Abstract
In earlier studies we identified a putative repressor of the human β-globin gene, termed beta protein 1 (BP1), which binds to two silencer DNA sequences upstream of the adult human β-globin gene and to a negative control region upstream of the adult δ-globin gene. Further studies demonstrated an inverse correlation between the binding affinity of the BP1 protein for the distal β-globin silencer sequence and the severity of sickle cell anemia, suggesting a possible role for BP1 in determining the production of hemoglobin S. We have now cloned a cDNA expressing the BP1 protein. Sequencing revealed that BP1 is a member of the homeobox gene family and belongs to the subfamily called Distal-less (DLX), genes important in early development. Further analysis showed that BP1 is an isoform of DLX4. BP1 protein has repressor function towards the β-globin promoter, acting through the two β-globin DNA silencers, demonstrated in transient transfection assays. Strong BP1 expression is restricted to placenta and kidney tissue, with no expression in 48 other human tissues. BP1 exhibits regulated expression in the human erythroid cell line MB-02, where its expression decreases upon induction of the β-globin gene. BP1 is thus the first member of the DLX family with known DNA binding sites and a function in globin gene regulation.
Expression of globin genes in the β-globin cluster is restricted to erythroid cells, with five different genes expressed during embryonic (ɛ), fetal (Gγ and Aγ), and adult (δ and β) development. Transcriptional activation of globin genes occurs not only by the binding of transcriptional activator proteins to the promoter of the gene being activated but also by a regulatory element located 6 to 18 kb upstream of the β-globin cluster, the locus control region (LCR) (23, 30, 44). It was believed that the LCR accomplishes this by opening the chromatin, but recently this idea has been called sharply into question (16). Instead, the LCR may serve strictly as an enhancer. Sequential activation of the β-globin cluster genes during ontogeny must be countered by repression of the globin genes inactive during a given developmental stage. Repression is caused by the binding of repressor proteins to promoter or upstream DNA and, in the case of the adult β-globin gene, may also be due to lack of activation by the LCR (10). In contrast to the identification and understanding of the role of transcriptional activators that bind to DNA sequences near the β-globin gene, little is known about the proteins that repress its transcription.
In earlier studies, two silencer regions upstream of the β-globin gene and a common protein that binds to both of them, termed beta protein 1 (BP1), were identified (3). BP1 binds within silencer I at −530 bp and within silencer II at −300 bp relative to the cap site (+1). Interestingly, it has been found that high mobility group HMG-I (Y), an architectural protein, binds to and bends the DNA at or near the BP1 binding site in silencer I but not in silencer II (5). HMG-I (Y) may facilitate the binding of BP1 and possibly other repressor proteins in this region. Similarly, HMG1, another high mobility group protein, binds to and bends the DNA near the BP1 binding site in silencer II (13).
An inverse correlation between the binding affinity of BP1 and the severity of sickle cell anemia (SCA) has been observed (15). In SCA, the mutated β-globin gene (βS) is in strict linkage disequilibrium with five haplotypes (26, 34). There are differences in the clinical severity of SCA among the haplotypes, with the Indian-Arabo (hereafter referred to as the Indian haplotype) being the mildest and the Bantu being the most severely affected (35). A sequence polymorphism within the −530 bp binding site for BP1 in silencer I is also in linkage disequilibrium with these five SCA haplotypes (6). Using competition electrophoretic mobility shift assays (EMSAs), it was found that BP1 binds five to six times more tightly to the Indian haplotype sequence than to the normal sequence (2, 15, 47). On the other hand, BP1 binds two to three times more weakly to the Bantu haplotype sequence than to the normal sequence (15), which may allow more transcription of βS, resulting in an increased concentration of hemoglobin S. This correlates with the increased severity of SCA observed in Bantus.
Taken together, the above data strongly support the hypothesis that BP1 is a repressor of the β-globin gene. We now report the cloning of BP1 cDNA and demonstrate that BP1 overexpression represses a reporter gene linked to the β-globin promoter and can act through either silencer I DNA or silencer II DNA. BP1 expression is regulated during erythroid differentiation. It is found in the fetal liver, the site of fetal erythropoiesis, and is gradually extinguished during erythroid differentiation of the human cell line MB-02. Sequencing revealed that BP1 contains a homeobox (HB), placing it in an important family of conserved genes encoding transcription factors which regulate each other and unrelated genes as well (19). It maps near a cluster of other HB genes on chromosome 17q21-22 (18). BP1 belongs to the Distal-less (DLX) family of HB genes, which are expressed during early development (7, 11, 39, 42).
MATERIALS AND METHODS
Cloning of BP1.
A λgt11 K562 cell cDNA expression library (Clontech, Inc.) was screened with an oligonucleotide probe containing the BP1 binding site located in silencer II DNA, which included sequences between −336 and −278 bp upstream of the β-globin gene. Screening was as described previously (46) with several modifications. Filters were prehybridized in binding buffer (3) containing 5% Carnation instant dry milk at 4°C for 30 min and then hybridized in the same buffer for 2 to 12 h at 4°C with shaking. Positive plaques were purified and subjected to three additional rounds of screening. Rapid amplification of cDNA ends was performed with a kit from Clontech.
EMSA.
The binding reaction mixture included 500 pg of fusion protein or 250 ng of nuclear extract added to binding buffer (3). Protein extracts from lysogens were prepared as described previously (9). Competitors were preincubated with nuclear extract for 20 min prior to the addition of the probe. Oligonucleotide sequences of probes and competitors for EMSAs were as follows: −530 normal, TGTATATATACACATATATATATATATTTTTTTTCTTTTCTTACCAGAAGGTTT; −530 Indian, TGTACATATACACATATATATATATATATATTTTTTCTTTTCTTACCAGAAGGTTT; −300 beta, TTCTTATTTGTGTAATAAGAAAATTGGGAAAACGATCTTCAATATGCTTACCAAGCTG; −530 delta, TTCTTTTAATGGATATTTATTTCAATATAATAAAAAATTAGAGTTTTA; and nonspecific, TGCATATATATGTATATGTATGTGTGTATA.
The supershift assay was performed by using a polyclonal antibody raised against an N-terminal polypeptide of BP1 (Research Genetics, Inc.). The probe and nuclear extract were incubated for 20 min prior to the addition of 10 μg of the antibody for an additional 20 min. Samples were then immediately loaded onto a polyacrylamide gel for analysis.
Plasmid construction.
To construct a plasmid expressing the BP1 open reading frame (ORF), cDNA was amplified by reverse transcriptase (RT) PCR from K562 cell mRNA. The resulting 1,013-bp product was cloned into the pGEM7 vector (Promega, Inc.), and its identity was confirmed by sequencing. This plasmid was cleaved with HindIII and XbaI to release the BP1 ORF, which was directionally cloned into pRc/RSV (Invitrogen, Inc.).
Transfections and CAT assays.
Transient-transfection assays were performed in K562 cells with DMRIE-C reagent and OPTI-MEM serum-free medium (Gibco-BRL, Inc.). Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (3).
Northern blot analysis.
An RNA dot blot containing mRNA from 50 normal human tissues was purchased (Clontech, Inc.); the amounts of RNA loaded on the blot were normalized by the manufacturer against eight different housekeeping genes, obviating the need to normalize the samples. The probe was obtained by PCR amplification of BP1 cDNA cloned in pBluescript with subsequent EcoRI digestion to release a 3′ BP1 cDNA of 630 bp lacking HB sequences. It was labeled by random priming and hybridized overnight at 65°C. Blots were washed and exposed to X-ray film overnight.
RT-PCR.
Total RNA was isolated by using Trizol reagent (Gibco-BRL) according to the manufacturer's specifications. One microgram of RNA was reverse transcribed with SuperScript II RT (Gibco-BRL) in a total reaction volume of 20 μl. PCR was performed with 1 μl of reverse transcription product in 25 μl. Based on our linearity assay results, the following PCR conditions were used for BP1: each PCR cycle consisted of a denaturation step (94°C, 1 min), an annealing step (62°C, 1 min), and an elongation step (72°C, 1.5 min) for 27 cycles followed by an additional extension (72°C, 5 min). BP1 primers, designed to amplify a product of 588 bp and specific for BP1, were 5′-CACCTCCTGTCTTACCCCTACACC-3′ (forward) and 5′-GCCCTTCCCCAGATTCACATCATC-3′ (reverse). PCR products were electrophoresed on a 2% agarose gel and visualized with ethidium bromide. The product was verified by cleavage with restriction enzymes and hybridization with an internal probe. Primers designed for amplification of β-globin were 5′-GGATCTGTCCACTCCTGATG-3′ (forward) and 5′-TTGAGGTTGTCCAGGTGAGC-3′ (reverse). The product size is 108 bp. β-Actin primers were used as an internal control for all samples.
Growth and induction of MB-02 cells.
The cell line MB-02 was maintained in continuous suspension culture as described previously (32). All cytokines were purchased from R&D Systems. Briefly, stock cultures were grown in nutrient medium RPMI 1640 supplemented with 10% human serum and 5 ng of granulocyte-macrophage colony-stimulating factor/ml. For erythroid differentiation, granulocyte-macrophage colony-stimulating factor depletion was achieved by washing the cells twice in nutrient medium. The cells were reseeded in serum-containing medium supplemented with 25 ng of stem cell factor/ml and 4 U of erythropoietin/ml.
In vitro transcription and translation.
BP1 cDNA was subcloned from pRc/RSV into pGEM-7Zf at the HindIII and XbaI sites. pGEM-7Zf-BP1 was used for in vitro transcription and translation with the TNT coupled reticulocyte or wheat germ lysate system (Promega) according to the manufacturer's protocol. Five micrograms of the reaction mixture was analyzed by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis. A kaleidoscope prestained standard was used as a molecular weight marker (Bio-Rad).
Western blotting.
A polyclonal antibody was raised in rabbits against the N-terminal BP1 peptide SYPYTEPANPGDSYLSCQQ (Research Genetics). Freshly prepared K562 cell protein lysate or cold in vitro-translated protein product was separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis. The proteins were electrophoretically transferred to nitrocellulose membranes. BP1 antibody was diluted in phosphate-buffered saline (1:2,500) and added for 2 h at room temperature, and then the anti-rabbit secondary antibody was added. Detection of the signal was accomplished by using an ECL kit according to the recommended protocol (Amersham Pharmacia Biotech).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been published in the GenBank/EBI Data Bank with accession number AF254115.
RESULTS
Cloning of BP1 cDNA.
A λgt11 cDNA expression library made from human K562 cells was probed with a multimerized oligonucleotide containing the −300 bp BP1 binding site. Two million plaques were screened, and one positive plaque was isolated which expressed a protein that recognized the −300 bp BP1 binding site but not a negative control sequence.
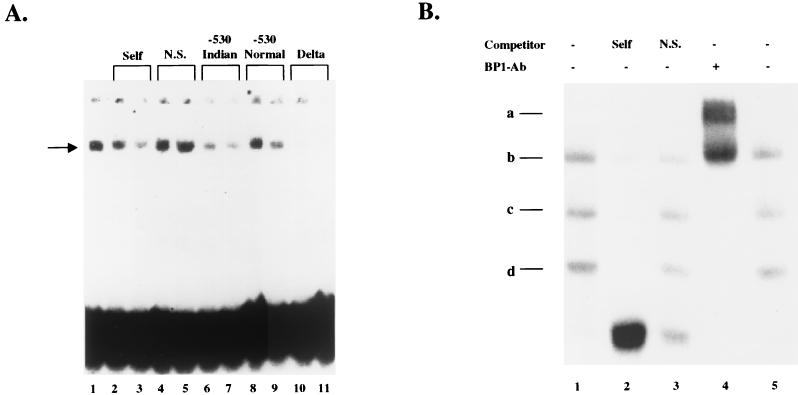
The presence of BP1 protein was originally defined based on its binding sites: it bound to silencers I and II upstream of the β-globin gene and it exhibited extremely tight binding to the Indian haplotype DNA sequence of silencer I (2, 15, 47). Furthermore, BP1 binds in a negative regulatory region located −530 bp upstream of the δ-globin gene (P. Berg, unpublished data). To verify that our cDNA encoded BP1, as defined by its binding site specificity, a competition EMSA was performed. The putative BP1 protein, fused to β-galactosidase, was expressed from a λ lysogen as described in Materials and Methods. This fusion protein was partially purified and incubated with the silencer II probe used for screening the library, resulting in a single shifted band shown in Fig. 1A, lane 1. Cold competitors were added at a 100× and 200× molar excess to determine specificity. The shifted band was competed by self DNA (unlabeled −300 binding site) (Fig. 1A, lanes 2 to 3), the BP1 binding site at −530 bp in silencer I containing either the normal or Indian haplotype sequences (Fig. 1A, lanes 6 to 9), and the BP1 binding site found upstream of the δ-globin gene (Fig. 1A, lanes 10 to 11) but not by a nonspecific DNA (Fig. 1A, lanes 4 to 5). Note that the Indian sequence is a stronger competitor than the normal sequence, indicating tighter binding of BP1 to the Indian sequence, as was previously observed with K562 nuclear extracts (15). The BP1 site upstream of the δ-globin gene is also a strong competitor for BP1 binding. A negative control, pure β-galactosidase protein, did not exhibit binding activity towards the probe (data not shown). The mobility shift analysis thus demonstrated that the binding properties of the fusion protein are identical to those of unpurified BP1.
FIG. 1.
Verification of the cloning of BP1. (A) Binding characteristics of the BP1 protein. The probe was incubated with fusion protein, as described in Materials and Methods, in the absence or presence of cold competitor oligonucleotide. The band at the top marks the position of the wells, and the band at the bottom contains the free probe. The arrow indicates the position of the shifted band. Per reaction, 10,000 cpm of probe was used, and the amount of cold competitor added was based on the specific activity of the probe. Lanes 2, 4, 6, 8, and 10 contain a 100× excess of the cold competitor indicated, and lanes 3, 5, 7, 9, and 11 contain a 200× excess of cold competitor. (B) Antibody against the cloned BP1 protein (BP1-Ab) recognizes the BP1 protein in a nuclear extract. Bands c and d represent specific shifted bands, and bands a and b are supershift bands. The amount of competitors were 500× self DNA (lane 2) and 500× nonspecific (N.S.) DNA (lane 3). +, present; −, absent.
To further demonstrate that the BP1 cDNA encodes the same protein identified as BP1 in crude extracts, a supershift assay was performed with a polyclonal antibody against BP1. K562 nuclear extract was incubated with the oligonucleotide probe containing the Indian haplotype sequence (Fig. 1B, lanes 1 and 5). Competition with cold self DNA (Fig. 1B, lane 2) compared with nonspecific DNA (Fig. 1B, lane 3) demonstrated two specific bands, c and d, and a nonspecific band near position b; the free probe is at the bottom. The presence of more than one specific shifted band may represent homo- or heterodimers of BP1, or BP1 and a partner protein binding to the probe, and is consistent with previous gel shift experiments that identified BP1 (3). When BP1 antibody was added, both bands c and d largely disappeared and new bands appeared in positions a and b, a characteristic of supershifting. Thus, the antibody raised against our cloned protein recognizes the specific shifted bands from a nuclear extract of K562 cells, showing that we have cloned BP1.
Verification of BP1.
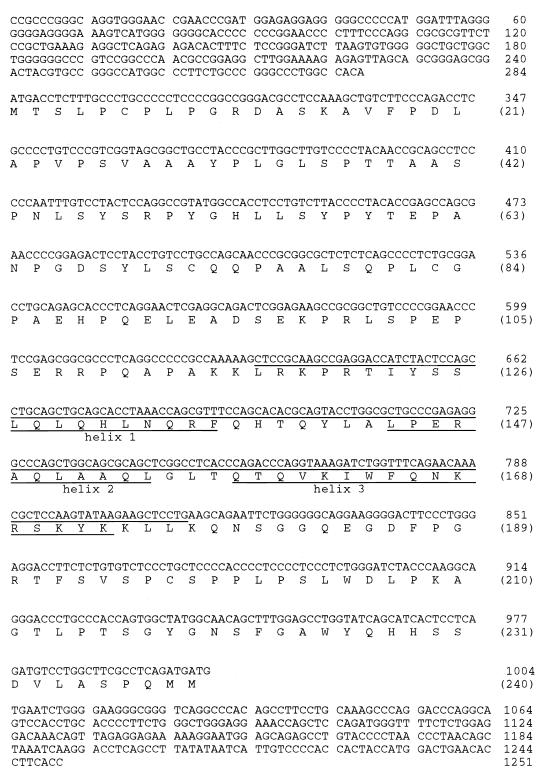
The initial BP1 cDNA was 630 bp in length. When part of this sequence was used as a probe in a Northern blot containing K562 RNA, a single RNA band of 2.1 kb was recognized, indicating that the cloned BP1 sequence was incomplete (data not shown). To extend the cDNA sequence, we used rapid amplification of cDNA ends. Additional sequences in both the 5′ and 3′ directions were obtained, giving a cDNA of 1,251 bp. The predicted ORF, determined by computer analysis and identification of a Kozak consensus sequence (25), is shown in Fig. 2.
FIG. 2.
BP1 cDNA sequence and predicted ORF. Numbers at the right without parentheses indicate nucleotide numbers, and numbers with parentheses indicate amino acid numbers. The nucleotides included in the HB are underlined, and the three predicted α helices are indicated by underlining of the appropriate amino acids.
Computer analysis with BLAST (1) indicated that BP1 contains an HB, placing it in a family of genes known to be important in development. The HB in Fig. 2 is underlined, and the amino acids comprising the three predicted α helices found in homeodomains (HDs) (19) are also indicated. The second and third helices comprise the helix-turn-helix motif which is characteristic of HDs. BP1 belongs to the DLX subfamily based on DNA sequence homology to the HB of these genes.
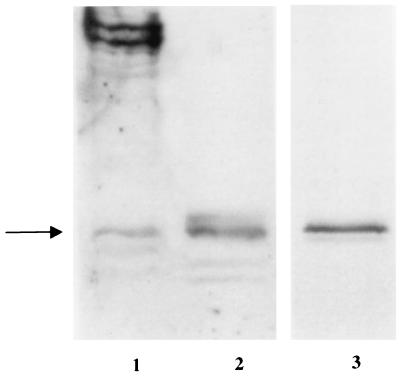
To verify that we cloned the complete BP1 ORF, we compared the sizes of in vitro-translated BP1 protein with proteins from K562 cells by using Western blot analysis (Fig. 3, lanes 1 to 2). Proteins from K562 cells are in lane 1, and the in vitro-translated protein product is in lane 2. Controls with rabbit reticulocyte lysate only and with preimmune serum only did not show any bands (data not shown). There is a protein band in common to lanes 1 and 2. The higher molecular mass bands in lane 1 are 80 to 100 kDa, too large to be encoded by BP1 unless there are extensive posttranslational modifications. The presence of a common band recognized by BP1 antibody in both K562 cells and in vitro-translated BP1 protein indicates that the cloned BP1 ORF is complete. The band size is approximately 32 kDa. The same size band was seen after 35S labeling of the in vitro BP1 protein product (Fig. 3, lane 3), confirming that we have cloned the complete BP1 ORF. The predicted size of the BP1 protein is 26 kDa, suggesting that the BP1 protein may undergo posttranslational modifications.
FIG. 3.
Identification of the BP1 protein by Western blot analysis and in vitro transcription and translation. Western blot analysis was performed with protein from K562 cells (lane 1) and in vitro-translated BP1 protein (lane 2) with polyclonal BP1 antibody. Lane 3 shows 35S-labeled in vitro-translated BP1 protein after autoradiography. The arrow indicates the position of the 32-kDa BP1 band.
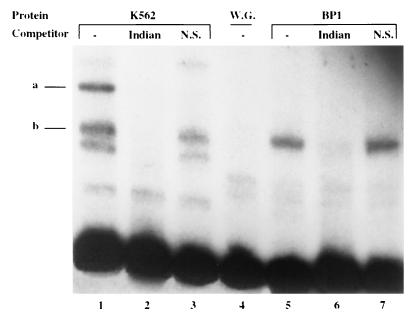
Using an EMSA, we also compared the binding of BP1 from K562 extracts with the binding of in vitro-translated BP1 (Fig. 4). There is a specific shifted band at position b in both protein extracts which is competed by the Indian sequence (Fig. 4, lanes 2 and 6) but not by DNA lacking a BP1 binding site (Fig. 4, lanes 3 and 7). Band a, also a specific shifted band, is found only with K562 extract and could be the result of the binding of BP1 and another protein(s). These results further demonstrate that the in vitro BP1 protein expressed from the cloned cDNA is full length.
FIG. 4.
Proteins from K562 cells and BP1 in vitro protein give a common shifted band with the EMSA. Proteins from K562 cells or BP1 in vitro protein translated in a wheat germ system were incubated with a probe containing the BP1 binding site in silencer I. The free probe is at the bottom. Cold DNA competitors, added at 1,000× excess, were the Indian haplotype sequence (lanes 2 and 6) or nonspecific (N.S.) DNA (lanes 3 and 7). Lanes 1 and 5 are controls lacking competitors, and lane 4 contains only wheat germ extract (W.G.) as a control.
BP1 represses the β-globin promoter.
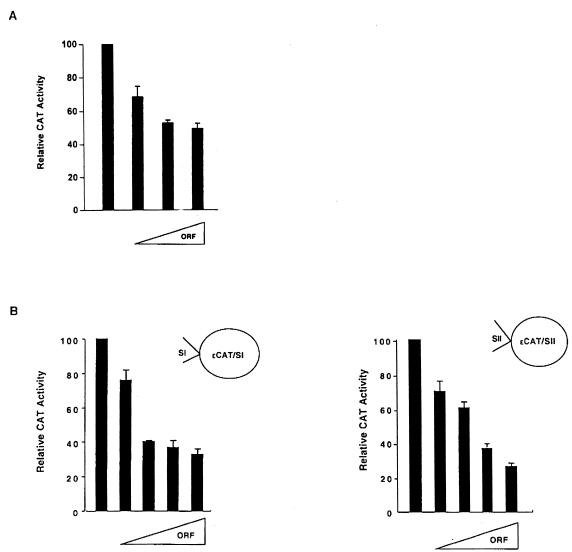
To determine the function of BP1 protein, transient transfection assays were performed in K562 cells. K562 cells are frequently used as a model to study β-globin expression (for examples, see references 28 and 48). They were transiently cotransfected with increasing amounts of either an empty vector or pRSV/BP1-ORF, which encodes the BP1 protein driven by the Rous sarcoma virus (RSV) promoter, along with a target plasmid (Fig. 5). The target was a construct which fuses the CAT reporter gene downstream of the β-globin promoter, which extends to −338 bp and includes silencer II (3). However, CAT levels are barely detectable with this plasmid in K562 cells (3), so the simian virus 40 (SV40) enhancer was added to boost expression. This plasmid is called pβD338/SV40. CAT activity was normalized against a cotransfected internal standard, pCMV/β-gal. Relative CAT activity was calculated as the ratio of CAT activity in cells receiving pRSV/BP1-ORF to the CAT activity of cells receiving only the empty vector. Experiments were repeated two to five times, with at least four replicates for each point. When increasing amounts of pRSV/BP1-ORF DNA were added to pβD338/SV40 (Fig. 5A), CAT activity was repressed. The level of repression was 2.0-fold with 10 μg of pRSV/BP1-ORF; repression leveled off at 5 μg. This relatively low level of repression may be due to the fact that the BP1 protein must overcome the activation of the β-globin promoter by the SV40 enhancer before repression is detectable. Nonetheless, it is clear that the BP1 protein exhibits repressor activity towards the β-globin promoter.
FIG. 5.
Repressor activity of BP1. (A) Expression of pβD338/SV40 was measured in the presence of 2, 5, and 10 μg of pRSV/BP1-ORF. (B) Expression of pɛCAT/SI was measured in the presence of 2, 3, 4, and 5 μg of pRSV/BP1-ORF, and pɛCAT/SII expression was measured with 1, 2, 3, and 4 μg of pRSV/BP1-ORF. Increasing amounts of pRSV/BP1-ORF are indicated with wedges.
To test repression without the interference of an enhancer and to determine whether BP1 protein represses through the two silencers we previously identified, additional experiments were performed. Either silencer I (pɛCAT/SI) or silencer II (pɛCAT/SII), cloned into an expression vector containing the ɛ-globin promoter fused to the CAT reporter gene, was used as a target of the BP1 protein (Fig. 5B). These plasmids express well in K562 cells without the addition of an enhancer (3). There was also a plateau in CAT expression when silencer I was the target, beginning at 3 μg of pRSV/BP1-ORF and giving about a threefold level of repression. Likewise, adding increasing amounts of pRSV/BP1-ORF DNA relative to pɛCAT/SII caused increasing repression of the CAT reporter gene. Even at the lowest amount, 1 μg of pRSV/BP1-ORF, there was a sharp decrease in CAT expression. Maximal repression was about fivefold. A control, an ɛCAT plasmid lacking silencer DNA, was unresponsive to the addition of pRSV/BP1-ORF (data not shown). Thus, BP1 protein represses the β-globin promoter and can act through silencer I or silencer II DNA.
Expression analysis of BP1.
To analyze expression in a broad spectrum of tissues, a dot blot containing normal human RNA from 50 tissues was hybridized with a BP1 cDNA probe (Fig. 6 and Table 1). A signal was observed for only two tissues, placenta and kidney. RNA from the remaining 48 tissues showed no hybridization, even after 5 days of autoradiography (data not shown). BP1 expression in placenta and kidney tissue was verified by RT-PCR with BP1-specific primers (data not shown).
FIG. 6.
Expression of BP1 in normal tissues. An RNA Northern dot blot was screened with a BP1 probe and exposed to X-ray film overnight. Position E1 contains kidney RNA, and position F4 contains placental RNA. Sources of RNA are given in Table 1.
TABLE 1.
Tissue-specific expression of BP1
| Tissue source | BP1 expression result |
|---|---|
| Placenta | + |
| Kidney | + |
| Whole brain | − |
| Amygdala | − |
| Caudate nucleus | − |
| Cerebellum | − |
| Hippocampus | − |
| Frontal lobe | − |
| Medulla oblongata | − |
| Occipital lobe | − |
| Putamen | − |
| Substantia nigra | − |
| Temporal lobe | − |
| Subthalamic nucleus | − |
| Thymus | − |
| Heart | − |
| Aorta | − |
| Skeletal muscle | − |
| Colon | − |
| Bladder | − |
| Uterus | − |
| Prostate | − |
| Stomach | − |
| Testis | − |
| Ovary | − |
| Pancreas | − |
| Adrenal gland | − |
| Thyroid gland | − |
| Salivary gland | − |
| Pituitary gland | − |
| Trachea | − |
| Peripheral leukocyte | − |
| Lymph node | − |
| Bone marrow | − |
| Appendix | − |
| Thalamus | − |
| Liver | − |
| Spinal cord | − |
| Mammary gland | − |
| Small intestine | − |
| Spleen | − |
| Cerebral cortex | − |
| Lung | − |
| Fetal kidney | − |
| Fetal spleen | − |
| Fetal thymus | − |
| Fetal brain | − |
| Fetal heart | − |
| Fetal liver | − |
| Fetal lung | − |
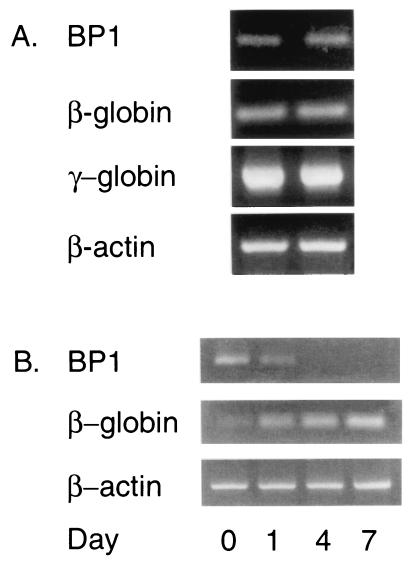
If BP1 represses the β-globin gene, it may be expressed in the fetal liver, the site of fetal erythropoiesis. Although analysis of the dot blot did not reveal expression of BP1 in fetal liver tissue, Northern analysis is relatively insensitive. For this reason, RT-PCR was performed on two 20-week human fetal liver samples. Using this approach, expression of BP1 mRNA was observed (Fig. 7A). Expression of the fetal γ-globin gene was high, as expected. β-Globin gene transcription has begun by 20 weeks (29) and was detectable in both fetal liver tissues.
FIG. 7.
Expression of BP1 during erythroid differentiation. (A) Expression in fetal liver tissue. RT-PCR was used to analyze expression of BP1, β-globin, and γ-globin in fetal liver tissue at 20 weeks. (B) Expression during terminal differentiation of MB-02 cells. Expression of BP1 and β-globin was compared on days 1, 4, and 7 after erythroid induction. β-Actin was an internal control in both panels A and B.
BP1 expression was examined during erythroid differentiation by using the cell line MB-02, a human erythroid progenitor cell line derived from a patient with megakaryoblastic leukemia (32). Although MB-02 cells have not been successfully used in transient transfection assays or stably transfected (D. Morgan, unpublished data), they serve as an excellent model of fetal and adult erythroid differentiation since, upon treatment with erythropoietin, they show increased expression of fetal globin genes and induction of the β-globin gene (32). We compared BP1 and β-globin expression by RT-PCR after induction of MB-02 cells with erythropoietin (Fig. 7B). β-Globin mRNA expression increased from day 0 (no inducer) to days 1, 4, and 7 postinduction. Simultaneously, BP1 mRNA decreased on day 1 and was undetectable by day 4 or 7. Thus, BP1 expression is gradually extinguished upon induction of β-globin mRNA.
DISCUSSION
In this paper, we report the isolation of a cDNA expressing a new HD protein, BP1, and establish its function as a β-globin gene repressor. Two approaches were used to demonstrate that we successfully cloned BP1. First, cloned and expressed BP1 protein recognizes the BP1 binding sites in silencers I and II of the β-globin gene (3) and a DNA site in a negative control region upstream of the δ-globin gene. These common BP1 binding sites near the β- and δ-globin genes may be a point of coordinate regulation of the adult β-like globin genes. The cloned BP1 protein also retains the increased binding affinity for the Indian haplotype DNA sequence that was demonstrated for BP1 in crude protein extracts (15). Furthermore, a supershift assay with an antibody raised against the expressed protein recognizes BP1 protein in K562 nuclear extracts, demonstrating that we have cloned the same protein we called BP1 in nuclear extracts.
As predicted by earlier studies, BP1 exhibits repressor function towards the β-globin gene. Using increasing amounts of a plasmid containing the BP1 ORF causes increasing repression of three target reporter genes, one including the β-globin promoter and two constructs with silencer I or II DNA cloned separately. These experiments demonstrate that BP1 can function as a repressor through either silencer DNA (Fig. 5). Expression of the reporter gene was not completely extinguished, indicating a possible requirement for binding of additional, low abundance repressor proteins or the lack of partner proteins.
The binding site recognized by the BP1 protein has been determined by DNase I footprint analysis in both silencers (3), and a consensus BP1 binding site, (A/T)T(A/C)(A/T)ATATG, can be deduced. This site, like HOX binding sites, is AT rich (45). Based on our footprint analysis, site-directed mutagenesis of the BP1 binding sites in both silencers I and II was performed in a β-globin vector containing the CAT reporter gene (13, 14). When these constructs were transfected into K562 cells, repression was relieved, shown by a four- to sixfold increase in CAT expression. Furthermore, the BP1 protein no longer bound to the mutated BP1 binding site from either silencer. Those experiments demonstrated that it is the BP1 binding site within each silencer that is a repressor binding site and are supportive of the data presented here.
BP1 is an HB gene and a member of the Distal-less family. HB genes are known as master regulator genes during development because, upon mutation, vast morphological abnormalities occur. Distal-less genes are found in organisms as diverse as mice, Xenopus, zebra fish, and humans (43). In Drosophila, Distal-less (called Dll) is required for the normal development of larval sensory organs of the head and thorax. In larval and adult flies, Dll plays an important role in the development of the distal regions of the limbs (7, 8). There is a single Dll gene in Drosophila, but there are multiple Distal-less genes in other organisms. In mammals, seven Distal-less (called Dlx in mice and DLX in humans) genes have been identified so far. During murine development, Dlx genes are expressed in branchial arches, forebrain, limbs, eyes, teeth, bones, and facial mesenchyme (11, 39, 42).
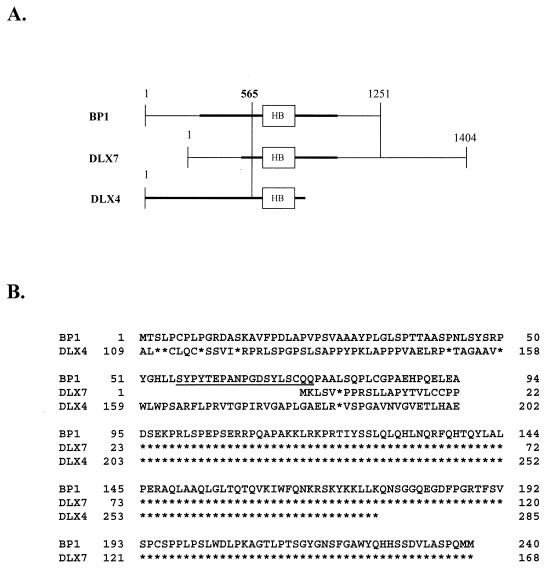
Interestingly, BP1 and two recently cloned human cDNAs, DLX4 and DLX7 (33, 38), appear to be isoforms (splice variants), since their HBs are identical (Fig. 8A) and they exhibit extensive additional areas of amino acid identity (Fig. 8B). There is limited sequence available for DLX4 (38), and to the best of our knowledge, its function is not yet defined. BP1 and DLX4 diverge upstream of nucleotide 565 of BP1 (Fig. 8A), and they share amino acid sequence identity downstream of that site (Fig. 8B). BP1 and DLX7 (33, 37) share almost complete sequence identity from nucleotides 565 to 1251 of BP1, differing by only a few base pairs 3′ to their respective ORFs. Further upstream of nucleotide 565, there is no homology, indicating that nucleotide 565 may be a splice site. A murine Dlx7 has been cloned which shares 88% DNA sequence homology with BP1 and only 46% homology with DLX7 (33), raising the possibility that the murine Dlx7 actually corresponds to BP1. In addition, the murine ORF and the predicted BP1 ORF share the same translational start site, and the following 8 bp are identical. Regarding function, abrogation of DLX7 expression leads to reduced expression of c-myc and GATA-1 and then to apoptosis (41). A consensus binding site for DLX7 has not yet been determined, but DLX7 protein binds to silencer I DNA, although it lacks repressor function (18). Since the sequence of DLX4 was the first to appear in GenBank (accession no. U31762), we consider BP1 and DLX7 to be splice variants of DLX4.
FIG. 8.
BP1 and DLX7 are isoforms of DLX4. (A) cDNA sequence homology. Thick black lines denote predicted ORFs. Number 565 indicates the nucleotide of BP1 where divergence occurs among BP1, DLX4, and DLX7. HB is the HB region. The regions between the two vertical lines indicate the regions of homology. (B) Amino acid sequence homology. Asterisks indicate the same amino acid. The amino acids comprising the peptide of BP1 used to generate a polyclonal antibody are underlined.
A striking feature of BP1 is its restricted expression. BP1 mRNA expression was observed in only 2 of 50 tissues, placenta and kidney (Table 1 and Fig. 6). Although the probe would recognize BP1, DLX4, or DLX7, the expression of BP1 in placenta and kidney tissue was confirmed by RT-PCR (data not shown). Expression in the placenta is consistent with a role for BP1 in early development, a feature of HB genes. The significance of high expression in adult kidney tissue is not known. However, it is intriguing that the expression of erythropoietin, a cytokine that controls the differentiation, proliferation, and survival of erythroid progenitors, also occurs in the kidney (reviewed in reference 31). These data suggest that BP1 may have multiple regulatory roles during development.
BP1 expression is regulated during erythroid differentiation. It is expressed in the fetal liver, the site of fetal hematopoiesis, and a tissue in which β-globin expression is low. In MB-02 cells, BP1 is down-regulated during erythroid maturation and expression of the β-globin gene. These findings are consistent with the repressor function of BP1. Interestingly, BP1 expression has also been detected in CD34-negative cells (22), early hematopoietic progenitors. We propose a model in which BP1 is expressed in primitive hematopoietic cells and down-regulated during erythroid differentiation. Potentially, BP1 could be used in the therapy of SCA to repress transcription of the βs-globin gene. Expression of BP1 could be placed under the control of a β-globin promoter lacking silencers I and II and targeted to erythroid cells by using a vector such as the one described by Kasahara et al. (24). Current treatments of SCA focus on chemical reactivation of fetal globin genes, which may have adverse side effects. Repression of hemoglobin S may reduce or eliminate the need for such chemicals. Reduction of βs-globin gene expression may also indirectly reactivate fetal globin genes. This idea is based on well-established observations, both in vivo and in vitro, which demonstrate reciprocal regulation of fetal and adult globin genes (12, 17, 36).
Few direct molecular targets of human homeotic proteins are known. In vitro, human homeotic proteins HOXD9 and HNF-1 bind to and regulate the liver cell adhesion molecule gene from chickens (20), and several human HOX genes have been shown to self regulate or to regulate other HOX genes (45). For example, the human HOXB3 gene transactivates the rat thyroid transcription factor-1 (TTF-1) gene, which is also an HB-containing gene, making the human TTF-1 gene a likely target of HOXB3 (21). Interestingly, Lavelle et al. have shown that a homeotic protein, the product of HOXB2, binds to DNA sequences in the β-globin locus control region, the γ-globin promoter and the Aγ-globin enhancer (27); transient transfection assays show that HOXB2 has a negative regulatory effect on transcription of a γ-globin-luciferase construct (4). Also, overexpression of HOX 2.2 is associated with reduced erythroid features in K562 cells, although the mechanism for this effect is unknown (40). In mice, Hox genes have been shown to regulate not only cell adhesion molecules and integrins but also transcription factors involved in the maturation of thymocytes and genes involved in B-cell development and thyroid increase (45). The identification of BP1 as a homeotic protein with the ability to bind to two β-globin silencer sequences and repress the β-globin gene makes this one of the few human HB genes with a defined target and the first with a known function in erythropoiesis.
Acknowledgments
We thank Allan Goldstein, Valerie Hu, Allen Eskenazi, and Christopher Frantz for helpful discussions and suggestions and Christine Obriecht for excellent technical assistance.
This work was supported by National Institutes of Health grant R01 DK53533 (P.B.).
M.B.C. and S.F. contributed equally to this work.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, P. E., M. Mittelman, J. Elion, D. Labie, and A. N. Schechter. 1991. Increased protein binding to a −530 mutation of the human β-globin gene associated with decreased β-globin synthesis. Am. J. Hematol. 36:42-47. [DOI] [PubMed] [Google Scholar]
- 3.Berg, P. E., D. M. Williams, R.-L. Qian, R. B. Cohen, S. X. Cao, M. Mittelman, and A. N. Schechter. 1989. A common protein binds to two silencers 5′ to the human β-globin gene. Nucleic Acids Res. 17:8833-8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Case, S. S., P. Huber, and J. A. Lloyd. 1999. The gamma PE complex contains both SATB1 and HOXB2 and has positive and negative roles in human gamma-globin gene regulation. DNA Cell Biol. 18:805-817. [DOI] [PubMed] [Google Scholar]
- 5.Chase, M. B., S. Haga, W. D. Hankins, D. M. Williams, Z. Bi, J. W. Strovel, C. Obriecht, and P. E. Berg. 1999. Binding of HMG-I(Y) elicits structural changes in a silencer of the human β-globin gene. Am. J. Hematol. 60:27-35. [DOI] [PubMed] [Google Scholar]
- 6.Chebloune, Y., J. Pagnier, G. Trabuchet, C. Faure, G. Verdier, D. Labie, and V. M. Nigon. 1988. Structural analysis of the 5′ flanking region of the β-globin gene in African sickle cell anemia patients: further evidence for three origins of the sickle cell mutation in Africa. Proc. Natl. Acad. Sci. USA 85:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. M., G. Bronner, F. Kuttner, G. Jurgens, and H. Jackle. 1989. Distal-less encodes a homeodomain protein required for limb development in Drosophila. Nature 338:432-434. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, S. M., and G. Jurgens. 1989. Proximal-distal pattern formation in Drosophila: cell autonomous requirement for Distal-less gene activity in limb development. EMBO J. 8:2045-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowell, I. G., and H. C. Hurst. 1994. Cloning transcription factors from a cDNA expression library, p. 120-122. In D. S. Latchman (ed.), Transcription factors: a practical approach. IRL Press, New York, N.Y.
- 10.Crossley, M., and S. H. Orkin. 1993. Regulation of the β-globin locus. Curr. Opin. Genet. Dev. 3:232-237. [DOI] [PubMed] [Google Scholar]
- 11.Dolle, P., M. Price, and D. Duboule. 1992. Expression of the murine Dlx-1 homeobox gene during facial, ocular, and limb development. Differentiation 49:93-99. [DOI] [PubMed] [Google Scholar]
- 12.Dover, G. J., and S. H. Boyer. 1987. Fetal hemoglobin-containing cells have the same mean corpuscular hemoglobin as cells without fetal hemoglobin: a reciprocal relationship between gamma and beta-globin gene expression in normal subjects and in those with high fetal hemoglobin production. Blood 69:1109-1113. [PubMed] [Google Scholar]
- 13.Drew, L., D. C. Tang, P. E. Berg, and G. P. Rodgers. 2000. The role of trans-acting factors and DNA-bending in the silencing of human beta-globin gene expression. Nucleic Acids Res. 28:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebb, D., D. C. Tang, L. Drew, K. Chin, P. E. Berg, and G. P. Rodgers. 1998. Identification of regulatory elements that repress adult beta-like globin genes. Blood Cells Mol. Dis. 24:356-369. [DOI] [PubMed] [Google Scholar]
- 15.Elion, J., P. E. Berg, C. Lapoumeroulie, G. Trabuchet, M. Mittelman, R. Krishnamoorthy, A. N. Schechter, and D. Labie. 1992. DNA sequence variation in a negative control region 5′ to the β-globin gene correlates with the phenotypic expression of the βS mutation. Blood 79:787-792. [PubMed] [Google Scholar]
- 16.Epner, E., A. Reik, D. Cimbora, A. Telling, M. A. Bender, S. Fiering, T. Enver, D. I. K. Martin, M. Kennedy, G. Keller, and M. Groudine. 1998. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol. Cell 2:447-455. [DOI] [PubMed] [Google Scholar]
- 17.Fibach, E., P. Prasanna, G. P. Rodgers, and D. Samid. 1993. Enhanced fetal hemoglobin production by phenylacetate and 4-phenylbutyrate in erythroid precursors derived from normal donors and patients with sickle cell anemia and β-thalassemia. Blood 82:2203-2209. [PubMed] [Google Scholar]
- 18.Fu, S., H. Stevenson, J. W. Strovel, S. B. Haga, J. Stamberg, K. Do, and P. E. Berg. 2001. Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene 278:131-139. [DOI] [PubMed] [Google Scholar]
- 19.Gehring, W. J. 1993. Exploring the homeobox. Gene 135:215-221. [DOI] [PubMed] [Google Scholar]
- 20.Goomer, R. S., B. D. Holst, I. C. Wood, F. S. Jones, and G. M. Edelman. 1994. Regulation in vitro of an L-CAM enhancer by homeobox genes HoxD9 and HNF-1. Proc. Natl. Acad. Sci. USA 91:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guazzi, S., R. Lonigro, L. Pintonello, E. Boncinelli, R. DiLauro, and F. Mavilio. 1994. The thyroid transcription factor-1 gene is a candidate target for regulation by Hox proteins. EMBO J. 13:3339-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga, S., S. Fu, J. E. Karp, D. D. Ross, D. M. Williams, W. D. Hankins, F. Behm, F. W. Ruscetti, M. Chang, B. D. Smith, D. Becton, S. C. Raimondi, and P. E. Berg. 2000. BP1, a new homeobox gene, is frequently expressed in acute leukemias. Leukemia 14:1867-1875. [DOI] [PubMed] [Google Scholar]
- 23.Hardison, R., J. L. Slightom, D. L. Gumucio, M. Goodman, N. Stojanovic, and W. Miller. 1997. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205:73-94. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara, N., A. M. Dozy, and Y. W. Kan. 1994. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science 266:1373-1376. [DOI] [PubMed] [Google Scholar]
- 25.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulozik, A. E., J. S. Wainscoat, G. R. Serjeant, B. D. Kar, B. Al-Awamy, G. J. F. Essan, A. G. Falusi, S. K. Haque, A. M. Hilali, S. Kate, W. A. E. P. Ranasinghe, and D. J. Weatherall. 1986. Geographical survey of βs-globin gene haplotypes: evidence for an independent Asian origin of the sickle-cell mutation. Am. J. Hum. Genet. 39:239-244. [PMC free article] [PubMed] [Google Scholar]
- 27.Lavelle, D., J. Ducksworth, E. Eves, G. Gomes, M. Keller, P. Heller, and J. DeSimone. 1991. A homeodomain protein binds to gamma-globin gene regulatory sequences. Proc. Natl. Acad. Sci. USA 88:7318-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. S., H. Ngo, D. Kim, and J. H. Chung. 2000. Erythroid Kruppel-like factor is recruited to the CACCC box in the beta-globin promoter but not to the CACCC box in the gamma-globin promoter: the role of the neighboring promoter elements. Proc. Natl. Acad. Sci. USA 97:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley, T. J., K. A. Maloney, J. I. Gordon, and A. L. Schwartz. 1989. Globin gene expression in erythroid human fetal liver cells. J. Clin. Investig. 83:1032-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, D., S. Fiering, and M. Groudine. 1996. Regulation of beta-globin gene expression: straightening out the locus. Curr. Opin. Genet. Dev. 6:488-495. [DOI] [PubMed] [Google Scholar]
- 31.Maxwell, P., and P. Ratcliffe. 1998. Regulation of expression of the erythropoietin gene. Curr. Opin. Hematol. 5:166-170. [DOI] [PubMed] [Google Scholar]
- 32.Morgan, D. A., D. L. Gumucio, and I. Brodsky. 1991. Granulocyte-macrophage colony-stimulating factor-dependent growth and erythropoietin-induced differentiation of a human cell line MB-02. Blood 78:2860-2871. [PubMed] [Google Scholar]
- 33.Nakamura, S., D. W. Stock, K. L. Wydner, J. A. Bollekens, K. Takeshita, B. M. Nagai, S. Chiba, T. Kitamura, T. M. Freeland, Z. Zhao, J. Minowada, J. B. Lawrence, K. B. Weiss, and F. H. Ruddle. 1996. Genomic analysis of a new mammalian Distal-less gene: Dlx-7. Genomics 38:314-324. [DOI] [PubMed] [Google Scholar]
- 34.Pagnier, J., J. G. Mears, O. Dunda-Belkhodja, K. E. Schaefer-Rego, C. Beldjord, R. L. Nagel, and D. Labie. 1984. Evidence of the multicentric origin of the hemoglobin S gene in Africa. Proc. Natl. Acad. Sci. USA 81:1771-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrine, R. P., M. E. Pembrey, S. Perrine, and F. Shoup. 1978. Natural history of sickle cell anemia in Saudi Arabs. A study of 270 subjects. Ann. Intern. Med. 88:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Perrine, S. P., B. A. Miller, D. V. Faller, R. A. Cohen, E. P. Vichinsky, D. Hurst, B. H. Lubin, and T. Papayannopoulou. 1989. Sodium butyrate enhances fetal globin gene expression in erythroid progenitors of patients with HbSS and β thalassemia. Blood 74:454-459. [PubMed] [Google Scholar]
- 37.Price, J. A., D. W. Bowden, J. T. Wright, M. J. Pettenati, and T. C. Hart. 1998. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum. Mol. Genet. 7:563-569. [DOI] [PubMed] [Google Scholar]
- 38.Quinn, L. M., B. V. Johnson, J. Nicholl, G. R. Sutherland, and B. Kalionis. 1997. Isolation and identification of homeobox genes from the human placenta including a novel member of the Distal-less family, DLX4. Gene 187:55-61. [DOI] [PubMed] [Google Scholar]
- 39.Robinson, G. W., and K. Mahon. 1994. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mech. Dev. 48:199-215. [DOI] [PubMed] [Google Scholar]
- 40.Shen, W.-R., K. Detmer, C. H. E. Mathews, F. M. Hack, D. A. Morgan, C. Largmen, and H. J. Lawrence. 1992. Modulation of homeobox gene expression alters the phenotype of human hematopoietic cell lines. EMBO J. 11:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimamoto, T., S. Nakamura, J. Bollekenn, F. H. Ruddle, and K. Takeshita. 1997. Inhibition of DLX-7 homeobox gene causes decreased expression of GATA-1 and c-myc genes and apoptosis. Proc. Natl. Acad. Sci. USA 94:3245-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simeone, A., D. Acampora, M. Pannese, M. D'Esposito, A. Stornaiuolo, M. Gulisano, A. Mallamaci, K. Kastury, T. Druck, and K. Huebner. 1994. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA 91:2250-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock, D. W., D. L. Ellies, Z. Zhao, M. Ekker, F. H. Ruddle, and K. M. Weiss. 1996. The evolution of the vertebrate Dlx family. Proc. Natl. Acad. Sci. USA 93:10858-10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuan, D., W. Soloman, Q. Li, and I. M. London. 1985. The “β-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. USA 82:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Oostveen, J. W., J. J. Bijl, F. M. Raaphorst, J. J. M. Walbooners, and C. J. L. M. Miejer. 1999. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia 13:1675-1690. [DOI] [PubMed] [Google Scholar]
- 46.Vinson, C. R., K. L. LaMarco, P. F. Johnson, W. H. Landschulz, and S. L. McKnight. 1988. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 2:801-806. [DOI] [PubMed] [Google Scholar]
- 47.Zeng, F.-Y., G. P. Rodgers, S.-Z. Huang, A. N. Schechter, M. Salamah, S. Perrine, and P. E. Berg. 1994. Sequence of the −530 region of the beta globin gene of sickle cell anemia patients with the Arabian haplotype. Hum. Mutat. 3:163-165. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, S. Z., Q. Li, G. Stamatoyannopoulos, and A. Srivastava. 1996. Adeno-associated virus 2-mediated transduction and erythroid cell-specific expression of a human beta-globin gene. Gene Ther. 3:223-229. [PubMed] [Google Scholar]