Abstract
The E2F1, -2, and -3 transcription factors are key downstream targets of the retinoblastoma protein (pRB) tumor suppressor that drive expression of proliferation-associated genes. Here we use mutant mouse strains to investigate E2F3's role in vivo. We show that E2F3 is essential for embryonic viability in the pure 129/Sv background but the presence of C57BL/6 alleles yields some adult survivors. Although growth retarded, surviving E2f3−/− animals are initially healthy. However, they die prematurely, exhibiting no obvious tumor phenotype but with the typical signs of congestive heart failure. The defects are completely distinct from those arising in E2f1 mutant mice (S. J. Field et al., Cell 85:549-561; 1996; L. Yamasaki et al., Cell 85:537-548, 1996), supporting the prevailing view that these E2Fs must have some unique biological functions in vivo. To test this model, we examined the phenotypes of E2f1 E2f3 compound mutant mice. Almost all of the developmental and age-related defects arising in the individual E2f1 or E2f3 mice were exacerbated by the mutation of the other E2f. Thus, E2F1 and E2F3 appear to play critical, overlapping roles in the development and maintenance of a variety of tissues. Importantly, this study did identify one major difference in the properties of E2F1 and E2F3: either alone or in combination with E2F1 loss, E2f3 mutation did not increase the incidence of tumor formation. These data strongly suggest that tumor suppression is a specific property of E2F1 and not E2F3.
The E2F transcription factors control the cell cycle-dependent expression of genes that are essential for cellular proliferation, including key components of the DNA replication and cell cycle control machinery (reviewed in reference 13). E2F activity is regulated by the retinoblastoma protein (pRB), a tumor suppressor that is functionally inactivated in most, if not all, human tumors (reviewed in reference 42). The hypophosphorylated form of pRB binds to E2F during the G1 phase of the cell cycle (5). This association inhibits activation of transcription by E2Fs (15, 21), and the pRB-E2F complex has the capacity to actively repress E2F-responsive genes by recruiting histone-modifying enzymes to the promoter (2, 35, 52). In response to mitogenic signaling, pRB is phosphorylated by the cyclin-dependent kinases (3, 6, 10) and transcriptionally active E2F is released at the G1/S transition (reviewed in reference 13). In this manner, E2F can contribute to either the repression or activation of E2F-responsive genes.
To date, eight genes encoding components of E2F have been cloned (reviewed in reference 13). The protein products are subdivided into two groups, the E2Fs (1 through 6) and DRTF proteins 1 and 2 (DPs), that heterodimerize to generate functional E2F activity (22, 27). Although the DP subunit is critical for activity, the functional specificity of the E2F-DP complex is determined by the E2F subunit. The E2F family can be divided into three distinct subgroups based on differences in both sequence homology and functional properties.
Two of the three E2F subgroups appear to be primarily involved in the repression of E2F-responsive genes. In the case of E2F4 and E2F5, this is thought to occur via recruitment of the pRB family members and their associated histone-modifying enzymes (reviewed in reference 19). In contrast, E2F6 lacks the sequences required for pRB binding (4, 18, 39, 48) but instead associates with the mammalian polycomb complex, a known transcriptional repressor (49). Mutant mouse models have been generated for E2F4 and E2F5 (17, 24, 32, 45). Analysis of these mice indicates that E2F4 and E2F5 are fully dispensable for cellular proliferation but are required for cell cycle arrest and the terminal differentiation of specific cell lineages. The role of E2F6 in development and/or tumorigenicity has yet to be determined.
The final E2F subclass contains E2F1, E2F2, and E2F3. When overexpressed, these three E2Fs are potent activators of E2F-responsive genes (28) and their ectopic expression is sufficient to induce quiescent cells to enter S phase (26, 34). The endogenous E2F1-DP, E2F2-DP, and E2F3-DP complexes are specifically regulated by pRB, not p107 or p130, and the timing of their release from pRB correlates with the timing of the activation of E2F-responsive genes (28, 38). These observations strongly suggested that the tumor-suppressive properties of pRB are at least partially dependent upon its ability to block the induction of proliferation by E2F1, E2F2, and E2F3. This hypothesis is directly supported by the finding that E2F1 loss reduces tumor development in Rb+/− mutant mice (56).
Recent studies have identified a second form of E2F3, called E2F3b, in which the N-terminal 121 amino acids of the original E2F3 variant (now called E2F3a) are replaced by 6 novel amino acids (20, 30). The E2f3a and E2f3b mRNAs are transcribed from distinct promoters at different stages of the cell cycle (1, 30). The biological properties of E2F3b have yet to be described.
Given the importance of E2F1, E2F2, and E2F3, considerable attention has been focused on understanding their precise roles in vivo. Mutant mouse strains have been used to assess the phenotypic consequences of E2F1 deficiency. E2f1−/− mice are fully viable but develop several tissue-specific defects (14, 16, 57, 58). There is a decrease in the apoptotic potential of E2f1−/− thymocytes that results in increased lymphoid proliferation. E2f1−/− males develop testicular atrophy. Finally, E2f1+/− and E2f1−/− mutant mice have an increased susceptibility to a variety of tumor types, including histiocytic sarcomas, hemangiosarcomas, lymphomas, hepatocarcinomas, and lung tumors. This increase in cancer susceptibility might seem paradoxical in light of E2F1's purported role in proliferation. However, E2F1's tumor suppressive role is believed to be due to its known ability to trigger cells to undergo p53-dependent apoptosis in response to inappropriate cell cycle entry (11, 23, 44, 46, 55).
We have previously generated an E2f3 mutant mouse strain that is deficient for both E2F3a and E2F3b (25). Since we cannot attribute the resulting phenotypes to the loss of one or both of the two E2F3 variants, we refer to this strain using the general term “E2F3 deficient.” Using mouse embryonic fibroblasts (MEFs) derived from these animals, we have shown that E2F3 acts in a dose-dependent manner to control the mitogen-induced transcriptional activation of many known E2F-responsive genes (25). As a result, E2F3 controls the rate of proliferation of both primary and transformed MEFs. Consistent with this observation, E2F3 makes a major contribution to both the inappropriate proliferation and the apoptosis that results from the loss of the pRB (60). Given its key proliferative role, we have focused our attention on establishing the in vivo consequences of E2F3 loss. We show that E2F3 is essential for full embryonic viability in a strain-specific manner. The small proportion of animals that survive to adulthood exhibit growth retardation that is dependent on the dosage of E2F3. In addition, this study reveals an unexpected and critical role for E2F3 in the adult heart. Significantly, the phenotypes of the E2f3 mutant mice are completely distinct from those of E2f1 mutants. Through the generation and analysis of E2f1 E2f3 compound mutant mice, we also show that the role of E2F3 in cardiac function, as well as the regulation of many other developmental processes, is a shared property of E2F1 and E2F3 but that E2F1 has a specific function in tumorigenesis.
MATERIALS AND METHODS
Animal maintenance and histological analysis.
The E2f1 and E2f3 mutant mouse strains were genotyped using previously described PCR protocols (25, 57). The window of lethality of E2f3−/− (or E2f1−/− E2f3−/−) embryos was determined by establishing timed pregnancies through the detection of vaginal plugs (embryonic day 0.5 [e0.5]). Embryos were harvested by cesarean section, and viability was assessed by detection of a heartbeat. For histology, embryos and tissues were fixed in 10% formalin or Bouin's solution and embedded in paraffin blocks and 6-μm sections were stained with either hematoxylin and eosin or Gomori's one-step trichrome stain (Poly Scientific). All conclusions about statistical significance were derived using Student's t test.
RESULTS
E2f3 is essential for normal embryonic development.
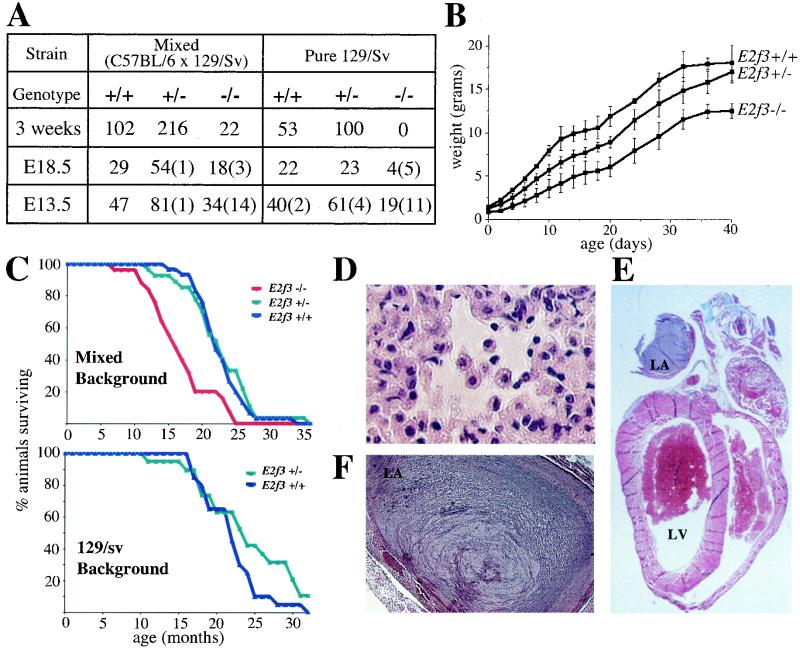
We have previously shown that E2F3 is required for mitogen-induced cell cycle entry and that viable E2f3−/− mice are underrepresented at weaning (Fig. 1A) (25). Given these observations, we wished to determine the developmental consequences of E2F3 deficiency. Upon close examination of the newborn litters, we found that a significant number of the mixed (C57BL/6 × 129/Sv) background E2f3−/− mice were born alive but failed to thrive and died within 48 h of birth. Although we were able to successfully foster wild-type and E2f3+/− littermate controls, the use of foster mothers did not increase the proportion of surviving E2f3−/− neonates. Thus, the high incidence of perinatal lethality results from an intrinsic defect in the E2f3−/− mice rather than the E2f3+/− mothers.
TABLE 1.
Genotypes of progeny from E2f1 E2f3 compound mutant animals
| Progenyb | No. with E2f1 E2f3 status
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| +/+ +/+ | +/− +/+ | −/− +/+ | +/+ +/− | +/− +/− | −/− +/− | +/+ −/− | +/− −/− | −/− −/−a | |
| 3 Wk (455) | |||||||||
| Observed | 36 | 82 | 35 | 69 | 149 | 72 | 12 | 0 | 0 |
| Expected | 28 | 57 | 28 | 57 | 114 | 57 | 28 | 57 | 28 |
| e18.5 (52) | |||||||||
| Observed | 3 | 8 | 5 | 9 | 17 | 8 | 2 | 0 | 0 |
| Expected | 3 | 7 | 3 | 7 | 13 | 7 | 3 | 7 | 3 |
| e13.5 (72) | |||||||||
| Observed | 3 | 15 | 7 | 8 | 19 | 9 | 4 | 7 | 0 |
| Expected | 5 | 9 | 5 | 9 | 18 | 9 | 5 | 9 | 5 |
| e11.5 (50) | |||||||||
| Observed | 4 | 6 | 7 | 5 | 8 | 11 | 4 | 3 | (2) |
| Expected | 3 | 6 | 3 | 6 | 13 | 6 | 3 | 6 | 3 |
| e10.5 (123) | |||||||||
| Observed | 10 | 13 | 4 | 18 | 30 | 13 | 9 | 23 | 1 (2) |
| Expected | 7 | 15 | 7 | 15 | 31 | 15 | 7 | 15 | 7 |
| e9.5 (130) | |||||||||
| Observed | 13 | 19 | 8 | 10 | 42 | 15 | 4 | 11 | 8 |
| Expected | 8 | 16 | 8 | 16 | 32 | 16 | 8 | 16 | 8 |
Numbers in parentheses indicate numbers of nonviable embryos.
Numbers in parentheses are the total number of animals.
To determine whether the reduced viability of the E2f3−/− mutant animals was entirely attributable to perinatal lethality, we set up intercrosses of the mixed background E2f3+/− animals and monitored embryonic development using timed pregnancies (Fig. 1A). The E2f3−/− animals were present immediately prior to birth (e18.5) but their weight (0.77 ± 0.12 g; n = 16) was significantly lower (P < 1.2 × 10−11) than that of either their wild-type (1.17 ± 0.146 g; n = 28) or E2f3+/− (1.06 ± 0.106 g; n = 57) littermate controls. Moreover, these embryos seemed to be underrepresented. Analysis of earlier developmental stages confirmed that some of the E2f3−/− animals die in utero. E2f3−/− embryos were present at the expected frequency at e13.5, but 29% of these were already dead. Dead E2f3−/− embryos were also present at subsequent developmental stages (Fig. 1A and data not shown), indicating that the window of lethality extends from e13.5 to the postpartum stages. Apart from the significant growth retardation, histopathological examination did not reveal any obvious defects in either the E2f3−/− embryos or neonates.
FIG. 1.
E2F3 is essential for full embryonic viability, promotes growth in a dose-dependent manner, and is critical for normal cardiac function. (A) Progeny arising from E2f3+/− intercrosses at various stages of embryonic development. Numbers in parentheses indicate numbers of nonviable embryos found at each developmental stage. (B) Representative weight curve of female littermate animals. Similar data was obtained for other litters and for males; however, the number of animals examined was insufficient to allow statistical analysis. (C) Kaplan-Meier plots of wild-type (n = 30), E2f3+/− (n = 27), and E2f3−/− (n = 25) littermates with the mixed (C57/BL6 × 129/Sv) genetic background and wild-type (n = 20) and E2f3+/− (n = 19) littermates with the pure 129/Sv background. (D) Hematoxylin- and eosin-stained sections of the lungs (×40 magnification) of an E2f3−/− animal that died from congestive heart failure, displaying macrophages in the alveolar spaces. (E and F) Sections of hearts (×1 and ×5 magnification, respectively) from E2f3−/− mice, depicting atrial thrombi stained with Gomori's one-step trichrome stain. LA, left atrium. LV, left ventricle.
The deleterious nature of E2F3 deficiency was increased in the pure 129/Sv genetic background. There was a significant increase in the ratio of dead to viable E2f3−/− embryos at e13.5, and viable E2f3 homozygotes were present at only 30% of the expected frequency at e18.5 (Fig. 1A). Finally, heterozygous intercrosses generated no viable E2f3−/− neonates. Only four E2f3−/− pups were recovered at birth, all of which were dead. We therefore conclude that E2F3 is essential for viability in the pure 129/Sv background. Thus, the survival of the mixed background E2f3−/− animals must depend upon the presence of one or more of the C57BL/6 alleles.
E2f3 mutant mice suffer premature death that results from congestive heart failure.
We monitored the development of the surviving mixed (C57BL/6 × 129/Sv) background E2f3−/− animals to determine whether E2F3 is required postnatally. Consistent with the growth retardation in utero, we saw a significant reduction in the weight of the E2f3−/− animals relative to that of the littermate controls that was maintained throughout their life span (Fig. 1B and data not shown). Furthermore, the growth of the E2f3+/− animals was found to be intermediate between that of their wild-type and E2f3−/− littermates. Thus, in addition to playing a key role in embryonic development, E2F3 acts in a dose-dependent manner to promote growth in the adult animal.
Previous studies have shown that E2f1+/− and E2f1−/− mice arise at the expected frequency but develop a variety of age-related defects, including an increased predisposition to tumors (14, 57). We therefore established and monitored colonies of both the mixed and pure 129/Sv background E2f3 mutant mice to determine whether E2f3 mutation had any influence on aging animals (Fig. 1C). The life span and cause of death of the mixed background and the pure 129/Sv E2f3+/− mice were indistinguishable from those of their littermate controls. In contrast to the E2F1-deficient mice, there was no difference in either the incidence or spectrum of tumors. The E2f3−/− (mixed background) animals began dying at 7 months of age, and their average life span was significantly shorter that that of the wild-type controls (16.6 ± 4.8 months versus 22.6 ± 3.6 months; P < 3.6 × 10−6) (Fig. 1C). By visual inspection, the only evidence of ill health in these E2f3−/− animals was a mildly ruffled coat in the last 1 to 2 months of life and dyspnea (shortness of breath) in the last few days of life.
Full necropsy showed that, like the E2f3+/− animals, the E2f3−/− animals were predominantly tumor free. Thus, E2F3 was not acting as a tumor suppressor in a manner similar to that of E2F1. Instead, greater than 85% of the animals (23 of 27) showed indications of congestive heart failure. Specifically, we observed bilateral pleural effusions as well as pulmonary edema. Histological examination of the lungs revealed the presence of macrophages in the alveolar spaces (Fig. 1D). Strikingly, histological analysis also revealed the presence of massive thrombi within the left atria of the E2f3−/− hearts (Fig. 1E and F). In several cases, the clots nearly completely filled the atrium and were also present in the left ventricle. The thrombi were all focally organized, indicating that they were formed at least one day prior to death (Fig. 1F). However, there was no evidence of pulmonary embolism in these animals. We also detected a large atrial thrombus in one of the four pure 129/Sv E2f3−/− pups that was recovered dead immediately after birth, indicating that a cardiac etiology was likely responsible for this preperinatal death. Importantly, fibrin thrombi were consistently detected in the heart but not other organs of the E2F3 deficient mice, indicating that clot formation does not result from a general hypercoagulable state. Also, there was no evidence of bacterial infection that could lead to the development of a septic thrombus. Thus, we believe that the congestive heart failure and the formation of atrial thrombi likely resulted from a primary defect of the heart. Although the underlying mechanistic basis for this phenotype is still unclear, the high penetrance of heart failure in these animals demonstrates that E2F3 plays a critical role in cardiac function.
E2F1 and E2F3 play overlapping roles in a variety of developmental processes.
The activating E2Fs, E2F1, -2, and -3, are key downstream targets of the pRB tumor suppressor, and therefore, considerable attention has focused on understanding their relative roles in vivo. Significantly, the defects arising in the E2f3−/− mice are completely distinct from the reported phenotypes of the E2f1−/− mice, which include full neonatal viability, testicular atrophy, defective thymocyte apoptosis, and increased tumor susceptibility (14, 16, 57, 58). This finding supports the prevailing view that E2F1 and E2F3 have distinct roles in vivo. To test this hypothesis, we crossed the E2f1 and E2f3 mutant mouse strains to assess whether their specific developmental defects of each were exacerbated by the mutation of the other E2F. This analysis was conducted in the mixed (C57BL/6 × 129/Sv) background to enable generation of E2f1−/− and E2f3−/− control animals.
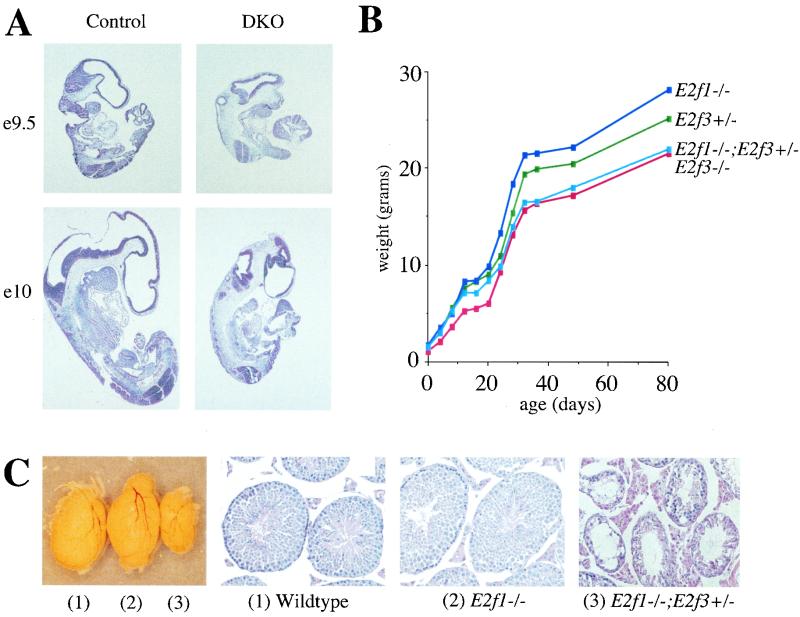
We first tested whether E2f1 mutation influences the viability of the E2f3−/− mice by assessing the frequency of progeny arising from E2f1+/− E2f3+/− intercrosses (Table 1). E2f1+/− E2f3+/− and E2f1−/− E2f3+/− compound mutants were generated at the expected frequency. Viable E2f3−/− mice were present at a reduced frequency, as described above. The additional mutation of one or both alleles of E2f1 progressively shortened the life span of these E2f3−/− mice. No viable E2f1+/− E2f3−/− mice were detected at weaning or immediately prior to birth (e18.5) but these were present at reasonable numbers at e13.5. Moreover, the E2f1−/− E2f3−/− embryos died between e9.5 and e10.5. At e9.5, the E2f1−/− E2f3−/− embryos appeared essentially normal but the single viable E2f1−/− E2f3−/− embryo detected at e10 was substantially smaller than the wild-type controls (Fig. 2A). Thus, E2F1 contributes to the viability of E2F3 deficient embryos and neonates in a dose-dependent manner.
FIG. 2.
E2F1 and E2F3 have overlapping functions in a variety of tissue types. (A) Hematoxylin- and eosin-stained sections (×4 magnification) of E2f1−/− E2f3−/− embryos and littermate controls at embryonic days 9.5 and 10. (B) Weight curve from a representative litter of sex-matched (male) mice of the indicated genotypes. Similar data were obtained for other litters; however, the number of animals examined was insufficient to allow statistical analysis. (C) External view and hematoxylin- and eosin-stained sections (×20 magnification) of testes from representative four-month-old wild-type, E2f1−/−, and E2f1−/− E2f3+/− littermates.
We next examined same-sex littermate controls for evidence of growth retardation, a specific defect of the E2f3+/− and E2f3−/− (this study) but not the E2f1−/− (14, 57) mice. Significantly, the E2f1−/− E2f3+/− mice consistently displayed a degree of growth retardation that was similar to that of the E2f3−/− rather than that of the E2f3+/− littermates (Fig. 2B and data not shown). Thus, E2F1 loss potentiates the growth retardation that results from the heterozygous mutation of E2f3.
We also screened single and compound mutant animals for the presence of testicular atrophy, a specific defect of E2f1−/− (57) but not E2f3−/− (data not shown) mice. Yamasaki et al. (57) showed that E2f1−/− mice display testicular atrophy by 8 months of age. We therefore screened 4- to 5-month-old wild-type, E2f1−/−, and E2f1−/− E2f3+/− littermate males for this defect (Fig. 2C). At this age we were able to detect mild testicular atrophy in some but not all of the E2f1−/− males, as evidenced by a reduction in testicular weight and the number of sperm in the seminiferous tubules. The severity of this defect was exacerbated in the E2f1−/− E2f3+/− compound mutants. The testes were considerably smaller than those of the E2f1−/− littermate controls, and they contained empty and degenerated seminiferous tubules with no apparent loss of the Leydig cells. Importantly, these changes were indistinguishable from those seen in older E2f1−/− males, indicating that the reduction in E2F3 dosage increases severity but does not alter the fundamental nature of the phenotype.
Together, these studies show that we can increase the severity of developmental defects that are specific to either the E2f1 (testicular atrophy) or the E2f3 (viability and growth retardation) mutant mouse strains by reducing or abolishing the levels of the second E2F family member. This strongly suggests that E2F1 and E2F3 have one or more shared biochemical properties that are essential for the development and/or maintenance of a variety of embryonic and adult tissues.
Phenotypes of aging E2f1 E2f3 compound mutant mice.
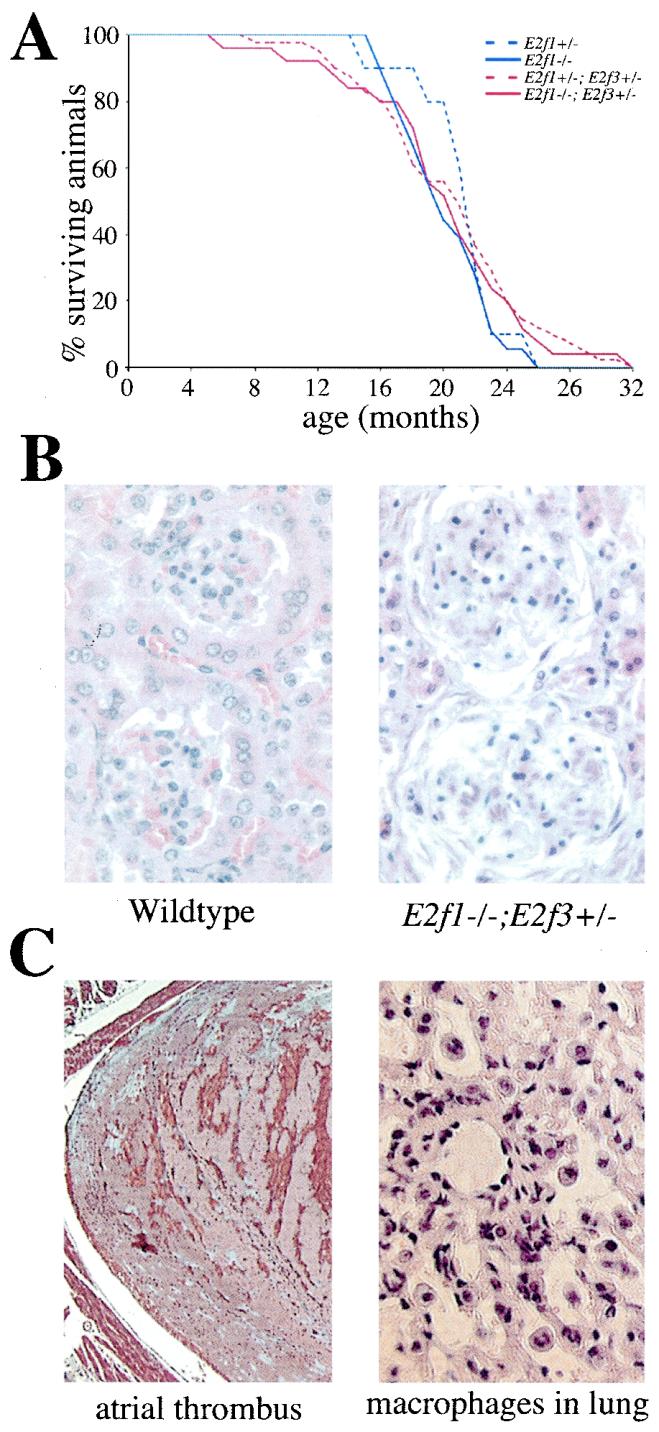
The average life spans of E2f1+/− and E2f1−/− mutant mice are shortened by an increased tumor predisposition (57), while E2f3−/− mice die of congestive heart failure as described above. We therefore wished to establish the incidence of these two defects in the viable compound E2f1 E2f3 mutant mice. To address this, we monitored aging cohorts of E2f1+/− E2f3+/− and E2f1−/− E2f3+/− mice versus those of E2f1+/− and E2f1−/− littermate controls. Statistical analysis did not detect any significant differences in the life spans of these four genotypes (Fig. 3A). Whenever possible, the cause of death was established by necropsy and histological analysis (Table 2).
FIG. 3.
Aging phenotypes of E2f1 E3f3 compound mutant animals. (A) Relative life spans of mixed (C57BL/6 X129/Sv) E2f1+/− (n = 10), E2f1−/− (n = 18), E2f1+/− E2f3+/− (n = 41), and E2f1−/− E2f3+/− (n = 27) mice. (B) Hematoxylin- and eosin-stained sections of kidneys from a control wild-type animal and from an E2f1−/− E2f3+/− animal that died at 6 months of age with severe glomerulonephritis. (C) Signs of congestive heart failure in an E2f1+/− E2f3+/− animal that died at 8 months of age. The histological section of the left atrium of the heart (×5 magnification) was stained with Gomori's one step trichrome stain, and the section of the lung (×40 magnification) was stained with hematoxylin and eosin.
TABLE 2.
Phenotypes and time of death of E2f1 E2f3 compound mutant animals
| Phenotype | Age at death (mo)
|
||
|---|---|---|---|
| E2f1+/− or E2f1−/− (n = 24) | E2f1+/−E2f3+/− (n = 37) | E2f1−/−E2f3+/− (n = 16) | |
| Tumor | |||
| Histiocytic sarcoma | 16, 17, 17.5, 19, 19.5, 22.5, 23, 23.5 | 12.5, 18, 18, 20.5, 21.5, 21.5, 27.5, 32 | 13, 18, 18.5, 21, 26.5 |
| Hemangiosarcoma | 19, 19.5, 22.5 | 11.5, 14.5, 18, 20.5, 21.5, 23, 24.5, 26, 29 | 13, 17.5 |
| Hepatocarcinoma | 18 | 17, 23.5, 27 | 18.5, 31.5 |
| Adenocarcinoma | 15.5, 20, 22, 22.5 | 20.5, 24, 29 | 31.5 |
| Lymphoma-leukemia | 21, 22, 22, 22.5 | 15, 17.5, 18, 18, 19, 21, 27 | |
| Pheochromocytoma | 16.5 | 22.5 | |
| Other | |||
| Glomerulonephritis | 14.5, 21 | 8, 14, 15, 16.5, 19 | 6, 10, 16, 18, 19 |
| Polyarteritis nodosa | 14.5, 21, 21 | 10, 12.5, 19 | |
| Congestive heart failure | 8, 14, 17, 17 | 20, 22.5, 24.5 | |
First, we considered the tumor phenotype of these animals. The incidence and spectrum (histiocytic sarcomas, hemangiosarcomas, hepatocarcinomas, and lung tumors) of tumors arising in the E2f1+/− and E2f1−/− controls were similar to those previously described (57). The one difference was that the histiocytic sarcomas were primarily detected in the reproductive tract by Yamasaki et al. (57) (and therefore were classified as reproductive tract sarcomas) but were found in many different tissues in our study. Importantly, there was no significant difference in the incidence, spectrum, or time of onset of tumors in the E2f1+/− E2f3+/− and E2f1−/− E2f3+/− mice versus the E2f1+/− and E2f1−/− controls. This finding, along with the absence of any increased tumor predisposition in the E2f3+/− and E2f3−/− mutant mice, suggests that E2F3 does not make a significant contribution to the suppression of tumor formation in a manner analogous to that of E2F1.
Although the difference was not statistically significant, we observed that a fraction of the E2f1+/− E2f3+/− and E2f1−/− E2f3+/− mice died several months earlier than the E2f1+/− and E2f1−/− controls (Fig. 3A). This was due to the increased incidence, and severity, of age-related lesions that had previously been observed in the E2f1+/−, E2f1−/−, and E2f3−/− mice rather than the appearance of any novel phenotypes (Table 2). First, a larger proportion of the E2f1+/− E2f3+/− (21.6%) and E2f1−/− E2f3+/− (31.3%) animals died with severe polyarteritis nodosa and/or glomerulonephritis (Fig. 3B), defects that arise at a low frequency (12.5%) in both the E2f1+/− and E2f1−/− mice (reference 57 and this study). This suggests that the mutation of a single E2f3 allele exacerbates the inflammatory defects that arise in the E2f1 mutant mice. Most importantly, we discovered that four of the E2f1+/− E2f3+/− and three of the E2f1−/− E2f3+/− animals but no E2f1+/− or E2f1−/− controls died prematurely, with indications of cardiac failure including large organized atrial thrombi, bilateral pleural effusions, and pulmonary edema (Table 2; Fig. 3C.) Although wild-type and E2f3+/− animals do develop heart failure at a similar, low frequency (approximately 1 of 40), this only occurs in older (>18 months) animals (our unpublished data). Notably, two of the E2f1+/− E2f3+/− animals died at 8 and 14 months, well before the control animals are susceptible to this defect. Although the incidence of heart failure in the E2f1 E2f3 compound mutants is relatively low, these data suggest that mutation of E2f1 can potentiate the effects of E2f3 mutation and that a reduction in dosage of these two activating E2Fs is sufficient to induce congestive heart failure.
Since mutation of Rb is known to increase the amount of free E2F (37, 43, 50, 56, 60), we were interested in establishing whether Rb mutation would modulate the incidence of the congestive heart failure in the E2F3-deficient animals. We therefore examined a cohort of E2f3−/− and Rb+/− E2f3−/− littermate animals that had been independently generated for a tumor study (U. Ziebold and J. A. Lees, unpublished data). The Rb+/− E2f3−/− animals died as a result of tumor formation, but their life spans (252 to 541 days) encompassed the timeframe in which the E2f3 mutant mice die from heart failure (Table 3). Despite this fact, none of the Rb+/− E2f3−/− mice (n = 25) showed any evidence of congestive heart failure. In contrast, 7 of the 10 E2f3−/− littermate controls, including 5 that were within the critical age range for the Rb+/− E2f3−/− animals (213, 304, 317, 383, and 428 days of age), exhibited signs of cardiac failure (Table 3). The high incidence of congestive heart failure in these younger E2f3−/− littermates strongly suggests that the suppression of this defect that results from Rb mutation is not merely a consequence of their shortened life span. These results, combined with the incidence of heart failure in the E2f1 E2f3 compound mutant animals, strongly suggest that normal heart function is dependent on a critical dosage of activating E2Fs.
TABLE 3.
Rb mutation rescues heart failure of E2f3−/− mice
| Age at death (days) | Deaths from heart failure/total no. of deaths of mice of genotype:
|
|
|---|---|---|
| E2f3−/− | Rb+/−E2f3−/− | |
| 200-250 | 1/1 | |
| 250-300 | 0/4 | |
| 300-350 | 2/3 | 0/6 |
| 350-400 | 1/1 | 0/6 |
| 400-450 | 1/1 | 0/5 |
| >450 | 2/4 | 0/4 |
| Total | 7/10 | 0/25 |
DISCUSSION
E2F3 is required for embryonic, neonatal and adult viability.
The endogenous E2F transcriptional activity is derived from the combined action of multiple E2F-DP complexes (reviewed in reference 13). Since the biological properties of the individual E2F-DP species are determined by the E2F moiety, considerable attention has focused on understanding the precise roles of the individual E2F family members. In this study, we used mouse models to investigate the phenotypic consequences of E2F3 deficiency in vivo. First, we have shown that E2F3 is critical for normal embryonic development. This distinguishes E2F3 from all other members of the E2F family examined to date (E2F1, E2F2, E2F4, and E2F5), which are fully dispensable for embryonic viability (14, 24, 32, 41, 57, 59).
Importantly, the incidence of embryonic lethality differed in different genetic backgrounds, indicating the presence of strain-specific modifiers of E2F3, and was fully penetrant in the pure 129/Sv strain. The large window of lethality (from e13 to birth) and the absence of any detectable tissue-specific defect(s) have made it difficult to establish the cause of death. However, we have detected a strong correlation between the penetrance of the embryonic lethality and that of the proliferation defect in MEFs. Specifically, MEFs derived from mixed background E2f3−/− embryos show a range of proliferation defects from significantly impaired to near normal (25), while those derived from pure 129/Sv E2f3−/− embryos consistently display a substantial proliferation defect (J. E. Cloud and J. A. Lees, unpublished observations). This shows that the strain-specific modifier(s) influences E2F3's role in both viability and cellular proliferation, and it suggests that the viability of the individual E2f3−/− embryos may be at least partially determined by their proliferative capacity.
Our ability to generate viable E2f3−/− animals allowed us to assess the requirement for E2F3 in the adult. This analysis revealed an unexpected role for E2F3 in cardiac function. Specifically, histopathological analysis of E2F3-deficient mice demonstrated a high incidence (greater than 85%) of atrial thrombus formation and pulmonary edema resulting from congestive heart failure. Importantly, there was no evidence of coagulation defects in other tissues or of a bacterial infection that could produce a septic thrombus. We therefore conclude that the phenotype is most likely due to a primary heart defect. There are two opposing mechanisms for the onset of heart failure in these animals. One possibility is that a problem in the heart in utero could set up a defect that does not manifest itself as heart failure until adulthood. For example, subtle changes in the geometry of the heart could affect cardiac function in the adult mouse. Intriguingly, our analyses showed that the E2f1−/− E2f3−/− mice failed around day 10, a time point that is known to be critical for heart development. To date, we have not detected any structural abnormalities in the hearts of either the E2f3−/− or E2f1−/− E2f3−/− embryos, but it remains possible that subtle defects exist. An alternative possibility is that the congestive heart failure occurs late in the lives of the animals because E2F3 is required for the maintenance of the heart. There are many potential mechanisms for late-onset heart problems, including either perturbations in the electrical conduction system or deficiencies in the contractile properties of the heart muscle itself (7). A clearer understanding of the physiology of the cardiac dysfunction will help to elucidate the underlying molecular basis for the role of E2F3 in the development and/or maintenance of the heart.
The absence of both E2F3a and E2F3b proteins in these animals further complicates our understanding of the underlying basis of the heart failure. Although E2F3a is known to be a potent transcriptional activator controlling cellular proliferation, the properties of E2F3b are not well understood. An E2F3a deletion mutant (E2F3ΔN) that is identical to E2F3b, apart from six novel N-terminal amino acids, behaves as a transcriptional activator (28), suggesting that E2F3b may have an activating function similar to that of E2F3a. Consistent with this observation, the proliferation defect arising in E2f mutant MEFs can be suppressed by expression of either E2F3a or E2F3b (54). However, the high levels of E2f3b mRNA in quiescent cells (20, 30) have led a number of groups to propose that E2f3b functions in an analogous manner to the repressive E2Fs, E2F4 and E2F5. This latter model would be consistent with the idea that the majority of the mass of the heart is comprised of terminally differentiated nonproliferative cardiomyocytes.
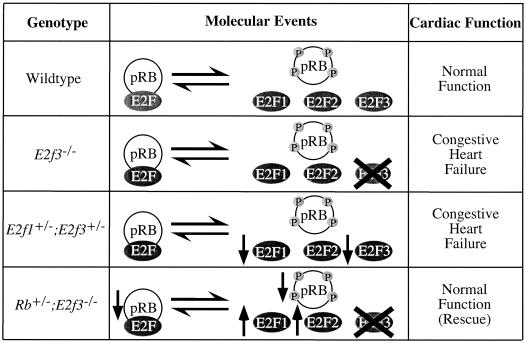
Although our data do not address the issue of E2F3a versus E2F3b function, the analysis of the E2f1 E2f3 and Rb E2f3 compound mutant mice strongly suggests that the congestive heart failure results from the loss of an activating function of E2F3 (Fig. 4). Specifically, cardiac failure can be triggered by either the complete loss of E2F3 or a reduction in the dosages of both E2F1 and E2F3. Moreover, Rb mutation, which is known to induce the release of its associated E2Fs, completely rescues the congestive heart failure resulting from E2F3 deficiency. These data strongly suggest that normal cardiac function is dependent on a critical dosage of activating E2Fs.
FIG. 4.
Model of E2F action in cardiac function. As demonstrated by the comparison of wild-type, E2f3−/−, E2f1+/− E2f3+/−, and Rb+/− E2f3−/− mice, the dosage of free activating E2F is critical for normal cardiac function.
The relative roles of E2F1 and E2F3 in vivo.
The analysis of E2f1 E2f3 compound mutant mice yields additional insight into the relative roles of E2F1 and E2F3 in vivo. This is a major issue in the field, and there are currently two opposing models. The first model predicts that E2F1, -2, and -3 regulate distinct subsets of target genes and thus have distinct biological properties in vivo . Support for this model comes from a number of studies. First, ectopic expression of E2F1, E2F2, or E2F3 has been reported to mediate the differential activation of known E2F-responsive genes (11, 12). Second, microinjection assays and the analysis of E2F-deficient MEFs support the notion that E2F3, but not E2F1, is critical for activation of E2F-responsive genes and therefore cellular proliferation (25, 29). Finally, it is widely believed that E2F1, but not other members of the E2F family, induces p53- and p73-dependent apoptosis (11, 33). Our finding that the phenotypic spectrum of the E2f3−/− mice is completely nonoverlapping with that of the E2f1 mutant animals appeared to be consistent with this specificity model of E2F action.
The alternative model of E2F action proposes that E2F1, -2, and -3 have similar molecular properties. There is also considerable evidence to support this second model. Several groups have shown that E2F1, -2, and -3 are similarly capable of inducing either ectopic S-phase entry (40) or programmed cell death (51) (S. Lowe, personal communication; A. Aslanian and J. A. Lees, unpublished data). Consistent with these findings, mutant mouse models confirm that the endogenous E2F1 and E2F3 proteins act in a similar manner to induce the inappropriate proliferation and apoptosis that arises in pRB-deficient mouse embryos (50, 60). Finally, in vivo chromatin immunoprecipitation assays have not detected any specificity for the association of individual E2F-DP complexes to a variety of known E2F-responsive promoters (47, 53). One limitation of this model is that it does not immediately explain the distinct phenotypes of the E2f1 and E2f3 mutant mice. However, in situ hybridization studies reveal clear differences in the expression levels of the individual E2F proteins in different tissues (8, 9). Thus, we speculated that E2F1 and E2F3 deficiency could reduce the total level of free E2F activity below a critical threshold in a defined, but distinct, subset of tissues, giving rise to a nonoverlapping spectrum of tissue-specific defects. Importantly, the phenotypes of the compound E2f1 E2f3 mutant mice provide strong support for the dosage model of E2F action as opposed to the specificity model. Specifically, we found that all of the developmental and age-related lesions of the E2f1 and E2f3 mutant mouse strains, including embryonic lethality, growth retardation, testicular atrophy, polyarteritis nodosa, glomerulonephritis, and congestive heart failure, were exacerbated by loss or reduction in the levels of the other E2F. We therefore conclude that the development of a wide variety of tissues shows a critical dependence on the dosage of E2F1 and E2F3.
Importantly, our analysis does suggest one major difference in the properties of E2F1 and E2F3 in vivo. Whereas E2f1+/− and E2f1−/− mice are susceptible to tumors, E2f3+/− and E2f3−/− mice do not have any increased tumor incidence relative to that of wild-type littermate controls. This difference could have been attributed to early death from heart failure in the E2f3−/− animals. However, mutation of a single E2f3 allele also failed to alter the incidence, spectrum, or timing of tumor development in the E2f1+/− or E2f1−/− mice. Interestingly, analysis of DNA derived from the tumors of E2f1+/− mice indicates that tumor formation occurs without loss of the wild-type E2f1 allele, explaining why E2f1+/− and E2f1−/− mice develop tumors with similar kinetics (57, 59) (J. E. Cloud and J. A. Lees, unpublished observations). Thus, a reduction in the dosage of E2F1 seems sufficient to induce tumorigenesis, while E2F3 does not appear to behave as a tumor suppressor in vivo.
There has been considerable debate about the underlying basis of E2F1's tumor-suppressive properties. These were originally thought to reflect E2F1's ability to participate in the repression of E2F-responsive genes via recruitment of pRB and its associated histone-modifying enzymes (57). However, the absence of any tumor phenotype in mice deficient for E2F4 (24), the major repressive E2F in vivo, was inconsistent with this model. The prevailing view is that E2F1's tumor-suppressive properties are dependent upon its ability to induce apoptosis when cells enter the cell cycle inappropriately. Since we have shown that E2F3 also induces apoptosis in vivo (60) but does not to act as a tumor suppressor (this study), this model also seems in doubt. Interestingly, two recent studies suggest a link between E2F1 and the DNA damage response pathway. Lin et al. showed that E2F1, but not E2F2 or E2F3, is stabilized by phosphorylation by the ATM and ATR kinases after DNA damage (31). The second study suggests why an accumulation of E2F1 might be beneficial to a cell with DNA damage: E2F1 was also found to associate with Nbs1 and the Mre11 recombination/repair complex (36). Maser et al. hypothesized that E2F1 is required to target the Mre11 complex near the origins of DNA replication and suppress origin firing when DNA is damaged. Although it has yet to be shown that Nbs1 binds specifically to E2F1 and not E2F3, this link to DNA damage could be the elusive explanation for E2F1's tumor-suppressive properties.
Acknowledgments
We thank Lili Yamasaki and Tyler Jacks for kindly providing the E2f1 and Rb mutant mouse strains. We are grateful to Tyler Jacks, Terry Orr-Weaver, Bob Weinberg, and members of the Lees lab for helpful discussions during this study and the preparation of the manuscript.
U.Z. was supported by a fellowship from the Deutsche Forschungsgemeinschaft. This work was supported by grants from the ACS (RPG-99-094-01) and NIH (PO1-CA42063) to J.A.L.
REFERENCES
- 1.Adams, M. R., R. Sears, F. Nuckolls, G. Leone, and J. R. Nevins. 2000. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol. Cell Biol. 20:3633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 3.Buchkovich, K., L. A. Duffy, and E. Harlow. 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097-1105. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright, P., H. Muller, C. Wagener, K. Holm, and K. Helin. 1998. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17:611-623. [DOI] [PubMed] [Google Scholar]
- 5.Chellappan, S. P., S. Hiebert, M. Mudryj, J. M. Horowitz, and J. R. Nevins. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 65:1053-1061. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P. L., P. Scully, J. Y. Shew, J. Y. Wang, and W. H. Lee. 1989. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell 58:1193-1198. [DOI] [PubMed] [Google Scholar]
- 7.Chien, K. R. 2000. Genomic circuits and the integrative biology of cardiac diseases. Nature 407:227-232. [DOI] [PubMed] [Google Scholar]
- 8.Dagnino, L., C. J. Fry, S. M. Bartley, P. Farnham, B. L. Gallie, and R. A. Phillips. 1997. Expression patterns of the E2F family of transcription factors during mouse nervous system development. Mech. Dev. 66:13-25. [DOI] [PubMed] [Google Scholar]
- 9.Dagnino, L., C. J. Fry, S. M. Bartley, P. Farnham, B. L. Gallie, and R. A. Phillips. 1997. Expression patterns of the E2F family of transcription factors during murine epithelial development. Cell Growth Differ. 8:553-563. [PubMed] [Google Scholar]
- 10.DeCaprio, J. A., J. W. Ludlow, D. Lynch, Y. Furukawa, J. Griffin, H. Piwnica-Worms, C. M. Huang, and D. M. Livingston. 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58:1085-1095. [DOI] [PubMed] [Google Scholar]
- 11.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirks, P. B., J. T. Rutka, S. L. Hubbard, S. Mondal, and P. A. Hamel. 1998. The E2F-family proteins induce distinct cell cycle regulatory factors in p16-arrested, U343 astrocytoma cells. Oncogene 17:867-876. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 14.Field, S. J., F. Y. Tsai, F. Kuo, A. M. Zubiaga, W. G. Kaelin, Jr., D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549-561. [DOI] [PubMed] [Google Scholar]
- 15.Flemington, E. K., S. H. Speck, and W. G. Kaelin, Jr. 1993. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 90:6914-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia, I., M. Murga, A. Vicario, S. J. Field, and A. M. Zubiaga. 2000. A role for E2F1 in the induction of apoptosis during thymic negative selection. Cell Growth Differ. 11:91-98. [PubMed] [Google Scholar]
- 17.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 18.Gaubatz, S., J. G. Wood, and D. M. Livingston. 1998. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. USA 95:9190-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., M. K. Armanious, M. J. Thomas, and W. D. Cress. 2000. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene 19:3422-3433. [DOI] [PubMed] [Google Scholar]
- 21.Helin, K., E. Harlow, and A. Fattaey. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 13:6501-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7:1850-1861. [DOI] [PubMed] [Google Scholar]
- 23.Hiebert, S. W., G. Packham, D. K. Strom, R. Haffner, M. Oren, G. Zambetti, and J. L. Cleveland. 1995. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol. Cell. Biol. 15:6864-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humbert, P. O., C. Rogers, S. Ganiatsas, R. L. Landsberg, J. M. Trimarchi, S. Dandapani, C. Brugnara, S. Erdman, M. Schrenzel, R. T. Bronson, and J. A. Lees. 2000. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell 6:281-291. [DOI] [PubMed] [Google Scholar]
- 25.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. G., J. K. Schwarz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 27.Krek, W., D. M. Livingston, and S. Shirodkar. 1993. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262:1557-1560. [DOI] [PubMed] [Google Scholar]
- 28.Lees, J. A., M. Saito, M. Vidal, M. Valentine, T. Look, E. Harlow, N. Dyson, and K. Helin. 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 13:7813-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leone, G., J. DeGregori, L. Jakoi, J. G. Cook, and J. R. Nevins. 1999. Collaborative role of E2F transcriptional activity and G1 cyclin-dependent kinase activity in the induction of S phase. Proc. Natl. Acad. Sci. USA 96:6626-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone, G., F. Nuckolls, S. Ishida, M. Adams, R. Sears, L. Jakoi, A. Miron, and J. R. Nevins. 2000. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 20:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, W. C., F. T. Lin, and J. R. Nevins. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15:1833-1844. [PMC free article] [PubMed] [Google Scholar]
- 32.Lindeman, G. J., L. Dagnino, S. Gaubatz, Y. Xu, R. T. Bronson, H. B. Warren, and D. M. Livingston. 1998. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 12:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lissy, N. A., P. K. Davis, M. Irwin, W. G. Kaelin, and S. F. Dowdy. 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407:642-645. [DOI] [PubMed] [Google Scholar]
- 34.Lukas, J., B. O. Petersen, K. Holm, J. Bartek, and K. Helin. 1996. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 16:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 36.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCaffrey, J., L. Yamasaki, N. J. Dyson, E. Harlow, and A. E. Griep. 1999. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell. Biol. 19:6458-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moberg, K., M. A. Starz, and J. A. Lees. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morkel, M., J. Wenkel, A. J. Bannister, T. Kouzarides, and C. Hagemeier. 1997. An E2F-like repressor of transcription. Nature 390:567-568. [DOI] [PubMed] [Google Scholar]
- 40.Muller, H., M. C. Moroni, E. Vigo, B. O. Petersen, J. Bartek, and K. Helin. 1997. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol. Cell. Biol. 17:5508-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murga, M., O. Fernandez-Capetillo, S. J. Field, B. Moreno, L. R. Borlado, Y. Fujiwara, D. Balomenos, A. Vicario, A. C. Carrera, S. H. Orkin, M. E. Greenberg, and A. M. Zubiaga. 2001. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 15:959-970. [DOI] [PubMed] [Google Scholar]
- 42.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 43.Pan, W., T. Sun, R. Hoess, and R. Grafstrom. 1998. Defining the minimal portion of the retinoblastoma protein that serves as an efficient substrate for cdk4 kinase/cyclin D1 complex. Carcinogenesis 19:765-769. [DOI] [PubMed] [Google Scholar]
- 44.Qin, X. Q., D. M. Livingston, W. G. Kaelin, Jr., and P. D. Adams. 1994. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 91:10918-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rempel, R. E., M. T. Saenz-Robles, R. Storms, S. Morham, S. Ishida, A. Engel, L. Jakoi, M. F. Melhem, J. M. Pipas, C. Smith, and J. R. Nevins. 2000. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol. Cell 6:293-306. [DOI] [PubMed] [Google Scholar]
- 46.Shan, B., and W. H. Lee. 1994. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol. Cell. Biol. 14:8166-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 48.Trimarchi, J. M., B. Fairchild, R. Verona, K. Moberg, N. Andon, and J. A. Lees. 1998. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. USA 95:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai, K. Y., Y. Hu, K. F. Macleod, D. Crowley, L. Yamasaki, and T. Jacks. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2:293-304. [DOI] [PubMed] [Google Scholar]
- 51.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 19:6379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weintraub, S. J., C. A. Prater, and D. C. Dean. 1992. Retinoblastoma protein switches the E2F site from positive to negative element. Nature 358:259-261. [DOI] [PubMed] [Google Scholar]
- 53.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 55.Wu, X., and A. J. Levine. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA 91:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamasaki, L., R. Bronson, B. O. Williams, N. J. Dyson, E. Harlow, and T. Jacks. 1998. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(±) mice. Nat. Genet. 18:360-364. [DOI] [PubMed] [Google Scholar]
- 57.Yamasaki, L., T. Jacks, R. Bronson, E. Goillot, E. Harlow, and N. J. Dyson. 1996. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85:537-548. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, J. W., D. DeRyckere, F. X. Li, Y. Y. Wan, and J. DeGregori. 1999. A role for E2F1 in the induction of ARF, p53, and apoptosis during thymic negative selection. Cell Growth Differ. 10:829-838. [PubMed] [Google Scholar]
- 59.Zhu, J. W., S. J. Field, L. Gore, M. Thompson, H. Yang, Y. Fujiwara, R. D. Cardiff, M. Greenberg, S. H. Orkin, and J. DeGregori. 2001. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol. Cell. Biol. 21:8547-8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziebold, U., T. Reza, A. Caron, and J. A. Lees. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15:386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]