Abstract
We have found that the Dna2 helicase-nuclease, thought to be involved in maturation of Okazaki fragments, is a component of telomeric chromatin. We demonstrate a dynamic localization of Dna2p to telomeres that suggests a dual role for Dna2p, one in telomere replication and another, unknown function, perhaps in telomere capping. Both chromatin immunoprecipitation (ChIP) and immunofluorescence show that Dna2p associates with telomeres but not bulk chromosomal DNA in G1 phase, when there is no telomere replication and the telomere is transcriptionally silenced. In S phase, there is a dramatic redistribution of Dna2p from telomeres to sites throughout the replicating chromosomes. Dna2p is again localized to telomeres in late S, where it remains through G2 and until the next S phase. Telomeric localization of Dna2p required Sir3p, since the amount of Dna2p found at telomeres by two different assays, one-hybrid and ChIP, is severely reduced in strains lacking Sir3p. The Dna2p is also distributed throughout the nucleus in cells growing in the presence of double-strand-break-inducing agents such as bleomycin. Finally, we show that Dna2p is functionally required for telomerase-dependent de novo telomere synthesis and also participates in telomere lengthening in mutants lacking telomerase.
Saccharomyces cerevisiae Dna2p is a highly conserved helicase-nuclease essential for DNA replication (3, 10, 12-14, 30, 35, 38). It is also essential for DNA replication in Schizosaccharomyces pombe and in Xenopus laevis (34, 38). Mounting evidence indicates that Dna2p participates in the processing of Okazaki fragments, either compensating for or cooperating with the RAD27-encoded FEN-1 nuclease (4, 10, 12, 25, 33). Unlike most DNA replication mutants, dna2 mutants are also defective in repair of double-strand breaks (DSBs) by the postreplication repair pathway (11, 25). dna2 mutants require rad9 for cell cycle arrest at the restrictive temperature (24, 25). However, the double mutants have greater viability than dna2 mutants at semipermissive temperatures. In this work, we describe an unprecedented type of interaction of this or any other DNA replication protein with telomeres.
Telomeres are specialized structures at the ends of chromosomes, important both for facilitating complete DNA replication and for stabilizing the ends by preventing end-to-end fusions. Yeast telomeres contain about 300 bp of heterogeneous C1-3A/TG1-3 repeats at the extreme termini. Subtelomeric repeats, called Y′, are found at some but not all of the yeast telomeres, and a second set of repeats, X, are found at all telomeres (51). The tandem array of C1-3A/TG1-3 repeats binds a set of specific proteins that nucleate a higher order chromatin structure that leads to silencing of genes up to several kilobase pairs internal to the terminal repeats. Such silencing is called telomere position effect and requires the RAP1, SIR2, SIR3, and SIR4 genes (53). Many of the proteins are also components of the telomere capping complex that protects against fusions (6). The majority of the chromosome terminus replicates late in S phase due to late activation of autonomously replicating sequences (ARSs) within 40 kb from the telomere (18, 22, 60).
The replisome emanating from this region replicates the subtelomeric repeats and some of the C1-3A/TG1-3 repeat sequences. Recent evidence obtained from in vitro replication of a linear simian virus 40 chromosome suggests that the eukaryotic replisome can completely copy the leading-strand (C1-3A template strand in S. cerevisiae), but that the lagging strand terminates gradually within ≈50 bp from the 3′ end, producing single-stranded ends (47). This is a reflection of the so-called end replication problem, the inability to repair the gaps left by removal of the last primer or to prime the terminal Okazaki fragment on the lagging-strand. Perhaps related to the observations in the simian virus 40 system in vitro, transient single-stranded TG1-3 tails, 30 to 150 nucleotides long, arise late in S phase at yeast telomeres (61). Their appearance requires passage of the replication fork (20) and may involve nucleolytic processing, in addition to failure to complete the lagging strand, since both leading- and lagging-strand telomeres have TG1-3 tails (19, 61).
The end replication problem is in part solved because TG1-3 tails are extended by a specialized DNA polymerase, telomerase. Without telomerase, telomeres gradually shorten. The products of the yeast TLC1, EST1, EST2 (the catalytic subunit of telomerase), EST3, and CDC13 (also known as EST4) genes are required for TG1-3 tail extension (46). TG1-3 tails disappear at G2/M, presumably by C1-3A strand fill-in, but the source of the lagging-strand machinery (which we will now call the primosome, in analogy to bacterial terminology) in the absence of a standard replication fork is unknown (50). It seems unlikely that it is the same primosome assembled at the subtelomeric ARSs (50).
Which of the enzymes involved in normal lagging-strand DNA replication are required to fill in the complementary strand, how are they recruited to telomeres, and how do they interact with telomerase (50)? Compared to the intensive efforts to study telomerase mechanism, little attention has been aimed at the interaction between telomerase extension and lagging-strand fill-in. Several mutants affecting lagging-strand polymerases exhibit telomere length deregulation and effects on telomere position effect (1, 15, 41). Mutants lacking RAD27, an Okazaki fragment processing enzyme, have long-lived single-stranded TG tails (48). However, it has been pointed out that these phenotypes could be an indirect effect due to slower replication forks or the inherent instability of repeats in rad27Δ mutants (18, 48).
One elegant study showed directly that polymerase α, polymerase δ, and primase are required for telomere synthesis, whereas polymerase ɛ is not (18). A second important study revealed that polymerase α interacts with Cdc13p, a telomere binding protein (52). Based on this and other recent work (16, 49), it appears that synthesis of the TG1-3-rich strand by telomerase and the C1-3A-rich strand by the lagging-strand enzymes is highly coordinated. Though such coregulation might occur through formation of a large complex of both enzymatic machines at the chromosome ends (21), evidence for such a complex is lacking.
Using a set of assays and specially marked yeast strains previously validated in laboratories devoted to the study of telomere biology (7, 18, 28, 37, 39, 54), in addition to chromatin immunoprecipitation (ChIP) assays and indirect immunofluorescence, we show directly both that Dna2p is required for telomere biosynthesis and that Dna2p is dynamically sequestered in an apparently large complex at telomeres. This work expands the known repertoire of primosomal proteins needed for C-strand fill-in to processing enzymes, in addition to the known polymerases. It also answers the question of when and where at least one component of the telomere primosome assembles at telomeres, and the answer is unexpected. Our findings suggest that Dna2p is part of a dynamic telomeric chromatin-based complex that we propose not only functions in telomere replication but also serves as a store for replication and DSB repair proteins, a telomere cap, and perhaps a telomere checkpoint.
MATERIALS AND METHODS
Telomere position effect and HML silencing assay.
Yeast strains UCC3505, with the URA3 gene inserted at the left telomere of chromosome VII, and UCC3515 (Table 1), with URA3 inserted at HML, were transformed with the pTRP-based plasmid, which overexpresses DNA2 under the GAL1,10 promoter, or with pTRP with no insert (54). A 6-kb EcoRI-XhoI fragment carrying the entire DNA2 gene (13, 14) was subcloned into pTRP cleaved with EcoRI-XhoI, placing DNA2 under GAL1,10. Transformants were plated on SD minus tryptophan plates to select for colonies containing the TRP-bearing plasmids. After 3 days of growth, colonies were restreaked on plates containing 2% galactose and 1.5% raffinose to induce DNA2 expression.
TABLE 1.
Strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| YM701HIS-Tel | MATaura3-52 his3-200 ade2-101 lys2-801 trp1-901 tyr1 adh4::URA3-TG1-3 promoter-defective HIS, 20 kb Δ of DNA distal to adh4 | 7 |
| YM701HIS-INT | MATaura3-52 his3-200 ade2-101 lys2-801 trpl-901 tyr1 adh4::URA3 promoter-defective HIS | 7 |
| YM701HIS-TELsir3Δ | MATaura3-52 his3-200 ade2-101 lys2-801 trp1-901 tyr1 adh4::URA3-TG1-3 promoter-defective HIS, 20 kb Δ of DNA sir3::LYS2 distal to adh4 | 7 |
| UCC3505 | MATaade2-101 his3-200 leu2-1 lys2-801 trp1-63 ura3-52 ppr1::HIS3 adh4::URA3-TEL-VIIL DIA5-1 | 54 |
| UCC3515 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp1-63 ura3-52 hml::URA3 | 54 |
| UCC5706 | MATa-inc ura3-52 lys2-801 ade2-101 ochre trp1-63 his3-200 leu2-1::GAL1-HO-LEU2 TELVIIL::ADE2-TG1-3-Hosite-LYS2 rad52::HISG | 18 |
| UCC5706dna2-2 | MATa-inc ura3-52 lys2-801 ade2-101 ochre trp1-63 his3-200 leu2-1::GAL1-HO-LEU2 TELVIIL::ADE2-TG1-3-Hosite-LYS2 rad52::HISG dna2::dna2-2-URA3 | This work |
| est2 | MATaleu2Δ0 his3Δ1 met15Δ ura3Δ0 est2::KANMX4 | Research Genetics |
| est2dna2-2 | MATaleu2Δ0 his3Δ1 met15Δ ura3Δ0 est2::KANMX4 dna2-2 sol3Δ::LEU2 | This work |
| TF1087-10-2 | MATα trp1 leu2 ura3 his3 dna2-2 sol3-Δ(::LEU2) | 25 |
| TF7610-2-1 | MATα trp1 leu2 ura3 his3 DNA2 sol3-Δ(::LEU2) | 25 |
| est1 | MATaleu2Δ0 his3Δ1 met15Δ ura3Δ0 est1::KANMX4 | Research Genetics |
| JCYrad52 | MATarad52-8Δ::TRP1 ura3 trp1 leu2 his3 gal2 | 11 |
| DNA2TMTH | MATacan1-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 pep4::TRP1 bar1::LEU2 dna2::DNA2-TEV-9MYC-HIS3 | This work |
| DNA2TMTH Sir3 Δ | MATacan1-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 pep4::TRP1 bar1::LEU2 dna2::DNA2-TEV-9MYC-HIS3 sir3::KANR | This work |
| est1dna2-2 | MATaleu2Δ0 his3Δ1 met15Δ ura3Δ0 est1::KANMX4 dna2-2 sol3Δ::LEU2 | This work |
In order to assay the ability to grow in the absence of uracil, samples (5 μl) of 10-fold serial dilutions of the same numbers of each transformant were spotted onto 2% galactose-1.5% raffinose plates minus uracil. To control for the number of cells plated, the serial dilutions were also plated on the 2% galactose-1.5% raffinose plus uracil medium at the same time. Plates were photographed after 4 days of incubation.
Telomere one-hybrid assay.
The telomere one-hybrid assay was carried out using strains and plasmids as described (7). For the telomere one-hybrid assay, the DNA2 open reading frame (ORF) (EcoRI-XhoI) was cloned in frame into the EcoRI-XhoI sites of pJG4-5 (7) to make pJG4-5DNA2. When pJG4-5 carries DNA2, it expresses a polypeptide that is a fusion of DNA2 with the B42 activation domain, the hemagglutinin (HA) tag, and a nuclear localization signal. B42 is an Escherichia coli sequence that activates transcription in S. cerevisiae. DNA2 was also cloned in the same vector without the activation domain, as a control.
Isogenic strains (YM701HIS-TEL and YM701HIS-TELsir3Δ) (Table 1 and Fig. 2) carrying plasmids expressing the relevant fusion proteins under GAL1,10 promoter control were grown at 30°C for 2 days in SC liquid medium lacking tryptophan and containing 3% raffinose to avoid glucose repression. The vector, Rap1-B42, and Cdc13-B42 plasmids have been described (7). To screen for expression of HIS3, 5 μl of cells was spotted in 10-fold dilutions onto 2% galactose-2% raffinose minus histidine plates containing 3-aminotriazole (3-AT) or 2% galactose-2% raffinose plus histidine control plates. It was necessary to use different concentrations of 3-AT to reduce the background of the His3+ phenotype of each transformant, as described (7). The growth of each transformant was measured after 3 days of incubation on 2% galactose-2% raffinose plus histidine and after 10 days on the same medium minus histidine. The assay was repeated three times on different days.
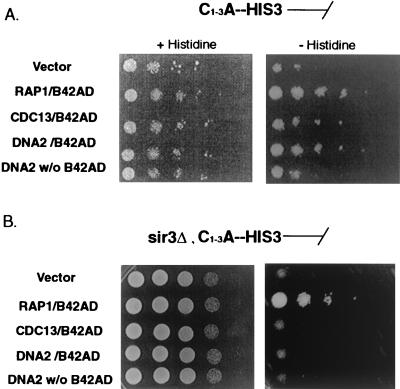
FIG. 2.
Telomere one-hybrid assay of DNA2. The genes indicated were fused to the bacterial B42 transcriptional activation domain as described in Materials and Methods and overexpressed under the GAL1,10 promoter. The promoterless HIS3 reporter gene was integrated at the telomere (A) (7). In panel B, the HIS3 reporter was integrated at the telomere in an isogenic strain carrying sir3Δ. See Materials and Methods and Table 1 for strain descriptions. The experimental results are shown in the panels on the right in panel A and panel B. On the left are loading controls. The loading control in panel A was plated on galactose-raffinose plus histidine and in panel B on YPGR (YP plus galactose and raffinose).
Indirect immunofluorescence.
DNA2 was fused at its C terminus to 9 Myc tag peptides and inserted into the chromosome, replacing the endogenous copy of DNA2. For this construct, PCR was performed on plasmid pJSVM53H (pRS303-based plasmid containing 9 Myc tags adjacent to a HIS3 marker, gift of R. Deshaies, California Institute of Technology, Pasadena) using an upstream primer, P1, carrying 45 bases of DNA2 C-terminal coding sequence and 25 bases of Myc tag sequence (AGCGATAAACCTATCATAAAGGAAATTCTACAAGAGTATGAAAGTGAGAACCTGTACTTCCAGGG) and downstream primer P2 containing 25 bases of HIS3 and 45 bases of the DNA2 3′ untranslated region (UTR) (TGTGATAGCTTTCCTGTTATGGAGAAGCTCTTCTTATTCCCCCTGACAGCAGTATAGCGACCAGC). The PCR product was used to transform strain W303 pep4bar1, with selection for growth on histidine, resulting in replacement of the endogenous gene under the control of its native promoter, as verified by PCR and Western blotting with the 9E10 anti-Myc tag monoclonal antobody. The resulting strain showed the same growth rates as the parental strain. This strain, designated DNA2TMTH (Table 1), was used for indirect immunofluorescence.
Indirect immunofluorescence was performed as described (28). Briefly, cells were fixed by incubation in 3.7% formaldehyde. Fixed cells were washed with 1 ml of YPD containing 1 M sorbitol and resuspended in sorbitol buffer. Spheroplasts of fixed cells were obtained by treatment with zymolyase for 30 min at 30°C. Spheroplasts were deposited on lysine-coated multiwell slides. Cells were stained sequentially with anti-Myc 9E10 primary antibody and then with anti-Sir3p antibody, followed by sequential staining with fluorescent secondary donkey anti-mouse and goat anti-rabbit immunoglobulin antibody to visualize the signal. Finally, cells were stained with 4′,6′-diamidino-2phenylindole (DAPI) to reveal DNA.
The 9E10 anti-Myc monoclonal antibody from Babco Company and polyclonal antibodies against Sir3p (a kind gift from L. Guarente) were used. Secondary antibodies were goat tetramethyl rhodamine thiocyanate (TRITC)-labeled anti-rabbit and donkey fluorescein isothiocyanate (FITC)-labeled anti-mouse immunoglobulin (Jackson Immuno Research, West Grove, Pa.). As controls, immunofluorescence was performed without primary antibody or without secondary antibody and the possibility of cross-reaction between antibodies was ruled out. Digital images were obtained using a Hamatu digital camera using a Nikon Eclipse TE300 inverted microscope. The camera was controlled by Metaphore image acquisition software (Universal Image Corp., patent 4558302).
ChIP.
The chromatin immunoprecipitation assay (ChIP) assay used in this study is based on the methods described by Tanaka et al. (56). Four sets of primers were used for the PCR amplification step: Tel3500.fwd, TGATTCTGCTTTATCTACTTGCGTTTC; Tel3500.rev, AGAGTAACCATAGCTATTTACAATAGG; Tel300.fwd, GGATATGTCAAAATTGGATACGCTTATG; Tel-300.rev, CTATAGTTGATTATAGATCCTCAATGTC (45); Cup1-1, GGCATTGGCACTCATGACCTTCAT; Cup1-2, TCTTTTCCGCTGAACCGTTCCAGCA (29); rDNA-1, CCACCCGATCCCTAGTCGCATAG; and rDNA-2, GTGTACTGGATTTCCAACGGGGCC.
PCR products were separated on a 2% agarose gel and stained with 0.2 μg of ethidium bromide/ml. Gels were photographed, and the negatives were scanned and quantitated by densitometry. The DNA2TMTHin Δ strain was constructed in the DNA2TMTH strain (Table 1 and above). The SIR3 gene was deleted by a one-step PCR method using the sir3 del fw oligomer (GCAGCCCTTTCATCACCTTCCTTACAGGGGTTTAAGAAAGTTGTTTTG) and sir3 del rev (ATTAAGGAATACAGAAGAGACTGCATGTGTACATAGGCATATCTATGGCGG) in the same way as for the DNA2TMTH strain (see above).
Telomerase-mediated telomere addition assay.
The telomere addition assay was carried out as described (18). Strain UCC5706 dna2-2 was made in three steps. Strain UCC5706 (a kind gift from D. Gottschling, Fred Hutchinson Cancer Center, Seattle, Wash.) was transformed with pSD190 containing RAD52 to create UCC5706RAD52. Linearized pRS306DNA2C containing the dna2-2 gene was integrated into UCC5706RAD52 to form UCC5706RAD52dna2-2. To eliminate the pSD190-encoded RAD52 from UCC5706RAD52dna2-2, the strain was incubated in YPD rich medium overnight and plated on SD minus tryptophan. To confirm the UCC5706 dna2-2 rad52Δ genotype, temperature sensitivity was checked at 37°C (11).
The assay was performed exactly as described (18). Briefly, cells of wild-type UCC5706 rad52Δ and the temperature-sensitive strain UCC5705 dna2-2 rad52Δ were grown in complete medium lacking lysine (2% glucose), diluted into YEP-2.5% raffinose for four generations, and arrested in the cell cycle with nocodazole at 23°C until 95% of cells showed the appropriate dumbbell cell morphology. Nocodazole was added to a log-phase culture from 10-mg/ml stock in dimethyl sulfoxide (DMSO) to a final concentration of 10 μg/ml. Cell cycle arrest (2C DNA content) was confirmed by flow cytometry (not shown).
For galactose induction, cells were then centrifuged and resuspended in warmed YEP-3% galactose, and incubation was continued at 37°C. At various times, cells were harvested, genomic DNA was prepared and digested with enzyme, and 0.8% agarose gel electrophoresis was carried out. Hybridization with the 32P-labeled ADE2 gene fragment was carried out after blotting the gel to nylon membrane (Zeta-probe GT genomic membrane) by vacuum transfer. Telomere sequence addition efficiency at each time point was normalized based on the amount of signal in the band containing ADE2 from its endogenous chromosomal location (band labeled INT in Fig. 6B) using phosphorimager analysis.
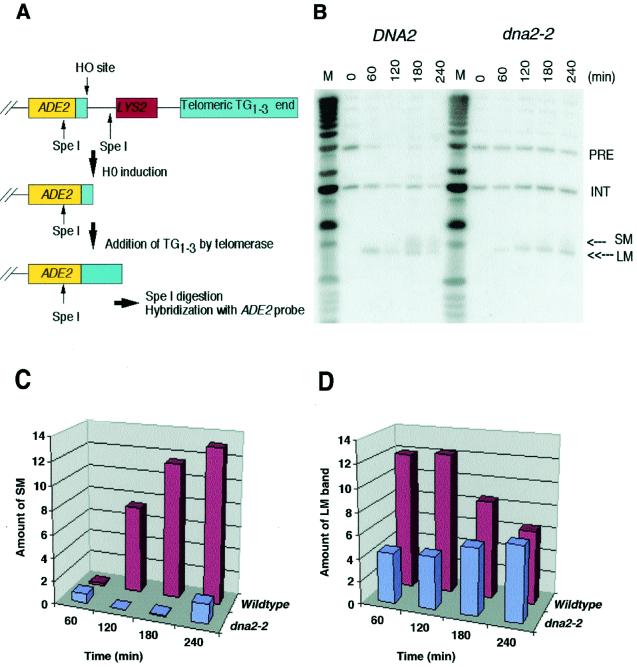
FIG. 6.
Dna2p plays a role in telomerase-dependent telomere replication. (A) Schematic representation of the telomere addition assay (18). Briefly, a strain carrying an ectopic copy of ADE2 adjacent to a stretch of TG1-3 repeats and an HO endonuclease site is arrested at G2/M with nocodazole, a time when telomere addition has been shown to be active. The HO endonuclease is induced in the strain by incubation in galactose. At various times after induction, samples are taken; DNA is prepared and digested with SpeI, generating the fragments indicated. These fragments are separated by gel electrophoresis and blotted and probed with an ADE2 probe. Note that the strain carries an additional copy of ADE2 at its endogenous chromosomal location, not shown in the diagram. See Materials and Methods for details. (B) Hybridization of ADE2 probe to SpeI-digested DNA samples taken at the indicated times after induction of HO endonuclease by the addition of galactose. PRE, parental DNA uncut by HO; LM, HO-cut DNA; SM, telomeric addition; INT, SpeI fragment containing the ADE2 gene at its endogenous chromosomal location as an internal hybridization control. (C) Quantitation of telomere addition onto TG1-3/HO ends after normalization of SM to the INT fragment. (D) Quantitation of disappearance of the LM band after normalization of LM to the INT fragment. The y axis in panels C and D represents normalized phosphorimager readings in arbitrary units.
RESULTS
Telomere position effect and Dna2p.
dna2 mutations have been reported to affect telomeres in at least three different ways. First, overexpression of 300 amino acids of the N terminus of DNA2 reduces telomere position effect (54). Second, telomeres in dna2 mutants are slightly longer than in the wild type, although not as long as in other lagging-strand mutants (1, 15, 25, 41). Third, overexpression of DNA2 leads to rapid appearance of single-stranded TG1-3 tails (48). Dna2p is a candidate for a component of the putative telomeric end replication complex, especially because of the unexpected and as yet unexplained observation that DNA2 overexpression, just like overexpression of TLC1, the RNA component of telomerase, reduces telomere position effect (54).
To increase confidence that the effect of the N-terminal fragment of DNA2 on telomere position effect had relevance to the function of Dna2p in the cell, we asked whether overexpression of intact DNA2 also reduces telomere position effect (54). Strain UCC3505 has URA3 next to the left telomere of chromosome VII (VII-L). The URA3 gene located at the telomere normally switches between transcriptional states. However, deleting its transcriptional activator gene PPR1 causes the telomere-adjacent URA3 gene to be completely silenced in UCC3505, and the strain grows poorly on medium lacking uracil.
As shown in the serial dilution assay in Fig. 1A, overexpression of DNA2 increased the efficiency of growth on medium lacking uracil. No difference in the number of Ura+ colonies was observed between strains containing the vector and DNA2 constructs on glucose medium, where DNA2 was not overexpressed. Overexpression of the full-length DNA2, as reported previously for the N-terminal portion of DNA2 (54), also reduced silencing of URA3 integrated at the silent mating type locus HML, as evidenced by increased ability to form colonies without uracil (Fig. 1A). Thus, the full-length DNA2 gene behaves like the N terminus in reducing silencing, especially the telomere position effect.
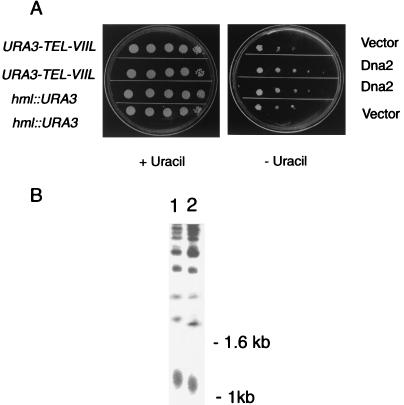
FIG. 1.
(A) Telomere position effect is reduced by overexpression of DNA2. UCC3505 and UCC3515, containing URA3 at the telomere or at HML as described in Materials and Methods, were transformed with a plasmid expressing DNA2 under GAL1,10 control or with the plasmid vector alone as a control. Tenfold serial dilutions of the indicated transformants were plated on medium containing galactose, to induce DNA2, and plus or minus uracil, to monitor expression of the URA3 gene. (B) Telomere length is reduced by overexpression of DNA2. Dna2p was overproduced in strain W303 RAD5 using a plasmid with DNA2 under control of the GAL1,10 promoter as described in Materials and Methods. The same strain containing vector alone was used as a control. A single colony was picked, and after three restreaks (about 100 generations) on YPGal plates, telomere length was determined as described (48). DNA was digested with XhoI, which cleaves near the telomere-proximal end of Y′ repeats, leaving primarily tracts of C1-3A repeats that migrate in agarose gels around the size of the 1-kb marker. Telomere length was assessed by gel electrophoresis and Southern blotting using hybridization with a 32P-labeled 22-mer of the telomere sequence 5′-CCCACCACACACACCCACACCC-3′ (referred to as the CA oligonucleotide) as described (39). Each lane contains equal amounts of DNA. Lane 1, strain carrying plasmid lacking DNA2 insert; lane 2, overexpression of GAL-DNA2. The diffuse band around 1 kb represents the Y′ telomeres.
The same strains used to test the effect of overexpression of DNA2 on telomere position effect were also used to measure the effect of overproduction on telomere length. As shown in Fig. 1B, overexpression of DNA2 reduced telomere length. Three independent induction experiments showed the same shortening compared to wild type. Thus, overproduction of Dna2p reduces both telomere position effect and telomere length.
Telomere one-hybrid assay.
To test whether reduction in telomere position effect and telomere length might be a consequence of physical association of Dna2p with telomeric chromatin, we carried out a telomere one-hybrid assay that monitors association of telomeric proteins with telomeres in vivo (7). This assay uses a promoter-defective HIS3 reporter gene inserted adjacent to C1-3A tracts at the telomere of chromosome VI (7). This telomere was shown to reflect organization at a natural telomere and to be fully functional (7). Since the HIS3 gene lacks upstream activation sequences, HIS3 can only be efficiently activated by binding of a transcriptional activator to the telomere. A similar construct was used to monitor interaction with C1-3A repeats at an internal position in the chromosome (not shown).
DNA2 was fused to the B42 transcriptional activation domain, a bacterial transcriptional activation domain that functions in S. cerevisiae. The ability of this Dna2-B42 fusion protein to activate HIS3 transcription when expressed in S. cerevisiae was compared to proteins known to interact with telomeric DNA and telomeric chromatin, Rap1p-B42 and Cdc13p-B42, by scoring growth on medium lacking histidine. Dna2p-B42 was able to activate the telomeric C1-3A-HIS3 reporter gene as efficiently as Rap1p-B42, and activation required the B42 transcriptional activation domain (Fig. 2A).
Since the B42 activation domain is required, it does not appear that overexpression of Dna2p is simply causing titration of factors that silence the HIS3 gene, but rather that it is bound at the telomeric repeats and is actually activating transcription. (Note that in Fig. 2A, the panel on the left constitutes a loading control and shows less growth than in the experiment on the right because these control plates were incubated for only 3 days while the experimental plates were incubated for 10 days, as in the original description of this assay [7].)
Since overexpression of DNA2 caused derepression of silencing at HML as well as at telomeres (Fig. 1A), it seemed plausible that the apparent association of Dna2p with telomeres (Fig. 2A) might require components of silent chromatin shared between HML and telomeres. SIR3 encodes such a protein (53). We therefore asked if the ability to activate transcription in the one-hybrid assay depended on SIR3 by repeating the assay in an isogenic sir3Δ strain, as previously demonstrated for Cdc13 (7). As shown in Fig. 2B, right, the Rap1-B42AD fusion protein activated the telomeric promoter-defective HIS3 gene, consistent with the fact that Rap1p is known to bind directly to the C1-3A repeats. Dna2p-B42 failed to express the gene, however, suggesting that binding of Dna2p at telomeres requires a Sir3-dependent chromatin structure (see also Fig. 5D). (Note that the loading control on the left in Fig. 2B was plated on YPGR [yeast extract, peptone, galactose, and raffinose], accounting for the higher efficiency of growth compared to controls in Fig. 2A.)
FIG. 5.
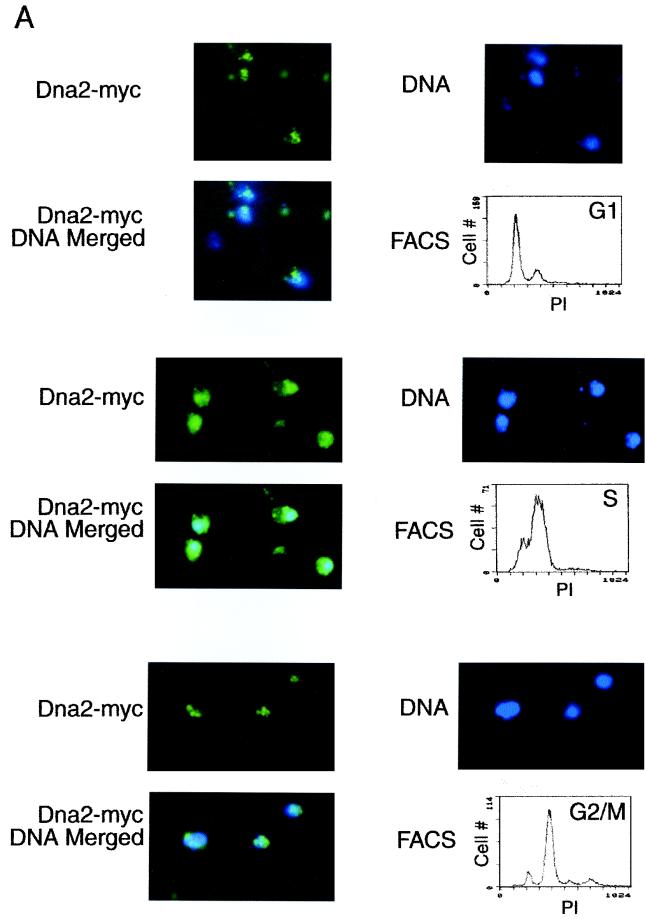
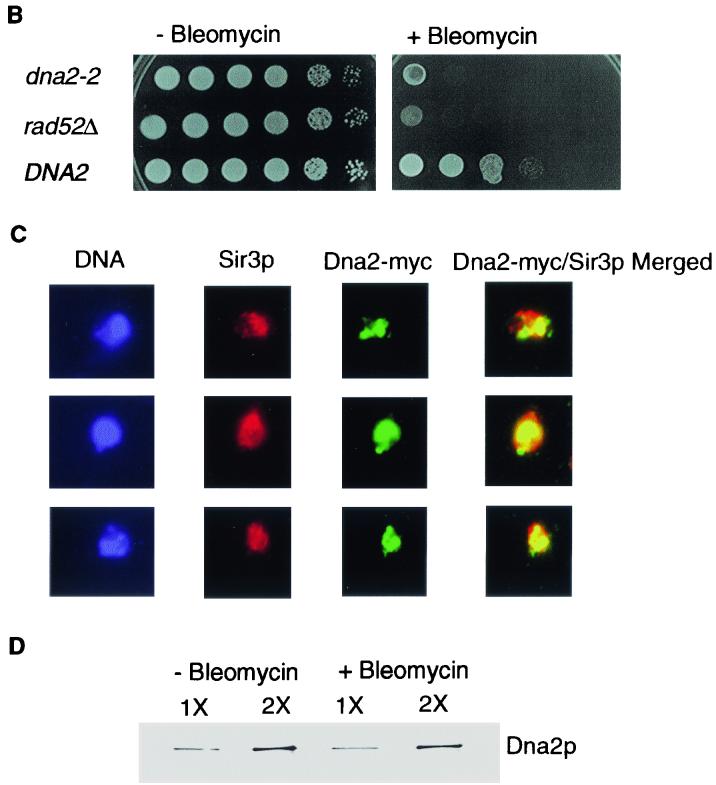
Dna2p localization is dynamic. (A) Dna2p relocalizes in the nucleus during the cell cycle. Cells arrested by α-factor were released in YPD medium and harvested at various times after release as described in Materials and Methods. Cells in G1, S, and G2/M were identified by flow cytometry analysis of each sample, as indicated, and Dna2-Myc localization was examined by indirect immunofluorescence with anti-Myc antibody (green stain) at the cell cycle stage indicated. Cells were costained with DAPI for DNA (blue). Preparation of cells for flow cytometry was performed as described (5). (B) Bleomycin sensitivity of strain TF1087-10-2dna2-2, TF7610-2-1 DNA2, and JCY rad52Δ (see Table 1). Tenfold serial dilutions of log-phase cultures were plated on YPD medium with or without bleomycin (15 mU/ml). (C) Indirect immunofluorescence after treatment with bleomycin-Sir3p (red) and Dna2-Myc (green), merged; yellow indicates colocalization. Costaining and visualization were carried out as described in Materials and Methods and the legend to Fig. 3. (D) Dna2p levels do not rise after DNA damage. Extracts were prepared from untreated and bleomycin-treated cells, and the level of Dna2-Myc was determined by Western blotting as described previously (14). 2X refers to loading of twice as much extract as in the 1X lane, to ensure the blot was in a linear range of Dna2p concentration. A loading control using Cdc28 and anti-Cdc28 antibody showed equal loading of the 1X lanes from bleomycin-treated and untreated cells (not shown).
Dna2p can be cross-linked to telomeric and subtelomeric regions and localization fluctuates during the cell cycle.
As a direct test for association of Dna2p with telomeric DNA, we determined the localization of Dna2p using a ChIP protocol (56). Since Dna2p is not abundant, in order to detect Dna2p without overproduction, the chromosomal DNA2 coding sequence was precisely replaced, as described in Materials and Methods, with an epitope-tagged DNA2 gene (DNA2-myc) under control of its native promoter. We found no differences in growth rates between cells carrying DNA2 and DNA2-myc.
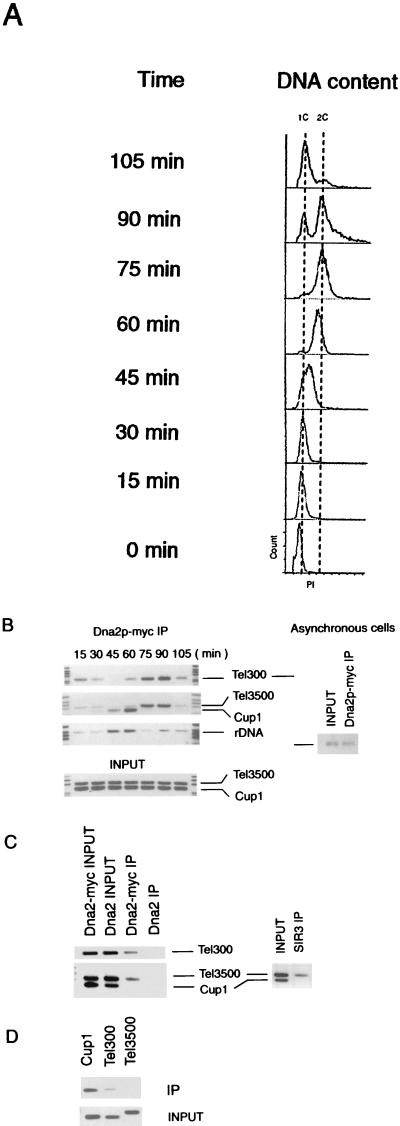
ChIP assays were performed on both asynchronous cells and cells progressing through a synchronous cell cycle. For the synchrony experiments, cells were arrested with α-factor and then released into a synchronous cell cycle. S phase begins about 40 min after release under these conditions (Fig. 3A). Samples of either asynchronous cells (Fig. 3B, right) or synchronized cells at the indicated times after release from α-factor arrest (Fig. 3B, left) were cross-linked using formaldehyde, and extracts were prepared at the indicated times. DNA in the extracts was sonicated to about 500 bp, followed by immunoprecipitation with 9E10 anti-Myc antibody. After reversing the cross-links, quantitative PCR was performed using primers complementary to a subtelomeric region 3,500 bp from the right telomere of chromosome VI (Tel3500), and to a region 300 bp from the same telomere (Tel300) (45).
FIG. 3.
Chromatin immunoprecipitation assay of Dna2p localization. (A) Flow cytometric analysis of experiment shown in panel B. Propidium iodide (PI) is a measure of DNA content. (B) Cross-linking of Dna2p to various DNA sequences in asynchronous (right two lanes) and synchronized cells (panel on the left) as indicated. An experiment using Dna2p-Myc immunoprecipitates is shown. Cells were synchronized as in Materials and Methods and shown in panel A. ChIP assays were preformed on samples collected at the time indicated after release from pheromone arrest. The regions amplified by the primers are described in the text. Tel refers to telomeric and subtelomeric primers, while CUP1 and rDNA (RDN1) primers are nontelomeric sequences. The Tel300 primers give a PCR product identical in length to the CUP1 PCR product and therefore represents a separate PCR, as does the rDNA PCR series. PCRs were in the linear range, as determined by monitoring different numbers of cycles. Input indicates that PCRs were preformed on the extracts from which the immunoprecipitates were prepared, using the primers indicated, providing a loading control as well as a control that the primers used amplified the respective DNAs equally. The lanes at the left and right of each strip contain the size markers. For asynchronous cells, only Tel300 primers were used for the PCR. (C) Binding of both Sir3p and Dna2-Myc protein to telomeric sequences in nocodazole-arrested (G2/M) phase cells. A Dna2-Myc-tagged (strain DNA2TMTH) was arrested with nocodazole as described in Materials and Methods. ChIP assays were performed using anti-Myc to localize Dna2-Myc and anti-Sir3 antibody for Sir3p. The left lanes represent the Dna2-Myc immunoprecipitates (IP). Right two lanes represent a Sir3p immunoprecipitate from the same extracts. Primers used to generate the PCR products are as indicated. Input is PCR using the extract from which the immunoprecipitate was prepared. Lanes labeled untagged DNA2 represent anti-Myc immunoprecipitates from strains synchronized in the same way but lacking a Dna2-Myc protein, as a control for the specificity of the antibody. (D) Localization of Dna2p from G2 phase DNA2TMTH sir3Δ cells (see Table 1). The chromatin immunoprecipitation experiment was identical to that in panel C. The sir3Δ strain used was isogenic with the strain used in panel C and was constructed by standard PCR techniques (see Materials and Methods and Table 1).
The Tel3500 and Tel300 primers amplify unique sequences. Sir3p has been shown to cross-link efficiently to DNA amplified by these probes (45), and yeast Ku (yKu) has been shown to be associated with similar probes (42). Primers to a centromere-proximal region around the CUP1 gene (29) and to another internal region in the ribosomal DNA (rDNA) (RDN1) locus were also used (see Materials and Methods). The results shown in Fig. 3B indicate that Dna2-Myc binds preferentially to telomeric DNA compared to centromere-proximal sequences in G1. Tel300 and Tel3500 were enriched 22-fold and 9.2-fold, respectively, compared to the CUP1 sequences in the Dna2-Myc immunoprecipitates from G1 phase cells. Controls show that CUP1 probes are amplified with similar efficiency to Tel3500 in extracts (lanes labeled Input), even though some strains contain repeated copies of CUP1.
In S phase, the pattern of DNA associated with Dna2-Myc was dramatically reversed (Fig. 3A). The two telomeric PCR products were virtually undetectable in the Dna2-Myc precipitates (45-min time point, cells greater than 90% budded), and the CUP1 and RDN1 bands, which should now correspond to replicating DNA, were prominent. The timing of association of Dna2p with CUP1 and RDN1 is similar to the timing of association of another replication elongation factor, DNA polymerase Ε, with CYC1, another centromere-proximal locus (43), and serves as an independent marker of S phase. The redistribution of the bulk of Dna2p from Tel3500 and Tel300 to internal DNA sequences at S phase suggests that the Dna2p is sequestered at telomeres in G1 and that this may, in part, reflect storage of Dna2p or a protective capping role.
The preferential telomeric localization pattern was observed again as cells exited S phase or as they entered G2 (75 and 90 min, 100% large budded cells) and remained at a maximum throughout G2. This coincided with the time that telomerase was active in vivo. The final time point in the experiment (105 min, 90% unbudded cells) represents the next G1 phase. The fluctuation in localization is probably not due to changes in expression of DNA2 during the cell cycle, since its mRNA expression is constant in the cell cycle (17, 55). Western blotting also showed that Dna2 protein was expressed throughout the cell cycle, though there may have been a slight reduction in levels during α-factor arrest in G1 (data not shown).
It has previously been shown by ChIP assay that Sir3p interacts with both Tel300 and Tel3500 DNA and even with a probe 5,800 bp internal to the telomere (45). To insure that the ChIP assays for Dna2-Myc were valid, extracts were also tested for Sir3p localization (Fig. 3C). Cells were arrested in our experiment with nocodazole, and arrest was monitored by flow cytometry (not shown). Extracts were cross-linked, and immunoprecipitations were carried out on parallel samples from the same extract with anti-Myc antibody and anti-Sir3 antibody. We found the same enrichment of telomeric DNA compared to CUP1 in the anti-Dna2-Myc immunoprecipitates as we found in cells in G2 phase after release from α-factor arrest (compare Fig. 3B and Fig. 3C), and Sir3p showed the same occupancy as Dna2-Myc in the nocodazole-arrested cells (Fig. 3C). Thus, there is a correlation between the sequences to which Sir3p binds and those to which Dna2-Myc binds.
The ChIP experiments shown in Fig. 3B and 3C provide strong evidence that Dna2p is bound to both telomeric and subtelomeric chromatin and that this localization changes with progression through the cell cycle. The presence of Dna2p at telomeres and subtelomeric regions in G1 phase was surprising and implies that sequestration is not due solely to the enzymatic role of Dna2p in Okazaki fragment processing at these chromosomal sites (see below), neither of which replicates in G1 (18, 22, 23, 40). This is the first DNA replication protein that has been found to show preferential subnuclear localization to telomeric chromatin in G1 phase, where there is no DNA replication. The G1 localization is further shown by immunofluorescence studies below (see Fig. 5A).
Amount of Dna2p at telomeres in G2 phase is dramatically reduced in a strain lacking SIR3.
Two results suggested that Sir3p might be required, either directly or indirectly, for the association of Dna2p with telomeres. The one-hybrid assay presented in Fig. 2 showed that Dna2p failed to associate with C1-3A repeats in a sir3Δ strain. Furthermore, the ChIP assay presented in Fig. 3C showed Sir3p was present on the same subtelomeric sequence (Tel3500) as Dna2-Myc. To test for Sir3p involvement in Dna2p localization, ChIP assays were repeated in an isogenic sir3Δ strain (see Materials and Methods).
As shown in Fig. 3D, Dna2p localization to telomeres (Tel300) in cells arrested in G2 with nocodazole was indeed severely reduced in a sir3Δ strain compared to the isogenic SIR3 strain shown in Fig. 3C. Association of Dna2-Myc with subtelomeric chromatin (Tel3500) was undetectable in the sir3Δ strain. Interestingly, in contrast to the SIR3 strain, in the sir3Δ strain, CUP1 sequences were now found in the Dna2-Myc immunoprecipitates (compare the CUP1 bands in Fig. 3C and Fig. 3D). This may suggest that Dna2-Myc associates with internal DNA when not sequestered at the telomere, although much further work would be required to verify this. Although the apparent Sir3 dependence of Dna2-Myc localization was virtually identical to the Sir3 dependence of yKu localization to telomeric and subtelomeric chromatin (42), it is unexpected for a DNA replication protein.
The residual Dna2p associated with Tel300 is consistent with the fact that DNA2 may be required for their telomerase-dependent replication (see below), which occurs actively in nocodazole-arrested cells (18). The Tel3500 DNA is most likely replicated by standard forks moving out from the subtelomeric ARS, and this replication is likely complete long before cells arrest in nocodazole. Therefore, the Sir3-dependent localization of Dna2p to Tel3500 in G2 is not likely due to the function of Dna2p in subtelomeric replication, which does not require Sir3p. The SIR3 requirement strongly suggests that the telomeric association of Dna2-Myc observed in the ChIP assays is physiologically significant, as well as identifying at least one factor that is important for efficient Dna2p recruitment/and or retention at telomeres.
Localization of Dna2p by indirect immunofluorescence.
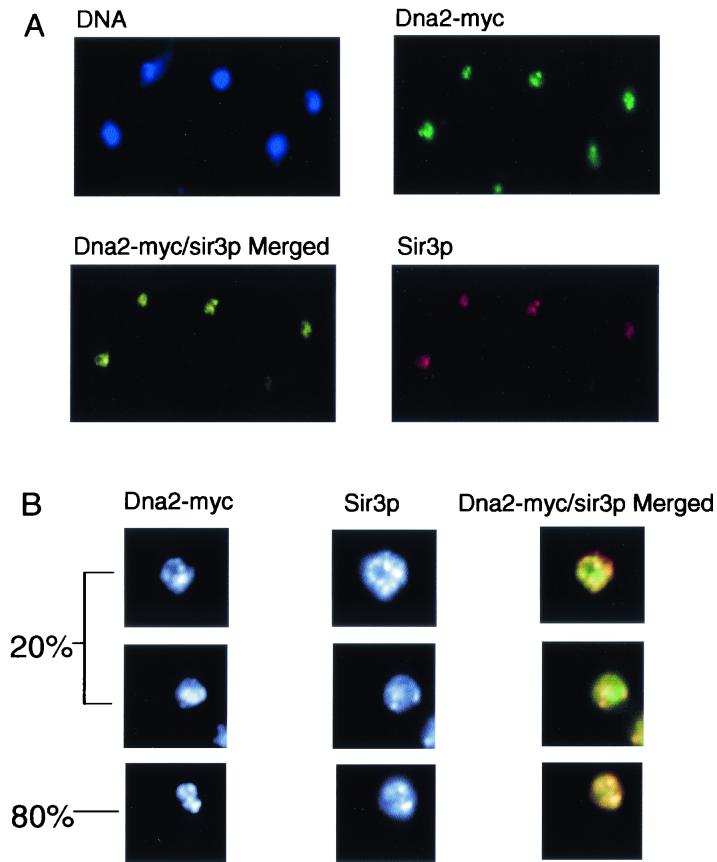
To obtain an overview of the localization of the entire complement of Dna2p in the cell, we used indirect immunofluorescence microscopy. The same strain, carrying Dna2-Myc under the control of its natural promoter, was used for these experiments. Telomeres have been found to cluster in several large foci around the nuclear periphery in S. cerevisiae (28). Cells from an asynchronous culture were fixed and costained with 9E10 anti-Myc antibody, to detect Dna2-Myc, and anti-Sir3p antibody, the latter serving as a marker for telomeres (36). Sir3p showed large foci (Fig. 4A), and these were near the nuclear periphery (Fig. 4B), as expected for telomeric foci. Strikingly, Dna2-Myc protein also showed a focal staining pattern (Fig. 4A). The merged fluorescence of the anti-Myc and anti-Sir3p antibodies showed overlap of Dna2-Myc foci with Sir3p in almost 80% of the cells (Fig. 4A). Thus, Dna2-Myc is abundant at telomeres.
FIG. 4.
Dna2p is concentrated in telomeric domains. Indirect immunofluorescence was carried out as described in Materials and Methods. (A) DNA in nuclei was stained with DAPI and is shown in blue. Staining of Sir3p (red) and Dna2-Myc (green) and costaining (yellow) is indicated. (B) Left column, black and white images of anti-Myc-stained cells. Center column, same cells showing the anti-Sir3p stain. Right column, high-resolution color image of the merged anti-Sir3p- and anti-Myc-stained images of three different cells. Yellow indicates colocalization. The bottom cell represents 80% of the cells in the culture. The remaining 20% appear similar to the top and middle cells, indicated by brackets. About 50 cells were evaluated. (C) Confocal microscopy of six sections through a single cell stained with Sir3p antibody (red), Dna2-Myc (green), and the bottom row shows the merged images in each section of the Z stack shown. Locations of foci within the nucleus are diagrammed to the left.
Although 75 to 80% of the cells showed most of the Dna2-Myc in the Sir3-containing foci, 20 to 25% of the cells clearly showed more dispersed Dna2-Myc. The top two cells in Fig. 4B represent the 20% with clearly dispersed Dna2-Myc. In the top cell, there appear to be some clumps of Dna2-Myc, but there is also a more dispersed Dna2-Myc staining that fails to overlap with Sir3p. The middle cell shows Sir3p foci and even more clearly nonoverlapping dispersed Dna2-Myc. The bottom cell represents the major class in which the two proteins are almost entirely colocalized. A likely explanation for the heterogeneity, given the results of the ChIP assays, is that the two patterns, focal and diffuse, represent cells at different points in the cell cycle and/or cells in transition (see Fig. 5).
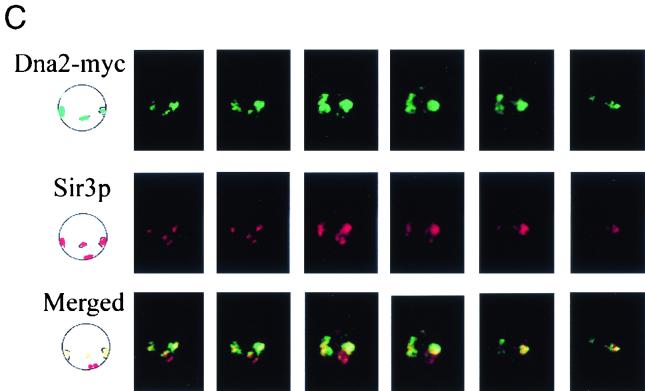
To ensure that the foci overlapped in three dimensions, cells were observed in the confocal microscope. In Fig. 4C, the first picture in each row is an interpretation of the real images of four foci in the first of the six Z-sections of a single cell shown in the six adjacent photographs in the same row. The first row shows Dna2p in green, the second row shows Sir3p in red, and the extent of colocalization of the two proteins is shown in the merged images in the third row (yellow/orange). Three out of four Dna2p foci overlap Sir3p foci.
Redistribution of Dna2p during the cell cycle as shown by immunofluorescence.
To demonstrate that the focal and diffuse staining patterns for Dna2-Myc were the result of dynamic localization to telomeres, synchronization studies were carried out. As shown in Fig. 5A, Dna2-Myc localized in nuclear foci in G1 phase (peripheral, bright green foci on the blue background of DAPI-stained DNA). This, like the ChIP assays (Fig. 3), suggests that Dna2p is localized to telomeres even at a point in the cell cycle when the telomere is clearly not replicating (18, 40). In contrast to G1 phase, Dna2-Myc showed diffuse nuclear staining in S phase, appearing as an aqua stain over almost the entire DAPI-stained area. This pattern is different from that reported for Sir3p, which is found in foci during S phase as well as in the other cell cycle phases (36).
The difference in Sir3p and Dna2-Myc localization in S phase likely explains our observation that there was not a direct one-to-one correspondence between Sir3p foci and Dna2-Myc foci in every cell in an asynchronous culture (Fig. 4B). The diffuse staining pattern in S phase is probably not due to an increase in expression of DNA2 during S phase, since its expression is constant in the cell cycle (17, 55). Furthermore, the ChIP assays confirm the reduction of Dna2p at telomeres during S phase (Fig. 3), and Western blots show the same amounts of Dna2p in G1, S, and G2 phase cells (data not shown). We propose that much of the Dna2p found at telomeres in G1 delocalized during S phase to associate with replication forks for Okazaki fragment processing or to repair DSBs arising due to replication errors (62).
Dna2p redistributes in response to DNA damage.
The yKu and Sir proteins are normally enriched in telomeric foci, but redistribute throughout the nucleus in S phase upon treatment of cells with agents that cause DSBs (42, 45). dna2 mutants are sensitive to such agents (9, 11, 25). Bleomycin causes DSBs by a concerted free radical attack on sugar moieties, preferentially attacking adjacent residues on opposite strands. A serial dilution growth assay shows that dna2-2 mutant strains are roughly 1,000 times more sensitive than the wild type to bleomycin and almost as sensitive as the rad52Δ recombination repair-defective control (Fig. 5B).
We examined the localization of Dna2-Myc after treatment of wild-type cells with a low level (15 mU/ml) of bleomycin for 3 h. Dna2-Myc showed a diffuse staining pattern in all cells, as did the Sir3p control (Fig. 5C), rather than the focal pattern shown in Fig. 4A. Like yKu and Sir3p after DNA damage (42, 45), Dna2p and Sir3p do not completely colocalize in damaged cells, but both are clearly dispersed. Also as in the case of Sir3p and yKu, some foci remain. When a ChIP assay was carried out on extracts prepared from cells treated identically with bleomycin, there was a significant reduction of the amount of Dna2p located at telomeres (O. Imamura, W. Choe, and J. L. Campbell, unpublished data).
The changes in the Dna2-Myc focal staining pattern are likely due to relocalization rather than to changes in expression of Dna2p, since Western blotting showed equal amounts of Dna2-Myc before and after bleomycin treatment (Fig. 5D). This relocalization of Dna2-Myc could be due to some change in telomere structure or to a checkpoint-regulated mobilization, such as Mec1-dependent yKu and Sir3p redistribution after DNA damage (27, 42). Alternatively, since Dna2p relocalizes in S phase without DNA damage (Fig. 3B and 5A), the Dna2, damage-dependent pattern could be due to an extension of S phase due to engagement of the S phase checkpoint.
DNA2 functions in telomerase-dependent telomere replication.
dna2 mutants synthesize a 2C DNA content, but the DNA is fragmented. They are thus deficient in processing of the newly synthesized DNA rather than in synthesis per se (10, 24). To assess whether Dna2p functioned in telomere DNA replication and whether it was required for synthesis and/or processing, a physical assay that allows monitoring of telomere addition at the fully nonpermissive temperature was adopted (Fig. 6A) (18). The experiment measures addition of telomeric repeats onto a chromosomal end generated by induction of the HO endonuclease and terminating in C1-3A repeats (Fig. 6A). In this assay, rad52 must be deleted to prevent healing of broken telomeres by recombination. Like many other mutations causing defects in lagging-strand DNA replication, such as rad27Δ, most dna2 alleles cause severe growth defects and reduced maximum permissive temperature when combined with rad52Δ mutations. It is possible, however, to isolate viable dna2-2 rad52Δ mutants. These double mutants grow well at 30°C (9) but become dependent on RAD52 for survival and fail to grow at 37°C.
As shown in Fig. 6B, dna2-2 showed reduced efficiency of telomere addition at the restrictive temperature. In this figure, the band labeled LM is the product of HO cutting of the fragment labeled PRE, and telomere addition is represented by the SM smear of elongating double-stranded DNA just above LM. Quantitation of the appearance of the SM smear and disappearance of the LM band after normalization by the internal hybridization control (band labeled INT) shows that dna2 mutants are at least six times less efficient in telomere synthesis than the wild type at the 240-min point (Fig. 6C). In DNA2 wild type, telomere sequence addition is more efficient than HO cutting, such that LM was rapidly converted to SM. In the dna2 mutant, telomere sequence addition is less efficient than HO cutting, such that LM accumulated instead of converting to SM (Fig. 6D). This pattern is striking because formation of LM itself is slower in the dna2 mutant than in the wild type; that is, HO cutting is less efficient in the dna2 mutant at the nonpermissive temperature than in the wild type (see Fig. 6B, band labeled PRE, and quantitation in Fig. 6D). Thus, LM accumulation in the mutant cannot be due to more rapid production of LM (Fig. 6D), but must be due to failure of telomere sequence addition. Similar slow digestion with HO was seen in the replication mutants studied previously (18). We conclude that DNA2 is required for telomerase-dependent telomere synthesis and not just for subsequent processing (see Discussion).
There is some residual elongation of the HO cut end (conversion of LM to SM, Fig. 6) in the dna2-2 mutant. Since the DNA2 gene is essential for viability, dna2-2 mutants, which are viable at all temperatures (in the presence of RAD52), are by definition leaky. Thus, the small amount of synthesis observed may be due to residual dna2 activity, as was found for polymerase δ mutants (pol3), in the study that originally used this assay (18). However, further work is required to distinguish leakiness from the participation of a backup mechanism.
dna2 mutations accelerate the senescence of mutants lacking telomerase.
Since the results in Fig. 6 are most simply interpreted as a coordination between telomerase and polymerase α, polymerase δ, primase, and Dna2p, perhaps akin to the coordination of leading and lagging-strands at normal replication forks, we investigated the phenotypes of dna2 mutants lacking telomerase. EST2 encodes the catalytic subunit of telomerase. EST1, a telomerase RNA binding protein, also affects telomerase activity in vivo, since est1 mutants show progressive telomere shortening (32).
To test the interaction between DNA2 and telomerase function, strains heterozygous for both DNA2 and EST2 or DNA2 and EST1 were sporulated, and tetrads were dissected. When otherwise isogenic dna2-2 EST2/DNA2 est2Δ or EST2/est2Δ heterozygotes were sporulated, the dna2-2 est2Δ and est2Δ spores germinated efficiently (Fig. 7A and Table 1). When the spores were restreaked, however, there was a large reduction in viability in the dna2-2 est2Δ compared to the est2Δ colonies (Fig. 7A). The same was true for spores with the dna2-2 est1Δ genotype compared to est1Δ; that is, a much smaller percentage of the double mutant cells remained viable after the germinating spore grew into a colony and was restreaked (data not shown). This suggests that the double mutants senesce more rapidly than the single est mutants (Fig. 7A).
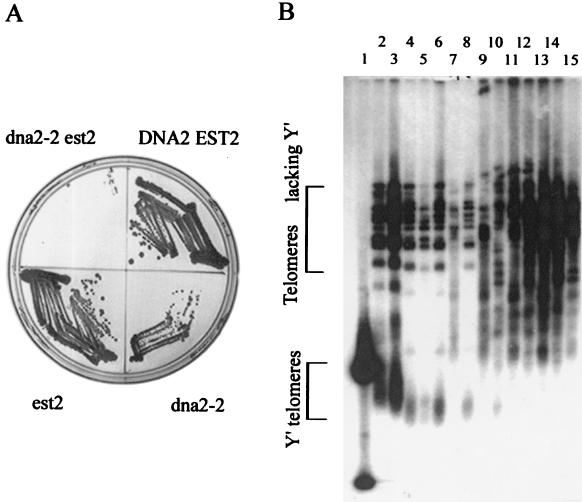
FIG. 7.
Telomerase-deficient mutant deficient in dna2 shows accelerated senescence. (A) Increased senescence in dna2-2 est2Δ mutants. A diploid strain (MATα est2::KanMX4 leu2Δ met15Δ ura3Δ/MATa dna2-2::LEU2 KANMX4 leu2Δ met15Δ ura3Δ) heterozygous at both the EST2 and DNA2 loci was sporulated and dissected. After 2 days (about 25 generations), the spores were replica plated onto G418-containing medium lacking leucine. Individual spores from a tetrad with the tetratype genotype were then restreaked on rich medium and grown to single colonies (about 25 divisions). DNA2+ EST2+, dna2-2 est2, DNA2 +est2Δ, and dna2-2 EST2+ cells were restreaked on a YPD plate. The same experiment was performed with a dna2-2 est1Δ heterozygous diploid (not shown, but see panel B). (B) Telomere maintenance in senescing estΔ and dna2 est1Δ mutants. A diploid strain (MATα est1::KanMX4 leu2Δ met15Δ ura3Δ/MRTa dna2-2::LEU2 KANMX4 leu2Δ met15Δ ura3Δ) heterozygous at both the EST1 and DNA2 loci was sporulated and dissected, and the genotype of the spore products determined, as in panel A. A tetrad with the tetratype genotype was then streaked onto a YPD plate at 30°C as in panel A. From this plate, est1Δ and dna2-2 est1Δ colonies were inoculated into 5 ml of YPD medium. After the cultures grew to saturation, 2.5 μl of each culture was added to 5 ml of fresh YPD medium. The inoculations were repeated six times. The type II survivors appeared in the dna2-2 est1Δ strain after two inoculations and in the est1Δ strain after four inoculations. Telomere maintenance was assessed by preparation of DNA from each culture, digestion with XhoI, gel electrophoresis, blotting, and hybridization of the telomere blot with a C1-3A telomere probe as described (39). Lane 1, marker; large spot in the Y′ telomere bracketed region is about 1 kb; lane 2, DNA2+, asynchronous culture; lane 3, dna2-2, asynchronous culture; lane 4, est1Δ after one passage; lane 5, dna2-2 est1Δ after one passage; lane 6, est1Δ after two passages; lane 7, dna2-2 est1Δ after two passages; lane 8, est1Δ after three passages; lane 9, dna2-2 est1Δ after three passages; lane 10, est1Δ after four passages; lane 11, dna2-2 est1Δ after four passages; lane 12, est1Δ after five passages; lane 13, dna2-2 est1Δ after five passages; lane 14, est1Δ after six passages; lane 15, dna2-2 est1Δ after six passages. The smear beginning above the bracket denoting Y′ telomeres and the larger rearranged bands represent the putative type II intermediates. These experiments were performed before publication of a higher-resolution assay for type I and type II (58, 59).
Dna2p may participate in telomerase-independent telomere elongation.
Although most cells in the double mutants die, survivors do appear. Telomere maintenance was monitored in both the dna2-2 est1Δ and est1Δ strains as they senesced. Although initially the small percentage of cells remaining viable in the dna2-2 est1Δ spores grew slowly, the dna2-2 est1Δ survivors showed much larger colonies than the est1Δ survivors after only 60 generations (data not shown). This suggested that so-called type II survivors, which have a growth advantage over type I survivors, might arise more rapidly in the dna2-2 est1Δ in the est1Δ mutant (39, 59). Type I survivors arise through RAD51-dependent homologous recombination between Y′ elements, whereas type II survivors acquire very long C1-3A tracts due to some as yet uncharacterized, RAD51-independent, RAD50-dependent mechanism (37, 39, 58, 59). Type I telomeres have multiple, tandem copies of Y′ elements terminating in a short segment of C1-3A tracts and are slightly longer than normal telomeres. Type II telomeres arise from very short telomeres and contain up to 12 kb of C1-3A tracts. This is the type of survivor seen in alternative methods of maintaining telomeres (ALT) in mammalian tumor cells (8).
Single colonies arising from the first restreak of the germinated est1Δ and dna2-2 est1Δ spores were serially passaged, and the structure of the telomeres was monitored during this outgrowth (39). DNA from survivors isolated after various numbers of passages was digested with XhoI, electrophoresed, blotted, and probed with a C1-3A repeat probe (Fig. 7B). After the first passage, telomeres seemed to have shortened in both dna2-2 est1Δ and est1Δ strains to a similar degree. However, patterns characteristic of fast-growing type II survivors appeared much earlier in the double mutant (two passages, or about 40 generations; Fig. 7B, lanes 7 and 9) than in the single est1Δ mutant (four passages; Fig. 7B, lane 10). In lanes 7 and 9, the dna2-2 est1Δ double mutant, a smear of new higher-molecular-weight bands appeared concomitant with a decrease in intensity in the broad telomeric XhoI bands characteristic of the early survivors or wild type (marked Y′ in lanes 2 to 5). In lanes 6 and 8, the est1Δ mutant after the same number of passages, neither the disappearance of the major XhoI band nor the smear of longer bands was seen.
The pattern in lanes 7 and 9, dna2-2 est1Δ, corresponds to that described as type II (see Fig. 1B, lane 5, in Lundblad and Blackburn [39] and Le et al. [37]). In Fig. 7B, lanes 4 and 6, est1Δ, the faint band running just above the 1 kb marker may represent type I survivors that have acquired a Y′ element in the est1Δ strain. Since type II grows more rapidly than type I, type II eventually appears in est1Δ and predominates in the later passages from both dna2-2 estΔ and est1Δ colonies (lanes 10 to 15). Lanes 2 and 3 contain controls showing that telomeres are more heterogeneous in length in dna2-2 than in an isogenic DNA2 strain, though not exclusively longer as reported previously in a different genetic background (25). Premature senescence and early appearance of type II survivors were also seen in rad51 tlc1Δ double mutants (37).
DISCUSSION
Roles of Dna2p localization and relocalization in telomere replication.
Although dynamic association/disassociation of telomere factors and their relation to the functions of telomeres is an active area of current research (16, 49), the studies generally fail to directly address the interaction of telomerase with the putative telomeric primosome. Yet, given the evidence, albeit indirect that there is coordination of telomerase with components of the lagging-strand apparatus, a full understanding of the end-replication problem will only be possible by understanding lagging-strand functions at telomeres as well as telomerase and its components (21).
Our localization results are just the beginning, but the details are surprising and suggest much more complex functions for replication proteins than heretofore proposed. We have established that, in addition to lagging-strand DNA polymerases, Dna2p, a processing enzyme, has a function in de novo telomere synthesis (Fig. 6 and 7). What is unanticipated is that Dna2p does not appear to function solely in processing, but that it is required for synthesis itself (Fig. 6). (We reject the alternative idea that 100-bp Okazaki fragments are made and destroyed as inconsistent with previous work on discontinuous synthesis.) This is the first direct evidence that an entire primosome, that is, one containing both synthesis and maturation functions, is essential for proper end replication.
In support of a telomeric primosome complex containing Dna2p is that dna2 mutants are synthetically lethal with ctf4 mutants. Ctf4p is a polymerase α binding protein, and ctf4 mutations dramatically reduce the elongated telomeres observed in polymerase 1 mutants (25). We propose that, as in bacteria, there may be at least two ways to assemble a eukaryotic primosome: one mediated by Orc-dependent assembly of primosomes at unwound replication origins, and a second mediated perhaps by the telomerase machine at single-stranded chromosomal termini.
The data in Fig. 6 not only show that Dna2p is required for synthesizing a double-stranded telomere but also imply that Dna2p must interact with or is dependent on previous action of telomerase in order to synthesize this double strand (18). The fact that dna2-2 mutants show slightly longer telomeres than the wild type (25) yet are deficient in the telomere elongation assay (this work) suggests that completion of the lagging-strand involving Dna2p at telomeres may require tight coordination of the lagging-strand replisome with telomerase activity. The synergistic effect on senescence of introducing the dna2-2 mutation into an est2Δ or est1Δ mutant background (Fig. 7A) is also consistent with integration of leading-strand synthesis by telomerase and lagging-strand maturation by a primosome (if one interprets synthetic lethality as indicating interaction). One might envision that, just as the standard primosome coordinates with the leading-strand replication machinery at a conventional fork, a similar primosome coordinates with telomerase for concomitant 3′ extension of the G-rich strand and 5′-to-3′ fill-in of the C-rich strand.
Role in telomere capping?
It was during our probing of the spatial and temporal pattern of Dna2p interaction with telomeres that we encountered evidence suggesting that Dna2p played additional roles at telomeres. We propose that one of these is in the telomere capping or end protection process. First, the relative fraction of the cellular Dna2p sequestered at telomeres (Fig. 4) is unexpectedly large for the synthesis of what would be anticipated to constitute a small fraction of the total number of Okazaki fragments. Second, the timing of association-dissociation is unexpected for a protein having a role exclusively in end-replication. Namely, Dna2p is at telomeres in G1 phase (Fig. 3 and Fig. 5), when there is no DNA replication in any part of the chromosome (18, 40). At this time, telomerase is present and catalytically active, a state that has been shown to have a protective effect on telomeres (44), and transcriptional activators cannot activate transcription at telomeres, suggesting a closed structure (21).
Elsewhere in the chromosome, ORC, Cdc6, and the Mcms, replication proteins that will later recruit the replication fork primosome, are associating with ARSs. Dna2p (Fig. 3), however, like other elongation factors, is undetectable in the bulk chromatin (2, 56, 57). This timing and several additional findings in our work lead to the interpretation that the G1 telomeric Dna2p may be involved capping of the telomere. First, while localization of Dna2p at C1-3A/TG1-3 repeats in G2 might be expected, since their terminal portion is replicated by telomerase, enrichment in G1 is unexplained. Second, enrichment in sequences 3,500 bp away from the telomere in G1 is unexpected since this DNA is replicated by the standard replication fork, which is not present in G1. Association with subtelomeric chromatin probably accounts for the effects of overproduction on telomere position effect (Fig. 1). Telomere position effect in turn may reflect telomere capping, since several of the same proteins affect both (6). Perhaps an imbalance in one putative component, Dna2p, may lead to disruption of the capping-silencing complexes. Our observation that telomeres shorten when Dna2p is overproduced (Fig. 1B) is also more consistent with disruption of a protective cap than with an effect of overproduction on Okazaki fragments. A capping role is also supported by the fact that the phenotype of the dna2-2 est1Δ (Fig. 7) mutant is the same as that of yKu tlc1Δ double mutants (31). yKu is a well-known component of the telomere cap in S. cerevisiae (44).
We were at first surprised at the dramatic change in Dna2p localization at the G1/S phase transition. Instead of associating with telomeres for their replication at this time, Dna2p spreads out throughout the entire nucleus (Fig. 3 and 5) and is now found associated with internal sequences replicating from activated ARSs (Fig. 3). In fact, Dna2p is not even detectable at the telomere in the ChIP assay at the first S phase time point, when it is already associated with Cup1 and the rDNA. We propose now that this relocalization is associated with transient uncapping of telomeres and preparation for their replication later in the cell cycle. This may represent the “judiciously regulated degree of uncapping” proposed by Blackburn, during which the telomere has to actually transiently resemble a break for telomerase to act (6).
A role for Dna2p in capping would also explain why Sir3p, required for telomere position effect but not for DNA replication, is apparently required for Dna2p association with telomeres (Fig. 2B and Fig. 3). The Sir3p requirement for association with subtelomeric chromatin, in addition to the ends, would be especially surprising if the only role of Dna2p were for DNA replication, since these sequences should obtain their replicative complement of Dna2p from standard replication forks and not need Sir3p to recruit Dna2p for Okazaki fragment processing.
An alternative ad hoc model is that Dna2p does not function in capping but is merely stored at telomeres and that its release from storage at telomeres is an S phase regulatory mechanism. It is noteworthy that Dna2p is an unusual DNA replication protein in that DNA2 contains no MCB cell cycle-regulated upstream promoter element and accordingly is not transcriptionally regulated in the cell cycle (55). Thus, Dna2p may be regulated differently from other replication proteins. There is, however, no precedent for such a model.
In late S phase and G2 phase, Dna2p is recruited again to telomeres. This correlates with the time at which Dna2p is required for telomerase-dependent telomere elongation (Fig. 6) and when subtelomeric regions replicate (22, 60). The large amount of Dna2p now associated with telomeres might also have a capping role in stabilizing the newly formed telomeres. Supporting this additional role, again Sir3p is required for localization in G2, for at least some of the Dna2p (Fig. 3C).
Checkpoint role for Dna2p at telomeres-unifying capping, replication, and repair.
Sir proteins and yKu can relocalize from telomeres to DSBs elsewhere in the chromosome (42, 45, 53). The change in the Dna2-Myc staining pattern in nuclei after DNA damage (Fig. 5) could analogously suggest that Dna2p is stored at telomeres and targeted away from telomeres to the sites of damage. If the only role for Dna2p at telomeres were C-rich strand fill-in, then relocalization after DNA damage would not be necessary. Release could occur in several different ways. Damage at telomeres might lead to an alteration of telomeric structure that releases Dna2p, or the DNA damage checkpoint might induce relocalization, as has been shown for Sir3p and yKu (26, 42, 45). It is not yet known if RAD9 or MEC1 is required for relocalization of Dna2p, as they are for Sir3p and yKu (42, 45). This response to damage may also reflect the telomerase-independent role of Dna2p in telomerase biogenesis (Fig. 6). Interpretation of the redistribution seen in our damage experiments will require further work, however, since the effect of DNA damage is superimposed on the cell cycle distribution seen in undamaged cells.
An appealing alternative model for telomeric sequestration of proteins suggests that assembly of Sir proteins, yKu, and now Dna2p at telomeres signals the completion of both DNA replication and repair of DSBs through a telomere chromatin checkpoint (27). When these proteins are free in the nucleus, it triggers a checkpoint to block entry into mitosis until DNA replication and repair are complete and the proteins return to telomeric chromatin, releasing cells from the checkpoint. The involvement of Dna2p in a checkpoint has been proposed to account for the fact that dna2 mec1 and dna2 rad9 double mutants are viable, though they fail to arrest mitosis (24, 25). In a mec1 mutant, Dna2p might, like yKu and the Sir proteins, also remain associated with telomeres, allowing passage through mitosis and a second chance to repair damage in the next cell cycle.
In summary, the localization study, supported by genetics and biochemistry, suggest a dual role for Dna2p at telomeres. We provide the first evidence of when and where the lagging-strand proteins associate with telomeres. Taken together with the demonstration by others that polymerase α is found associated with Cdc13p, a telomere binding protein, it also suggests that a primosome complex forms that may account for coordination between telomerase and lagging-strand maturation. In addition, we provide evidence for a novel capping-checkpoint function for a lagging-strand DNA replication protein.
Acknowledgments
We thank D. Gottschling, V. Zakian, and L. Guarente for generously providing the yeast strains for telomeric assays and Sir3p antibody described in Materials and Methods. We thank the Caltech-ERATO center for use of the Nikon microscope and image processing.
This research was supported by NSF POWRE grant MCB9805943 and support from the Pomona College Research Committee to L.L.M.H. and NIH GM25508 and NSF MCB9985527 to J.L.C.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, O. M., D. M. Weinstein, and S. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 3.Bae, S., E. Choi, K. Lee, J. Park, S. Lee, and Y. Seo. 1998. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273:26880-26890. [DOI] [PubMed] [Google Scholar]
- 4.Bae, S. H., K.-H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, E. H. 2000. Telomere states and cell fates. Nature 408:53-56. [DOI] [PubMed] [Google Scholar]
- 7.Bourns, B. D., M. K. Alexander, A. M. Smith, and V. A. Zakian. 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18:5600-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 9.Budd, M. E., and J. L. Campbell. 2000. Interrelationships between DNA repair and DNA replication. Mutat. Res. 451:241-255. [DOI] [PubMed] [Google Scholar]
- 10.Budd, M. E., and J. L. Campbell. 1995. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budd, M. E., and J. L. Campbell. 2000. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459:173-186. [DOI] [PubMed] [Google Scholar]
- 12.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out Its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budd, M. E., W.-C. Choe, and J. L. Campbell. 1995. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270:26766-26769. [DOI] [PubMed] [Google Scholar]
- 14.Budd, M. E., W.-C. Choe, and J. L. Campbell. 2000. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275:16518-16529. [DOI] [PubMed] [Google Scholar]
- 15.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in S. cerevisiae. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 16.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, R., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conay, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 18.Diede, S. G., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerase alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 19.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dionne, I., and R. J. Wellinger. 1998. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 26:5365-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuBois, M. L., S. J. Diede, A. E. Stellwagen, and D. E. Gottschling. 2000. All things must end: telomere dynamics in S. cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 65:281-296. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson, B. M., B. J. Brewer, A. E. Reynolds, and W. L. Fangman. 1991. A yeast origin of replication is activated late in S-phase. Cell 65:507-515. [DOI] [PubMed]
- 23.Ferguson, B. M., and W. L. Fangman. 1992. A position effect on the time of replication origin activation in S. cerevisiae. Cell 68:333-339. [DOI] [PubMed] [Google Scholar]
- 24.Fiorentino, D. F., and G. R. Crabtree. 1997. Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S Phase. Mol. Biol. Cell 8:2519-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formosa, T., and T. Nitiss. 1999. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full DNA2 helicase activity is not essential for growth. Genetics 151:1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galy, V., J.-C. Olivo-Marin, H. Scherthan, V. Doyes, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403:108-112. [DOI] [PubMed] [Google Scholar]
- 27.Gasser, S. 2000. A sense of the end. Science 288:1377-1378. [DOI] [PubMed] [Google Scholar]
- 28.Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Sherthan, and S. M. Gasser. 1996. The clustering of telomeres and colocalization with rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy, M. Grunstein, and S. M. Gasser. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould, K. L., C. G. Burns, A. Feoktistiva, C. Hu, S. G. Pasion, and S. L. Forsburg. 1998. Fission yeast cdc24+ encodes a novel replication factor required for chromosome integrity. Genetics 149:1221-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravel, S., M. Larrivé, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed]
- 32.Hughes, T. R., D. K. Morris, A. Salinger, N. Walcott, C. I. Nugent, and V. Lundblad. 1998. The role of the EST genes in S. cerevisiae telomere replication. Ciba Found. Symp. 211:41-47. [DOI] [PubMed] [Google Scholar]
- 33.Ireland, M. J., S. S. Reinke, and D. M. Livingston. 2000. The impact of lagging-strand replication mutations on the stability of CAG repeat tracts in S. cerevisiae. Genetics 155:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang, J.-Y., E. Choi, S.-H. Bae, K.-H. Lee, B.-S. Gim, H.-D. Kim, C. Park, S. A. MacNeill, and Y.-S. Seo. 2000. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics 155:1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo, C.-L., C.-H. Huang, and J. L. Campbell. 1983. Isolation of yeast DNA replication mutants using permeabilized cells. Proc. Natl. Acad. Sci. USA 80:6465-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laroche, T., S. G. Martin, M. Tsai-Pflugfelder, and S. Gasser. 2000. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 129:159-174. [DOI] [PubMed] [Google Scholar]
- 37.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Q., W.-C. Choe, and J. L. Campbell. 1999. Identification of the Xenopus laevis homolog of Saccharomyces cerevisiae DNA2 and its role in DNA replication. J. Biol. Chem. 275:1615-1624. [DOI] [PubMed] [Google Scholar]
- 39.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 40.Marcand, S., V. Brevet, C. Mann, and E. Gilson. 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10:487-490. [DOI] [PubMed] [Google Scholar]
- 41.Martin, A. A., I. Donne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and Sir proteins in response to DNA strand breaks in S. cerevisiae. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 43.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerases alpha and varepsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 45.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 97:609-620. [DOI] [PubMed] [Google Scholar]
- 46.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: Components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 47.Ohki, R., T. Tsurimoto, and F. Ishikawa. 2001. In vitro reconstitution of the end replication problem. Mol. Cell. Biol. 21:5753-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in S. cerevisiae mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 50.Price, C. M. 1997. Synthesis of the telomeric C-strand. A review. Biochemistry (Moscow) 62:1216-1223. [PubMed] [Google Scholar]
- 51.Pryde, F. E., H. C. Gorham, and E. J. Louis. 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7:822-827. [DOI] [PubMed] [Google Scholar]
- 52.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 53.Shore, D. 2000. The Sir2 protein family: a novel deacetylase for gene silencing and more. Proc. Nat. Acad. Sci. USA 97:14030-14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Nahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes in the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an MCM protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka, T., and K. Nasmyth. 1998. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teng, S.-C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 59.Teng, S.-C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wellinger, R., A. Wolf, and V. Zakian. 1993. Origin activation and formation of single-strand TG1-3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13:4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wellinger, R. J., K. Ethier, P. Labrecque, and V. A. Zakian. 1996. Evidence for a new step in telomere maintenance. Cell 85:423-433. [DOI] [PubMed] [Google Scholar]
- 62.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]