Abstract
p107 functions to control cell division and development through interaction with members of the E2F family of transcription factors. p107 is phosphorylated in a cell cycle-regulated manner, and its phosphorylation leads to its release from E2F. Although it is known that p107 physically associates with E- and A-type cyclin/cyclin-dependent kinase 2 (Cdk2) complexes through a cyclin-binding RXL motif located in the spacer domain, the mechanisms underlying p107 inactivation via phosphorylation remain poorly defined. Recent genetic evidence indicates a requirement for cyclin D1/Cdk4 complexes in p107 inactivation. In this work, we provide direct biochemical evidence for the involvement of cyclin D1/Cdk4 in the inactivation of p107's growth-suppressive function. While coexpression of cyclin D1/Cdk4 can reverse the cell cycle arrest properties of p107 in Saos-2 cells, we find that p107 in which the Lys-Arg-Arg-Leu sequence of the RXL motif is replaced by four alanine residues is largely refractory to inactivation by cyclin D/Cdk4, indicating a role for this motif in p107 inactivation without a requirement for its tight interaction with cyclin D1/Cdk4. We identified four phosphorylation sites in p107 (Thr-369, Ser-640, Ser-964, and Ser-975) that are efficiently phosphorylated by Cdk4 but not by Cdk2 in vitro and are also phosphorylated in tissue culture cells. Growth suppression by p107 containing nonphosphorylatable residues in these four sites is not reversed by coexpression of cyclin D1/Cdk4. In model p107 spacer region peptides, phosphorylation of S640 by cyclin D1/Cdk4 is strictly dependent upon an intact RXL motif, but phosphorylation of this site in the absence of an RXL motif can be partially restored by replacement of S643 by arginine. This suggests that one role for the RXL motif is to facilitate phosphorylation of nonconsensus Cdk substrates. Taken together, these data indicate that p107 is inactivated by cyclin D1/Cdk4 via direct phosphorylation and that the RXL motif of p107 plays a role in its inactivation by Cdk4 in the absence of stable binding.
Progression through the G1/S transition is regulated by D- and E-type cyclins. These proteins function as essential activators of two distinct classes of cyclin dependent kinases: cyclin-dependent kinase 4 (Cdk4)/6 and Cdk2, respectively (48, 56). D-type cyclins link mitogenic signaling pathways with the basic cell division apparatus and are thought to promote progression through G1 by both catalytic and noncatalytic mechanisms (57). Cyclin D1 expression is induced during G1 in response to growth factors, and this induction is known to require the Ras pathway (4, 35, 57). Once synthesized, cyclin D1 assembles with Cdk4 in a manner that requires members of the p21Cip1 class of CDK inhibitors (8, 14, 33, 50). Activated cyclin D1/Cdk4 promotes the G1/S transition via phosphorylation of the retinoblastoma (Rb) protein (6, 19, 23, 56). Two major lines of evidence support a role for D-type cyclins in Rb inactivation. First, overexpression of the Cdk4/6 inhibitor p16 leads to G1 arrest in Rb-positive but not in Rb-negative cells, indicating that Cdk4/6 activity is indispensable in cells containing Rb (40, 45). Second, cyclin D1 can phosphorylate Rb on particular sites, which when removed by mutation to nonphosphorylatable residues, block the ability of cyclin D1/Cdk4 to promote inactivation of Rb (15). Other kinases, including cyclin E/Cdk2 and Cdc2, also contribute to Rb inactivation by direct phosphorylation (22, 37).
Rb functions, in part, to regulate E2F transcriptional activity. E2F transcription factors are composed of a DNA binding DP subunit and one of five E2F family members. Association of Rb with E2F1 to E2F4 leads to inhibition of E2F transcriptional activity either by blocking the E2F transactivation domain or by active repression via recruitment of histone deacetylase (19, 23). Phosphorylation of Rb by Cdks is thought to result in the release of Rb and its associated chromatin remodeling enzymes from E2F, thereby facilitating transcriptional activation of numerous target genes (49).
p107 and p130 are members of the Rb family of proteins and contain two domains, the A and B pockets, which are responsible for binding to transforming oncogenes such as E1A. Like Rb, p107 and p130 block transcription of E2F-regulated genes but function exclusively through interactions with E2F4 and E2F5 (19, 39). Analysis of mouse embryo fibroblasts lacking either p107/p130 or Rb indicate that p107/p130 and Rb regulate the expression of different sets of E2F-responsive genes (28). Until recently, it was thought that the major, if not exclusive substrate of cyclin D1/Cdk4 was Rb (40, 45). However, the finding that mouse embryo fibroblasts lacking p107/p130 or E2F4/E2F5 are insensitive to growth arrest by p16 expression challenges this view (11, 20). These observations indicate that Rb family members function combinatorially to control cell cycle transitions (16, 53) and that cyclin D1/Cdk4 may participate in the functional inactivation of all three family members. In fact, earlier biochemical studies had placed cyclin D1/Cdk4 upstream of p107 inactivation: p107 becomes hyperphosphorylated as cells traverse the G1/S-phase transition and ectopic expression of cyclin D1/Cdk4 induces p107 hyperphosphorylation, as well as inactivation of its growth-suppressive and E2F-binding activities (7, 63). However, these experiments did not address whether the effects of cyclin D/Cdk4 on p107 were direct or reflect CKI sequestration and Cdk2 activation.
Other lines of evidence suggest a role for cyclin A and E/Cdk2 in the regulation of p107. In contrast to D-type cyclins, cyclins E and A/Cdk2 form stable complexes with p107 during late-G1 and S phases (36). Both cyclins interact with p107 and the related p130 protein, at least in part, through a conserved RXL-type cyclin/Cdk2 binding motif within the spacer region (2, 12, 21, 62, 67, 68). This cyclin-binding motif is present in a number of cyclin/Cdk2 binding proteins, including substrates, such as Rb, E2F-1, Skp2, Cdh1, human papillomavirus (HPV) replication protein E1, p220NPAT, and Cdc6, and inhibitor proteins, such as p21Cip1, p27Kip1, and p57Kip2 (2, 3, 13, 17, 18, 21, 34, 42, 43, 51, 52, 54, 60, 61). The RXL motif binds to a conserved “hydrophobic patch” on cyclins A and E (10, 52, 55). This RXL-dependent interaction of substrates with cyclins A and E is thought to facilitate their phosphorylation by the Cdk2 subunit (1). However, the functional significance of the interaction between p107 and cyclin/Cdk2 complexes is unclear since p107 growth suppression activity does not appear to be regulated by these kinases in vivo (7).
Although several key residues in the “hydrophobic patch” to which the RXL motif binds are conserved in the D-type cyclins, it is not clear whether D-type cyclins depend upon RXL motifs for the recognition of p107 or any other substrate. Dutta and coworkers (61) showed that an RXL-containing peptide derived from p21Cip1 competitively inhibits cyclin/Cdk2 kinase activity but does not affect cyclin D1/Cdk4 kinase activity. On the other hand, when the E2F1-derived RXL was fused to the C terminus of an otherwise nonphosphorylatable mutant of Rb that contains phosphoacceptor sites but lacks RXL sequences, in vitro phosphorylation by cyclin D/Cdk4 was restored (3).
In this study we provide the first biochemical evidence that cyclin D/Cdk4 directly phosphorylates and regulates p107 in vivo. Moreover, we show that, while p107 and cyclin D1/Cdk4 do not form a stable complex, the ability of cyclin D1/Cdk4 to overcome p107-mediated growth arrest is enhanced by the presence of an intact RXL motif, presumably for a transient enzyme:substrate interaction. In vitro, this interaction allows more efficient phosphorylation of four sites in p107 whose modification is required to inactivate p107's growth-suppressive activity in tissue culture cells.
MATERIALS AND METHODS
Cell culture and transfection.
C33A, U2OS, and Saos-2 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. The cells were maintained in a humidified 37°C incubator with 5% CO2. Sf9 and Hi5 insect cells were grown in Graces media at 27°C. Transfection was performed by using FuGENE 6 (Roche). Briefly, the indicated quantities of p107, cyclin, and Cdk expression plasmids were added to 10 μl of FuGENE 6 previously diluted into 100 μl of Opti-MEM1 (Gibco). This mixture was incubated for 30 min and then dispersed into a 60-mm dish containing the indicated cells at 50 to 60% confluence. For immunostaining, the cells were seeded on sterilized glass coverslips.
Plasmids.
pcDNA3-p107HA was used as a template for site-directed mutagenesis to generate pcDNA-p107A4HA in which K657-L660 were replaced by alanine. To construct baculoviruses, BamHI/XbaI fragments containing p107HA or p107A4HA were inserted into pBlueBac4.5 (Invitrogen) prior to cotransfection with Bac-N-Blue DNA into Sf9 cells. To generate EYFP-p107 fusion constructs, SacII/XbaI fragments containing p107HA or p107A4HA were cloned into pEYFP-C1 (Clontech). pGEX-2TK/p107(618-672) and pGEX-2TK/p107A4(618-672) were generated by PCR amplification by using pcDNA/p107HA or pcDNA3/p107A4HA as templates with the 5′ primer (CCATATGCACCCAAGAG) and the 3′ primer (CCGAATTCAGATCTTGTCC). The PCR products were cloned into pTOPO2.1 (Invitrogen). p107(618-672) and p107A4(618-672) as NdeI/BamHI fragments were then ligated into the NdeI/BamHI site of pGEX-2TK. Phosphorylation site mutations were generated by conventional PCR based mutagenesis by using high-fidelity Taq polymerase (Roche). All constructs were sequenced in their entirety to rule out spurious mutations. The sequences of oligonucleotides employed here are available upon request.
Protein purification.
Recombinant p107HA and p107A4HA were purified from infected Sf9. Briefly, cells were lysed in NETN buffer (0.5% Nonidet P-40, 20 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA, 5 mM NaF, 30 mM p-nitrophenylphosphate, 1 mM phenylmethylsulfonyl fluoride) containing freshly added protease inhibitors (1 μg/ml of antipain, leupeptin and aprotinin). Cleared lysates were incubated with anti-HA monoclonal antibody (BAbCO 16B12; dilution 1:150) and protein A/G plus agarose (Santa Cruz) at 4°C for 2 h. Cyclin D1/glutathione S-transferase (GST)-Cdk4, GST-cyclin A/Cdk2, and GST-cyclin E/Cdk2 complexes were expressed in Hi5 cells and purified by using GSH-Sepharose (Pharmacia), followed by elution with 50 mM Tris, 150 mM NaCl, 20% glycerol, and 20 mM glutathione (pH 8.0) as described previously (15). For purification of p107 spacer mutants, the indicated pGEX2TK-p107(618-672) constructs were transformed into BL21(DE3) cells. For each construct, 100 ml of culture was induced with 400 nM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. Cells were collected and resuspended in NETN buffer with protease inhibitors, followed by sonication. Cleared lysates were incubated with GSH-Sepharose for 2 h at 4°C. Immobilized GST-p107(618-672) proteins were eluted with 50 mM Tris, 150 mM NaCl, 20% glycerol, and 20 mM glutathione (pH 8.0). In some experiments, p107(618-672) (100 μg) was cleaved from GST by using thrombin (0.1 U) at room temperature for 6 h. Thrombin activity was terminated by adding phenylmethylsulfonyl fluoride at a final concentration of 1 mM. The pGEX-2TK plasmid places a protein kinase A phosphorylation site at the N terminus of p107(618-672) that is retained after thrombin cleavage. Protein concentrations were determined by Bradford assays (Bio-Rad). In some cases, the relative concentrations of p107(617-672) proteins were determined by measuring the [γ-32P]ATP incorporation after incubation with protein kinase A (Sigma) in 20 mM HEPES (pH 7.5)-15 mM MgCl2 in the presence of 50 μM ATP and 25 μCi of [γ-32P]ATP (37°C for 30 min).
For immunoprecipitation of p107 from C33A cells, asynchronous cells from four 150-mm dishes were collected and lysed in 0.5% NETN with freshly added protease inhibitors. Anti-p107 monoclonal antibodies (Santa Cruz SC-250; dilution 1:100) and protein A/G plus agarose (Santa Cruz) were added to the cleared lysate and rotated at 4°C for 2 h. Immune complexes were washed three times with NETN and two times with 1× kinase buffer (20 mM HEPES [pH 7.5], 15 mM MgCl2) before kinase assays were performed.
In vitro kinase assays.
In vitro kinase assays were performed in 20 mM HEPES (pH 7.5)-15 mM MgCl2 in the presence of 50 μM ATP and 5 μCi of [γ-32P]ATP. The reaction was incubated at 37°C for 30 min and terminated by adding 2× sodium dodecyl sulfate (SDS) loading buffer.
In vivo labeling.
U2OS cells were transfected with pcDNA3-p107HA by using FuGENE 6. At 24 h after transfection, cells were labeled with [32P]orthophosphate (1 mCi/ml) for 6 h before being harvested. Cells were lysed in NETN buffer supplemented with 5 mM NaF, 30 mM p-nitrophenylphosphate, 20 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, and a protease inhibitor cocktail (Roche). The cleared lysate was subjected to immunoprecipitation by using a mixture of antibodies (anti-HA, anti-p107 [SD9], and anti-p107 [C-18]) to facilitate efficient capture of p107. The immune complex was washed three times with NETN and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 4 to 12% gel prior to autoradiography and peptide mapping.
Phosphopeptide mapping.
Phosphorylated p107 was fractionated by SDS-PAGE and gels transferred to nitrocellulose prior to autoradiography. Radiolabeled p107 was excised, and filters were then treated with 10 μg of sequencing-grade trypsin (Roche) in 50 mM ammonium bicarbonate at 37°C overnight. The resulting tryptic peptides were oxidized with performic acid and processed as previously described (9). Approximately 4,000 cpm were subjected to two-dimensional phosphopeptide mapping on a thin-layer cellulose glass plate (VWR). Electrophoresis was carried out in a solution composed of 88% formic acid-acetic acid-H2O (50:156:1,794 [vol/vol/vol]; pH 1.9) for 45 min at 1,024 V by using the Hunter Thin Layer Electrophoresis System (HTLE-7000). The second dimension was developed by ascending chromatography in n-butanol-pyridine-acetic acid-H2O (75:50:15:60 [vol/vol/vol/vol]) for 12 h. Phosphopeptides were visualized by autoradiography.
Matrix-assisted laser desorption-ionization mass spectrometry with delayed extraction (Voyager-DE; Perspective Biosystems) was used for the identification of phosphopeptides, as described previously (65). An electrospray ion trap mass spectrometer (LCQ, Finnigan) coupled on-line with a capillary high-pressure liquid chromatograph (Magic 2002) was used for identification of phosphorylation sites. A MAGICMS C18 column (5 micron particle diameter) was used for liquid chromatography-tandem mass spectrometry analysis.
Phosphoamino acid analysis and phosphopeptide sequencing.
For phosphoamino acid analysis, peptides were scraped off the electrophoresis plate and eluted from the cellulose support by using 88% formic acid-acetic acid-H2O (50:156:1,794 [vol/vol/vol/vol]; pH 1.9) three times with 200 μl each time, followed by a single elution with 200 μl of H2O. Pooled elutions were dried and hydrolyzed in 6 N HCl at 110°C for 1 h. The sample was dried and dissolved in 20 μl of 88% formic acid-acetic acid-H2O (50:156:1,794 [vol/vol/vol]; pH 1.9) prior to being mixed with phosphoamino acid standards. The sample was applied to a thin-layer cellulose glass plate, followed by electrophoresis as previously described (9). The positions of phosphoamino acid standards were visualized by 0.25% ninhydrin dissolved in acetone and marked with a pencil. Radiolabeled phosphoamino acids were visualized by autoradiography. For phosphopeptide sequencing, eluted phosphopeptides were subjected to automated Edman degradation (Baylor Protein Core Facility), and the radioactivity in each cycle was determined by liquid scintillation.
Pulse bromodeoxyuridine (BrdU) labeling and immunofluorescence.
At 24 h after transfection, cells were incubated with 50 μM BrdU for 6 to 10 h prior to fixation with 4% paraformaldehyde. Cells were washed two times with phosphate-buffered saline (PBS) and permeablized with 0.1% Triton X-100-PBS for 10 min at room temperature. Cells were washed twice with PBS and incubated with anti-BrdU monoclonal antibodies (Amersham RPN 202) at 37°C for 4 h, followed by washing with PBS. Anti-BrdU antibodies were detected by using Alexa 594-conjugated goat anti-mouse antibodies (1:200 in 50% goat serum-PBS). The cells were washed two times with PBS and incubated in DAPI (4′,6′-diamidino-2-phenylindole) solution (a 1:2,000 dilution in PBS from a stock of 2 mg/ml) for 5 min at room temperature. Coverslips were mounted with 20 μl of Vectashield (H-1000; Vector Laboratories, Inc.) and observed under a fluorescence microscope (Olympus).
Sequence analysis and modelling.
Sequence conservation analysis was performed after the sequences of A-, D-, and E-type cyclins were aligned by using CLUSTALW 1.6. The sequences used were human cyclin E1 (A40270), mouse cyclin E1 (X75888), frog cyclin E2 (Q91780), human cyclin E2 (AF106690), mouse cyclin E2 (Q9Z238), fly cyclin E1 (S41756), zebra fish cyclin E1 (P47794), human cyclin A2 (X68303), cow cyclin A2 (X68321), fly cyclin A (JC1390), clam cyclin A (A26328), mouse cyclin D1 (S78355), human cyclin D3 (M92287), zebra fish cyclin D1 (S62730), human cyclin D1 (A38977), fly cyclin D (U41808), and rat cyclin D2 (D16308). The resulting alignment was analyzed in the program Aesop (M. Noble, unpublished data) to identify conserved residues that contribute to the cyclin molecular surface. Amino acids with similar properties (L, I, and V; S and T; D and E; Y and F; and R and K) were treated as identical. Residues 100% conserved by this criterion are indicated in Fig. 1.
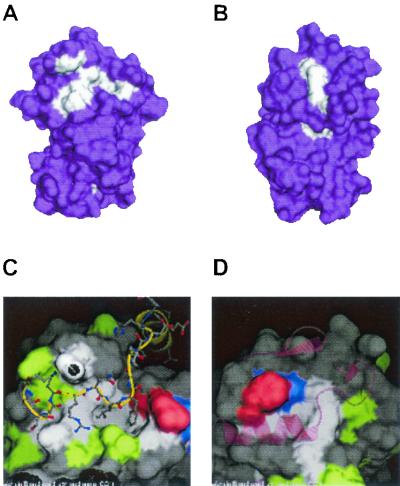
FIG. 1.
The Cdk interacting surface and the hydrophobic patch represent the major surfaces conserved in A-, E-, and D-type cyclins. Multisequence alignments for 17 vertebrate A-, D-, and E-type cyclins were generated and used to project on the structure of cyclin A. Residues that are conserved in character are shown in white. Residues that are not conserved among all three cyclins appear in purple. (A) Face of the projection containing the hydrophobic surface that interacts with RXL motifs. (B) Face of the projection containing the Cdk interacting surface. (C) Pairwise surface representation of the hydrophobic pocket bound to the RXL motif of p27 (stick model, backbone in yellow, side chains in gray). Residues conserved in all three proteins are shown in white. Residues conserved in cyclins A and D are shown in red, residues conserved in cyclins D and E are shown in green, and residues conserved in cyclins A and E are shown in blue. Surface residues that are not conserved are shown in gray. (D) Pairwise surface representation of the Cdk interaction surface in the composite cyclin. The PSTAIRE helix in Cdk2 is modeled onto the cyclin surface and is shown in the pink ribbon trace. Color coding is as described for panel C.
RESULTS
Efficient inactivation of p107's growth-suppressive function by cyclin D1/Cdk4 requires an intact cyclin-binding motif in the spacer region.
Cyclin E/Cdk2 and cyclin A/Cdk2 complexes interact with p107 via a cyclin-binding motif (Arg-Arg-Leu-Phe-Gly) located in the spacer region of p107, and both cyclin A and cyclin E form stable complexes with p107 in extracts from tissue culture cells (12, 24, 36, 58, 62, 67). However, neither cyclin E nor cyclin A complexes can inactivate growth suppression by p107 (7, 63). In contrast, available data have failed to reveal a stable interaction between D-type cyclins and p107 (24; unpublished data). Given the genetic evidence of a role for Cdk4 complexes in p107/p130 regulation, we sought to directly address the role of cyclin D1/Cdk4 in p107 regulation and to determine whether the RXL motif played a role in this process. The involvement of RXL motifs in targeting cyclin D/Cdk complexes remains poorly understood. In vitro experiments suggest that phosphorylation of Rb by cyclin D1/Cdk4 can involve both RXL-independent and RXL-dependent events (3). However, other work indicates that RXL-containing peptides do not function as inhibitors of cyclin D1/Cdk4, unlike the situation with cyclin A/Cdk2 (61), implying that cyclin D1/Cdk4 kinases bind poorly to certain RXL-containing peptides. Therefore, we initially performed modeling studies to determine the extent to which cyclin D's hydrophobic pocket might resemble that found in A-type cyclins, for which a structure exists. We constructed alignments of 17 vertebrate A-, E-, and D-type cyclins present in GenBank. We then sought to determine which residues are conserved in character between all cyclins. Two areas of extremely high conservation were observed: the region in cyclins known to interact with the PSTAIRE helix in Cdks (Fig. 1B and D) and the region corresponding to the hydrophobic pocket that interacts with RXL motifs (Fig. 1A and C). Overall, we found that residues comprising the hydrophobic pocket in these cyclins are highly conserved. Pairwise comparisons revealed extended conservation on surfaces surrounding the hydrophobic pocket, which could potentially contribute to specificity (Fig. 1C). However, we were unable to identify other extended regions of surface conservation among individual classes of cyclins that could potentially reflect novel substrate-binding sites. Because the sequence of the RXL motif in p107 (RRLFG) is very similar to that of p27 (RNLFG), we reasoned that cyclin D1 could interact with p107 through this motif, but possibly with an affinity that is too weak to be visualized in coprecipitation assays. Such a situation appears to exist with cyclin E, which phosphorylates Rb in an RXL-dependent manner without forming stable complexes (3, 22).
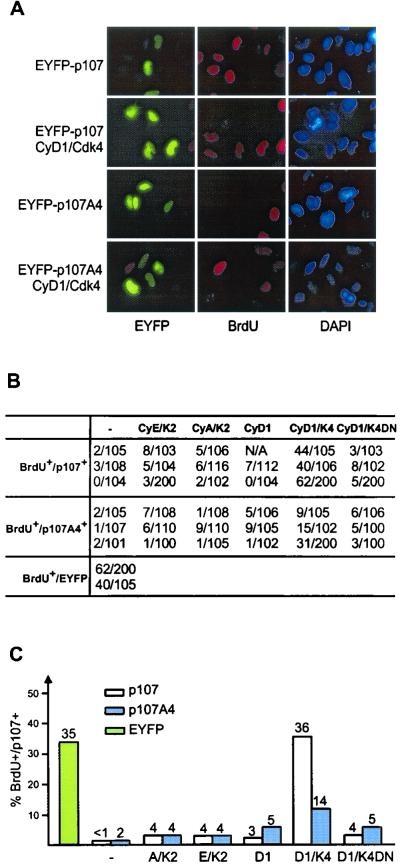
To test this possibility, we generated a p107 mutant (p107A4) in which the cyclin binding motif (Lys-Arg-Arg-Leu) at residues 657 to 660 was replaced by four alanine residues. Similar mutations in other RXL motifs have been shown to abolish association with A- or E-type cyclins (32, 43). Coding sequences for this p107 mutant, as well as for wild-type p107, were fused at the N terminus with enhanced yellow fluorescent protein (EYFP) and placed under control of the cytomegalovirus (CMV) promoter. p107A4 retained the ability to interact with E2F4 in insect cell coinfection experiments (data not shown) and, as such, this mutant protein was expected to block cell proliferation via repression of E2F target genes. Wild-type p107 and p107A4 were transiently expressed in p107-sensitive Saos-2 cells, and the number of p107-expressing S-phase cells was assessed by measuring BrdU incorporation. Fewer than 2% of cells expressing EYFP-p107A4 were in S phase 36 h after transfection versus 35% of cells transfected with pCMV-EYFP alone (Fig. 2). The extent of growth suppression by p107A4 was similar to that seen with wild-type p107 (Fig. 2). Consistent with previous studies, cotransfection of vectors expressing cyclin D1/Cdk4 led to a dramatic reversal of growth suppression by p107, with 36% of cells in S phase (Fig. 2), indicating a virtually complete reversal of G1 arrest mediated by p107. In contrast, cotransfection of cyclin D1/Cdk4 with EYFP-p107A4 resulted in a much smaller increase in S-phase cells (up to 14%) (Fig. 2). Reversal of growth suppression required catalytically active Cdk4, since cotransfection of cyclin D1 and a catalytically inactive Cdk4 mutant (D158N) led to only 5% of the cells entering S phase (Fig. 2). These results indicate that the RXL motif in the spacer region of p107 contributes to its inactivation by cyclin D1/Cdk4.
FIG. 2.
The cyclin-binding RXL motif in the spacer of p107 is dispensable for p107-induced growth arrest, but it is required for full inactivation of p107-mediated growth suppression by cyclin D1/Cdk4. (A) Saos-2 cells were transfected with vectors expressing EYFP-p107 or EYFP-p107A4 (2 μg) alone or together with the indicated cyclin/Cdk (1 μg each). At 24 h after transfection, cells were labeled with BrdU for 6 h before immunostaining. p107 expression was visualized as EYFP fluorescence. BrdU incorporation was detected by using anti-BrdU antibodies (red). Nuclei were stained with DAPI (blue). (B) Determination of the number of EYFP-p107 positive cells that are also BrdU positive from multiple experiments. (C) Summary of results in panel B depicting the proportions of p107-expressing cells that incorporated BrdU.
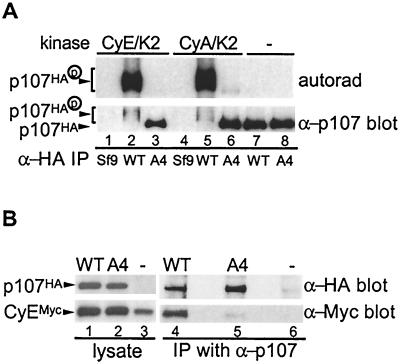
Previous studies have implicated both an N-terminal CRK motif and the spacer RXL motif in the association of E- and A-type cyclins with p107 (12). The available data would suggests that these two domains function together to allow tight association with the cyclin/Cdk complex, since mutation of either abolishes tight binding to either cyclin A or cyclin E (12, 68). We examined whether disruption of the RXL motif affected the ability of cyclin E or cyclin A to overcome p107-mediated growth arrest. In principle, the ability of p107 to inhibit Cdk2 complexes could negate the ability of these kinases to promote S-phase entry, and loss of the interaction could therefore allow for S-phase entry. As shown in Fig. 2, neither kinase was able to overcome growth suppression by the p107A4 mutant under conditions where RbΔCdk is overcome by cyclin E/Cdk2 (38; data not shown). To verify that the p107A4 mutant was defective in association with cyclin E, Saos-2 cells expressing cyclin EMyc, together with p107HA or p107A4HA, were subjected to immunoprecipitation with anti-p107 antibodies and blotted for cyclin EMyc (Fig. 3B). While cyclin EMyc was found to associate with p107 (lane 4), no association was seen with p107A4 (lane 5). Similar results were obtained in binding experiments performed with purified proteins from insect cells (Fig. 3A). These results suggest that the inability of cyclins E and A to overcome p107-mediated growth arrest is not simply a result of its sequestration and/or inhibition by p107 via the RXL motif.
FIG. 3.
The RXL motif in the spacer of p107 is required for stable interaction with cyclin E and cyclin A. (A) A total of 500 ng of immunopurified p107HA and p107A4HA (in which the KRRL motif is replaced by AAAA) was incubated with purified GST-cyclin E/Cdk2 or GST-cyclin A/Cdk2 (limiting subunit, ∼500 ng) for 2 h at 4°C to allow for complex assembly. Free Cdk complexes were removed by washing complexes with binding buffer, and the remaining complexes were washed with kinase buffer. Complexes were incubated with [γ-32P]ATP (5 μCi) and ATP (50 μM) at 37°C for 30 min, followed by SDS-8% PAGE. The proteins were transferred to NC membrane, followed by autoradiography and Western blotting. (B) pcDNA3-p107HA or p107A4HA (5 μg) were cotransfected with vectors expressing Myc-tagged cyclin E (cyclin EMyc) and Cdk2 (0.5 μg each) into U2OS cells, and the cell lysates were subjected to immunoprecipitation with anti-p107 antibodies. Immune complexes and crude lysates were fractionated by SDS-8 to 16% PAGE, followed by Western blotting with anti-HA and anti-Myc antibodies.
Site-specific phosphorylation of p107 by cyclin D1/Cdk4.
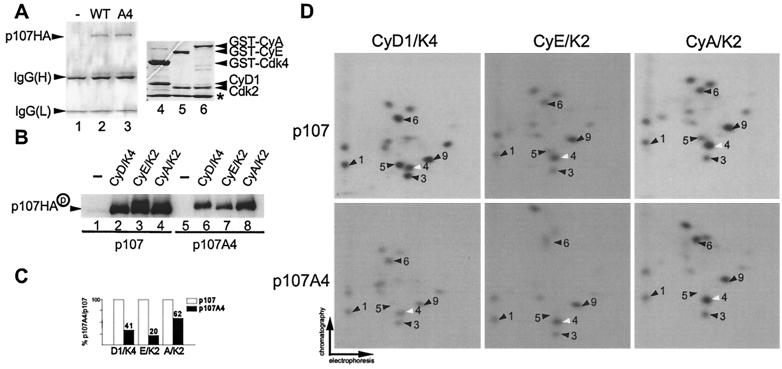
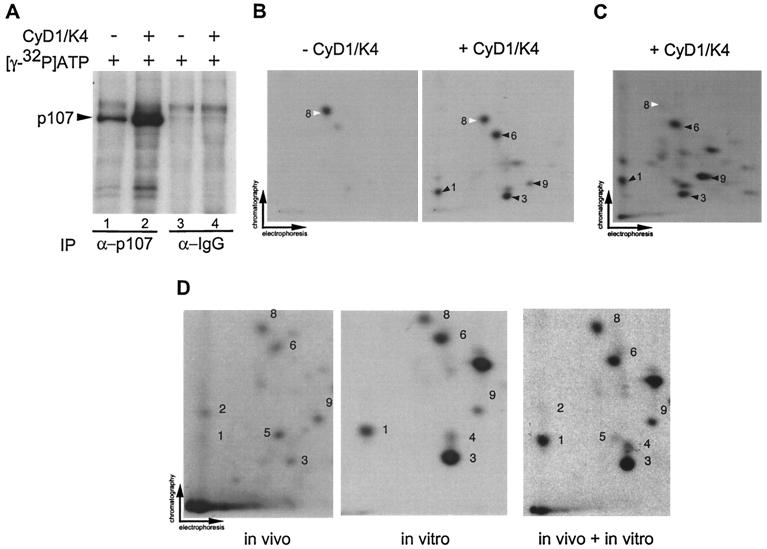
To understand the mechanistic basis of p107 inactivation by cyclin D1/Cdk4 and to establish that Cdk4's role is direct, we sought to determine the sites of phosphorylation of p107 by Cdks. p107 contains 17 potential Cdk phosphorylation sites, conforming to the minimal consensus sequence of serine or threonine followed by proline (26, 47, 59). We first examined the phosphorylation of p107 and p107A4 in vitro. Hemagglutinin (HA) epitope-tagged p107 proteins were produced in insect cells and purified by immunoaffinity chromatography (Fig. 4A). These proteins (500 nM) were then treated with purified cyclin/Cdk complexes (20 nM) (Fig. 4A and B) without a prebinding step, and the phosphorylated p107 was subjected to two-dimensional phosphopeptide mapping (Fig. 4D). Overall, cyclin A/Cdk2 and cyclin E/Cdk2 produced virtually identical phosphopeptides, with phosphorylation of peptides 4 and 9 being enhanced relative to other peptides. In contrast, cyclin D1/Cdk4 produced a distinct pattern with peptides 1, 3, 5, 6, and 9 being enhanced relative to other phosphopeptides (Fig. 4D). The pattern of phosphopeptides observed for Cdk2-phosphorylated p107 was unaffected by prebinding of the cyclin/Cdk2 complex to p107 (data not shown). To examine the contribution of the spacer RXL motif to phosphorylation, assays were performed with purified p107A4HA. Although p107A4HA was phosphorylated by Cdk4 and Cdk2, the efficiency was reduced two- to fivefold compared to wild-type p107 (Fig. 4B and C). Inactivation of the RXL motif did not result in dramatic changes in the phosphopeptide patterns observed with Cdk2 complexes. In contrast, the preference for residues preferred by Cdk4 (phosphopeptides 1, 3, 5, 6, and 9) was largely abolished upon mutation of the RXL motif (Fig. 4D).
FIG. 4.
Selective phosphorylation of recombinant p107 by cyclin D1/Cdk4 and Cdk2 kinase complexes in vitro. (A) Purified proteins used in this study. p107HA and p107A4HA were produced in insect cells and purified by immunoaffinity chromatography by using anti-HA antibodies. Proteins were separated by SDS-PAGE and stained with Coomassie blue (lanes 2 and 3). GST-cyclin A/Cdk2 (lane 6), GST-cyclin E/Cdk2 (lane 5), and GST-cyclin D1/Cdk4 (lane 4) were purified as described previously (15) and subjected to SDS-PAGE prior to staining with Coomassie blue. The asterisk indicates the position of insect cell GST which copurifies with the recombinant GST proteins. (B) Immunopurified p107HA and p107A4HA (500 nM) were subjected to in vitro kinase assays with purified cyclin D1/GST-Cdk4, GST-cyclin E/Cdk2, and GST-cyclin A/Cdk2 (20 nM) without a prebinding step. Reaction products were fractionated by SDS-8% PAGE and transferred to nitrocellulose; 32P-incorporation was then visualized by autoradiography. (C) Normalization of 32P incorporation from panel B. (D) Two-dimensional tryptic phosphopeptide mapping of phosphorylated p107HA or p107A4HA derived from panel B. Spot 4 (white arrow) is preferentially phosphorylated by Cdk2 complexes relative to Cdk4 complexes. Phosphopeptides 1, 3, 6, and 9 are preferentially phosphorylated by Cdk4. Among these phosphopeptides, spot 9 is also prominent in Cdk2 peptide maps.
To complement our analysis of recombinant p107 phosphorylation, three additional peptide-mapping experiments were performed. In one experiment, endogenous p107 was isolated from C33A cells by immunoprecipitation and incubated with [γ-32P]ATP in the presence or absence of cyclin D1/Cdk4 (20 nM). As shown in Fig. 5A, p107 was phosphorylated by an associated kinase activity (lane 1) but the extent of phosphorylation was greatly increased upon addition of cyclin D1/Cdk4 (lane 2). Peptide mapping revealed that phosphopeptide 8 was prominent in samples lacking exogenous cyclin D1/Cdk4, whereas multiple phosphopeptides, including peptides 1, 3, 6, and 9, were formed in the presence of cyclin D/Cdk4 (Fig. 5A and B). These peptides were among those found to be efficiently phosphorylated by cyclin D1/Cdk4 by using recombinant p107. In a second experiment, we expressed p107HA in Saos-2 cells. We reasoned that p107 isolated from these cells, which are arrested in G1, would lack phosphorylation events important for p107 inactivation and, therefore, that these sites would be available for modification by cyclin D1/Cdk4 in vitro. We found that phosphopeptides 1, 3, 6, and 9 were efficiently phosphorylated, while phosphopeptide 8 was weakly phosphorylated (Fig. 5C). These data suggested that phosphopeptides 1, 3, 6, and 9 might be important for p107 inactivation by cyclin D1/Cdk4. In a third experiment, we addressed the in vivo phosphorylation status of p107. p107-insensitive U2OS cells were transfected with pCMV-p107HA and metabolically labeled with orthophosphate prior to immunoprecipitation and peptide mapping (Fig. 5D). Phosphopeptides 1, 2, 3, 5, 6, 8, and 9 were detected in the in vivo-labeled p107, a finding consistent with the peptides observed with in vitro modified p107 isolated from mammalian cells or recombinant p107 from insect cells (Fig. 4D and 5D).
FIG. 5.
Visualization of cyclin D1/Cdk4-specific phosphorylation events in p107. (A) Anti-p107 or control immune complexes from asynchronous C33A cells were incubated with [γ-32P]ATP in the presence or absence of purified cyclin D1/Cdk4 (40 nM). Reaction mixtures were separated by SDS-8% PAGE and transferred to nitrocellulose prior to autoradiography. (B) Two-dimensional tryptic phosphopeptide mapping of samples derived from panel A. In the absence of Cdk4, the single major phosphopeptide 8 was found (white arrow). In the presence of Cdk4, four additional phosphopeptides (peptides 1, 3, 6, and 9) were observed (black arrow). (C) Phosphorylation of p107 immune complexes from Saos-2 cells by cyclin D1/Cdk4 in vitro. pcDNA-p107HA was transiently expressed in Saos-2 cells, and anti-HA immune complexes were then used in kinase assays with cyclin D1/Cdk4 as described in panel B. Phosphorylated p107HA was subjected to tryptic peptide mapping. The major phosphopeptides seen with Cdk4 in panel B were found in panel C (peptides 1, 3, 6, and 9) as indicated by the black arrows. Peptide 8 (white arrow) was weakly phosphorylated in this setting. (D) In vivo phosphorylation of p107. p107HA was expressed in U2OS cells by transient transfection prior to metabolic labeling and analysis as described in Materials and Methods. Labeled p107 (600 cpm) was subjected to peptide mapping in the presence or absence of in vitro-phosphorylated p107HA obtained by immunoprecipitation from U2OS cells.
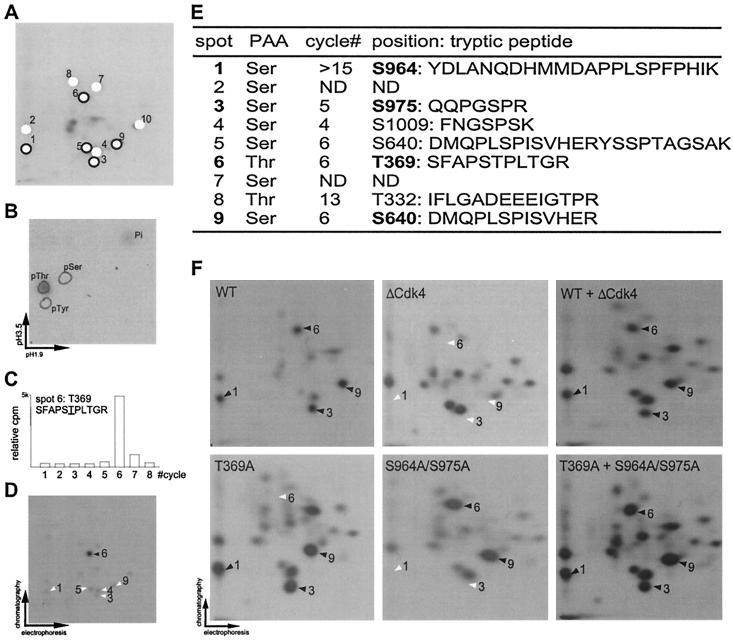
Identification of p107 phosphorylation sites.
Multiple strategies were used to identify cyclin D1/Cdk4 phosphorylation sites in p107. The results for phosphopeptide 6 are shown in Fig. 6B to D, and the results of all identified phosphopeptides are summarized in Fig. 6E. First, phosphoamino acid analysis was carried on each major phosphopeptide (Fig. 6A, B, and E). Phosphopeptides 1 to 5, 7, and 9 were found to contain serine, while phosphopeptides 6 and 8 contain threonine. Second, phosphopeptides were subjected to Edman degradation, and the radioactivity released at each cycle was determined (Fig. 6C and E). Third, synthetic peptides corresponding to predicted tryptic peptides 3, 4, 5, 6, 8, and 9 were phosphorylated in vitro and subjected to two-dimensional electrophoresis in the presence or absence of tryptic peptides derived from full-length p107 in order to assess comigration with predicted sites (Fig. 6D). Based on these data and the predicted tryptic peptides in p107, we assigned the major phosphorylation sites as follows: peptide 1, S964 or S515; peptide 3, S975; peptide 4, S1009; peptide 5, S640; peptide 6, T369 or T997; peptide 8, T332; and peptide 9, S640 (Fig. 6E). Peptides 5 and 9 were both found to contain phosphoserine 640 but differed in the position of tryptic cleavage, presumably reflecting the presence of a glutamic acid preceding the cleavage site at R647 which reduces the efficiency of trypsin cleavage. To unequivocally determine the identities of phosphorylation sites in the major Cdk4-phosphorylated peptides 1, 3, 6, and 9, we generated combinatorial point mutations in full-length p107HA. Residues S964, S975, T369, and S640 were replaced with alanine. Wild-type and mutant proteins were expressed in U2OS cells, isolated by using anti-HA antibodies, and subjected to phosphorylation by using cyclin D/Cdk4. As shown in Fig. 6F, mutation of all four sites (p107ΔCdk4) led to the absence of phosphopeptides 1, 3, 6, and 9, as demonstrated by mixing experiments. Similarly, single (T369A) and double (S964A/S975A) mutants displayed loss of peptide 6 or peptides 1 and 3, respectively (Fig. 6F). Therefore, we conclude that phosphopeptide 1 derives from phosphorylation of S964 and that phosphopeptide 6 derives from phosphorylation of T369 (Fig. 6E). We found that p107ΔCdk4 contains several prominent phosphopeptides in addition to those that we identified (Fig. 6F). The majority of these peptides are present in wild-type p107 peptide maps but at low stoichiometry. The simplest explanation for these results is that mutation of preferred sites leads to enhanced phosphorylation of sites that are normally phosphorylated but with low efficiency.
FIG. 6.
Determination of in vitro Cdk phosphorylation sites in p107. (A) Designation of p107 phosphopeptides. Major phosphopeptides (designated 1 to 10) are indicated schematically with open circles for Cdk2 preferred sites and with closed circles for Cdk4 preferred sites. (B) Phosphoamino acid analysis of phosphopeptide 6. Tryptic phosphopeptide 6 from Cdk4 phosphorylated p107 (Fig. 4D) was eluted from a two-dimensional electrophoresis plate and subjected to phosphoamino acid analysis. The positions of unlabeled phosphoamino acids used as standards are circled. The phosphorylated amino acid was visualized by autoradiography. (C) Edman degradation of phosphopeptide 6. Residues released at each Edman cycle were subjected to liquid scintillation counting, and the radioactivity at each cycle is presented as a histogram. Based on p107 tryptic peptides, phosphopeptide 6 was predicted to correspond to SFAPS TPLTGR, where the underlined residue is phosphorylated. (D) A synthetic tryptic peptide corresponding to SFAPS TPLTGR was phosphorylated in vitro with cyclin D1/Cdk4, purified, and then subjected to peptide mapping with a limiting amount of p107HA previously phosphorylated by cyclin D1/Cdk4. The phosphorylated synthetic peptide comigrated with phosphopeptide 6 from p107, as indicated by the solid arrow. (E) Summary of Cdk phosphorylation sites in p107 identified in this study. Cyclin D1/Cdk4-specific sites are indicated in boldface type. (F) Major cyclin D1/Cdk4 phosphorylation sites are absent in the p107ΔCdk4 mutant in which T369, S640, S964, and S975 are replaced by alanine. HA-tagged p107, p107(T369A), p107(S964A/S975A), and p107ΔCdk4 were expressed in U2OS cells, purified by using anti-HA antibodies, and phosphorylated by cyclin D1/Cdk4 (40 nM) in vitro. Proteins were subjected to two-dimensional peptide mapping either alone or in various combinations. The positions of Cdk4 phosphorylation sites are shown by black arrows when present, and their expected positions are shown by white arrows in maps derived from mutants containing nonphosphorylatable residues.
Additional analysis of phosphorylation sites in p107 modified by cyclin A/Cdk2 was performed by using mass spectrometry (Table 1). By this more sensitive assay, we detected phosphorylation of several sites identified as Cdk4 sites (residues T332, S640, S964, and S975) and, in addition, phosphorylation of T385, S762, S988, and T997 was also observed.
TABLE 1.
Analysis of cyclin A/Cdk2-phosphorylated p107 by mass spectrometrya
| Peptide (residues) | Mass
|
|
|---|---|---|
| Measured | Theoretical | |
| IFLGADAEEEIGTPR (320-334) | 1,617.80 | 1,617.77 |
| EKEAVITPVASATQSVSR (379-396) | 1,873.10 | 1,873.09 |
| DMQPLSPISVHER (635-647) | 1,508.70 | 1,508.72 |
| VKSPVSLTAHSLIGASPK (747-764) | 1,792.30 | 1,792.10 |
| YDLANQHMMDAPPLSPFPHIK (949-970) | 2,537.90 | 2,537.90 |
| QQPGSPR (971-977) | 769.00 | 768.82 |
| RISQQHSIYISPHKNGSGLTPR (978-999) | 2,476.80 | 2,476.78 |
| SALLYKFNGSPSK (1000-1012) | 1,411.60 | 1,411.62 |
The positions of phosphorylated residues are underlined.
Phosphorylation of p107 on major Cdk4 phosphorylation sites is required to overcome cell cycle arrest.
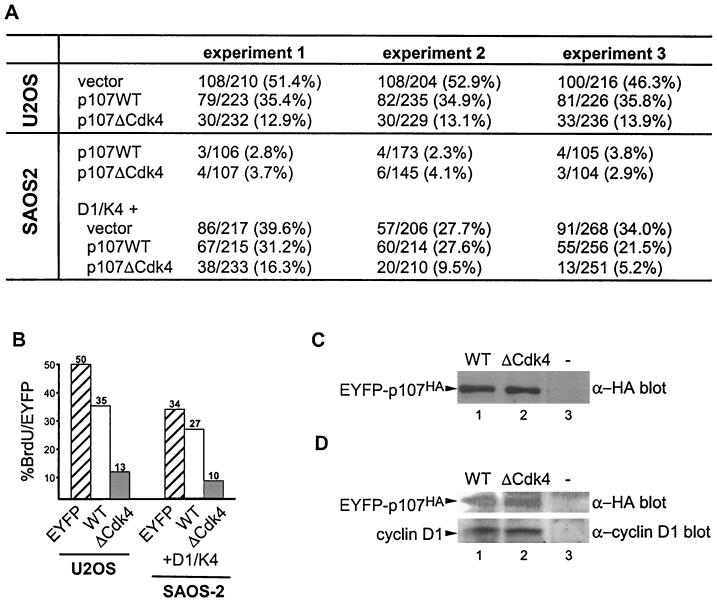
Having identified four major Cdk4 phosphorylation sites (Fig. 6) and found that these sites were among those whose phosphorylation is enhanced by the RXL motif in p107 (Fig. 4), we sought to determine whether p107 lacking these sites is resistant to inactivation. Two established assays were used. Previous studies indicate that U2OS cells are resistant to the cell cycle arrest properties of p107 in transient-transfection assays, presumably as a result of active cyclin D/Cdk4 complexes in these cells (66). Thus, the prediction is that if the p107ΔCdk4 protein is resistant to inactivation, it would induce growth arrest in these cells. Therefore, we expressed EYFP-p107 and EYFP-p107ΔCdk4 (containing alanine at T369, S640, S964, and S975) in U2OS cells and determined the number of EYFP-positive S-phase cells by BrdU incorporation. EYFP-p107 had a modest effect of BrdU incorporation relative to EYFP alone (35% versus 50% BrdU-positive, EYFP-positive cells). In contrast, only 13% of EYFP-p107ΔCdk4-expressing cells were BrdU positive, when expressed at levels similar to that found with wild-type EYFP-p107 (Fig. 7A, B, and C).
FIG. 7.
Mutation of Cdk4 phosphorylation sites in p107 abolishes the ability of cyclin D1/Cdk4 to reverse growth suppression. (A) p107 insensitive U2OS cells or p107-sensitive Saos-2 cells were transfected with vectors expressing EYFP-p107 or EYFP-p107ΔCdk4 (3 μg) in the presence or absence of vectors expressing cyclin D1 and Cdk4 (1 μg each). After 24 h, cells were labeled with BrdU. EYFP-p107-positive cells incorporating BrdU were determined by immunofluorescence. The numbers of BrdU-positive and EYFP-p107-positive cells are shown for multiple independent experiments. (B) Histogram of data presented in panel A. (C) The levels of p107 and p107ΔCdk4 expressed in U2OS cells were similar, as determined by immunoblotting with anti-p107 antibodies. (D) Immunoblots of cyclin D1 and EYFP-p107 proteins revealed similar expression levels in Saos-2 cells.
A second assay employed p107-sensitive Saos-2 cells, together with cotransfection of p107 and cyclin D/Cdk4-expressing plasmids. As expected, p107ΔCdk4 blocked cell division to an extent similar to that found with wild-type p107 (Fig. 7A). However, in contrast with wild-type p107, coexpression of cyclin D1/Cdk4 was unable to reverse growth arrest mediated by the p107ΔCdk4 mutant (Fig. 7A, B, and D). Thus, the results of these two assays are consistent with the idea that phosphorylation of p107 on key sites modified preferentially by cyclin D1/Cdk4 is required to reverse the growth-suppressive function of p107.
RXL-mediated phosphorylation on nonconsensus Cdk phosphorylation sites.
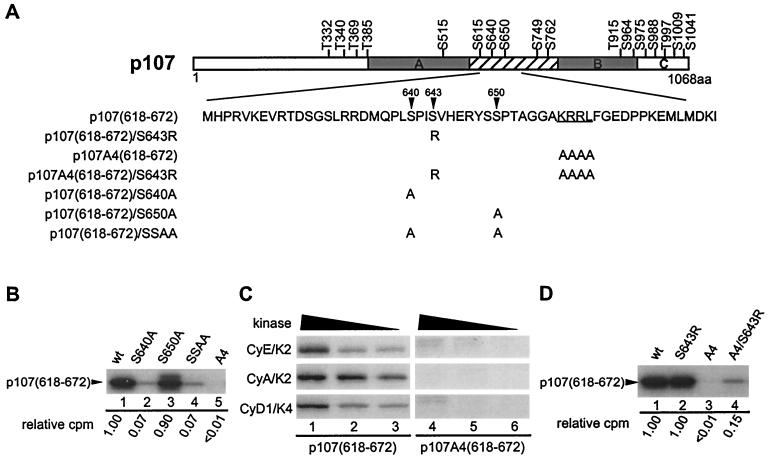
Structure-function studies with peptide substrates have defined S/T-P-X-K/R as a consensus sequence for Cdk2 and Cdc2 (26, 47, 59). The specificity of cyclin D1/Cdk4 has not been studied as intensely but in Rb, several Cdk4 phosphorylation sites conform to this consensus (15, 30, 64). Interestingly, the majority of preferred Cdk4 phosphorylation sites in p107 (S964, S975, T369, and S640) did not conform to the Cdk consensus. This, coupled with the apparent decrease in preference for these sites in the p107A4 mutant lacking the cyclin interaction motif in the spacer region (Fig. 4), led us to examine the role of the RXL motif in facilitating phosphorylation of S640 in greater detail. This residue is located near the RXL motif beginning at residue 658. We produced a GST fusion protein containing p107 (residues 618 to 672), i.e., p107(618-672), with or without the RXL motif (Fig. 8A). To aid in the analysis, we generated nonphosphorylatable point mutations at the two potential Cdk phosphorylation sites in this region (S640A and S650A). p107 fragments were then released from GST by treatment with thrombin. In vitro kinase assays with these p107 spacer fragments revealed that the vast majority of phosphorylation occurs on S640, since its mutation to alanine largely abolished phosphorylation by Cdk2 and Cdk4 (Fig. 8B and data not shown). The extent of phosphorylation observed was 7% of wild-type p107(618-672) (lanes 1 and 2). In fact, this level of phosphorylation was indistinguishable from a mutant lacking both S640 and S650 (lane 4). In contrast, mutation of S650 had little effect on the extent of phosphorylation (90% relative to wild-type p107) (lane 3). Importantly, the extent of phosphorylation of the RXL mutant (A4) was less than 1%, indicating a strict requirement for the RXL motif in phosphorylation of S640 in this p107 fragment (Fig. 8B, lane 5, and C). Interestingly, we found that mutation of S643 to arginine, which generates a more typical Cdk consensus site at S640, led to a 15-fold increase in phosphorylation at S640 in the context of a mutant lacking the RXL motif (Fig. 8D, lanes 1, 3, and 4). In contrast, this S643R mutation had no effect on S640 phosphorylation in the context of an intact RXL motif (Fig. 8D, lanes 1 and 2). Previous crystallographic studies indicate that basic residues in peptide substrates form hydrogen bonds with phosphothreonine 160 in Cdk2 and also with the main-chain oxygen of I270 in cyclin A3, thereby contributing to substrate binding (10). Thus, our data suggest that one role of the RXL motif may be to allow for efficient phosphorylation of substrate sequences lacking a basic residue at the +3 position. This may be important in the case of cyclin D1/Cdk4 since the majority of the sites modified in p107 and p130 lack basic residues at position +3 (Fig. 9).
FIG. 8.
Phosphorylation of S640 is RXL dependent in p107(618-672). (A) Schematic representation of spacer mutants used in this study. (B) S640 is the Cdk target site in p107(618-672). The indicated proteins were phosphorylated by cyclin A/Cdk2 (20 nM) in vitro. Reaction products were separated on 8 to 20% Tricine gels, followed by autoradiography and quantitation with a PhosphorImager. (C) Phosphorylation of S640 is RXL dependent. GST-p107(618-672) and GST-p107A4(618-672) were incubated with the indicated kinases and [γ-32P]ATP, and the reaction products were separated by SDS-PAGE prior to autoradiography. (D) Cdk phosphorylation on S640 is abolished in the absence of RXL motif, but it is partially restored by replacing the +3 residue with arginine. Reactions were performed as in panel B. Quantitation by PhosphorImager analysis is indicated below each lane.
FIG. 9.
(A) Conserved organization of Cdk4 phosphorylation sites in pocket proteins. Major Cdk4 phosphorylation sites are indicated by black arrows. The position of the RXL motif in p107 and p130 is indicated by an asterisk. (B) Alignment of Cdk4 phosphorylation sites in p107 (the present study), p130 (21), and Rb (15, 30, 64).
DISCUSSION
Work from several laboratories has revealed that cyclins not only serve as Cdk activating subunits but also function in substrate recognition through direct interaction with substrates (2, 3, 18, 25, 27, 29, 30, 32, 44, 46, 60). Our understanding of this property of cyclins has been extended primarily through analysis of A- and E-type cyclins. These cyclins interact with substrates containing a conserved RXL motif through a hydrophobic surface composed of elements within the cyclin box (10, 55). There appear to be two distinct classes of RXL motifs that differ in their strength of association with the cyclin. In one extreme, the cyclin interacts tightly with the RXL-containing substrate and remains tightly bound after phosphorylation in vitro. Examples of this class include the p107/cyclin A or E complex and the cyclin E/HPV E1 complex. In the case of p107, the pocket protein becomes the obligate substrate for the kinase and blocks access of exogenous substrates to either the Cdk, the cyclin box, or both (62, 67). In the case of the cyclin E/HPV E1 complex, substrates that do not require association with the cyclin to be phosphorylated, such as histone H1, are still accessible (42). On the other extreme are substrates that contain RXL motifs but nevertheless do not bind tightly to the cyclin. For example, phosphorylation of several sites in Rb by A- and E-type cyclin/Cdk complexes requires an RXL motif located in the C pocket of Rb, but neither of these cyclins forms stable complexes with Rb (3). Presumably, the cyclin associates transiently with the RXL motif to facilitate Rb phosphorylation.
In this study, we have addressed the question of how cyclin D1/Cdk4 controls the activity of p107 and the role of the RXL motif in p107 in this process. Several facts were established. First, although expression of cyclin D1/Cdk4 readily overcame growth suppression by p107 in a Saos-2 cotransfection assay, mutation of the RXL motif located in the spacer domain in p107 substantially reduced the efficiency of reversal by cyclin D1/Cdk4. This effect required catalytically active Cdk4, since mutation of the active site D145 essentially abolished the ability of cyclin D1/Cdk4 to overcome p107-mediated growth arrest (Fig. 2). This is consistent with a direct role for cyclin D1/Cdk4 as opposed to a role in CKI sequestration (57). Second, cyclin D1/Cdk4 phosphorylates p107 with a specificity distinct from that seen with cyclin A and cyclin E/Cdk2 (Fig. 4). The efficiency of Cdk4 preferred phosphorylation events is reduced when the RXL motif is abolished by mutation, a finding consistent with the effects of this mutation on reversal of growth suppression in transfected cells (Fig. 2). Third, we identified four phosphorylation sites that are preferentially targeted by cyclin D1/Cdk4 in vitro, and these sites are phosphorylated in tissue culture cells (Fig. 5 and 6). Fourth, replacement of these four major cyclin D1/Cdk4 phosphorylation sites in p107 by nonphosphorylatable alanine residues (p107ΔCdk4) leads to a dominantly acting p107 protein. Expression of p107ΔCdk4 in p107-insensitive U2OS cells leads to a dramatic reduction in BrdU-positive cells relative to wild-type p107 (Fig. 7). This effect is reminiscent of that recently observed with a nonphosphorylatable p130 protein (21). Moreover, growth suppression by p107ΔCdk4 in Saos-2 cells is not efficiently overcome by coexpression of cyclin D1/Cdk4, indicating that phosphorylation of one or more of these sites is required for p107 inactivation and for cyclin D1/Cdk4 to promote proliferation in the face of p107 expression (Fig. 7). These data are consistent with the recent finding that cells lacking p107 and p130 are insensitive to the growth suppressive effects of p16 overexpression (11, 20). Thus, there is now both genetic and biochemical data directly linking Cdk4 function with inactivation of both p107 and p130. This further supports the idea that the p16/cyclin D signaling pathway is not a simple linear pathway but instead has two branches that independently regulate E2F-1, E2F-2, E2F-3, and E2F-4 family members through Rb and E2F-4 and E2F-5 through p107 and p130 (11).
The domain structures and the positions of Cdk4 phosphorylation sites in Rb (15, 30, 64), p107 (this work), and p130 (21) are summarized in Fig. 9. The data indicate that Cdk4-mediated phosphorylation of these proteins occurs primarily in three locations: (i) near the border between the N terminus and the A pocket, (ii) in the spacer region located between the A and B pockets, and (iii) in the C pocket. This overall conservation of modification events suggests that the biochemical mechanism of pocket protein inactivation is conserved in all three family members.
A total of 13 phosphorylation sites preferred by cyclin D1/Cdk4 have now been identified in three pocket proteins (15, 21, 30, 31, 37, 64). Among these sequences, the −2 position was found to contain proline in nine cases and two other phosphorylation sites contained proline at −3. Thus, these data would indicate that cyclin D/Cdk4 complexes strongly prefer substrates containing proline residues at −2 or −3. Previous studies with peptide substrates have suggested a preference for basic residues at position +3 for Cdk2 and Cdc2 (26, 59). At this position, arginine or lysine side chains make salt bridges with a phosphate group present on Thr-160 in Cdk2, the site of modification by Cdk activating kinase. This interaction likely increases the affinity of substrates for the kinase and may also help orient the substrate (10). Although the majority of Cdk4 sites in Rb contain a basic residue at the +3 position, this situation is rare in p107 and p130. One such site in p107 is S640, which contains a serine at the +3 position (S643). We have found that phosphorylation of S640 in the context of a spacer fragment of p107 is fully dependent upon the presence of an intact RXL motif with all three cyclin/Cdk complexes examined (Fig. 8 and data not shown). Interestingly, phosphorylation at S640 in the absence of an intact RXL motif could be partially restored upon replacement of S643 by arginine (Fig. 8D). These data suggest that RXL motifs can facilitate the phosphorylation of substrate sequences that lack a basic residue at +3. In principle, the presence of an RXL motif in a substrate can greatly expand the types of sequences available within Cdk phosphorylation sites because the loss of binding energy in substrates with suboptimal sequences can be partially compensated for by RXL-mediated substrate recruitment.
It has been proposed that cyclin D1/Cdk4 mediated phosphorylation of Rb allows access of cyclin E/Cdk2 to the pocket region to phosphorylate sites that complete the inactivation process (22). What is the role of cyclin E and cyclin A/Cdk complexes in p107 regulation? Although it is clear that these kinases assemble with p107 and p130 in a sequential manner during cell division, the precise roles for these interactions are unclear (36, 58, 67). Previous studies have provided evidence that interaction of p107 with cyclin A/Cdk2 may serve an inhibitory function, much like Cdk inhibitors, while simultaneously leading to p107 phosphorylation (62, 67). However, it appears that E- and A-type cyclin/Cdk complexes are not sufficient to inactivate the growth-suppressive function of p107, presumably because they do not efficiently target sites that are required for p107 inactivation (7, 63; the present study). We also note that coexpression of cyclin E/Cdk2 cannot reverse the growth-suppressive effects of a mutant p107 protein lacking the spacer RXL motif despite the fact that this mutant p107 protein does not stably associate with cyclin E (Fig. 2 and 3). Thus, it would appear that, in contrast to the situation with a nonphosphorylatable Rb protein, active cyclin E/Cdk2 complexes are unable to bypass p107 growth arrest (5, 38, 41). This reinforces the notion that growth arrest by p107 and Rb occur via distinct mechanisms.
Acknowledgments
We thank Louise Johnson and Jane Endicott for stimulating discussions.
This work was supported by National Institutes of Health grant GM54137 and the Welch Foundation to J.W.H. A portion of this work was performed with the support of a Vallee Visiting Professorship at Oxford University to J.W.H.
REFERENCES
- 1.Adams, P. D., and J. W. Harper. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511-2526. [DOI] [PubMed] [Google Scholar]
- 2.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-Cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed]
- 3.Adams, P. D., X. Li, W. R. Sellers, K. B. Baker, X. Leng, J. W. Harper, Y. Taya, and W. G. Kaelin, Jr. 1999. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-Cdk complexes. Mol. Cell. Biol. 19:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aktas, H., H. Cai, and G. M. Cooper. 1997. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol. Cell. Biol. 17:3850-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alevizopoulos, K., J. Vlach, S. Hennecke, and B. Amati. 1997. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 16:5322-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartek, J., J. Bartkova, and J. Lukas. 1997. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 237:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Beijersbergen, R. L., L. Carlee, R. M. Kerkhoven, and R. Bernards. 1995. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 9:1340-1353. [DOI] [PubMed] [Google Scholar]
- 8.Blain, S. W., E. Montalvo, and J. Massague. 1997. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J. Biol. Chem. 272:25863-25872. [DOI] [PubMed] [Google Scholar]
- 9.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 10.Brown, N. R., M. E. Noble, J. A. Endicott, and L. N. Johnson. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1:438-443. [DOI] [PubMed] [Google Scholar]
- 11.Bruce, J. L., R. K. Hurford, Jr., M. Classon, J. Koh, and N. Dyson. 2000. Requirements for cell cycle arrest by p16INK4a. Mol. Cell 6:737-742. [DOI] [PubMed] [Google Scholar]
- 12.Castano, E., Y. Kleyner, and B. D. Dynlacht. 1998. Dual cyclin-binding domains are required for p107 to function as a kinase inhibitor. Mol. Cell. Biol. 18:5380-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J., P. Saha, S. Kornbluth, B. D. Dynlacht, and A. Dutta. 1996. Cyclin-binding motifs are essential for the function of p21CIP1. Mol. Cell. Biol. 16:4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, M., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connell-Crowley, L., J. W. Harper, and D. W. Goodrich. 1997. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell 8:287-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmolino, L. M., P. Saha, and A. Dutta. 2001. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276:26947-26954. [DOI] [PubMed] [Google Scholar]
- 18.Dynlacht, B. D., K. Moberg, J. A. Lees, E. Harlow, and L. Zhu. 1997. Specific regulation of E2F family members by cyclin-dependent kinases. Mol. Cell. Biol. 17:3867-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 20.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, K., T. Farkas, J. Lukas, K. Holm, L. Ronnstrand, and J. Bartek. 2001. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 20:422-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbour, J. W., R. X. Luo, S. A. Dei, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 23.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 24.Hauser, P. J., D. Agrawal, B. Chu, and W. J. Pledger. 1997. p107 and p130 associated cyclin A has altered substrate specificity. J. Biol. Chem. 272:22954-22959. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann, I., G. Draetta, and E. Karsenti. 1994. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E-dependent phosphorylation at the G1/S transition. EMBO J. 13:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, J. K., and M. J. Solomon. 1996. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 271:25240-25246. [DOI] [PubMed] [Google Scholar]
- 27.Huang, D., J. Moffat, W. A. Wilson, L. Moore, C. Cheng, P. J. Roach, and B. Andrews. 1998. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol. Cell. Biol. 18:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurford, R. K., Jr., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa, M., H. Higashi, I. Suzuki-Takahashi, K. Segawa, S. K. Hanks, Y. Taya, S. Nishimura, and A. Okuyama. 1995. Phosphorylation of E2F-1 by cyclin A-cdk2. Oncogene 10:229-236. [PubMed] [Google Scholar]
- 30.Kitagawa, M., H. Higashi, H. K. Jung, I. Suzuki-Takahashi, M. Ikeda, K. Tamai, J. Kato, K. Segawa, E. Yoshida, S. Nishimura, and Y. Taya. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15:7060-7069. [PMC free article] [PubMed] [Google Scholar]
- 31.Knudsen, E. S., and J. Y. Wang. 1996. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J. Biol. Chem. 271:8313-8320. [DOI] [PubMed] [Google Scholar]
- 32.Krek, W., M. E. Ewen, S. Shirodkar, Z. Arany, W. G. Kaelin, Jr., and D. M. Livingston. 1994. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78:161-172. [DOI] [PubMed] [Google Scholar]
- 33.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 34.Lacy, S., and P. Whyte. 1997. Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes. Oncogene 14:2395-2406. [DOI] [PubMed] [Google Scholar]
- 35.Lavoie, J. N., G. l'Allemain, A. Brunet, R. Muller, and J. Pouyssegur. 1996. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 271:20608-20616. [DOI] [PubMed] [Google Scholar]
- 36.Lees, E., B. Faha, V. Dulic, S. I. Reed, and E. Harlow. 1992. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 6:1874-1885. [DOI] [PubMed] [Google Scholar]
- 37.Lees, J. A., K. J. Buchkovich, D. R. Marshak, C. W. Anderson, and E. Harlow. 1991. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 10:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leng, X., L. Connell-Crowley, D. Goodrich, and J. W. Harper. 1997. S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr. Biol. 7:709-712. [DOI] [PubMed] [Google Scholar]
- 39.Lipinski, M. M., and T. Jacks. 1999. The retinoblastoma gene family in differentiation and development. Oncogene 18:7873-7882. [DOI] [PubMed] [Google Scholar]
- 40.Lukas, J., D. Parry, L. Aagaard, D. J. Mann, J. Bartkova, M. Strauss, G. Peters, and J. Bartek. 1995. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503-506. [DOI] [PubMed] [Google Scholar]
- 41.Lukas, J., T. Herzinger, K. Hansen, M. C. Moroni, D. Resnitzky, K. Helin, S. I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 11:1479-1492. [DOI] [PubMed] [Google Scholar]
- 42.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsushime, H., M. E. Ewen, D. K. Strom, J. Y. Kato, S. K. Hanks, M. F. Roussel, and C. J. Sherr. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71:323-334. [DOI] [PubMed] [Google Scholar]
- 45.Medema, R. H., R. E. Herrera, F. Lam, and R. A. Weinberg. 1995. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl. Acad. Sci. USA 92:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyerson, M., and E. Harlow. 1994. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14:2077-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minshull, J., R. Golsteyn, C. S. Hill, and T. Hunt. 1990. The A- and B-type cyclin-associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 9:2865-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 49.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parry, D., D. Mahony, K. Wills, and E. Lees. 1999. Cyclin D-CDK subunit arrangement is dependent on the availability of competing INK4 and p21 class inhibitors. Mol. Cell. Biol. 19:1775-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersen, B. O., J. Lukas, C. S. Sorensen, J. Bartek, and K. Helin. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 53.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saha, P., Q. Eichbaum, E. D. Silberman, B. J. Mayer, and A. Dutta. 1997. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol. Cell. Biol. 17:4338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherr, C. J. 1994. G1 phase progression: cycling on cue. Cell 79:551-555. [DOI] [PubMed] [Google Scholar]
- 57.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 58.Shirodkar, S., M. Ewen, J. A. DeCaprio, J. Morgan, D. M. Livingston, and T. Chittenden. 1992. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell 68:157-166. [DOI] [PubMed] [Google Scholar]
- 59.Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms, and L. C. Cantley. 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4:973-982. [DOI] [PubMed] [Google Scholar]
- 60.Takeda, D. Y., J. A. Wohlschlegel, and A. Dutta. 2001. A bipartite substrate recognition motif for cyclin-dependent kinases. J. Biol. Chem. 276:1993-1997. [DOI] [PubMed] [Google Scholar]
- 61.Wohlschlegel, J. A., B. T. Dwyer, D. Y. Takeda, and A. Dutta. 2001. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol. Cell. Biol. 21:4868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo, M. S., I. Sanchez, and B. D. Dynlacht. 1997. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol. Cell. Biol. 17:3566-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao, Z. X., D. Ginsberg, M. Ewen, and D. M. Livingston. 1996. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc. Natl. Acad. Sci. USA 93:4633-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zarkowska, T., and S. Mittnacht. 1997. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J. Biol. Chem. 272:12738-12746. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, X., C. J. Herring, P. R. Romano, J. Szczepanowska, H. Brzeska, A. G. Hinnebusch, and J. Qin. 1998. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal. Chem. 70:2050-2059. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, L., S. van den Heuvel, K. Helin, A. Fattaey, M. Ewen, D. Livingston, N. Dyson, and E. Harlow. 1993. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7:1111-1125. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, L., E. Harlow, and B. D. Dynlacht. 1995. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 9:1740-1752. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, L., G. Enders, J. A. Lees, R. L. Beijersbergen, R. Bernards, and E. Harlow. 1995. The pRB-related protein p107 contains two growth suppression domains: independent interactions with E2F and cyclin/cdk complexes. EMBO J. 14:1904-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]