Abstract
The yeast [PSI+], [URE3], and [PIN+] genetic elements are prion forms of Sup35p, Ure2p, and Rnq1p, respectively. Overexpression of Sup35p, Ure2p, or Rnq1p leads to increased de novo appearance of [PSI+], [URE3], and [PIN+], respectively. This inducible appearance of [PSI+] was shown to be dependent on the presence of [PIN+] or [URE3] or overexpression of other yeast proteins that have stretches of polar residues similar to the prion-determining domains of the known prion proteins. In a similar manner, [PSI+] and [URE3] facilitate the appearance of [PIN+]. In contrast to these positive interactions, here we find that in the presence of [PIN+], [PSI+] and [URE3] repressed each other's propagation and de novo appearance. Elevated expression of Hsp104 and Hsp70 (Ssa2p) had little effect on these interactions, ruling out competition between the two prions for limiting amounts of these protein chaperones. In contrast, we find that constitutive overexpression of SSA1 but not SSA2 cured cells of [URE3], uncovering a specific interaction between Ssa1p and [URE3] and a functional distinction between these nearly identical Hsp70 isoforms. We also find that Hsp104 abundance, which critically affects [PSI+] propagation, is elevated when [URE3] is present. Our results are consistent with the notion that proteins that have a propensity to form prions may interact with heterologous prions but, as we now show, in a negative manner. Our data also suggest that differences in how [PSI+] and [URE3] interact with Hsp104 and Hsp70 may contribute to their antagonistic interactions.
The Saccharomyces cerevisiae [PSI+] and [URE3] non-Mendelian genetic elements are prions (infectious isoforms) of the Sup35 and Ure2 proteins, respectively (39). The prion isoforms aggregate in the cytoplasm and propagate by converting the soluble form of the protein into the same misfolded form as it joins the aggregate. Purified Sup35p and Ure2p spontaneously aggregate into amyloid fibers (12, 14, 37), suggesting that the aggregates in vivo are amyloid in nature and indicating that generation and replication of the amyloid forms of these proteins do not require other cellular factors. Spontaneously arising [PSI+] or [URE3] clones in cell populations are rare, however, indicating that prion appearance is either hindered by molecular interactions of the native proteins in the cell or actively repressed by cellular processes. Additionally, in vivo propagation of all yeast prions tested is dependent upon Hsp104, a protein chaperone that plays a major role in recovery from stress by solubilizing protein aggregates (26).
The protein chaperones Hsp104 and Hsp70 both play important roles in the stable propagation of [PSI+] and [URE3] (2, 13, 23). Hsp104's ability to modify protein conformation has been suggested to aid in inducing structural changes in proteins that could promote propagation of prions (4, 27, 32). Hsp104 may also break established aggregates into smaller particles that are more efficiently transmitted as prion “seeds” (16, 28). Overexpressing Hsp104 under growth conditions where it is not normally induced causes [PSI+] to be lost. Simultaneous overexpression of the HSP70 isoform SSA1 moderates this [PSI+]-curative effect (24). Additionally, a specific SSA1 mutation (SSA1-21) causes [PSI+] to become unstable (13). HSP104 overexpression does not cure cells of [URE3] (23), indicating that there is a difference in the nature of the two prions, perhaps reflecting how their replicative forms are recognized as substrates by these chaperones.
Although it is believed to require nucleation, or seeding, what triggers the initial appearance of prions is unknown. Overexpressing the protein determinant of a prion leads to more frequent appearance of the prion, presumably by increasing the number of molecules that can undergo the apparently stochastic change into the prion conformation. The [PIN+] state (for Psi inducibility), reflecting the capability to be induced to become [PSI+] by overexpression of Sup35p, was originally shown to be dependent upon a cytoplasmic element and subsequently shown to be conferred by an aggregated prion form of Rnq1p (5, 6). It was further shown that [URE3], and overexpression of proteins that share stretches of polar residues similar to those found in known yeast prion proteins, can provide [PIN+] function. [URE3] and [PSI+] can also act as [PIN+] factors for induction of the aggregated form of Rnq1p (5). The mechanism underlying this dependence on heterologous prions for prion induction is unknown. It has been suggested that it may be due to some level of cross-seeding of prion polymers or to the sequestration by a resident prion of cellular factors that would otherwise repress generation of heterologous prions (5, 25). Amyloid propagation is thought to be similar to crystal growth (18), so negative effects due to interactions of heterologous subunits should also be expected. Here we find negative interactions between prions that may be explained by such a mechanism.
In preparing strains with both [PSI+] and [URE3], we discovered that [URE3PSI+] appearance was less frequent in cells harboring [URE3]. Further characterization of interactions between [PSI+] and [URE3] led to our findings that in our strains, which are [PIN+], [PSI+] and [URE3] repressed each other's de novo appearance and that [URE3] and [PSI+] negatively affected each other's propagation when both were replicating in the same cell. We thus provide genetic evidence of antagonistic interactions between yeast prion proteins with heterologous prion-determining domains. Our additional finding that overproduction of Ssa1p but not Ssa2p cures cells of [URE3] uncovers a functional distinction between these nearly identical Hsp70 isoforms. Differences in how [URE3] and [PSI+] interact with Hsp70 and Hsp104 may thus contribute to their mutual antagonism.
MATERIALS AND METHODS
Strains and media.
Strains 403-3A (MATakar1-1 ade2-1 SUQ5 arg− ura2) and 628-4B (MATα kar1-1 ade2-1 SUQ5 his3Δ202 ura2) were constructed in our laboratory and are related but not isogenic. Strain 628-4BL was made by replacing the LEU2 coding DNA of strain 628-4B with that of KanMX (38). Strain 628-4BMC was made by cotransforming a [PSI+] variant of 628-4B with pDCM66 (see below) and a DNA fragment containing the SUP35 gene lacking codons 2 to 124. Plasmid transformants that had become [psi−] were verified by PCR and sequencing to have the chromosomal SUP35 replaced by the truncated allele. One was selected for further use. Consistent with their [PIN+] phenotype, all 628-4B variants display the aggregated (prion) form of Rnq1p detected as foci of fluorescence in cells expressing a green fluorescent protein-Rnq1 fusion protein (34). The source of [URE3] ([URE3-1] [17]) and [PSI+] was strain 3347 (39), obtained from Reed Wickner (National Institutes of Health, Bethesda, Md.).
The four variants of 628-4B (see Fig. 1), which were derived from the same [PSI+][URE3] strain, were used throughout this study. Reconstructed [psi−] variants were assayed as controls. Reconstructed variants were made by reciprocal cytoplasmic transfer (20) to ensure that all strains had the same complement of known and possibly unknown prions. Cytoplasm from [PSI+][URE3] and [PSI+][ure-o] variants of 628-4B was first transferred to 403-3A. These recipients were then used as cytoplasm donors to 628-4BMC, which is incapable of propagating [PSI+] because it lacks the SUP35 N-terminal prion-determining domain. Cytoplasm from these recipients was then returned to 403-3A and subsequently to 628-4B and 628-4BL, which regenerated the [psi−] variants with and without [URE3]. Essentially identical results were obtained in experiments using 628-4B variants cured of [PSI+] by transient elevation of Hsp104 or by cytoductions using the 628-4BMC intermediate. The 628-4BMC variants themselves also produced similar results (see Results and data not shown). This indicates that in the 628-4B [PSI+][URE3] strain, transient overexpression of Hsp104 caused elimination of [PSI+] and did not affect [PIN+] or any other, unknown elements that may have influenced our observations.
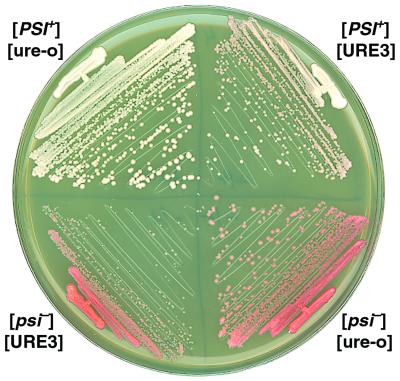
FIG. 1.
Growth and color phenotypes of cells propagating different prions. Cells with the indicated combinations of [PSI+] and [URE3] were incubated at 30°C on YPD for 2.5 days.
Rich medium (0.5×YPD, referred to hereafter as YPD) contains 0.5% yeast extract, 2% peptone, and 2% dextrose. YPAD is similar but contains 1% yeast extract and 400 mg of adenine per liter, which represses the adenine biosynthetic pathway and prevents red pigment accumulation in ade2-1 strains (see below). In all experiments where color was not relevant, media were supplemented with 400 mg of adenine per liter. Synthetic dextrose medium (SD) contains 2% dextrose, 0.7% yeast nitrogen base, and required nutritional supplements. Synthetic galactose medium (SG) is similar but has 2% galactose in place of dextrose and contains 2% raffinose. Ureidosuccinate selection medium (USA) is supplemented SD with 40 mg of USA per liter in place of uracil. Solid media are identical but contain 2% agar.
Plasmids.
Single-copy (pRS313) and multicopy (pRS423) plasmids have been described previously (33). Plasmid pH218 (23), from Herman Edskes (National Institutes of Health), is pRS313 with HSP104. Plasmid pDCM64 is pRS313 with the SSA2 gene at the BamHI site. SSA2 was PCR amplified from strain 628-3A (13) by using primers 5′-CGGGATCCCGCCGCTTAAGCGGTGCCCC-3′ and 5′-CGGGATCCCGGTTGACCGGTATCCCCGC-3. Plasmid pDCM65 contains HSP104 under control of the Gal1,10 promoter and was made by ligating the ClaI-SacI fragment from pYSGal104 (19) to pRS423 digested with the same enzymes. Plasmid pDCM66 contains ure2Δ151-158 under control of the Gal1,10 promoter and was made by blunting the ends of the NheI-BglII fragment from pH 377 (9) by using DNA polymerase Klenow fragment (New England Biolabs, Beverly, Mass.) and then ligating it to SmaI-cut pRS423. Plasmid pDCM71 contains SUP35 under control of the Gal1,10 promoter and was made by replacing the BamHI fragment of pDCM66 with a BamHI-digested SUP35 gene spanning nucleotides −25 to +2219 (where +1 is the A of the translation initiator), which was PCR amplified from strain 628-3A by using primers 5′-CCGGATCCATCTATATCTGCCCACTAG-3′ and 5′-CCGGATCCTTGTTTATGGTATATGGTAC-3′. Plasmid pDCM74 has the Escherichia coli β-galactosidase gene under control of the SSA1 promoter and was made by ligating the SalI fragment of pZFO (36) to pRS313 cut with SalI. Plasmid pC2D5, which contains the DAL5 promoter fused to E. coli β-galactosidase, was used to monitor Ure2p activity. It was constructed by first destroying the PstI site in the β-lactamase gene of plasmid pRR29 (31) by site-directed mutagenesis, creating pRR29-P, and then replacing the XbaI-PstI fragment of pRR29-P with the XbaI-PstI fragment containing HIS3 from pRS423. Plasmids pUKC815-L and pUKC817-L (35), used for measuring nonsense suppression, were gifts from M. Tuite (Canterbury, United Kingdom).
SSA1 was put under control of the strong, constitutive SSA2 promoter by first making NdeI sites at the initiator ATGs of SSA1 on plasmid pJ120 (13) and of SSA2 on pDM64, making pN-1 and pN-2, respectively. The BamHI-NdeI fragment of pN-1, containing the SSA1 promoter, was then replaced by the same fragment from pN-2, containing the SSA2 promoter, creating plasmid pC210. The BamHI-SphI fragment of pC210 was then used to replace the BamHI-SphI fragment of pJ130, creating plasmid pC211. Plasmid pJ130 is pRS313 with SSA1 on the BamHI-ApaI fragment from pJ120.
Assays for [PSI+] and [URE3].
[PSI+], an inactive aggregated form of the translation release factor Sup35p, was monitored and its presence was verified as described previously (13). The presence of [PSI+] causes nonsense suppression by depleting the cytosol of functional Sup35p. In ade2-1 cells this suppression reduces accumulation of red pigment and confers adenine prototrophy. In [PSI+] cells, the abundance of active (soluble) Sup35p increases proportionally with temperature (13), so in some experiments [PSI+] cells grown on medium lacking adenine were incubated at 25 rather than 30°C. This is a compromise between reducing growth due to the lower temperature and increasing it due to more efficient nonsense suppression of ade2-1, but it aids growth of cells with a weakened [PSI+] phenotype. [URE3] was monitored by its ability to allow ura2 mutants to grow on USA selection medium, and its presence was confirmed by its curability when grown in the presence of 3 to 5 mM guanidine hydrochloride. In most instances, verification of [URE3] presence or loss included cytoduction using a representative number of clones. In the experiment described in Table 4, the difference in loss of [URE3] from [PSI+] cells with and without induced Hsp104 expression was 4.3 and 7%, respectively. By the Student's t test, there is less than a 7% chance that this difference was due to chance fluctuations alone.
TABLE 4.
[URE3] does not affect curing of [PSI+] by transient overexpression of Hsp104a
| Starting strain | HSP104 induction | No. (%) of colonies
|
|||
|---|---|---|---|---|---|
| [PSI+][URE3] | [PSI+][ure-o] | [psi−][URE3] | [psi−][ure-o] | ||
| [PSI+][URE3] | − | 1,256 | 86 (7) | 0 | 0 |
| + | 497 | 28 (1.7) | 1,057 (65) | 43 (2.6) | |
| [PSI+][ure-o] | − | 0 | 1,760 | 0 | 0 |
| + | 0 | 619 | 0 | 1,347 (69) | |
| [psi−][URE3] | − | 0 | 0 | 697 | 0 |
| + | 0 | 0 | 915 | 0 | |
Large inocula of 628-4B cells carrying pDCM65 (galactose-inducible Hsp104) and the indicated prions were taken from plates lacking histidine and grown at 30°C for 2 days to confluent masses of cells on SG (induction) or SD (no induction) plates supplemented with uracil and adenine. Cells from the SD and SG plates were then suspended in water and spread onto YPD plates. The resulting colonies from two independent experiments were scored and verified for prions as indicated in Table 2.
Assays for prion appearance were done as indicated (see Results). For quantitative assays of [URE3] appearance (9), titers of cultures were determined by plating dilutions onto YPAD, and USA plates with between 50 and 200 colonies were scored after incubation for 4 days at 30°C.
Other methods.
Western analyses and cell lysate fractionation assays were performed as indicated or described previously (13), using cells grown in YPAD to an optical density at 600 nm (OD600) of 0.6 to 1.0.
RESULTS
[PSI+] and [URE3] can stably exist in the same cell but, each weakens the other.
After mating with [URE3-1] strain 3347, cytoduction recipients of [psi−][ure-o] strain 628-4B were found to be both [URE3] and [PSI+]. We cannot directly monitor the [PSI+] status of strain 3347 because of its genotype, but this result indicated that 3347 was both [PSI+] and [URE3]. Although the 628-4B recipients stably propagated both [URE3] and [PSI+], on YPD they were slightly pink (Fig. 1). The pink color was due to a reduced ability of [PSI+] to mediate nonsense suppression (Table 1), which reflects an increase in the relative abundance of functional Sup35p. [URE3] did not reduce nonsense suppression in the absence of [PSI+]. The [PSI+][URE3] cells also grew more slowly than isogenic [ure-o] cells (Fig. 1 and Table 1). When these cells were grown nonselectively, there was a noticeable frequency of appearance of faster-growing white colonies. Analysis of these white clones showed that they remained [PSI+] but had spontaneously become [ure-o] (Table 2). Thus, loss of [URE3] from cells with both prions restored the normal [PSI+] phenotype and wild-type growth, indicating that [URE3] impaired [PSI+]-mediated nonsense suppression and was inhibitory to growth.
TABLE 1.
Effect of [PSI+] and [URE3] on nonsense suppression, DAL5 and SSA1 promoter activity, and growth rate
| Strain | Nonsense suppressiona
|
Promoter activityb
|
Growth rate (min/cell division)c | |||
|---|---|---|---|---|---|---|
| UGG | UAA | % Readthrough | DAL5 | SSA1 | ||
| [PSI+][URE3] | 108 ± 3 | 8.0 ± 0.5 | 7.5 | 202 ± 26 | 216 ± 9 | 104 ± 2 |
| [PSI+][ure-o] | 137 ± 7 | 20 ± 3 | 15 | 6.6 ± 0.5 | 197 ± 7 | 90 ± 3 |
| [psi−][URE3] | 89 ± 11 | 2.0 ± 0.4 | 2.2 | 370 ± 44 | 160 ± 23 | 134 ± 7 |
| [psi−][ure-o] | 301 ± 32 | 4.8 ± 0.2 | 1.6 | 5.6 ± 0.4 | 117 ± 12 | 87 ± 2 |
Nonsense suppression is expressed as units of β-galactosidase (22). Data are averages and standard deviations of duplicate measurements of at least three cultures from at least two independent experiments. Suppression was measured in 628-4BL transformants with plasmids pUKC815-L and pUKC817-L, which have leader peptides with an in-frame UGG or UAA, respectively, fused to the E. coli β-galactosidase gene (35). Percent readthrough = UAA/UGG × 100.
Expressed as units of β-galactosidase (22). Data are averages and standard deviations of duplicate measurements of at least three cultures from at least two independent experiments. Activities of DAL5 and SSA1 promoters were determined in 628-4B transformants with plasmids pC2D5 and pDCM74, respectively.
Data are averages and standard deviations of results from six log-phase 628-4B cultures in YPAD at 30°C.
TABLE 2.
[PSI+] reduces [URE3] mitotic stabilitya
| Starting strain | % of cells from:
|
|||
|---|---|---|---|---|
| YPAD plates
|
Plates without adenine
|
|||
| [URE3] | [ure-o] | [URE3] | [ure-o] | |
| [PSI+][URE3] | 99.1 | 0.9 | 96.7 | 3.3 |
| [psi−][URE3] | 100 | 0 | NDb | ND |
[URE3] cells from USA plates were grown to colonies on YPAD plates, suspended in water, and spread onto YPAD plates or plates lacking adenine. At least three times, three 1-mm colonies from each of the latter plates were pooled, suspended in water, and spread onto YPD. The resulting colonies were scored for [PSI+] and [URE3] on the basis of color and size (see text) and verified as described in Materials and Methods. For [PSI+] and [psi−] starting strains grown on each medium, about 1,200 and 100,000 colonies were scored, respectively.
ND, not determined.
A [psi−][URE3] variant was obtained by selectively eliminating [PSI+] from the [PSI+][URE3] strain by transient overexpression of Hsp104 (2) (see below). The resulting [psi−][URE3] cells grew even more slowly than the [PSI+][URE3] cells (Fig. 1 and Table 1), indicating that the growth-inhibitory effect of [URE3] was diminished by the presence of [PSI+]. In contrast to the [PSI+] cells, [URE3] was completely stable in isogenic [psi−] cells. We have not observed spontaneous loss of [URE3] from the [psi−][URE3] variant in routine handling of cells stored for various times on rich medium at 4 to 25°C. Upon quantification, we saw no loss of [URE3] among 100,000 [psi−][URE3] cells screened (Table 2).
Together, these results show that both prions could propagate in the same cell, but the presence of [PSI+] impaired [URE3] propagation and [URE3] modestly impaired [PSI+] propagation and inhibited growth independently of [PSI+]. The effect of the two prions on colony color and size provided a useful way of monitoring both prions on nonselective medium. On YPD plates [psi−][ure-o] cells are red and large, [PSI+][ure-o] cells are white and large, [PSI+][URE3] cells are pink and small, and [psi−][URE3] cells are red and smaller (Fig. 1). These and all strains used throughout this study are also [PIN+] (see Materials and Methods).
To further analyze the apparent competition between the two prions, we tested prion stability when [PSI+][URE3] cells were grown under conditions selecting for phenotypes conferred by the prions. Since prion phenotypes depend upon reduced protein activity, optimal growth under such conditions might be expected to require depletion of more of the active form of the protein than growth under nonselective conditions. If so, then this may enhance the negative interactions between [PSI+] and [URE3] by promoting formation of the selected prion. Indeed, when [PSI+][URE3] cells were grown on medium lacking adenine, which selects for [PSI+], [URE3] was lost three- to fourfold more frequently (Table 2).
As expected, when [PSI+][URE3] cells were grown on USA, which selects for [URE3], [URE3] was not lost. Growth on USA did not affect the mitotic stability of [PSI+]. However, when cells were grown on medium selecting for both prions, incubation for over a week at 25°C was required to form 0.5-mm colonies, and no colonies formed after two weeks at 30°C, a temperature suboptimal for [PSI+]-mediated nonsense suppression (13) (see Materials and Methods). Although the growth inhibition on the double-selection plates may be due at least partially to the effects of [PSI+] on [URE3], these results are consistent with enhanced weakening of [PSI+]-mediated nonsense suppression by [URE3] when cells are grown on USA. In agreement with this explanation, unlike what was seen under nonselective conditions, the presence of [PSI+] did not affect growth of [URE3] cells on USA (see Fig. 5).
FIG. 5.
Growth of the four prion variant strains expressing elevated levels of Hsp70 or Hsp104. Plasmids are pRS313 (lanes -, vector control), pDCM64 (lanes 70, extra copy of SSA2), or pH218 (lanes 104, extra copy of HSP104). All plates lacked histidine for plasmid selection. The media select for no prions (left), [PSI+] (medium lacking adenine [-ade], center), or [URE3] (USA, right). Cells from stock plates without histidine were suspended in water, and 300 to 500 cells were spread onto the indicated selection plates. Shown are representative 2-cm squares from selection plates. Incubation times and temperatures are indicated.
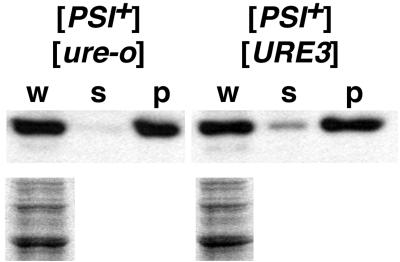
The weakened [PSI+] phenotypes caused by [URE3] were reflected in an increase in the detectable amount of soluble Sup35p in [PSI+] cells when [URE3] was present (Fig. 2). This indicates that [URE3] was reducing nonsense suppression in [PSI+] cells by impairing propagation of the aggregated prion form of Sup35p. Although it was not discussed, this same result was obtained in a similar experiment done by others (see Fig. 4 of reference 11).
FIG. 2.
Solubility of Sup35p in [PSI+] cells with or without [URE3]. (Upper panels) Western analysis of samples equivalent to 10 μg of the whole lysate was performed using polyclonal anti-Sup35p as described in Materials and Methods. Whole lysates (lanes w) of cells with the indicated prions grown in YPAD were separated by centrifugation at 10,000 × g for 30 min into soluble (lanes s) and pellet (lanes p) fractions. (Lower panels) Coomassie blue-stained loading control of duplicate samples of the whole lysates.
FIG. 4.
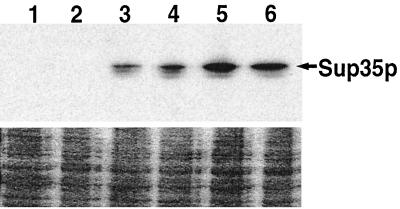
Galactose-induced expression of Sup35p. The strains used were the [psi−] 628-4B transformants from one replicate of the experiment described in Table 3. Immediately (lanes 1 and 2), 2 h (lanes 3 and 4), and 6 h (lanes 5 and 6) after transfer to SG medium, cells were removed and assayed for Sup35p abundance as described in Materials and Methods. Cells are [URE3] (lanes 1, 3, and 5) and [ure-o] (lanes 2, 4, and 6)
The antagonistic effect of [PSI+] on [URE3] was reflected in the level of derepression of the DAL5 promoter. DAL5 expression is repressed by Ure2p when a good source of nitrogen is present (3, 31), such as in standard growth media, and the activity of its promoter reflects the level of active Ure2p in the cell. The reduced activity of Ure2p caused by the presence of [URE3] resulted in a 60-fold increase in DAL5 promoter activity (Table 1). This derepression was reduced twofold by [PSI+], indicating that there was more functional Ure2p in [URE3] cells when [PSI+] was present.
[PSI+] and [URE3] repress each other's genesis.
Transient overexpression of Sup35p or Ure2p in yeast significantly increases the frequency of appearance of [PSI+] and [URE3], respectively (1, 39). We examined induction of [URE3] by using galactose-induced overexpression of a deletion derivative of URE2 known to cause [URE3] to appear at high frequencies (ure2Δ151-158) (21). The frequency of induced [URE3] appearance was significantly reduced when [PSI+] was present (Fig. 3). Unexpectedly, [URE3] appearance was elevated in strains with the inducing plasmid without galactose induction. This is likely due to incomplete repression of the galactose promoter, resulting in low-level expression of the strongly [URE3]-inducing Ure2Δ151-158 protein. The inhibitory effect of [PSI+] on [URE3] appearance was also seen under this condition. Spontaneous [URE3] appearance, measured as the frequency of USA+ cells arising in 628-4B cultures grown to saturation in YPAD, was reduced twofold from 16 × 10−6 to 7.6 × 10−6 in [PSI+] cells. Thus, the presence of [PSI+] decreased the frequency of both inducible and spontaneous de novo appearance of [URE3].
FIG. 3.
Frequency of induced appearance of [URE3] is reduced by [PSI+]. Stationary-phase [ure-o] cells, from stock plates lacking histidine, carrying pDCM66 (galactose-inducible ure2Δ151-158) or pRS313 (control plasmid) were suspended in water and inoculated to the same density onto SD (dextrose) and SG (galactose) plates selecting for the plasmids. After incubation for 2 h at 30°C, cells were replica plated onto USA selection plates containing histidine. The USA plates were photographed after incubation for 3 days at 30°C followed by 2 days at 25°C. [PSI+] and [psi−], 628-4B; MC, is 628-4BMC.
It was possible that the transiently elevated Hsp104 used to cure [PSI+] had an effect on other, unknown prions that may be present in our strains and that this contributed to the difference we saw in [URE3] induction. To ensure that this was not the case, we eliminated [PSI+] from the [PSI+][ure-o] variant of strain 628-4B by replacing SUP35 with an allele lacking the N-terminal region, which is necessary for [PSI+] propagation, creating the [psi−][ure-o] strain 628-4BMC. The relative efficiency of [URE3] induction by overexpression of Ure2Δ151-158 in 628-4BMC was similar to that of the [psi−] variant obtained by HSP104 overexpression (Fig. 3). This indicates that the presence of [PSI+] alone was responsible for the decrease in [URE3] appearance.
Induction of [PSI+] by Sup35p overexpression was reduced 14-fold when [URE3] was present (Table 3). This reduction in [PSI+] appearance was not due to a reduction in induced Sup35p expression as determined by Western analysis (Fig. 4). Also, among more than 1,800 [psi−][URE3] cells that remained [psi−], none lost [URE3]. In contrast, [URE3] was lost from 5 of the 17 [psi−][URE3] cells that became [PSI+]. Thus, [URE3] reduced the frequency of de novo appearance of [PSI+], and there was a significant correlation between appearance of [PSI+] and loss of [URE3].
TABLE 3.
[URE3] reduces induced [PSI+] appearancea
| Starting strain | No. of colonies
|
% [PSI+] | |||
|---|---|---|---|---|---|
| [PSI+][URE3] | [PSI+][ure-o] | [psi−][URE3] | [psi−][ure-o] | ||
| [psi−][ure-o] | 0 | 159 | 0 | 1,221 | 12 |
| [psi−][ure-o]b | 0 | 101 | 0 | 439 | 19 |
| [psi−][URE3] | 6 | 2 | 1,223 | 0 | 0.7 |
| [psi−][URE3]b | 6 | 3 | 610 | 0 | 1.5 |
| Total | |||||
| [psi−][ure-o] | 0 | 260 | 0 | 1,660 | 14 |
| [psi−][URE3] | 12 | 5 | 1,833 | 0 | 0.9 |
Stationary-phase 628-4B [psi−] cells carrying pDCM71 (galactose-inducible SUP35) from stock plates lacking histidine were inoculated into SG liquid medium without histidine at an OD600 of 0.25, incubated for 6 h at 30°C (at which time both cultures were at an OD600 of 0.28), diluted, and spread onto YPD plates. [PSI+] and [URE3] were scored and verified as indicated in Table 2. Sup35p induction in [URE3] and [ure-o] strains was similar (Fig. 4). [PSI+] did not appear in any of about 2,000 [URE3] or [ure-o] cells grown in SD (dextrose) medium or with either carbon source when pRS313 transformants were used. One colony (0.05%) among [URE3] pRS313 transformants grown in each carbon source was [ure-o].
628-4B variant derived from cytoductions using the 628-4BMC strain to eliminate [PSI+] (see Materials and Methods).
[URE3] does not affect curing of [PSI+] by Hsp104 overexpression.
Transient overexpression of Hsp104 efficiently cures [PSI+] but does not cure [URE3] (2, 23). We overexpressed HSP104 in [PSI+][URE3] cells to test if the apparently weakened [PSI+] was more susceptible to this curing (Table 4). In the absence of induced Hsp104 overexpression, [PSI+] was completely stable regardless of the presence of [URE3]. After transient induction of Hsp104 expression, about 70% of both [URE3] and [ure-o] cells lost [PSI+]. Thus, the presence of [URE3] did not affect the ability of Hsp104 to cure [PSI+]. Consistent with previously reported results (23), Hsp104 overexpression did not cure [URE3] in [PSI+] or [psi−] cells. Loss of [URE3] was observed only in [PSI+] cells, again showing that [URE3] propagation was impaired by [PSI+]. Consistent with the known weakening of [PSI+] by increased Hsp104, [URE3] was slightly stabilized in [PSI+] cells when Hsp104 was elevated (4.3 versus 7% [ure-o] colonies [see Materials and Methods]).
[URE3] and [PSI+] do not compete for limiting amounts of Hsp104 or Hsp70.
Since yeast prion propagation is critically dependent upon properly functioning Hsp104 and Hsp70, we tested to see if the negative effects between [PSI+] and [URE3] were due to competition for limiting amounts of these proteins (Fig. 5). An extra copy of the HSP70 isoform SSA2, which has a strong constitutively active promoter and normally produces most of the cytosolic Ssa protein in the cell (10), had no effect on growth of any strain under any condition. An extra copy of HSP104 only affected growth of [PSI+] cells, and in every instance effects were consistent with the known weakening of [PSI+] caused by elevated Hsp104.
In the absence of selection for prions, [PSI+] did not affect growth of [ure-o] cells but increased the growth rate of [URE3] cells (Fig. 5, left panels; see also Table 1 and Fig. 1). Consistent with its presence impairing [URE3]-mediated DAL5 expression and reducing mitotic stability of [URE3], [PSI+] likely relieved the growth inhibition caused by [URE3] by compromising [URE3] propagation. In agreement with this, weakening of [PSI+] in these cells by an extra copy of HSP104 restored the growth inhibition.
When cells were grown on medium lacking adenine to select for [PSI+], similar but accentuated effects were seen. In the absence of [URE3], elevated Hsp104 predictably weakened the ability of [PSI+] cells to grow without adenine. [URE3] also significantly slowed growth, and when it was combined with Hsp104, growth was further reduced. In contrast, when cells were grown on USA to select for [URE3], [PSI+] did not improve growth. Moreover, elevated Hsp104 did not improve growth of [PSI+][URE3] cells on USA. These two observations indicate that growth conditions selecting for [URE3] overcome the inhibitory effect of [PSI+] on [URE3]. Together, these results further illustrate the antagonistic interactions between [PSI+] and [URE3] and indicate that Hsp70 or Hsp104 is not limiting for prion propagation when [PSI+], [URE3], or both are present.
SSA1 overexpression cures [URE3].
Since our earlier results suggested that Ssa1p and Ssa2p might have nonoverlapping roles in the cell, and because all effects of Ssap on yeast prions described to date involve Ssa1p, we examined the effects of Ssa1p overexpression. When the normally regulated SSA1 gene was expressed in an extra copy from the strong constitutively active SSA2 promoter (plasmid pC211), [URE3] was mitotically unstable. Eighty percent of 628-4B [psi−][URE3] transformants of pC211 did not grow on USA when replica plated from the primary transformation selection plates. When transformants that remained USA+ were subsequently grown overnight in YPAD and then spread onto YPD plates, about 60% of colonies from all cultures were USA− (Table 5). These were verified as having lost [URE3]. Of these [ure-o] clones, 10 to 30% had lost the plasmid, indicating that excess Ssa1p was not simply altering the USA phenotype. In contrast, [URE3] was completely stable in [psi−] transformants carrying the SSA2 or control plasmids. In [PSI+] cells, loss of [URE3] was enhanced by excess Ssa2p to a level similar to that seen when [PSI+][URE3] cells were grown without adenine (Table 2). In contrast, the curing effect of Ssa1p, while still greater than that of Ssa2p, was reduced when [PSI+] was present.
TABLE 5.
[URE3] stability is reduced by overexpressed SSA1 but not SSA2a
| Elevated Hsp70 | Strain variant | No. of colonies
|
% [ure-o] | |||
|---|---|---|---|---|---|---|
| [URE3]
|
[ure-o]
|
|||||
| With plasmid | Without plasmid | With plasmid | Without plasmid | |||
| Ssa1p | [psi−] | 538 | 101 | 932 | 143 | 63 |
| [PSI+] | 698 | 324 | 73 | 35 | 10 | |
| Ssa2p | [psi−] | 579 | 162 | 0 | 0 | 0 |
| [PSI+] | 1,663 | 390 | 48 | 27 | 3.5 | |
| None | [psi−] | 1,311 | 282 | 0 | 0 | 0 |
| [PSI+] | 336 | 195 | 4 | 0 | 0.8 | |
Primary selection plates (lacking histidine) with 628-4B transformants of pC211 (Ssa1p), pDCM64 (Ssa2p), or pRS313 (none) were replica plated onto secondary plates lacking histidine and onto USA plates. Cells from USA+ colonies, taken from the secondary plates lacking histidine, were grown in YPAD overnight and spread onto YPAD plates. The resulting colonies were scored for the presence of [URE3] and plasmids. Data are from 8 to 10 independently tested USA+ clones from each transformation.
After cells were grown to colonies twice on medium selecting for the plasmid, very similar results were seen. All colonies of both [psi−] and [PSI+] transformants with the SSA2 plasmid remained [URE3], but among transformants with the SSA1 plasmid, all [psi−] colonies had lost [URE3], and about 80 to 90% of [PSI+] colonies retained [URE3]. Thus, overproduction of Ssa1p, but not Ssa2p, eliminated [URE3] in [psi−] cells, and [PSI+] partially protected [URE3] from the effects of overproduced Ssa1p. [PSI+] stability was unaffected by overexpression of either Ssa2p or Ssa1p regardless of the presence of [URE3].
SSA1 promoter activity and abundance of Hsp104 in prion variant strains.
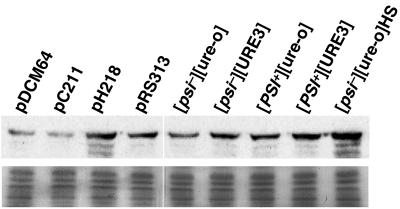
We previously showed that the presence of [PSI+], which presumably causes a stress to cells, elevates Hsp104 abundance and SSA1 promoter activity (13). We tested the four prion variant strains to see if [URE3] or the combination of prions had similar effects. [URE3] activated the SSA1 promoter, but less so than [PSI+], and there was a further increase in promoter activity when both prions were present (Table 1). Thus, both prions induced SSA1 expression, and SSA1 promoter activity was higher in [URE3] cells when [PSI+] was present. Hsp104 abundance was elevated to a similar level when either [URE3] or [PSI+] was present (Fig. 6). When cells had both [URE3] and [PSI+], the increase in Hsp104 abundance was roughly additive. Overexpression of either Ssa1p or Ssa2p repressed Hsp104 expression, presumably by reducing accumulation of Hsp104 substrates. As expected, Hsp104 abundance was higher in cells with the HSP104 plasmid.
FIG. 6.
Hsp104 abundance in 628-4B transformants and prion variant strains. Western analysis (upper panel) was done as described previously (13) from cells grown to mid-log phase in SD lacking histidine (transformants) or in YPAD. All plasmid transformants are [psi−][ure-o]; pDCM64, pC211, and pH218 are the pRS313 control plasmid with SSA2, SSA1, and HSP104, respectively. Strains with different combinations of [PSI+] and [URE3] are indicated. [psi−][ure-o] and [psi−][ure-o]HS are from the same culture, but those labeled HS were heat shocked at 39°C for 30 min before preparation of lysates. The lower panel shows a Coomassie blue-stained loading control of the same samples.
DISCUSSION
We demonstrate that different yeast prions replicating in the same cell interfere with each other's propagation. In our [PIN+] strains, [PSI+] considerably destabilized [URE3] and increased the relative amount of active Ure2p in [URE3] cells. When [URE3] was present in [PSI+] cells, there was an increase the detectable amount of soluble Sup35p, which correlated with reduced [PSI+]-mediated nonsense suppression. These mutual antagonistic effects were likely due to impaired propagation of the prion forms of Ure2p and Sup35p. [PSI+] and [URE3] also reduced each other's de novo appearance. Expression of Hsp70 and Hsp104 chaperones was elevated when either of these prions was present and was further elevated when both were present. The mitotic stability of [PSI+] and [URE3] was sensitive to elevated levels of different chaperones. Together, our data suggest that the antagonistic interactions between [PSI+] and [URE3] involve other cellular factors, but they do not rule out direct interactions between Sup35p and Ure2p and their counterpart prions.
It was previously shown that in a general way, the presence of one yeast prion potentiates the de novo appearance of other prions. Induction of [PSI+] by overproduction of Sup35p requires the presence of [PIN+], an aggregated prion form of Rnq1p (5, 6). Yeast [URE3] and the New1p-Sup35p hybrid [NU+] also confer [PSI+] inducibility by overproduced Sup35p in the absence of [PIN+]. Moreover, [URE3] and [PSI+] were shown to be capable of acting like [PIN+], facilitating appearance of Rnq1 aggregates (5, 25). The yeast proteins that can form prions all have a region enriched in polar amino acids, which is believed to allow hydrogen bonding between like molecules that confers prion-forming ability. One suggestion to explain the positive effects that prions have on biogenesis of heterologous prions is that established prion aggregates may, at a frequency much less efficient than that seen for the homologous protein, interact with similar but nonidentical prion-determining domains of heterologous proteins (5). This would convert the heterologous prion protein into its prion conformation, initiating formation of the heterologous prion.
It might be expected that such direct interactions could not only fail to provide the surface necessary for seeding the heterologous prion but also eliminate or reduce the continued growth of resident prion polymers. Indeed, this is believed to be how expression of the PNM2 allele of SUP35 causes loss of [PSI+] (7, 15). Additionally, [URE3] propagation can be disrupted by overexpressing various Ure2p fragments containing the [URE3] prion-determining domain fused to green fluorescent protein (8). Various degrees of interference of prion propagation by expression of heterologous prion proteins have also been observed in mammalian cells (29, 30). In all of these studies, the interacting proteins shared identical or near-identical prion-determining amino acid sequences. Here, we see similar negative effects but between proteins with significantly less identity in their prion-determining regions. Nevertheless, the mutual interference between [URE3] and [PSI+] prions could be explained if Sup35p and Ure2p interacted with their counterpart prions in an analogous manner.
Another explanation for the positive effects that yeast prions have on the appearance of other prions is that limiting concentrations of cellular factors that normally prevent prion appearance are sequestered by preexisting prions, thus allowing more frequent appearance of heterologous prions (5, 25). It follows, then, that an excess of such factors would have a negative effect on prion appearance. Obvious candidates for these factors are Hsp104 and Hsp70, protein chaperones known to be involved in yeast prion propagation.
Our data show that Hsp104 and Hsp70 are not independently limiting for prion propagation when [PSI+], [URE3], or both are present. In contrast, because of the different interactions that [PSI+] and [URE3] have with Hsp104 and Hsp70, the antagonistic effects that we observed between the prions may be due at least partially to elevated levels of these chaperones when both prions are present. [PSI+] is easily eliminated by elevating Hsp104 expression, but [URE3] is insensitive to large increases in Hsp104 abundance (Table 4) (2, 23). Since the relative abundance of Hsp104 in [PSI+] cells is higher when [URE3] is present, this may explain or contribute to the observed weakening of [PSI+] by [URE3]. We now show that overexpression of SSA1 eliminated [URE3] but had no noticeable effect on [PSI+], which is exactly the opposite effect for [PSI+] and [URE3] with regard to overproduction of Hsp104. The increased expression of SSA1 in [URE3] cells when [PSI+] is present may explain or contribute to the inhibition of [URE3] propagation. In this instance, however, the mechanism may be more complicated, since the presence of [PSI+] also diminished the curative effect of elevated Ssa1p on [URE3].
In [psi−] cells, overexpression of SSA1, but not SSA2, cured cells of [URE3]. In [PSI+] cells the ability of Ssa1p to cure [URE3] was compromised, but overexpression of Ssa2p moderately cured [URE3]. Elevated expression of either Ssa1p or Ssa2p reduced Hsp104 expression, which would be expected to strengthen [PSI+]. This may at least partially explain the earlier observation that excess Ssa1p impairs Hsp104-mediated basal thermotolerance and was suggested to promote the [PSI+] phenotype (24). The enhanced loss of [URE3] from [PSI+] cells overexpressing SSA2 was similar to that seen when [PSI+] cells were grown with selection for the [PSI+] phenotype. Both conditions may similarly enhance [URE3]-inhibiting effects of [PSI+] by promoting [PSI+] propagation. Excess Ssa2p would do so by reducing Hsp104 abundance, and selection for [PSI+] would do so by requiring reduced amounts of active Sup35p for optimal growth. Since the ability of Ssa1p to cure [URE3] was reduced by [PSI+], Ssa1p must act in an additional or different way than Ssa2p. Perhaps Ssa1p is preferentially recruited by Sup35p aggregates, which diverts it from acting on [URE3]. Alternatively, Ssa1p may have a specific role in regulating the activity or expression of other stress response factors, which is altered by [PSI+]. [URE3] was previously shown to be cured by overexpression of Ydj1p, a known Hsp40 cochaperone for Ssa1p (23), and it is possible that curing of [URE3] by Ydj1p is mediated through a specific interaction with Ssa1p.
In addition to uncovering a specific interaction between [URE3] and Ssa1p, the difference in the abilities of Ssa1p and Ssa2p to cure [URE3] clearly demonstrates that these nearly identical proteins have nonoverlapping functions. Further characterization of Ssa1p and Ssa2p interactions with yeast prions will aid in identifying these functions. This result also provides new evidence that there are differences in the nature of the replicative forms of [PSI+] and [URE3] and highlights the importance of the relationship between the activity of the Hsp70 and Hsp104 chaperones and optimal propagation of yeast prions. To better understand their roles in yeast prion propagation, it will be important to systematically test at what level Hsp104 and Hsp70 act in the de novo appearance and propagation of these different prions.
.
Acknowledgments
We thank Herman Edskes and Mick Tuite for plasmids.
REFERENCES
- 1.Chernoff, Y. O., I. L. Derkatch, and S. G. Inge-Vechtomov. 1993. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24:268-270. [DOI] [PubMed] [Google Scholar]
- 2.Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov, and S. W. Liebman. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880-884. [DOI] [PubMed] [Google Scholar]
- 3.Cox, K. H., R. Rai, M. Distler, J. R. Daugherty, J. A. Coffman, and T. G. Cooper. 2000. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem. 275:17611-17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DebBurman, S. K., G. J. Raymond, B. Caughey, and S. Lindquist. 1997. Chaperone-supervised conversion of prion protein to its protease-resistant form. Proc. Natl. Acad. Sci. USA 94:13938-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derkatch, I. L., M. E. Bradley, J. Y. Hong, and S. W. Liebman. 2001. Prions affect the appearance of other prions. The story of [PIN+]. Cell 106:171-182. [DOI] [PubMed] [Google Scholar]
- 6.Derkatch, I. L., M. E. Bradley, P. Zhou, Y. O. Chernoff, and S. W. Liebman. 1997. Genetic and environmental factors affecting the de-novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doel, S. M., S. J. McCready, C. R. Nierras, and B. S. Cox. 1994. The dominant PNM2− mutation which eliminates the Ψ factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137:659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edskes, H. K., V. T. Gray, and R. B. Wickner. 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA 96:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edskes, H. K., and R. B. Wickner. 2000. A protein required for prion generation: [URE3] induction requires the Ras-regulated Mks1 protein. Proc. Natl. Acad. Sci. USA 97:6625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellwood, M. S., and E. A. Craig. 1984. Differential regulation of the 70K heat shock gene and related genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Bellot, E., E. Guillemet, F. Ness, A. Baudin-Baillieu, L. Ripaud, M. Tuite, and C. Cullin. 2002. The [URE3] phenotype: evidence for a soluble prion in yeast. EMBO Rep. 3:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover, J. R., A. S. Kowal, E. C. Schirmer, M. M. Patino, J.-J. Liu, and S. Lindquist. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89:811-819. [DOI] [PubMed] [Google Scholar]
- 13.Jung, G., G. Jones, R. D. Wegrzyn, and D. C. Masison. 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, C. Y., P. Tittmann, H. Gross, R. Gebert, M. Aebi, and K. Wuthrich. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochneva-Pervukhova, N. V., S. V. Paushkin, V. V. Kushnirov, B. S. Cox, M. F. Tuite, and M. D. Ter-Avanesyan. 1998. Mechanism of inhibition of Psi+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 17:5805-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushnirov, V. V., and M. D. Ter-Avanesyan. 1998. Structure and replication of yeast prions. Cell 94:13-16. [DOI] [PubMed] [Google Scholar]
- 17.Lacroute, F. 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106:519-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lansbury, P. T., Jr., and B. Caughey. 1995. The chemistry of scrapie infection: implications of the ′ice 9′ metaphor. Chem. Biol. 2:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Lindquist, S., and G. Kim. 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. USA 93:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masison, D. C., M.-L. Maddelein, and R. B. Wickner. 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. USA 94:12503-12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masison, D. C., and R. B. Wickner. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 273:93-95. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. F. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Moriyama, H., H. K. Edskes, and R. B. Wickner. 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20:8916-8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newnam, G. P., R. D. Wegrzyn, S. L. Lindquist, and Y. O. Chernoff. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19:1325-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osherovich, L. Z., and J. S. Weissman. 2001. Multiple gln/asn-rich prion domains confer susceptibility to induction of the yeast. Cell 106:183-194. [DOI] [PubMed] [Google Scholar]
- 26.Parsell, D. A., A. S. Kowal, M. A. Singer, and S. Lindquist. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475-478. [DOI] [PubMed] [Google Scholar]
- 27.Patino, M. M., J.-J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622-626. [DOI] [PubMed] [Google Scholar]
- 28.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 29.Priola, S. A., B. Caughey, R. E. Race, and B. Chesebro. 1994. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prusiner, S. B., M. Scott, D. Foster, K. M. Pan, D. Groth, C. Mirenda, M. Torchia, S. L. Yang, D. Serban, G. A. Carlson, et al. 1990. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63:673-686. [DOI] [PubMed] [Google Scholar]
- 31.Rai, R., F. S. Genbauffe, R. A. Sumrada, and T. G. Cooper. 1989. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serio, T. R., A. G. Cashikar, A. S. Kowal, G. J. Sawicki, J. J. Moslehi, L. Serpell, M. F. Arnsdorf, and S. L. Lindquist. 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289:1317-1321. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondheimer, N., N. Lopez, E. A. Craig, and S. Lindquist. 2001. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 20:2435-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone, D. E., and E. A. Craig. 1990. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1622-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, K. L., N. Cheng, R. W. Williams, A. C. Steven, and R. B. Wickner. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283:1339-1343. [DOI] [PubMed] [Google Scholar]
- 38.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 39.Wickner, R. B. 1994. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 264:566-569. [DOI] [PubMed] [Google Scholar]