Abstract
In the U12-dependent spliceosome, the U4atac/U6atac snRNP represents the functional analogue of the major U4/U6 snRNP. Little information is available presently regarding the protein composition of the former snRNP and its association with other snRNPs. In this report we show that human U4atac/U6atac di-snRNPs associate with U5 snRNPs to form a 25S U4atac/U6atac.U5 trimeric particle. Comparative analysis of minor and major tri-snRNPs by using immunoprecipitation experiments revealed that their protein compositions are very similar, if not identical. Not only U5-specific proteins but, surprisingly, all tested U4/U6- and major tri-snRNP-specific proteins were detected in the minor tri-snRNP complex. Significantly, the major tri-snRNP-specific proteins 65K and 110K, which are required for integration of the major tri-snRNP into the U2-dependent spliceosome, were among those proteins detected in the minor tri-snRNP, raising an interesting question as to how the specificity of addition of tri-snRNP to the corresponding spliceosome is maintained. Moreover, immunodepletion studies demonstrated that the U4/U6-specific 61K protein, which is involved in the formation of major tri-snRNPs, is essential for the association of the U4atac/U6atac di-snRNP with U5 to form the U4atac/U6atac.U5 tri-snRNP. Subsequent immunoprecipitation studies demonstrated that those proteins detected in the minor tri-snRNP complex are also incorporated into U12-dependent spliceosomes. This remarkable conservation of polypeptides between minor and major spliceosomes, coupled with the absence of significant sequence similarity between the functionally analogous snRNAs, supports an evolutionary model in which most major and minor spliceosomal proteins, but not snRNAs, are derived from a common ancestor.
The major or U2-dependent spliceosome, which catalyzes the removal of the vast majority of introns from nuclear pre-mRNA molecules, is a large 50S-60S ribonucleoprotein complex composed of the so-called major snRNPs U1, U2, U4/U6, and U5 and numerous non-snRNP-associated proteins (reviewed in references 4 and 27). It assembles on the pre-mRNA via an ordered pathway that involves dynamic interactions between the different snRNPs and also between snRNPs and the pre-mRNA (27, 28, 33). First, the U1 and the U2 snRNPs recognize the 5′ splice site and the branch site, respectively, forming the prespliceosome. After the integration of the U4/U6 and U5 snRNPs as a preformed 25S U4/U6.U5 tri-snRNP complex and subsequent rearrangements, a mature spliceosome is formed.
More recently, a second spliceosome has been identified. This minor or U12-dependent spliceosome recognizes a distinct set of very rare nuclear pre-mRNA introns (so-called U12-type introns) that contain unique 5′ splice site, branch site, and 3′ splice site sequences (5, 11). The minor spliceosome contains a different set of snRNPs, namely, U11, U12, and U4atac/U6atac, but shares the U5 snRNP with the major spliceosome (12, 34-36). As shown both in vitro and in vivo, the U11, U12, and U4atac/U6atac snRNPs represent functional counterparts of the major U1, U2, and U4/U6 snRNPs, respectively (12, 15, 17, 34, 35, 43). During assembly of the minor spliceosome, the U11 and U12 snRNPs, in the form of a stable 18S U11/U12 complex, recognize cooperatively the 5′ splice site and the branch point sequence of the pre-mRNA, respectively (9). Subsequently, the U4atac/U6atac and U5 snRNPs associate with the pre-mRNA; after numerous structural rearrangements, a catalytically active spliceosome is formed.
In general, the minor and major snRNAs are engaged in analogous snRNA/snRNA and snRNA/pre-mRNA interactions such that a similar, dynamic RNA network is formed. Within the U4atac/U6atac di-snRNP, the U4atac and U6atac snRNAs undergo base pairing with each other like the major snRNAs U4 and U6 within the U4/U6 snRNP (35). Although the overall sequence conservation levels are not very high between these minor and major snRNAs, the structures of the U4atac/U6atac and U4/U6 snRNA duplexes are very similar (35). Upon recruitment of U5 and either the U4atac/U6atac or the U4/U6 snRNP into the spliceosome, the base pairing interaction in both di-snRNPs is disrupted and U5 interacts with exon sequences at the 5′ and 3′ splice sites (10, 39). U6atac base pairs with the 5′ splice site, displacing U11, and also interacts with U12 to form what is thought to be the catalytic core of the minor spliceosome (15, 17, 35, 43), whereas in the major spliceosome, U6 replaces U1 at the 5′ splice site and base pairs with U2.
Each of the major spliceosomal snRNPs consists of one or (in the case of U4/U6) two snRNAs, a common set of core proteins (the Sm proteins), and snRNP-specific polypeptides. The U4/U6 di-snRNP interacts with U5 to form a U4/U6.U5 tri-snRNP particle which contains at least 29 different polypeptides, many of which are essential for splicing. Some of these proteins are also present in either the 13S U4/U6 or 20S U5 snRNPs, while others are only found in the 25S trimeric particle (21, 40). In addition to the Sm core proteins, seven U5-associated proteins, with molecular masses of 220, 200, 116, 102, 100, 40, and 15 kDa, have been identified in the U4/U6.U5 tri-snRNP. Several of these proteins, namely, the 220K protein (hPrp8), the 200K protein (a DEXH box RNA helicase), the 116K protein (a putative GTPase that is homologous to the ribosomal elongation factor EF-2), and the 40K protein (a WD repeat protein), interact with each other, forming an RNA-free complex (1). In addition to these U5 proteins, the major tri-snRNP particle contains several U4/U6-specific polypeptides. These include the 61K protein, which is essential for tri-snRNP formation (22); the 15.5K protein, which interacts with the 5′ stem-loop of both U4 and U4atac snRNAs (29); and three proteins, 20K (a cyclophilin), 60K (a WD repeat protein), and 90K, which form a complex with each other (14, 37). Three tri-snRNP-specific SR-related proteins, 27K (8), 65K, and 110K (21), are also present. Although the 110 and 65K proteins are loosely associated with 20S U5 snRNP particles, they are predominantly found in 25S tri-snRNPs and mediate the recruitment of the tri-snRNP to the prespliceosome (21). Recently, a novel class of proteins, namely, that comprising the human Sm-like proteins 2 through 8 (hLSm2 through hLSm8), was shown to be present in human U4/U6.U5 tri-snRNPs and to interact with U6 snRNA by recognizing the oligo(U) tail at its 3′ end (2, 23).
Although much has been learned about the snRNA components of the minor spliceosome, less is known about its protein composition. At least two protein families, namely, those of the Sm and the SR proteins, are shared between the major and minor spliceosomes (13, 26). Elucidation of the protein composition of the 18S U11/U12 di-snRNP particle revealed that it contains both unique proteins and four subunits of the heteromeric splicing factor SF3b (SF3b-155, -145, -130, and -49) which are also associated with the U2 snRNP (42). More recently, p14, a branch site interacting protein that is also an SF3b component, was shown to be shared by U2 and U11/U12 snRNPs (41). Previous studies have shown that the U5-specific 220K protein (hPrp8) is a component of both spliceosomes (19), raising the questions of whether other U5-associated proteins are also shared and which of them are involved in mediating the interaction of the U5 snRNP with the U4atac/U6atac di-snRNP particle.
Here, we have biochemically characterized the human U4atac/U6atac snRNP in more detail. We first demonstrated that U4atac/U6atac snRNPs associate with U5 snRNPs to form a minor 25S U4atac/U6atac.U5 tri-snRNP particle. We next investigated whether this complex contains not only U5-specific but also U4/U6- or major tri-snRNP-specific proteins. For this purpose, we performed immunoprecipitation experiments with antibodies against a variety of proteins that are present in the major tri-snRNP. These investigations revealed that the protein composition of the minor U4atac/U6atac.U5 tri-snRNP is remarkably similar to that of the major tri-snRNP. Moreover, the U4/U6 snRNP-specific 61K protein was shown to mediate the association of the U4atac/U6atac snRNP with U5. Finally, immunoprecipitation experiments revealed that most of those proteins associated with both the major and minor tri-snRNP particles were also present in U2- and U12-dependent spliceosomes. The extensive conservation of proteins between the two spliceosomes strongly supports the idea that one of the spliceosomes evolved in the presence of the other.
MATERIALS AND METHODS
Preparation of snRNPs from HeLa nuclear extract.
Splicing-active nuclear extracts were prepared from HeLa cells (Computer Cell Culture, Mons, Belgium) according to the procedures described in reference 7. Total U snRNP particles were purified from nuclear extract by anti-m3G affinity chromatography at 250 mM NaCl and then fractionated by centrifugation on a 10 to 30% glycerol gradient containing 150 mM KCl as described previously (18). The positions of the U snRNPs were determined either by silver staining (25) or Northern blotting (see below).
Immunoprecipitation and Northern blotting of snRNAs.
Twenty microliters of antiserum was incubated with 20 μl of preswollen protein A-Sepharose beads (PAS; Pharmacia) in 400 μl of phosphate-buffered saline (PBS) (130 mM NaCl, 20 mM KPO4 [pH 8.0]) for 2 h at room temperature. The beads were first washed three times with PBS containing 0.1% Nonidet P-40 and then once with PBS and subsequently were resuspended in 300 μl of PBS. A total of 100 μl of snRNP particles from the 25S or 13S region of a 10 to 30% glycerol gradient (see above and Fig. 1) was added to the PAS-bound antibodies. The mixture was then incubated for 3 h at 4°C with end-over-end rotation. The PAS beads were washed five times with 500 μl of IPP150 (20 mM Tris-HCl [pH 8.0]-0.1% Nonidet P-40 containing 150 mM NaCl). Alternatively, to determine the salt stability of U4atac/U6atac.U5 tri-snRNP, the last wash was performed by incubation for 1 h at 4°C in IPP buffer containing NaCl at the indicated concentrations (Fig. 2). Bound RNAs were recovered by incubating the beads with 100 μl of PK buffer (100 mM Tris-HCl [pH 7.5], 12.5 mM EDTA [pH 8.0], 150 mM NaCl, 1% [wt/vol] sodium dodecyl sulfate [SDS]) containing 100 μg of proteinase K for 30 min at 37°C, extracting with phenol-chloroform, and precipitating with 2 volumes of ethanol. RNAs were then separated on a 10% polyacrylamide-7 M urea gel and electroblotted on a nylon membrane (Biodyne B membrane; Pall).
FIG. 1.
U4atac/U6atac snRNPs cosediment with U5 snRNPs as a 25S complex in glycerol gradients. Sedimentation behavior of anti-m3G affinity-purified U snRNP particles on a 10 to 30% glycerol gradient is shown. snRNAs present in each fraction were isolated and separated on a 10% polyacrylamide-7 M urea gel. The sedimentation coefficients of the major snRNP peaks are indicated at the top, and the identities of the RNAs are marked on the left. Major snRNAs were visualized by silver staining (upper panel), whereas the minor U4atac and U6atac snRNAs were detected by Northern blotting (lower panel).
FIG. 2.
Major and minor tri-snRNP particles exhibit similar salt stabilities. Tri-snRNP particles from the 25S region of a 10 to 30% glycerol gradient (see Fig. 1) were immunoprecipitated with preimmune serum (lanes 2, NIS) or an antibody against the U5-specific 116K protein (lanes 3 to 8). Bound particles were incubated with buffer containing 150 to 600 mM NaCl as indicated. Coprecipitated RNAs were separated on a 10% polyacrylamide-7 M urea gel, and Northern blotting was performed with probes against the major U4, U6, and U5 snRNAs (upper panel) or the minor U4atac and U6atac snRNAs (lower panel). Lanes 1 show 100% of the input material.
Following a 6-h prehybridization at 42°C in hybridization buffer (50% [vol/vol] formamide, 0.5% [wt/vol] SDS, 5× Denhardt's solution, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 25 mM NaPO4 [pH 6.5], 0.1 mg of salmon sperm DNA/ml), the membrane was first hybridized with 32P-labeled DNA probes specific for the minor U4atac and U6atac snRNAs for 40 h at 42°C. The membrane was washed twice with 2× SSC-0.5% SDS, twice with 2× SSC-0.1% SDS at room temperature, and then once with 2× SSC-0.1% SDS at 50°C. snRNAs were subsequently visualized by autoradiography. The DNA probes were removed by boiling the membrane in DH buffer (15 mM NaCl, 1.5 mM Na citrate, 0.1% SDS) for 10 min, and the membrane was subsequently hybridized with probes against the major U4, U5, and U6 snRNAs under the same conditions. All probes were generated by random primer labeling of plasmids with a Prime It II kit (Stratagene).
Immunodepletion and Western blotting of nuclear extracts.
For immunodepletion of HeLa nuclear extracts, antiserum against a C-terminal peptide of the U4/U6-specific 61K protein was affinity purified on a SulfoLink column (Pierce) containing the immobilized peptide according to the instructions of the manufacturer. The salt concentration of 400 μl of nuclear extract in buffer C (7) was adjusted to 700 mM NaCl. The extract was then incubated twice with 60 μl of PAS charged with the affinity-purified antibodies for 2 h at 4°C with end-over-end rotation and, after removal of PAS, dialyzed for 4 h against buffer D (20 mM HEPES-KOH [pH 7.9], 20% [vol/vol] glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol) in a Slide-A-Lyzer 3.5 K apparatus (Pierce). For complementation studies, the bound 61K protein was eluted from the PAS in 200 μl of buffer D containing 0.5 mg of the antigenic peptide/ml and then added back to the depleted extract. Mock-depleted extracts were treated in an identical manner, except that the PAS beads were either preblocked with bovine serum albumin and tRNA or incubated with preimmune serum. The two methods yielded essentially identical results. Western blotting analysis with the antiserum against the 61K protein (diluted 1:1,000) was performed as previously described (20).
For sedimentation analysis, depleted and mock-depleted extracts were diluted 1:1 with gradient buffer (20 mM HEPES-KOH [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithioerythritol) after dialysis and loaded on a linear 4-ml 10 to 30% (wt/vol) glycerol gradient. After centrifugation for 14 h at 29,000 rpm in a Beckman SW60 rotor, the gradient was harvested manually from the top. RNAs were isolated from the gradient fractions by phenol-chloroform extraction followed by ethanol precipitation and separated on a 10% polyacrylamide-7 M urea gel, and the U4atac and U6atac snRNAs were detected by Northern blotting. Immunoprecipitation of snRNPs from mock-depleted, 61K-depleted, or 61K-complemented extract was performed with antibodies directed against the Sm proteins (monoclonal antibody Y12, kindly provided by Iain Mattaj, European Molecular Biology Laboratory, Heidelberg, Germany), the U5-specific 116K or the U4/U6-specific 60K proteins, as previously described (20). The precipitated snRNAs were recovered and analyzed by Northern blotting as described above.
In vitro pre-mRNA splicing.
U12-dependent splicing was performed with in vitro-transcribed 32P-labeled P120 pre-mRNA (34), with a specific activity of 2.5 × 106 cpm/pmol, as previously described (41). Standard 12.5-μl splicing assays contained 7.5 μl of nuclear extract with final concentrations of 12% (vol/vol) glycerol, 0.12 mM EDTA (pH 8.0), 0.3 mM dithioerythritol, 44 mM HEPES-KOH (pH 7.6), 60 mM KCl, 2.5 mM MgCl2, 0.48 mM ATP, 20 mM creatine phosphate, 2.6 μM anti-U2 deoxyoligonucleotide (5′-AGA TAC TAC ACT TGA TC- 3′) and 3 × 104 to 5 × 104 cpm of 32P-labeled P120 pre-mRNA and were incubated for 6 h at 30°C. Splicing reactions were preincubated for 10 min at 30°C to promote RNase H degradation of targeted U2 snRNA prior to the addition of the P120 pre-mRNA substrate. U2-dependent splicing was performed for 60 min at 30°C with 32P-labeled pre-mRNA derived from the adenovirus major late transcription unit (44). The splicing conditions were the same as those described for the U12-dependent pathway except that no antisense oligonucleotide was added.
Immunoprecipitation of U2- and U12-dependent spliceosomes.
Following in vitro splicing, heparin was added to each reaction to a final concentration of 0.5 mg/ml to minimize nonspecific interactions. After incubating for an additional 5 min at 30°C, PAS-bound antibodies were added to the splicing reactions and the mixture was incubated with end-over-end rotation for 16 h at 4°C. All washing and RNA recovery steps were identical to those described above. Precipitated RNA species were separated on 14% polyacrylamide-8 M (MINX) or 8% polyacrylamide-8 M (P120) urea gels, respectively, and visualized by autoradiography.
RESULTS
Sedimentation behavior of U4atac/U6atac snRNPs in glycerol gradients.
As the U4atac/U6atac snRNP is the functional counterpart of the U4/U6 snRNP and the U5 snRNP appears to be shared by both spliceosomes (34), we first investigated whether U4atac/U6atac and U5 snRNPs also interact to form a U4atac/U6atac.U5 complex. For this purpose we separated anti-m3G immunoaffinity-purified snRNPs on a 10 to 30% glycerol gradient. Figure 1 shows the distribution of the snRNAs throughout such a gradient. The upper panel shows the sedimentation behavior of the major snRNPs U1, U2, U4/U6, and U5 as detected by silver staining. While the U1 and U2 snRNPs sedimented predominantly in the 12S region, the majority of U5 and U4/U6 snRNPs cosedimented as a 25S U4/U6.U5 complex. In addition, 20S U5 snRNPs, as well as 13S U4/U6 snRNPs, were observed.
To detect the positions of the less abundant U4atac and U6atac snRNAs in the gradient, Northern blot analysis was performed (Fig. 1, lower panel). Consistent with results demonstrating that the U4atac and U6atac snRNAs interact with each other and thus form di-snRNP particles (35), a fraction of snRNPs containing these snRNAs peaked in the 13S region of the gradient. Significantly, most of the complexes containing U4atac and U6atac snRNAs sedimented in the 25S region, suggesting that U4atac/U6atac snRNPs also interacted with U5 snRNPs to form a 25S U4atac/U6atac.U5 tri-snRNP complex like that of their counterparts in the major spliceosome. Note that the apparent underrepresentation of the U6atac snRNA compared to that of U4atac is most likely due to differences in the hybridization efficiencies of the two probes. Of the peak fractions of U4/U6 and U4atac/U6atac snRNAs in the 25S region, a higher percentage of the U4atac/U6atac snRNAs appears to exhibit the faster sedimentation behavior, suggesting that the minor particle may contain additional proteins or that its structural organization might be different.
The minor and major tri-snRNPs exhibit similar levels of salt stability.
To investigate whether the proposed minor 25S tri-snRNP particle has a structural organization similar to that of the major 25S tri-snRNP particle, we analyzed the association behavior of U5 snRNPs with either U4atac/U6atac or U4/U6 snRNPs at increasing salt concentrations. For this purpose, we immunoprecipitated particles from the 25S region of glycerol gradients with an antibody against the U5 snRNP-specific 116K protein and then subjected the PAS-bound particles to increasing concentrations of NaCl (Fig. 2). The snRNAs of the precipitated particles were isolated and then separated by gel electrophoresis and analyzed by Northern blotting by using 32P-labeled probes specific for either U4, U6, and U5 snRNAs (Fig. 2, upper panel) or U4atac and U6atac snRNAs (lower panel). Significantly, both U4/U6 and U4atac/U6atac snRNAs were precipitated with the antibody against the U5-116K protein at lower salt concentrations (Fig. 2, lanes 3 and 4). These results clearly confirm that the U4atac and U6atac snRNPs indeed associate with U5 snRNPs, forming a U4atac/U6atac.U5 tri-snRNP complex. When the bound particles were washed with increasing concentrations of NaCl, U5 snRNP remained stably associate with the PAS-bound antibody whereas both U4/U6 and U4atac/U6atac snRNAs dissociated (Fig. 2, lanes 5 to 8). Interestingly, both tri-snRNPs began to dissociate at about 350 mM NaCl (Fig. 2, lanes 6), demonstrating that they exhibit very similar salt stabilities. This result suggests that the structural organizations of these particles may be similar. Taken together, these data provide further evidence that the U4atac/U6atac snRNPs integrate into the spliceosome as part of a preformed U4atac/U6atac.U5 complex. The existence of a minor tri-snRNP was suggested previously by in vitro spliceosome assembly studies, which demonstrated that U4atac and U6atac associate with the prespliceosome at the same time as the U5 snRNP (35).
Minor and major tri-snRNP particles share several proteins
To determine the protein composition of the minor U4atac/U6atac.U5 tri-snRNP, we first tried to affinity select this complex from 25S gradient fractions using biotinylated antisense oligonucleotides directed against U4atac or U6atac snRNA. However, due to consistently high background precipitation, it was not possible to obtain sufficiently pure minor tri-snRNPs. Therefore, as an alternative approach, we performed immunoprecipitation experiments with antibodies against proteins of the major tri-snRNP and in each case compared the precipitation patterns of the minor and major snRNPs (Fig. 3). For this purpose, the snRNAs of the precipitated particles were separated by gel electrophoresis and analyzed by Northern blotting as shown in Fig. 2.
FIG. 3.
Antibodies against several major tri-snRNP proteins also precipitate U4atac/U6atac.U5 tri-snRNP particles. Immunoprecipitations were performed with fractions from the 25S region of a 10 to 30% glycerol gradient (see Fig. 1) and antibodies against the following proteins: (A) lanes 2 to 5, the U5-specific proteins 220, 116, 40, and 100K, respectively; lanes 6,7, 9, and 10, the tri-snRNP specific proteins 110, 65, 61, and 27K, respectively; lanes 8, the U4/U6-specific protein 60K; lanes 11, preimmune serum (background precipitation); (B) lanes 2, a second antiserum against the U5-100K protein; lanes 3, preimmune serum (background precipitation); (C) lanes 2 and 3, antibodies against the hLSm2 and hLSm4 proteins; lanes 4, the SmF protein; lanes 5, preimmune serum (background precipitation). Precipitated minor (upper panel) or major (lower panel) RNAs were analyzed as for Fig. 2. Lanes 1 of panels A and C show 100% of the input material, and lanes 1 of panel B show 15% of the input material.
We first tested all available antisera against U5-associated proteins, using fractions from the 25S region of the gradient. As expected, antibodies against the U5-specific 220, 116, 100 (hPrp28p), and 40K proteins precipitated major tri-snRNPs, albeit with differing efficiencies (Fig. 3A, lower panel, lanes 2 to 5). Since the precipitation efficiency of the anti-100K antiserum (Fig. 3A, lane 5) was very low, we used a second antibody against this protein to confirm its presence in minor tri-snRNP particles (Fig. 3B). Interestingly, tri-snRNPs containing the U4atac/U6atac snRNAs were precipitated by each of these antibodies with efficiencies similar to that of the major tri-snRNP (Fig. 3A, upper panel, lanes 2 to 5). Only minimal background precipitation of both minor and major snRNPs (less than 1% of the input material) was observed when immunoprecipitations were performed with nonimmune serum (Fig. 3A, lanes 11). These results provide strong evidence that the U5-specific 220, 116, 100, and 40K proteins are components of the minor tri-snRNP particle. Since the 220, 116, and 40K proteins are known to form a stable, RNA-independent complex with the U5-specific 200K protein (1), it is very probable that this protein is also present in the minor tri-snRNP. Unfortunately, our antiserum, which is specific for the U5-200K protein in Western blotting experiments, did not recognize native U5-containing particles (not shown). The presence of U5-220 in the minor tri-snRNP is consistent with previous studies demonstrating that the U5-220/hPrp8p protein is common to both spliceosomes (19). These data confirm that the majority of known U5-specific proteins are intrinsic components of the minor tri-snRNP particle.
We next investigated whether the minor tri-snRNP also contains major tri-snRNP-specific proteins. Thus, we first tested antibodies against the 110 and 65K proteins (Fig. 3A, lanes 6 and 7), which are only present in low amounts, if at all, in isolated 20S U5 snRNP particles but are very abundant in 25S major tri-snRNPs (21). These antisera precipitated both minor and major tri-snRNPs with similar efficiencies. Antibodies against the tri-snRNP-specific 27K protein also precipitated both minor and major tri-snRNPs (Fig. 3A, lanes 10). However, while most of the tested antibodies precipitated minor and major particles to comparable extents, the anti-27K antibody precipitated major tri-snRNPs significantly more efficiently (51% of the input material) than minor tri-snRNPs (3% of the input material). A likely explanation for this difference is that the 27K protein is less stably associated in the minor particle or that the epitope for the antibody is not accessible in the minor tri-snRNP, suggesting that its protein components may either be arranged in a different way or that it may contain additional, novel polypeptides (see Discussion).
The finding that tri-snRNP proteins are shared by the major and minor complexes prompted us to test whether U4/U6 snRNP-associated proteins were also present in the minor tri-snRNP complex. Indeed, antibodies against the U4/U6-specific 60 and 61K proteins also precipitated both minor and major tri-snRNP particles (Fig. 3A, lanes 8 and 9). Unfortunately, our antibodies against the U4/U6-specific 20 and 90K proteins did not recognize native U4/U6- or U4atac/U6atac-containing particles (data not shown). However, given that the 20K and 90K proteins form a stable complex with the 60K protein (14, 37), the former proteins most likely are also present in U4atac/U6atac.U5 tri-snRNPs.
Since U6atac snRNA contains a 3′-terminal oligo(U) tail similar to that of the U6 snRNA and this region is essential for binding of the so-called Sm-like proteins hLSm2 through hLSm8 to U6 (2), we next investigated whether hLSm proteins are also present in minor U4atac/U6atac.U5 tri-snRNPs. For this purpose, we performed immunoprecipitation experiments with antibodies against two hLSm proteins (hLSm2 and hLSm4) and snRNPs from the 25S region of the gradient (Fig. 3C). For comparison, we also tested an antibody against the SmF protein. Interestingly, antibodies against hLSm2 and hLSm4 as well as the canonical SmF protein also recognized both minor and major particles (Fig. 3C, lanes 2 to 4). Since hLSm and Sm proteins form higher-ordered complexes even in the absence of RNA (2, 16, 30), it is highly likely that, as is the case with the major tri-snRNP, not just the investigated proteins but rather the complete sets of Sm and hLSm proteins are present in the minor tri-snRNP. In sum, these data suggest that the majority of proteins present in the major tri-snRNP are also found in the minor U4atac/U6atac.U5 complex.
To determine whether proteins which are associated with major tri-snRNPs are also present in the U4atac/U6atac di-snRNP, we performed immunoprecipitations with U snRNPs migrating in the 13S region of the gradient (Fig. 4). Indeed, 13S U4atac/U6atac di-snRNP particles were precipitated above background level with an antibody against the U4/U6-60K protein (compare lanes 2 and 6 of Fig. 4). Thus, the 60K protein and very likely its interaction partners, the U4/U6-20 and 90K proteins, were present (see above). U4atac/U6atac or U4/U6 snRNPs were precipitated only weakly, if at all, by anti-61K antibodies (Fig. 4, lanes 3), suggesting that the protein dissociated from the particle during either gradient centrifugation or the precipitation procedure (see below). Interestingly, antibodies specific for the hLSm2 and SmF proteins also precipitated 13S U4atac/U6atac snRNPs (Fig. 4, lanes 4 and 5), demonstrating that hLSm proteins as well as Sm proteins are components of U4atac/U6atac di-snRNPs. The latter result is consistent with previous studies demonstrating the presence of Sm proteins in the U4atac/U6atac di-snRNP particle (35). These data demonstrate that U4/U6 snRNP proteins are present not only in the minor tri-snRNP complex but also in the 13S U4atac/U6atac particle.
FIG. 4.
Antibodies against U4/U6 snRNP proteins also precipitate 13S U4atac/U6atac di-snRNP particles. Immunoprecipitations were performed with fractions from the 13S region of a 10 to 30% glycerol gradient (see Fig. 1) and antibodies against the following proteins: lanes 2, the U4/U6-specific 60K protein; lanes 3, the U4/U6-specific 61K protein; lanes 4, the U6-specific hLSm2 protein; and lanes 5, the SmF protein. Lanes 6 show the background precipitation with preimmune serum. Precipitated minor (upper panel) or major (lower panel) RNAs were analyzed as for Fig. 2. Lanes 1 show 12.5% of the input material.
The 61K protein is essential for the formation of 25S U4atac/U6atac.U5 tri-snRNPs.
Recent results from our laboratory have revealed that the 61K protein is essential for U4/U6.U5 tri-snRNP formation (22). To investigate whether the 61K protein plays a similar role in minor tri-snRNP formation, we prepared 61K-immunodepleted HeLa nuclear extracts by using an anti-peptide anti-61K antibody that was coupled to protein A Sepharose. The 61K protein was efficiently removed (>95%), as demonstrated by immunoblotting (Fig. 5A). To ensure that only the 61K protein, and not snRNPs, was removed, the depletion reaction was performed at 700 mM salt. Under these conditions, the 61K protein was completely dissociated from both major and minor snRNP particles, as evidenced by glycerol gradient centrifugation and immunoprecipitation experiments (reference 20 and data not shown). Consistent with this result, material eluted from the PAS-bound 61K antibodies with an excess of the antigenic peptide, subsequent to depletion, contained the 61K protein almost exclusively (data not shown).
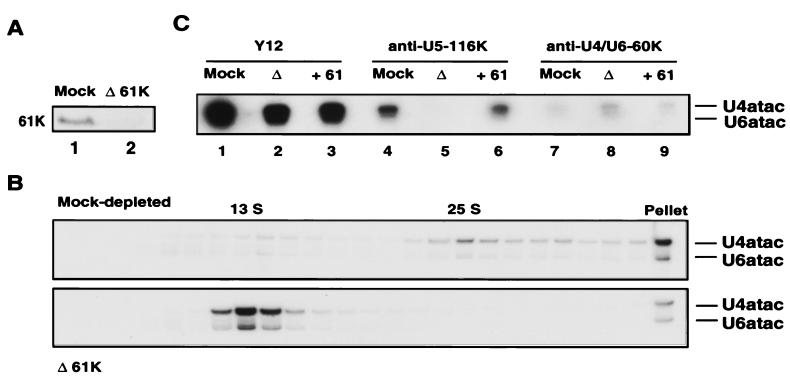
FIG. 5.
The U4/U6-specific 61K protein is essential for the formation of 25S U4atac/U6atac.U5 particles. (A) Western blot analysis was performed with either mock-depleted (Mock) (lane 1) or 61K-immunodepleted (Δ61K) (lane 2) HeLa nuclear extract by using antiserum against the U4/U6-specific 61K protein. (B) Mock-depleted (upper panel) or 61K-immunodepleted (Δ61K) (lower panel) HeLa nuclear extract was fractionated on a 10 to 30% glycerol gradient. The RNA from each fraction was extracted and separated on a 10% polyacrylamide-7 M urea gel, and the sedimentation behavior of the U4atac and U6atac snRNAs in the gradient was determined by Northern blotting. The sedimentation coefficients of the snRNP peaks are indicated at the top. (C) Immunoprecipitation experiments were performed with mock-depleted (Mock) (lanes 1, 4, and 7) or 61K-depleted (Δ) HeLa nuclear extract (lanes 2, 5, and 8) or 61K-depleted extract complemented with the 61K protein (+61) (lanes 3, 6, and 9) and the following antibodies: Y12 (anti-Sm) (lanes 1 to 3), anti-U5-116K (lanes 4 to 6), and anti-U4/U6-60K (lanes 7 to 9).
After the depletion reaction was performed, the extract was dialyzed against buffer containing 100 mM KCl to allow reassembly of snRNP particles. To elucidate the effect of the removal of the 61K protein on the formation of minor tri-snRNP particles during dialysis, we first performed glycerol gradient centrifugation with both mock-depleted and 61K-depleted nuclear extract. RNAs from the gradient fractions were first separated by denaturing gel electrophoresis, and the U4atac and U6atac snRNAs were detected by Northern blotting. As shown in Fig. 5B (upper panel), the majority of the U4atac/U6atac snRNPs in the mock-depleted extract were present as intact 25S tri-snRNP particles, whereas a smaller amount (∼25%) peaked in the 13S region of the gradient. In contrast, virtually all of the U4atac/U6atac snRNPs migrated as 13S di-snRNP particles in the 61K-depleted extract (Fig. 5B, lower panel). This result demonstrates that the minor tri-snRNP particle does not reassemble in the absence of the 61K protein.
Subsequent to dialysis, we also performed immunoprecipitation experiments with the mock-depleted and depleted extracts to test for tri-snRNP formation. In these experiments we also investigated the effect of adding back 61K protein that had been eluted from the PAS-bound anti-61K antibodies under native conditions (see above). An antibody that is directed against the Sm proteins (Y12) served as a positive control. As shown in Fig. 5C, lanes 1 to 3, this antibody precipitated U4atac/U6atac snRNPs from the mock-depleted, depleted, and complemented extracts, demonstrating that they were not co-depleted during the depletion procedure. Consistent with the results obtained by glycerol gradient centrifugation, U4atac/U6atac snRNPs were strongly coprecipitated from mock-depleted extract with anti-U5-116K antibodies (Fig. 5C, lane 4), demonstrating that they were associated with the U5 snRNP in a tri-snRNP particle. In contrast, no coprecipitation was observed in the depleted extract (Fig. 5C, lane 5), suggesting that the tri-snRNP was completely dissociated. Addition of the 61K protein to the depleted extract restored the coprecipitation of the U4atac/U6atac snRNP (Fig. 5C, lane 6), demonstrating that the tri-snRNP could be reassembled in the presence of the 61K protein. In contrast, when we used an antibody against the U4/U6-specific 60K protein, which is present not only in minor tri-snRNPs but also in 13S U4atac/U6atac di-snRNP particles, U4atac/U6atac snRNPs were precipitated from all three extracts (Fig. 5C, lanes 7 to 9). Consistent with the data shown in Fig. 5, this result additionally confirms that removal of the 61K protein did not affect the stability of 13S U4atac/U6atac di-snRNPs.
Antibodies against major U4/U6.U5 tri-snRNP proteins precipitate U2- and U12-dependent spliceosomes.
As shown in the previous sections, antibodies raised against proteins associated with major tri-snRNPs also recognized minor tri-snRNP particles. Thus, we next investigated whether the tested proteins are also present in the U2- and U12-dependent spliceosomes. For this purpose, we performed in vitro splicing with a 32P-labeled template containing either a U2- (MINX) or a U12-dependent (P120) intron and then carried out immunoprecipitations with various antisera. Precipitated RNAs were separated by gel electrophoresis and visualized by autoradiography (Fig. 6). To prevent the formation of U2-dependent spliceosomes on the P120 pre-mRNA, splicing was performed after inactivation of the U2 snRNP with an oligonucleotide directed against the U2 snRNA.
FIG. 6.
Major tri-snRNP proteins are present in both U2- and U12-dependent spliceosomes. U2- or U12-dependent spliceosomes were assembled on 32P-labeled MINX (A) or P120 (B) pre-mRNA, respectively, and immunoprecipitations were performed with the same subset of antibodies which was used to precipitate 25S tri-snRNP particles (see Fig. 3 and 4). The identity of each antibody is indicated above each lane. (A) Lane 1 shows 100% of the input material. Precipitated RNAs (pre-mRNA, splicing intermediates, and products), as designated on the left, were separated on a 14% polyacrylamide-8 M urea gel and detected by autoradiography. Lane 13 shows background precipitation using protein A Sepharose without antiserum. (B) Lanes 1 and 13 show 50 and 25% of the input material, respectively. Precipitated RNAs were separated on 8% polyacrylamide-8 M urea gels and detected by autoradiography. Lanes 12 and 15 show background precipitation using protein A Sepharose without antiserum.
After incubating the MINX or the P120 pre-mRNA under splicing conditions for 1 or 6 h, respectively, both products and intermediates were formed (Fig. 6A, lane 1, and 6B, lanes 1 and 13). Interestingly, several of the tested antibodies precipitated the U2- and the U12-dependent pre-mRNA, as well as the splicing intermediates and products, above background (Fig. 6A and B [left and right panels], lanes PAS), albeit with very distinct precipitation efficiencies. These include antibodies directed against the U5-specific 220K, 116K, and 40K proteins (Fig. 6A, lanes 2, 3, and 6, and 6B, lanes 2, 3, and 5) and the tri-snRNP-associated 110K and 65K proteins (Fig. 6A, lanes 4 and 8, and 6B, lanes 14 and 7), as well as an antiserum against the SmF protein (Fig. 6A, lane 12, and 6B, lane 11). Each of these precipitated the two spliceosomes to similar extents. In contrast, antibodies against the U4/U6-specific 60K and 61K proteins and the tri-snRNP-specific 27K proteins precipitated a low level of the MINX splicing complexes but none of the P120 complexes (Fig. 6A, lanes 7, 9, and 10, and 6B, lanes 6, 8, and 9). Moreover, the antibody against the U5-100K protein, which recognizes both major and minor tri-snRNP particles (Fig. 3A and B), completely failed to precipitate U2- and U12-dependent spliceosomes (Fig. 6A, lane 5, and 6B, lane 4), suggesting that the epitope for the antibody may not be accessible in either spliceosome. Alternatively, U5-100K may dissociate during spliceosome assembly or as a result of the addition of heparin prior to immunoprecipitation. Interestingly, some of the antisera, especially those against the U5-220K, the U4/U6-60K, and the hLSm2 proteins, clearly precipitated a larger amount of mRNA than the pre-mRNA or splicing intermediates. A notable difference between the two spliceosomes was observed with anti-hLSm2 antibodies, which precipitated the P120 pre-mRNA and splicing products and intermediates (Fig. 6B, lane 10) with a significantly higher efficiency than that of MINX (Fig. 6A, lane 11). Taken together, these data show clearly that most of the tested minor and major tri-snRNP proteins are also present in U12- and U2-dependent spliceosomes and remain associated throughout the catalytic steps of splicing.
DISCUSSION
The protein composition of major and minor tri-snRNPs is largely conserved.
Characterization of the protein composition of the U4atac/U6atac.U5 tri-snRNP complex by immunoprecipitation experiments revealed that most, if not all, proteins are shared between minor and major tri-snRNPs. Consistent with the fact that the U5 snRNP is part of both particles, all of the tested U5-specific polypeptides were present in minor tri-snRNPs. Interestingly, our data revealed that U4/U6- and major-tri-snRNP-specific proteins were also present in U4atac/U6atac.U5 particles. Moreover, U4/U6-specific proteins were also shown to associate with 13S U4atac/U6atac snRNPs (Fig. 4). Significantly, according to the results of database searches, none of the tested U4/U6- or tri-snRNP-specific proteins appears to belong to a larger protein family. For this reason, it is not very likely that the minor tri-snRNP possesses variants of the major proteins, which are still recognized by our antibodies.
The predicted secondary structures of U4atac and U6atac are remarkably similar to those of the U4/U6 snRNAs (35), supporting the idea that major and minor snRNAs may associate with the same proteins. Interestingly, it could be shown that the intramolecular stem-loop structure of U6 snRNA can indeed functionally replace the U6atac snRNA stem-loop (32). Despite the fact that the U4/U6 and U4atac/U6atac snRNA duplexes exhibit analogous secondary structures, the overall sequence similarity between the corresponding snRNAs is not very high (approximately 40% between both U6 and U6atac and U4 and U4atac snRNAs) (35). Conserved nucleotides are mostly clustered in regions that are involved in RNA-RNA interactions within the spliceosome (i.e., between U6atac and U4atac, U12, or the pre-mRNA substrate). However, the 5′ stem-loops of U4atac and U4, which protrude from the two intermolecular helices formed with U6atac and U6, respectively, are very similar in sequence and structure. Previous studies, including RNase H protection assays, have shown that the loop region of the 5′ stem-loop of the U4 snRNA is tightly associated with proteins in HeLa nuclear extracts (3). Recently, it has been demonstrated that the 15.5K protein of the human U4/U6.U5 tri-snRNP binds directly and specifically to the 5′ stem-loops of both U4 and U4atac snRNAs in vitro (29). Unfortunately, we were not able to precipitate either major or minor tri-snRNP particles with an antibody against the 15.5K protein, and thus we could not confirm that the 15.5K protein is also present in the U4atac/U6atac snRNP and/or minor tri-snRNP complex. Significantly, studies in our laboratory have shown that the 61K protein, which is also present in major and minor particles (see Fig. 3A, lane 9), could be cross-linked to both U4 and U4atac snRNAs near the 5′ stem-loop and that its association was dependent on the presence of the 15.5K protein (S. Nottrott, H. Urlaub, and R. Lührmann, unpublished data), supporting the idea that the 15.5K protein may serve as a nucleation factor, mediating the association of several identical proteins with both the U4/U6 and U4atac/U6atac snRNPs.
As our immunoprecipitation studies were limited to a subset of tri-snRNP proteins, it is presently not clear whether the complete set of major tri-snRNP proteins was present in the minor complex. In addition, these studies did not allow us to determine whether unique proteins also associate with the U4atac/U6atac.U5 tri-snRNP. Affinity-purified human 18S U11/U12 snRNPs, for example, have been shown to contain several novel proteins in addition to polypeptides (i.e., subunits of SF3b) that are also found in the U2 snRNP (41, 42). However, at least one result of our immunoprecipitation studies suggests that some attributes of the major and minor particles, aside from the sequences of their snRNAs, could be distinct. Using an antibody against the major tri-snRNP-specific, SR domain-containing 27K protein (8), we precipitated the major tri-snRNP very well while the minor tri-snRNP was only poorly recognized (Fig. 3A, lanes 10). This differential precipitation could reflect differences in the association behavior of the 27K protein; i.e., under the same salt conditions it may be stably associated with major tri-snRNPs but largely dissociated from minor ones. It is possible that the minor particle is lacking U4/U6.U5 tri-snRNP proteins that help stabilize binding of the 27K protein. An alternative explanation for the weak precipitation of minor particles is that there may be a reduced accessibility of the epitope recognized by the anti-27K serum. This reduction in accessibility could arise if the same set of tri-snRNP proteins were arranged in different ways in each particle or if the minor particle contained additional U4atac/U6atac.U5-specific proteins. Finally, it is conceivable that the 27K protein is simply not present in U4atac/U6atac.U5 tri-snRNPs but is recruited to the U12-dependent spliceosome by other means. Unfortunately, our attempts to isolate U4atac/U6atac.U5 tri-snRNPs using biotinylated antisense oligonucleotides and streptavidin agarose beads were not successful. A clear answer to the open question as to whether unique proteins are present in the minor tri-snRNPs will depend on the future isolation of this snRNP complex or the U12-dependent spliceosome itself.
The U4/U6 snRNP-specific 61K protein is essential for minor and major tri-snRNP formation.
Since U5 and U4/U6 snRNPs associate via protein-protein interactions, the identification of apparently identical proteins in the minor tri-snRNP suggests that similar molecular mechanisms are responsible for the formation of the major and minor tri-snRNPs. Indeed, the salt stability of the minor tri-snRNP mirrored that of the major tri-snRNP complex (Fig. 2), consistent with the idea that U5 and either U4atac/U6atac or U4/U6 di-snRNPs interact with each other via the same protein-protein interactions. One of the proteins shared by both complexes, the U4/U6-61K protein, was shown by immunodepletion studies to be required for the association of U4atac/U6atac snRNPs with the U5 snRNP. Significantly, this protein has also been shown to mediate the interaction of the U4/U6 snRNP with U5 and, moreover, to be essential for the splicing of a U2-dependent pre-mRNA substrate (22). Thus, at least one essential bridging interaction appears to be conserved in both types of tri-snRNPs. Unfortunately, we were not able to determine whether the 61K protein is also required for U12-dependent splicing; attempts to splice the P120 pre-mRNA with either 61K- or mock-depleted nuclear extract failed due to a severe reduction in U12-dependent splicing activity stemming from the immunodepletion procedure.
A large number of proteins are shared by the major and minor spliceosomes.
Immunoprecipitation studies further revealed that most of those proteins which we detected in the U4atac/U6atac.U5 tri-snRNP were also present in the U12-dependent spliceosome. Significantly, a very similar precipitation pattern was obtained with splicing complexes formed on a U2- or U12-type pre-mRNA. In addition to the pre-mRNAs, splicing intermediates and products were also coprecipitated by several of the anti-sera tested. A similar result was previously reported for the U5-220K (hPrp8) protein (19); antibodies against this protein also precipitated both spliceosomes, including those that had undergone the first and second catalytic steps of splicing. Several of the tested antisera also precipitated intermediates, in particular sera against U5-116K, U5-40K, the tri-snRNP-specific 65K protein, and SmF. Due to their generally lower precipitation efficiencies, it was not clear if the remaining antibodies also precipitated splicing intermediates to a significant extent. Interestingly, although the precipitation efficiencies are comparable, there was a clear difference between the precipitation patterns of the sera specific for the 116 and 40K proteins in both spliceosomes (Fig. 6A, lanes 3 and 6, and Fig. 6B, lanes 3 and 5); with the latter serum, significantly more excised intron relative to mRNA was precipitated, suggesting that the 40K protein could be part of intron-containing postspliceosomal complexes. Several antibodies, such as anti-U5-220 and hLSm2, also appeared to precipitate higher amounts of the MINX mRNA relative to the other RNA species (Fig. 6A, lanes 2 and 11). However, additional experiments are required to clarify whether certain tri-snRNP proteins are preferentially found in the mRNP or excised intron complex in either spliceosome. Nevertheless, our data indicate that most of the tested proteins remain associated with U12- and U2-dependent spliceosomes during the catalytic steps of splicing.
Data presented here, together with those of several other previous studies, indicate that both a subset of U2 and most tri-snRNP-associated proteins, as well as non-snRNP proteins of the SR family, are present in both spliceosomes (13, 19, 42). The identification of such a large number of shared proteins between minor and major spliceosomes suggests that similar protein-protein and protein-RNA networks are formed in both splicing machineries. Several of the shared proteins, such as components of SF3b and U5-220 (hPrp8p), appear to be core components of the major spliceosome, consistent with the idea that the structure of the catalytic core of the minor spliceosome is conserved. Furthermore, several proteins thought to be involved in structural rearrangements during spliceosome assembly and splicing catalysis also appear to be shared, suggesting that the dynamics of the splicing reactions are achieved by similar mechanisms. Of particular interest is the apparent presence of a putative GTPase, namely the U5-116K protein, and several RNA unwindases, including the U5-100K protein (Prp28p in Saccharomyces cerevisiae), a DEAD box protein involved in rearranging snRNP interactions at the 5′ splice site during spliceosome assembly. In S. cerevisiae, Prp28p has been shown to be involved in the displacement of U1 from the 5′ splice site (6). The presence of its human orthologue, U5-100K, in 25S U4atac/U6atac.U5 snRNP particles suggests that it may also be involved in the displacement of the U11 snRNP from the 5′ splice site.
How are major and minor tri-snRNPs distinguished during spliceosome formation?
The formation of a mixed spliceosome, consisting of U1 and U2 snRNPs and the minor U4atac/U6atac.U5 tri-snRNP or U11/U12 snRNP and the major U4/U6.U5 tri-snRNP, has not been observed to date. Given that the minor and major tri-snRNPs differ minimally, if at all, in terms of their protein compositions, what mechanisms ensure that the correct tri-snRNP is recruited to the major or minor prespliceosome?
During prespliceosome formation, specific base pairing interactions between the U11 snRNA and the U12-type 5′ splice site and the U12 snRNA and the U12-type branch site favor the association of these minor snRNPs over U1 and U2 snRNPs. Proteins uniquely associated with the former snRNPs may also contribute to their specific interactions with U12-type introns. In addition, U11 and U12 snRNPs form a stable 18S complex and interact with the pre-mRNA as such, thereby ensuring the pairing of the proper splice sites and apparently preventing the formation of a mixed prespliceosome. Similarly, discrimination of the subsequently interacting tri-snRNP likely relies on specific interactions between the U6atac snRNA and the U12 snRNA and/or the U12-type 5′ splice site (or conversely, between U6 and U2 and the U2-type 5′ splice site in the major spliceosome). However, the initial docking of the tri-snRNP complex may involve protein-protein contacts. Indeed, in the major spliceosome, the U2 snRNP-associated protein SPF30 has been shown to tether the major tri-snRNP to the major prespliceosome, apparently via interactions with the tri-snRNP 90K protein (24, 31). Whether a similar protein-protein contact also contributes to tri-snRNP addition in the minor spliceosome is not known; to date, it is not clear whether SPF30 is also present in the U12-dependent spliceosome. On the other hand, our data clearly show that the major tri-snRNP-specific 110 and 65K proteins were present in the minor particle. These SR-related proteins are indispensable for the recruitment of the major tri-snRNP to the prespliceosome and potentially mediate tri-snRNP addition through interaction with other SR proteins (21). Since SR proteins are also required in the minor splicing pathway (13), and since other SR domain-containing proteins, such as the tri-snRNP-specific 27K protein (8, 38), are present in the minor spliceosome, the 110 and 65K proteins may also mediate the addition of the tri-snRNP to the minor prespliceosome via a similar mechanism. The association of the major tri-snRNP with minor prespliceosomes via similar protein-protein interactions may initially block the assembly of a functional spliceosome, due to the inability of U6 snRNA to base pair with the 5′ splice site and the U12 snRNA. Nonproductive interactions of the major tri-snRNP during minor spliceosome assembly, which block the association of the less abundant minor tri-snRNP, may contribute to the relatively slow kinetics of U12-dependent splicing in vitro. Conceivably, such interactions may be monitored, and ultimately disrupted, in some way by a yet-to-be-identified proofreading mechanism, thereby enhancing the assembly of functional minor spliceosomes.
Implications for the evolutionary relationship of the major and minor spliceosomes.
The recent identification of the minor spliceosome has led to intriguing questions regarding its origin and evolutionary relationship to the major spliceosome. Three potential models for their relationship have been proposed: the so-called fission-fusion, codivergence, and parasitic invasion models (5). In the first two models, both types of spliceosomes are homologous; i.e., they are derived from a common ancestor. Thus, the present differences in the snRNAs, snRNP, and non-snRNP proteins are the result of the divergence of the two spliceosomes during the course of evolution. Proteins carrying out very important, identical functions in both spliceosomes may have diverged little or not at all, thus giving rise to the phenomenon of shared spliceosomal proteins. However, the absence of sequence similarities between U1 and U11 snRNAs and U2 and U12 snRNAs, as well as the very limited homologies between U4 and U4atac snRNAs and U6 and U6atac snRNAs, favors a hypothesis of nonhomologous origins for these snRNAs. In this respect the parasitic invasion model is more attractive. In this model, a parasitic group II intron invaded a number of genes of a common ancestor of animals and plants which had a preexisting spliceosome. Subsequent fragmentation of the intron gave rise to novel snRNAs, and the utilization of many of the proteins of the preexisting spliceosome led to the formation of novel snRNPs with distinct snRNAs but many shared proteins. That such a large number of spliceosomal proteins are conserved, including some that do not appear to perform essential functions, is nonetheless a very unexpected finding. The future characterization of the minor U12-dependent spliceosome may reveal additional surprises and help to further elucidate the evolutionary relationship of both spliceosomes.
Acknowledgments
We thank Axel Badouin, Gabi Heyne, Peter Kempkes, and Winfried Lorenz for excellent technical assistance. We gratefully acknowledge Joan Steitz for providing plasmid encoding the P120 pre-mRNA.
This work was supported by the Gottfried Wilhelm Leibniz Program and a grant from the Deutsche Forschungsgemeinschaft (SFB 397/A6) to R.L.
REFERENCES
- 1.Achsel, T., K. Ahrens, H. Brahms, S. Teigelkamp, and R. Lührmann. 1998. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18:6756-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. L., and J. A. Steitz. 1986. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell 46:697-704. [DOI] [PubMed] [Google Scholar]
- 4.Burge, C., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. Gesteland, T. R. Cech, and J. Atkins (ed.), RNA world. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 5.Burge, C. B., R. A. Padgett, and P. A. Sharp. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2:773-785. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, Jr., L. J. Latus, and T. H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7:227-232. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fetzer, S., J. Lauber, C. L. Will, and R. Lührmann. 1997. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA 3:344-355. [PMC free article] [PubMed] [Google Scholar]
- 9.Frilander, M. J., and J. A. Steitz. 1999. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 13:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frilander, M. J., and J. A. Steitz. 2001. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell 7:217-226. [DOI] [PubMed] [Google Scholar]
- 11.Hall, S. L., and R. A. Padgett. 1994. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 239:357-365. [DOI] [PubMed] [Google Scholar]
- 12.Hall, S. L., and R. A. Padgett. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271:1716-1718. [DOI] [PubMed] [Google Scholar]
- 13.Hastings, M. L., and A. R. Krainer. 2001. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz, D. S., R. Kobayashi, and A. R. Krainer. 1997. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA 3:1374-1387. [PMC free article] [PubMed] [Google Scholar]
- 15.Incorvaia, R., and R. A. Padgett. 1998. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA 4:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Lührmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375-387. [DOI] [PubMed] [Google Scholar]
- 17.Kolossova, I., and R. A. Padgett. 1997. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA 3:227-233. [PMC free article] [PubMed] [Google Scholar]
- 18.Laggerbauer, B., J. Lauber, and R. Lührmann. 1996. Identification of an RNA-dependent ATPase activity in mammalian U5 snRNPs. Nucleic Acids Res. 24:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, H. R., G. A. Moreau, N. Levin, and M. J. Moore. 1999. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5:893-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarov, E. M., O. V. Makarova, T. Achsel, and R. Lührmann. 2000. The human homologue of the yeast splicing factor Prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 298:567-575. [DOI] [PubMed] [Google Scholar]
- 21.Makarova, O. V., E. M. Makarov, and R. Lührmann. 2001. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 20:2553-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova, O. V., E. M. Makarov, S. B. Liu, H. P. Vornlocher, and R. Lührmann. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21:1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayes, A. E., L. Verdone, P. Legrain, and J. D. Beggs. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meister, G., S. Hannus, O. Plottner, T. Baars, E. Hartmann, S. Fakan, B. Laggerbauer, and U. Fischer. 2001. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 20:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merril, C. R., D. Goldman, and M. L. Van Keuren. 1983. Silver staining methods for polyacrylamide gel electrophoresis. Methods Enzymol. 96:230-239. [DOI] [PubMed] [Google Scholar]
- 26.Montzka, K. A., and J. A. Steitz. 1988. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. USA 85:8885-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, M., C. C. Query, and P. A. Sharp. 1993. Splicing of precursors to mRNA by the spliceosome, p. 303-357. In R. Gesteland and J. Atkins (ed.), RNA world. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 28.Nilsen, T. W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing, p. 279-307. In R. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 29.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raker, V. A., G. Plessel, and R. Lührmann. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256-2269. [PMC free article] [PubMed] [Google Scholar]
- 31.Rappsilber, J., P. Ajuh, A. I. Lamond, and M. Mann. 2001. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem. 276:31142-31150. [DOI] [PubMed] [Google Scholar]
- 32.Shukla, G. C., and R. A. Padgett. 2001. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA 7:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 34.Tarn, W. Y., and J. A. Steitz. 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84:801-811. [DOI] [PubMed] [Google Scholar]
- 35.Tarn, W. Y., and J. A. Steitz. 1996. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273:1824-1832. [DOI] [PubMed] [Google Scholar]
- 36.Tarn, W. Y., and J. A. Steitz. 1997. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 22:132-137. [DOI] [PubMed] [Google Scholar]
- 37.Teigelkamp, S., T. Achsel, C. Mundt, S. F. Gothel, U. Cronshagen, W. S. Lane, M. Marahiel, and R. Lührmann. 1998. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA 4:127-141. [PMC free article] [PubMed] [Google Scholar]
- 38.Teigelkamp, S., C. Mundt, T. Achsel, C. L. Will, and R. Lührmann. 1997. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3:1313-1326. [PMC free article] [PubMed] [Google Scholar]
- 39.Umen, J. G., and C. Guthrie. 1995. The second catalytic step of pre-mRNA splicing. RNA 1:869-885. [PMC free article] [PubMed] [Google Scholar]
- 40.Will, C. L., and R. Lührmann. 1997. snRNP structure and function, p. 130-173. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 41.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Lührmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20:4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Will, C. L., C. Schneider, R. Reed, and R. Lührmann. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284:2003-2005. [DOI] [PubMed] [Google Scholar]
- 43.Yu, Y. T., and J. A. Steitz. 1997. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl. Acad. Sci. USA 94:6030-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zillmann, M., M. L. Zapp, and S. M. Berget. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]