Abstract
The adeno-associated virus type 2 (AAV) large Rep proteins can act to increase the ratio of spliced to unspliced AAV RNA when they are targeted to the transcription template via a Rep binding element. The required Rep binding site is both location and orientation independent; however, Rep enhancement decreases as the distance between the promoter and the intron of the affected transcription unit increases. Only the AAV intron and an extended polyadenylation site must remain for the AAV transcription unit to manifest responsiveness to Rep. A number of promoters, when driving the AAV capsid gene transcription unit, were responsive to targeted Rep, though to various degrees. Transactivation of transcription initiation is not sufficient for the enhancement of RNA processing, because activation of the P40 transcription unit by other activators targeted to this transcription template did not result in enhancement of the ratio of spliced to unspliced AAV RNA. These results suggest that Rep may act as a trans regulator of RNA processing by modulating such functions coupled to RNA polymerase II (RNA pol II) transcription, perhaps by affecting the composition of the transcription complex either prior to or during elongation. These results reveal another way in which gene expression can be regulated by trans-acting proteins and help explain an important feature of the parvovirus life cycle.
It has become increasingly clear that various features of pre-mRNA cap formation, splicing, and polyadenylation as well as mRNA export are coupled to the transcription of these RNAs (reviewed in references 3, 11, 12, 16, 31, 36, and 40). The C-terminal domain (CTD) of RNA polymerase II (RNA pol II) has been implicated in the coordination of these events (17, 26, 48), and most recently, domains of the CTD which can independently stimulate pre-mRNA processing events have been identified (14). It has also been shown that the same RNA, when transcribed from different promoters, can undergo different levels of processing (10, 20) and, in addition, that the function of certain SR proteins in splicing is dependent upon promoter structure (9).
One way in which trans-acting transcriptional activators can affect the transcriptional activity of RNA pol II is by changing the composition or the activity of the transcription machinery binding at the promoter (28, 47). That trans-acting factors may affect RNA processing by inducing changes in the transcription machinery binding at the promoter or during elongation has so far only been suggested (16, 36, 40).
The human parvovirus adeno-associated virus type 2 (AAV) is a single-stranded DNA virus which utilizes a helper virus (typically adenovirus or herpesvirus) in the establishment of productive infection (reviewed in references 4 and 5). The genetic map (Fig. 1A) and the temporal profile of the appearance of AAV RNA (27) have been determined. The genome of approximately 4.7 kb is flanked by palindromic inverted terminal repeats (ITRs) that are required for viral replication and site-specific integration into human chromosome 19 in the absence of helper virus or cellular stress. RNAs are generated from three viral promoters (P5, P19, and P40), and each pre-mRNA class contains the same intron, which, when excised, utilizes a single splice donor site and either one of two splice acceptor sites (Fig. 1A). The ratio at which these two acceptors are used for each class of AAV RNA has not yet been determined. All AAV RNAs have been shown to be very stable (27), and all spliced and unspliced RNAs are exported at similar rates (J. Qiu and D. J. Pintel, unpublished data). Both the transcription and splicing of AAV RNAs are enhanced by adenovirus coinfection (5, 27, 41). Interestingly, the ratio of steady-state spliced to unspliced P40-generated RNAs is much greater than that of RNAs generated from the upstream promoters (27, 41).
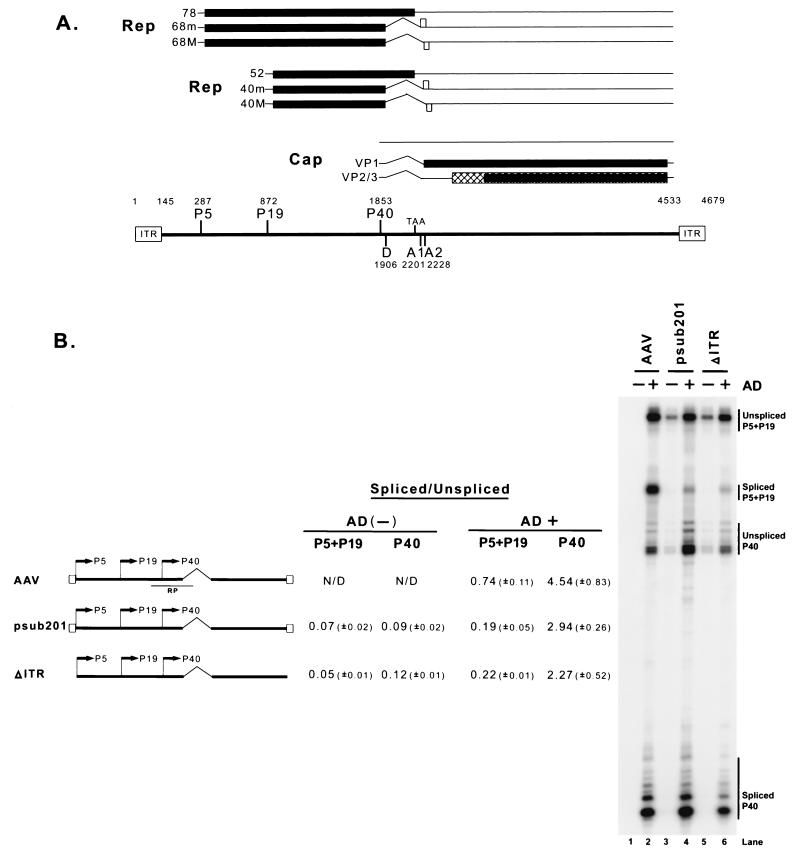
FIG. 1.
(A) Genetic map of AAV. Transcripts and protein-encoding regions for the three AAV promoters are shown. The promoters, intron donor (D) and acceptors (A1 and A2), and ITRs are mapped with their nucleotide designations. (B) The ratios of spliced to unspliced RNAs generated from the different AAV promoters are different during both wild-type AAV infection and transfection of full-length AAV plasmid constructs. 293 cells were either infected with AAV2 (MOI, 10) or transfected with either psub201 or ΔITR in the absence (−) or presence (+) of adenovirus (AD) (MOI, 10). A map of these constructs (not to scale) is shown. Ten micrograms of total RNA from infected or transfected cells was protected by the RP probe, which is depicted in relation to the AAV genome. A representative experiment is shown, and the AAV-specific RNA bands are indicated on the right. Multiple bands protected by P40 unspliced and spliced RNA likely reflect the use of multiple initiation sites within these regions (27). Quantitations of the ratio of spliced to unspliced RNAs specifically derived from the P5+P19 and P40 promoters are shown. All of the values are averages of the results of at least three separate experiments, with standard deviations in parentheses. Previous work has shown (27) that the ratios of spliced to unspliced AAV RNA increase through the course of infection; lane 2 shows the maximal levels of spliced products seen at late times during infection. N/D, not determined
The AAV large Rep proteins (Rep 78 and 68), translated from P5-generated mRNAs, are site-specific DNA-binding transcriptional regulators that have been reported to both stimulate and repress the AAV P5 promoter and stimulate the P40 promoter in the presence of adenovirus (34, 35). We show here that the large Rep proteins, in the presence of adenovirus, can also enhance the ratio of spliced to unspliced RNAs generated from transcription templates to which they have been targeted. This effect is dependent upon the presence of both the AAV intron and polyadenylation sites.
This observation reveals another way in which trans-acting proteins can regulate gene expression.
MATERIALS AND METHODS
Cells and viruses.
Human 293 cells were maintained as described previously (27). AAV was a gift from Kevin Brown, Hematology Branch, National Institutes of Health, Bethesda, Md. Adenovirus type 5 (dl309) was a gift from R. J. Samulski, University of North Carolina, Chapel Hill. Viral coinfections were done at multiplicities of infection (MOI) of 10 for both AAV and adenovirus, as previously described (27).
Transfections.
Two micrograms of DNA per 60-mm dish was transfected into 293 cells with Lipofectamine and Plus reagents from Gibco BRL (Gaithersburg, Md.) as previously described (27). When additional plasmids were used for cotransfection, 0.5 μg of DNA per 60-mm dish was used. Where indicated, adenovirus was immediately added at an MOI of 10 following transfections. Total RNA was isolated 36 to 40 h later as previously described (30).
RNase protection assay and quantitation.
RNA was isolated with guanidine isothiocyanate, and RNase protection assays were performed as previously described (27, 30, 39). Hybridizations were done in substantial probe excess, and RNase protection assay (RPA) signals were quantified by using Molecular Imager FX and Quantity One (version 4.2.2) image software (Bio-Rad, Hercules, Calif.). The relative molar ratios of individual species of RNA were determined after adjusting for the number of 32P-labeled uridines in each protected fragment as previously described (39). In experiments in which total RNA levels were being compared (Fig. 4), a plasmid expressing enhanced green fluorescent protein (EGFP) (pEGFP) was added as an internal transfection control and activation levels were standardized to green fluorescent protein (GFP) RNA levels.
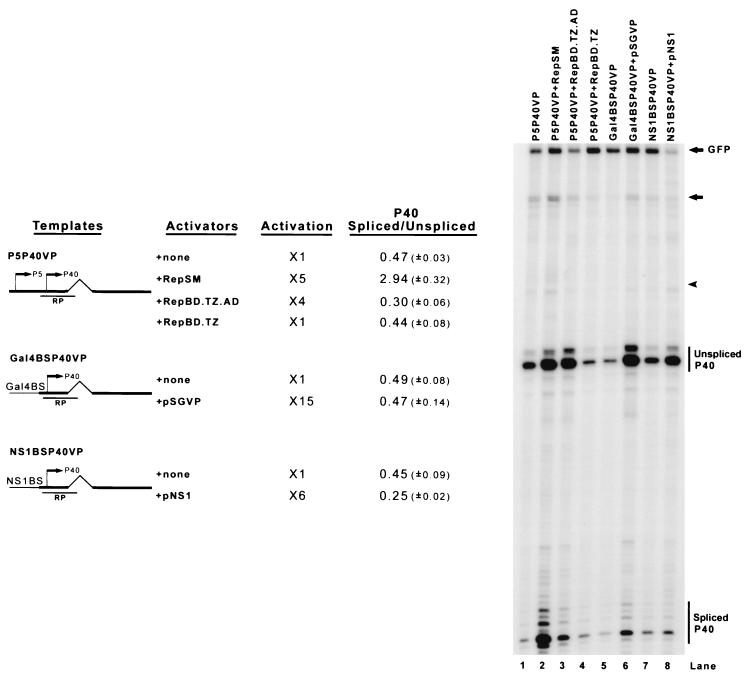
FIG. 4.
Activation of the P40 promoter is not enough to enhance the ratio of spliced to unspliced AAV RNA. 293 cells were cotransfected with minimal AAV capsid gene constructs bearing either the native P5 promoter (P5P40VP), three repeated Gal4 binding sequences (Gal4BSP40VP), or the MVM NS1 binding sequence (NS1BSVP) as described in the text, along with the activator plasmids shown. RepBD.TZ expressed only the oligomerized Rep binding domain (Rep aa 1 to 244), RepBD.TZ.AD expressed a Rep oligomerized binding domain/VP16 activation domain chimera, pSGVP expressed the chimeric Gal4 binding domain/VP16 activation domain fusion, and pNS1 expressed the MVM NS1 protein. To evaluate the levels of promoter activation, an EGFP-expressing plasmid (pEGFP) was cotransfected at 0.04 μg/60-mm dish in all groups as an internal control. Total RNAs were protected with the RP probe and a GFP probe. Bands specifically protected by P40-generated spliced and unspliced RNA and GFP RNA from a representative experiment are shown and labeled on the right. The fold activation and quantitations of the ratio of spliced to unspliced RNAs from at least three separate experiment are shown. Activation levels were standardized to that of the GFP internal control and compared to the unactivated background, which was set to 1 (lanes 1, 5, and 7). The ratios of spliced to unspliced RNAs include standard deviations in parentheses. For lanes 1 to 4, the unlabeled arrow and arrowhead denote a combination of unspliced (arrow) and spliced (arrowhead) P5 plus a low level of read-around transcription RNA; in the other lanes, these bands are assumed to derive from plasmid read-around transcription.
Plasmid constructs: full-length constructs.
psub201 was a gift from R. J. Samulski (38). ΔITR, which was created by the deletion of both ITRs in psub201, was a gift from Greg Tullis, University of Missouri, Columbia, Mo. pSK45 was created by the insertion of the AAV sequence from nucleotides (nt) 145 to 4492 (nucleotide numbers refer to the AAV sequence with GenBank accession number AF 043303) from pIM45 (24) into the polylinker of pBluescript SK(+) (SK+) (Stratagene, La Jolla, Calif.). To create pSK45mP5ATG, the ATG for Rep 68 and 78 was mutated to GGC. pSK45mP19TA was obtained by the mutation of both P19 TATA boxes (TAATAT→CGCGGG and TATTTAA→GCGCGGG) in pSK45mP5ATG.
AAV minimal constructs.
P40VP contains AAV sequences (nt 1700 to 4492) which include the AAV P40 promoter (nt 1700 to 1880) (24), intron, capsid gene sequences, and polyadenylation site cloned into the polylinker of SK+. The native distance of the P40 RNA initiation site from the AAV intron is 53 nt. P5VP contains the AAV P5 promoter (nt 146 to 320) (34) adjacent to AAV sequences from nt 1882 to 4492. The distance between the P5 RNA initiation site and the AAV intron in this construct is 58 nt. P5P40VP was created by inserting the AAV P5 promoter (nt 146 to 320) at nt 1700 upstream of the P40 promoter in P40VP. To generate P5(mMLTF)P40VP, P5(mYY1-60)P40VP, P5(ExmTATA)P40VP, P5(mRBE)P40VP, and P5(mYY1 + 1)P40VP, previously described mutant plasmids (gifts from N. Muzyczka, University of Florida) (34) were used as PCR templates and mutations were transferred into P5P40VP. P5(mTATA)P40VP was created by changing the TATTT in the TATA box of the P5 promoter in P5P40VP to CCCGG. MiniP5P40VP was created by inserting AAV sequences 255 to 299, which contain the TATA box, RBE, and (+1)YY1 domains, in front of the P40 promoter in P40VP at AAV nt 1700. ITRP40VP was obtained by inserting the ITR sequences from nt 1 to 145 in front of the P40 promoter in P40VP at nt 1700. P40VPP5 was created by insertion of the P5 promoter (nt 146 to 320) into the PvuII site of SK+, 245 nt downstream of the capsid gene sequences in P40VP. (−)P5P40VP was created by inserting the AAV P5 promoter (nt 146 to 320) in reverse orientation at nt 1700, in front of the P40 promoter in P40VP.
Rep protein supplementation plasmids.
Silent mutations were inserted into all of the Rep supplementation plasmids within the region covered by the RP protection probe. This allowed the specific detection of expression from the reporter plasmid.
The plasmid RepSM, which was used to supply all of the Rep proteins, was generated from pHIVRep (1) by insertion of silent mutations across the RP probe protection region. Rep78/52SM was mutated so that the splice acceptor in RepSM was destroyed (this mutation has been described elsewhere [15]). Rep68/40SM was altered so that the AAV intron in RepSM was removed (this mutation has been previously described [15]). Rep78SM was mutated so that the Rep52 ATG in Rep78/52SM was mutated to GGG (this mutation has been previously described [7]). To create Rep68SM, Rep52ATG in Rep 68/40SM was mutated to GGG. To create Rep52SM, silent mutations in the RP probe regions were created in the previously characterized pHIVRep52 (15). To create Rep40SM, silent mutations within the RP probe regions were created in the previously described pHIVRep40 (15). Plasmid RepSMP19SM, which was designed for supplementation of Rep in trans for analyzing the splicing of P5-generated RNA produced from pSK45mP19TA, contained additional silent mutations in the P19 promoter region covered by the SB probe. All of the Rep supplementation parent plasmids were gifts from R. J. Samulski, University of North Carolina.
Activation plasmids.
The GalBSP40VP and NS1BSP40VP plasmids were constructed by fusing three Gal4 binding site repeats (CGGAGTACTGTCCTCCG) (44) or the minute virus of mice (MVM) NS1 binding site (TTGGTTGGTAAAGAATGGTTACCAATCTACCA) (8), respectively, in front of the P40 promoter in P40VP. The Gal4VP16 chimeric expression plasmid (pSGVP) was described elsewhere (37), and the MVM NS1 expression plasmid pNS1, previously called pCMV1989, has also been previously described (29). The RepBD.TZ.AD chimera contained the AAV Rep 78/68 DNA-binding domain (amino acids [aa] 1 to 244), an oligomerization signal from p53, and a VP16 activation domain; the RepBD.TZ plasmid contained the AAV Rep 78/68 DNA-binding domain (aa 1 to 244) and the same oligomerization signal. Both constructs were gifts from Matt Weitzman (Salk Institute, San Diego, Calif.) (6).
Plasmids for determining cis-acting requirement.
HIVVP was obtained by inserting the human immunodeficiency virus (HIV) long terminal repeat (LTR) promoter (nt 573 to 1095 from pHIVRep) (1) in the HindIII site at AAV nt 1882. This plasmid also contains an 80-nt linker from pBR322 (nt 376 to 455) at this site, which was found to increase the efficiency of transcription of this promoter. The distance between the HIV RNA initiation site and the AAV intron in this construct was approximately 176 nt. MVMP4VPP5 was created by inserting the MVM P4 promoter (MVM nt 116 to 470) in the HindIII site at AAV nt 1882. The distance from the RNA initiation site to the intron in this construct was 310 nt. The AAV P5 promoter was inserted at the PvuII site in SK+, 245 nt downstream of the AAV sequence in the opposite orientation. CMVVPP5 was created by inserting the cytomegalovirus (CMV) immediate-early (IE) promoter (CMV nt 206 to 888 from pcDNA3; Invitrogen) into the HindIII site at AAV nt 1882. The distance from the putative RNA initiation site to the intron was 81 nt. The AAV P5 promoter was inserted 245 nt downstream of AAV sequences in the opposite orientation. P5HIV was created by replacing the simian virus 40 promoter region (PstI-XbaI) of pDM128 (18) (gift of Bryan Cullen, Duke University, Durham, N.C.) with the AAV P5 promoter region (nt 146 to 320). P5AAVHIV contains the AAV P5 promoter, the AAV intron, and the HIV polyadenylation signal and was created by replacing P5VP AAV nt 2233 to 4492 with nt 6083 to 6896 of pDM128, which contains the HIV polyadenylation site. P5HIVAAV contains the AAV P5 promoter, the HIV intron, and the AAV capsid sequences and was created by replacing the AAV intron in P5VP (nt 1882 to 2233) with the HIV intron from pDM128 (nt 3291 to 6082). P5sPA contains the P5 promoter, the AAV intron, and the core AAV polyadenylation signal (nt 4326 to 4492). P5mPA contains the P5 promoter, the AAV intron, and the extended AAV polyadenylation signal (nt 3941 to 4492). P5 (400)sPA and P5 (1470)sPA contain the AAV P5 promoter, the AAV intron, and either a 403 (nt 29 to 432)- or a 1,466 (nt 29 to 1495)-nt fragment from pBR322 DNA upstream of the core polyadenylation signal (nt 4326 to 4492).
Plasmids for determining distance requirement.
P5 (180)VP, P5 (390)VP, P5 (780)VP, and P5 (1560)VP were created by inserting a 182 (nt 2376 to 558)-, a 390 (nt 376 to 766)-, a 780 (nt 376 to 1156)-, and a 1,561 (nt 376 to 1937)-nt DNA fragment from pBR322, respectively, at the HindIII site at nt 1882 between the AAV P5 promoter and the AAV intron in P5VP. This made the distance between the P5 initiation site and the AAV intron in these constructs 215, 423, 813, and 1,594 nt, respectively. P40 (180)VP, P40 (390)VP, P40 (780)VP, and P40 (1560)VP were created by inserting the same 182-, 390-, 780-, and 1,561-nt DNA fragments from pBR322 as described above, respectively, at the HindIII site at nt 1882 between the AAV P40 promoter and the AAV intron in P40VP. This made the distance between the P40 initiation site and the AAV intron in these constructs 211, 419, 809, and 1,590 nt, respectively. The AAV P5 promoter was inserted 245 nt downstream of the AAV sequence in the opposite direction to provide a fully functional Rep binding element.
Plasmids for probe generation.
Plasmids for RP and SB probes were described previously (27). Briefly, the RP probe protected bands of 191 and 139 nt for unspliced and spliced P5+P19-generated RNA, respectively, and bands of 105 and 53 nt for unspliced and spliced P40-generated RNA, respectively. The SB probe protected a band of 155 nt for P5-generated RNA and 92 nt for P19-generated RNA. The antisense probe-generating plasmids RRP5, RPHIV, and RPCMV were created by cloning, from the relevant reporter plasmid, a DNA fragment from a position upstream of the transcription initiation site to AAV nt 1960, which lies within the intron, into pGEM3Z (Promega, Madison, Wis.). The RPP5 probe protected bands of 112 and 58 nt for unspliced and spliced RNA, respectively, which were generated from P5 promoter in the constructs in which P5 was linked directly to the AAV intron. The RPHIV probe protected bands of 230 and 176 nt for unspliced and spliced HIV LTR-generated RNA, respectively, from plasmid transfection of HIVVP. The RPCMV probe protected bands of 135 and 81 nt, based on the putative initiation site on the CMV IE promoter, for unspliced and spliced CMV IE-generated RNA, respectively, from CMVVP plasmid transfection. RPP4 was created by cloning from MVMP4VP a 160-nt fragment beginning at nt 160 and spanning the MVM-AAV fusion up to AAV nt 1960, which lies within the AAV intron, into pGEM3Z. The RPP4 probe protected bands of 160 and 106 nt for unspliced and spliced MVM P4-generated RNA, respectively, from MVMP4VP transfection. The probe plasmid RRPP5HIV was created by cloning a 370-nt DNA fragment from a position upstream of the P5 initiation site to nt 3349 (pDM128), which lies within the HIV intron, into pGEM4Z (Promega). The RPP5HIV probe detected bands of 288 and 237 nt for unspliced and spliced P5-generated RNA, respectively, following P5HIV and P5HIVAAV transfection. Plasmid RPPBR was created by cloning a 140-nt DNA fragment beginning at nt 437 on pBR322 DNA and ending at AAV nt 1960, which lies in the AAV intron, into pGEM3Z. The RPPBR-generated probe detected bands of 140 and 86 nt for unspliced and spliced RNA, respectively, in all of the plasmids designed to analyze promoter-intron distance effects. Plasmid RPGFP was created by cloning a 245-nt fragment from the GFP gene (nt 613 to 858 of pEGFP; Clontech, Palo Alto, Calif.) into pGEM3Z. The RPGFP probe detected a band of 245 nt for GFP mRNA.
All plasmids were sequenced to confirm that only the desired mutations were generated.
RESULTS
AAV Rep proteins can enhance the ratio of spliced to unspliced AAV RNAs.
All AAV RNAs share the same intron; however, analysis of the steady-state levels of AAV RNA generated after AAV infection of 293 cells in the presence of adenovirus (which is known to stimulate both the expression and splicing of AAV-generated RNAs [5, 27, 41]) showed that the ratio of spliced to unspliced RNAs generated by the intron-proximal P40 promoter was considerably greater than that for RNA generated by the upstream P5 and P19 promoters (Fig. 1B, lane 2). This was also the case following transfection of replicating (psub201) or essentially nonreplicating, ITR-lacking (ΔITR) plasmid clones of AAV (Fig. 1B, lanes 4 and 6, respectively).
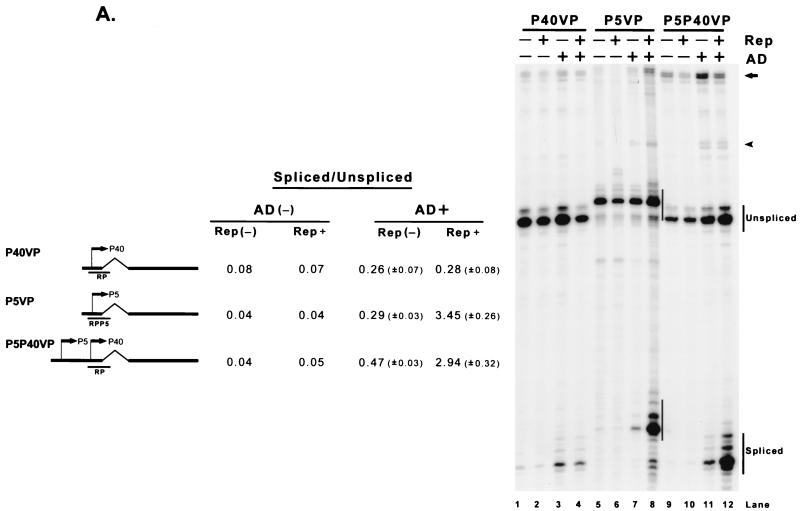
Although the P40-generated RNAs, when expressed in the context of the complete viral genome, are normally spliced very efficiently, the ratio of spliced to unspliced RNA was found to be poor in 293 cells when generated from minimal constructs in which the capsid-coding transcription unit was driven by either P40 (P40VP) or P5 (P5VP) in either the absence or presence of adenovirus (Fig. 2A, lanes 1 and 3 and lanes 5 and 7, respectively). Since the minimal constructs lacked the ability to express Rep proteins, the splicing levels of these constructs were then determined after the addition in trans of the Rep proteins. Surprisingly, the addition of the Rep proteins could enhance, in the presence of adenovirus, the ratio of spliced to unspliced RNAs generated from the P5-driven minimal construct (Fig. 2A, compare lanes 7 and 8) to levels reached by P40-driven RNA generated from full-length constructs. The P40-driven construct, however, was unaffected by Rep addition (Fig. 2A, compare lanes 3 and 4). Linkage of P5 sequences upstream of P40 in these constructs, however, allowed the generation of enhanced ratios of spliced to unspliced RNA initiated at the authentic P40 initiation site in the presence of Rep (Fig. 2A, lanes 9 to 12). Similar results were obtained following transfection of these constructs into HeLa cells in the presence of adenovirus (data not shown).
FIG. 2.
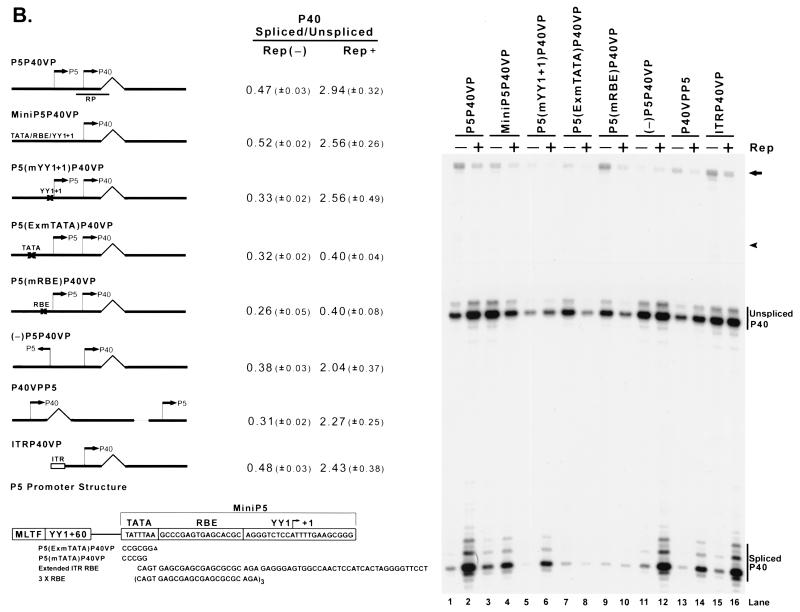
The AAV Rep proteins, when targeted to the transcription template, can enhance the ratio of spliced to unspliced AAV RNAs. (A) 293 cells were transfected with plasmid P40VP (lanes 1 to 4), P5VP (lanes 5 to 8), and P5P40VP (lanes 9 to 12) with (Rep+) or without [Rep(−)] cotransfection of the Rep supplementation plasmid RepSM in the absence [AD(−)] or presence (AD+) of adenovirus (MOI, 10). Ten micrograms of total RNA from transfected cells was protected by the homologous probes RP (lanes 1 to 4 and 9 to 12) and RPP5 (lanes 5 to 8), which are depicted under their respective plasmid diagrams on the left. Spliced and unspliced RNA specifically initiated from either the P40 (lanes 1 to 4 and 9 to 12) or P5 (lanes 5 to 8) promoters in a representative experiment is shown and labeled on the right. Quantitations of the ratio of spliced to unspliced RNAs for the intron-proximal promoters are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. For quantitations in the absence of adenovirus, all standard deviations were <0.01. For the P5P40VP construct, a portion of the bands shown by the arrow and arrowhead reflect unspliced and spliced RNA generated from the P5 promoter, respectively (lanes 9 to 12); in the other lanes, these bands are assumed to derive from low levels of plasmid read-around transcription. (B) Mutant constructs, as described in the text, are shown. An X in the plasmid diagrams is used to show the location of some of these mutations. 293 cells were transfected with (+) or without (−) Rep supplementation by RepSM in trans in the presence of adenovirus. Harvested total RNA samples were protected by the RP probe, and the unspliced and spliced RNA bands specifically initiated at the P40 promoter of a representative experiment are indicated on the right. Quantitations of the ratio of spliced to unspliced P40-initiated RNAs are shown and are the averages of the results of at least three separate experiments, with standard deviations in parentheses. For the experiment shown in lanes 1 to 10, the portions of the bands shown by the arrow and arrowhead reflect the unspliced and spliced RNAs generated from the P5 promoter, respectively; in the other lanes, the bands are assumed to derive from low levels of plasmid read-around transcription. The structure of the AAV P5 promoter is shown at the bottom of the diagram. The sequence shows the mini P5 element (used in MiniP5P40VP), which confers Rep responsiveness to the transcription template (lanes 3 and 4). The boxed region indicates sequence elements previously characterized to bind YY1, MLTF, and TBP and also the RBE. TATA mutations that either abolish Rep responsiveness [P5(ExmTATA)P40VP; lanes 7 and 8] or have no effect on this function [P5(mTATA)P40VP; see text] are depicted.
Enhancement of the ratio of spliced to unspliced RNA depends on targeting Rep to the transcription template, but the Rep binding site is position and orientation independent.
One of the most striking differences between P40 and P5 is the presence of a Rep binding site (RBE) in P5. Our analysis showed that an extended Rep binding site was both required and sufficient to confer Rep-dependent enhancement of the ratio of spliced to unspliced RNA to the P40-driven minimal construct (Fig. 2B). First, mutation of the RBE abolished the help from Rep proteins added in trans (Fig. 2B, lanes 9 and 10). In addition, a previously characterized (34) debilitating mutation of the P5 (+1)YY1 site (Fig. 2B, lanes 5 and 6), a mutation of the major late transcription factor (MLTF) and (−60)YY1 binding sites (data not shown), or a mutation of the P5 TATA sequence predicted to abolish TATA-binding protein binding [P5(mTATA)P40VP; TATTTAA to CCCGGAA, diagrammed in Fig. 2B] had no deleterious effect on Rep enhancement of the ratios of spliced to unspliced RNA (data not shown).
The addition of the Rep binding site-containing ITR to the transcription template (Fig. 2B, lanes 15 and 16) was also able to confer Rep responsiveness; however, the addition of a synthetic oligonucleotide containing three repeats of the ITR core RBE (6) (also diagrammed in Fig. 2B) was unable to do so (data not shown). This was apparently because additional sequences that extend beyond the core RBE were required; more extensive mutation of the region that impinged upon the P5 TATA sequences [P5(ExmTATA)P40VP; Fig. 2B, lanes 7 and 8] and the (+1)YY1 site (data not shown) which flank the RBE abolished the enhancement of the ratio of spliced to unspliced RNA that had been caused by added Rep protein. A small sequence of the P5 promoter region (diagrammed in Fig. 2B), comprising the TATA motif, the RBE, and the (+1)YY1 site (nt 255 to 299) (Fig. 2B, lanes 3 and 4), or an extended ITR RBE (also diagrammed in Fig. 2B) which contains the core RBE but is otherwise dissimilar (containing additional downstream sequences spanning nt 112 to 145) was found to be enough, when linked to P40 in minimal constructs, to confer Rep-dependent enhancement of the ratio of spliced to unspliced P40-driven RNA (data not shown).
These results demonstrated that the AAV Rep proteins could enhance the ratios of spliced to unspliced RNA generated from AAV P40, but only when they were targeted to the DNA molecule carrying that promoter. The P5 region that mediated this effect could function in either orientation (Fig. 2B, lanes 11 and 12) and at various positions on the plasmid (Fig. 2B, lanes 13 and 14). Thus, the RBE exhibited orientation and position independence similar to those of the classical transcription factor-binding enhancer element.
Enhancement of the ratio of spliced to unspliced AAV RNA is specific to the large Rep proteins.
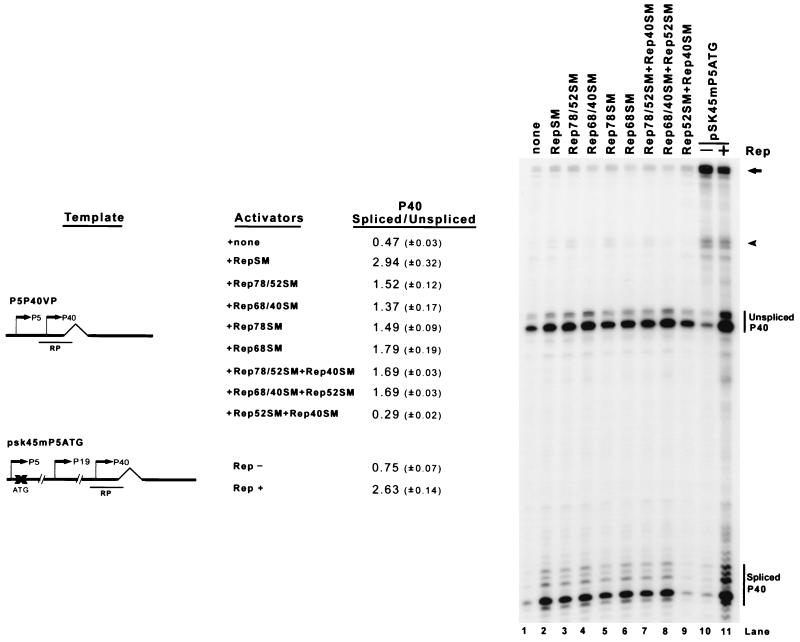
The large Rep proteins, Rep 78 and Rep 68, supplied in trans were required and sufficient to achieve Rep-dependent enhancement of splicing from the P5 and P40 minimal constructs (Fig. 3). Constructs that individually expressed Rep 78 or Rep 68, either alone (Fig. 3, lanes 5 and 6) or in the presence of Rep 52 or 40, respectively (Fig. 3, lanes 3 and 4), enhanced the ratio of spliced to unspliced P40-generated RNA, though not quite to the level of a construct that expressed all of the Rep proteins (Fig. 3, lane 2). The enhancement by either of the individual large Rep proteins was not significantly increased when both small Rep proteins were added (Fig. 3, lanes 7 and 8). As expected, the small Rep proteins alone had no effect, since they lack the Rep DNA-binding domain (Fig. 3, lane 9). As predicted from these results, the destruction of the initiating AUG for the large Rep proteins in the full-length ITR(−) AAV clone pSK45 resulted in a decreased ratio of spliced to unspliced P40-generated RNAs (Fig. 3, lanes 10 and 11).
FIG. 3.
The large Rep proteins are required and sufficient to enhance the ratio of spliced to unspliced AAV RNA. 293 cells were transfected with the minimal P5P40VP reporter plasmid in the presence of adenovirus, and different Rep proteins were provided in trans by cotransfection. For the sample shown in lane 11, Rep was supplied by the plasmid RepSM. pSK45mP5ATG, a full-length AAV construct in which the initiating AUG for the large Rep proteins has been mutated, is described in the text. Total RNA was protected by the RP probe, and bands specifically protected by P40-initiated spliced and unspliced RNAs from a representative experiment are designated. Quantitations of the ratio of spliced to unspliced P40-initiated RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. All of the Rep-supplying plasmids contained silent mutations in the region protected by the RP probe and are described in Materials and Methods. The arrow and arrowhead denote the unspliced and spliced RNAs, respectively, from the P5+P19 promoters (lanes 10 and 11) and from the P5 promoter (lanes 1 to 9), with some low-level contribution from plasmid read-around (lanes 1 to 9).
Activation of P40 transcription is not sufficient to enhance the ratio of spliced to unspliced RNA.
In all situations so far described, the enhancement of splicing by Rep was concomitant with the activation of the transcriptional initiation event. However, when the transcription of the P40 promoter was activated by either Gal4/VP16 (pSGVP, 15-fold) or the transcriptional activator NS1 produced by the autonomous parvovirus MVM (43) (pNS1, 6-fold) targeted to the transcription template, the ratio of spliced to unspliced AAV RNAs was not enhanced (Fig. 4, lanes 5 and 6 and lanes 7 and 8, respectively). Interestingly, neither a chimera of the Rep DNA-binding domain (Rep aa 1 to 244) fused to the VP16 activation domain (Rep BD.TZ.AD) (6), which activated P40 transcription in minimal constructs fourfold (Fig. 4, lane 3), nor an oligomerized Rep DNA-binding domain alone (Rep BD.TZ; Fig. 4, lane 4) (6) was able to enhance the ratio of spliced to unspliced RNAs. These results indicated that mere activation of P40 transcription initiation was not sufficient to cause an increased ratio of spliced to unspliced RNAs. This effect was either specific to Rep activation or dependent upon an additional Rep activity. Binding by the Rep DNA-binding domain was not sufficient for enhancement of the spliced product, suggesting that a feature of the C-terminal domain of the molecule was also required.
Enhancement by Rep requires the AAV intron and an extended polyadenylation site.
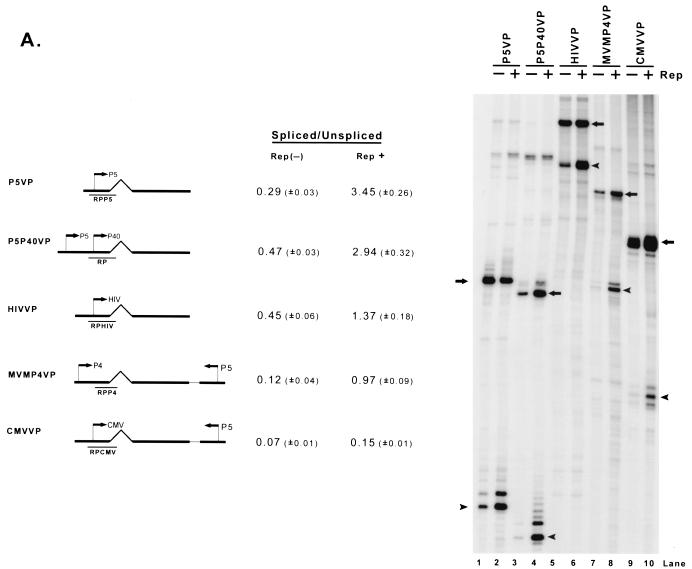
Which features of the P40 transcription unit confer responsiveness to Rep enhancement of the spliced-to-unspliced RNA ratio? Both the constitutive relative levels of spliced RNA and the ability of Rep, when targeted to the transcription template, to enhance the ratio of spliced to unspliced AAV RNA varied depending upon the promoter used to generate that RNA (Fig 5A ). The HIV LTR (which contains a Rep binding site [1, 25]) generated approximately the same ratio of spliced to unspliced RNA in the absence of Rep as was seen for the constructs driven by AAV P5 or P40 (HIVVP; Fig. 5A, lane 5), and the addition of Rep enhanced the level of spliced RNA threefold (Fig. 5A, lane 6), compared to the six- to sevenfold enhancement seen when the capsid gene transcription unit was driven by AAV promoters. When the AAV capsid-coding region was driven by either the MVM P4 or CMV IE promoters (MVMP4VP and CMVVP, respectively), the ratios of spliced to unspliced RNA in the absence of Rep were much lower than those of RNAs generated from either AAV P5, AAV P40, or the HIV LTR (Fig. 5A, compare lanes 7 and 9 to lanes 1, 3 and 5). The addition of Rep enhanced the ratio of spliced to unspliced MVM P4-driven AAV RNA approximately eightfold (Fig. 5A, lane 8), an increase similar to that seen when the AAV promoters were used, although the final spliced-to-unspliced ratios were considerably lower. By comparison, targeting of Rep to the CMV-driven AAV transcription unit enhanced the ratio of spliced to unspliced AAV RNA twofold. That the basal ratios of spliced to unspliced AAV RNA differed depending on the promoter used to drive their expression may have been expected from results obtained in other systems (10, 20); however, in addition, the ability of Rep to enhance the levels of the final spliced product was also promoter specific.
FIG. 5.
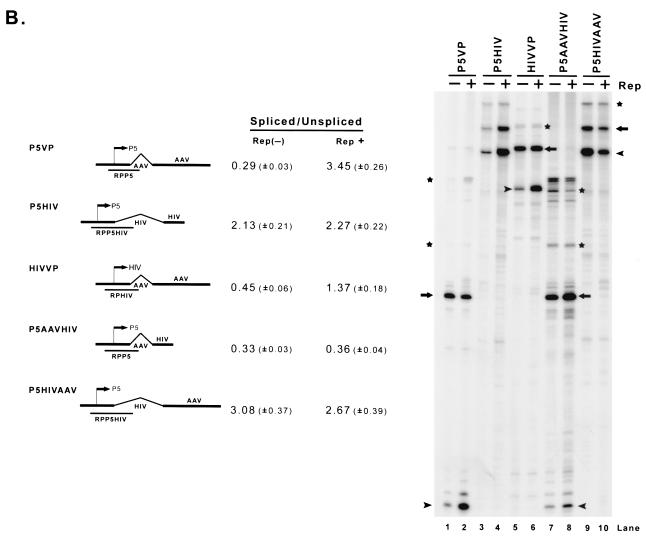
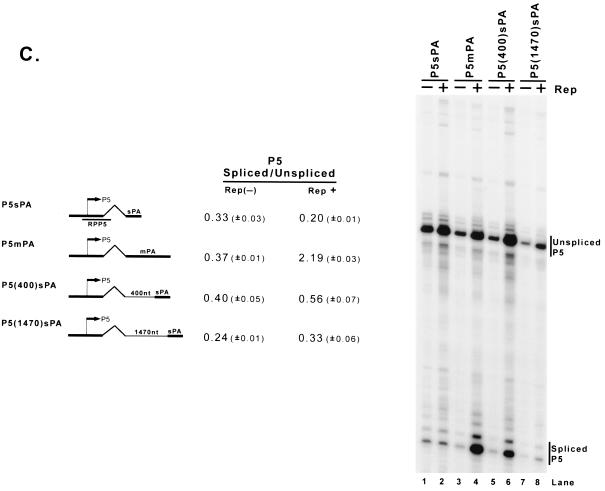
Enhancement of the ratio of spliced to unspliced RNA levels by Rep is promoter specific and requires the AAV intron and polyadenylation site. (A) 293 cells were transfected with AAV capsid gene sequences driven by the native P5 or P40 promoters (P5VP or P5P40VP), the HIV LTR (HIVVP), the MVM P4 promoter (MVMP4VP), or the CMV IE promoter (CMVVP). P5 sequences were inserted into MVMP4VP and CMVVP downstream of the AAV capsid gene in the opposite orientation to provide a fully functional Rep binding element as shown in Fig. 2B; the HIV LTR contains an RBE that has been previously characterized (1, 25). Cells were either cotransfected with RepSM (Rep+) or not [Rep(−)] in the presence of adenovirus. Total RNAs were protected with their respective homologous probes (RPP5, RP, RPHIV, RPP4, and RPCMV), and bands representing authentically initiated spliced (arrowhead) or unspliced (arrow) RNA from a representative sample are shown (except for P4-generated RNA, which uses an internal probe). Quantitations of the ratio of spliced to unspliced RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. (B) Plasmids diagrammed at left and described in the text were transfected into 293 cells either with (Rep+) or without [Rep(−)] cotransfection of the Rep-supplementing plasmid RepSM in the presence of adenovirus. Total RNAs were protected by the homologous probes (RPP5, RPP5HIV, and RPHIV) indicated under the plasmid diagrams, and bands protected by specifically initiated spliced (arrowhead) or unspliced (arrow) RNA from a representative experiment are shown. Bands labeled with an asterisk are likely due to low levels of read-around transcription and were not considered in the quantitation. Quantitations of the ratio of spliced to unspliced RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. (C) Plasmids P5sPA (containing only the AAV P5 promoter, the AAV intron, and the minimal AAV polyadenylation site [nt 4326 to 4492]), P5mpA (containing the AAV P5 promoter, the AAV intron, and the extended AAV polyadenylation site [nt 3941 to 4492]), or P5 (400)sPA and P5 (1470)sPA (which contain 403 and 1,466 nt of pBR322 DNA sequences inserted into P5sPA, respectively) were cotransfected into 293 cells either with (Rep+) or without [Rep(−)] cotransfection of the Rep-supplying plasmid RepSM in the presence of adenovirus. Total RNAs were protected by the RPP5 probe, which is depicted under the diagram of P5sPA. Bands protected by spliced and unspliced RNA from a representative experiment are shown and so labeled. Quantitations of the ratio of P5-specifically initiated spliced to unspliced RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses.
Further analysis of the cis-acting requirement for Rep responsiveness demonstrated that both the AAV intron and the extended polyadenylation site were required (Fig. 5B). Rep, when targeted to the transcription template, could enhance the ratio of spliced to unspliced RNA generated from the AAV capsid gene driven by either the AAV (P5VP) or HIV (HIVVP) promoter (Fig. 5A and B, lanes 1, 2, 5, and 6). In contrast, neither a complete HIV transcription unit (pDM128, data not shown) nor a minimal HIV transcription unit containing the HIV intron and polyadenylation site driven by AAV P5 was similarly responsive to Rep (P5HIV; Fig. 5B, lanes 3 and 4). This suggested that at least some responding element resided within the transcribed AAV pre-mRNA. (Although the basal ratio of spliced to unspliced RNAs generated from this construct was high, these levels would still allow the detection of Rep-dependent enhancement.) Rep was unable to enhance the ratio of spliced to unspliced RNA generated from a construct in which the normal AAV polyadenylation site of P5VP had been replaced with that of HIV (P5AAVHIV; Fig. 5B, lanes 7 and 8), and perhaps not unexpectedly, the AAV polyadenylation site alone was unable to confer Rep responsiveness to the HIV intron (P5HIVAAV; Fig. 5B, lanes 9 and 10). The most efficient enhancement by Rep occurred when an extended sequence including AAV sequences upstream of the canonical polyadenylation site (nt 3941 to 4492 [m]) was included in the transcription unit (P5mPA; Fig. 5C, lanes 3 and 4); the core AAV polyadenylation sequence (nt 4326 to 4492 [s]), either adjacent to (P5sPA; Fig. 5C, lanes 1 and 2) or at various distances downstream of the intron, was unable to sustain Rep responsiveness [P5 (400)sPA and P5 (1470)sPA; Fig. 5C, lanes 5 to 8]. These results demonstrated that the effect of the Rep protein in influencing the ratio of spliced to unspliced RNA was not universal and that it was most effective when targeted to an AAV transcription unit containing the AAV intron paired to its natural extended polyadenylation site or, for maximum effect, driven by the AAV P5 or P40 promoters.
Distance between the promoter and the AAV intron affects the level of Rep enhancement.
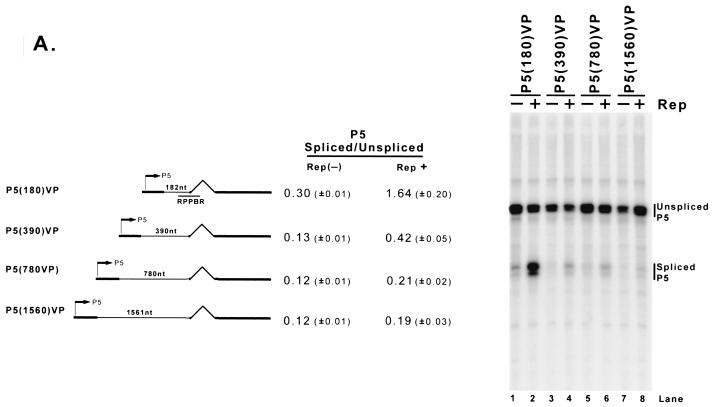
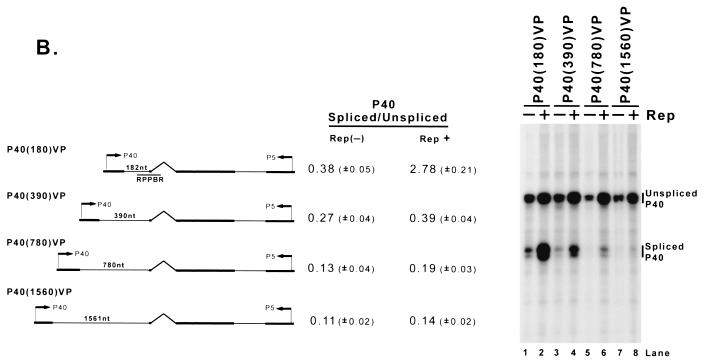
Although the Rep binding element required for enhancement of the ratio of spliced to unspliced P40-generated RNA could function at various locations in the transcription template-expressing molecule (Fig. 2B), the distance between the promoter and the intron of the RNAs affected was critical. As the distance in minimal constructs between P5 and the intron increased, the basal ratio of spliced to unspliced RNA decreased and the effect of Rep enhancement was diminished (Fig. 6A ). This was also the case in minimal constructs containing the P5 region downstream of P40 (Fig. 6B).
FIG. 6.
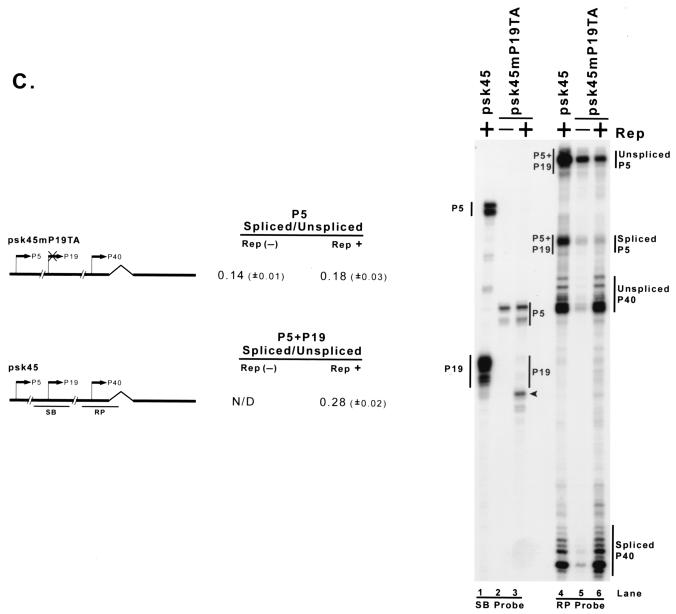
As the distance between the promoter and the AAV intron is increased, the level of Rep enhancement is diminished. (A) Plasmids with differently sized insertions of pBR322 DNA between the AAV P5 promoter and the AAV intron (as described in the text) were transfected into 293 cells with (Rep+) or without [Rep(−)] cotransfection of the Rep-supplementing plasmid RepSM in the presence of adenovirus. Total RNA was protected by the RPPBR probe, which is depicted under the diagram of P5 (180)VP. Bands protected by spliced and unspliced RNA from a representative experiment are shown and labeled on the right. The insertion in P5 (1560)VP (lanes 7 and 8) put the P5 promoter at approximately its native position relative to the AAV intron. Quantitations of the ratio of P5-specifically initiated spliced to unspliced RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. (B) Plasmids described in the text and similar to those described in panel A, except utilizing the AAV P40 promoter and bearing P5 sequences downstream in the opposite orientation (which serves as a fully functional Rep binding element [see Fig. 2B]), were transfected into 293 cells either with (Rep+) or without [Rep(−)] cotransfection of the Rep-supplementing plasmid RepSM in the presence of adenovirus. Total RNA was protected by the RPPBR probe, which is depicted under the diagram of P40 (180)VP. Bands protected by spliced and unspliced RNA from a representative experiment are shown and labeled on the right. Quantitations of the ratio of P40-specifically initiated spliced to unspliced RNAs are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. (C) pSK45 and pSK45mP19TA, in which the P19 promoter was destroyed (see text for details), were transfected into 293 cells in the presence of adenovirus and, in the case of pSK45mP19TA, with (Rep+) or without [Rep(−)] cotransfection of the Rep-supplying plasmid RepSMP19SM, which contained a series of silent mutations within both the RP and SB probe regions (see text for details). Total RNAs were individually protected by the SB (to distinguish P5 from P19 products) and RP (to distinguish P5+P19 from P40 products) probes, and bands protected by these probes from a representative experiment are shown and labeled accordingly. The arrowhead denotes an RNA breakdown band created from protection of P5+P19 RNA by cotransfected RepSMP19SM. Inspection of results with the SB probe (lanes 2 and 3) confirms that pSK45mP19TA no longer generates detectable P19-generated RNA, and so the spliced and unspliced RNAs labeled as being P5 generated in lanes 5 and 6 are uncontaminated by P19 products. Quantitations of the ratio of P5-specifically initiated spliced to unspliced RNAs for the pSK45mP19TA construct and of P5+P19-generated spliced to unspliced RNAs for pSK45 are shown and are averages of the results of at least three separate experiments, with standard deviations in parentheses. N/D, not determined.
Consistent with these results, the ratio of spliced to unspliced P5-generated RNA expressed from its naturally occurring position in a full-length construct (in which P19 activity was destroyed [as confirmed in Fig. 6C, lanes 2 and 3] to allow specific detection of the P5 products alone) was found to be similarly low (Fig. 6C, lanes 5 and 6).
DISCUSSION
In this report, we show that the AAV Rep protein can act to enhance the ratio of spliced to unspliced RNA when Rep is targeted to the transcription template on transfected plasmids. The binding site requirement for this activity of Rep is both location and orientation independent relative to the transcription start site, as in the cases of classical transcription enhancers. Our experiments suggest that Rep can function as a trans-regulator of RNA processing after binding to the transcription template, perhaps via promoter interactions. A number of RNA processing events are known to be cotranscriptional, and our results suggest that Rep may act to influence the composition of factors associated with the RNA pol II CTD.
The AAV Rep proteins are known to be transactivators of transcription initiation of both the AAV P5 and P40 promoters when targeted to the transcription template. Our results, however, suggest that neither activation of P40 nor mere binding to an extended RBE is sufficient to affect RNA processing. Activation of P40 by either Gal4/VP16, a Rep DNA-binding domain/VP16 activation domain chimera, or the NS1 protein of MVM did not result in an increase in the ratio of spliced to unspliced RNA (in the case of NS1, the ratio was significantly decreased). While transcriptional activation may be intimately related to the enhancement of RNA processing, it is not sufficient; this enhancement requires an additional function of the Rep protein. The minimal domain of Rep competent for transcription activation has not yet been defined. We were unable, however, to show enhancement of either transcriptional initiation or RNA processing of the minimal P40-driven AAV transcription unit by a targeted Gal4/Rep52 fusion protein (data not shown), leaving the definition of the domain required for the enhancement of the ratio of spliced to unspliced RNA unresolved. The large Rep proteins have been shown only to bind the AAV genome at the P5 promoter and the ITR sequences, and current models of P40 activation involve looping out models in which distally bound Rep affects the transcription complex downstream (35). The ability of Rep to enhance the ratio of spliced to unspliced P40 RNAs might also utilize such a looping out model, although it is not clear whether this mechanism is possible in the plasmid system used here.
Splicing of AAV-generated RNAs is enhanced by adenovirus coinfection, although the mechanism of this enhancement is not yet known (5, 27, 41). It is in this background that Rep increases the ratio of spliced to unspliced AAV RNAs, and this effect of Rep may require the participation of adenovirus gene products. It is known that during viral infection, AAV targets adenovirus replication centers at late times during coinfection (45), and that during coinfection, the splicing of AAV RNA reaches, at late times, levels not seen for RNA generated during transfection of AAV plasmids (27) (Fig. 1B). For the following reasons, however, it is unlikely that in our system, Rep acts merely to target template plasmids to adenovirus replication centers. Rep does not localize to adenovirus replication centers when expressed from an HIV-driven expression vector in adenovirus-infected cells (45). In addition, the ratio of Rep proteins derived from spliced AAV RNAs to Rep proteins derived from unspliced RNAs generated by an integrated copy of AAV is high (22). Finally, Rep can increase the ratio of spliced to unspliced AAV RNAs when a plasmid supplying only a minimal set of adenovirus helper genes, but not replicating adenovirus, is used as a helper (Qiu and Pintel, unpublished).
Rep enhancement of the ratio of spliced to unspliced RNA requires that targeting of Rep to the transcription template must be via an extended Rep binding site. This is consistent with recent suggestions that the binding of Rep to an extended site is qualitatively different from binding to an RBE site alone, which derived from the observation that the RBE alone is required but not sufficient for specific nicking of the AAV terminal resolution site by Rep (46).
While Rep was shown to enhance the ratio of spliced to unspliced RNA driven by various promoters to different degrees, the AAV intron and an extended AAV polyadenylation site were required to mediate Rep's effect on AAV RNA. This makes it unclear, as yet, what the primary effect of Rep is in this regard. Although the steady-state ratios of spliced to unspliced RNAs are detected to be different, it is possible that the primary effect of Rep is on either the capping or polyadenylation processes, which subsequently determine the final spliced-to-unspliced ratios. AAV RNA has only a single intron separating a large 5′-terminal exon and a 3′-terminal exon. Influences of both 5′ capping (19, 21) and 3′ polyadenylation (2, 13, 23, 32, 33, 42) have been shown to have effects on the splicing of adjacent introns. All three of these RNA processing events have recently been linked to the transcription process (see reference 14 and references therein). The observation that the effect of Rep is decreased as the distance between the RNA start site and the intron is increased, however, suggests that whatever the primary effect of Rep is, this effect must be sustained until the intronic region is reached by polymerase. Most recently, a role for RNA pol II elongation in the splicing process has been suggested, based on the observation that alternative splicing of the same pre-mRNA was different when transcription of that RNA was activated by two activators that have reportedly opposite effects on transcription elongation (20).
It is unlikely that in our system, differences in RNA export determine the effects we see. P40-generated spliced and unspliced RNAs are exported to the cytoplasm with similar kinetics during either viral infection or transfection (as determined by measuring the localization of pulse-labeled AAV RNA) (Qiu and Pintel, unpublished).
It has been suggested that if RNA processing factors function cotranscriptionally, one might find an effect of distance relative to the initiation site, i.e., factors so bound may disassociate at a given rate (36, 40). We do in fact observe that the effect of Rep on the ratio of spliced to unspliced RNA decreases as the distance between the promoter and intron increases, and this may also help explain why the same intron is excised at different final levels from P5- versus P40-generated AAV RNAs during infection. The splicing of the AAV intron may be constitutively poor (it has poor 5′ donor and 3′ acceptor sites), and perhaps some component of regulation is achieved because help by Rep is only effective when the promoter is close to 5′ splice sites. The increase in the concentration of Rep proteins that is seen as infection progresses would also help ensure the maximum accumulation of spliced P40-generated mRNAs required for capsid protein production. Also, since the relative A1 to A2 usage for each class of AAV RNA is not known, it remains possible that the effect of Rep on splicing to these acceptors is different.
In summary, we have demonstrated that the AAV Rep proteins can influence the steady-state ratio of spliced to unspliced RNA after targeting to the transcription template on transfected plasmids. Our results suggest that Rep may act to modulate processing functions coupled to RNA pol II transcription, perhaps by affecting the composition or function of factors associated with the CTD either prior to or during elongation. Although the mechanism of this activity has not yet been characterized, this observation identifies another way in which trans-acting proteins can regulate gene expression and helps explain an important feature of the AAV life cycle.
Acknowledgments
We thank R. Jude Samulski (University of North Carolina, Chapel Hill), Nick Muzyczka (University of Florida), Bryan Cullen (Duke University), Matthew Weitzman (Salk Institute), and Kevin Brown (NIH) for supplying plasmids, virus, and antibodies used in this work. We thank Greg Tullis (University of Missouri) for plasmids, helpful discussion, and critical reading of the manuscript. We are particularly indebted to Matthew Mouw, who initiated the AAV work in this lab and whose involvement helped form the initial direction of this project. We also thank Lisa Burger for excellent technical assistance.
Jianming Qiu was supported by a University of Missouri Molecular Biology Program fellowship. This work was supported by Public Health Service grant ROI A14658 and ROI A121302 from NIAID to D.J.P.
REFERENCES
- 1.Antoni, B. A., A. B. Rabson, I. L. Miller, J. P. Trempe, N. Chejanovsky, and B. J. Carter. 1991. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J. Virol. 65:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauren, G., S. Belikov, and L. Wieslander. 1998. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 12:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, D. L. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347-351. [DOI] [PubMed] [Google Scholar]
- 4.Berns, K. 1996. Parvoviridae: the viruses and their replication, p. 1017-1041. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 5.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 6.Cathomen, T., D. Collete, and M. D. Weitzman. 2000. A chimeric protein containing the N terminus of the adeno-associated virus Rep protein recognizes its target site in an in vivo assay. J. Virol. 74:2372-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., J. Christensen, J. P. F. Nüesch, and P. Tattersall. 1995. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J. Virol. 69:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramer, P., J. F. Cáceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2000. Connections between the processing and nuclear export of mRNA: evidence for an export license? Proc. Natl. Acad. Sci. USA 97:4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3:371-378. [DOI] [PubMed] [Google Scholar]
- 14.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segment of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavin, D. K., S. M. Young, Jr., W. Xiao, B. Temple, C. R. Abernathy, D. J. Pereira, N. Muzyczka, and R. J. Samulski. 1999. Charge-to-alanine mutagenesis of the adeno-associated virus type 2 Rep78/68 proteins yields temperature-sensitive and magnesium-dependent variants. J. Virol. 73:9433-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear event. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 17.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue, K., M. Ohno, H. Sakamoto, and Y. Shimura. 1989. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev. 3:1472-1479. [DOI] [PubMed] [Google Scholar]
- 20.Kadener, S., P. Cramer, G. Nogues, D. Cazalla, M. de la Mata, J. P. Fededa, S. Werbajh, A. Srebrow, and A. R. Kornblihtt. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20:5759-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. Meguigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1-snRNP with the cap-proximal 5′ splice-site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., F. Vaulgaropoulou, R. Chen, P. R. Johnson, and K. R. Clark. 2000. Selective Rep-Cap gene amplification as a mechanism for high-titer recombinant AAV production from stable cell lines. Mol. Ther. 2:394-403. [DOI] [PubMed] [Google Scholar]
- 23.Lutz, C. S., and J. C. Alwine. 1994. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 8:576-586. [DOI] [PubMed] [Google Scholar]
- 24.McCarty, D. M., M. Christensen, and N. Muzyczka. 1991. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J. Virol. 65:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarty, D. M., D. J. Pereira, I. Zolotukhin, X. Zhou, J. H. Ryan, and N. Muzyczka. 1994. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 68:4988-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 27.Mouw, M., and D. J. Pintel. 2000. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 74:9878-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 29.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of MVM is required for efficient DNA replication and infectious virus production in a cell-type specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6:1107-1111. [DOI] [PubMed] [Google Scholar]
- 31.Neugebauer, K. M., and M. B. Roth. 1997. Transcription units as RNA processing units. Genes Dev. 11:3279-3285. [DOI] [PubMed] [Google Scholar]
- 32.Niwa, M., and S. M. Berget. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 5:2086-2095. [DOI] [PubMed] [Google Scholar]
- 33.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 34.Pereira, D. J., D. M. McCarty, and N. Muzyczka. 1997. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 71:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira, D. J., and N. Muzyczka. 1997. The adeno-associated virus type 2 p40 promoter requires a proximal Sp1 interaction and a p19 CArG-like element to facilitate Rep transactivation. J. Virol. 71:4300-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot, N. J. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 37.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563-564. [DOI] [PubMed] [Google Scholar]
- 38.Samulski, R. J., L. S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 40.Steinmetz, E. J. 1997. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell 89:491-494. [DOI] [PubMed] [Google Scholar]
- 41.Trempe, J. P., and B. J. Carter. 1988. Regulation of adeno-associated virus gene expression in 293 cells: control of mRNA abundance and translation. J. Virol. 62:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 43.Vanacker, J.-M., and J. Rommelaere. 1995. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin. Virol. 6:291-297. [Google Scholar]
- 44.Wang, Y., B. W. O'Malley, Jr., S. Y. Tsai, and B. W. O'Malley. 1994. A regulatory system for use in gene transfer. Proc. Natl. Acad. Sci. USA 91:8180-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, J., M. D. Davis, and R. A. Owens. 2001. A rep recognition sequence is necessary but not sufficient for nicking of DNA by adeno-associated virus type-2 rep proteins. Biochem. Biophys. Res. Commun. 389:271-277. [DOI] [PubMed] [Google Scholar]
- 47.Zaman, Z., A. Z. Ansari, L. Gaudreau, J. Nevado, and M. Ptashne. 1998. Gene transcription by recruitment. Cold Spring Harbor Symp. Quant. Biol. 63:167-171. [DOI] [PubMed] [Google Scholar]
- 48.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]