Abstract
Transcriptional regulation of the Saccharomyces cerevisiae ARG1 gene is controlled by positive and negative elements. The transactivator Gcn4p is required for activation in minimal medium, while arginine repression requires the ArgR/Mcm1 regulatory complex, which binds to two upstream arginine control elements. We have found that the coordinated regulation of ARG1 requires components of the SAGA chromatin-remodeling complex. Using gcn5 deletion strains and a Gcn5 protein carrying the E173Q mutation in the histone acetyltransferase (HAT) region, we show that the HAT activity of Gcn5p is required for repression of ARG1 in rich medium. Similar increases in expression were seen upon deletion of other SAGA components but not upon deletion of the ADA-specific component, Ahc1p. Chromatin immunoprecipitations using antibodies to acetylated H3 confirmed that a decrease in the level of acetylated histones at the ARG1 promoter correlated with increased ARG1 expression. Up-regulation of ARG1 in the absence of Gcn5p also correlated with increased binding of TATA-binding protein to the promoter. The analysis of promoter deletions showed that Gcn5/Ada repression of ARG1 was mediated through the action of the ArgR/Mcm1 regulatory complex. In addition, studies with minimal medium demonstrated a requirement for the Ada proteins in activation of ARG1. This suggests that SAGA has a dual role at ARG1, acting to repress transcription in rich medium and activate transcription in minimal medium.
Transcriptional regulation can occur by alteration of the recruitment or activity of the transcriptional machinery (2, 34, 70, 85). In eucaryotic cells the packaging of DNA into chromatin plays an important role in regulating the accessibility of transcription factors (84). Nucleosomes, the principal packaging element of DNA in the nucleus, may be the primary determinant of accessibility for gene-specific regulatory proteins and the basal transcriptional machinery (26, 83). One mechanism by which chromatin structure can be changed is through the reversible acetylation of lysines within the amino-terminal domains of the histones (34, 78, 85). Often, enhanced gene expression corresponds with increased acetylation of nucleosomes (33, 70); however, there are cases where histone acetylation appears to be unchanged upon activation (18, 56) and others where increased acetylation correlates with repression (3, 10, 19, 50, 54, 68, 75).
The importance of histone acetylation in transcription has been confirmed by the identification of histone acetyltransferase (HAT) activities within coregulatory complexes (79). Saccharomyces cerevisiae GCN5 was originally described as a gene encoding a transcriptional adaptor (28) and was found to be functionally linked to a group of genes whose disruption resulted in reduced toxicity from the overexpression of VP16 (5, 12, 39, 48, 58). Subsequently Gcn5p was identified as the catalytic component of two multicomponent HAT complexes, the SAGA and ADA complexes (29, 66), both of which preferentially acetylate nucleosomal histone H3 and to a lesser extent histone H2B. SAGA is a 1.8-MDa complex composed of distinct classes of transcription factors: Ada proteins (Ada1p, Ada2p, Ngg1p/Ada3p, and Ada4p/Gcn5p) (5, 8, 40, 58), TATA-binding protein (TBP)-related SPT proteins (Spt3p, Spt7p, Spt8p, and Spt20p/Ada5p) (24, 27, 47, 62, 82), and TBP-associated factors or (TAFIIs) (TAFII90, TAFII68/61, TAFII60, TAFII25/23, and TAFII17) (30). Also included in the complex is the DNA-dependent protein kinase related molecule Tra1p (31, 67). As well as displaying HAT activity, components of the complex have interactions with activators and with TBP (4, 69). The ADA complex is approximately 900 kDa and contains Gcn5p, Ada2p, and Ngg1p/Ada3p (29, 66) as well as the ADA complex-specific component Ahc1p (23).
The SAGA complex is generally regarded as a coactivator complex, with the HAT activity of Gcn5p contributing a key function in a multistep pathway leading to gene activation (14, 45, 80). However, the Ada proteins have been linked with transcriptional repression. Disruption of NGG1/ADA3 and ADA2 leads to enhanced transcription of Gal4p-, Pdr1p-, and Pdr3p-activated genes (8, 49). Furthermore, the genome-wide analysis of Holstege et al. (37) showed that of the 5.2% of the yeast genes regulated twofold or more by Gcn5p, approximately one-third of them were negatively regulated.
When yeast cells were grown in rich medium (yeast extract-peptone-dextrose [YPD]), mRNA for the gene encoding arginosuccinate synthetase (ARG1), the enzyme involved in the penultimate step in the biosynthesis of arginine, was found to be elevated 8.2-fold upon disruption of gcn5 (37). This implicates acetylation as having a negative role in the expression of this gene. ARG1 is controlled by two regulatory pathways. It is induced under amino acid starvation through the general control pathway, culminating in activation by the transcription factor, Gcn4p (15, 17). ARG1, like the arginine biosynthetic genes ARG5,6, ARG8, and ARG3, is repressed by arginine (16, 20, 42, 51). Arginine repression operates at the level of transcription through a complex of proteins including Arg80p (ArgRI), Arg81p (ArgRII), and Mcm1p (the ArgR/Mcm1complex) (21, 52) and is further regulated by Arg82p (ArgRIII) (60). In addition to repressing the anabolic genes, the ArgR/Mcm1 complex is required for the arginine-specific induction of the catabolic genes CAR1 and CAR2 (22, 52). ARG1 contains two sites of binding for ArgR/Mcm1 termed arginine boxes or arginine control elements (ARC elements) (15). The mechanism of repression by ArgR/Mcm1 is unknown but does not involve steric inhibition of Gcn4p binding (15).
In this study we show that the Ada and Spt components of the SAGA complex are involved in the repression of ARG1 in rich medium. The repression by SAGA requires the HAT activity of Gcn5p. The increased expression of ARG1 in the absence of GCN5 correlates with decreased acetylation of histone H3 at the promoter and enhanced binding of TBP. Interestingly, the corresponding studies performed for cells grown in minimal medium demonstrate a requirement for Gcn5p and the Ada proteins in the activation of ARG1. This suggests that acetylation mediated by the SAGA complex can lead to activation or repression of ARG1, depending on the signals impinging on the promoter.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The strains used in this study are shown in Table 1. FY86 and FY1370 (61) were kindly provided by F. Winston. ARY96 is isogenic to BY249 but contains a TRP1 disruption of NGG1 (8). ARY144 and ARY146 are isogenic to FY86 and FY1370, respectively, but contain a Tn10LUK disruption of the entire ARG80 coding region, using the oligonucleotide pairs 5′-GGAGGTCCTGTGTTCGATCC-3′ with 5′-CGGGATCCGTCGCTATTCGACGTCA-3′ (N terminal) and 5′-GGGGATCCGGCAAACTGAACATGCG-3′ with 5′GCTCTAGAAAAATTACTCAAAGGGGGA-3′ (C terminal) to construct the disrupting allele.
TABLE 1.
Yeast strains and genotypes
| Strain | Genotype | Reference or source |
|---|---|---|
| FY86 | matα his3Δ200 leu2Δ1 ura3-52 | 61 |
| FY1370 | Isogenic to FY86 except gcn5Δ::HIS3 | 61 |
| BY4741 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742 | matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| BY4282 | Isogenic to BY4741 except ada2Δ0 | Research Genetics |
| BY3534 | Isogenic to BY4741 except ngg1Δ0 | Research Genetics |
| BY249 | Isogenic to BY4741 except gcn4Δ0 | Research Genetics |
| ARY96 | Isogenic to BY249 except ngg1Δ::TRP3 | This study |
| BY4228 | Isogenic to BY4741 except spt3Δ0 | Research Genetics |
| BY3218 | Isogenic to BY4741 except spt7Δ0 | Research Genetics |
| BY1799 | Isogenic to BY4741 except ahc1Δ0 | Research Genetics |
| BY1114 | Isogenic to BY4741 except rpd3Δ0 | Research Genetics |
| BY11695 | Isogenic to BY4742 except sin3Δ0 | Research Genetics |
| BY12666 | Isogenic to BY4742 except spt8Δ0 | Research Genetics |
| BY17285 | Isogenic to BY4742 except gcn5Δ0 | Research Genetics |
| ARY144 | Isogenic to FY86 except arg80Δ0::URA3 | This study |
| ARY146 | Isogenic to FY1370 except arg80Δ0::URA3 | This study |
Yeast strains were grown at 30°C in YPD broth (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [vol/vol] glucose) or minimal medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [vol/vol] glucose; supplemented with amino acids as required).
DNA constructs.
To generate Myc-epitope tagged GCN5, the complete GCN5 coding sequence flanked by NotI and SacI restriction sites was generated by PCR using oligonucleotides 5′-ATAAGAATGCGGCCGCCATGGTCACAAAACATCAGATTGA-3′ and 5′-CGAGCTCAATTGATCACATCGTCTC-3′. The NotI-SacI fragment was ligated into NotI-SacI-digested YCp88-myc (9) to allow expression of Myc-tagged Gcn5p from the DED1 promoter. To generate GCN5E173Q, the EcoRI site of YCp88-myc was removed by EcoRI digestion and backfilling with the Klenow fragment of DNA polymerase I (YCp88-mycΔRI). An EcoRI-SacI fragment containing the E173Q mutation (boldfaced) in GCN5 was synthesized by PCR using oligonucleotides 5′-GGAATTCGCACAAATTGTTTTCTGTGCCATC-3′ and 5′-CGAGCTCAATTGATCACATCGTCTC-3′. This fragment and the 506-bp NotI-EcoRI 5′-terminal fragment of YCp88-mycGCN5 were triple-fragment ligated into NotI-EcoRI-digested YCp88-mycΔRI.
The hemagglutinin (HA)-tagged TBP molecule was constructed by inserting three copies of the HA epitope (HA3) after codon 3 of the TBP open reading frame. This was achieved by using oligonucleotides 5′-CGGGATCCATGGCCGATTGAGGGCGGCCGCGAACGTTTAAAGGAGTTTAAAAGAG-3′ and 5′-CGGGAATTCGTCTACTCCTTCCCCATCA-3′ to synthesize the complete TBP-coding sequence with a NotI site placed after the third codon, flanked by BamHI and EcoRI. The BamHI-EcoRI fragment was ligated into YCp88 (38). An oligonucleotide encoding a triple HA tag (kindly provided by M. Manolson) was cloned directly into the engineered NotI site.
lacZ reporter constructs were cloned as his3-lacZ fusions into the LEU2 centromeric plasmid YCp87 (8). ARG1-lacZ was isolated by PCR using oligonucleotides 5′-CCCCAAGCTTCCTTGGCGGCATCGAAATC-3′ and 5′-CCCGGATCCCCCAACCAATTTATCATTATAC-3′, creating an 855-bp fragment encompassing positions −640 to +213, relative to the transcriptional start site (15), flanked by BamHI and HindIII restriction sites. After BamHI-HindIII digestion, this fragment was cloned directly into YCp87. ARG1ΔABF1 was similarly constructed using an upstream oligonucleotide, 5′-CCCGGATCCTCTAGAGCTCTCCAGTCATTTATGTG-3′, that incorporates ARG1 sequences downstream of −301.
ARG1ΔARC-lacZ was constructed by inserting SalI and XbaI restriction sites between positions −175 and −197 (ARC1) and positions −214 and −236 (ARC2). Initially, a SalI restriction site was placed in ARC1 by PCR using oligonucleotides 5′-ACGCGTCGACAGGAGAGAGAGATTACTCATC-3′ and 5′-CCCCAAGCTTCCTTGGCGGCATCGAAATC-3′ to create a SalI-HindIII fragment and oligonucleotides 5′-ACGCGTCGACAAATGACTCTTCCATACGGCA-3′ and 5′-CCCGGATCCCCCAACCAATTTATCATTATAC-3′ to generate a SalI-BamHI fragment. The two PCR fragments were digested and then triple-fragment ligated into YCp87. This molecule was used as the template to disrupt ARC2 by using the strategy outlined above and the oligonucleotides 5′-GCTCTAGACATTAACGGTGCCGTATGG-3′ and 5′-GCTCTAGAGGTTGCGACAGCGGAAAAA-3′. ARG1ΔABF1ΔARC was constructed by PCR using ARG1ΔARC-lacZ as a template and the primers used for construction of ARG1ΔABF1.
β-Galactosidase assays.
For the analysis of ARG1-lacZ fusion reporters, saturated cultures grown in minimal medium were inoculated at a 1/100 dilution in YPD or minimal medium (supplemented with the required amino acids), and grown at 30°C to an A600 of 1.0 to 1.5. Cells were pelleted, washed in LacZ buffer, and resuspended in 500 μL to 1 ml of LacZ buffer. β-Galactosidase activity was determined with o-nitrophenyl-β-d-galactosidase (ONPG) as the substrate as described previously, standardizing to cell density (1).
Chromatin immunoprecipitation (ChIP) assays.
Chromatin was prepared as described previously (36) with the following modifications. A 150-ml portion of a yeast culture grown to an A600 of approximately 1.5 was treated with 1% formaldehyde for 20 min at room temperature with occasional swirling. Cells were collected, washed twice with cold phosphate-buffered saline (140 mM NaCl, 2.5 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.5) and resuspended in 1.2 ml of ice-cold lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] sodium deoxycholate) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 0.05 mg of N-tosyl-l-phenylalanine chloromethyl ketone per ml, 0.01 mg of aprotinin [prepared in 10 mM HEPES-KOH, pH 8.0] per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml). The suspension was aliquoted to 400 μl into chilled 1.5-ml Microfuge tubes containing an equal volume of glass beads (0.5-mm diameter). Cross-linked chromatin was isolated, pooled, and fragmented as described previously (36). The chromatin solution (400 μl) was incubated with antibody (2 μl of anti-acetylated histone H3 [anti-AcH3] [Upstate Biotechnology], 15 μl of ascites fluid derived from the Myc1-9E10 cell line [25], or 15 μl of an ascites fluid from the 12CA5 cell line [anti-HA]) for 4 h to overnight at 4°C. Protein A (for anti-AcH3 immunoprecipitation) or G Sepharose (for anti-HA and anti-Myc) was added and left for an additional hour at 4°C. Washes were for 5 min at room temperature. These washes included the following: for immunoprecipitations using anti-AcH3 and -HA antibodies, twice with 1 ml of lysis buffer, once with wash buffer 1 (lysis buffer containing 500 mM NaCl), once with wash buffer 2 (10 mM Tris-HCl [pH 8.0], 0.25 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and once with TE (10 mM Tris-HCl [pH 8.0], 2 mM EDTA); for immunoprecipitations with anti-Myc antibody, three times with 1 ml of lysis buffer, once with wash buffer (lysis buffer containing 100 mM NaCl), and once with TE.
Immunoprecipitated material was eluted from the beads at 65°C for 30 min in 100 μl of TE-1% sodium dodecyl sulfate followed by centrifugation at 10,000 × g. Cross-links were reversed by incubation at 65°C for 6 h to overnight. DNA was extracted with phenol-chloroform-isoamyl alcohol and chloroform, precipitated in 3 volumes of ethanol (containing 20 μg of glycogen, 1/10 volume 5 M LiCl, and 50 mM Tris-HCl, pH 8) at −20°C, and then washed with 70% ethanol and resuspended in TE.
Precipitated DNA was analyzed by quantitative PCR using primers from ARG1 positions −304 to −79 (5′-ATACTATTGAGACAGTGCCAGT-3′ and 5′-ACGGCTCTCCAGTCATTTATG-3′) (for anti-AcH3 and anti-Myc immunoprecipitation), ARG1 positions −968 to −649 (upstream region) (5′-GCAGATCTGATCACCACCTG-3′ and 5′-GGCAACAGGAAAGATCAGAG-3′) (for anti-AcH3 and anti-Myc immunoprecipitation), ARG1 positions −201 to +26 (5′-GAACAATGTTCCTTATCGCTGCACA-3′ and 5′-GAGTCATTTAAAGGCAGGAGAGAGAGA-3′) (for HA3TBP immunoprecipitation), TDH3 (5′-CTACCGAAA CCGTTAATAGCAAC-3′ and 5′-GTTCATAGGTCCATTCTCTTAGC-3′), and PGK1 (5′-GGACAGACAACTTTGAAGATAAAG-3′ and 5′-GAATCGTGTGACAACAACAGCGTG-3′). The linear range for primer pairs was determined using decreasing amounts of template. Approximately 1/50 of precipitated DNA and 1/13,000 of total DNA input were used. PCRs were carried out in 50 μl containing 50 pmol of primers, 0.2 mM deoxynucleoside triphosphates, 1× PCR buffer (Promega), 1.5 mM MgCl2, 1 mg of glycogen per ml, and 2 U of Taq polymerase (Promega). The standard cycling program was 2 min at 95°C, followed by 25 to 28 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C and a final extension step of 5 min at 72°C. PCR products were separated by electrophoresis on 6 to 8% native acrylamide gels, stained with ethidium bromide (0.1 μg/ml), and photographed during UV exposure on a gel documentation system (Alpha Innotech Corporation). Band intensities were quantitated using the alpha image spot densitometer function. Background and no-antibody readings were subtracted from all those for samples.
RESULTS
Gcn5p HAT activity is required for ARG1 repression.
The genome-wide analysis of Holstege et al. (37) indicated that Gcn5p was involved directly or indirectly in the regulation of approximately 5% of all yeast genes. Approximately one-third of these genes were negatively regulated by Gcn5p. ARG1 is of particular interest because its expression was enhanced 8.2-fold in the absence of GCN5, suggesting that acetylation may play a key role in the repression of this promoter. An ARG1-lacZ fusion which included sequences from bp −614 to +213 relative to the transcriptional start site was constructed (Fig. 1). This fragment includes all of the cis-acting elements known to influence expression of the promoter (15). ARG1-lacZ on a centromeric plasmid was introduced into the yeast strains used in the analysis by Holstege et al. (37), FY86 and the isogenic gcn5 disruption strain FY1370 (kindly provided by Fred Winston) (61). Cultures were grown in YPD medium for analysis in β-galactosidase assays. As shown in Table 2 there was a 5.6-fold increase in expression of ARG1-lacZ in the gcn5 deletion background, consistent with the results of Holstege et al. (37). To verify that the derepression due to the gcn5 disruption was a result of a loss of acetyltransferase activity, a variant of Gcn5p containing an E173Q mutation in the HAT domain (72, 80) was introduced into FY1370 on a centromeric plasmid and compared to a similarly introduced wild-type Gcn5p (both Myc epitope tagged). The wild-type Gcn5p reduced expression of ARG1-lacZ to levels seen in FY86 (GCN5). In contrast, the strain with the E173Q mutation showed ARG1-lacZ expression similar to that seen in the null background (Table 2). Since Gcn5pE173Q is expressed at levels comparable to those in the wild-type (80), we conclude that acetylation by Gcn5p is required for the full repression of ARG1 transcription when cells are grown in rich medium.
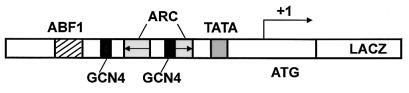
FIG. 1.
The ARG1 promoter. An ARG1 promoter region encompassing positions −640 to +213 (relative to the principal transcriptional start site [15]) was cloned as a BamHI-HindIII fragment into the LEU2 centromeric plasmid YCp87-lacZ. cis elements are shown as follows: TATA element (grey) centered at bp −73 relative to the start site of transcription, ARC elements (arrows) at bp −197 to −175 and −236 to −214, Gcn4p binding sites (black) at bp −201 to −195 and −272 to −265), and the Abf1p consensus sequence (hatched) at bp −314 to −302.
TABLE 2.
Gcn5p is required for repression of ARG1 in rich mediuma
| Strain | GCN5 plasmid | β-Galactosidase activity
|
|
|---|---|---|---|
| LacZ unitsb | Ratio (FY1370/FY86) | ||
| FY86 | None | 7.9 ± 0.4 | |
| FY1370 | None | 44.2 ± 0.6 | 5.6 |
| FY1370 | Myc-Gcn5p | 12.6 ± 0.3 | 1.6 |
| FY1370 | Myc-Gcn5pE173Q | 43.1 ± 0.6 | 5.5 |
Saturated cultures of FY86 (wild type), FY1370 (gcn5), FY1370 expressing Myc-GCN5, and FY1370 expressing Myc-GCN5E173Q, each containing YCp87-ARG1-lacZ, were grown in minimal medium and then diluted 1/100 into YPD and grown to an OD600 of approximately 1.5.
β-Galactosidase activity was determined using ONPG as the substrate, standardizing to cell density. Numbers represent the means and standard errors of the means from 10 independent experiments.
Disruption of GCN5 leads to increased binding of TBP to the ARG1 promoter.
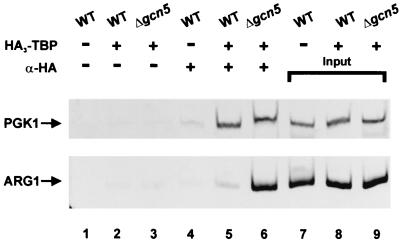
The increased expression of ARG1-lacZ in the GCN5 deletion background was fully consistent with the gene array data of Holstege et al. (37) yet could have arisen from enhanced stability of the mRNA in the absence of Gcn5p. To support a direct involvement of Gcn5p in the transcription of ARG1, we analyzed binding of TBP to the ARG1 promoter in the presence and absence of Gcn5p. Yeast strains FY86 (GCN5) and strain FY1370 (gcn5) were transformed with a centromeric plasmid expressing HA3 epitope-tagged TBP, grown in YPD medium and assayed by ChIP for the binding of TBP at the ARG1 promoter. As shown in Fig. 2, ChIP assays indicated that there was enhanced TBP binding to the ARG1 promoter in the gcn5 deletion background but not to the PGK1 promoter (Fig. 2; compare lanes 5 and 6). Densitometric analysis of three independent experiments indicated a 3.8-fold increase in TBP binding to ARG1 in the gcn5 disruption strain. This result supports the view that Gcn5p is required for the transcriptional repression of the ARG1 promoter and suggests that this repression is at a stage prior to or involving the binding of TBP.
FIG. 2.
Increased TBP binding at the ARG1 promoter in the absence of Gcn5p. Yeast strains FY86 (wild type [WT]), FY86 expressing HA3TBP, and FY1370 (gcn5) expressing HA3TBP were grown in 150 ml of rich medium and cross-linked with 1% formaldehyde. Extracts from these strains were subjected to mock immunoprecipitation (lanes 1 to 3) and immunoprecipitation with anti-HA (α-HA) antibody (lanes 4 to 6). Immunoprecipitated DNA and input DNA (lanes 7 to 9) were analyzed by PCR using primers specific to ARG1 and the PGK1 promoter (see Materials and Methods). Purified extended products were analyzed on 6% native polyacrylamide gels and stained with ethidium bromide. The data are representative of those from a minimum of three independent whole-cell extracts and ChIP assays.
Components of the SAGA complex are required for the repression of ARG1 in rich medium.
Gcn5p is part of the two multisubunit complexes ADA and SAGA. Components of these complexes enhance the ability of Gcn5p to acetylate nucleosomal histones (29, 40, 64, 66). The smaller ADA complex shares components with SAGA (Ada2p, Ngg1p/Ada3p, and Gcn5p/Ada4p), with the exception of the more recently described Ahc1p (23). SAGA contains the SPT proteins (Spt3p, Spt7p, and Spt20p/Ada5p) and Ada1p (40, 81). Also found within SAGA are a subset of the TBP-associated factors (30) and the 430-kDa Tra1p (31, 66). To determine the relationship between ARG1 repression and the different Gcn5p-containing complexes, we analyzed ARG1-lacZ expression in strains disrupted for genes encoding ADA and SAGA complex components. This was of particular interest since Chen et al. (13) recently reported that deletion of AHC1 results in enhanced transcription mediated by a unique Gal4p-σ54 fusion protein. This suggested that the ADA complex may mediate transcriptional repression (13). ARG1-lacZ expression was analyzed in yeast strains BY4741 (wild type), BY17285 (gcn5), BY3534 (ngg1/ada3), BY4282 (ada2), and BY1799 (ahc1) after growth of cells in YPD medium (Fig. 3A). Loss of Gcn5p, Ngg1p/Ada3p, or Ada2p resulted in increased expression of ARG1-lacZ by 4.8-, 6.2-, and 8.1-fold, respectively. In contrast to the findings with gcn5, ada2, and ngg1/ada3, deletion of ahc1 did not result in a change in expression of ARG1-lacZ, indicating that repression at ARG1 is distinct from the Ahc1p-dependent repression seen for Gal4p-σ54 (13).
FIG. 3.
The SAGA complex is required for ARG1 repression in rich medium. (A) β-Galactosidase analysis of ARG1-lacZ expression in strains BY4741 (wild type [WT]), BY17285 (gcn5), BY3534 (ngg1/ada3), BY4282 (ada2), and BY1799 (ahc1) grown to saturation in minimal medium and then diluted 1/100 in YPD medium. Cells were harvested at an optical density at 600 nm (OD600) of 1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The data are presented as ratios of the LacZ activity of the null mutant to that of the wild type. Error bars represent standard errors of the means for five independent experiments. (B) β-Galactosidase analysis of ARG1-lacZ expression in strains BY4741 (WT), BY4742 (WT), BY4228 (spt3), BY3218 (spt7), BY12666 (spt8) grown to saturation in minimal medium and then diluted 1/100 in YPD medium. Cells were harvested at an OD600 of 1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The data are presented as ratios of the LacZ activity of the null mutant to that of the wild type. Error bars represent standard errors of the means for a minimum of five independent experiments.
SAGA-specific components were also analyzed for their effect on ARG1-lacZ expression (Fig. 3B). Disruption of either spt7 or spt8 resulted in increased expression of ARG1-lacZ (5.2- and 9.1-fold, respectively) (Fig. 3B). The effect noted upon disruption of spt3 was markedly reduced from that for disruption of the other SPT genes (1.8-fold) (Fig. 3B) but still consistent with a role for the SAGA complex in the repression of ARG1.
Repression of the ARG1 promoter by SAGA components correlates with increased acetylation of histone H3.
SAGA has been shown to acetylate nucleosomal histone H3 and, to a lesser extent, H2B. To determine if the enhanced expression of ARG1 seen upon disruption of SAGA components coincided with deacetylation of histones at the promoter, ChIP analyses were performed. FY86 (GCN5), FY1370 (gcn5), and FY1370 containing centromeric plasmids expressing Gcn5pE173Q or functional Gcn5p were grown in rich medium, and after cross-linking and chromatin fragmentation, protein-DNA complexes were immunoprecipitated with anti-AcH3. As shown in Fig. 4, there was a decrease in the level of acetylated H3 in the strain with gcn5 deleted as detected using a region encompassing positions −304 to −79 of ARG1 (compare lanes 5 and 6), which parallels the increased expression of ARG1-lacZ in the gcn5 disruption background (Table 2). Quantitation of results from six independent experiments indicated a 2.4-fold decrease in histone H3 acetylation between the wild-type and gcn5 disruption strains. This same decrease in acetylated histone H3 was not seen at a promoter unaffected by gcn5 deletion, TDH3 (37, 44). The activity of Gcn5p at ARG1 was further demonstrated by decreased histone H3 acetylation in the strain expressing the acetylase-deficient mutant (E173Q) (compare lanes 7 and 8 of Fig. 4; 2.3-fold effect as determined by densitometry), which also has a high level of lacZ expression (Table 2). These data demonstrate that histone H3 acetylation by Gcn5p correlates with repression of ARG1 transcription.
FIG. 4.
Histone H3 acetylation status of ARG1 in wild-type and gcn5 deletion strains. FY86 (wild type [WT]), FY1370 (gcn5), FY1370 expressing Myc-GCN5, and FY1370 expressing Myc-GCN5E173Q were grown overnight in 150 ml of rich medium, and cross-linked with 1% formaldehyde, followed by immunoprecipitation with no antibody (lanes 1 to 4) or anti-AcH3 (α-AcH3) (lanes 5 to 8) antibody. Immunoprecipitated DNA and input DNA (lanes 9 to 12) were analyzed by PCR using primers specific to ARG1 and the TDH3 promoter (see Materials and Methods). Purified extended products were analyzed on 8% native polyacrylamide gels and stained with ethidium bromide. The data are representative of ChIP analyses of six independent whole-cell extracts and PCRs carried out in triplicate.
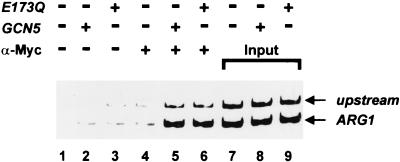
We have used ChIP analysis to determine if Gcn5p is found at the ARG1 promoter. Anti-Myc antibody was used in extracts of FY1370 (gcn5) containing Myc epitope-tagged wild-type Gcn5p or Gcn5pE173Q (Fig. 5). Two regions upstream of the ARG1 coding sequences were analyzed. The first encompassed the promoter region from position −304 to −79 relative to the start site of translation (labeled ARG1 in Fig. 5). The second was centered an additional 345 bp upstream from position −968 to −649 (labeled upstream). Gcn5p was found to associate with the ARG1 promoter region (∼7-fold above that seen in a strain lacking Gcn5p; compare lane 4 with lane 5). The ChIP assay also indicated an association between Gcn5p and the upstream region (compare lanes 4 and 6) but at a level twofold below that of the promoter region as determined by densitometry. Since the ChIP assay is not expected to fully discriminate the boundary of association, this suggests that Gcn5p is preferentially recruited to the promoter region. Interestingly, the HAT activity of Gcn5p is not required for its localization to the ARG1 promoter. The association of Gcn5pE173Q with the promoter region was equivalent to that of wild-type Gcn5p (compare lanes 5 and 6). Together these results support the view that the changes in acetylation and transcription attributed to Gcn5p at ARG1 arise from its direct action at the promoter.
FIG. 5.
Gcn5p is present at the ARG1 promoter. Yeast strain FY1370 (gcn5) containing no plasmid, Myc-GCN5, or Myc-GCN5E173Q was grown in 150 ml of rich medium and cross-linked with 1% formaldehyde. Extracts from these strains were subjected to mock immunoprecipitation (lanes 1 to 3) and immunoprecipitation with anti-Myc (α-Myc) antibody (lanes 4 to 6). Immunoprecipitated DNA and input DNA (lanes 7 to 9) were analyzed by PCR using primers specific to the ARG1 promoter (positions −304 to −79 relative to the start site of translation [ARG1]) or an upstream region (positions −968 to −649 [upstream]) (see Materials and Methods). Purified extended products were analyzed on 8% native polyacrylamide gels and stained with ethidium bromide. The data are representative of those from three ChIP analyses with independent whole-cell extracts and PCRs carried out in duplicate.
Sin3p and Rpd3 are not required for the repression of ARG1.
In many instances, the deacetylation of histones correlates with transcriptional repression. Components of the Sin3p-Rpd3p deacetylase complex have thus been implicated in transcriptional repression (43, 65). Indeed, the CAR1 and CAR2 arginine-regulated promoters are repressed by a mechanism involving Sin3p-Rpd3p (53). The involvement of Gcn5p with repression at ARG1 implied that Rpd3p-Sin3p would not have a similar role at this promoter. A recent genome-wide analysis by Bernstein et al. (6) indicated a 2- to 2.5-fold decrease in ARG1 expression upon disruption of RPD3-SIN3. We have similarly analyzed the expression of ARG1-lacZ in sin3 and rpd3 deletion backgrounds when cells are grown on YPD. ARG1-lacZ expression was down regulated 1.7-fold and 1.9-fold in BY1114 (rpd3) and BY11695 (sin3), respectively, supporting a role for Rpd3p and Sin3p in the expression of ARG1 (data not shown).
Gcn5p-dependent repression of ARG1 is dependent upon the ARC elements and ArgR/Mcm1 complex.
The ARG1 promoter has been extensively studied to identify cis-acting elements that are involved in both the activation and repression of transcription (15, 17). The ARG1 promoter contains an upstream binding site for Abf1p (bp −314 to −302) and two Gcn4p binding sites (bp −272 to −265 and −201 to −195), with the downstream site sharing 3 bp with one of two ARC elements (bp −236 to −214 and −197 to −175). The ARC elements are the binding site for the ArgR/Mcm1 regulatory complex (52). A TATA element is positioned from bp −81 to −73 relative to the start site of transcription (15) (Fig. 1).
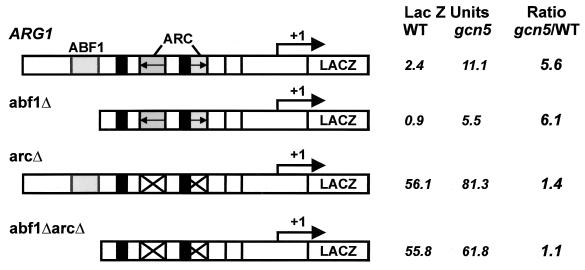
To analyze the role of Abf1p and the ArgR/Mcm1 proteins, ARG1ΔABF1-lacZ and ARG1ΔARC-lacZ, which lacked the Abf1p binding site and ARC elements, respectively, were engineered. While deletion of the Abf1p binding site (ARG1ΔABF1) resulted in a ∼2-fold decrease in the expression of the ARG1 promoter when cells were grown in rich medium, the Abf1p binding site was not required for the Gcn5p effect, since β-galactosidase activity was elevated 5.6-fold in FY1370 (gcn5) compared to FY86 (GCN5) (Fig. 6). As expected for loss of the ARC elements, expression of the ARGΔARC promoter increased relative to that of the intact promoter when cells were grown in rich medium. Interestingly, only a 1.4-fold increase in β-galactosidase activity was seen when this allele was present in the gcn5 deletion strain, suggesting that the ARC elements have a role in repression by Gcn5p. A similar 1.1-fold increase in expression was seen for ARG1ΔABF1ΔARC-lacZ, which lacks both the Abf1p binding site and ARC elements
FIG. 6.
Mapping of ARG1 promoter elements involved in Gcn5/Ada-mediated repression of ARG1. Yeast strains FY86 (wild type [WT]) and FY1370 (gcn5) transformed with YCp87-ARG1-lacZ (−614 to +213), YCp87-ARG1ΔABF1 (−302 to +213), YCp87-ARG1ΔARC (mutated for both ARC elements), or YCp87-ARG1ΔABF1ΔARC, were grown to saturation in minimal medium and then diluted 1/100 in YPD medium. Cells were harvested at an OD600 of 1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The means from three independent experiments are shown.
To verify the role of the ARC elements, we assayed the expression of ARG1-lacZ in an arg80 gcn5 double deletion background (Table 3). In this experiment deletion of gcn5 resulted in a 5.5-fold increase in ARG1-lacZ expression compared to that with wild-type GCN5 (compare FY86 with FY1370). Expression of ARG1-lacZ was increased upon deletion of arg80; however, similar to the result with ARGΔARC-lacZ, deletion of gcn5 in this arg80 background resulted in only a further 1.8-fold increase in expression (compare ARY144 with ARY146). This result confirms that arginine control proteins are required for full SAGA-mediated repression of ARG1.
TABLE 3.
SAGA-dependent ARG1 repression in gcn4 and arg80 deletion strainsa
| Strain | LacZb
|
|
|---|---|---|
| U | ΔadaADA ratio | |
| BY4741 (WT) | 8.6 ± 1.2 | 6.3 |
| BY3534 (ngg1) | 54.2 ± 3.1 | |
| BY249 (gcn4) | 3.6 ± 0.8 | 10.5 |
| ARY96 (gcn4 ngg1) | 37.8 ± 1.1 | |
| FY86 (WT) | 8.4 ± 0.5 | 5.5 |
| FY1370 (gcn5) | 46.6 ± 0.6 | |
| ARY144 (arg80) | 94.9 ± 8.2 | 1.8 |
| ARY146 (gcn5 arg80) | 176.4 ± 8.4 | |
Saturated cultures of BY4741 (wild type [WT]), BY3534 (ngg1), BY249 (gcn4), and ARY96 (gcn4ngg1) containing YCp87-ARG1-lacZ were grown in minimal medium and then diluted 1/100 in YPD and grown to an OD600 of approximately 1.5.
β-Galactosidase activity was determined using ONPG as the substrate and standardizing to cell density. The means and standard errors of the means from five independent experiments are shown.
To determine if Gcn4p was required for SAGA-mediated repression, we assayed ARG1-lacZ expression in a gcn4 ngg1/ada3 double deletion background. Deletion of ngg1/ada3 results in a 6.3-fold increase in ARG1-lacZ expression compared to that with wild-type NGG1/ADA3 (Table 3). Expression was slightly reduced in the gcn4 deletion background (BY249 gcn4), but the strain with disruptions of both gcn4 and ngg1/ada3 showed an increase in ARG1-lacZ expression similar to that found for the ngg1 deletion strain, indicating that Gcn4p is not required for the SAGA-dependent repression.
Gcn5p/Ada proteins are required for activation of ARG1 in minimal medium.
We next examined the effects of SAGA components on the expression of ARG1 under conditions of promoter activation. FY86 (wild type), FY1370 (gcn5), BY4741 (wild type), BY4282 (ada2), BY3534 (ngg1), BY4228 (spt3), BY3218 (spt7), and BY12666 (spt8) containing the ARG1-lacZ reporter were grown in minimal medium lacking arginine, conditions in which general control is induced and arginine repression minimized. In contrast to the results in rich medium (Fig. 3; Table 2), strains lacking the Ada proteins and Spt7p had levels of ARG1-lacZ expression below those of the wild-type strains. The Ada proteins and Spt7p are therefore required for full activation of the ARG1 promoter in minimal medium (Table 4). Interestingly, disruption of spt3 and spt8 resulted in a threefold increase in ARG1-lacZ expression in minimal medium (Table 4). Spt3p and Spt8p thus serve to repress transcription in both minimal and rich media. Furthermore, this finding demonstrates a dichotomy of function between Spt3p and Spt8p and the other components of the SAGA complex.
TABLE 4.
Effect of SAGA components on ARG1 expression in minimal mediuma
| Strain | Relevant genotype | β-Galactosidase activityb
|
|
|---|---|---|---|
| LacZ units | Ratio (null/wildtype) | ||
| FY86 | Wild type | 43.96 ± 0.8 | |
| FY1370 | Δgcn5 | 18.9 ± 0.4 | 0.4 |
| BY4741 | Wild type | 30.07 ± 1.2 | |
| BY4742 | Wild type | 34.07 ± 4.1 | |
| BY4282 | Δada2 | 21.33 ± 1.3 | 0.7 |
| BY3534 | Δngg1 | 23.54 ± 3.5 | 0.8 |
| BY4228 | Δspt3 | 90.6 ± 4.3 | 3.0 |
| BY3218 | Δspt7 | 15.3 ± 1.0 | 0.5 |
| BY12666 | Δspt8 | 99.1 ± 1.8 | 2.9 |
Yeast strains FY86 (wild type), FY1370 (gcn5), BY4741 (wild type), BY4742 (wild type), BY4282 (ada2), BY3534 (ngg1/ada3), BY4228 (spt3), BY3248 (spt7), and BY12666 (spt8) containing YCp87-ARG1-lacZ were grow in minimal medium to an OD600 of approximately 1.5.
β-Galactosidase activity was determined using ONPG as the substrate and standardizing to cell density. Means and standard errors of the means for five independent experiments are indicated.
DISCUSSION
Posttranslational modification of histones represents an important mechanism by which the interaction of histones and DNA can be regulated to allow specific changes in gene expression. The acetylation and deacetylation of core histones have been extensively investigated, with a general pattern of acetylation correlating with actively transcribed chromatin (35, 85). There is, however, a growing body of evidence to suggest that for any individual gene, more complex relationships between transcription and acetylation exist (3, 10, 19, 50, 54, 57, 59, 68, 71). In the present work, we show that the HAT activity of the SAGA complex is required for the repression of the ARG1 gene in rich medium. Histone acetylation results in decreased expression of ARG1 in a manner that is likely shared with the approximately 80 yeast genes whose expression is elevated in the absence of Gcn5p (37). Furthermore, we show that the role of SAGA in the regulation of ARG1 is dynamic, with a subset of the components of SAGA required for the activation of the promoter in minimal medium.
Acetylation by Gcn5p is required for the repression of ARG1 in rich medium.
Through the analysis of an ARG1-lacZ promoter fusion, we have found that Gcn5p is required for the repression of ARG1 when cells are grown in YPD medium. The fold increase in expression we observed for the lacZ fusion is consistent with the ∼8-fold effect observed in the gene array analysis by Holstege et al. (37) and with the increased recruitment of TBP upon disruption of gcn5 (Fig. 2). The repression due to Gcn5p is largely dependent on its HAT activity, since an E173Q mutation, which almost completely abolishes HAT activity (72, 80), showed a corresponding decrease in its ability to repress ARG1. ChIP assays revealed a decrease in H3 acetylation in the absence of gcn5 and the acetylase-deficient Gcn5pE173Q mutant. While we cannot exclude the possibility that other relevant substrates for acetylation by Gcn5p exist, these results are fully consistent with the view that acetylation of histone H3 at the ARG1 promoter contributes to its repression in rich medium. Also in agreement with acetylation leading to repression is the finding that deletion of the deacetylase components Rpd3p and Sin3p leads to hyperrepression of ARG1.
Regulation of ARG1 expression by the SAGA complex.
Gcn5p is found in the ADA and SAGA complexes. Compositional differences between the complexes have been established (23, 32), which has provided the opportunity to define distinct functions for SAGA. We have shown that repression of ARG1 in rich medium requires the SAGA components Ada2p, Ngg1p/Ada3p, Spt7p, Spt8p, and, to a lesser extent, Spt3p. The Ada complex-specific Ahc1p did not have an effect on repression. This suggests that the SAGA complex is the active factor in the repression of ARG1. With the exception of Spt3p and Spt8, these same components are required for the activation of ARG1 in minimal medium (Table 4). Activation in minimal medium, like repression in rich medium, correlated with increased acetylation of histone H3 at the promoter (data not shown) (44).
Recruitment of SAGA to the ARG1 promoter.
Our ability to locate Myc-tagged Gcn5p at the ARG1 promoter by ChIP supports its direct modification of promoter bound nucleosomes. The ARG1 promoter contains binding sites for the general factor Abf1p, the activator protein Gcn4p, and the ArgR/Mcm1 regulatory complex that potentially could be involved in SAGA function. Through analysis of promoter derivatives, we found that the ARC elements are required for ARG1 to be fully sensitive to Gcn5p-mediated repression in rich medium (Fig. 6). Furthermore, a strain with the ArgR/Mcm1 component Arg80p deleted showed correspondingly reduced levels of Gcn5p-mediated repression, thus confirming the role of this complex.
Although we have not demonstrated a direct dependence or interaction between SAGA and the ArgR/Mcm1 complex, the simplest model to explain the requirement for ArgR/Mcm1 and the localization of Gcn5p at the ARG1 promoter is that ArgR/Mcm1 is capable of recruiting SAGA. This would be similar to the case for other yeast regulatory factors that interact with the SAGA complex, such as Gal4p (7, 46), VP16 (11, 76), and Gcn4p (55, 74). In agreement with a recruitment model is the finding that Gcn5p-dependent histone H3 acetylation occurs preferentially in the promoter region compared to upstream sequences (data not shown). Currently, we cannot exclude an alternative model whereby the action of SAGA facilitates the binding of ArgR/Mcm1 in a promoter-specific fashion.
The ArgR/Mcm1 complex is required for the repression of the arginine biosynthetic genes and the induction of the arginine catabolic genes. The general role for histone acetylation in the function of ArgR/Mcm1p shows a high degree of promoter variability. For example, neither of the promoters for the arginine biosynthetic genes ARG5,6 (1.9-fold increase) or ARG8 (no change) is as greatly affected by loss of Gcn5p as is ARG1 (37). Expression of the catabolic genes CAR1 and CAR2 shows a 1.2-fold and 2.1-fold dependency on Gcn5p, respectively (37). The explanation of how acetylation differentially affects these promoters probably reflects the specific positioning and spacing of promoter elements. At ARG1, rotational or translational repositioning of nucleosomes, caused by histone deacetylation, may impart a repressive state on the promoter by sterically blocking recruitment of the basal transcriptional machinery or other regulatory proteins. It should be noted that repression by the ArgR/Mcm1complex also involves mechanisms in addition to those requiring the SAGA complex, since removal of ArgR/Mcm1 results in a greater relief of repression than that seen upon removal of the SAGA components.
A role for chromatin structure in the regulation of ARG1 has also been suggested by our recent finding that the E2 ubiquitin conjugase Rad6p is required for repression of ARG1 (73). Rad6p ubiquitinates histone H2B in vivo (63), and H2B is the probable target for Rad6p at ARG1 (73). Interestingly, analysis of a rad6 ada2 double deletion strain indicates that SAGA components and Rad6p are acting in a common pathway (73). This common regulation may involve signaling between the two chromatin modifications or perhaps the requirement for both modifications to establish the repressive chromatin structure.
In the absence of the ArgR/Mcm1 complex, the ARG1 promoter does retain a small degree of sensitivity to the action of the SAGA complex in rich medium (ranging from 1.1- to 1.8-fold repression in three experiments). This may result from nontargeted acetylation of the ARG1 promoter by SAGA. Nontargeted acetylation would be consistent with the results of Vogelauer et al. (77), who found that histone acetylation by Gcn5p is dispersed throughout coding and noncoding regions. Kuo et al., (44) suggested that Gcn5p is capable of acetylating the yeast genome in a random fashion but that upon recruitment by specific activators there can be a further promoter-selective targeted hyperacetylation. Consideration of the exact contribution of any one HAT complex to the pattern of global acetylation is complicated by the possibility of redundant functions, for example, histone H3 acetylation by both SAGA and the SAS acetyltransferase (41).
It is intriguing that SAGA can lead to either repression or activation at ARG1. While nucleosome acetylation contributes to the reduced expression of ARG1 in rich medium, it is also required for the response of the promoter to activating conditions. This is consistent with the suggestion of Vogelauer et al. (77) that a base level of acetylation creates a condition whereby a gene can be turned on and off rapidly. As noted by Crabeel et al. (15), the change from activation to repression of ARG1 in large part reflects the recruitment of Gcn4p to the promoter. Gcn4p induction of ARG1, like that of other Gcn4p-activated promoters, would thus seem to require Gcn5p function (28, 44).
Spt8p in ARG1 regulation.
It is interesting that Spt8p had the most dramatic effect on ARG1 in rich medium and, with Spt3p, a distinct effect in minimal medium. Belotserkovskaya et al. (4) found that disruption of spt8 led to depression of Gcn4p-activated promoters, in contrast to the effects seen for Spt7p and Gcn5p. Spt8p is involved with Spt3p in the interaction between SAGA and TBP (4, 69) and, unlike other Ada and Spt proteins, has not been found to influence the HAT activity of Gcn5p. Our results thus suggest that repression of ARG1 by SAGA may be a multistep process, involving the HAT activity of Gcn5p and perhaps a subsequent step involving the action of Spt8p.
Acknowledgments
We thank Ilona Skerjanc, David Litchfield, George Chaconas, Carol Hannam, and Suzanne Turner for helpful comments on the manuscript. Strains FY86 and FY1370 were kindly provided by Fred Winston.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to C.J.B. A.R.R was supported by a National Sciences and Engineering Research Council of Canada (NSERC) studentship.
REFERENCES
- 1.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Protocols in molecular biology, p. 13.6.2-13.6.4. John Wiley & Sons, New York, N.Y.
- 2.Barberis, A., and L. Gaudreau. 1998. Recruitment of the RNA polymerase II holoenzyme and its implications in gene regulation. Biol. Chem. 379:1397-1405. [DOI] [PubMed] [Google Scholar]
- 3.Bartsch, J., M. Truss, J. Bode, and M. Beato. 1996. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc. Natl. Acad. Sci. USA 93:10741-10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl, C. J., A. M. Furlanetto, J. A. Martens, and K. S. Hamilton. 1993. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 12:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandl, C. J., J. A. Martens, A. Margaliot, D. Stenning, A. M. Furlanetto, A. Saleh, K. S. Hamilton, and J. Genereaux. 1996. Structure/functional properties of the yeast dual regulator protein NGG1 that are required for glucose repression. J. Biol. Chem. 271:9298-9306. [DOI] [PubMed] [Google Scholar]
- 10.Bresnick, E. H., S. John, D. S. Berard, P. LeFebvre, and G. L. Hager. 1990. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc. Natl. Acad. Sci. USA 87:3977-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 12.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 13.Chen, B. S., Z. W. Sun, and M. Hampsey. 2001. A Gal4-sigma 54 hybrid protein that functions as a potent activator of RNA polymerase II transcription in yeast. J. Biol. Chem. 276:23881-23887. [DOI] [PubMed] [Google Scholar]
- 14.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 15.Crabeel, M., M. de Rijcke, S. Seneca, H. Heimberg, I. Pfeiffer, and A. Matisova. 1995. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast 11:1367-1380. [DOI] [PubMed] [Google Scholar]
- 16.Crabeel, M., R. Lavalle, and N. Glansdorff. 1990. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol. Cell. Biol. 10:1226-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crabeel, M., S. Seneca, K. Devos, and N. Glansdorff. 1988. Arginine repression of the Saccharomyces cerevisiae ARG1 gene. Comparison of the ARG1 and ARG3 control regions. Curr. Genet. 13:113-124. [DOI] [PubMed] [Google Scholar]
- 18.Crane-Robinson, C., T. R. Hebbes, A. L. Clayton, and A. W. Thorne. 1997. Chromosomal mapping of core histone acetylation by immunoselection. Methods 12:48-56. [DOI] [PubMed] [Google Scholar]
- 19.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delforge, J., F. Messenguy, and J. M. Wiame. 1975. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR− mutations and the general control of amino-acid biosynthesis. Eur. J. Biochem. 57:231-239. [DOI] [PubMed] [Google Scholar]
- 21.Dubois, E., and F. Messenguy. 1991. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol. Cell. Biol. 11:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubois, E., and F. Messenguy. 1997. Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p, and Ume6p. Mol. Gen. Genet. 253:568-580. [DOI] [PubMed] [Google Scholar]
- 23.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates 3rd, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenmann, D. M., C. Chapon, S. M. Roberts, C. Dollard, and F. Winston. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenfeld, G. 1992. Chromatin as an essential part of the transcriptional mechanism. Nature 355:219-224. [DOI] [PubMed] [Google Scholar]
- 27.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgakopoulos, T., and G. Thireos. 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 30.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates 3rd, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 31.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates 3rd, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 32.Grant, P. A., D. E. Sterner, L. J. Duggan, J. L. Workman, and S. L. Berger. 1998. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8:193-197. [DOI] [PubMed] [Google Scholar]
- 33.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 34.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 37.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 38.Hope, I. A., and K. Struhl. 1986. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46:885-894. [DOI] [PubMed] [Google Scholar]
- 39.Horiuchi, J., N. Silverman, G. A. Marcus, and L. Guarente. 1995. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horiuchi, J., N. Silverman, B. Pina, G. A. Marcus, and L. Guarente. 1997. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol. Cell. Biol. 17:3220-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs, P., J. C. Jauniaux, and M. Grenson. 1980. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J. Mol. Biol. 139:691-704. [DOI] [PubMed] [Google Scholar]
- 43.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo, M. H., E. von Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 45.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcus, G. A., J. Horiuchi, N. Silverman, and L. Guarente. 1996. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol. Cell. Biol. 16:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martens, J. A., J. Genereaux, A. Saleh, and C. J. Brandl. 1996. Transcriptional activation by yeast PDR1p is inhibited by its association with NGG1p/ADA3p. J. Biol. Chem. 271:15884-15890. [DOI] [PubMed] [Google Scholar]
- 50.McKnight, G. S., L. Hager, and R. D. Palmiter. 1980. Butyrate and related inhibitors of histone deacetylation block the induction of egg white genes by steroid hormones. Cell 22:469-477. [DOI] [PubMed] [Google Scholar]
- 51.Messenguy, F. 1976. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J. Bacteriol. 128:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messenguy, F., F. Vierendeels, B. Scherens, and E. Dubois. 2000. In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J. Bacteriol. 182:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montecino, M., B. Frenkel, A. J. van Wijnen, J. B. Lian, G. S. Stein, and J. L. Stein. 1999. Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry 38:1338-1345. [DOI] [PubMed] [Google Scholar]
- 55.Natarajan, K., B. M. Jackson, E. Rhee, and A. G. Hinnebusch. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2:683-692. [DOI] [PubMed] [Google Scholar]
- 56.O'Neill, L. P., and B. M. Turner. 1995. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14:3946-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ormandy, C. J., A. de Fazio, P. A. Kelly, and R. L. Sutherland. 1992. Transcriptional regulation of prolactin receptor gene expression by sodium butyrate in MCF-7 human breast cancer cells. Endocrinology 131:982-984. [DOI] [PubMed] [Google Scholar]
- 58.Pina, B., S. Berger, G. A. Marcus, N. Silverman, J. Agapite, and L. Guarente. 1993. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol. Cell. Biol. 13:5981-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plesko, M. M., J. L. Hargrove, D. K. Granner, and R. Chalkley. 1983. Inhibition by sodium butyrate of enzyme induction by glucocorticoids and dibutyryl cyclic AMP. A role for the rapid form of histone acetylation. J. Biol. Chem. 258:13738-13744. [PubMed] [Google Scholar]
- 60.Qui, H. F., E. Dubois, and F. Messenguy. 1991. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol. Cell. Biol. 11:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 64.Ruiz-Garcia, A. B., R. Sendra, M. Pamblanco, and V. Tordera. 1997. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 403:186-190. [DOI] [PubMed] [Google Scholar]
- 65.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 66.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 67.Saleh, A., D. Schieltz, N. Ting, S. B. McMahon, D. W. Litchfield, J. R. Yates 3rd, S. P. Lees-Miller, M. D. Cole, and C. J. Brandl. 1998. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J. Biol. Chem. 273:26559-26565. [DOI] [PubMed] [Google Scholar]
- 68.Sheldon, L. A., M. Becker, and C. L. Smith. 2001. Steroid hormone receptor-mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J. Biol. Chem. 11:11.. [DOI] [PubMed] [Google Scholar]
- 69.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi, I., H. Miyaji, T. Yoshida, S. Sato, and T. Mizukami. 1996. Selective inhibition of IL-2 gene expression by trichostatin A, a potent inhibitor of mammalian histone deacetylase. J. Antibiot. (Tokyo) 49:453-457. [DOI] [PubMed] [Google Scholar]
- 72.Trievel, R. C., J. R. Rojas, D. E. Sterner, R. N. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, and R. Marmorstein. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96:8931-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turner, S. D., A. R. Ricci, H. Petropoulos, J. Genereaux, I. S. Skerjanc, and C. J. Brandl. 2002. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22:4011-4019. [DOI] [PMC free article] [PubMed]
- 74.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 75.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 76.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 78.Wade, P. A., D. Pruss, and A. P. Wolffe. 1997. Histone acetylation: chromatin in action. Trends Biochem. Sci. 22:128-132. [DOI] [PubMed] [Google Scholar]
- 79.Wade, P. A., and A. P. Wolffe. 1997. Histone acetyltransferases in control. Curr. Biol. 7:R82-R84. [DOI] [PubMed]
- 80.Wang, L., L. Liu, and S. L. Berger. 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12:640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 82.Winston, F., K. J. Durbin, and G. R. Fink. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675-682. [DOI] [PubMed] [Google Scholar]
- 83.Wolffe, A. P. 1994. Transcription: in tune with the histones. Cell 77:13-16. [DOI] [PubMed] [Google Scholar]
- 84.Wolffe, A. P., J. Wong, and D. Pruss. 1997. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells 2:291-302. [DOI] [PubMed] [Google Scholar]
- 85.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]