Abstract
Surfaces of human TATA box-binding protein (hsTBP) required for activated transcription in vivo were defined by constructing a library of surface residue substitution mutations and assaying them for their ability to support activated transcription in transient-transfection assays. In earlier work, three regions were identified where mutations inhibited activated transcription without interfering with TATA box DNA binding. One region is on the upstream surface of the N-terminal TBP repeat with respect to the direction of transcription and corresponds to the TBP surface that interacts with TFIIA. A second region on the stirrup of the C-terminal TBP repeat corresponds to the TFIIB-binding surface. Here we report that the third region where mutations inhibit activated transcription in mammalian cells, the convex surface of the N-terminal repeat, corresponds to a surface on TBP that interacts with hsTAF1, the major scaffold subunit of TFIID. Since mutations at the center of the hsTAF1-interacting region inhibit the ability of the protein to support activated transcription in vivo, these results are consistent with the conclusion that an interaction between hsTBP and TAFIIs is required for activated transcription in mammalian cells.
Transcription of protein-coding genes by RNA polymerase II (Pol II) requires general transcription factors TFIIA, -B, -D, -E, -F, and -H in addition to the polymerase (54). TFIID is a multisubunit factor that includes the TATA box-binding subunit (TBP) and TBP-associated factors (TAFIIs). TBP is sufficient for in vitro basal transcription from a promoter with a high-affinity TATA box, but TAFIIs are required for in vitro transcription from promoters lacking a TATA box (69), where they bind to initiator (11) and downstream promoter elements (9). Initial studies indicated that TAFIIs were required for activator proteins to stimulate transcription in in vitro reactions (12, 31, 53). However, another early report (25) as well as several more recent studies have shown that TBP alone can support activated transcription in vitro (28, 32, 46, 73).
The question of whether TAFIIs are generally required for activated transcription in vivo has been controversial. Initial studies with Saccharomyces cerevisiae utilizing engineered depletion (42) or temperature inactivation of TAF temperature-sensitive mutants (70) argued that TAFIIs were not generally required for Pol II transcription in vivo or for activation by most activators. However, subsequent studies indicated that S. cerevisiae TAF6 (scTAF6) and scTAF12 (40, 44), scTAF9 (2, 40, 43), and scTAF10 (57) are in fact required for most, if not all, Pol II transcription in yeast. (Pol II TAFs are named according to reference 66a.) The interpretation of these experiments was complicated by the finding that many of the yeast TAFIIs are also subunits of the SAGA histone acetylase complex (19), just as many of the mammalian TAFIIs are subunits of the P/CAF histone acetylase complex (47). Consequently, the phenotypes of these TAF mutants could be due to defects in both TFIID and SAGA complexes. However, studies with scTAF11 temperature-sensitive mutants, which were free of this complication since scTAF11 is not associated with the SAGA complex, indicated that it is also generally required for Pol II transcription in vivo (33).
Further support for the model that transcription from most yeast promoters does not require TAFIIs came from chromatin immunoprecipitation experiments. They indicated that yeast TAFIIs are underrepresented at promoters that show resistance to TAFII depletion compared to promoters where TAFIIs are required (34, 35).
The requirement for TAFIIs in activated transcription in vivo in mammalian cells has not been addressed. The biochemistry of TBP in mammalian cells is quite different from that in yeast cells. Whereas TBP in yeast extracts is largely monomeric (7), free TBP is not observed in extracts of mammalian cells (63, 66, 75). In HeLa cell extracts, TBP is tightly associated with other polypeptides in TFIID as well as with the Pol I factor SL1 (TFID/TIF-IB), with the Pol III factor TFIIIB (reviewed in reference 21), or in complex with human BTAF1 (hsBTAF1), a homolog of yeast Mot1 (13, 67). Consequently, whereas yeast TBP might associate with some promoters in the absence of TAFIIs (34, 35), this seems unlikely in mammalian cells.
In an earlier study, we generated a large set of human TBP mutants with single amino acid substitutions of surface residues. These mutants were analyzed for their ability to support activated transcription in vivo in a transient-transfection assay (5) and for their ability to bind TFIIA and TFIIB in gel shift assays. Mutations in three discrete regions on the surface of TBP inhibited the ability of the protein to support activated transcription. One of these corresponded to the region that was found to interact with TFIIA, and one corresponded to the region that interacts with TFIIB. Mutations in a third region on the convex surface of the N-terminal repeat of hsTBP (referred to here as the top of the hsTBP N-terminal repeat) also strongly inhibited activated transcription in vivo, although they had no effect on basal transcription in vitro. This region did not correspond to the binding site for any known factor.
In this study, we analyzed the ability of this set of hsTBP mutants to bind full-length hsTAF1 in vitro. hsTAF1 is the largest hsTAFII and makes the principal contact between hsTBP and hsTAFIIs in TFIID (61, 75). The Drosophila homolog of hsTAF1, dmTAF1, is required for the assembly of partial TFIID complexes in vitro (12). Similarly, partial human TFIID complexes can be assembled by using hsTBP, hsTAF1 and additional hsTAFIIs (20). Consequently hsTAF1 appears to be a major scaffold with which TBP and other TAFIIs interact in the assembly of TFIID. Our results indicate that the top of the N-terminal hsTBP repeat corresponds to a principal hsTAF1 binding site. Together with an earlier study (5), these results imply that an interaction between hsTBP and hsTAF1 is required for activated transcription in vivo in mammalian cells.
MATERIALS AND METHODS
hsTAF1 binding assay.
Wild-type and mutant hsTBPs were transcribed and translated in vitro (TNT coupled reticulocyte lysate system; Promega) and labeled with Tran35S-label (ICN; 20 μCi/25-μl reaction mixture). In vitro transcription-translation reaction mixtures (25 μl) were diluted to 175 μl in D buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 5 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 0.1% NP-40) containing 0.15 M KCl and incubated with 25 μl of protein G-Sepharose beads for 1 h at room temperature. After 10 s of centrifugation at 14,000 rpm in an Eppendorf microcentrifuge, the precleared supernatant was removed, and 25 μl was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and PhosphorImager analysis (Molecular Dynamics) of the dried gel. Volumes of hemagglutinin (HA)-tagged mutant TBP approximately equal in counts to 25 μl of wild-type TBP or human TBP with an m3 triple mutation (hm3TBP; see below) were mixed and diluted to a total volume of 225 μl with 0.15 M KCl D buffer plus 0.2% NP-40. A 25-μl portion of hsTAF1 matrix was added, and the suspension was incubated with constant mixing overnight at 4°C (∼16 h). After centrifugation for 10 s at 15,000 × g, the supernatant was aspirated, and the beads were washed twice with 250 μl of 1.0 M KCl D buffer and twice with 250 μl of 0.15 M D buffer. Bound proteins were eluted in Laemmli sample buffer and subjected to SDS-PAGE, followed by PhosphorImager analyses (Molecular Dynamics) of the dried SDS gels.
Preparation of the hsTAF1 matrix.
Recombinant full-length HA-tagged hsTAF1 was expressed in Hi5 cells (Invitrogen) with the recombinant baculovirus (pbHAX-TAFII250) described previously (55). Cells were infected at a multiplicity of infection of 10 and harvested 48 h postinfection, and nuclear extract was prepared (15). A 25-μl portion of protein G-Sepharose (Pharmacia) was loaded with 25 μg of anti-hsTAF1 monoclonal antibody (anti-TAFII250 monoclonal antibody; catalog no. sc-735 [Santa Cruz]) by incubation in 25 μl of 0.5× phosphate-buffered saline for 1.5 h at room temperature. After centrifugation for 10 s at 15,000 × g, the supernatant was removed and the beads were washed three times with 250 μl of phosphate-buffered saline. A 25-μl portion of nuclear extract was incubated with 25 μl of protein G-Sepharose loaded with anti-hsTAF1 antibody for 2 h at room temperature, the beads were centrifuged, the supernatant was removed, and the beads were washed three times with 250 μl of 0.15 M KCl D buffer plus 50 μg of benzamidine per ml, 1 μg of pepstatin A per ml, and 1 μg of leupeptin per ml.
Binding to GST-hsTAF1(1172-1344).
A fusion of glutathione S-transferase (GST) to hsTAF1 residues 1172 to 1344 was prepared by PCR amplification of the corresponding coding region of an hsTAF1 cDNA (55) and cloning between the EcoRI and XhoI sites of pGEX-5X-1 (Pharmacia) to generate pGEX 5X-hsTAF1-1172-1344. The fusion protein was expressed in Escherichia coli strain BL21 and purified on glutathione Sepharose according to Pharmacia procedures. GST was purified similarly following expression from pGEX-5X-1. Glutathione Sepharose beads (25 μl) containing prebound GST-hsTAF1(1172-1344) or GST were incubated with equal counts of 35S-labeled epitope-tagged mutant hm3TBP and hm3TBP as described above for the hsTAF1 binding assay, except that all washes were in 0.15 M KCl D buffer plus 0.2% NP-40, unless otherwise noted.
Purification of TFIIA.
The TFIIA αβ and γ subunits were expressed in E. coli BL21 from plasmids pQIIA-αβ and pQIIA-γ, respectively, and purified by Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen) chromatography as described previously (49). Equimolar amounts of the αβ and γ subunits in Ni-NTA elution buffer B (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl [pH 8.0]) containing 200 mM imidazole were diluted to 0.1 mg/ml in the same buffer and dialyzed sequentially against 8, 2, 1, 0.5, and 0 M urea, all in D buffer for 2 h each at 4°C in a Slide-A-Lyzer cassette (Pierce). Following gradient dialysis, precipitated protein was removed by centrifugation at 15,000 rpm for 20 min at 4°C in a Sorvall SA-600 rotor.
In vivo analysis of mutant hsTBP function in Pol II transcription.
One microgram of hm3TBP mutant expression vector described previously (5), 1 μg of p4xGAL c-fosTGTluc (65), 0.2 μg of pGAL4-E1A (38) or 0.02 μg of pG4VP16 (56), and 0.02 μg of pRLTK (Promega) were transiently transfected with Superfect reagent (Qiagen) into one well of a 24-well dish of COS cells that had been plated at 6 × 104 cells/well the previous day. Cells were harvested 48 h later with 100 μl of passive lysis buffer (dual luciferase assay system; Promega) and incubated for 15 min at room temperature on a shaker. Firefly luciferase activity was measured with a Monolight 2010 luminometer (Analytical Luminescence Laboratory) with 10-s measurements with the Promega firefly luciferase assay substrate. Renilla luciferase activity was then measured with the Promega Renilla luciferase assay substrate for 10 s. Firefly luciferase assays were normalized by dividing firefly luciferase units by Renilla luciferase units. The background control was from a transfection in which the TBP expression vector was replaced by the same vector expressing an inactive, truncated form of hm3 (deleted at the StuI site to produce hm3TBP terminated after amino acid residue 271, 22 residues into the second TBP repeat). Reported units are normalized firefly luciferase units minus background as a percentage of the activity observed with hm3TBP.
RESULTS
Binding of hsTBP mutants to hsTAF1.
Human TBP (hsTBP) mutants that had been studied earlier for their interactions with TFIIA and TFIIB in vitro and their activities for in vitro basal and in vivo activated transcription (5) were assayed for their interaction with hsTAF1. These mutations were each constructed in the background of the m3 triple mutation in residues on the concave DNA-binding surface of the hsTBP saddle. The m3 mutations allow the protein (hm3TBP) to bind to a TGTAAA variant TATA box to which wild-type TBP cannot bind (60). The use of the m3 background allowed the additional mutations constructed on the convex surface of hm3TBP to be assayed for their influence on in vivo transcription by using reporter genes with TGTAAA boxes. These reporters are poorly transcribed by the endogenous wild-type TBP but are responsive to hm3TBP (26, 60, 65).
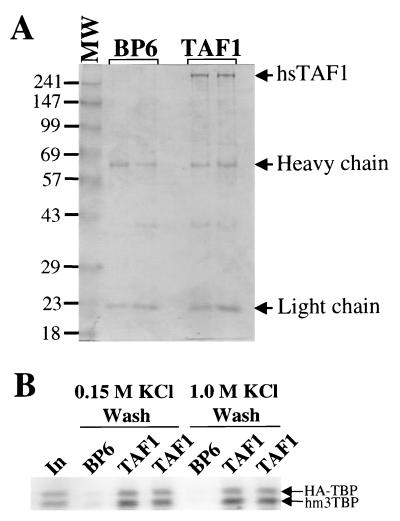
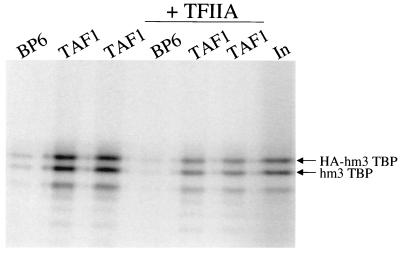
N-terminal peptides from the Drosophila and S. cerevisiae homologs of hsTAF1, dmTAF1 and scTAF1, respectively, bind to the concave DNA binding surface of scTBP (30, 37, 48). Therefore, we analyzed whether the m3 mutations engineered into the concave surface of human TBP (hm3TBP) influenced the binding of hsTBP to full-length hsTAF1. An hsTAF1 affinity matrix was constructed by expressing full-length hsTAF1 from a baculovirus vector in insect cells (55) and binding the protein to anti-hsTAF1 monoclonal antibody bound to protein G Sepharose (Fig. 1A). Both 35S-labeled epitope-tagged wild-type hsTBP and untagged hm3TBP were incubated with the hsTAF1 matrix, washed with a buffer containing 1 M KCl, eluted, and analyzed by PAGE (Fig. 1B). Since the lower-mobility epitope-tagged hsTBP could be distinguished from the hm3TBP lacking an epitope tag, the binding of the two proteins to hsTAF1 could be directly compared in the same reaction. The hm3TBP mutant bound to hsTAF1 at least as well as wild-type hsTBP, demonstrating that the m3 mutations do not affect hsTAF1 binding.
FIG. 1.
The m3 mutations in hm3TBP do not inhibit binding to hsTAF1. (A) Proteins eluted from the hsTAF1 matrix and the control BP6 matrix. Anti-hsTAF1 monoclonal antibody immobilized on protein G Sepharose was incubated with nuclear extract from insect cells infected with a hsTAF1 baculovirus expression vector (TAF1), or from insect cells infected with the empty baculovirus vector (BP6). Proteins from independently prepared, duplicate matrices were eluted with SDS, resolved by SDS-PAGE, and stained with Coomassie blue. MW, molecular weight markers (molecular weights are in thousands). (B) The control BP6 matrix and duplicate hsTAF1 matrices were incubated with 35S-labeled, in vitro-translated, HA epitope-tagged wild-type hsTBP and hm3TBP in a buffer with 0.15 M KCl and then washed with 0.15 or 1.0 M KCl buffer. Bound proteins were eluted, resolved by SDS-PAGE, and analyzed with a PhosphorImager. Input (In) represents 10% of the labeled proteins added to the binding reaction mixture.
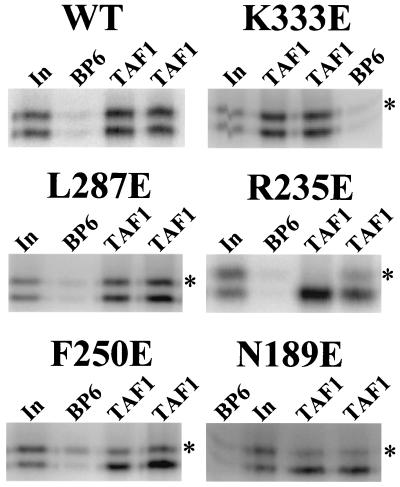
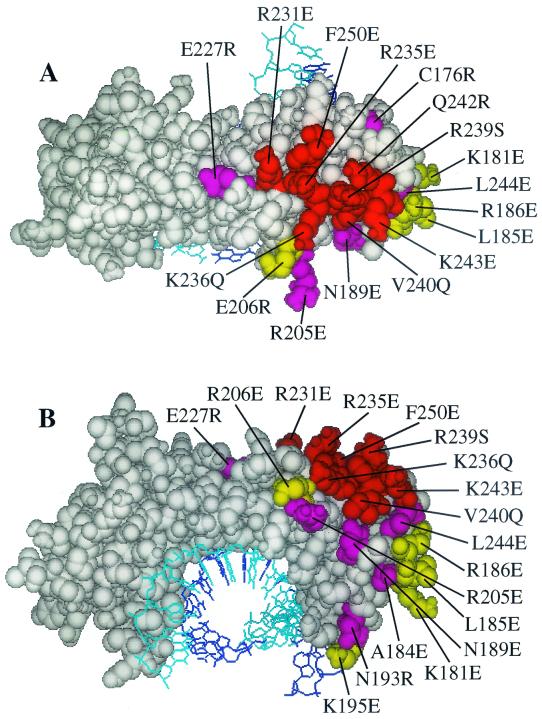
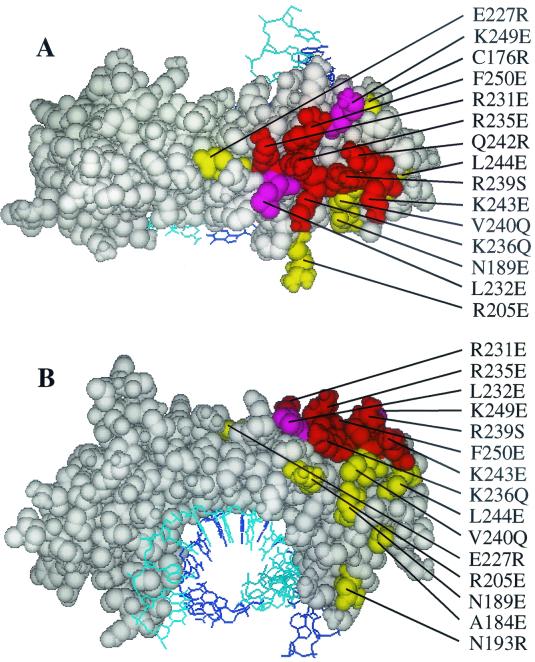
Seventy-five hsTBP mutants in the m3 background (5) were assayed for binding to the hsTAF1 matrix as described above. Each of these mutants binds to TATA box DNA in vitro and thus is correctly folded (5). Mutants Q242R and K243E were also constructed on the hm3TBP background and analyzed. Fifteen of the hm3TBP mutations bound hsTAF1 less than 50% as well as hm3TBP (Fig. 2; Table 1). Ten of the residues where mutations reduce hsTAF1 binding cluster into a continuous region on the top of the hsTBP saddle within the N-terminal repeat (Fig. 3A). The remaining inhibitory mutations map to the upstream surface (relative to the transcription start site (23, 27, 45) of the N-terminal repeat (Fig. 3B). We infer that these residues delineate the surface of hsTBP that interacts with hsTAF1.
FIG. 2.
Representative hsTAF1 binding reactions. Epitope-tagged hm3TBP (WT) or the indicated hsTBP mutants (in the m3 background, marked by an asterisk) were mixed with equal amounts (counts per minute) of untagged hm3TBP and incubated in duplicate with a matrix containing bound hsTAF1 (T250) or the control BP6 matrix. After washing with 1 M KCl, eluted proteins were resolved by SDS-PAGE. Input (In) represents 10% of the counts added to the binding reactions.
TABLE 1.
Binding of hsTBP mutants to the hsTAF1 matrix and to GST-hsTAF1(1172-1344)a
| TBP mutant | % Binding b
|
TBP mutant | % Bindingb
|
|||
|---|---|---|---|---|---|---|
| hsTAF1 | HMG | hsTAF1 | HMG | |||
| S156E | 64 ± 1.9 | |||||
| S157E | 92 ± 5.3 | |||||
| S158E | 76 ± 2.6 | |||||
| S159E | 72 ± 1.8 | |||||
| I161R | 96 ± 2.2 | |||||
| V162R | 98 ± 3.1 | |||||
| N173E | 71 ± 2.3 | |||||
| G175R | 103 ± 3.8 | 110 ± 0.1 | ||||
| C176R | 40 ± 0.1 | 83 ± 11 | ||||
| K177E | 62 ± 1.2 | |||||
| D179R | 100 ± 3.1 | |||||
| K181E | 66 ± 18 | |||||
| A184E | 8 ± 1.0 | 74 ± 0.3 | ||||
| L185E | 96 ± 5.1 | |||||
| R186E | 59 ± 7.1 | |||||
| N189E | 20 ± 1.8 | 78 ± 1.8 | ||||
| E191R | 125 ± 16 | |||||
| N193R | 25 ± 0.1 | 103 ± 1.0 | ||||
| P194E | 55 ± 6.6 | |||||
| K195E | 60 ± 0.8 | |||||
| R205E | 10 ± 1.2 | 60 ± 6.6 | ||||
| E206R | 112 ± 4.1 | |||||
| R208E | 92 ± 1.8 | |||||
| S215E | 61 | |||||
| S216R | 90 ± 0.2 | |||||
| E227R | 43 ± 0.8 | 79 ± 1.8 | ||||
| R231E | 48 ± 1.5 | 53 ± 2.5 | ||||
| R231E, R235E, R239S | 2 ± 18 | 4.9 ± 0.5 | ||||
| L232E | 58 ± 1.1 | 51 ± 0.4 | ||||
| R235E | 13 ± 7.2 | 51 ± 1.0 | ||||
| K236Q | 16 ± 7.9 | 45 ± 1.0 | ||||
| R239S | 27 ± 9.7 | 58 ± 3.2 | ||||
| V240Q | 19 ± 5.9 | 94 ± 11 | ||||
| Q242R | 22 ± 1.8 | 107 ± 3.6 | ||||
| K243E | 39 ± 7.2 | 51 ± 0.1 | ||||
| L244E | 25 ± 0.6 | 78 ± 13 | ||||
| G245E | 86 ± 0.1 | |||||
| P247E | 82 ± 6.1 | |||||
| K249E | 59 ± 7.1 | 52 ± 1.4 | ||||
Boldface indicates ≤33% (hsTAF1) or <60% (HMG).
Binding of hsTBP mutants to the hsTAF1 matrix and GSt-hsTAF1(1172-1344) (HMG) was calculated as a percentage of binding of hm3TBP in the same binding reaction. Data are means and standard deviations from 4 to 10 assays, with the exception of S215E, which was assayed once.
FIG. 3.
Residues where mutations inhibit hsTAF1 and BRF binding to hsTBP. A space-filling model of Arabidopsis thaliana TBP bound to TATA box DNA (wire model) is shown, looking down on the top of the TBP saddle (A) and at the surface facing upstream from the TATA box (B), with the N-terminal repeat on the right. Residues equivalent to those in hsTBP where the indicated mutations decrease hsTAF1-binding to less than 50% of that observed with the wild type and decrease BRF-binding to less than 30% of wild-type binding as measured in a gel shift assay (59) are in red. Residues where mutations inhibit hsTAF1-binding to <50% but do not inhibit BRF binding to 30% or less of wild-type TBP are in magenta (A184, N189, N193, R205, E227, and L244). Residues where mutations inhibit BRF binding to <30% of wild-type TBP but do not inhibit binding of hsTAF1 to less than 50% of wild-type TBP are in yellow.
A subdomain of hsTAF1 containing an HMG box-like region interacts with hsTBP.
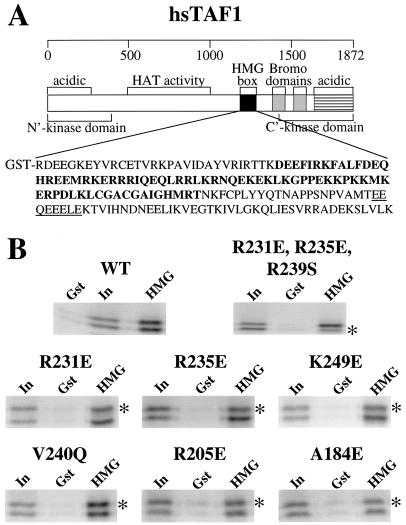
The hsTAF1-interacting region described above was also found to constitute a large portion of the hsTBP binding site for BRF, the Pol III TAF that makes the principal TAFIII-TBP interaction in TFIIIB (59) (Fig. 3). This interaction is through a C-terminal domain of BRF (59) that includes an HMG box-related sequence (71). An HMG box-related sequence was also identified within the C-terminal half of hsTAF1 (58). The HMG-like regions are the only regions in hsTAF1 and BRF that appear to be even distantly related in sequence, even though these two proteins interact with a similar surface of TBP. These considerations suggested to us that hsTAF1 and BRF might bind to the top of the N-terminal hsTBP repeat through structurally similar domains that include their respective HMG-like regions. To test this hypothesis, we analyzed the binding of hm3TBP to hsTAF1 residues 1172 to 1344, which includes the HMG-like region (Fig. 4A). In a binding reaction with both untagged and epitope-tagged hm3TBP, equivalent and substantial binding of both was observed (Fig. 4B). When complexes formed in 0.15 M KCl were exhaustively washed with increasing salt concentrations, approximately half of the hm3TBP washed off the matrix at 0.3 M KCl. Thus, the GST-hsTAF1(1172-1344)-hsTBP interaction was less stable than the interaction of full-length hsTAF1 with hsTBP, suggesting that additional hsTAF1-hsTBP interactions contribute to the stability of the complex formed with the full-length TAF. Recombinant hsTBP expressed in and purified from E. coli also bound to GST-hsTAF1(1172-1344) (data not shown).
FIG. 4.
hsTBP binds to the region of hsTAF1 that includes the HMG box homologous sequence. (A) Diagram of the primary sequence of hsTAF1 showing the N- and C-terminal regions required for its protein kinase activity (16), the region with histone acetylase activity (41), the HMG box-like region (58), and the double bromodomains (22). The sequence from 1172 to 1344 that was fused to GST is shown with the HMG-like region highlighted, and a highly acidic region C-terminal to it is underlined. (B) Representative results. Both untagged and epitope-tagged hm3TBP lacking any additional mutations on the convex surface (WT) are shown at the upper left. The triple mutant R231E, R235E, R231S was untagged (∗) and compared to epitope-tagged hm3TBP. For each point mutant, the epitope-tagged mutant (∗) was compared to untagged hm3TBP. Gst, binding to GST; HMG, binding to GST-hsTAF1(1172-1344); In, 10% of the input counts.
A triple mutant of hm3TBP (R231E, R235E, R239S) that has severely reduced binding to BRF and support of Pol III in vitro transcription (59) also had severely reduced binding to full-length hsTAF1 (Table 1). We found that this mutant also had severely reduced binding to GST-hsTAF1(1172-1344) (Fig. 4B; Table 1). Several single point mutants of hsTBP were also tested for binding to GST-hsTAF1(1172-1344) (Fig. 4B; Table 1). As observed for binding to full-length hsTAF1, mutation of several residues on the top of the N-terminal hsTBP repeat inhibited binding to GST-hsTAF1(1172-1344) (Fig. 5). However, mutations of residues on the upstream surface of the N-terminal hsTBP repeat that inhibited binding of full-length hsTAF1 had much less effect on binding to GST-hsTAF1(1172-1344). These results confirm the presence of a hsTAF1-interaction surface on the top of the hsTBP N-terminal repeat and lead us to suggest that this region interacts with hsTAF1 residues within the region from 1172 to 1344.
FIG. 5.
Relation of the hsTBP binding surface for hsTAF1(1172-1344) to the surface involved in binding full-length hsTAF1. Residues where mutations reduce binding to GST-hsTAF1(1172-1344) to less than 60% of that observed for wild-type TBP and reduce binding of full-length hsTAF1 to less than 50% are in red. Mutations where binding to GST-hsTAF1(1172-1344) is reduced to <60% of wild-type TBP but binding to full-length hsTAF1 is >50% (L232 and K249) are in magenta. Residues where mutations reduce binding of full-length hsTAF1 to <50% of wild-type TBP but do not reduce binding to GST-hsTAF1(1172-1344) to <60% of wild-type TBP are in yellow.
TFIIA competes with hsTAF1 for binding to TBP.
Some of the mutations found to inhibit hsTAF1 binding (A184E, N189E, N193R, and R205E) also correspond to the interface between TFIIA and TBP in the crystal structures of TBP-TFIIAΔ-TATA box complexes (18, 62). These results suggest that hsTAF1 and TFIIA bind to overlapping regions on the surface of hsTBP. To test this possibility, we examined whether a high concentration of TFIIA could inhibit hm3TBP binding to the hsTAF1 matrix. Binding of in vitro-translated hm3TBP and epitope-tagged hm3TBP was performed in the absence and presence of recombinant hsTFIIA at a threefold molar excess over the hsTAF1. This high concentration of TFIIA inhibited binding to 25% of that observed in the absence of hsTFIIA (Fig. 6). These results indicate that the TFIIA-binding site on hsTBP does overlap with the hsTAF1-binding site, at least in the complex between hsTBP and hsTAF1 formed in vitro in the absence of other TAFIIs. Results consistent with this have also been reported by Kokubo et al. (30) and Bagby et al. (3) for N-terminal fragments of dmTAF1 and scTAF1. However, the in vivo significance of this competition has yet to be determined. As discussed below, TFIIA readily binds to a TFIID-DNA complex, even though the TFIID complex contains hsTAF1 (36). TFIIA did not interfere with the binding of hsTBP to GST-hsTAF1(1172-1344) (data not shown).
FIG. 6.
Inhibition of hsTBP binding to hsTAF1 by a threefold molar excess of TFIIA over hsTAF1.
Reduced in vivo stability of mutants defective in hsTAF1 binding.
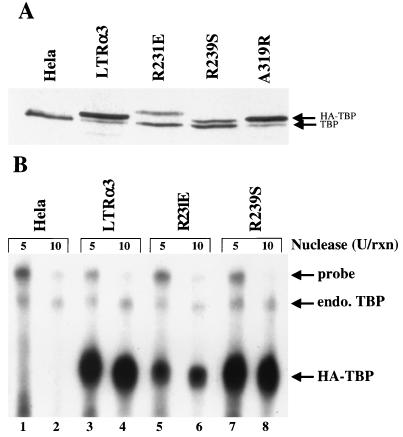
In earlier studies, a HeLa cell line (LTRα3) was constructed by using a retrovirus vector that expresses an epitope-tagged version of hsTBP, facilitating the purification of the TFIID complex (76). To analyze the association of mutant hsTBPs with TAFs in vivo, the same retrovirus vector system was used to transduce genes for the epitope-tagged mutant TBPs into HeLa cells. These were single-amino-acid-substitution mutants that did not also contain the m3 mutations. The expression of tagged mutant and untagged endogenous TBPs in cloned transduced cell lines was analyzed by Western blotting with an anti-TBP monoclonal antibody (Fig. 7A). TBP mRNA from these cells also was analyzed by S1 protection analysis of cytoplasmic RNA using a 5′-end-labeled, single-stranded probe (Fig. 7B). This generated S1-protected fragments of ∼80 nucleotides from the endogenous hsTBP mRNA in HeLa cells (Fig. 7B, lane 2) and ∼48 nucleotides from the hsTBP mRNA expressed from the retrovirus vector (HA-TBP) (Fig. 7B, lanes 3 to 8), consistent with the sequence of hsTBP cDNA included in the vector.
FIG. 7.
In vivo expression of mutant hsTBPs in retrovirus vector-transduced HeLa cells. (A) Western blot of hsTBPs in nuclear extract from cells transduced with a retrovirus vector expressing the indicated hsTBP mutants. LTRα3 cells express epitope-tagged wild-type hsTBP from the same vector. The anti-hsTBP monoclonal antibody used detects both untagged and tagged hsTBPs. (B) S1 nuclease protection assay of hsTBP mRNAs. The 5′-end-labeled single-stranded DNA probe (from −15 to +80 of the endogenous hsTBP gene) yielded S1-protected fragments of 80 and 48 nucleotides, respectively, from the endogenous hsTBP mRNA in HeLa cells and the hsTBP mRNA transcribed from the retrovirus vector.
In LTRα3 cells expressing tagged wild-type hsTBP from the vector, vector hsTBP mRNA was present at far higher levels than the endogenous hsTBP mRNA (Fig. 7B, lanes 3 and 4). However, the level of total hsTBP protein in these cells was only modestly increased over the level of hsTBP observed in nontransduced HeLa cells (Fig. 7A, compare lanes 1 and 2), and most of it was epitope tagged. In addition, the level of endogenous, untagged TBP protein was greatly decreased in LTRα3 cells compared to that in nontransduced HeLa cells. We infer from these results that the epitope-tagged wild-type hsTBP probably was translated at much higher levels than untagged, endogenous TBP in LTRα3 cells due to the high level of vector-derived hsTBP mRNA. We postulate that the decrease in the level of untagged endogenous hsTBP in these cells resulted from competition between the epitope-tagged TBP and endogenous TBP for binding to class I, II, and III TAFs and hsBTAF1. TBP molecules produced in excess of the supply of TAFs appear to be unstable and rapidly degraded, since the overall level of hsTBP in the transduced cells was only modestly increased over that in nontransduced HeLa cells. Also, we did not observe significant levels of free hsTBP (i.e., TBP that is not associated with TAFs) in nuclear extracts prepared from LTRα3 cells (74).
Mutant A319R was not defective for binding to hsTAF1 (Table 1) or for supporting activated transcription in vivo (5). When this mutant was expressed in HeLa cells from the retrovirus vector, the protein accumulated to high levels similar to epitope-tagged wild-type hsTBP, and its expression also resulted in a decrease in the level of endogenous TBP (Fig. 7A, lane 5). These results imply that A319R competes effectively with wild-type TBP for binding to TAFs. A319R accumulated to steady-state levels similar to those of the tagged wild-type TBP in LTRα3 cells, consistent with the model that association with TAFs stabilized the mutant TBP. We postulate that the decreased level of endogenous TBP in A319R cells resulted from its instability in the absence of TAFs that were sequestered by A319R.
Results with mutants R231E and R239S, which have a reduced capacity to bind hsTAF1 in vitro (Table 1) and to support activated transcription in vivo (5), contrasted with those for A319R. Even though vector mRNAs encoding mutants R231E and R239S were also expressed at far higher levels than the endogenous hsTBP mRNA (Fig. 7B, lanes 5 to 8), these tagged mutants did not accumulate to the high levels observed for tagged wild-type hsTBP in LTRα3 cells or mutant A319R (Fig. 7A, lanes 3 and 4). Their expression also did not result in a significant decrease in the level of endogenous wild-type TBP compared to HeLa cells. The low steady-state levels of R231E and R239S in stably transformed HeLa cells were not due to inherent instability of these mutants compared to wild-type TBP or A319R or inefficient translation of the mRNAs encoding them, because they were expressed at levels comparable to those of wild-type hm3TBP from the same vectors in transient-transfection assays (5). We infer from these observations that mutants R231E and R239S did not compete with endogenous TBP for binding to endogenous TAFs as well as epitope-tagged wild-type hsTBP or mutant A319R. Consequently, they were not efficiently stabilized by their association with TAFs and therefore did not accumulate to the levels observed for epitope-tagged wild-type hsTBP and mutant A319R. Their expression did not lead to a significant decrease in the level of endogenous TBP because endogenous TBP preferentially bound to TAFs. Consequently, the endogenous TBP was stabilized and accumulated to the same extent as in untransduced HeLa cells. Thus, the results of these in vivo expression studies are consistent with the in vitro binding assays showing that the R231E and R239S mutations inhibit binding of hsTBP to both hsTAF1 (Table 1) and BRF (59).
Isolation and characterization of epitope-tagged TFIID from cells expressing mutants R231E and R239S indicated that the mutant proteins that did accumulate in these cells assembled into TFIID complexes containing the complete set of hTAFIIs (data not shown). However, based on the arguments above, we infer that these mutants were incorporated into TFIID complexes much less efficiently than wild-type hsTBP. The TFIID complexes that did form with TBP mutants R231E and R239S were stable to 1 M KCl and to purification by immunoaffinity chromatography. However, we were not able to isolate these mutant complexes in sufficient quantity to study their activity in in vitro transcription assays. We presume that the R231E and R239S mutant TBPs that were incorporated into TFIID formed stable complexes because of TAF-TAF and TAF-TBP interactions in addition to the TBP-hsTAF1 interaction we map in the present study.
In vivo function of mutants Q242R and K243E.
Most hm3TBP mutants with significantly reduced hsTAF1 binding (Table 1) had severely reduced abilities to support activated transcription in vivo (5). Two mutants (Q242R and K243E) with reduced hsTAF1 binding had not been assayed previously for their in vivo activities. We performed transient-transfection assays with these mutants in the presence of a Gal4-responsive promoter and Gal4-E1A or Gal4-VP16 expression vectors. Q242R and K243E exhibited only 10 to 15% and 5 to 10%, respectively, of the activity of “wild-type” hm3TBP. Therefore, as for other hsTBP mutants with significantly reduced hsTAF1 binding in vitro, the ability of Q242R and K243E to support activated transcription in vivo was severely reduced.
DISCUSSION
The surface of hsTBP that interacts with hsTAF1.
We identified a putative surface of hsTBP that interacts with hsTAF1 by measuring the binding of 75 hsTBP point mutants to a hsTAF1 affinity matrix. Together, these 75 mutants cover much of the surface of hsTBP aside from the underside of the TBP saddle that interacts with TATA box DNA (5, 23, 27, 45). Mutations in 10 residues that form a continuous surface on the top of the N-terminal hsTBP repeat (Fig. 3A) inhibited hsTAF1 binding twofold or more (Table 1). There were no mutations of residues in the C-terminal hsTBP repeat that inhibited hsTAF1 binding significantly. These results are consistent with those of others who reported that triple mutations of arginines or lysines in the H2 helix on the top of the TBP N-terminal repeat inhibited binding to hsTAF1 (65) and an N-terminal peptide of scTAF1 (30). Also, mutation of two lysine residues in scTBP that lie immediately adjacent to the region highlighted in Fig. 3A result in temperature-sensitive association with the yeast TAFIIs in vivo (51). In light of these data and the results presented here, it seems likely that hsTAF1 interacts directly with the surface of hsTBP defined by the highlighted residues in Fig. 3A. In addition to this region on the top of the N-terminal hsTBP repeat, mutations in residues on the upstream surface of the N-terminal hsTBP repeat (R205E, N189E, A184E, and N193R) also inhibited hsTAF1 binding (Fig. 3B).
The RNA Pol III TAF BRF also binds to the top of the hsTBP N-terminal repeat (59) (Fig. 4). BRF (71) and hsTAF1 (58) both contain a region with weak homology to the HMG box DNA-binding domain. Since both BRF and hsTAF1 bind to a similar surface of hsTBP, and the domain of BRF that makes tight contact with hsTBP contains the HMG box-related region (59, 71), we analyzed the interaction of hsTBP with a fragment of hsTAF1 containing its HMG box-related region (residues 1172 to 1344). We found that this domain of hsTAF1 also bound specifically to hsTBP. Moreover, mutations on the top of the first TBP repeat inhibited this interaction, whereas mutations on the upstream surface of the first TBP repeat did not. Based on these results, we suggest that the hsTAF1(1172-1344) region interacts with the positively charged surface on the top of the first TBP repeat and that another region of hsTAF1 interacts with residues on the upstream surface of TBP, further stabilizing the hsTBP-hsTAF1 interaction in the complex formed in the absence of other TAFIIs. Based on the nuclear magnetic resonance (NMR) studies of Bagby et al. (3), it is likely that the upstream region of the first TBP repeat interacts with an N-terminal region of hsTAF1. The complex formed between TBP and hsTAF1 in the absence of other TAFIIs is also likely stabilized by an interaction between the extreme N terminus of hsTAF1 and the concave DNA-binding surface of TBP (3, 4, 30, 31, 37). We postulate that the HMG box-related regions of hsTAF1 and BRF fold similarly and make similar interactions with the top of the N-terminal TBP repeat. This may occur through highly acidic regions in hsTAF1 (Fig. 5A) and hsBRF that lie just C-terminal to their respective HMG box-related regions. The SL1 subunit hTAFI 48 also may bind to the top of the N-terminal TBP repeat, since it competes with hsTAF1 for binding to TBP (14).
The conclusion that hsTAF1 interacts with hsTBP through the hsTAF1(1172-1344) region contrasts with results indicating that the very N-terminal regions of dmTAF1 (residues 11 to 77) and scTAF1 (residues 10 to 64) bind to the DNA-binding concave surface of scTBP (30, 37). In addition, Bagby et al. (3) reported that a region of dmTAF1 from residues 78 to 156 interacts with the upstream surface and top of the scTBP N-terminal repeat. Nonetheless, other studies showed that an N-terminal deletion of dmTAF1 lacking these sequences (deletion of positions 1 to 659) does bind dmTBP (72) and assembles into transcriptionally active partial TFIID complexes (12, 72). Consequently, although the very N-terminal regions of hsTAF1 homologs can bind to TBP as isolated peptides, other regions of these hsTAF1 homologs also bind to TBP. Mencia and Struhl (39) observed that overexpressed scTAF1 truncation mutants lacking the HMG-like region are incorporated into complexes with scTBP and other TAFIIs in vivo. Bai et al. (4) made similar observations for a scTAF1 mutant with most of the HMG box homologous region deleted. However, these mutant complexes were not transcriptionally active (4, 39) and did not associate with Pol II promoters in vivo or bind to DNA in vitro (39). It may be that when these truncation mutants are expressed at high levels, the N-terminal scTAF1 region of the truncated protein associates with the scTBP DNA-binding surface as observed for peptides in vitro (30). This would explain the inhibition of scTBP DNA-binding activity in these mutant complexes (39). In contrast, a stable interaction between the N-terminal region of hsTAF1 and the DNA-binding surface of hsTBP in wild-type hsTFIID is unlikely, since purified hsTFIID binds to TATA box DNA (68, 76).
Conformational changes in hsTAF1 implied by differences between the TBP-hsTAF1 complex and TFIID.
The interactions between TBP and hsTAF1 differ comparing the isolated proteins and the TFIID complex containing the full set of TAFIIs. When the isolated proteins interact, the DNA-binding activity of TBP is blocked (29, 48). In contrast, the TFIID complex, which includes hsTAF1, binds specifically to TATA box DNA (68, 76). In the interaction between the isolated proteins, hsTAF1 appears to interact with the upstream surface of the N-terminal TBP repeat, since mutations on this surface inhibit hsTAF1 binding (Table 1; Fig. 3B). This is the same region of TBP that interacts with TFIIA in TBP-ΔTFIIA-DNA crystal structures (18, 62). As expected from these results, TFIIA inhibits the binding of TBP to hsTAF1 (Fig. 6). In contrast, the TBP in a TFIID complex appears to interact with TFIIA on TATA box DNA in the same way as in the TBP-ΔTFIIA-DNA crystal structures, since TFIIA can join a TFIID-TATA box DNA complex, producing a footprint in the same region observed in the TBP-TFIIA-DNA complex (6, 36). These considerations imply that interactions of hsTAF1 with other TAFIIs in the TFIID complex alter the conformation of hsTAF1 compared to its conformation in the TBP-hsTAF1 complex so that the TBP DNA-binding surface is free to bind to TATA box DNA and the upstream surface of the TBP N-terminal repeat is free to interact with TFIIA. Indeed, Guermah et al. (20) recently reported that whereas a complex of hsTBP with hsTAF1 is unable to bind to a TATA box in the presence or absence of TFIIA, an hsTBP-hsTAF1-hsTAF4 complex binds to a TATA box cooperatively with TFIIA. Since the fragment of hsTAF1 containing the HMG box-like region (residues 1172 to 1344) interacts with the top of the first TBP repeat and not the upstream surface that interacts with TFIIA (Fig. 5), we propose that it is this region of hsTAF1 that interacts with hsTBP in the native TFIID complex.
It has been suggested that one mechanism of transcriptional activation may be the relief of hsTAF1 inhibition of TBP DNA binding through a competition for TBP binding between activation domains and hsTAF1 (30, 37, 48). Another possible function in vivo for the ability of hsTAF1 to inhibit hsTBP binding in the absence of other hTAFIIs may be to inhibit partially assembled TFIID complexes from binding to promoters so that only fully assembled TFIID is incorporated into a preinitiation complex. This would be analogous to the inhibition of E. coli σ70 DNA binding activity before σ70 associates with E. coli RNA polymerase, forming a fully functional holoenzyme (17).
TAFIIs appear to be required for activated transcription in vivo in mammalian cells.
Mutations R231E, R235E, R239S, Q242R, and K243E all greatly inhibited activated transcription in vivo in response to the VP16 and E1A activation domains (5; see above). These mutations also inhibited hsTAF1 binding to TBP in vitro (Table 1) and lie at the center of the hsTAF1-binding region on the top of the N-terminal repeat (Fig. 3A). Consequently, these results suggest that an interaction between TBP and hsTAF1 is required for activated transcription in vivo. However, this issue is complicated by reports that the same region of TBP interacts with TFIIA (3, 8, 64) and hsBTAF1 (50), the human homolog of yeast Mot1 (13, 50), which also interacts with this region of scTBP (1, 10). Consequently, mutations in this region of the TBP surface might inhibit activated transcription by interfering with TFIIA or hsBTAF1 binding rather than hsTAF1 binding or because of a combination of effects on interactions with hsTAF1, TFIIA, and hsBTAF1. We argue, however, that the inhibition of activated transcription by these mutations is principally a consequence of reduced binding to hsTAF1 for the following reasons.
First, although the top of the TBP N-terminal repeat interacts with TFIIA in the complex formed in the absence of DNA as analyzed in the NMR study of Bagby et al. (3) and in our own column binding assays (5), this interaction is much less significant in the far more stable complex formed between TBP, TFIIA, and TATA box DNA as analyzed in a gel shift assay (5). We observed that the R231E, R235E, and R239S mutations inhibited TBP binding to a TFIIA affinity column in the absence of DNA. However, these same mutations had no significant effect on the formation of a hsTBP-hTFIIA-TATA box DNA complex assayed by gel shift (5). This apparent discrepancy between the results of the NMR and column binding assays on the one hand and gel shift assays on the other may be explained by differences in the stability of TBP-TFIIA complexes formed in the presence and absence of DNA. Ranish and Hahn (52) reported a Kd of ∼6 × 10−4 M for yTFIIA binding to scTBP in the absence of DNA compared to a Kd of 2 × 10−11 M for yTFIIA binding to a scTBP-TATA box DNA complex. The many TFIIA-DNA and TBP-DNA interactions observed in the crystal structures of the TBP-ΔTFIIA-DNA complexes (18, 62) provide an explanation for the increased stability of the complex in the presence of DNA. In the absence of DNA, interactions between the highly positively charged surface on the top of the TBP N-terminal repeat and a negatively charged region of the hsTFIIA large subunit may make significant contributions to the stability of the weak TBP-TFIIA complex, as discussed by Bagby et al. (3). However, with the additional interactions of TFIIA and TBP with DNA, such interactions between TFIIA and the top of the N-terminal repeat of TBP are less significant, since the single-residue mutations have little effect in the gel shift assay.
A second reason for concluding that the R231E, R235E, R239S, Q242R, and K243E mutations inhibit activated transcription by interfering with TAFII binding is that free hsTBP is not detectable in extracts of mammalian cells (63, 66, 74). Consequently, TFIID complexes, rather than free hsTBP, probably function in virtually all Pol II transcription in mammalian cells. The results presented here indicate that the hsTAF1 subunit of the TFIID complex binds to the top of the N-terminal hsTBP repeat (Fig. 3A). Because of this, the top of the hsTBP N-terminal repeat would be sterically blocked from interacting with TFIIA during preinitiation complex assembly in vivo in mammalian cells. Steric blocking of the top of the TBP N-terminal domain by hsTAF1 and BRF may also explain why hsBTAF1 inhibits transcription in reactions with TBP but does not inhibit transcription in reactions with TFIID or TFIIIB (13). NC2 is yet another factor that interacts with TBP. However, recent genetic (10) and crystallographic (24) studies indicate that NC2 interacts with the top and downstream surface of the second TBP repeat in a region on the surface of TBP that is far from the hsTAF1-binding surface mapped in the present study.
Based on these considerations, we conclude that the inability of hsTBP mutants R231E, R235E, and R239S to support activated transcription in vivo in transient-transfection assays is most likely due to their reduced interaction with hsTAF1 and not to a reduced interaction with TFIIA or hsBTAF1. The finding that these residues required for activated transcription are at the center of the hsTAF1 binding surface on hsTBP (Fig. 3A) argues strongly that TAFIIs are required for activated Pol II transcription in vivo in mammalian cells.
R231E, R235E, R239S103 ± 3.810 ± 0.1F250E22 ± 2.355 ± 10L251E50 ± 0.775 ± 9.9 D252R85 ± 4.5K254E75 ± 0.6S261E108 ± 3.8D263R76 ± 3.4R265E78 ± 1.0P267E58 ± 0.4R269E71 ± 0.9E271R93 ± 2.5L275R79 ± 1.0H277E82 ± 3.4Q278E75 ± 0.3A279E110 ± 14F280E97 ± 3.8S282E77 ± 4.6E284R95 ± 1.3E286R84 ± 1.1L287E74 ± 1.5K297E64 ± 1.7R299E116 ± 9.5F305A86 ± 6.1V306E90 ± 4.5S307F58 ± 0.7K316E88 ± 2.1V317E66 ± 2.2R318E133 ± 11A319R96 ± 1.7E320R89 ± 0.0Y322E75 ± 1.9E323R90 ± 4.4E326A84 ± 5.7N327E84 ± 4.1Y329E73 ± 0.4P330R124 ± 0.4K333E84 ± 2.8R336E81 ± 1.3K337E92 ± 2.4T338R80 ± 2.3
Acknowledgments
This work was supported by grant CA25235 from the National Cancer Institute. L.S.M. was supported by NRSA CA-09056 from the National Cancer Institute.
We thank R. Tjian for supplying baculovirus vector pbHAX-TAFII250, Rod Hori for the recloned baculoviral stock and his advice on use of the hsTAF1 expression vector, and Mark Surby for TFIIA.
REFERENCES
- 1.Adamkewicz, J. I., K. E. Hansen, W. A. Prud'homme, J. L. Davis, and J. Thorner. 2001. High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J. Biol. Chem. 276:11883-11894. [DOI] [PubMed] [Google Scholar]
- 2.Apone, L. M., C. A. Virbasius, F. C. Holstege, J. Wang, R. A. Young, and M. R. Green. 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2:653-661. [DOI] [PubMed] [Google Scholar]
- 3.Bagby, S., T. K. Mal, D. Liu, E. Raddatz, Y. Nakatani, and M. Ikura. 2000. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 468:149-154. [DOI] [PubMed] [Google Scholar]
- 4.Bai, Y., G. M. Perez, J. M. Beechem, and P. A. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, G. O., L. S. Martel, S. K. Burley, and A. J. Berk. 1996. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 10:2491-2504. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski, S., S. Hahn, P. A. Sharp, and L. Guarente. 1988. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334:37-42. [DOI] [PubMed] [Google Scholar]
- 8.Buratowski, S., and H. Zhou. 1992. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science 255:1130-1132. [DOI] [PubMed] [Google Scholar]
- 9.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cang, Y., D. T. Auble, and G. Prelich. 1999. A new regulatory domain on the TATA-binding protein. EMBO J. 18:6662-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 13.Chicca, J. J., D. T. Auble, and B. F. Pugh. 1998. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol. Cell. Biol. 18:1701-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comai, L., J. C. Zomerdijk, H. Beckmann, S. Zhou, A. Admon, and R. Tjian. 1994. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266:1966-1972. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 17.Dombroski, A. J., W. A. Walter, M. T. Record, D. A. Siegele, and C. A. Gross. 1992. Polypeptides containing highly conserved regions of transcription initiation factor sigma-70 exhibit specificity of binding to promoter DNA. Cell 70:501-512. [DOI] [PubMed] [Google Scholar]
- 18.Geiger, J. H., S. Hahn, S. Lee, and P. B. Sigler. 1996. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272:830-836. [DOI] [PubMed] [Google Scholar]
- 19.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 20.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 23.Juo, Z. S., T. K. Chiu, P. M. Leiberman, I. Baikalov, A. J. Berk, and R. E. Dickerson. 1996. How proteins recognize the TATA box. J. Mol. Biol. 261:239-254. [DOI] [PubMed] [Google Scholar]
- 24.Kamada, K., F. Shu, H. Chen, S. Malik, G. Stelzer, R. G. Roeder, M. Meisterernst, and S. K. Burley. 2001. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106:71-81. [DOI] [PubMed] [Google Scholar]
- 25.Kao, C. C., P. M. Lieberman, M. C. Schmidt, Q. Zhou, R. Pei, and A. J. Berk. 1990. Cloning of a transcriptionally active human TATA binding factor. Science 248:1646-1650. [DOI] [PubMed] [Google Scholar]
- 26.Keaveney, M., A. Berkenstam, M. Feigenbutz, G. Vriend, and H. G. Stunnenberg. 1993. Residues in the TATA-binding protein required to mediate a transcriptional response to retinoic acid in EC cells. Nature 365:562-566. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y., J. H. Geiger, S. Hahn, and P. B. Sigler. 1993. Crystal structure of a yeast TBP/TATA-box complex. Nature 365:512-520. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 29.Kokubo, T., D. W. Gong, S. Yamashita, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1993. Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 7:1033-1046. [DOI] [PubMed] [Google Scholar]
- 30.Kokubo, T., M. J. Swanson, J. I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokubo, T., R. Takada, S. Yamashita, D. W. Gong, R. G. Roeder, M. Horikoshi, and Y. Nakatani. 1993. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J. Biol. Chem. 268:17554-17558. [PubMed] [Google Scholar]
- 32.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466-469. [DOI] [PubMed] [Google Scholar]
- 33.Komarnitsky, P. B., B. Michel, and S. Buratowski. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13:2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 35.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman, P. M., and A. J. Berk. 1994. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 8:995-1006. [DOI] [PubMed] [Google Scholar]
- 37.Liu, D., R. Ishima, K. I. Tong, S. Bagby, T. Kokubo, D. R. Muhandiram, L. E. Kay, Y. Nakatani, and M. Ikura. 1998. Solution structure of a TBP-TAFII230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94:573-583. [DOI] [PubMed] [Google Scholar]
- 38.Martin, K. J., J. W. Lillie, and M. R. Green. 1990. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature 346:147-152. [DOI] [PubMed] [Google Scholar]
- 39.Mencia, M., and K. Struhl. 2001. Region of yeast TAF 130 required for TFIID to associate with promoters. Mol. Cell. Biol. 21:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michel, B., P. Komarnitsky, and S. Buratowski. 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2:663-673. [DOI] [PubMed] [Google Scholar]
- 41.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 42.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 43.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 44.Natarajan, K., B. M. Jackson, E. Rhee, and A. G. Hinnebusch. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2:683-692. [DOI] [PubMed] [Google Scholar]
- 45.Nikolov, D. B., H. Chen, E. D. Halay, A. Hoffman, R. G. Roeder, and S. K. Burley. 1996. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc. Natl. Acad. Sci. USA 93:4862-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oelgeschlager, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 47.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 48.Ozer, J., K. Mitsouras, D. Zerby, M. Carey, and P. M. Lieberman. 1998. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J. Biol. Chem. 273:14293-14300. [DOI] [PubMed] [Google Scholar]
- 49.Ozer, J., P. A. Moore, A. H. Bolden, A. Lee, C. A. Rosen, and P. M. Lieberman. 1994. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 8:2324-2335. [DOI] [PubMed] [Google Scholar]
- 50.Pereira, L. A., J. A. van der Knaap, V. van den Boom, F. A. J. van den Heuvel, and H. T. M. Timmers. 2001. TAFII170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Mol. Cell. Biol. 21:7523-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranallo, R. T., K. Struhl, and L. A. Stargell. 1999. A TATA-binding protein mutant defective for TFIID complex formation in vivo. Mol. Cell. Biol. 19:3951-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranish, J. A., and S. Hahn. 1991. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 266:19320-19327. [PubMed] [Google Scholar]
- 53.Reese, J. C., L. Apone, S. S. Walker, L. A. Griffin, and M. R. Green. 1994. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature 371:523-527. [DOI] [PubMed] [Google Scholar]
- 54.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 55.Ruppert, S., E. H. Wang, and R. Tjian. 1993. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature 362:175-179. [DOI] [PubMed] [Google Scholar]
- 56.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563-564. [DOI] [PubMed] [Google Scholar]
- 57.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 58.Sekiguchi, T., Y. Nohiro, Y. Nakamura, N. Hisamoto, and T. Nishimoto. 1991. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol. Cell. Biol. 11:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, Y., G. A. Kassavetis, G. O. Bryant, and A. J. Berk. 1998. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol. 18:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strubin, M., and K. Struhl. 1992. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68:721-730. [DOI] [PubMed] [Google Scholar]
- 61.Takada, R., Y. Nakatani, A. Hoffmann, T. Kokubo, S. Hasegawa, R. G. Roeder, and M. Horikoshi. 1992. Identification of human TFIID components and direct interaction between a 250-kDa polypeptide and the TATA box-binding protein (TFIID tau). Proc. Natl. Acad. Sci. USA 89:11809-11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan, S., Y. Hunziker, D. F. Sargent, and T. J. Richmond. 1996. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381:127-151. [DOI] [PubMed] [Google Scholar]
- 63.Tanese, N., B. F. Pugh, and R. Tjian. 1991. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 5:2212-2224. [DOI] [PubMed] [Google Scholar]
- 64.Tang, H., X. Sun, D. Reinberg, and R. H. Ebright. 1996. Protein-protein interactions in eukaryotic transcription initiation: structure of the preinitiation complex. Proc. Natl. Acad. Sci. USA 93:1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tansey, W. P., S. Ruppert, R. Tjian, and W. Herr. 1994. Multiple regions of TBP participate in the response to transcriptional activators in vivo. Genes Dev. 8:2756-2769. [DOI] [PubMed] [Google Scholar]
- 66.Timmers, H. T., R. E. Meyers, and P. A. Sharp. 1992. Composition of transcription factor B-TFIID. Proc. Natl. Acad. Sci. USA 89:8140-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a.Tora, L., et al. A unified nomenclature for TATA box binding protein (TBP) associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., in press. [DOI] [PubMed]
- 67.van der Knaap, J. A., J. W. Borst, P. C. van der Vliet, R. Gentz, and H. T. Timmers. 1997. Cloning of the cDNA for the TATA-binding protein-associated factor II170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc. Natl. Acad. Sci. USA 94:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Dyke, M. W., R. G. Roeder, and M. Sawadogo. 1988. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science 241:1335-1338. [DOI] [PubMed] [Google Scholar]
- 69.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 70.Walker, S. S., J. C. Reese, L. M. Apone, and M. R. Green. 1996. Transcription activation in cells lacking TAFIIs. Nature 383:185-188. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Z., and R. G. Roeder. 1995. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc. Natl. Acad. Sci. USA 92:7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinzierl, R. O., B. D. Dynlacht, and R. Tjian. 1993. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature 362:511-517. [DOI] [PubMed] [Google Scholar]
- 73.Wu, S. Y., E. Kershnar, and C. M. Chiang. 1998. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 17:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou, Q., and A. J. Berk. 1995. The yeast TATA-binding protein (TBP) core domain assembles with human TBP-associated factors into a functional TFIID complex. Mol. Cell. Biol. 15:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, Q., T. G. Boyer, and A. J. Berk. 1993. Factors (TAFs) required for activated transcription interact with TATA box-binding protein conserved core domain. Genes Dev. 7:180-187. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]