Abstract
Histone deacetylases (HDACs) are important for gene regulation and the maintenance of heterochromatin in eukaryotes. Schizosaccharomyces pombe was used as a model system to investigate the functional divergence within this conserved enzyme family. S. pombe has three HDACs encoded by the hda1+, clr3+, and clr6+ genes. Strains mutated in these genes have previously been shown to display strikingly different phenotypes when assayed for viability, chromosome loss, and silencing. Here, conserved differences in the substrate binding pocket identify Clr6 and Hda1 as class I HDACs, while Clr3 belongs in the class II family. Furthermore, these HDACs were shown to have strikingly different subcellular localization patterns. Hda1 was localized to the cytoplasm, while most of Clr3 resided throughout the nucleus. Finally, Clr6 was localized exclusively on the chromosomes in a spotted pattern. Interestingly, Clr3, the only HDAC present in the nucleolus, was required for ribosomal DNA (rDNA) silencing. Clr3 presumably acts directly on heterochromatin, since it colocalized with the centromere, mating-type region, and rDNA as visualized by in situ hybridization. In addition, Clr3 could be cross-linked to mat3 in chromatin immunoprecipitation experiments. Western analysis of bulk histone preparations indicated that Hda1 (class I) had a generally low level of activity in vivo and Clr6 (class I) had a high level of activity and broad in vivo substrate specificity, whereas Clr3 (class II) displayed its main activity on acetylated lysine 14 of histone H3. Thus, the distinct functions of the S. pombe HDACs are likely explained by their distinct cellular localization and their different in vivo specificities.
Heterochromatic regions of the chromosomes, such as the centromeres and telomeres, are specialized noncoding structures with important functions for genomic stability. Euchromatic regions, on the other hand, are the chromosomal domains in which a majority of the transcriptionally active genes are located. In general, heterochromatin contains underacetylated histone amino-terminal tails compared to euchromatin (22). The acetylation of histones is carried out by histone acetyl transferases (HATs) (5) at several conserved lysine residues of the N-terminal tails of histone H3 and H4. Important positions are K9 and K14 of histone H3 and K5, K8, K12, and K16 of histone H4. There are two protein families with histone deacetylase (HDAC) activity: the SIR2 family of NAD-dependent HDACs, the enzymatic activity of which was only recently discovered (20, 28), and the “classical” HDAC family. The “classical” HDAC family members fall into two different phylogenetic classes, namely class I and class II (14, 29). The HDAC enzyme was first purified by binding to trapoxin, an inhibitor that irreversibly bind to the active site of the enzyme (39). Subsequently, HDACs were shown to participate in large multiprotein complexes as corepressors requiring the HDAC enzymatic activity for repression of gene transcription (18, 19, 23, 27, 34). Generally HDAC enzymes work on acetylated histone substrates, but there are cases in which other proteins, such as p53, become deacetylated by HDAC1 (30). The structure of a sequence homologue of the HDAC family of proteins from the archaebacterium Aquifex aeolicus has been determined. It revealed that the conserved regions in HDACs form an active site with a Zn2+ atom in the center and, furthermore, that this site can be blocked by TSA (11).
The Schizosaccharomyces pombe putative HDACs are encoded by the hda1+, clr3+, and clr6+ genes (with clr for cryptic loci regulator) (13, 25, 36). The hda1+ gene was discovered due to its homology to RPD3 from Saccharomyces cerevisiae, while clr3+ and clr6+ were identified in screens aimed at identifying mutants deficient in mating-type silencing in fission yeast. Hda1 has been shown to have an HDAC activity (25). Genetic analysis of the S. pombe HDACs indicates functional divergence of the three proteins. The clr6+ gene is essential, while hda1Δ cells are viable (13, 25). Strains with mutations in clr3+ or clr6+ are silencing deficient, while hda1Δ results in strengthened silencing (10, 13, 36, 41). In addition, clr6+ is necessary for genomic stability, since a conditional mutation in the gene results in chromosome loss (13). This phenotype is not displayed by mutations in clr3+ or hda1+ (1, 36). It is tempting to seek mechanistic explanations for the observed and sometimes counterintuitive phenotypic differences within this enzyme family by using a simple model system.
In this study, we have compared the properties of three HDACs in S. pombe. We have investigated the localization of the proteins in the cell and established their in vivo enzymatic activities. Their importance for silencing of polymerase II (Pol II) marker genes inserted into the ribosomal DNA (rDNA) was examined. A more profound investigation of one of the enzymes, Clr3, reveals a major activity in Western analysis of bulk histone preparations for lysine K14 of histone H3. Clr3 acts directly on the silent mating-type chromatin, since it colocalizes with mat2/3 and can be cross-linked to the mating-type region in chromatin immunoprecipitation (ChIP) experiments.
MATERIALS AND METHODS
Strains and media.
The characteristics of the S. pombe strains used in this study are listed in Table 1 . Media were prepared according to reference 40. The hda1+, clr3+, and clr6+ genes were tagged at their endogenous locus, and the clr3+ gene was deleted by the method described in reference 2. Pom152-green fluorescent protein (GFP) was constructed according to reference 26. The resulting PCR products were integrated into the genome of the haploid strain FY367 or FY1180 or of the diploid strain Hu114. PCR and subsequent sequencing of the PCR products confirmed correct integration.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| FY367 | h+ura4-D18 leu1-32 ade6-M210 | 8 |
| FY648 | h+otr1R(SphI)::ura4+ura4-DS/E leu1-32 ade6-M210 | 1 |
| FY1180 | h+otr1R(SphI)::ade6+ura4-D18 leu1-32 ade6-M210 | 8 |
| Hu114 | h+/h−ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/M216 | This study |
| Hu268 | h−hda1-HA::kanMX6 ura4-D18 leu1-32 ade6-M210 | This study |
| Hu056 | h+clr3-myc::kanMX6 ura4-D18 leu1-32 ade6-M210 | This study |
| Hu397 | h+clr3-myc::kanMX6 pom152-GFP::kanMX6 ura4-D18 leu1-32 ade6-M210 | This study |
| Hu401 | h−clr6-HA::kanMX6 pom152-GFP::kanMX6 ura4-D18 leu1-32 ade6-M210 | This study |
| Hu067 | h−clr3-myc::kanMX6 clr6-HA::kanMX6 ura4-D18 leu1-32 ade6-M210 | This study |
| Hu415 | h−hdal-HA::kanMX6 clr3-myc::kanMX6 leu1-32 ura4-D18 ade6-M210 | This study |
| Hu386 | h+hda1-myc::kanMX6 clr6-HA::kanMX6 otr1R(SphI)::ade6+leu1-32 ura4-D18 ade6-M210 | This study |
| FY597 | h90 mat3::ura4+ura4-D/SE leu1-32 ade6-M210 | 1 |
| FY2606 | h90 hda1::LEU2+mat3-M::ura4+ura4-DS/E leu1-32 ade6-M210 | 36 |
| Hu427 | h90 clr3Δ::kanMX6 mat3-M::ura4+ura4-DS/E leu1-32 ade6-M210 | This study |
| Hu460 | h90 clr6-1 mat3-M::ura4+ura4-DS/E leu1-32 ade6-210 | This study |
| Hu393 | h+leu1/Ylp2.4pUCura4+-7 ura4-DS/E leu1-32 ade6-M216 | This study |
| Hu434 | h+hda1Δ::LEU2+leu1/Ylp2.4 pUCura4+-7 ura4DS/E leu1-32 ade6-M210 | This study |
| Hu395 | hA clr3Δ::kanMX6 leu1/Ylp2.4pUCura4+-7 ura4-DS/E leu1-32 ade6-M210 | This study |
| Hu451 | h+clr6-1 leu1/YIp2.4 pUCura4+-7 ura4DS/E leu1-32 ade6-M210 | This study |
| FY498 | h+imr1R(NcoI)::ura4+ura4-D/SE leu1-32 ade6-M210 | 1 |
| Hu383 | h+hda1::LEU2+imr1R(NcoI)::ura4+ura4-DS/E leu1-32 ade6-M210 | This study |
| Hu379 | hA clr3::kanMX6 imr1R(NcoI)::ura4+ura4-DS/E leu1-32 ade6-M210 | This study |
| SPG141 | h+clr6-1 imr1R(NcoI)::ura4+ura4-DS/E leu1-32 ade6-M210 | 13 |
| FY520 | h+ Ch16 m23::ura4+-TEL[72] ura4-D/SE leu1-32 ade6-M210 (Ch16 ade6-M216) | 1 |
| FY2602 | h+hda1::LEU2+ Ch16 m23::ura4+-TEL[72] ura4-D/SE leu1-32 ade6-M210 (Ch16 ade6-M216) | 36 |
| Hu377 | hA clr3Δ::kanMX6 Ch16 m23::ura4+-TEL[72] ura4-D/SE leu1-32 ade6-M210 (Ch16 ade6-M216) | This study |
| Hu454 | h−clr6-1 Ch16 m23::ura4+-TEL[72] ura4-D/SE leu1-32 ade6-M210 (Ch16 ade6-M216) | This study |
| Hu619 | h90 clr3-myc::kanMX6 mat3-M::ura4+ura4-D/SE leu1-32 ade6-M216 | This study |
(hA), mating type ambiguous because silent mating-type loci are deregulated in this background.
Construction of a phylogenetic tree.
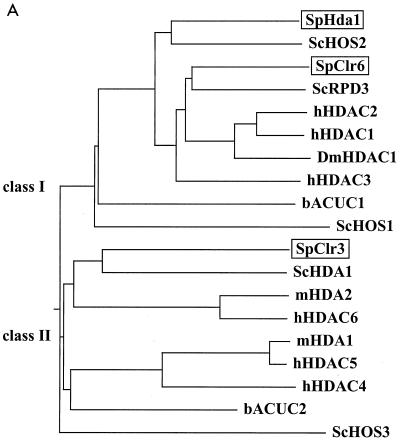
A ClustalW multiple alignment of HDACs was performed with Mac Vector 6.5 (Genetics Computer Group, Inc.) by using the “blosum” matrix and a gap penalty of 50.0. A total of 19 HDAC amino acid sequences were aligned. The resulting dendrogram showing the phylogenetic relationships is depicted in Fig. 1A, and the sequence alignment is presented in Fig. 1B. The following protein amino acid sequences (species and accession numbers follow in parentheses) were used: SpHda1 (S. pombe, AL021046), ScHOS2 (S. cerevisiae, X91837), SpClr6 (S. pombe, AFO64206), ScRPD3 (S. cerevisiae, P32561), hHDAC2 (Homo sapiens, NP_001518), hHDAC1 (H. sapiens, Q13547), DmHDAC1 (Drosophila melanogaster, AF026949), hHDAC3 (H. sapiens, AF039703), bACUC1 (A. aeolicus, D70388), ScHOS1 (S. cerevisiae, Z49219_23), SpClr3 (S. pombe, AFO64207), ScHDA1 (S. cerevisiae, Z71297), mHDA2 (Mus musculus, AF006603), hHDAC6 (H. sapiens, AF132609), mHDA1 (M. musculus, AF006602), hHDAC5 (H. sapiens, AF132608), hHDAC4 (H. sapiens, AF132607), bACUC2 (A. aeolicus, A70481), and ScHOS3 (S. cerevisiae, U43503).
FIG. 1.
S. pombe HDACs are members of both phylogenetic classes. (A) A dendrogram representing the phylogenetic relationships of the HDAC family was constructed by ClustalW alignment (see Materials and Methods). The species is indicated immediately before the protein name: for example, ScHDA1 (S. cerevisiae HDA1). Abbreviations: Hs, H. sapiens; Dm, D. melanogaster; b, A. aeolicus; Sc, S. cerevisiae; Sp, S. pombe; m, Mus musculus. The two phylogenetic classes are indicated in the figure. (B) Part of the sequence alignment showing the enzymatic core region, which is the conserved part of the HDAC proteins. Class I and class II are indicated. L3 to L7 are the loops that are known to form a pocket domain around the active site. The amino acid residues coordinating the Zn2+ ion in the catalytic site are indicated (solid diamonds). β4 is a β-sheet stretch in close proximity to the active pocket.
Coimmunoprecipitations (co-IPs) were carried out according to reference 17. Lysis of log-phase cells with glass beads and subsequent co-IPs were performed in radioimmunoprecipitation assay (RIPA) buffer or a nonionic general lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, Complete Protease Inhibitor Cocktail from Boehringer Mannheim). Preclearing and co-IP were conducted with protein A coupled to Sepharose beads. Immunoprecipitates were obtained with rabbit anti-myc (Research Laboratories), mouse antihemagglutinin (anti-HA) (Boehringer Mannheim), or rabbit anti-HA (Upstate) antibodies. The immunoprecipitated material was subjected to Western analysis (17).
Immunofluorescence microscopy.
S. pombe cells were prepared for immunofluorescence microscopy according to the formaldehyde fixation procedure with some modifications (16). Log-phase cultures were incubated for 5 to 30 min in yeast extract with supplements (YES) plus 1.2 M sorbitol before harvest. In most cases, PEMAL (PEM [100 mM PIPES, 1 mM EGTA, 1 mM MgSO4, pH 6.9] plus 5% or 0.03% milk, 0.1 M l-lysine HCl, cleared by 30 min of centrifugation at 20,000 × g) was used instead of PEMBAL (PEM plus 1% bovine serum albumin [essential fatty acids and globulin-free sigma] and 100 mM lysine hydrochloride, pH 6.9). The following primary antibodies were used: rabbit anti-myc, mouse anti-myc (Sigma), mouse anti-HA, mouse anti-GFP (Molecular Probes), and rabbit anti-Nop1 (21). Clr6 antibodies were raised in rabbits against the N-terminal peptide MGFGKKKVSYFYD and C-terminal peptide IAKEEFTIMDERV. The sera were affinity purified against the peptides, mixed 1:1, and used for immunostaining. Fluorescein isothiocyanate- or Texas red-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories or Sigma. Fluorescence in situ hybridization (FISH) experiments were carried out with the pRS314 probe to detect centromeres and the probe for rDNA loci according to reference 9. The c1555 probe was used to detect the entire mat2/3 region in S. pombe (33). Cells were visualized with a Zeiss Axioskop II microscope equipped with a Hamamatsu C4742-95 charge-coupled device camera. A z-series digital confocal deconvolution analysis with Openlab software, version 2:09 (Improvision), was performed with 0.2- to 0.3-μm sample z-series spacing and nearest-neighbor deconvolution. An object magnification of ×100 and a lens aperture of 1.4 were used.
Western analysis of bulk histones.
S. pombe strains were grown at 36°C overnight, and crude histones were prepared, separated, and Western blotted according to reference 8. Antibody incubation was performed according to manufacturer's (Upstate) instructions. Antibodies against histone H3 acetylated at position K9 or K14 or histone H4 acetylated at position K8 or K12 were purchased from Upstate. Antibodies against histone H4 acetylated at positions K5 and K16 were as described by reference 43 or purchased from Serotec. An antibody against the C-terminal part of histone H3 was used (A.Verreault, unpublished data). The band intensity was quantified with Fujifilm Image Gauge software. The signal was normalized to the loading control anti-histone H3 or Coomassie staining of the histones.
ChIPs.
Chromatin immunoprecipitations (ChIPs) were performed according to reference 9 with antibodies against histone H3 acetylated at position K9 or K14 or against histone H4 acetylated at position K8 or K12 (Upstate) or with mouse antibodies directed against the myc epitope (Sigma). The immunoprecipitated DNA was amplified and labeled with 5′-GAGGGGATGAAAAATCCCA-3′ and 5′-TTCGACAACAGGATTACGAC-3′ as ura4 primers in a PCR with [α-32P]dCTP. PCR products were separated on 4% polyacrylamide gels, detected with Fujifilm Phosphorimager FLA3000, and quantified with Fujifilm Image Gauge software.
RESULTS
Phylogenetic relationships between HDACs.
A database search of the S. pombe genome (www.sanger.ac.uk/Projects/S_pombe/) was performed to investigate the total number of putative HDACs in the organism. The entire S. pombe genome (pompep) protein database including 4,952 protein sequences was BLAST-searched with hHDAC1. The search identified the three previously described HDAC homologues of S. pombe, Hda1, Clr3, and Clr6, and did not reveal any additional S. pombe members of this HDAC family. The three amino acid sequences were aligned together with HDAC proteins from other organisms, and a phylogenetic tree was constructed. As expected from previous studies (29), the resulting dendrogram of HDAC sequences formed two major groups, class I and class II (Fig. 1A). It was clear that the two classes were older than the divergence into three kingdoms (Archae, Eubacteria, and Eukarya), since the two archaebacterial sequences from the same species (A. aeolicus) were present with one in each class (see bACUC1 and bACUC2). The S. pombe HDAC homologues also appeared in both phylogenetic classes. Hda1 and Clr6 belong together with RPD3 of S. cerevisiae and HDAC1, -2, and -3 from humans in class I, while Clr3 belonged to class II together with HDA1 from S. cerevisiae and HDAC4, -5, and -6 from humans. The sequence alignment (Fig. 1B) revealed drastic conserved differences between the class I and class II enzymes. The differences were particularly clear in the L3, L4, L5, L6, and L7 loops and in the adjacent β4 region, which form a pocket domain where the active site has been determined to be, in the HDAC homologue bACUC1 from the archaebacterium A. aeolicus (11).
Distinct localization patterns of the S. pombe HDACs
In order to further investigate the three S. pombe HDACs, their gene products were epitope tagged at their respective loci under the control of their own endogenous promoters. Hda1 and Clr6 were tagged with the HA epitope, and Clr3 was tagged with the myc epitope. Western analysis of the epitope-tagged strains showed that the migration of the proteins corresponded to their predicted sizes: the class I proteins, Hda1 and Clr6, with smaller sizes of 48 and 45 kDa, respectively, and the larger class II protein, Clr3, of 76 kDa (data not shown).
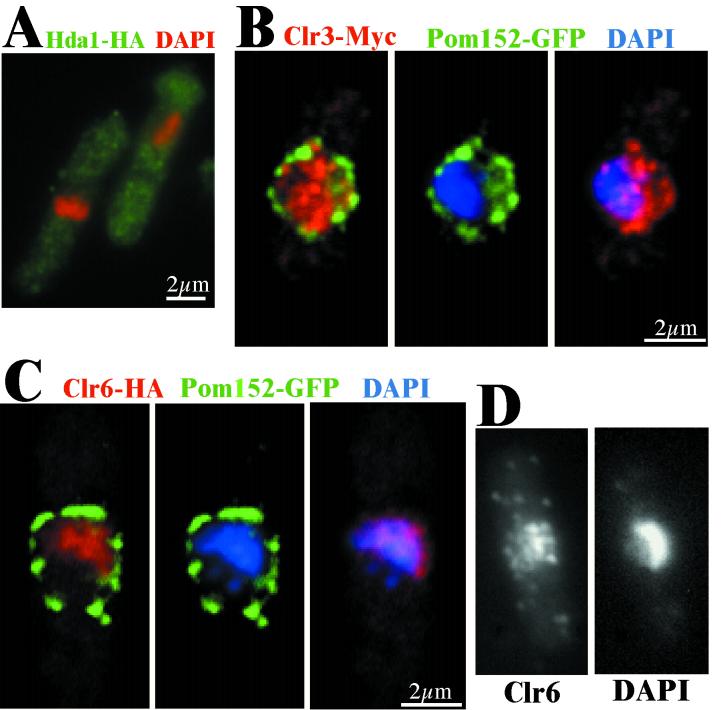
The tagged strains were used to investigate the subcellular localization of the three S. pombe HDAC proteins by indirect immunofluorescence microscopy. The immunofluorescence analysis clearly showed that Hda1-HA was a cytoplasmatic protein throughout the cell cycle (Fig. 2A) (data not shown), while Clr3 and Clr6 localized to the nucleus. To facilitate a more detailed localization study of the nuclear proteins, the nuclear membrane was labeled by tagging the Pom152 protein with GFP. Pom152 is an S. pombe homologue of S. cerevisiae POM152, which is a previously described component of the nuclear pore complex (6, 44). Double immunofluorescence labeling and z-series digital confocal deconvolution analysis of Clr3-myc and Pom152-GFP revealed that Clr3-myc was predominantly nuclear with Clr3-myc foci occupying the entire nuclear space, including the 4′,6′-diamidino-2-phenylindole (DAPI)-stained chromosomes and the nucleolus (Fig. 2B). The Clr3-myc signal was directly adjacent to and occasionally overlapping with the Pom152-GFP signal (Fig. 2B [Clr3-myc and Pom152-GFP merged]), indicating that Clr3-myc also occupied a region at the nuclear periphery. In addition, a subpopulation of Clr3-myc could be found in the cytoplasm of the cell (Fig. 2B; see Fig. 4B). A similar analysis was carried out with Clr6-HA and revealed that Clr6-HA was localized in the nucleus with foci exclusively on the DAPI-stained chromosomes (Fig. 2C). The deconvolution analysis revealed a Clr6-HA free space at the nuclear periphery close to the Pom152-GFP signal (Fig. 2C). The chromosomal localization of the HA-tagged Clr6 protein was confirmed by raising antibodies against the Clr6 protein and using them in indirect immunofluorescence (Fig. 2D). Staining of wild-type cells with the antibodies against Clr6 showed the same spotted localization pattern on the chromatin as the Clr6-HA strain did. As controls, an untagged wild-type strain was incubated with antibodies against the tags used. No signal was detected in these control experiments, indicating that the detected signals truly reflected the localization of the epitope-tagged proteins (data not shown).
FIG. 2.
Each of the HDACs in S. pombe has a distinct characteristic cellular localization pattern. Cells of the indicated strains were processed for immunofluorescence microscopy. (A) The strain Hu268 (hda1-HA) was stained with DAPI (red) and HA antibodies (green). Hda1-HA (green) is located primarily in the cytoplasm. (B) The strain Hu397 (clr3-myc pom152-GFP) was stained with anti-myc (red) and anti-GFP (green) antibodies and DAPI (blue). The Clr3-Myc pattern colocalizes with the DNA (blue), but is not restricted to the chromatin-containing regions of the nucleus. (C) The strain Hu401 (clr6-HA pom152-GFP) was stained with anti-HA (red) and anti-GFP (green) antibodies and DAPI (blue). The Clr6-HA (red) foci were restricted to the chromatin and thus excluded from the perinuclear region, surrounded by the Pom152-GFP counter stain. (D) The strain FY648 (wild type) was stained with anti-Clr6 antibodies and DAPI. (B, C, and D) The immunofluorescence microscopy images were deconvolved (z = 0.3 μm) and merged.
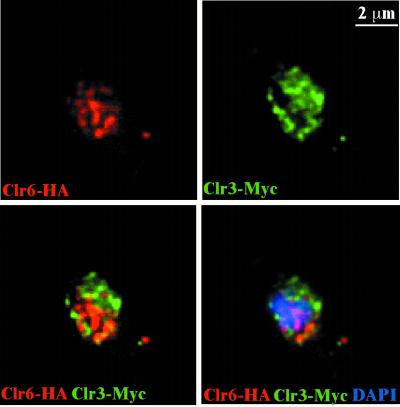
We also performed double immunofluorescence localization experiments and z-series digital confocal deconvolution analysis on a strain epitope tagged with both Clr3-myc and Clr6-HA. This three-dimensional analysis revealed little overlap between the Clr6 and Clr3 signals (Fig. 3). The localization data suggested that there was little or no physical interaction between the three S. pombe HDACs. To further test this possibility, strains carrying pairs of epitope-tagged S. pombe HDACs were subjected to co-IP experiments. If the HDACs were part of the same complexes, they would most likely be detected in reciprocal immunoprecipitates by using the myc and HA epitopes. None of the HDACs underwent co-IP with any of the other HDACs; instead they remained in the supernatants (data not shown). Taken together, these immunolocalization and co-IP experiments indicate that the S. pombe HDACs are not physically associated and, furthermore, they occupy different cellular compartments.
FIG. 3.
Clr6 and Clr3 nuclear foci do not coincide. The strain Hu067 (clr6-HA clr3-myc) was processed for immunofluorescence microscopy and stained with anti-HA (red) and anti-myc (green) antibodies and DAPI (blue). The spotted nuclear patterns of the Clr6-HA (red) and Clr3-Myc (green) rarely colocalized completely (yellow) after deconvolution (z = 0.3 μm).
Clr3 acts directly in mating-type silencing.
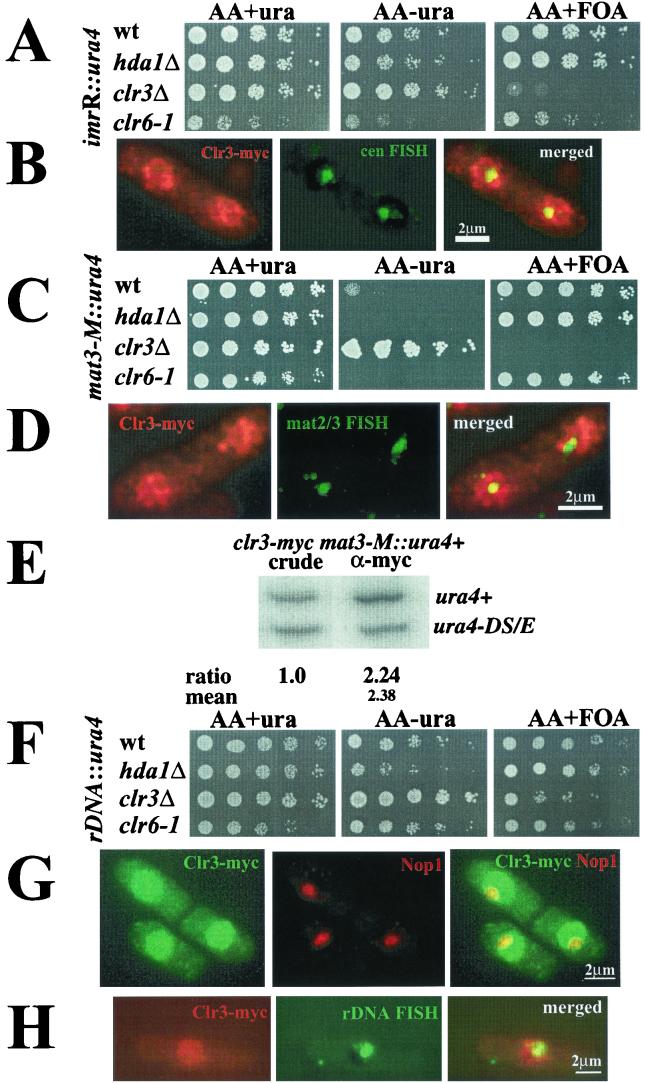
To further investigate the function of the clr3+ gene, a complete null allele was constructed. The resulting haploid clr3::kanMX6 (clr3Δ) cells were viable, showing that clr3+ is not an essential gene. To determine whether the clr3Δ null allele had the same effects on silencing as previously described for the clr3-E36 mutation (1), the clr3Δ marker was combined with a ura4+ reporter gene inserted into the different heterochromatic regions of S. pombe (centromere, telomere, and mating-type regions) (1). The strains were subjected to a silencing assay in which growth on plates with or without uracil and on plates containing the drug fluoroorotic acid (FOA), was compared. Strains whose sole functional copy of the ura4+ gene resides either in the centromere region, imrR(NcoI)::ura4+, or in the mating-type region, mat3-M::ura4+, grow poorly on medium lacking uracil and grow well on the FOA-containing plates due to the efficient repression of the ura4+ reporter gene at these locations (Fig. 4A and C, top row). In the clr3Δ genetic background, the imrR(NcoI)::ura4 marker gene was derepressed, so cells grew better on plates lacking uracil as compared to the wild type and could not form colonies on FOA (Fig. 4A, third row). In the clr3Δ genetic background, the mat3-M::ura4+ marker gene was strongly derepressed, resulting in full growth on plates lacking uracil, and no growth on FOA-containing plates (Fig. 4C, third row). Silencing of a telomeric marker gene was also investigated, and we found that the clr3Δ allele had a slight effect on telomeric silencing. All of these silencing effects were in accordance with the phenotype previously reported for the clr3-E36 allele (1). For comparison, silencing of the imrR(NcoI)::ura4 and mat3-M::ura4+ marker genes in strains with the hda1Δ or clr6-1 mutation were monitored on the same plates under semirestrictive conditions (30°C) for the clr6-1 temperature-sensitive allele. These strains displayed wild-type or slightly enhanced repression levels in agreement with phenotypes reported earlier (13, 36).
FIG.4.
Clr3 is necessary for silencing of the mating-type region and the rDNA. (A, C, and F) Silencing assays. Cell suspensions were fivefold serially diluted, and each dilution was spotted onto the indicated medium and incubated for 3 days at 30°C. (A) The strains FY498 (wild type [wt]), Hu383 (hda1Δ), Hu379 (clr3Δ), and SPG141 (clr6-1) have the ura4+ gene inserted into the outer repeats of the centromere [imrR(NcoI)::ura4+]. (C) The strains FY597 (wt), FY2606 (hda1Δ), Hu427 (clr3Δ), and Hu460 (clr6-1) have the ura4+ gene inserted next to the mating-type region (mat3-M::ura4+). (F) The strains Hu393 (wt), Hu434 (hda1Δ), Hu395 (clr3Δ), and Hu451 (clr6-1) have the ura4+ gene inserted into the rDNA repeats (rDNA::ura4+). (B, D, and H) FISH experiments with Clr3-myc. Clr3-myc were processed for immunofluorescence microscopy and subjected to FISH. (B) The pRS314 cosmid was used to detect the centromere (green); Clr3-myc appears red. (D) The c1555 probe was used to detect the mat2/3 region (green); Clr3-myc appears red. (H) The rDNA probe was used for FISH (green); Clr3-myc appears red. The pictures were deconvolved (z = 0.2 μm) and merged. Yellow color indicates colocalization. (E) ChIP of Clr3-myc to the mat3-M::ura4+ marker. The strain Hu619 was subjected to ChIP. (G) Hu56 cells were stained against Clr3-myc (green) and (G) Nop1 (red). Pictures were deconvolved (z = 0.2 μm) and merged.
To investigate whether the effects on centromeric and mating-type silencing correlated with the Clr3 protein directly localizing to these regions, fluorescence in situ hybridization (FISH) experiments were carried out. A centromere probe was used to detect the cen region (green) in cells stained for the Clr3-myc fusion protein (red). The Clr3-myc and cen-FISH double-labeled cells were subjected to z-series digital confocal deconvolution analysis (Fig. 4B). The yellow color in merged deconvolved images indicated that Clr3 indeed is present in the cen region. The cosmid c1555, which detects the mat2/3 region in S. pombe (33), was used on cells stained for the Clr3-myc fusion protein. These double-labeled cells were also subjected to z-series digital confocal deconvolution analysis (Fig. 4D). It was evident in the merged picture that the mat2/3-FISH signal (green) partially colocalized with Clr3-myc (red), resulting in a yellow color. To further test if Clr3 was bound to mat2/3 chromatin, a ChIP experiment was carried out (Fig. 4E). To this end, a strain with the ura4+ reporter gene inserted into the mating-type region, mat3-M::ura4, was used. This strain also had a second reporter gene consisting of a mini-ura4 allele, ura4-DS/E, at the endogenous, euchromatic ura4 locus. The ura4-DS/E allele functions as an internal control for the PCR analysis of the DNA recovered from each ChIP experiment (8, 9). In the first lane in Fig. 4E, the PCR products from the input of the crude chromatin lysate are shown. The larger band corresponded to the full-length ura4+ gene, and the smaller band corresponded to the internal control, ura4-DS/E. The ratio between the two bands in the crude extracts was set to 1.0. The ratios between the inserted ura4+ and ura4-DS/E genes were normalized to the true ratios of the input crude extract and therefore reflected the relative levels of chromatin immunoprecipitated with the myc antibody. The myc antibody selectively immunoprecipitated chromatin from the mating-type region, resulting in a difference in intensity greater than twofold between the PCR product made from the full-length heterochromatic band and the small euchromatic ura4 band. Taken together, these experiments demonstrate a direct role for Clr3 in the formation of the heterochromatin in the mating-type region.
Clr3 is necessary for rDNA silencing.
To explore whether any of the three HDACs were involved in rDNA silencing of Pol II-transcribed genes in S. pombe, mutations in the HDACs were separately combined with a ura4+ reporter gene integrated into the rDNA arrays (rDNA::ura4+) (42). The resulting strains were compared in the growth assay described above. The wild-type control strain displayed repression of rDNA::ura4+ (Fig. 4F, top row). The repression of rDNA::ura4+ was observed as a 5- to 25-fold-reduced growth on plates lacking uracil and strong growth on FOA-containing plates. The hda1Δ mutation resulted in enhanced silencing of rDNA::ura4+, as revealed by a fivefold-reduced growth on the plate lacking uracil in comparison to that of the wild type (Fig. 4F, first and second rows). This result was not surprising, since the hda1Δ allele had previously been reported to enhance telomeric, centromeric, and mating-type silencing (36). The clr6-1 mutation did not significantly affect expression of rDNA::ura4+ in comparison to wild-type cells (Fig. 4F, bottom lane). The clr3Δ mutation, however, resulted in a strong derepression of rDNA::ura4+, leading to increased growth on plates lacking uracil and a 25-fold reduction in growth on plates containing FOA compared to the wild-type control (Fig. 4F, first and third rows). Thus, Clr3 seemed to be the only one of the S. pombe HDACs tested that was required for silencing of Pol II-transcribed genes inserted into the rDNA repeats. This finding prompted us to investigate the localization of Clr3 in relation to the nucleolus. To this end, we used an antibody against Nop1 (21). The Nop1 protein had previously been described as a nucleolar marker in fission yeast (12). Cells were subjected to indirect immunofluorescence with double labeling of the Clr3-myc fusion protein and Nop1 and z-series digital confocal deconvolution analysis. Although there was less Clr3-myc in the nucleolus than in the rest of the nucleus, the merged deconvolved picture clearly demonstrated a nucleolar localization for a subpopulation of the Clr3-myc protein (Fig. 4G). The presence of Clr3 in the nucleolus was confirmed in a FISH experiment involving an rDNA probe together with Clr3-myc labeling, which resulted in a partial yellow colocalization signal in the merged picture (Fig. 4H). Thus, the silencing and localization studies showed that Clr3 was important for keeping the rDNA regions repressed for Pol II transcription, possibly by directly localizing to the rDNA in the nucleolus.
Differences in the in vivo deacetylation activity of the HDACs.
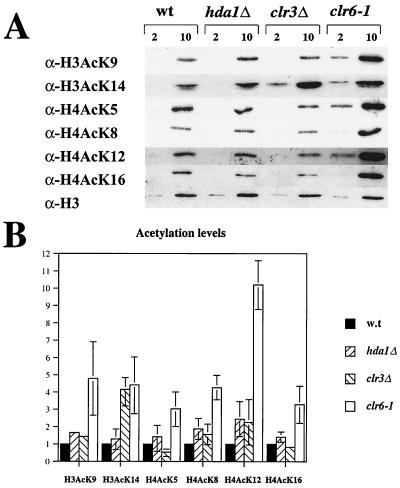
To determine whether the putative HDACs affect histone acetylation in vivo, a Western analysis of bulk histones was undertaken (8). This assay measures differences in acetylation levels of specific lysine residues for the entire genome in vivo. Bulk histones were prepared from wild-type cells and from strains carrying hda1Δ, clr3Δ, or clr6-1 mutations and submitted to a Western blot analysis with antibodies against specifically acetylated histone H3 or H4 (Fig. 5A). A mutated HDAC was expected to cause an increase, in comparison with wild type, of the signal corresponding to the acetyl group that the enzyme normally removes. The bands were either normalized to the signal from total histone H3, measured with an antibody against the histone H3 C terminus (Fig. 5A, bottom row), or normalized to Coomassie staining of the histones. The measurements were repeated two or three times, and the mean and range of the relative differences in intensity of the wild type and the mutants of each acetylated lysine position were determined (Fig. 5B). The hda1Δ mutant did not show a striking change in bulk acetylation levels compared to the wild-type control. In contrast, the clr6-1 temperature-sensitive mutant under restrictive conditions (36°C) gave strongly elevated acetylation levels of all lysines tested at the histone H3 and H4 amino-terminal tails (Fig. 5A, compare the first and the last lanes; and B, white bar). This observation was consistent with the in vitro activity of the purified Clr6 enzyme, which was found to be active on both acetylated histone H3 and histone H4 substrates (P. Bjerling, K. Ekwall, and S. Grewal, unpublished data). Histones prepared from a clr3Δ strain displayed little variation with respect to the wild-type strain. However, at the one position affected, namely lysine 14 of histone H3, a 4.5-fold increase in the acetylation level compared to that of the wild-type control was detected (Fig. 5A, second row, lanes 1 and 3; and B, third bar). This increase is statistically significant (P < 0.05, one-sample t test). Hence, we conclude that among the S. pombe HDACs, the class I enzyme, Clr6, displays the broadest in vivo specificity, while the class II enzyme, Clr3, displays the most narrow specificity, with a strong preference for acetylated K14 of histone H3.
FIG. 5.
The acetylation pattern in bulk histone preparations of the wild type and HDAC mutants. (A) Two and 10 μg of crude histone preparations from the strains FY498 (wild type [wt]), Hu383 (hda1Δ), Hu379 (clr3Δ), and SPG141 (clr6-1) were separated by SDS-PAGE, blotted and the blots were incubated with antibodies specific for the histone variants indicated to the left of the picture. At the bottom, a Western blot with an antibody against the C terminus of histone H3 is shown as a loading control. (B) The mean and range of the intensity of the bands in two to three experiments are shown.
Deletion of clr3+ results in increased acetylation levels at the mating-type region and the rDNA.
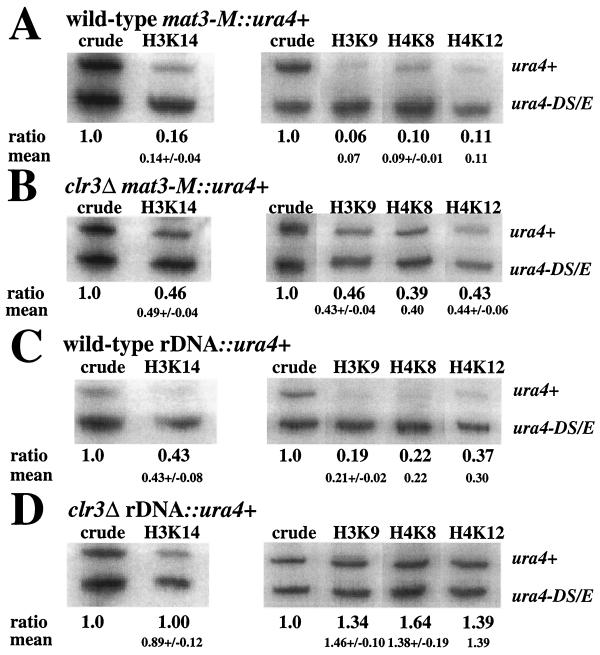
To investigate whether the mating-type region and the rDNA repeat were hypoacetylated and whether the enzymatic activity of Clr3 was necessary to maintain a low acetylation level in these regions, we utilized a ChIP assay, which measures acetylation of specific chromatin targets. The assay was performed with antibodies against histone H3 acetylated at K9 or K14 or antibodies against histone H4 acetylated at K8 or K12. Chromatin extracts were prepared from strains with the ura4+ reporter gene inserted into the chromosomal region of interest and a second reporter gene consisting of a mini-ura4 allele, ura4-DS/E, at the endogenous, euchromatic ura4 locus. Since the clr3Δ mutation was shown to mainly affect silencing at the mating-type region and at the rDNA (Fig. 4), the ChIP assays were performed with strains having the ura4+ reporter gene inserted into either of these domains. Figure 6 shows the results of the ChIP assays. In the first lanes, the PCR products from the input of the crude chromatin lysate are shown. Acetylation levels at all positions tested (histone H3 K9 and K14 and histone H4 K8 and K12) were low in mat3-M::ura4+ chromatin, resulting in ratios between the full-length ura4+ gene and the mini ura4-DS/E gene of 0.07 to 0.14 (Fig. 6A). In contrast, when mat3-M::ura4+ was combined with the clr3Δ mutation, the resulting acetylation levels increased to 0.40 to 0.49 (Fig. 6B). The rDNA region had a generally higher basal acetylation level, with the ChIP measurements of rDNA::ura4+ in a wild-type strain background giving ratios in the range 0.21 to 0.43 (Fig. 6C). The acetylation level of rDNA::ura4+ was further increased in the clr3Δ genetic background to ratios ranging from 0.89 to 1.46 (Fig. 6D). Thus, when the Clr3 enzyme is missing from the cells, the histone acetylation levels increase at least twofold in both the mating-type region and the rDNA repeats at all of the lysine residues tested.
FIG. 6.
clr3Δ cells exhibit elevated acetylation levels at several lysines in H3 and H4 in the mating-type region and at the rDNA regions. DNA from crude chromatin preparation (crude) and immunoprecipitated (ChIP) chromatin of the indicated strains was amplified by radioactive PCR. The normalized ratios between the two variants of the ura4+ gene (ura4+ and ura4-DS/E) are shown below each lane, and the mean value of duplicate or triplicate experiments is shown underneath. The full-length ura4+ gene was either inserted (A) next to the mating-type region (mat3-M::ura4+) in strains FY597 (wild type) and (B) Hu427 (clr3Δ) or (C) in the rDNA (rDNA::ura4+) in strains Hu393 (wild type) and (D) Hu395 (clr3Δ).
DISCUSSION
Distinct localization and in vivo specificity of HDACs in S. pombe
There are three members of the “classical” HDAC family in S. pombe. Previous studies indicated distinct functions for the three enzymes, since the mutants displayed different phenotypes (1, 13, 25, 36). In this study, we reveal some potential explanations for the functional divergence of S. pombe HDACs. First, we show that the S. pombe enzymes belong to evolutionarily distinct classes of HDACs and that these two classes have different conserved amino acid sequences in their L3-L7 loops and in their β4 regions, which together are known to form the active site (11). Next, we demonstrate that the enzymes have different cellular localization patterns. Clr3 and Clr6 were both observed in the nucleus, but in different subnuclear locations. Clr6 was found exclusively on the chromatin, while Clr3 occupied the entire nuclear space, including the chromatin, the nuclear periphery, and the nucleolus. Hda1, on the other hand, was found in the cytoplasm of the cell. In terms of the in vivo specificities, Western analysis of bulk histone preparations indicated that the class I enzyme Clr6 acted on both histones H3 and H4, whereas the class II enzyme Clr3 had a major effect on histone H3 acetylated at position K14. Finally, we demonstrate that clr3Δ is the only one of the three HDACs that is required for rDNA silencing. Thus, the different functions of the S. pombe HDACs can, at least in part, be explained by different cellular localizations and enzymatic activities.
The main in vivo activity of Clr3 is to deacetylate position K14 of histone H3.
We have presented a more detailed analysis of one of the HDACs, namely Clr3. First we showed that clr3+ was not an essential gene by deleting its entire open reading frame. Second, we established that Clr3 was indeed an HDAC, and not only a sequence homologue of known HDACs. Our evidence by Western analysis of bulk histone preparations clearly showed that the clr3Δ null allele caused elevated acetylation levels at histone H3 K14 (Fig. 5).
Furthermore, we demonstrated that the deacetylation of position K14 of histone H3 was crucial for maintaining the heterochromatin structure at the mating-type region and rDNA and that Clr3 was localized to these regions (Fig. 4). Taken together, these results strongly argue for a direct role of Clr3 in keeping the mating-type region and the rDNA repeats silenced by deacetylation of position K14 of histone H3.
ChIP analysis indicated that the acetylation of lysines other than K14 of histone H3 is also increased in the clr3Δ mutant, as observed in chromatin associated with the ura4+ reporter gene inserted into the rDNA repeats or the mat2/3 region (Fig. 6). The discrepancy between acetylation levels of these specific target genes and those of the bulk histone estimated by Western blot analysis in clr3Δ cells may be explained by assuming that the broad substrate specificity of Clr3 limited to small chromosomal regions that would not lead to an overall increase in acetylation in the bulk measurements. An alternative explanation is that Clr3 may have narrow substrate specificity for deacetylation of lysine K14 on histone H3 and that this activity would be required for subsequent deacetylation events in forming the heterochromatin of these regions. It has been shown that the clr6-1 mutant alone has a minor defect in mat2/3 silencing, whereas the clr3-735 and clr6-1 double mutant shows a strong synergistic reduction of silencing at mat2/3 (13). The synergism between clr3-735 and clr6-1 indicates that these two enzymes jointly keep the mat2/3 region underacetylated. This could explain the discrepancy between our analysis of the enzyme activities in bulk histone preparations (Fig. 5) and the ChIP data of mat2/3 chromatin in clr3Δ cells presented in Fig. 6. These results would also be consistent with a general model for formation of heterochromatin at mat2/3 suggested by reference 35. In this pathway, Clr3 would be required to deacetylate H3AcK14 prior to a more general deacetylation of H3 and H4 by Clr6 allowing for a methylation of H3K9 by Clr4 and subsequent binding of Swi6. The observed differences between the clr6-1 and clr3Δ mutants in the bulk histone preparations could thus be due to the fact that the regions of heterochromatin, where Clr3 and Clr6 activities may be linked, are relatively small as compared to the total S. pombe genome size. Presumably Clr6 and Clr3 functions are not dependent on each other in gene regulation in most other parts of the genome.
Silencing of the heterochromatic regions versus rDNA silencing of Pol II-transcribed genes.
Clr3 is shown to be necessary both for mating-type silencing and silencing of the rDNA. Studies in S. cerevisiae have shown that some of the factors needed for heterochromatic silencing are also important for rDNA silencing, while other factors are unique for one or the other chromatin (37). A similar picture is beginning to emerge in S. pombe. It has been reported previously that the chromo-domain proteins Swi6 and Chp2 and the methyltransferase Clr4 also influence silencing of both heterochromatin and rDNA (42). We show here that the function of Clr3 is also shared between the regions of heterochromatin and the rDNA repeats.
Clr3 localizes to the nucleolus, where it is involved in rDNA silencing. However, the function of rDNA silencing is not known in S. pombe. In S. cerevisiae, defects in rDNA silencing, for example, mutations in SIR2, lead to premature aging (15). In budding yeast, aging is easily monitored by the fraction of old cells with many bud scars in the cell population. In S. pombe, however, such an analysis is not feasible, since fission yeast divides by binary fission and bud scars are not easily recognizable.
A cytoplasmatic localization of Hda1.
The observed strengthening of silencing in an S. pombe hda1Δ strain background is not easily interpreted, since this mutation did not cause significant changes in the histone acetylation, as measured by ChIP at these loci (K. Ekwall, unpublished data). Hda1 could enhance silencing indirectly by affecting the transcription of a silencing factor or the formation of a specific acetylation pattern of histones in heterochromatin that was not detected in our bulk analysis. A recent paper discusses both of these mechanisms as a possible cause for obtaining enhanced silencing in an S. cerevisiae rpd3Δ strain (3). The unexpected cytoplasmic localization of Hda1 suggests a third possible explanation. Since Hda1 is found in the cytoplasm throughout the cell cycle, the protein is not in the same compartment as the heterochromatin, and thus the lack of Hda1 must indirectly influence the heterochromatin. Hda1 may counteract the cytoplasmic B-type HATs, creating a balance that optimizes the level and pattern of acetylation of the histones for nuclear import and nucleosome assembly. There is some evidence that the acetylation patterns are crucial for import and assembly. For example, when the N-terminal tail of histone H3 is removed, histone H4 needs to be acetylated at one or more of the positions K5, K8, or K12 for nucleosome deposition onto newly replicated DNA to occur (31). It is also possible that the cytoplasmic localization of Hda1 is not permanent, but that the protein is, perhaps transiently, translocated into the nucleus under certain conditions. Cytoplasmic localization of HDACs has also been reported for human HDAC4 and -5. These HDACs have been shown to move between cytoplasm and nucleus with the help of 14-3-3 proteins, where they repress MEF2-regulated genes (14, 32). It will be interesting to test whether Hda1 protein can be translocalized into the nucleus under particular conditions. Yet another possible function of Hda1 is that the enzyme might deacetylate nonhistone proteins similar to HDAC1 regulation of p53 in human cells by deacetylation (30).
Comparison of S. pombe and S. cerevisiae HDACs of class I and class II.
We have concluded that the main in vivo activity of Clr3 is to reduce acetylation level at histone H3 lysine 14. The closest relative to Clr3 in S. cerevisiae is HDA1 (Fig. 1A). ChIP assays on an S. cerevisiae strain lacking HDA1 shows a drastic increase in acetylation of histone H3 and H2B at the ENA1 promoter, while histone H4 is unaffected (45). Assuming that changes at the ENA1 promoter reflect the general activity of HDA1, it is possible that both of these class II enzymes primarily act on histone H3 in vivo.
In the clr6-1 mutant background, the in vivo acetylation levels were strongly increased at all positions examined in both histones H3 and H4. The closest homologue to Clr6 in S. cerevisiae is Rpd3. An rpd3 mutant strain exhibits increased acetylation levels at all lysine positions in histones H3, H4, H2A, and H2B at the INO1 promoter (38). Thus, both the class I HDAC enzymes SpClr6 and ScRPD3 seem to show a broad in vivo specificity. These differences in enzymatic activity between class I and class II enzymes might be the result of striking differences in their amino acid sequences observed between the two classes in the L3-L7 loops and the β4 region of the enzymatic core (Fig. 1B) (11). Perhaps these differences in the primary sequences reflect differences in the tertiary structure resulting in a more stringent substrate binding capacity for the class II enzymes.
It remains to be seen to what extent these similarities within the different classes of HDACs reflect conserved functions in the different organisms. Interestingly, the closest homologue to S. pombe Hda1 is S. cerevisiae HOS2. A deletion of S. pombe hda1Δ causes a defect in sporulation, a defect also displayed by an S. cerevisiae strain deleted for hos2Δ (4, 36). Thus, some regulated target genes of these two proteins might be shared between the two distantly related yeasts.
Nuclear foci for gene repression.
In this study, we demonstrated that both Clr3 and Clr6 HDAC had spotted localization patterns in the nuclei of S. pombe cells. This finding is not without precedent, since studies with mammalian cells also revealed such spotted localization patterns for HDAC1, HDAC5, and HDAC7 (7, 24). The HDAC1 protein forms heterochromatic foci in lymphocytes, which contain, among other proteins, the Ikaros protein and the Mi2 chromatin remodeling factor (24). In lymphocytes, developmental genes are regulated by selective recruitment to Ikaros foci leading to gene repression. Perhaps a similar mechanism with repression foci operates in the simple S. pombe system.
Taken together, our results indicate that HDACs have evolved highly divergent cellular functions. The distinct localization and specificity for different lysine residues of histone H3 and H4 of the different HDACs underlie the different phenotypes they display when mutated. Perhaps the explanation for the differences in enzymatic activity lies in the differences in amino acid sequences of the pocket domain between class I and class II HDACs.
Acknowledgments
We thank B. Turner for kindly providing us with antibodies against histone H4 acetyl K5 and K16. We also thank E. Hurt and A. Mutvei for sharing the Nop1 antibody and A. Verreault for kindly providing us with the antibody against the C terminus of histone H3.
Work in the K.E. laboratory was supported by the following grants: MFR K2000-31X-12562, NFR 0990-302, and CF 4284-B99. G.T. acknowledges support from the Novo Nordisk Foundation and from the Danish Natural Science Research Council. The S.G. laboratory was funded by a grant from NIH (RO1 GM 59772-01A1).
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilsland, E., M. Dahlen, and P. Sunnerhagen. 1998. Genomic disruption of six budding yeast genes gives one drastic example of phenotype strain-dependence. Yeast 14:655-664. [DOI] [PubMed] [Google Scholar]
- 5.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 6.Ding, D. Q., Y. Tomita, A. Yamamoto, Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5:169-190. [DOI] [PubMed] [Google Scholar]
- 7.Downes, M., P. Ordentlich, H. Y. Kao, J. G. Alvarez, and R. M. Evans. 2000. Identification of a nuclear domain with deacetylase activity Proc. Natl. Acad. Sci. USA 97:10330-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 9.Ekwall, K., and J. F. Partridge. 1999. Fission yeast chromosomal analysis: fluorescent in situ hybridisation (FISH) and chromatin immunoprecipitation (CHIP), p. 39-57. In W. A. Bickmore (ed.), Chromsome structural analysis: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 10.Ekwall, K., and T. Ruusala. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finnin, M. S., J. R. Donigian, A. Cohen, V. M. Richon, R. A. Rifkind, P. A. Marks, R. Breslow, and N. P. Pavletich. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188-193. [DOI] [PubMed] [Google Scholar]
- 12.Freeman-Cook, L. L., J. M. Sherman, C. B. Brachmann, R. C. Allshire, J. D. Boeke, and L. Pillus. 1999. The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol. Biol. Cell 10:3171-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grewal, S. I., M. J. Bonaduce, and A. J. Klar. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 16.Hagan, I. M., and K. R. Ayscough. 2000. Fluorescence microscopy in yeast, p. 179-206. In V. J. Allan (ed.), Protein localization by fluorescence microscopy: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 17.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 19.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 20.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 21.Jansen, R. P., E. C. Hurt, H. Kern, H. Lehtonen, M. Carmo-Fonseca, B. Lapeyre, and D. Tollervey. 1991. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 113:715-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeppesen, P., and B. M. Turner. 1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:281-289. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J., S. Sif, B. Jones, A. Jackson, J. Koipally, E. Heller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, R. Kingston, and K. Georgopoulos. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345-355. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y. B., A. Honda, M. Yoshida, and S. Horinouchi. 1998. phd1+, a histone deacetylase gene of Schizosaccharomyces pombe, is required for the meiotic cell cycle and resistance to trichostatin A. FEBS Lett. 436:193-196. [DOI] [PubMed] [Google Scholar]
- 26.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 28.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leipe, D. D., and D. Landsman. 1997. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 25:3693-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 31.Ma, X. J., J. Wu, B. A. Altheim, M. C. Schultz, and M. Grunstein. 1998. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. USA 95:6693-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizukami, T., W. I. Chang, I. Garkavtsev, N. Kaplan, D. Lombardi, T. Matsumoto, O. Niwa, A. Kounosu, M. Yanagida, T. G. Marr, et al. 1993. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell 73:121-132. [DOI] [PubMed] [Google Scholar]
- 34.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 36.Olsson, T. G., K. Ekwall, R. C. Allshire, P. Sunnerhagen, J. F. Partridge, and W. A. Richardson. 1998. Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res. 26:3247-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 38.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 39.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 40.Thon, G., K. P. Bjerling, and I. S. Nielsen. 1999. Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics 151:945-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thon, G., A. Cohen, and A. J. Klar. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thon, G., and J. Verhein-Hansen. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, B. M., A. J. Birley, and J. Lavender. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69:375-384. [DOI] [PubMed] [Google Scholar]
- 44.Wozniak, R. W., G. Blobel, and M. P. Rout. 1994. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 125:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]