methylate unmethylated imprinting regions, including the imprinted region of Igf2 and H19 (45). However, more extensive analysis of Dnmt1chip/− cells by Western blotting and methylation activity assays showed that the Dnmt1chip allele is a hypomorph with only low levels of Dnmt1 expression that are insufficient to achieve de novo methylation of the DMD imprinted region of Igf2 and H19. In contrast, overexpression of Dnmt1 as obtained by the integration of the Dnmt1 BAC in ES cells resulted in increased methylation levels at the imprinting box of Igf2 and H19 and biallelic Igf2 expression. Our results indicate that the unmethylated Igf2/H19 allele in wild-type or Dnmt1−/− cells is subject to postzygotic methylation under conditions of elevated Dnmt1 expression. We also detected that p57Kip2 is deregulated in Dnmt1-overexpressing cells. But due to the lack of a defined imprinted region of p57Kip2 and a complex gene structure, we were unable to detect the primary imprint with differential DNA methylation. However, analysis of the methylation status of the Lit1 CpG island might be informative, since a targeted deletion of the methylated maternal CpG island on a human chromosome results in the upregulation of p57Kip2 (13). In contrast to the imprinted genes Igf2 and p57Kip2, other imprinted genes such as Igf2r, Peg3, Snrpn, and Grf1 were resistant to de novo methylation even under conditions of elevated Dnmt1 expression. Igf2r was not expressed once the Igf2r imprinting box was demethylated, which is consistent with the model that methylation of the imprint region 2 of Igf2r is required for its transcription (50).
Genomic hypomethylation caused by the deletion of Dnmt1 MTase, though with no obvious effect on in vitro growth of undifferentiated ES cells, results in abnormal development and embryonic lethality (23). Similarly, overexpression of Dnmt1 and genomic hypermethylation had no obvious effect on ES cell proliferation. However, injection of the cells into blastocysts resulted in embryonic lethality of the chimeric embryos. Our results are consistent with the notion that DNA methylation has no obvious role in the survival of embryonic cells but is crucial for normal physiology of somatic cells (15). It has been shown that conditional deletion of Dnmt1 in fibroblasts leads to genomic demethylation and widespread ectopic gene activation, which may explain the cell death of Dnmt1 mutant cells (14). The mechanism of lethality induced by overexpression of Dnmt1, however, is unclear. It has been reported that Dnmt1 can be overexpressed in rare clones of tumor cells (47), suggesting that genomic hypermethylation is compatible with survival of transformed cells in contrast to that of primary cells. Our inability to long-term culture untransformed Dnmt1-overexpressing fibroblasts is consistent with that observation.
The susceptibility of Igf2 and H19 to postzygotic de novo methylation is of potential relevance for tumorigenesis because deregulation of Igf2 imprinting has been shown to occur in over 20 different tumor types, including Wilms' tumor (16, 27, 30, 35, 40). Igf2 is a potent cell survival factor that stimulates cell proliferation, and its overexpression leads to conditions that are favorable to increased cell proliferation and overgrowth. The oncogenic function of Igf2 has been confirmed by the experimental overexpression of Igf2 in mice leading to an increased probability of tumor development (2, 34, 38). The susceptibility of the imprinted region of Igf2 and H19 to postzygotic de novo methylation may be the basis for the high frequency of biallelic Igf2 expression observed in many cancers (27, 40). This appears to distinguish the Igf2 gene from other imprinted genes, such as Igf2r, Grf1, Snrpn, and Peg3, that were not susceptible to postzygotic de novo methylation. Consistent with this notion is the observation that tumor-specific alterations of these imprinted genes involve genetic mutations rather than epigenetic changes (16).
The results described in this paper suggest that Dnmt1 activity may be crucial for the final level of methylation of bulk genomic DNA and repetitive genes and for the imprinted region of Igf2 and H19. Our results imply that even a moderate increase in Dnmt1 expression in cells with low intrinsic de novo activity from Dnmt3 may be sufficient to shift the balance towards de novo methylation and activation of the silenced Igf2 allele. Thus, overexpression of Dnmt1 and activation of Dnmt3 as observed in many transformed cells may be of selective advantage for the incipient tumor cell (37, 51).
Acknowledgments
We thank H. J. Gross, Kerry Tucker, and W. M. Rideout III for stimulating discussions, Ruth Curry, Jessie Dausman, and Jan Loring for expert technical assistance, and C. Plass for providing the Grf1 probe.
This work was supported by NIH grant NIH/NCI 5-R35-CA44339 and in part by the ERC Program of the National Science Foundation under award EEC-9843342. D.B. was supported by the Deutsche Akademische Austauschdienst and the Fritz-Thyssen Stiftung. B.R. is supported by a Medical Research Council (United Kingdom) Clinician Scientist Fellowship.
REFERENCES
- 1.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. [DOI] [PubMed] [Google Scholar]
- 2.Bates, P., R. Fisher, A. Ward, L. Richardson, D. J. Hill, and C. F. Graham. 1995. Mammary cancer in transgenic mice expressing insulin-like growth factor II (IGF-II). Br. J. Cancer 72:1189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belinsky, S. A., K. J. Nikula, S. B. Baylin, and J. P. Issa. 1996. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc. Natl. Acad. Sci. USA 93:4045-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caspary, T., M. A. Cleary, C. C. Baker, X. J. Guan, and S. M. Tilghman. 1998. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 18:3466-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggan, K., H. Akutsu, J. Loring, L. Jackson-Grusby, M. Klemm, W. M. Rideout, R. Yanagimachi, and R. Jaenisch. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.el-Deiry, W. S., B. D. Nelkin, P. Celano, R. W. Yen, J. P. Falco, S. R. Hamilton, and S. B. Baylin. 1991. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc. Natl. Acad. Sci. USA 88:3470-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, R., T. Moore, J. Oswald, J. Walter, F. Sun, and W. Reik. 1997. The imprinted insulin-like growth factor 2 gene, p. 70-97. In W. Reik and A. Surani (ed.), Genomic imprinting. IRL Press, Oxford, England.
- 8.Feinberg, A. P., and B. Vogelstein. 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301:89-92. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg, A. P., and B. Vogelstein. 1983. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 111:47-54. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel, J. M., T. A. Gray, L. Stubbs, S. Saitoh, T. Ohta, and R. D. Nicholls. 1998. Structure and function correlations at the imprinted mouse Snrpn locus. Mamm. Genome 9:788-793. [DOI] [PubMed] [Google Scholar]
- 11.Gaudet, F., D. Talbot, H. Leonhardt, and R. Jaenisch. 1998. A short DNA methyltransferase isoform restores methylation in vivo. J. Biol. Chem. 273:32725-32729. [DOI] [PubMed] [Google Scholar]
- 12.Hogan, B., R. Beddington, F. Costantini, and F. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Horike, S., K. Mitsuya, M. Meguro, N. Kotobuki, A. Kashiwagi, T. Notsu, T. C. Schulz, Y. Shirayoshi, and M. Oshimura. 2000. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 9:2075-2083. [DOI] [PubMed] [Google Scholar]
- 14.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 15.Jaenisch, R. 1997. DNA methylation and imprinting: why bother? Trends Genet. 13:323-329. [DOI] [PubMed] [Google Scholar]
- 16.Jirtle, R. L. 1999. Genomic imprinting and cancer. Exp. Cell Res. 248:18-24. [DOI] [PubMed] [Google Scholar]
- 17.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163-167. [DOI] [PubMed] [Google Scholar]
- 18.Jouvenot, Y., F. Poirier, J. Jami, and A. Paldi. 1999. Biallelic transcription of Igf2 and H19 in individual cells suggests a post-transcriptional contribution to genomic imprinting. Curr. Biol. 9:1199-1202. [DOI] [PubMed] [Google Scholar]
- 19.Kautiainen, T. L., and P. A. Jones. 1986. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J. Biol. Chem. 261:1594-1598. [PubMed] [Google Scholar]
- 20.Kumar, S., X. Cheng, J. W. Pflugrath, and R. J. Roberts. 1992. Purification, crystallization, and preliminary X-ray diffraction analysis of an M.HhaI-AdoMet complex. Biochemistry 31:8648-8653. [DOI] [PubMed] [Google Scholar]
- 21.Leff, S. E., C. I. Brannan, M. L. Reed, T. Ozcelik, U. Francke, N. G. Copeland, and N. A. Jenkins. 1992. Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader-Willi syndrome region. Nat. Genet. 2:259-264. [DOI] [PubMed] [Google Scholar]
- 22.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 23.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 24.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 25.Li, L. L., I. Y. Szeto, B. M. Cattanach, F. Ishino, and M. A. Surani. 2000. Organization and parent-of-origin-specific methylation of imprinted peg3 gene on mouse proximal chromosome 7. Genomics 63:333-340. [DOI] [PubMed] [Google Scholar]
- 26.Lyko, F., B. H. Ramsahoye, H. Kashevsky, M. Tudor, M. A. Mastrangelo, T. L. Orr-Weaver, and R. Jaenisch. 1999. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 27.Moulton, T., W. Y. Chung, L. Yuan, T. Hensle, P. Waber, P. Nisen, and B. Tycko. 1996. Genomic imprinting and Wilms' tumor. Med. Pediatr. Oncol. 27:476-483. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, A., E. Gocza, E. M. Diaz, V. R. Prideaux, E. Ivanyi, M. Markkula, and J. Rossant. 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110:815-821. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, O., M. R. Eccles, J. Szeto, L. A. McNoe, K. Yun, M. A. Maw, P. J. Smith, and A. E. Reeve. 1993. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature 362:749-751. [DOI] [PubMed] [Google Scholar]
- 30a.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed]
- 31.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 32.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plass, C., H. Shibata, I. Kalcheva, L. Mullins, N. Kotelevtseva, J. Mullins, R. Kato, H. Sasaki, S. Hirotsune, Y. Okazaki, W. A. Held, Y. Hayashizaki, and V. M. Chapman. 1996. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 14:106-109. [DOI] [PubMed] [Google Scholar]
- 34.Pravtcheva, D. D., and T. L. Wise. 1998. Metastasizing mammary carcinomas in H19 enhancers-Igf2 transgenic mice. J. Exp. Zool. 281:43-57. [PubMed] [Google Scholar]
- 35.Rainier, S., L. A. Johnson, C. J. Dobry, A. J. Ping, P. E. Grundy, and A. P. Feinberg. 1993. Relaxation of imprinted genes in human cancer. Nature 362:747-749. [DOI] [PubMed] [Google Scholar]
- 36.Ramsahoye, B. H., D. Biniszkiewicz, F. Lyko, V. Clark, A. P. Bird, and R. Jaenisch. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 97:5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27:2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogler, C. E., D. Yang, L. Rossetti, J. Donohoe, E. Alt, C. J. Chang, R. Rosenfeld, K. Neely, and R. Hintz. 1994. Altered body composition and increased frequency of diverse malignancies in insulin-like growth factor-II transgenic mice. J. Biol. Chem. 269:13779-13784. [PubMed] [Google Scholar]
- 39.Sinsheimer, R. 1954. The action of pancreatic deoxyribonuclease. I. Isolation of mono- and dinucleotides. J. Biol. Chem. 208:445-459. [PubMed] [Google Scholar]
- 40.Steenman, M. J., S. Rainier, C. J. Dobry, P. Grundy, I. L. Horon, and A. P. Feinberg. 1994. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumour. Nat. Genet. 7:433-439. [DOI] [PubMed] [Google Scholar]
- 41.Stoger, R., P. Kubicka, C. G. Liu, T. Kafri, A. Razin, H. Cedar, and D. P. Barlow. 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73:61-71. [DOI] [PubMed] [Google Scholar]
- 42.Szabo, P., and J. R. Mann. 1994. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120:1651-1660. [DOI] [PubMed] [Google Scholar]
- 43.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008-1020. [DOI] [PubMed] [Google Scholar]
- 46.Tucker, K. L., D. Talbot, M. A. Lee, H. Leonhardt, and R. Jaenisch. 1996. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc. Natl. Acad. Sci. USA 93:12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vertino, P. M., R. W. Yen, J. Gao, and S. B. Baylin. 1996. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 16:4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed]
- 48.Warnecke, P. M., D. Biniszkiewicz, R. Jaenisch, M. Frommer, and S. J. Clark. 1998. Sequence-specific methylation of the mouse H19 gene in embryonic cells deficient in the Dnmt-1 gene. Dev. Genet. 22:111-121. [DOI] [PubMed] [Google Scholar]
- 49.Wu, J., J. P. Issa, J. Herman, D. E. Bassett, Jr., B. D. Nelkin, and S. B. Baylin. 1993. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA 90:8891-8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wutz, A., O. W. Smrzka, N. Schweifer, K. Schellander, E. F. Wagner, and D. P. Barlow. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745-749. [DOI] [PubMed] [Google Scholar]
- 51.Xie, S., Z. Wang, M. Okano, M. Nogami, Y. Li, W. W. He, K. Okumura, and E. Li. 1999. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 236:87-95. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., T. Shields, T. Crenshaw, Y. Hao, T. Moulton, and B. Tycko. 1993. Imprinting of human H19: allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am. J. Hum. Genet. 53:113-124. [PMC free article] [PubMed] [Google Scholar]