Abstract
While scaffold proteins are thought to be key components of signaling pathways, their exact function is unknown. By preassembling multiple components of signaling cascades, scaffolds are predicted to influence the efficiency and/or specificity of signaling events. Here we analyze a potential scaffold of the Ras/mitogen-activated protein kinase (MAPK) pathway, kinase suppressor of Ras (KSR), by generating KSR-deficient mice. KSR-deficient mice were grossly normal even though ERK kinase activation was attenuated to a degree sufficient to block T-cell activation and inhibit tumor development. Consistent with its role as a scaffold, high-molecular-weight complexes containing KSR, MEK, and ERK were lost in the absence of KSR. This demonstrates that KSR is a bona fide scaffold that is not required for but enhances signaling via the Ras/MAPK signaling pathway.
Sequential protein kinase cascades are highly conserved signaling pathways used by cells to respond to extracellular stimuli (18, 45, 65). While the kinase components of these pathways have been well described, how multiple kinases are regulated spatially or temporally is less well understood. A study of scaffold proteins, which by definition preassemble multiple components of a protein kinase cascade, can potentially explain how these signaling pathways are organized (5, 17, 32, 47, 48, 63, 64).
Scaffolds may serve different functions in signaling pathways. In some systems, scaffolds are integral components required for signaling events to occur. In budding yeast cells, inactivating the mitogen-activated protein kinase (MAPK) scaffold protein, STE5, blocks mating. Genetic studies demonstrate that STE5 is crucial for specifically directing the yeast MAPK components to activate the pheromone response pathway (reviewed by Elion [17]). In another example, the Drosophila photoreceptor scaffold protein InaD is not required but can dramatically influence the efficiency of signal transduction (59).
While scaffolds for the MAPK pathway are well documented in yeast, fly, and worm cells, deducing the physiologic importance and function of mammalian homologs has been less successful. Several proteins have been identified as potential mammalian scaffolds based on the ability of a particular protein to bind multiple kinases of a signaling pathway (10, 46, 70). To date, only one scaffold protein, JIP-1, has been examined in a physiologic context. JIP-1-deficient animals demonstrated defective JNK activation in response to excitotoxic and anoxic stress in the brain (64). There has not been, however, an in-depth analysis of candidate proteins in a physiologic context for the Ras/MAPK pathway. Here we analyze a putative MAPK scaffold, kinase suppressor of Ras (KSR). KSR was cloned in Caenorhabditis elegans and Drosophila melanogaster (31, 53, 55). Because the loss of KSR suppresses the phenotype conferred by an activated Ras molecule, KSR is thought to play a positive role in Ras/MAPK signaling. Genetic epistasis experiments position KSR after Ras and before MEK in the signaling pathway.
KSR remains an intriguing protein because its biological importance and precise role in the mammalian Ras/MAPK pathway are unclear. While the loss of KSR expression causes embryonic lethality in D. melanogaster, loss of KSR has little effect in C. elegans (31, 53, 55). While genetics support the idea that KSR is a positive regulator of the Ras/MAPK pathway, overexpression studies in mammalian cells suggests that KSR can both activate and inhibit Ras/MAPK signaling (6, 11, 26, 36, 52, 56, 71). One hypothesis that can reconcile these conflicting data proposes that KSR functions as a scaffold protein in the Ras/MAPK pathway since KSR can bind to both MEK and ERK (37, 38, 71). To support this, recent data demonstrate that KSR, when overexpressed in COS cells, can recruit MEK to the plasma membrane in response to epidermal growth factor (EGF) (39).
To test the role of KSR as a MAPK scaffold in vivo, KSR-deficient mice were generated. While KSR was not required for development, the loss of KSR decreased MAPK activation. The ability to study tissues and cells lacking KSR provided us with a unique opportunity to examine high-molecular-weight signaling complexes in the presence or absence of KSR, as well as its effect on MAPK signaling. We show here that KSR forms a ternary complex with both MEK and ERK and that the absence of KSR significantly diminishes MAPK signaling. These data support the idea that KSR is a MAPK pathway scaffold that is not required but functions mainly to facilitate MAPK activation.
MATERIALS AND METHODS
Construction of the KSR targeting vector and screening of mice.
Standard gene-targeting techniques and homologous recombination were used to generate KSR−/− mutant mice (57). The CA2 domain of the KSR protein was targeted and mice were screened by Southern blotting. Southern blotting confirmed the integrity of both the 5′ and 3′ targeting arms. Reverse transcription-PCR (RT-PCR) analysis showed that while the RNA machinery can splice out the neomycin cassette, it results in an out-of-frame transcript after amino acid 294. Heterozygous mice were bred and KSR−/− mice were screened with PCR primers specific for nucleotides 883 to 1008 in the KSR coding sequence. DO11.10 T-cell-receptor-positive (TCR+) mice were screened with primers specific for the alpha- and beta-chains of the DO11.10 TCR. In addition, a clonotypic monoclonal antibody, kJ1-26, was used to stain peripheral lymphocytes to detect T cells expressing the DO11.10 TCR. Mice transgenically expressing the polyomavirus middle T antigen (MT) were screened with primers generated to detect the splice A and splice B isoforms of the MT coding sequence.
Generation of MEFs.
Mouse embryonic fibroblasts (MEFs) were derived from 13.5-day-old embryos. After removal of the head and internal organs, embryos were rinsed with phosphate-buffered saline (PBS), minced, and digested with trypsin-EDTA (0.5% trypsin, 0.53 mM EDTA per embryo) at 37°C. Trypsin was inactivated by addition of Dulbecco minimal Eagle’s medium (DMEM) containing 10% fetal bovine serum, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were placed plates in T175 flasks and then incubated at 37°C in a 10% CO2 humidified chamber.
KSR protein analysis.
Lysates from spleen, thymus, and brain tissues were prepared from wild-type and KSR−/− mice. Tissues were homogenized in mammalian cell lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EDTA, 2 mM dithiothreitol [DTT], 0.5% NP-40, 100 μM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 20 μM pepstatin) by using prechilled glass homogenizers (2-ml capacity) and then passed three times through a 26-gauge needle. Lysates were cleared of insoluble debris by centrifugation at 4°C and 14,000 rpm for 10 min. The protein concentration was assessed by using a BCA Protein Assay Kit (Pierce). Studies were done either on immunoprecipitated KSR complexes or on cellular bulk lysates. The protein concentration was diluted to 1 mg/ml, and 1 ml was used for immunoprecipitation by using a goat anti-KSR (c-19) polyclonal antibody (Santa Cruz Biotechnology) and incubated overnight at 4°C. Protein A-Sepharose (Sigma) was then added for 1 h. For Western blot analysis, the immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylmide gel electrophoresis (SDS-PAGE) on 8% polyacrylamide. Immunoblotting was performed by using a rabbit polyclonal anti-KSR antibody generated to amino acids 118 to 248 of KSR (38) or a goat polyclonal anti-KSR antibody generated against amino acids 855 to 871 of KSR (c-19; Santa Cruz Biotechnology). Secondary donkey anti-rabbit horseradish peroxidase (HRP)-conjugated antibodies (Jackson Immunoresearch) were used at a 1:10,000 dilution. A Supersignal enhanced chemiluminescence detection kit (Pierce) was used to evaluate chemiluminescence on Kodak X-Omat AR film.
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis was performed on spleen, thymus, and bone marrow tissues homogenized by using complete DMEM placed on prefrosted glass slides and then cleared of debris by using cell strainers (Fisher). The cell suspensions were pelleted, washed twice with FACS staining buffer (PBS, 4% fetal bovine serum, 0.0002% sodium azide), and counted by using a hemacytometer. They were mixed with various antibodies conjugated to either fluorescein isothiocyanate (FITC) or phycoerythrin (PE). Antibodies from PharMingen included PE-conjugated anti-CD4, FITC-conjugated anti-CD8, PE-conjugated anti-B220, and biotin-conjugated anti-immunoglobulin M (IgM). Analyses were performed on a Becton Dickinson FACSCalibur instrument equipped with CellQuest software.
Proliferation assays.
Proliferation assays were performed with crude splenocytes. A total of 2 × 105 cells were plated in each well of a 96-well Costar tissue culture dish and incubated for 24, 48, or 72 h. T-cell mitogens used were 145-2C11 (monoclonal anti-CD3), ovalbumin OVA323-339 peptide, and phorbol myristate acetate (PMA) plus ionomycin or concavalin A (Sigma). All experiments were done in triplicate. The cells were incubated with 2 μCi of [3H]thymidine per well during the final 12 h of culture. The cells were harvested by using a Skatron Instruments Micro-96 Harvester and counted with a Beckman 6500 multipurpose scintillation counter.
IL-2 bioassays.
Interleukin-2 (IL-2) release was measured by using supernatants from proliferating T cells and adding them to the cytotoxic-T-cell line CTLL-2, which is IL-2 dependent for growth. After incubation with supernatants for 24 h, CTLL-2 cells were pulsed with 2 μCi of [3H]thymidine per well for 8 h. Cells were harvested in the same manner as for proliferation assays.
Recombinant baculovirus production.
For production of recombinant MEK baculovirus, MEK cDNA was subcloned into the vector pFastBac-HT (Life Technologies). Recombinant baculoviruses were then generated by using the Bac-to-Bac HT Baculovirus Expression System (Life Technologies) according to the manufacturer's instructions.
Immunoblotting.
MEFs, brain tissue, or T cells were isolated and used for immunoblots or in vitro kinase assays. For RasGTP analysis, 4 × 107 T cells were used per time point. After treatment with 100 nM PMA, cell lysates were incubated with glutathione S-transferase (GST)-RafRBD protein, and protein complexes were resolved by SDS-PAGE on 15% gels. For analysis of Raf kinase activity, 4 × 107 T cells were used per time point. After treatment with 100 nM PMA, Raf-1 was immunoprecipitated with polyclonal Raf-1 antibody as described previously (58) and then incubated in kinase buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 50 mM MgCl2, 2 mM EGTA, 150 μM ATP) with recombinant His-tagged MEK protein produced in Sf9 cells. Kinase assays were performed for 35 min at 37°C, and then proteins were analyzed by SDS-PAGE on 12% gels. For analysis of ERK activation, 2 × 105 to 10 × 105 cells were used per time point. Poststimulation, cell lysates were prepared, and proteins were resolved by SDS-PAGE on 12% gels. For detection of RasGTP, a Ras Activation Kit was used (catalog 17-218; Upstate Biotechnology). For immunoblotting of phosphorylated MEK, a polyclonal anti-phospho-MEK1/2 antibody (New England BioLabs) was used at a 1/1,000 dilution. For detection of phosphorylated ERK, a polyclonal anti-phospho-ERK1/2 antibody (New England BioLabs) was used at a 1:1,000 dilution. For immunoblotting of ERK1 and ERK2, ERK-1 (c-16) and ERK-2 (c-14) (Santa Cruz) were used at 1:500 dilutions. The secondary antibody was either goat anti-rabbit-HRP antibody (Santa Cruz) used at a 1:3,000 dilution or donkey anti-rabbit-HRP antibody (Jackson Immunoresearch) at a dilution of 1:10,000. For in vitro kinase assays, MEFs were deprived of serum, stimulated with either 10 nM EGF or 100 nM PMA for 5 min, and then lysed. Clarified cell lysates were immunoprecipitated with ERK-1 and ERK-2 antibodies, and in vitro kinase assays were performed with myelin basic protein as the substrate as described previously (3); reactions were quenched by the addition of sample buffer, denatured, and subjected to SDS-12% PAGE. Radiolabeled myelin basic protein was quantified by using phosphor storage technology (Molecular Dynamics).
Gel filtration chromatography.
Brain tissue was isolated from wild-type and KSR−/− animals. Whole brains were homogenized in 2-ml glass homogenizers in buffer B (i.e., 25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, and 2 mM EDTA supplemented with phenylmethylsulfonyl fluoride, leupeptin, pepstatin, and benzamidine). Homogenized tissue was then passed through a 26-gauge needle five times. Samples were spun down in sequential 15,000 × g centrifugation steps at 4°C. Clarified supernatants were then checked for protein concentration by using the BCA Protein Assay Kit (Pierce) and 1.5 mg of total protein was loaded onto a Superose 6 HR10/30 (Amersham Pharmacia Biotech) gel filtration column preequilibrated with buffer B at a flow rate of 0.25 ml/min as described previously (51, 62). Then, 35 1-ml fractions or 70 500-μl fractions were collected, subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted for MEK-1 (c-18; Santa Cruz), ERK1/2 (c-16; Santa Cruz), KSR (38), c-Raf-1 (clone 53 [Transduction Labs/PharMingen] or antibodies developed in our lab [58]), and B-Raf (c-19; Santa Cruz).
Th1/Th2 induction studies.
Splenic tissue from DO11.10 TCR+ wild-type or KSR−/− animals was isolated, homogenized, and subjected to red blood cell lysis. Ex vivo cultures were then set up with splenic lymphocytes and antigen-presenting cells, 1 μM ovalbumin peptide, and either (i) gamma interferon (IFN-γ), IL-12, and anti-IL-4 or (ii) IL-4, anti-IL-12, and anti-IFN-γ to induce Th1 or Th2 effector T-cell cultures. At day 3, cultures were split 1:3 and rested until day 7, when the cultures were restimulated with irradiated antigen-presenting cells and OVA323-339 peptide. Culture supernatants were then analyzed by enzyme-linked immunosorbent assay for IFN-γ and IL-4 production as described previously (24, 35).
Analysis of mammary tumors in KSR/MT+ mice.
KSR−/− mice were bred three generations onto the C57BL/6 background before being crossed to MT+ transgenic mice bred into the C57BL/6 background. F2 littermates were genotyped for KSR and MT. MT+ females were monitored daily starting at 3 weeks of age for the development of mammary tumors by palpating all five pairs of mammary glands as described previously (7, 22). Mice were sacrificed after tumors reached a size of 150 mm2 or larger.
RESULTS
Generation of KSR−/− animals.
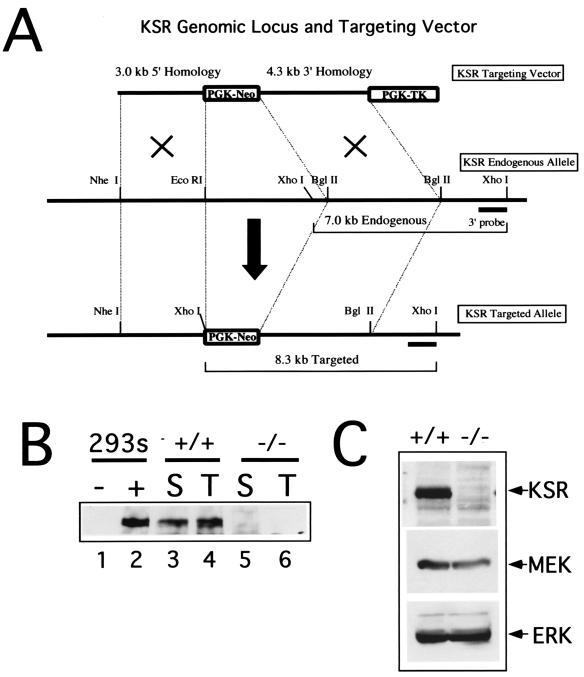
Standard gene-targeting methods were used to generate KSR−/− mice (57). Homologous recombination was used to replace the exon encoding the CA2 domain in the N terminus of the murine KSR gene locus with the neomycin resistance gene (Fig. 1A). Western blotting with both an N-terminal and a C-terminal specific antibody confirmed that full-length KSR, as well as any truncated form of KSR, was undetectable in the knockout animals (Fig. 1B and C). RT-PCR analysis showed that, while the RNA machinery can splice out the neomycin cassette, it results in an out-of-frame transcript (see Materials and Methods). Knockout animals were viable and phenotypically normal. The histology of all tissues was normal (data not shown). These results indicate that KSR is not required for development.
FIG. 1.
Generation of KSR−/− mice. (A) The targeting strategy is illustrated schematically. Exon 2 encoding the CA2 domain of KSR was targeted for homologous recombination. (B and C) Western blot analysis of KSR protein expression. Lysates prepared from spleen and thymus tissues (B) or brain tissue (C) were immunoprecipitated with goat anti-KSR antibodies and resolved by SDS-PAGE. These data were generated by immunoblotting with a polyclonal rabbit anti-KSR antibody.
Defective ERK activation in KSR−/− fibroblasts.
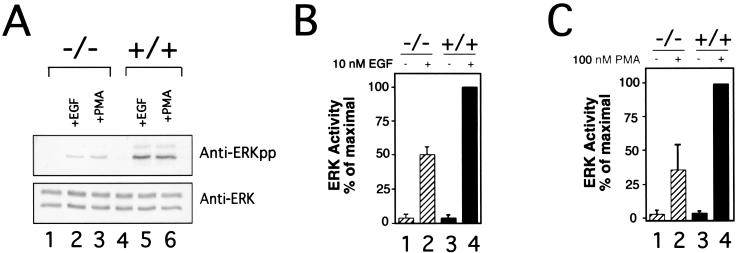
To test the function of KSR in the MAPK pathway, MEFs were generated from wild-type and KSR−/− animals. ERK activation was tested with two well-known ERK activators: EGF and PMA (4, 27). Both stimuli strongly induced ERK activity in wild-type cells, as detected by measuring ERK phosphorylation (Fig. 2A) or by in vitro kinase assays (Fig. 2B and C). In contrast, KSR−/− MEFs displayed a consistent 50% reduction in ERK activation (Fig. 2A) with decreased levels of phosphorylated ERK and decreased ERK activity detected. Wild-type and KSR−/− cells contain equivalent levels of MAPK. Thus, KSR potentiates ERK activation.
FIG. 2.
Impaired ERK activation in KSR−/− MEFs. (A) Defective ERK phosphorylation after EGF and PMA treatment. MEFs derived from wild-type (lanes 4 to 6) or KSR−/− (lanes 1 to 3) mice were treated with EGF (lanes 2 and 5) or PMA (lanes 3 and 6) or were left untreated (lanes 1 and 4). ERK1/ERK2 immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with antibodies to phospho-ERK (upper panel) or ERK (lower panel). (B and C) Diminished in vitro ERK kinase activity after EGF (B) or PMA (C) treatment. Wild-type or KSR−/− MEFs were treated with either 10 nM EGF or 100 nM PMA. ERK immunoprecipitates were used to phosphorylate myelin basic protein in vitro. The results are the means from four independent experiments.
Normal lymphocyte development in KSR−/− animals.
Given the normal phenotype of the KSR−/− animals, we focused on T-lymphocyte development and activation. The development of T lymphocytes is very well characterized and is known to be sensitive to changes in the signal transduction pathways. Furthermore, the Ras/MAPK pathway is thought to play an important role in the positive selection of T cells in the thymus (1, 2, 8, 43, 54).
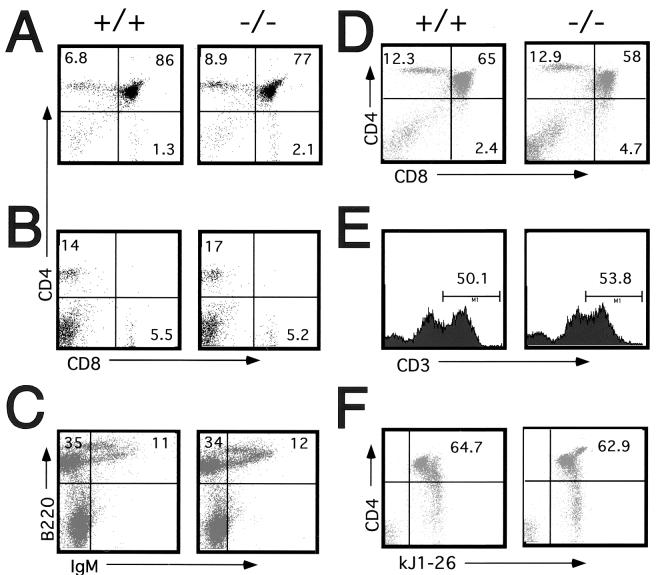
Thymocytes were isolated from 4- to 5-week-old wild-type or KSR−/− mice and analyzed. The cellularity and CD4/CD8 profiles of thymic T-cell populations in wild-type and KSR−/− mice were very similar (Fig. 3A). FACS analysis demonstrated there were no significant differences in percentages of thymocyte subsets as defined by expression of CD4 and CD8. Peripheral T-cell homeostasis was addressed by examining T cells in the spleen (Fig. 3B). The spleen had a normal cellularity with a normal distribution of mature T cells.
FIG. 3.
T-cell and B-cell development is normal in KSR−/− mice. (A) FACS analysis of thymocytes. Thymocytes from wild-type or KSR−/− mice were isolated and stained for CD4 and CD8 expression. (B) Peripheral T cells isolated from spleen tissues of wild-type or KSR−/− mice were stained for CD4 and CD8 expression. (C) Bone marrow cells isolated from wild-type or KSR−/− mice were stained for IgM and B220 expression. DO11.10 TCR+ thymocytes from wild-type or KSR−/− animals were stained for CD4 and CD8 expression (D), CD3 expression (E), and CD4 expression and kJ1-26 (DO11.10 TCR specific antibody) expression (F).
We analyzed T-cell development in greater detail by breeding KSR−/− mice to two different TCR transgenic mice, DO11.10 and H-Y. T cells expressing the DO11.10 TCR are activated by a peptide derived from ovalbumin (OVA323-339) (40), while the T cells expressing the H-Y TCR are activated by a male-specific peptide (29). The DO11.10 TCR is useful for studying CD4+ T-cell development, while the H-Y TCR is useful for studying CD8+ T-cell development. In both systems, thymic cellularity and T-cell development were not detectably impaired (Fig. 3D and data not shown). In repeated experiments, both wild-type and KSR−/− thymocytes expressed similar levels of TCR (Fig. 3E). Importantly, the presence of normal numbers of CD4+, DO11.10+ T cells and the presence of normal numbers of CD8+, H-Y TCR+ T cells demonstrated that positive selection was not affected by the loss of KSR (Fig. 3F and data not shown). Therefore, the loss of KSR did not detectably disrupt or inhibit the normal development of T cells.
We also examined B-cell development. Bone marrow-derived cells were stained for expression of CD43, B220, and IgM to analyze pro-B, pre-B, and immature B-cell populations (Fig. 3C and data not shown) (23). Normal populations of these developmental subsets were detected in KSR−/− mice. Thus, the loss of KSR did not detectably affect B-cell development.
Activation of MEK and ERK are impaired during T-cell activation.
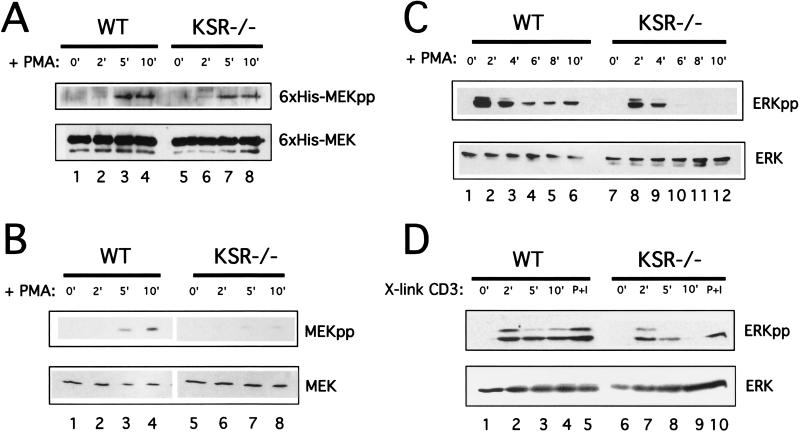
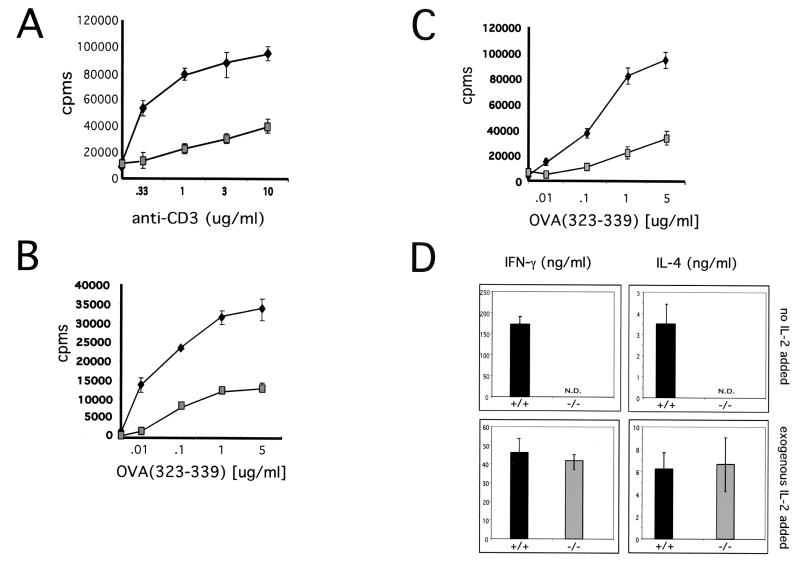
T-cell activation was analyzed first by CD3 cross-linking (Fig. 4A). T-cell proliferation was measured by [3H]thymidine incorporation. While wild-type and KSR−/− T cells proliferated equally well in response to PMA and ionomycin (data not shown), KSR−/− T cells exhibited a significant proliferation defect compared to wild-type T cells when stimulated by CD3 cross-linking. Given this proliferation defect, as well as the ERK activation defect observed in MEFs, the Ras/MAPK pathway was examined in greater detail. Since Ras can be activated by Ras-GRP in a diacylglycerol-dependent fashion (15, 16), PMA was used as the stimulus to examine Ras, Raf, MEK, and ERK activation kinetics in T cells.
FIG. 4.
Loss of KSR expression impairs MEK and ERK, but not Ras or Raf activation. (A) Raf activation and kinase activity are unaffected in KSR−/− T cells. (Top) Following treatment with PMA, immunoprecipitated Raf-1 was used for in vitro kinase assays with recombinant MEK protein produced from Sf9 cells. Phosphorylated MEK was detected by immunoblotting with an anti-phospho-MEK antibody. (Bottom) Total recombinant MEK protein was detected by using anti-MEK antibodies. (B) Defective MEK activation in response to PMA. (Top) PMA was used to stimulate T cells over a time course and lysates were used for immunoblotting. Active MEK was detected by using anti-phospho-MEK antibodies. (Bottom) Total endogenous MEK was detected by using an anti-MEK antibody. (C) Defective ERK activation in response to CD3 cross-linking. 2C11 was used to activate T cells from wild-type (lanes 1 to 4) or KSR−/− (lanes 6 to 9) mice. PMA and ionomycin were used as a positive control for MAPK activation (lanes 5 and 10). Cell lysates were immunoblotted with anti-phospho-ERK. (D) PMA was used to activate splenic T cells from wild-type (lanes 1 to 6) or KSR−/− (lanes 7 to 12) mice. Cell lysates were immunoblotted with anti-phospho-ERK (top) or with anti-ERK (bottom).
In response to PMA, wild-type and KSR−/− T cells demonstrated normal RasGTP formation with similar kinetics in wild-type and KSR−/− T cells (data not shown). Further, Raf-1 also displayed normal activation kinetics in the presence or absence of KSR. Raf-1 activation was examined by using an in vitro kinase assay. After PMA treatment, immunoprecipitated Raf-1 was used to phosphorylate recombinant MEK protein. Raf-1 activation was measured by immunoblotting for the level of phosphorylated recombinant MEK protein (Fig. 4B). Loss of KSR expression did not adversely affect Raf kinase activity in response to PMA (Fig. 4B, compare lanes 3 and 4 with lanes 7 and 8).
Unlike Ras and Raf, which display normal activation kinetics irrespective of the presence or absence of KSR, the loss of KSR expression results in impaired MEK activation (Fig. 4C, compare lanes 3 and 4 with lanes 7 and 8). ERK activation was also defective. In wild-type T cells (Fig. 4D, lanes 1 to 5) ERK activation in response to anti-CD3 cross-linking peaked at 2 min and was undetectable by 10 min. In comparison, ERK activation in KSR−/− T cells (Fig. 4D, lanes 6 to 10) was weak but exhibited similar kinetics. In response to PMA, ERK activation in wild-type T cells (Fig. 4E, lanes 1 to 6) peaked at 2 min and persisted over 10 min. In contrast, the pool of total phosphorylated ERK in KSR−/− T cells was significantly reduced, but the kinetics of activation were unchanged (Fig. 4E, lanes 7 to 12). These results are consistent with genetic epistasis experiments which place KSR between Ras and MEK in the Ras/MAPK pathway.
T-cell activation was next analyzed by using cognate antigen as the stimulus. DO11.10 transgenic T cells were isolated and stimulated by using a range of ovalbumin peptide concentrations. T-cell proliferation was measured at 24, 48, and 72 h after stimulation. KSR−/− transgenic T cells proliferated relatively poorly at all time points and at all ovalbumin peptide concentrations (Fig. 5A). Since secretion of the cytokine IL-2 is largely responsible for T-cell proliferation, we measured IL-2 secretion by using the IL-2-dependent cytotoxic-T-cell line CTLL-2 (20). IL-2 secretion from KSR−/− T cells was significantly decreased at all time points compared to that from wild-type T cells. This demonstrates that KSR−/− T cells are impaired in their ability to secrete IL-2 in response to antigen.
FIG. 5.
KSR−/− T cells display an activation defect ex vivo in response to physiologic antigen. (A) Defective T-cell proliferation in response to anti-CD3. 2C11 was used to stimulate splenic T cells from wild-type or KSR−/− mice. T-cell proliferation was measured by quantitation of [3H]thymidine incorporation. (B) KSR+/− or KSR−/− T cells transgenically expressing the DO11.10 TCR were used for proliferation assays in response to ovalbumin peptide (amino acids 323 to 339). After 24 h, cells were pulsed with [3H]thymidine for 12 h, and [3H]thymidine incorporation was measured. (C) IL-2 release was measured by using the IL-2-dependent cell line CTLL-2. After 24 h, supernatants from proliferating KSR+/− or KSR−/− T cells were used to stimulate CTLL-2 proliferation. (D) Th1/Th2 effector differentiation was studied by using wild-type and KSR−/− T cells under conditions which promote Th1 or Th2 development (top). Exogenous IL-2 was added to compensate for the proliferation defect of KSR−/− T cells (bottom).
When activated, naive T cells differentiate into cells of the Th1 or Th2 phenotype. Th1 cells secrete IFN-γ directing cellular responses during inflammatory immune challenges, while Th2 cells secrete IL-4, which upregulates antibody-dependent processes (21, 41). Since perturbation of MAPK signaling processes can affect Th1 and Th2 differentiation (14, 34), we were interested to test whether KSR played a role in Th1 and Th2 differentiation.
DO11.10+ T cells were placed under conditions promoting either Th1 or Th2 differentiation. Compared to wild-type controls, KSR−/− T cells did not produce appreciable amounts of IFN-γ or IL-4 under either of the induction conditions (Fig. 5C, upper panel). Since the lack of IL-2 secretion might explain this result, the Th1/Th2 experiment was repeated with the addition of exogenous IL-2 (Fig. 5C, lower panel). Supplemental IL-2 ameliorated the proliferative defect of the KSR−/− T cells, allowing normal Th1 and Th2 differentiation. In the presence of IL-2, KSR−/− Th1 and Th2 cells produced wild-type levels of cytokines. These data confirm that KSR is important for efficient T-cell proliferation and also demonstrate that KSR does not play an instructive role in Th1 or Th2 differentiation.
Loss of high molecular signaling complexes in KSR−/− mice.
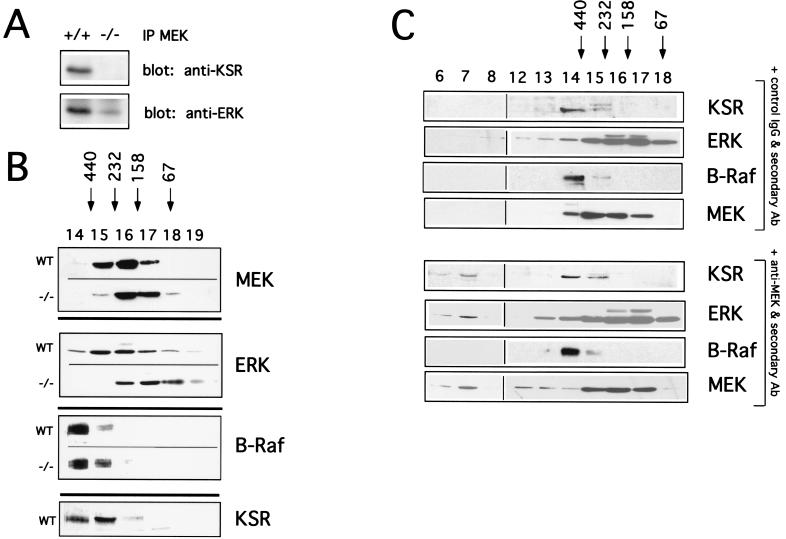
Recent studies demonstrate that KSR can coprecipitate with components of the Ras/MAPK pathways both in cell lines and in brain lysates (6, 11, 36, 38, 51, 56, 71) (Fig. 6A). Gel filtration studies demonstrate that when Raf is overexpressed, it can be detected in high-molecular-mass complexes of 400 to 500 kDa (62). We hypothesized that KSR, as a potential scaffold, might be required for nucleating a high-molecular-weight complex containing Raf, MEK, and ERK in vivo.
FIG. 6.
KSR, MEK, and ERK exist in high-molecular-weight complexes in vivo. (A) Coimmunoprecipitation of KSR, ERK, and MEK. Brain lysates were immunoprecipitated with anti-MEK antibodies and blotted for KSR and ERK (polyclonal antibodies). (B) Gel filtration analysis of brain tissue from wild-type and KSR−/− mice. Gel filtration fast-performance liquid chromatography fractions were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted for KSR, MEK, ERK, and B-Raf. Apparent-molecular-weight standards are indicated above. (C) Wild-type brain lysates were treated with anti-MEK antibody, followed by treatment with secondary anti-rabbit antibodies or control IgG antibodies, followed by treatment with secondary anti-rabbit antibodies. Each sample was then used for gel filtration. Fast-performance liquid chromatography fractions were immunoblotted for KSR, ERK, MEK, and B-Raf.
Cytoplasmic extracts were prepared from wild-type mouse brain and separated by gel filtration (Fig. 6B). We chose brain because it is known to express high levels of KSR (38). Fractions were immunoblotted for KSR, as well as components of the MAPK pathway. KSR, RAF, MEK, and ERK were all found to migrate in fractions containing high-molecular-mass complexes of 250 to 500 kDa (Fig. 6B). To our knowledge, this is the first demonstration that endogenous Raf, MEK, and ERK are contained in high-molecular-mass complexes in vivo.
We next used gel filtration to analyze brain cytoplasmic extracts from KSR−/− mice. Our finding, supporting a scaffolding role for KSR, was that the loss of KSR results in the shifting of complexes containing MEK and ERK to fractions of ca. 50 to 250 kDa. Immunoblotting for B-Raf (Fig. 6B) and c-Raf-1 (data not shown) showed no detectable shift, suggesting that Raf is not a component of the KSR-mediated scaffold.
While the data are consistent with the idea that KSR is stably associated with MEK and ERK in vivo, it remained unclear whether these three proteins form a ternary complex in vivo. To test this, we attempted to move complexes to the void volume of the gel filtration column by using antibody cross-linking. We reasoned that if KSR, MEK, and ERK existed in the same complex, aggregation with antibodies to MEK should also move KSR and ERK to the void volume.
Brain cytoplasmic extracts were treated with anti-MEK or a control antibody, followed by a secondary cross-linking antibody to generate large aggregates (Fig. 6C). In the presence of the anti-MEK, KSR and ERK shifted with MEK to the void volume of the gel filtration column. This was specific because the position of B-Raf was unchanged by treatment with the MEK antibody. In addition, the migration of MEK, ERK, and KSR complexes was unchanged when lysates were treated with control antibody. These data establish clearly that ternary complexes containing KSR, MEK, and ERK are stably present in vivo. Further, these data can potentially explain the activation defect in the Ras/MAPK signaling pathway in KSR−/− cells since MEK and ERK are inefficiently recruited to upstream activators such as Ras and Raf-1.
Attenuation of tumor growth in KSR−/− animals.
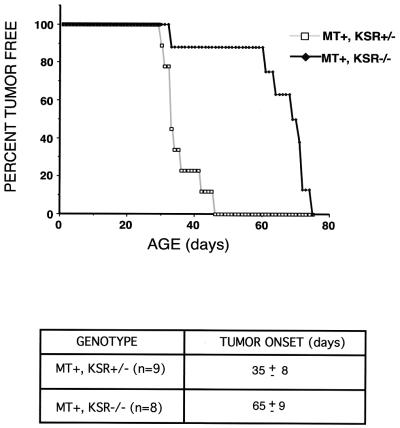
Since KSR was originally identified in a system in which Ras was constitutively active, we were interested in testing whether the loss of KSR affects the progression of Ras-dependent tumors. In female mice, the transgenic expression of the polyomavirus MT causes breast tumors to appear ca. 30 to 40 days after birth (22). MT is thought to be oncogenic in a Ras-dependent manner through the recruitment of the adaptor protein, Shc, and the lipid kinase, phosphatidylinositol 3-kinase (13, 60). KSR−/− mice were bred to MT+ transgenic mice, and the F2 littermate females were monitored. All mice used were bred at least four generations onto a C57BL/6 background.
MT+ KSR+/− and MT+ KSR−/− female littermate mice were caged separately after genotyping and checked daily for tumor formation by palpating each pair of mammary glands on each mouse. The onset of tumor formation in KSR+/− mice was found to be 35 ± 8 days (n = 9) (Fig. 7). In comparison, the onset of tumor formation in KSR−/− mice demonstrated slower kinetics since tumors were not detected until 65 ± 9 days (n = 8). Immunoblotting demonstrated that breast tumors from wild-type mice express KSR. These findings suggest that the loss of KSR expression can slow tumor progression in a well-characterized Ras-dependent tumor model. Further, this is also in agreement with original genetic screens in which the loss-of-function KSR mutants were able to suppress the effects caused by constitutively active Ras transgenes.
FIG. 7.
Loss of KSR expression inhibits tumor formation in mammary epithelial cells mediated by polyomavirus MT. Female littermates were genotyped for transgenic expression of polyomavirus MT (MT+) and KSR status (either KSR+/− or KSR−/−). These mice were examined for the development of mammary tumors starting at the age of 3 weeks. Tumors were detected by palpating all five pairs of mammary glands on each mouse daily. The onset of tumor formation in MT+ KSR+/− mice (n = 9) was 35 ± 8 days. The onset of tumor formation in MT+ KSR−/− mice (n = 8) was 65 ± 9 days.
DISCUSSION
Here we have established that KSR functions as a scaffold in the Ras/MAPK signaling pathway in vivo. While this role had previously been suggested by overexpression data (39, 51) or coimmunoprecipitation studies (38), our data show for the first time that KSR at endogenous expression levels exists in a high-molecular-weight complex that contains both MEK and ERK. Interestingly, we show that Raf also exists as in a high-molecular-weight complex, but these complexes are distinct from the ones containing KSR, MEK, and ERK. The advantage of this knockout approach is our ability to study the function of KSR by analyzing MAPK activation in cells expressing or lacking KSR. These studies showed that KSR positively regulates MAPK signaling but is not required for MAPK signaling. While the sequence of KSR is predicted to encode for a kinase and work from only one group has reported KSR kinase activity (67, 68, 72), KSR lacks several conserved residues found in virtually all kinases characterized to date (22a). A growing body of experimental data support the idea that KSR has no intrinsic kinase activity (36, 38, 39, 61).
KSR as a scaffold.
The prototypical MAPK scaffold is STE5 from Saccharomyces cerevisiae. Yeast cells use the same three MAPKKK/MAPKK/MAPK proteins—STE11, STE7, and FUS3—to respond to several different stimuli (as reviewed by Elion [17]). The function of these scaffolds is thought to be critical for the routing of specific extracellular signals to the MAPK pathway. The response to mating pheromone requires the scaffold protein, STE5, while the response to osmotic shock requires the scaffolds PBS2 and SHO1. The stimulus for invasive growth under starvation conditions does not appear to require a scaffold, but does use the conserved STE11/STE7/FUS3 triad. It is notable that, with the completion of the human genome project, homologs for STE5, PBS2, and SHO1 have not been identified in mammalian cells.
Based on binding to multiple components of a MAPK signaling pathway, several mammalian proteins have been suggested to function as MAPK scaffolds. For the Ras/MAPK signaling pathway, three proteins—KSR, MP-1, and RKIP (46, 70)—have been implicated. In the stress pathways that use JNK, JIP-1 and MEKK1 have been identified as potential scaffolds (9, 10, 12, 69). In general, they are defined as scaffolds based on their ability to bind to two or more components of the kinase pathway. However, with the exception of JIP-1 (64), the function of these proteins in vivo and whether or not they truly scaffold MAPK components in vivo remains largely unresolved.
Theoretically, scaffolds may have a large number of nonmutually exclusive functions (5, 30, 32). These properties include linking specific extracellular signals with the MAPK pathway, enhancing the kinetics of signaling (i.e., catalytic scaffolds), or preventing cross talk between MAPK pathways (i.e., insulators). STE5, PBS1, and SHO1 are good examples of scaffolds that are required to link specific extracellular signals with the MAPK signaling pathway. InaD, a scaffold important in Drosophila photoreceptor signal transduction, appears to function as a catalytic scaffold because signaling in its absence still occurs but with reduced efficiency and intensity (59).
Since ERK activation was not abolished and was attenuated in the KSR−/− mouse to the stimulation of multiple agonists, KSR does not appear to function as a specificity determinant in the MAPK pathway. Rather, KSR functions as a catalytic scaffold that enhances the kinetic efficiency of MAPK signaling. We suspect that KSR facilitates MAPK activation by preassembling components and/or by helping to deliver cytoplasmic MEK and ERK to Ras and Raf at the plasma membrane (37, 39). Since Raf did not appear to be bound to KSR, a specificity scaffold, if one exists, might be predicted to couple Raf with Ras. In fact, Sur-8, a potential adaptor/scaffold molecule identified in C. elegans, may function by preassembling Raf with Ras (33, 50). It is also interesting that STE5, PBS1, and SHO1 all bind to STE11, the ortholog of Raf-1, whereas our studies suggest that KSR does not bind to Raf-1 or B-Raf in vivo. Thus, KSR does not appear to function like STE5. It will be interesting to determine whether other proteins, such as MP-1 and RKIP, fulfill the criteria as scaffolds and whether they have functions that are distinct from those of KSR.
Phenotype of KSR-deficient mice.
Given the significant impairment of ERK activation to multiple stimuli, we were surprised that KSR-deficient mice develop normally. However, precedence for the plasticity of developmental processes can be found in several other mouse models wherein molecules central to the Ras/MAPK pathway were ablated. For example, development proceeds normally in mice deficient for N-Ras or H-Ras (19), and mice deficient for ERK1 exhibit only a moderate defect in T-cell development (44).
Alternatively, other mammalian KSR family members may exist that can replace KSR function during development. In C. elegans, another potential KSR family member protein, KSR2/Pex-1, was recently cloned. RNA-mediated interference of KSR2/Pex-1 in a KSR−/− background demonstrates that the loss of both isoforms is lethal in the worm (42). Lethality in the complete absence of KSR is consistent with the finding that the loss of the single KSR isoform in Drosophila is also lethal. It seems very likely, therefore, that other KSR family members will in turn be discovered in the mouse and human genomes. It is important to note, however, that if another form of KSR does exist in mouse, its expression is not sufficient to restore ERK activation nor to prevent the loss of high-molecular- weight signaling complexes in the brain tissue that we examined. It is possible that the residual level of ERK activity that we detected may be due to another isoform of KSR, but it is equally possible that other potential KSR family members will turn out to function in a tissue-specific fashion.
Since the Ras/MAPK pathway is thought to be critical for the positive selection of T cells (reviewed in references 28 and 49), it is also surprising that we were unable to detect any significant defects in T-cell development. While it is possible that the residual ERK activity in the KSR-deficient mice is sufficient to mediate positive selection, recent data support the idea that the Ras/MAPK pathway may not be as important for T-cell-positive selection as previously believed. The original studies were based on transgenic animals that express dominant-negative forms of Ras, Raf, and MEK (1, 2, 43). However, studies with dominant-negative MAPK proteins transduced by retroviruses (8) or with mice deficient for H-Ras or N-Ras (19) do not support the results of the original transgenic studies. We suspect that the Ras/MAPK signaling pathways may be more involved in mediating processes such as cell growth and proliferation and are less critical in instructive, developmental decisions.
In conclusion, we have established that KSR fulfills the generally accepted criteria of a scaffold (5, 25, 37). It participates in helping to generate high-molecular-weight multiprotein signaling complexes that simultaneously contain at least three components of the signaling pathway: KSR, MEK, and ERK. While it has been previously suggested that Raf might be a component of this signaling complex (56, 66), our results demonstrate that the high-molecular-weight complexes containing Raf are independent of the KSR/MEK/ERK complexes. It is possible that signaling may induce an association between the two complexes, but such experiments are technically not feasible at the present time. Lastly, our data are consistent with the role of KSR as a general positive mediator of Ras/MAPK signaling; ERK stimulation in the absence of KSR was attenuated in a variety of systems, including tumor growth and T-cell activation. Given that most components of the signaling pathways have been identified, the challenges for signal transduction in the next decade will be to determine how signaling pathways are regulated and modified in time and space. It is clear that understanding the function of scaffolds will play the central role in these processes.
Acknowledgments
We thank Andrew Chan, Alec Cheng, Osami Kanagawa, and Barry Sleckman for their insight and advice and Jeong Kim for critical reading of the manuscript.
This research was supported by the NIH (A.S.S.), an NIH training grant (A.N.), the Burroughs-Wellcome Fund (W.R.B.), NIH grants CA90400 and DK52809 (R.E.L.), and the American Diabetes Association (R.E.L. and R.K.).
REFERENCES
- 1.Alberola-Ila, J., K. A. Forbush, R. Seger, E. G. Krebs, and R. M. Perlmutter. 1995. Selective requirement for MAPK activation in thymocyte differentiation. Nature 373:620-623. [DOI] [PubMed] [Google Scholar]
- 2.Alberola-Ila, J., K. A. Hogquist, K. A. Swan, M. J. Bevan, and R. M. Perlmutter. 1996. Positive and negative selection invoke distinct signaling pathways. J. Exp. Med. 184:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehm, J. E., O. V. Chaika, and R. E. Lewis. 1998. Anti-apoptotic signaling by a colony-stimulating factor-1 receptor/insulin receptor chimera with a juxtamembrane deletion. J. Biol. Chem. 273:7169-7176. [DOI] [PubMed] [Google Scholar]
- 4.Bubenik, J., M. Indrova, J. Simova, H. Kypenova, D. Bubenikova, and K. Dolezalova. 1984. Phorbol myristate acetate-stimulated production of interleukin 2 by T-cell lymphoma and constitutive production by derived hybridomas. Neoplasma 31:497-505. [PubMed] [Google Scholar]
- 5.Burack, W. R., and A. S. Shaw. 2000. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 12:211-216. [DOI] [PubMed] [Google Scholar]
- 6.Cacace, A. M., N. R. Michaud, M. Therrien, K. Mathes, T. Copeland, G. M. Rubin, and D. K. Morrison. 1999. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol. Cell. Biol. 19:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, A. M., T. M. Saxton, R. Sakai, S. Kulkarni, G. Mbamalu, W. Vogel, C. G. Tortorice, R. D. Cardiff, J. C. Cross, W. J. Muller, and T. Pawson. 1998. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell 95:793-803. [DOI] [PubMed] [Google Scholar]
- 8.Crompton, T., K. C. Gilmour, and M. J. Owen. 1996. The MAPK pathway controls differentiation from double-negative to double-positive thymocyte. Cell 86:243-251. [DOI] [PubMed] [Google Scholar]
- 9.Cuenda, A., and D. S. Dorow. 1998. Differential activation of stress-activated protein kinase kinases SKK4/MKK7 and SKK1/MKK4 by the mixed-lineage kinase-2 and mitogen-activated protein kinase kinase (MKK) kinase-1. Biochem. J. 333:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 11.Denouel-Galy, A., E. M. Douville, P. H. Warne, C. Papin, D. Laugier, G. Calothy, J. Downward, and A. Eychene. 1997. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr. Biol. 8:46-55. [DOI] [PubMed] [Google Scholar]
- 12.Dickens, M., J. S. Rogers, J. Cavanagh, A. Raitano, Z. Xia, J. R. Halpern, M. E. Greenberg, C. L. Sawyers, and R. J. Davis. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693-696. [DOI] [PubMed] [Google Scholar]
- 13.Dilworth, S. M., C. E. Brewster, M. D. Jones, L. Lanfrancone, G. Pelicci, and P. G. Pelicci. 1994. Transformation by polyomavirus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature 367:87-90. [DOI] [PubMed] [Google Scholar]
- 14.Dong, C., D. D. Yang, C. Tournier, A. J. Whitmarsh, J. Xu, R. J. Davis, and R. A. Flavell. 2000. JNK is required for effector T-cell function but not for T-cell activation. Nature 405:91-94. [DOI] [PubMed] [Google Scholar]
- 15.Dower, N. A., S. L. Stang, D. A. Bottorff, J. O. Ebinu, P. Dickie, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317-321. [DOI] [PubMed] [Google Scholar]
- 16.Downward, J., J. D. Graves, P. H. Warne, S. Rayter, and D. A. Cantrell. 1990. Stimulation of p21ras upon T-cell activation. Nature 346:719-723. [DOI] [PubMed] [Google Scholar]
- 17.Elion, E. A. 1998. Routing MAP kinase cascades. Science 281:1625-1626. [DOI] [PubMed] [Google Scholar]
- 18.English, J., G. Pearson, J. Wilsbacher, J. Swantek, M. Karandikar, S. Xu, and M. H. Cobb. 1999. New insights into the control of MAP kinase pathways. Exp. Cell Res. 253:255-270. [DOI] [PubMed] [Google Scholar]
- 19.Esteban, L. M., C. Vicario-Abejon, P. Fernandez-Salguero, A. Fernandez-Medarde, N. Swaminathan, K. Yienger, E. Lopez, M. Malumbres, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillis, S., and K. A. Smith. 1977. Long term culture of tumour-specific cytotoxic T cells. Nature 268:154-156. [DOI] [PubMed] [Google Scholar]
- 21.Glimcher, L. H., and K. M. Murphy. 2000. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14:1693-1711. [PubMed] [Google Scholar]
- 22.Guy, C. T., R. D. Cardiff, and W. J. Muller. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family conserved features and deduced phylogeny of the catalytic domains. Science 241:42-62. [DOI] [PubMed] [Google Scholar]
- 23.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, C. S., S. E. Macatonia, A. O'Garra, and K. M. Murphy. 1995. T-cell genetic background determines default T helper phenotype development in vitro. J. Exp. Med. 181:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, M., K. Yoshioka, M. Akechi, S. Yamashita, N. Takamatsu, K. Sugiyama, M. Hibi, Y. Nakabeppu, T. Shiba, and K. I. Yamamoto. 1999. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol. Cell. Biol. 19:7539-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joneson, T., J. A. Fulton, D. J. Volle, O. V. Chaika, D. Bar-Sagi, and R. E. Lewis. 1998. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J. Biol. Chem. 273:7743-7748. [DOI] [PubMed] [Google Scholar]
- 27.Kamata, T., and J. R. Feramisco. 1984. Epidermal growth factor stimulates guanine nucleotide binding activity and phosphorylation of ras oncogene proteins. Nature 310:147-150. [DOI] [PubMed] [Google Scholar]
- 28.Kaye, J. 2000. Regulation of T-cell development in the thymus. Immunol. Res. 21:71-81. [DOI] [PubMed] [Google Scholar]
- 29.Kisielow, P., H. S. Teh, H. Bluthmann, and H. von Boehmer. 1988. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature 335:730-733. [DOI] [PubMed] [Google Scholar]
- 30.Kolch, W. 2000. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351(Pt. 2):289-305. [PMC free article] [PubMed] [Google Scholar]
- 31.Kornfeld, K., D. B. Hom, and H. R. Horvitz. 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83:903-913. [DOI] [PubMed] [Google Scholar]
- 32.Levchenko, A., J. Bruck, and P. W. Sternberg. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA 97:5818-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, W., M. Han, and K. L. Guan. 2000. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 14:895-900. [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, B., H. Yu, C. Chow, B. Li, W. Zheng, R. J. Davis, and R. A. Flavell. 2001. GADD45gamma mediates the activation of the p38 and JNK MAP kinase pathways and cytokine production in effector TH1 cells. Immunity 14:583-590. [DOI] [PubMed] [Google Scholar]
- 35.Macatonia, S. E., C. S. Hsieh, K. M. Murphy, and A. O'Garra. 1993. Dendritic cells and macrophages are required for Th1 development of CD4+ T cells from alpha beta TCR transgenic mice: IL-12 substitution for macrophages to stimulate IFN-gamma production is IFN-gamma-dependent. Int. Immunol. 5:1119-1128. [DOI] [PubMed] [Google Scholar]
- 36.Michaud, N. R., M. Therrien, A. Cacace, L. C. Edsall, S. Spiegel, G. M. Rubin, and D. K. Morrison. 1997. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl. Acad. Sci. USA 94:12792-12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison, D. K. 2001. KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci. 114:1609-1612. [DOI] [PubMed] [Google Scholar]
- 38.Muller, J., A. M. Cacace, W. E. Lyons, C. B. McGill, and D. K. Morrison. 2000. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell. Biol. 20:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, K. M., A. B. Heimberger, and D. Y. Loh. 1990. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science 250:1720-1723. [DOI] [PubMed] [Google Scholar]
- 41.O'Garra, A., and K. Murphy. 1996. Role of cytokines in development of Th1 and Th2 cells. Chem. Immunol. 63:1-13. [DOI] [PubMed] [Google Scholar]
- 42.Ohmachi, M., C. E. Rocheleau, D. Church, E. Lambie, T. Schedl, and M. V. Sundaram. 2002. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 12:427-433. [DOI] [PubMed] [Google Scholar]
- 43.O'Shea, C. C., T. Crompton, I. R. Rosewell, A. C. Hayday, and M. J. Owen. 1996. Raf regulates positive selection. Eur. J. Immunol. 26:2350-2355. [DOI] [PubMed] [Google Scholar]
- 44.Pages, G., S. Guerin, D. Grall, F. Bonino, A. Smith, F. Anjuere, P. Auberger, and J. Pouyssegur. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 286:1374-1377. [DOI] [PubMed] [Google Scholar]
- 45.Pawson, T., and T. M. Saxton. 1999. Signaling networks: do all roads lead to the same genes? Cell 97:675-678. [DOI] [PubMed] [Google Scholar]
- 46.Schaeffer, H. J., A. D. Catling, S. T. Eblen, L. S. Collier, A. Krauss, and M. J. Weber. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281:1668-1671. [DOI] [PubMed] [Google Scholar]
- 47.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schillace, R. V., and J. D. Scott. 1999. Organization of kinases, phosphatases, and receptor signaling complexes. J. Clin. Investig. 103:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sebzda, E., S. Mariathasan, T. Ohteki, R. Jones, M. F. Bachmann, and P. S. Ohashi. 1999. Selection of the T-cell repertoire. Annu. Rev. Immunol. 17:829-874. [DOI] [PubMed] [Google Scholar]
- 50.Sternberg, P. W., and J. Alberola-Ila. 1998. Conspiracy theory: RAS and RAF do not act alone. Cell 95:447-450. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, S., M. Sundaram, Y. Zhang, J. Lee, M. Han, and K. Guan. 1999. Kinase suppresor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 19:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugimoto, T., S. Stewart, M. Han, and K. Guan. 1998. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 17:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 54.Swan, K. A., J. Alberola-Ila, J. A. Gross, M. W. Appleby, K. A. Forbush, J. F. Thomas, and R. M. Perlmutter. 1995. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 14:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Therrien, M., H. C. Chang, N. M. Solomon, F. D. Karim, D. A. Wassarman, and G. M. Rubin. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83:879-888. [DOI] [PubMed] [Google Scholar]
- 56.Therrien, M., N. R. Michaud, G. M. Rubin, and D. K. Morrison. 1996. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 10:2684-2695. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, K. R., K. R. Folger, and M. R. Capecchi. 1986. High frequency targeting of genes to specific sites in the mammalian genome. Cell 44:419-428. [DOI] [PubMed] [Google Scholar]
- 58.Thorson, J. A., L. W. Yu, A. L. Hsu, N. Y. Shih, P. R. Graves, J. W. Tanner, P. M. Allen, H. Piwnica-Worms, and A. S. Shaw. 1998. 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell. Biol. 18:5229-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsunoda, S., and C. S. Zuker. 1999. The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell. Calcium 26:165-171. [DOI] [PubMed] [Google Scholar]
- 60.van der Geer, P., S. Wiley, V. K. Lai, J. P. Olivier, G. D. Gish, R. Stephens, D. Kaplan, S. Shoelson, and T. Pawson. 1995. A conserved amino-terminal Shc domain binds to phosphotyrosine motifs in activated receptors and phosphopeptides. Curr. Biol. 5:404-412. [DOI] [PubMed] [Google Scholar]
- 61.Volle, D. J., J. A. Fulton, O. V. Chaika, K. McDermott, H. Huang, L. A. Steinke, and R. E. Lewis. 1999. Phosphorylation of the kinase suppressor of ras by associated kinases. Biochemistry 38:5130-5137. [DOI] [PubMed] [Google Scholar]
- 62.Wartmann, M., and R. J. Davis. 1994. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J. Biol. Chem. 269:6695-6701. [PubMed] [Google Scholar]
- 63.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23:481-485. [DOI] [PubMed] [Google Scholar]
- 64.Whitmarsh, A. J., C. Y. Kuan, N. J. Kennedy, N. Kelkar, T. F. Haydar, J. P. Mordes, M. Appel, A. A. Rossini, S. N. Jones, R. A. Flavell, P. Rakic, and R. J. Davis. 2001. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev. 15:2421-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
- 66.Xing, H., K. Kornfeld, and A. J. Muslin. 1997. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr. Biol. 7:294-300. [DOI] [PubMed] [Google Scholar]
- 67.Xing, H. R., and R. Kolesnick. 2001. Kinase suppressor of Ras signals through Thr269 of c-Raf-1. J. Biol. Chem. 276:9733-9741. [DOI] [PubMed] [Google Scholar]
- 68.Xing, H. R., J. Lozano, and R. Kolesnick. 2000. Epidermal growth factor treatment enhances the kinase activity of kinase suppressor of Ras. J. Biol. Chem. 275:17276-17280. [DOI] [PubMed] [Google Scholar]
- 69.Yang, D., C. Tournier, M. Wysk, H. T. Lu, J. Xu, R. J. Davis, and R. A. Flavell. 1997. Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation, and defects in AP-1 transcriptional activity. Proc. Natl. Acad. Sci. USA 94:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeung, K., T. Seitz, S. Li, P. Janosch, B. McFerran, C. Kaiser, F. Fee, K. D. Katsanakis, D. W. Rose, H. Mischak, J. M. Sedivy, and W. Kolch. 1999. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401:173-177. [DOI] [PubMed] [Google Scholar]
- 71.Yu, W., W. J. Fantl, G. Harrowe, and L. T. Williams. 1998. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr. Biol. 8:56-64. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Y., B. Yao, S. Delikat, S. Bayoumy, X. H. Lin, S. Basu, M. McGinley, P. Y. Chan-Hui, H. Lichenstein, and R. Kolesnick. 1997. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89:63-72. [DOI] [PubMed] [Google Scholar]