Abstract
Transcription of the Saccharomyces cerevisiae ARG1 gene is under the control of both positive and negative elements. Activation of the gene in minimal medium is induced by Gcn4. Repression occurs in the presence of arginine and requires the ArgR/Mcm1 complex that binds to two upstream arginine control (ARC) elements. With the recent finding that the E2 ubiquitin conjugase Rad6 modifies histone H2B, we examined the role of Rad6 in the regulation of ARG1 transcription. We find that Rad6 is required for repression of ARG1 in rich medium, with expression increased ∼10-fold in a rad6 null background. Chromatin immunoprecipitation analysis indicates increased binding of TATA-binding protein in the absence of Rad6. The active-site cysteine of Rad6 is required for repression, implicating ubiquitination in the process. The effects of Rad6 at ARG1 involve two components. In one of these, histone H2B is the likely target for ubiquitination by Rad6, since a strain expressing histone H2B with the principal ubiquitination site converted from lysine to arginine shows a fivefold relief of repression. The second component requires Ubr1 and thus likely the pathway of N-end rule degradation. Through the analysis of promoter constructs with ARC deleted and an arg80 rad6 double mutant, we show that Rad6 repression is mediated through the ArgR/Mcm1 complex. In addition, analysis of an ada2 rad6 deletion strain indicated that the SAGA acetyltransferase complex and Rad6 act in the same pathway to repress ARG1 in rich medium.
In response to the role of Rad6/Ubc2 as an E2 ubiquitin conjugase, mutations in its gene affect multiple cellular processes. Rad6 acts with Rad18 in pathways of DNA repair (3-5, 46) and with the E3 ubiquitin ligase Ubr1 in the pathway leading to the degradation of multiubiquitinated protein substrates via the 26S proteasome (22, 48, 69). Independently of Rad18 and Ubr1, Rad6 is required for transcriptional silencing at telomeres and the HM loci (34). The ability of Rad6 to ubiquitinate histones H2A, H2B, and H3 in vitro (29, 30, 39) and H2B in vivo (59) has led to the suggestion that its ability to regulate gene expression results from changes in chromatin structure. This idea is supported by findings that disruption of rad6 results in changes in the sites of integration of retrotransposons (47, 56) and that a strain with a K123R mutation in the principal ubiquitination site of histone H2B has the same sporulation defect as a strain with rad6 deleted (59).
The ARG1 promoter provides a valuable system to study the role of factors involved in the activation and repression of transcription. ARG1 encodes argininosuccinate synthetase, which is required in a pathway that also includes ARG2, ARG5,6, ARG8, ARG3, and ARG4 for the biosynthesis of arginine. Transcription of this group of genes is subject to general amino acid control mediated by the activator protein Gcn4 (14, 20, 32). ARG1, as well as ARG5,6, ARG8, and ARG3, is also subject to repression by arginine (10, 14, 16, 20, 38, 50). Arginine repression requires a DNA binding complex of ArgR proteins, Arg80/ArgRI and Arg81/ArgRII, as well as Mcm1 (1, 8, 23-26, 51, 52, 57, 58). Interestingly, this same ArgR/Mcm1 complex is required for induction of CAR1 and CAR2, which are required for arginine catabolism (43, 52, 67). For both activation and repression, the ArgR/Mcm1 complex binds to upstream arginine control (ARC) elements (10, 17, 21, 24, 51). ARG1 contains two ARC elements, centered at −185 and −225 relative to the start site of transcription, that contribute to the arginine-specific repression.
The genomewide analysis of Holstege et al. (33) has shown that ARG1 regulation requires the histone acetyltransferase Gcn5. When grown in rich medium, cells with gcn5 deleted had an ∼8-fold increase in expression of ARG1 (33). We have recently found that repression of ARG1 in rich medium requires multiple components of the SAGA acetyltransferase complex and that this repression correlates with increased acetylation of histone H3 (58a). In the present work, we show that repression of ARG1 in rich medium also requires Rad6. Repression by Rad6 depends upon the active-site cysteine. Rad6-dependent repression is mediated by the ArgR/Mcm1 complex and acts through a pathway common to the SAGA components. Histone H2B is a likely target of Rad6 ubiquitination, since a K123R mutation that abolishes the principal ubiquitination site in H2B acts in the same pathway to regulate expression of ARG1. Unlike silencing at telomeres and the HM loci (34), a component of Rad6 repression at ARG1 also involves the E3 ubiquitin ligase Ubr1.
MATERIALS AND METHODS
Yeast strains.
All yeast strains were derivatives of BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (71) and were purchased from Research Genetics. They include BY4282 (ada2Δ0), BY4425 (rad6Δ0), BY618 (arg80Δ0), BY15787 (rad18Δ0), BY14814 (ubr1Δ0), BY249 (gcn4Δ0), BY13026 (htb2Δ0), and BY16148 (ubp3Δ0). URA3 disruptions of rad6 were made using a disruption allele synthesized by PCR with the oligonucleotides 5′-AAGATTATTTTTAGGCAGACAGAGACTAAAAGATAAAGCGTCATGAAGCTTTTCAATTCA-3′ and 5′-ATATCGGCTCGGCATTCATCATTAAGATTCTTTTGATTTTTCTCACCGAGATTCCCGGGTAATA-3′. This allele was integrated into BY249, BY618, BY14814, and BY4282 to generate the double-deletion strains CY1214 (gcn4 rad6), CY1251 (arg80 rad6), CY1304 (ubr1 rad6), and CY1215 (ada2 rad6), respectively. Strains CY1272 and CY1256 are derivatives of the htb2Δ0 strain (BY13026) which contain HIS3 insertions directly downstream of the htb1 allele. They were constructed by double-strand gene replacement in BY13026 using EcoRI-XbaI-digested CB1469 (wild-type HTB1) and CB1474 (HTB1K123R; see below). The strains were verified by PCR using oligonucleotides flanking the relevant alleles.
DNA constructs.
LacZ reporter constructs were cloned as his3-lacZ fusions into the LEU2 centromeric plasmid YCp87 (11). The ARG1 promoter constructs contain promoter sequences from −614 (ARG1-lacZ) or −302 (ARG1ΔABF1-lacZ), relative to the transcriptional start site and 213 bp of coding region fused to the first HindIII of HIS3 (Ricci et al., submitted). ARG1-lacZΔARC is a derivative of ARG1-lacZ in which the upstream ARC elements spanning nucleotides −175 to −197 and −214 to −239 have been replaced by SalI and BamHI restriction enzyme sites, respectively, using PCR-based mutagenesis strategies (58a).
The construction of C-terminally hemagglutinin (HA)-tagged TATA-binding protein (TBP) and its insertion into YCplac33 is described elsewhere (58a). The HIS3 centromeric plasmids expressing wild-type Rad6 (HH1) and Rad6-C88A (HH4) were generously provided by Susan Liebman (34).
The HTB1 integrating allele was constructed by PCR using oligonucleotides 5′-TTGAATTCTAAAAGAATTGGAATAAAAGTAC-3′ and 5′-GCTCTAGAGAATTGGCCTTAGTAGTGG-3′ and cloned as an EcoRI-to-XbaI fragment into pTZ18. The insert contains a unique BamHI site into which was inserted a 1.8-kb fragment that contains HIS3 to give CB1469. CB1474 with K123R was engineered by site-directed mutagenesis using CB1469 as the template and oligonucleotide 5′-GGTACTAGAGCTGTTACCAGGTACTCTTCCTCTACTC-3′ and its complement (68).
β-Galactosidase assays.
For the analysis of ARG1-lacZ fusion reporters, saturated cultures grown in minimal medium were inoculated at a 1/100 dilution into yeast-peptone-dextrose (YPD) or minimal medium (supplemented with the required amino acids) and grown at 30°C to an A600 of 1.0 to 1.5. Equal stability of the plasmids in the strains was verified by cell counts on rich- and minimal-medium plates. The cells were pelleted, washed in lacZ buffer, and concentrated 5- to 10-fold. β-Galactosidase activity was determined using O-nitrophenyl-β-d-galactosidase (ONPG) as a substrate, as described by Ausubel et al. (2), and standardizing to cell density.
RNA analyses.
Saturated cultures were grown in minimal medium and diluted 1/100 in YPD. The cells were grown at 30°C to an A600 of ∼1.3 (∼107 cells/ml; 10-ml total volume), and RNA was extracted by the hot acidic phenol method as described by Ausubel et al. (2). For Northern analysis, 12 μg of total RNA was separated by agarose-formaldehyde gel electrophoresis and probed with 32P-labeled DNA fragments for ARG1 and ACT1 as described by Skerjanc et al. (63). The ARG1 and ACT1 probes were each ∼700 bp long and were constructed by PCR using oligonucleotides 5′-GTTGGGTACCTCTTTGGCAA-3′ and 5′-GCCCAGAATGATGACGTTACCC-3′ for ARG1 and 5′-ACGAATTCAGAGTTGCCCCAGAAGAAC-3′ and 5′-CCCGGATCCACATTTGTTGGAAGGTA-3′ for ACT1. Following 16 h of hybridization at 42°C, the blots were washed five times for 5 min each time at 42°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate (SDS) and for 15 min at 65°C in 0.1× SSC-0.2% SDS. The blots were exposed to film for visualization, and densitometry was carried out by PhosphorImager analysis (ImageQuant version 1.11; Molecular Dynamics). Background was subtracted, and the ACT1 intensity was used to correct for loading. Primer extension analysis with 20 ng of ARG1 primer 5′-CCTTGGCGGCATCGAAATCTTC-3′ end labeled with [32P]ATP and 25 μg of total RNA was performed as described by Martens and Brandl (49).
Chromatin immunoprecipitation (ChIP).
Chromatin was prepared as described by Hecht and Grunstein (31) with the following modifications. One hundred fifty milliliters of cells grown to an A600 of ∼1.5 was treated with 1% formaldehyde for 20 min at room temperature with occasional swirling. The cells were pelleted, washed twice in phosphate-buffered saline (140 mM NaCl, 2.5 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.5), and then suspended in 1.2 ml of ice-cold lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] sodium deoxycholate with protease inhibitors [1 mM phenylmethylsulfonylfluoride; 1 mM benzamidine; 0.5 mg of N-tosyl-l-phenylalanine chloromethylketone/ml; 0.1 mg of aprotinin/ml in 10 mM HEPES-KOH, pH 8.0; 1.0 μg of leupeptin/ml; and 1.0 μg of pepstatin/ml]). The suspension was aliquoted into 400-μl volumes in 1.5-ml microcentrifuge tubes containing equal volumes of 0.5-nm-diameter glass beads (31). Cross-linked chromatin was isolated, pooled, and fragmented (31), and the chromatin solution (400 μl) was incubated with 15 μl of ascites fluid derived from the 12CA5 cell line for 4 h at 4°C. Protein G-Sepharose (Pharmacia Biotech, Inc.) was added for an additional hour, followed by 5-min washes as follows: once in lysis buffer containing 0.5 M NaCl; once in a solution of 10 mM Tris-HCl (pH 8.0), 0.25 mM LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, and 1.0 mM EDTA; and once in 10 mM Tris-HCl (pH 8.0)-2 mM EDTA (TE). The immunoprecipitated material was eluted from the beads by heating them at 65oC for 30 min in 100 μl of TE containing 1% SDS, followed by centrifugation at 10,000 × g. Cross-links were reversed by incubation at 65oC for 12 h. DNA was extracted with phenol-chloroform and then chloroform and precipitated in 3 volumes of ethanol containing 20 μg of glycogen, 0.1 volume of 5 M LiCl, and 50 mM Tris-HCl (pH 8.0) at −20°C. The DNA was pelleted, washed with 70% ethanol, and resuspended in TE. The precipitated DNA was analyzed by quantitative PCR using the ARG1 primers 5′ ATACTATTGAGACAGTGCCAGT-3′ and 5′-ACGGCTCTCCAGTCATTTATG-3′ and the ACT1 primers 5′ CATTCTTCCTTATCGGATCCTCA-3′ and 5′GGAAGGAAGAATACAAGAGAGAGG-3′. The linear range for each primer pair was determined using decreasing amounts of template. Approximately 1/50 of the precipitated DNA and 1/3,000 of the total DNA were used in a 50-μl volume containing 50 pmol of primers, 0.2 mM deoxynucleoside triphosphates, 1× reaction buffer (Promega, Inc.), 1.5 mM MgCl2, 1 mg of glycogen/ml, and 2 U of Taq polymerase. The cycling program was 2 min at 95°C, followed by 25 cycles of 30 s at 95oC, 30 s at 55oC, and 1 min at 72 oC, and a final extension at 72oC for 5 min. The products were analyzed on a 6% native polyacrylamide gel, stained with ethidium bromide, and photographed under UV on a gel documentation system (Alpha Innotech Corp.).
RESULTS
Rad6 represses ARG1 transcription.
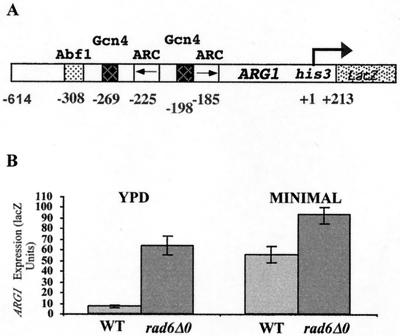
The yeast ARG1 promoter is subject to complex mechanisms of activation and repression. Our recent work has indicated that a component of this regulation requires chromatin modification mediated by the SAGA histone acetyltransferase complex (58a). Since the E2 ubiquitin-conjugating enzyme Rad6 has been implicated in transcriptional repression (34) and histone modification (59), we chose to examine its potential role in the repression of ARG1 transcription. A centromeric plasmid expressing a lacZ translational fusion containing ARG1 sequences from −614 to +213 (Fig. 1A; ARG1-lacZ) was introduced into the wild-type yeast strain BY4741 and the isogenic rad6 deletion strain, BY4425. Saturated cultures were grown in minimal medium and then diluted in rich medium (YPD). As shown in Fig. 1B, expression of ARG1-lacZ was increased 9.3-fold in the strain lacking RAD6, indicating that Rad6 acts to repress ARG1 expression in rich medium. A similar analysis was performed after cells were grown in minimal medium. As the result of Gcn4 activation and a loss of arginine repression, expression of ARG1-lacZ increased when BY4741 (RAD6) was grown in minimal medium compared to growth in YPD. However, in minimal medium the increase in expression in the rad6 deletion background was <1.7-fold, indicating that repression by Rad6 occurs predominately in rich medium.
FIG. 1.
Rad6 is required for the repression the ARG1 promoter in rich medium. (A) ARG1-lacZ reporter construct. ARG1 sequences from −640 to +213 were cloned as a BamHI-HindIII fragment into YCp87 to generate a his3-lacZ translational fusion on a LEU2 centromeric plasmid. Previously mapped regulatory sites (19) are shown as follows: ARC elements (centered at −175 and −214 relative to the principal transcriptional start site [16]), A TATA elements at −73 is not shown. Gcn4 binding sites (−195 and −265), and the Abf1 consensus sequence (−302). (B) β-Galactosidase analysis of ARG1-lacZ expression in wild-type (WT) and rad6 deletion backgrounds. Yeast strains BY4741 (wild type) and BY4425 (rad6Δ0) containing YCp87-ARG1-lacZ were grown to saturation in minimal medium and then diluted 1/100 in YPD medium or minimal medium (2% glucose supplemented with the required amino acids). The cells were harvested at an A600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean for two experiments performed in triplicate.
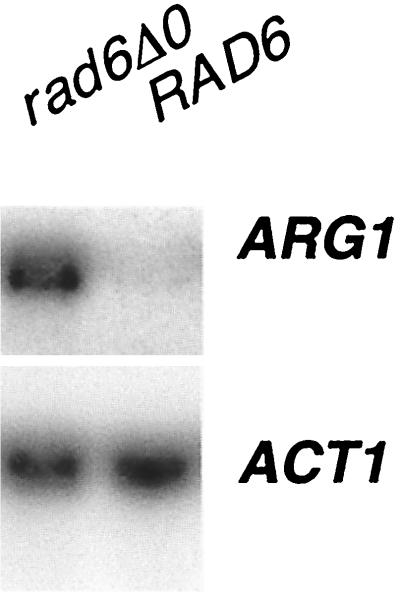
To verify that the increase in ARG1-lacZ expression was the result of an increase in RNA levels, Northern analysis was performed. BY4741 (RAD6) and BY4425 (rad6Δ0) strains were grown in rich medium. RNA was isolated from exponentially growing cells, separated by electrophoresis, and probed by Northern blotting with DNA fragments from ARG1 and ACT1 (Fig. 2). Under conditions in which ACT1 mRNA levels were unchanged, the level of ARG1 mRNA increased 10.5-fold (as determined from densitometry of three experiments). Primer extension analysis indicated that disruption of RAD6 did not result in changes to the principal mRNA start site for ARG1 (at −72 relative to the translational start site) (reference 16 and data not shown).
FIG. 2.
Northern analysis of ARG1 in a rad6 disruption strain. Yeast strains BY4741 (wild type) and BY4425 (rad6Δ0) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot shown is representative of analyses performed in triplicate.
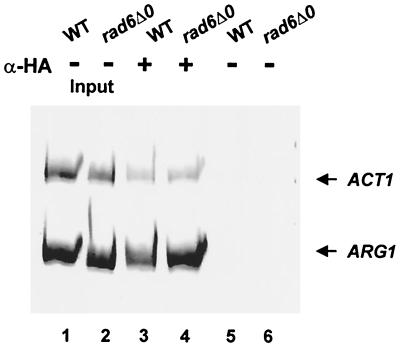
We next used a ChIP assay to determine if increased binding of TBP to the promoter paralleled the enhanced expression of ARG1. BY4741 and BY4425 (containing ARG1-lacZ) were transformed with a centromeric plasmid expressing HA epitope-tagged TBP. ChIP assays were performed on these RAD6 and rad6 extracts using anti-HA antibody. The PCR results for the ChIP analysis (Fig. 3) indicate that there was increased binding of TBP to the ARG1 promoter in the absence of Rad6. Densitometry of three independent experiments indicated that this increase was ∼2.4-fold. By comparison, the ACT1 promoter showed no change in the binding of TBP.
FIG. 3.
Rad6 inhibits the binding of TBP to the ARG1 promoter. BY4741 (wild type [WT]) and BY4425 (rad6Δ0) containing HA epitope-tagged TBP were grown in 150 ml of YPD medium and cross-linked with 1% formaldehyde followed by mock immunoprecipitation with no antibody (−; lanes 1 and 2) or immunoprecipitation with anti-HA (α-HA) antibody (+; lanes 3 and 4). Immunoprecipitated DNA and input DNA (lanes 5 and 6) were analyzed by PCR using primers specific for ARG1 and the ACT1 promoter. Linear ranges for PCR were determined by serial dilution. The purified extended products were analyzed on 6% native polyacrylamide gels and stained with ethidium bromide. The data are representative of three independent whole-cell extracts and ChIP assays.
Ubiquitin-conjugating activity of Rad6 is required for repression of ARG1 expression.
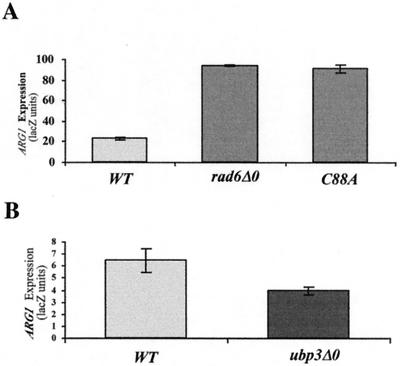
Ubiquitin conjugation by Rad6 involves the covalent attachment of ubiquitin to C88 of Rad6 through a thioester linkage (64). To determine if Rad6 represses the expression of ARG1 through a mechanism that requires ubiquitination, centromeric plasmids expressing wild-type Rad6 and Rad6 with a C88A mutation (kindly provided by Susan Liebman) were introduced into BY4425 (rad6Δ0) containing ARG1-lacZ. β-Galactosidase activity was determined after growth in rich medium. As shown in Fig. 4A, Rad6-C88A was unable to complement the null allele, suggesting that ubiquitination mediated by Rad6 is required for the repression of ARG1.
FIG. 4.
(A) Ubiquitin conjugase activity of Rad6 is required for its repression of ARG1 expression. BY4425 (rad6Δ0) containing YCp87-ARG1-lacZ and either no plasmid (rad6Δ0), a HIS3 centromeric plasmid expressing wild-type Rad6 (HH1; WT), or Rad6-C88A (HH4; C88A) (35) was grown to saturation in minimal medium and then diluted 1/100 in YPD medium. The cells were harvested at an A600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean of three samples. (B) Deletion of UBP3 results in reduced expression of ARG1 in rich medium. Yeast strains BY4741 and BY16148 (ubp3Δ0) containing ARG1-lacZ were grown, and β-galactosidase activity was determined as described above (n = 4; P = 0.04). The error bars represent the standard error of the mean.
If ubiquitin conjugation by Rad6 is required for repression of ARG1, then enhanced ubiquitination would be expected to lead to hyperrepression. The ubiquitin protease Ubp3 could act reciprocally with Rad6, removing ubiquitin moieties conjugated by Rad6. Such a role for UBP3 in the regulation of transcription is suggested by the findings that its deletion leads to hyperrepression at telomeres and HML (53) and that it is essential for growth in the absence of the transcription elongation factor TFIIS (18). We thus analyzed the expression of ARG1-lacZ in the ubp3 deletion strain, BY16148, after the growth of cells in YPD medium. As shown in Fig. 4B, deletion of ubp3 resulted in a decrease in expression of ARG1-lacZ to a level 60% of that seen in the wild-type background (P = 0.04). Although the effects of ubp3 disruption may be indirect, this result is consistent with a role for ubiquitination in the regulation of ARG1.
K123R mutations within histone H2B result in increased expression of ARG1.
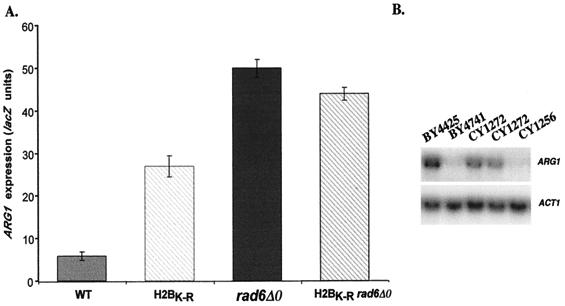
Histone H2B is a target for ubiquitination by Rad6 in vivo and in vitro (29, 30, 39, 59). Modification of histone H2B by Rad6 has been predicted to alter chromatin structure in a way that could influence transcription. To test this possibility, an allele of HTB1 containing a K123R mutation at the site of ubiquitination was introduced by gene replacement as the only cellular copy of histone H2B (yeast strain CY1272). As previously observed for the K123R mutation (59), CY1272 showed a reduced rate of growth in rich and minimal media (not shown). ARG1-lacZ expression was determined in this strain as well as the isogenic strain containing an integration of wild-type histone H2B (CY1256). As shown in Fig. 5A, H2BK123R resulted in a 4.6-fold increase in expression of ARG1. Since this was less than the 8.5-fold increase seen upon disruption of RAD6, to determine if mutations in htb1 and rad6 were acting through the same pathway, ARG1-lacZ expression was also determined in CY1284, which contains htbK-R in the rad6 disruption background. ARG1-lacZ expression in CY1284 was increased 7.5-fold compared to the wild type (CY1256). The nonadditive effects of the double deletion indicate that HTB and RAD6 are acting in the same pathway to regulate ARG1.
FIG. 5.
A K123R mutation within histone H2B results in relief of repression of the ARG1 promoter. (A) An allele of HTB1 containing a K123R mutation at the site of ubiquitination was introduced by gene replacement as the only cellular copy of histone H2B (yeast strain CY1272; H2BK-R). The isogenic strain CY1256 (WT) expressing wild-type H2B was similarly constructed. Yeast strain CY1284 (H2BK-R rad6Δ0) was constructed by disrupting rad6 in the CY1272 background. These strains, as well as BY4425 (rad6Δ0), all containing YCp87-ARG1-lacZ, were grown to saturation in minimal medium and then diluted 1/100 in YPD medium. The cells were harvested at an A600 of ∼1.5, and β-galactosidase activity was determined using ONPG as a substrate. Activities were standardized to cell density. The error bars represent the standard error of the mean of four samples. (B) Northern analysis. Yeast strains BY4425 (rad6Δ0), CY1256 (htb2Δ0), CY1272 (H2BK-R htb2Δ0), and BY4741 (wild type) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot is representative of analyses performed in triplicate. Two independent RNA preparations from CY1272 are shown.
Northern analysis was performed to ensure that the effect of htbK-R on expression in rich medium was at the level of ARG1 mRNA. As shown in Fig. 5B, htbK-R resulted in an increase in ARG1 mRNA levels (compare CY1272 to CY1256). This increase was 5.5-fold, as determined by densitometry of four experiments, compared to 10.5-fold upon disruption of RAD6 (for RAD6, compare BY4425 to BY4741). Together, these results suggest that ubiquitination of histone H2B by Rad6 plays a significant role in the repression of ARG1 in rich medium.
Ubr1 is required for a component of the Rad6-mediated repression.
The result described above indicated that approximately 5-fold of the 10-fold increase in ARG1 expression seen in the absence of rad6 was linked to histone H2B. Rad6 also interacts with Ubr1 and Rad18 for the multiubiquitination of amino-end rule proteolytic substrates and in mediating postreplication repair, respectively (3). To examine whether either of these interactions could account for the remaining Rad6-dependent repression of ARG1, expression of ARG1-lacZ was determined in BY15787 (rad18Δ0) and BY14814 (ubr1Δ0). As shown in Table 1, deletion of rad18 did not result in increased expression of ARG1-lacZ when cells were grown in YPD or in minimal medium. Disruption of ubr1 resulted in an ∼3-fold increase in expression of ARG1-lacZ when cells were grown in YPD and 2-fold for cultures grown in minimal medium. Rad6 and Ubr1 are acting in the same pathway to regulate ARG1, since the rad6 ubr1 double mutant showed the same level of expression as the disruption of rad6 alone. A component (∼25%) of the Rad6-dependent repression of ARG1 that is seen in rich medium is thus linked to Ubr1 and potentially proteolysis. This component can account for all of the Rad6-dependent repression seen in minimal medium.
TABLE 1.
ARG1-lacZ expression in rad6, rad18, and ubr1 deletion strains
| Strain | YPD medium
|
Minimal medium
|
||
|---|---|---|---|---|
| ARG1-lacZ expression | Null/WT ratio | ARG1-lacZ expression | Null/WT ratio | |
| rad6Δ0 | 80 | 12 | 149 | 1.7 |
| rad18Δ0 | 8.2 | 1.2 | 95 | 1.1 |
| ubr1Δ0 | 21 | 3.1 | 154 | 1.7 |
| ubr1Δ0 rad6Δ0 | 83 | 12 | 142 | 1.6 |
| WTa | 6.7 | 90 | ||
WT, wild type.
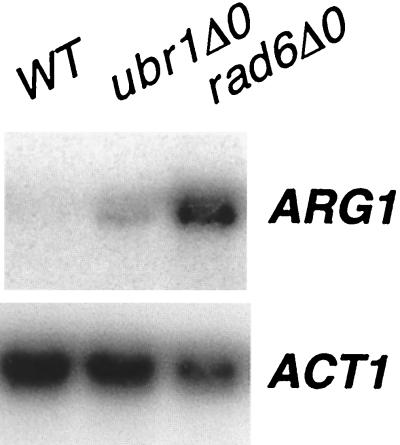
We performed a Northern analysis to verify that Ubr1 was affecting the level of ARG1 mRNA in rich medium. As shown in Fig. 6, disruption of ubr1 resulted in an increase of ARG1 mRNA (threefold, as determined by densitometric analysis of two experiments), in contrast to an ACT1 control, which was unaffected.
FIG. 6.
ARG1 expression in a UBR1 deletion strain . Yeast strains BY4741 (wild type [WT]), BY4425 (rad6Δ0), and BY14814 (ubr1Δ0) were grown to saturation in minimal medium and then diluted 1/100 in YPD. The cells were harvested, and total RNA was isolated. Twelve micrograms of total RNA was separated by electrophoresis on a 1.0% agarose formaldehyde gel and probed with 32P-labeled DNAs specific for ARG1 and ACT1. The blot was visualized by autoradiography and quantitated by PhosphorImager analysis (ImageQuant 1.11). The blot shown is representative of three experiments.
Repression by Rad6 at the ARG1 promoter requires components of arginine control.
The ARG1 promoter has been extensively analyzed to identify cis-acting elements that are involved in both the activation and repression of transcription (15, 17). As shown in Fig. 1A, the ARG1 promoter contains an upstream binding site for Abf1, two sites for Gcn4, and two sites for interaction of the ArgR/Mcm1 complex (ARC elements). Rad6 may repress transcription by influencing these trans-acting factors, or it could be recruited by these factors. To analyze the roles of Abf1 and the ArgR/Mcm1 proteins, ARG1ΔABF1-lacZ and ARG1ΔARC-lacZ, which lacked the Abf1 binding site and ARC elements, respectively, were engineered. While deletion of the Abf1 binding site (ARG1ΔABF1) resulted in a 2-fold decrease in the expression of the ARG1 promoter when cells were grown in rich medium, the Abf1 binding site was not required for the Rad6 effect, since β-galactosidase activity is elevated 12-fold in the rad6 deletion background (Fig. 6). As expected for loss of the ARC elements, expression of the ARGΔARC promoter increased relative to the intact promoter when cells were grown in rich medium. Interestingly, only a 1.8-fold increase in β-galactosidase activity was seen when this allele was present in the rad6 deletion strain. This was significantly less than the ∼12-fold increase that would be expected if Rad6 acted fully independently of the ARC elements, thus supporting the view that the ARC elements play a role in repression by Rad6. The 1.8-fold increase in expression of the ARGΔARC promoter in the rad6 deletion background can be attributed to the Ubr1 component of its repression, since twofold of the total threefold effect seen upon deletion of ubr1 is independent of the ARC elements (Fig. 7, compare ARG1 and Δarc promoters in the wild-type and ubr1Δ0 strains).
FIG. 7.
Recruitment of Rad6 to the promoter requires components of the ArgR/Mcm1 complex. Saturated cultures of BY4425 (rad6Δ0), BY14814 (ubr1Δ0), and BY4741 (wild type [WT]) containing YCp87-ARG1-lacZ, YCp87-ARG1ΔABF1-lacZ, or YCp87-ARG1ΔARC-lacZ were grown in minimal medium and then diluted 1/100 in YPD. The cultures were grown to an A600 of 1.0 to 1.5. β-Galactosidase activity was determined using ONPG as the substrate, standardizing to cell density. lacZ units represent the averages of the mean for an experiment performed in triplicate with a standard error of the mean under 10%. The ratio of lacZ expression in the null strain versus the wild-type (wt) strain is shown for each lacZ fusion construct. nd, not determined.
To verify the potential role of the ARC elements and to examine the roles of the two consensus binding sites for Gcn4, we assayed the expression of ARG1-lacZ in arg80 rad6 and gcn4 rad6 double-deletion backgrounds (Table 2). Disruption of gcn4 resulted in a decrease in expression of ARG1-lacZ relative to the wild type; however, deletion of rad6 in the gcn4 background resulted in a 10.6-fold increase in β-galactosidase activity, indicating that Gcn4 is not required for Rad6-dependent repression. Disruption of arg80 alone resulted in a significant increase in ARG1 expression compared to the wild type. Similar to the result with ARGΔARC-lacZ, deletion of rad6 in this arg80 deletion background resulted in only a 60% increase in expression. The finding that the fold increases in ARG1 expression as a result of deleting rad6 and arg80 were not additive suggests that the arginine control proteins are necessary for Rad6-mediated repression.
TABLE 2.
RAD6 repression of ARG1-lacZ in arg80 and gcn4 deletion strains
| Strain | ARG1 expression (lacZ units) | rad6Δ0/RAD6 ratio |
|---|---|---|
| rad6Δ0 | 53 | |
| 6.2 | ||
| Wild type | 8.5 | |
| arg80Δ0 rad6Δ0 | 135 | |
| 1.6 | ||
| arg80Δ0 | 86 | |
| gcn4Δ0 rad6Δ0 | 35 | |
| 10.6 | ||
| gcn4Δ0 | 3.3 |
Rad6 and Ada2 act through related pathways to repress ARG1 transcription in rich medium.
We have recently found that components of the SAGA coregulatory complex, including Gcn5, Ada2, and Ngg1/Ada3, are required for the repression of the ARG1 promoter in rich medium (58a). To determine if Rad6 and Ada-dependent repression of ARG1 are acting through a common pathway, we examined expression of ARG1-lacZ in an ada2 rad6 double-deletion strain (Table 3). In this experiment, deletion of rad6 resulted in an 11-fold increase in β-galactosidase activity when cells were grown in rich medium. This was compared with an ∼5-fold increase upon disruption of ada2 (BY4282). Expression of ARG1-lacZ in the double deletion (ada2 rad6; CY1215) was comparable (8.6-fold) to that found for the rad6 disruption. The nonadditive effects of ada2 and rad6 disruptions suggest that Rad6 and components of Ada/SAGA are acting through a shared pathway to regulate ARG1 expression.
TABLE 3.
ARG1-lacZ expression in rad6 and ada2 deletion strains
| Strain | ARG1 expression (lacZ units) | Deletion/WT ratio |
|---|---|---|
| WTa | 7.9 | 1 |
| rad6Δ0 | 87 | 11 |
| ada2Δ0 | 38 | 4.8 |
| ada2Δ0 rad6Δ0 | 68 | 8.6 |
WT, wild type.
DISCUSSION
There are a number of documented links between gene regulation and ubiquitination. Indeed, histones were the first proteins found to be ubiquitinated (27, 70), and this ubiquitination has been correlated with increased transcription (6, 19, 54). Ubiquitination of other targets has also been shown to influence transcription. These include the activation of NF-κB (40), the turnover of p53 (62), and the regulated turnover of the hypoxia-inducible factor 1α transcription factor by a ubiquitin ligase complex that includes the von Hippel-Lindau tumor suppressor protein (reviewed in reference 41). Notably, in some cases (for example, NF-κB and histones), ubiquitination does not lead directly to protein turnover. In yeast, the related hect-domain E3 ubiquitin ligases, Rsp5 and Tom1, have been implicated in transcriptional regulation. Rsp5 was identified as a suppressor of Spt3 (cited in reference 35), is required for activation by human steroid receptors in yeast (37), and modifies the carboxyl-terminal domain of the largest subunit of RNA polymerase II (7, 13, 36). Deletion of tom1 leads to transcriptional changes at the GAL10 and ADH2 promoters similar to those associated with components of the Ada complex (60). Also in agreement with the idea that targeted ubiquitination at a promoter can result in changes in transcription are the findings that UreB1, which contains a C-terminal hect-domain, was initially identified through its DNA binding activity (28), and that the TBP-associated factor TAFII250 possesses ubiquitin-activating and -conjugating activities (55). Furthermore, Salghetti et al. (61) have recently determined that the ubiquitn ligase Met30 is required for transcriptional activation by a LexA-VP16 fusion in yeast. Interestingly, a direct role for ubiquitination in the process was suggested by the fact that transcriptional activation was restored upon fusing ubiquitin to the amino terminus of LexA-VP16.
Rad6 is required for transcriptional repression of ARG1.
Previous work had shown that Rad6 was involved in transcriptional repression at telomeres and the HM loci (34). This suggested that Rad6 might play a more general role in gene regulation. We looked for possible effects of Rad6 on ARG1 expression because ARG1 is subject to distinct activation and repression mechanisms in which there is a marked requirement for gene-specific transcription factors and coactivators (15, 17, 33, 44). Analysis of ARG1-lacZ fusions revealed that Rad6 was required for the repression of ARG1 in rich medium. ARG1-lacZ expression was increased ∼10-fold when a rad6-disrupted strain was grown in YPD medium. Initially, we had concerns that a component of the increase in β-galactosidase levels in rad6 strains might result from reduced turnover of the reporter protein because of the involvement of Rad6 in the proteosomal degradation of proteins (22, 48, 69). However, Northern analysis confirmed that the increase was due to alterations in mRNA levels.
Histone H2B as a target for Rad6 in transcriptional regulation.
The work of Robzyk et al. (59) has shown that ubiquitinated histone H2B is not found in strains with rad6 disrupted and that a lysine-to-arginine change at the principal site of ubiquitination (K123R) within H2B confers defects in mitotic cell growth and meiosis similar to those caused by disruption of rad6. We have now shown that histone H2B is a likely target for Rad6 at the ARG1 promoter. A K123R mutation in H2B results in elevated levels of ARG1 expression. Through the analysis of a strain carrying htbK-R in the rad6 disruption background, we confirmed that Rad6 and histone H2B were acting in the same pathway to regulate expression of ARG1. Ubiquitination of histone H2B provides an obvious potential mechanism for Rad6 regulation of ARG1 expression through the modification of chromatin structure. How ubiquitination of H2B affects nucleosomal structure and function is unclear, but it is not likely to be related to direct turnover of histones.
As well as Rad6, the SAGA component proteins (including Ada2, Ngg1/Ada3, Gcn5, Spt7, and Spt8) are required for the full repression of ARG1 in rich medium (33, 58a). Double disruptions indicate that Rad6 and Ada2 are acting through a common pathway. Furthermore, disruption of gcn5 and rad6 both result in increased promoter binding of TBP, and repression by Rad6 and Ada2 require the ArgR/Mcm1 complex (58a). It is attractive to propose that SAGA and Rad6 are acting together to create a repressive chromatin structure or that one of the nucleosome modifications acts as the signaling event allowing the second chromatin modification to occur. This model is consistent with the finding that the effect of a K123R mutation within histone H2B on the expression of ARG1 closely parallels the effect of disrupting ada2. While our experiments do not address the order of events that facilitate repression, the ability of gene-specific regulators to interact with SAGA (9, 12, 45, 65, 66) suggests that targeting of SAGA by ArgR/Mcm1 and the resulting histone acetylation may initiate the process. Rad6 may then recognize acetylated nucleosomes and ubiquitinate histone H2B. Transcriptional repression could result from the rotational or translational repositioning of nucleosomes after their modification such that recruitment of the basal transcriptional machinery is sterically inhibited. We cannot exclude alternative models in which the initial ubiquitination of histone H2B facilitates the binding of the ArgR/Mcm1 complex or the preferential acetylation of nucleosomes by SAGA, although these mechanism do not provide an obvious means for promoter targeting.
The ARG1 promoter is subject to activation through the general control pathway involving Gcn4 and arginine repression involving the ArgR/Mcm1 regulatory complex. Kornitzer et al. (42) have shown that Gcn4 is subject to turnover by the ubiquitin pathway. This raised the possibility that the increase in ARG1 expression seen in the absence of Rad6 was due to increased levels of Gcn4-dependent activation. However, this is not the case, since increased ARG1 expression was observed in a strain with gcn4 deleted. Together with the finding that Rad6-dependent repression requires the ArgR/Mcm1 complex, this result implies that deletion of Rad6 leads to derepression of ARG1, not to enhanced activation. Based upon findings that expression of the ArgR/Mcm1-activated gene CAR1 is not stimulated in the absence of Rad6 (data not shown), an alternative view that Rad6 is required to convert ArgR/Mcm1 from an activator to a repressor seems unlikely.
A component of the Rad6-dependent repression of ARG1 is independent of histone H2B but requires the E3 ubiquitin ligase Ubr1. This repression is apparent whether cells are grown in rich or minimal medium. The involvement of Ubr1 (and Rad6) in N-end rule proteolysis suggests that the Ubr1 component of ARG1 regulation requires protein turnover. Our analyses to map the elements required for Ubr1-dependent repression do not clearly define a potential target for proteolysis that can account for the full effect; however, the increase in expression is reduced from approximately threefold to twofold upon removal of the ARC elements and in minimal medium, suggesting that turnover of ArgR/Mcm1 may partially contribute.
Acknowledgments
We thank Carol Hannam, David Litchfield, Greg Gloor, Colin Coros, and George Chaconas for their contributions to the manuscript. Plasmids and strains were kindly provided by Susan Liebman and Mary Ann Osley.
This work was supported by a grant from the Canadian Institutes of Health Research to C.J.B. A.R.R. was supported by an NSERC Studentship, and H.P. was supported by an Ontario Graduate Scholarship.
REFERENCES
- 1.Amar, N., F. Messenguy, M. El Bakkoury, and E. Dubois. 2000. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 20:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 4.Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. [DOI] [PubMed] [Google Scholar]
- 5.Bailly, V., S. Prakash, and L. Prakash. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barsoum, J., and A. Varshavsky. 1985. Preferential localization of variant nucleosomes near the 5′-end of the mouse dihydrofolate reductase gene. J. Biol. Chem. 260:7688-7697. [PubMed] [Google Scholar]
- 7.Beaudenon, S. L., M. R. Huacani, G. Wang, D. P. McDonnell, and J. M. Huibregtse. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechet, J., M. Grenson, and J. M. Wiame. 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in S. cerevisiae. Eur. J. Biochem. 12:31-39. [DOI] [PubMed] [Google Scholar]
- 9.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boonchird, C., F. Messenguy, and E. Dubois. 1991. Characterization of the yeast ARG5,6 gene: determination of the nucleotide sequence, analysis of the control region and of ARG5,6 transcript. Mol. Gen. Genet. 226:154-166. [DOI] [PubMed] [Google Scholar]
- 11.Brandl, C. J., A. M. Furlanetto, J. A. Martens, and K. S. Hamilton. 1993. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 12:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 13.Chang, A., S. Cheang, X. Espanel, and M. Sudol. 2000. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:20562-20571. [DOI] [PubMed] [Google Scholar]
- 14.Crabeel, M., R. Huygen, K. Verschueren, F. Messenguy, K. Tinel, R. Cunin, and N. Glansdorff. 1985. General amino acid control and specific arginine repression in Saccharomyces cerevisiae: physical study of the bifunctional regulatory region of the ARG3 gene. Mol. Cell. Biol. 5:3139-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabeel, M., S. Seneca, K. Devos, and N. Glansdorff. 1988. Arginine repression of the Saccharomyces cerevisiae ARG1 gene. Comparison of the ARG1 and ARG3 control regions. Curr. Genet. 13:113-124. [DOI] [PubMed] [Google Scholar]
- 16.Crabeel, M., R. Lavalle, and N. Glansdorff. 1990. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol. Cell. Biol. 10:1226-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crabeel, M., M. de Rijcke, S. Seneca, H. Heimberg, I. Pfeiffer, and A. Matisova. 1995. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast 11:1367-1380. [DOI] [PubMed] [Google Scholar]
- 18.Davie, J. K., and C. M. Kane. 2000. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol. Cell. Biol. 20:5960-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davie, J. R., and L. C. Murphy. 1990. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29:4752-4757. [DOI] [PubMed] [Google Scholar]
- 20.Delforge, J., F. Messenguy, and J. M. Wiame. 1975. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR-mutations and the general control of amino-acid biosynthesis. Eur. J. Biochem. 57:231-239. [DOI] [PubMed] [Google Scholar]
- 21.De Rijcke, M., S. Seneca, B. Punyammalee, N. Glansdorff, and M. Crabeel. 1992. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine. Mol. Cell. Biol. 12:68-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dohmen, R. J., K. Madura, B. Bartel, and A. Varshavsky. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois, E., J. Bercy, and F. Messenguy. 1987. Characterization of two genes, ARGRI and ARGRIII, required for specific regulation of arginine metabolism in yeast. Mol. Gen. Genet. 207:142-148. [DOI] [PubMed] [Google Scholar]
- 24.Dubois, E., and F. Messenguy. 1991. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol. Cell. Biol. 11:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois, E., and F. Messenguy. 1997. Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p, and Ume6p. Mol. Gen. Genet. 253:568-580. [DOI] [PubMed] [Google Scholar]
- 26.El Bakkoury, M., E. Dubois, and F. Messenguy. 2000. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1, by the pleiotropic factor ArgRIII is required for their stability. Mol. Microbiol. 35:15-31. [DOI] [PubMed] [Google Scholar]
- 27.Goldknopf, I. L., M. F. French, R. Musso, and H. Busch. 1977. Presence of protein A24 in rat liver nucleosomes. Proc. Natl. Acad. Sci. USA 74:5492-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu, J., K. Ren, R. Dubner, and M. J. Iadarola. 1994. Cloning of a DNA binding protein that is a tyrosine kinase substrate and recognizes an upstream initiator-like sequence in the promoter of the preprodynorphin gene. Mol. Brain Res. 24:77-88. [DOI] [PubMed] [Google Scholar]
- 29.Haas, A. L., P. M. Bright, and V. E. Jackson. 1988. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J. Biol. Chem. 263:13268-13275. [PubMed] [Google Scholar]
- 30.Haas, A. L., P. B. Reback, and V. Chau. 1991. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit homologs, E2(20K) and E2(32K). J. Biol. Chem. 266:5104-5112. [PubMed] [Google Scholar]
- 31.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 32.Hinnebusch, A. G. 1986. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. Crit. Rev. Biochem. 21:277-317. [DOI] [PubMed] [Google Scholar]
- 33.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 34.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huibregtse, J. M., J. C. Yang, and S. L. Beaudenon. 1997. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 94:3656-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imhof, M. O., and D. P. McDonnell. 1996. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol. Cell. Biol. 16:2594-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs, P., J. C. Jauniaux, and M. Grenson. 1980. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J. Mol. Biol. 139:691-704. [DOI] [PubMed] [Google Scholar]
- 39.Jentsch, S., J. P. McGrath, and A. Varshavsky. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131-134. [DOI] [PubMed] [Google Scholar]
- 40.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 41.Kondo, K., and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor gene. Exp. Cell Res. 264:117-125. [DOI] [PubMed] [Google Scholar]
- 42.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovari, L. Z., M. Fourie, H. D. Park, I. A. Kovari, H. J. Van Vuuren, and T. G. Cooper. 1993. Analysis of the inducer-responsive CAR1 upstream activation sequence (UASI) and the factors required for its operation. Yeast 9:835-845. [DOI] [PubMed] [Google Scholar]
- 44.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 45.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrence, C. W., and R. Christensen. 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82:207-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebman, S. W., and G. Newnam. 1993. A ubiquitin-conjugating enzyme, RAD6, affects the distribution of Ty1 retrotransposon integration positions. Genetics 133:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madura, K., R. J. Dohmen, and A. Varshavsky. 1993. N-recognin/Ubc2 interactions in the N-end rule pathway. J. Biol. Chem. 268:12046-12054. [PubMed] [Google Scholar]
- 49.Martens, J. A., and C. J. Brandl. 1994. GCN4p activation of the yeast TRP3 gene is enhanced by ABF1p and uses a suboptimal TATA element. J. Biol. Chem. 269:15661-15667. [PubMed] [Google Scholar]
- 50.Messenguy, F. 1976. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J. Bacteriol. 128:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messenguy, F., E. Dubois, and C. Boonchird. 1991. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol. Cell. Biol. 11:2852-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moazed, D., and A. D. Johnson. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86:667-677. [DOI] [PubMed] [Google Scholar]
- 54.Nickel, B. E., C. D. Allis, and J. R. Davie. 1989. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry 28:958-963. [DOI] [PubMed] [Google Scholar]
- 55.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 56.Picologlou, S., N. Brown, and S. W. Liebman. 1990. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol. Cell. Biol. 10:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu, H. F., E. Dubois, P. Broen, and F. Messenguy. 1990. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Gen. Genet. 222:192-200. [DOI] [PubMed] [Google Scholar]
- 58.Qiu, H. F., E. Dubois, and F. Messenguy. 1991. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol. Cell. Biol. 11:2169-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58a.Ricci, A. R., J. Genereaux, and C. J. Brandl. 2002. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 22:4033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 60.Saleh, A., M. Collart, J. A. Martens, J. Genereaux, S. Allard, J. Cote, and C. J. Brandl. 1988. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J. Mol. Biol. 282:933-946. [DOI] [PubMed] [Google Scholar]
- 61.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 62.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 63.Skerjanc, I. S., R. S. Slack, and M. W. McBurney. 1994. Cellular aggregation enhances MyoD-directed skeletal myogenesis in embryonal carcinoma cells. Mol. Cell. Biol. 14:8451-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sung, P., S. Prakash, and L. Prakash. 1990. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. USA 87:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 66.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viljoen, M., L. Z. Kovari, I. A. Kovari, H. D. Park, H. J. van Vuuren, and T. G. Cooper. 1992. Tripartite structure of the Saccharomyces cerevisiae arginase (CAR1) gene inducer-responsive upstream activation sequence. J. Bacteriol. 174:6831-6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChangeTM site-directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 69.Watkins, J. F., P. Sung, S. Prakash, and L. Prakash. 1993. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev. 7:250-261. [DOI] [PubMed] [Google Scholar]
- 70.West, M. H., and W. M. Bonner. 1980. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8:4671-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winzeler, E. A., et al. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed]