Abstract
Surprisingly, the contribution of defects in DNA replication to the determination of yeast life span has never been directly investigated. We show that a replicative yeast helicase/nuclease, encoded by DNA2 and a member of the same helicase subfamily as the RecQ helicases, is required for normal life span. All of the phenotypes of old wild-type cells, for example, extended cell cycle time, age-related transcriptional silencing defects, and nucleolar reorganization, occur after fewer generations in dna2 mutants than in the wild type. In addition, the life span of dna2 mutants is extended by expression of an additional copy of SIR2 or by deletion of FOB1, which also increase wild-type life span. The ribosomal DNA locus and the nucleolus seem to be particularly sensitive to defects in dna2 mutants, although in dna2 mutants extrachromosomal ribosomal circles do not accumulate during the aging of a mother cell. Several other replication mutations, such as rad27Δ, encoding the FEN-1 nuclease involved in several aspects of genomic stability, also show premature aging. We propose that replication fork failure due to spontaneous, endogenous DNA damage and attendant genomic instability may contribute to replicative senescence. This may imply that the genomic instability, segmental premature aging symptoms, and cancer predisposition associated with the human RecQ helicase diseases, such as Werner, Bloom, and Rothmund-Thomson syndromes, are also related to replicative stress.
Saccharomyces cerevisiae provides a promising model system for the identification and study of genetic pathways involved in determining longevity in more complex organisms (25). In yeast there is a spiral form of aging in which mother cells in each division increase in age by one generation, while each new bud is produced at age zero (33, 67). The total life span of yeast is counted as the number of times the mother cell is able to bud. This form of aging resembles mammalian cell replicative senescence (28), the finite number of divisions of a cell in culture, with two differences. Yeast cell division is asymmetric rather than symmetric with respect to age, and yeast cells lyse at the end of the life span rather than undergoing some form of differentiation. We have been interested in using yeast to study the mechanism underlying certain human premature aging and cancer susceptibility syndromes.
The yeast SGS1 gene encodes a homolog of the WRN, BLM, and RecQ4 helicases, members of the RecQ family. Mutations in these human genes cause disorders of premature aging and/or cancer susceptibility (16, 17, 45, 98). The helicase encoded by SGS1 is thought to be involved in the down-regulation of homologous recombination in the ribosomal DNA (rDNA) repeats. sgs1Δ mutants show a severely reduced average life span and are therefore of interest in understanding the molecular mechanisms of human diseases affecting homologous genes (80). Based on the phenotypes of aging that seemed to occur early in sgs1Δ mutants as well as on other findings, it was proposed early on that a reasonable model for yeast aging was that hyperrecombination in the rDNA produces extrachromosomal ribosomal circles (ERCs) that accumulate preferentially in mother cells and ultimately lead to cell death (79). The beauty of this model is that, if true, it would explain the asymmetry of yeast aging, since extrachromosomal plasmids lacking centromeres or 2μm circle replication origin and partition functions are inherited primarily by mothers and not by daughters (68). Evidence for early ERCs in sgs1 mutants compared to those of the wild type, however, has not been substantiated by later studies (29, 62). Also contradicting the model, genes that eliminate homologous recombination (RAD50, RAD51, RAD52, and RAD57) lead to shortened life span and do so without giving rise to ERCs (71). Furthermore, in cells recovering from stationary-phase arrest, life span is also shortened without the appearance of ERCs (2). Conversely, in cells in which the retrograde response (a process that involves signaling from the mitochondrion to the nucleus) is induced, life span is extended and yet ERCs appear and accumulate exponentially (43, 44; M. Jazwinski, personal communication). Thus, the contribution of ERCs to aging has become controversial (25). Furthermore, in organisms other than yeast, accumulation of rDNA copies does not correlate with replicative senescence (72).
Our studies were designed to test whether the hyperrecombination rDNA phenotype in aging wild-type and sgs1Δ mutant yeast could be secondary to some more general, systematic, endogenous DNA damage that increases recombination frequency to accomplish repair and leads to detrimental chromosomal rearrangements that accumulate with age. Mutations affecting the nucleotide excision repair, single-strand annealing repair, and transcription-coupled repair pathways do not affect life span (71). Recent studies with bacteria suggesting that DNA replication is more likely to be interrupted than previously supposed and to require recombinational repair (for reviews see references 48 and 51) led us to the hypothesis that DNA damage during DNA replication might be a factor contributing to finite life span of yeast. It is now known that replication forks assembled at origins encounter blocks during propagation, for instance, in the form of endogenous DNA damage, secondary structures, and protein complexes involved in other pathways. If the damage is a template strand interruption, fork encounter may result directly in a double-strand break. Forks encountering other blocks may be converted to four-way junctions by branch migration and reannealing of the newly synthesized strands (60, 61). Processing of this four-way junction in Escherichia coli by RuvABC may result in the generation of a recombinogenic double-strand break (DSB), while RecBCD processing leads to recombination-induced replication restart and removal of damage (75). Alternatively, paused forks may simply lead to disassembly of the replisome followed by replication restart (51). For bacteria, there is good evidence that replication mutations affecting helicases give rise to such damage and that recombination is involved in the rescue of the replication forks (63, 75). Failure of timely rescue leads to genetic instability.
Replication fork demise has also been shown to occur in yeast in response to replication inhibition by hydroxyurea (58). Such collapse may be recombinogenic, since recombination intermediates are known to increase spontaneously during DNA replication in yeast in the absence of any exogenous DNA damaging agent (100); also, specific replication mutants increase the frequency of replication-related recombination, especially at the nonpermissive temperature (27). In addition, several DNA replication mutants accumulate DSBs and require RAD52, essential for recombination or DSB repair, for viability, suggesting that recombination is required to repair the DSBs arising from replicational stress (30). Importantly, these phenomena are not limited to yeast. Early studies of higher eukaryotes showed that DNA damage is converted to DSBs and stimulates sister chromatid exchange only if damaged chromosomes are allowed to traverse S phase (93, 96).
We first investigated the life span of a known DNA replication mutant, dna2 (50), which contains a mutation that affects a yeast helicase/nuclease (4, 7, 9, 10). The helicase domain of DNA2 is closely related to WRN at the primary sequence level, though not as closely as is SGS1. What is striking, however, is that Dna2p, like WRN, contains both helicase and nuclease activities in the same polypeptide (76). Dna2p is thought to be an intrinsic component of the replication apparatus and to either compensate for or cooperate with the RAD27-encoded FEN-1 in the maturation of Okazaki fragments (5, 7, 9). Important in our choice of DNA2 as a test gene was the fact that inactivation of Dna2p leads to fragmentation of replicating chromosomal DNA (7, 38). Furthermore, dna2 mutants are defective in repair of DSBs caused by X rays or bleomycin, agents that mimic oxidative damage, or by methyl methane sulfonate (8, 20). Finally, overexpression of the N terminus of DNA2 reduces telomere position effect, the silencing of genes in telomere proximal positions, suggesting a role in silencing, a major aging pathway (81). In this work we show that dna2 and other replication mutants show premature aging, supporting our model that genomic instability due to replication defects can accelerate replicative senescence.
MATERIALS AND METHODS
Life span determination.
Life spans of virgin mothers were determined by dissection as described previously (33). Strain dna2-1 was grown at 23°C, and all other mutants were grown at 30°C. Statistical significance of differences in life spans was determined with a t test and was confirmed with a Mann-Whitney nonparametric test using StatMost software (Dataxiom Software Inc., Los Angeles, Calif.).
Nucleolar morphology of aging yeast cells.
Cultures of dna2-2 cells were labeled with biotin, grown, and then recovered after 8 to 9 generations with streptavidin paramagnetic beads as described by others (80). Sulfo-N-hydroxy succinamide (long chain)-biotin was from Pierce. Paramagnetic streptavidin-coated beads were from PerSeptive Biosystems (Framingham, Mass.) and were used at 5 mg/ml. An aliquot of the cells isolated was stained for 20 min in a 1 μg/ml-concentration of Calcofluor (Fluorescent Brightener 28; Sigma Chemical Company, St. Louis, Mo.) and was washed in phosphate-buffered saline, and the average number of bud scars was determined for 15 to 20 cells in UV epifluorescence to verify the age of the cells.
Immunofluorescence was performed by a slight modification of the method of Pringle et al. (74). Cells were fixed in formaldehyde, digested with Zymolyase (ICN) for 5 to 30 min until phase dark, washed, and attached to polylysine-coated slides. Slides were blocked for an hour, stained with primary antibody for an hour, rinsed five times, and finally stained with secondary antibody for an hour and rinsed five times. The primary antibody was A66 mouse monoclonal antibody to Nop1p or yeast fibrillarin, an abundant nucleolar protein (1). Donkey fluorescein isothiocyanate-labeled anti-mouse secondary antibodies were from Jackson Immunoresearch (West Grove, Pa.). Nuclei were stained for 5 s with 0.1 μg/ml-concentrations of DAPI (4′,6′-diamidino-2-phenylindole; Sigma) and rinsed for 5 s with deionized water. Cells were mounted in Vectashield (Vector Laboratory, Burlingame, Calif.). Stained cells were photographed with a Hamatsu Digital Camera with a Nikon Eclipse TE300 inverted microscope and Metaphore imaging software. All images are at the same magnification.
Analysis of rDNA from young and old yeast cells.
Young and old cells were prepared by the biotin method described above. DNA was isolated by gentle spheroplasting, and methods for one-dimension analysis to separate circular rDNA from total genomic rDNA (Fig. 2B) and for two-dimension gel electrophoresis in the presence of intercalators were similar to those described previously (79). Chloroquine gels were run in 1% (wt/vol) Tris-acetate-EDTA agarose at 1.3 V/cm for 39 h in 0.6 μg of chloroquine/ml in the first dimension and for 20 h in 3 μg of chloroquine/ml in the second dimension.
FIG. 2.
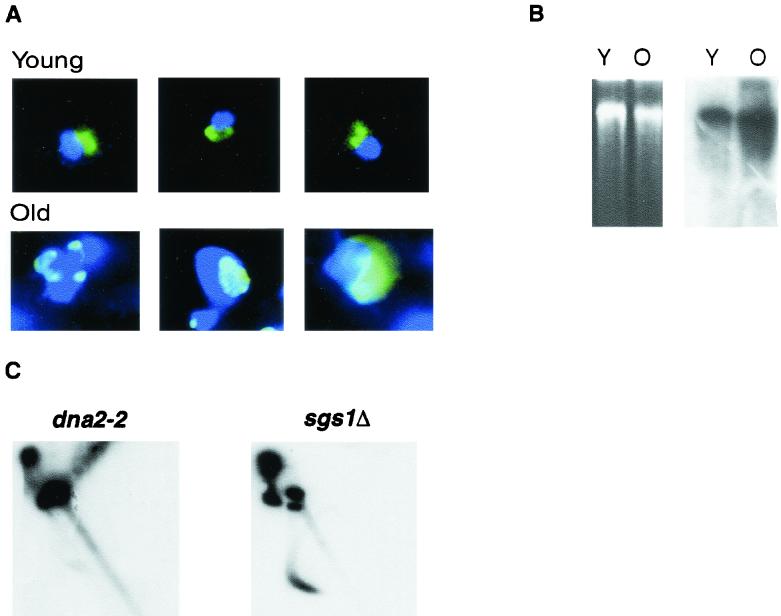
(A) Nucleolar enlargement and fragmentation accompanies aging in dna2-2 strains. Both young cells (labeled Young) and cells isolated 8 generations after being labeled for 1 generation with biotin (labeled Old) were examined (see Materials and Methods). Nuclei stained with DAPI (dark blue) and with anti-Nop1 antibody (green) were observed in young and in eighth-generation dna2-2 cells as indicated. The three cells shown are representative of more than 90 to 95% of the cells observed in each population and, as previously shown, do not simply represent dead cells (80). (B) rDNA is amplified in old cells derived from dna2-2 mutants. dna2-2 cells were prepared at 0 and 8 generations, and DNA was isolated as described previously (79). Total, undigested DNA was analyzed by conventional gel electrophoresis (left) and Southern blotting (right). The ethidium bromide-stained gel is shown on the left, with DNA from equal numbers of generation 0 (Y) and generation 8 (O) cells (as determined by hemocytometer counting). As shown on the right, this gel was blotted to nitrocellulose and hybridized to an rDNA probe labeled by PCR of a plasmid. The upper band (greater than 15 kb) is composed of total genomic DNA. Circular rDNA (5 kb) would migrate within or below the smear of degraded DNA. The wild type did not show amplification after 8 generations (data not shown). (C) rDNA isolated from dna2-2 mutants (7 to 8 generations) contain extrachromosomal rDNA circles but contain fewer than do age-matched sgs1Δ cells. Chromosomal DNA was prepared as described for panel B. Agarose gel electrophoresis was carried out in two dimensions, with each dimension containing chloroquine as described in Material and Methods. Southern blotting was carried out as described for panel B. The DNA on the diagonal represents linear DNAs. DNA forming an arc to the left and below the diagonal is composed of closed circular topoisomers.
ChIP assays.
Cells were synchronized with mating pheromone and released as described previously (14). Samples were taken at the indicated times, and chromatin cross-linking/immunoprecipitation (ChIP) assays were performed as described by Tanaka et al. (86). DNA2 was tagged with 9 myc epitopes and integrated at the DNA2 locus in strain W3110 pep4bar1(RAD5+). Anti-myc monoclonal antibody 9E10 was used for immunoprecipitation. One set of PCR primers is from the 35S RNA transcription region, as indicated in the diagram in Fig. 2D, and one spans the replication fork barrier (RFB) (47). The RFB site was chosen as the starting point for designing the probe with the Saccharomyces Data Base (SDB; Stanford University) and was assigned the nucleotide number 5000. The RFB probe spanned nucleotides 4971 to 5320. PCRs were carried out for different numbers of cycles to ensure that results were in the linear range, as described previously (87).
RESULTS
dna2 mutants have truncated life spans.
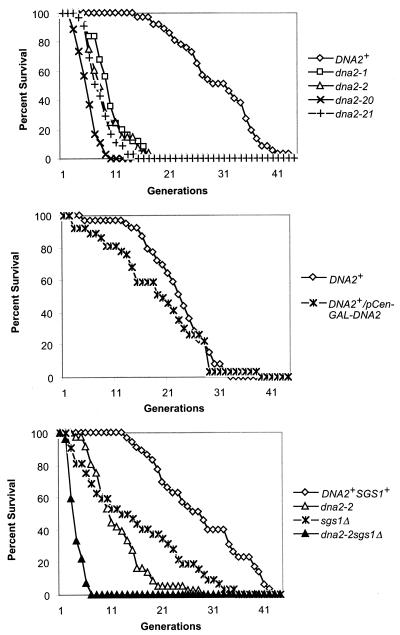
As shown in Fig. 1A, dna2 mutants do have severely shortened life spans. We observed a variety of mutants, including the temperature-sensitive dna2-1 mutant, at a permissive temperature, and dna2-20 and dna2-21, which require osmotic support (19). We also used dna2-2, which is not temperature sensitive but has a slightly reduced doubling time compared to that of the wild type. dna2-2 was isolated as a synthetic lethal mutation with ctf4Δ and is also synthetically lethal with chl12Δ. Ctf4 is a protein that binds tightly to DNA polymerase. ctf4 mutations are synthetically lethal with pol1 mutations, and dna2-2 pol1 mutations show synthetic effects. Chl12/Ctf8 is involved in an alternative clamp-loading complex. dna2-2 mutations also show synthetic effects with rfc1 mutations, which encode the major subunit of the replicative clamp loader. dna2-2 is also defective in postreplication repair but not in homologous recombination (8, 20). The average life span for dna2-1 was 8.1 ± 2.5 generations, that for dna2-2 was 8.2 ± 2.5 generations, that for dna2-20 was 4.9 ± 1.9, and that for dna2-21 was 7.2 ± 2.6 generations. Thus, for all four mutants, even the allele that had no lethal defect in young cells showed substantially shorter average life spans than their wild-type parent strains (30.4 ± 5.4 generations for the parent of dna2-1 and 26.55 ± 13.91 generations for the parent of dna2-2). All dna2 strains differed significantly from the wild type at P < 0.001. (The average life spans of the mutants used in this study are summarized below in tabular form.) We conclude that Dna2p is required for maximum life span in yeast. Since a regulatory effect on aging is more convincing if life span can be extended, we studied overproduction of Dna2p. Overproduction of Dna2p, however, has little or no effect on life span (Fig. 1B). In fact, overproduction leads to a small life span reduction, as might be expected, since overproduction of Dna2p can result in lethality (70).
FIG. 1.
(A) dna2 mutants have reduced life span. Average life spans (the numbers of cells dissected are given in parentheses) of dna2 mutants: dna2-1, 8.1 ± 2.5 generations (n = 25); dna2-2, 8.2 ± 2.5 generations (n = 25); dna2-20, 4.9 ± 1.9 generations (n = 36). The DNA2+ curve shown is for strain SS111 (wild-type parent of dna2-1), 30.4 ± 5.4 generations (n = 37). The isogenic wild-type strain for dna2-20 is not available (G. R. Crabtree, personal communication) (19). The isogenic parent of strain dna2-2, strain 7585-2-2, (20), is not shown for the sake of simplicity, but the average life span was determined and is 26.55 ± 13.91 generations (n = 31). (B) Dna2p overexpression strains have normal or slightly shortened life spans. Average life spans: UCC3515, DNA2+, 22.9 ± 5.9 generations (n = 39); UCC3515 DNA2+/p Gal-CEN-DNA2, 18.9 ± 8.5 generations (n = 37). Life span was determined in medium containing raffinose and galactose. (C) DNA2 and SGS1 are not epistatic. The strain background for this experiment was W303(RAD5), and all strains are isogenic (except for those with the dna2 and sgs1 alleles). Average life spans were the following: W303 DNA2 (wild type), 24.3 ± 10.2 generations (n = 35); dna2-2, 7.3 ± 3.8 generations (n = 22); sgs1Δ, 14.7 ± 10 generations (n = 22); dna2sgs1Δ, 3.2 ± 1 generations (n = 22).
Since SGS1 and DNA2 both encode helicases, it was of interest to test their relationship in life span determination. We investigated whether an sgs1Δdna2-2 double mutant strain had a more severe defect in life span than either single mutant alone. We found that sgs1Δdna2-2 double mutants were synthetically lethal in the genetic background that we chose but had the same doubling time as single mutants when osmotically supported with 0.5 M sorbitol, similar to data for dna2-20 strains (19). Thus, life spans of the double mutants were determined in medium containing sorbitol. Figure 1C shows that the life span of the double mutant sgs1Δdna2-2 differed significantly (P = 0.03 by Mann Whitney test; P = 0.02 by t test) from that of dna2-2 and even more so from that of sgs1Δ. To ensure that this observation was also true in genetic backgrounds where the two mutations were not synthetically lethal, we measured life span in double mutants that were viable without addition of sorbitol (8). The life span of the dna2-2 mutant is 8.2 ± 2.5 generations, and that of the sgs1Δ mutant is 12.6 ± 6.3 generations in this background. The double mutant again had a significantly shorter (P = 0.03) average life span than either single mutant, 6.5 ± 2.9 generations. Thus, the aging phenotype of the double mutants is not dependent on genetic background. Taken together with differences in phenotype of the single mutants documented previously, the fact that dna2 and sgs1Δ mutations clearly act additively leads us to propose that the two helicases have different substrates, accounting for their independent contributions, or act synergistically.
dna2 mutants show a segmental aging pattern.
Werner and Rothmund-Thomson syndromes have been described as segmental aging syndromes, since human patients suffering from these diseases show some, but not all, of the symptoms of normal human aging. In that sense, dna2 shows segmental aging in yeast. Old yeast cells show a characteristic set of phenotypes (78). Prematurely old dna2 mother cells showed almost all of the phenotypes of old wild-type mother cells, and by definition show them after fewer generations than the wild type, since the average life span is reduced. dna2 mother cells near the end of their life span became enlarged (Fig. 2A and data not shown) and showed a two- to threefold increase in cell cycle length (data not shown), as do wild-type cells near the end of their normal life span (33, 42). Daughters of old dna2 mother cells returned to normal cell size and cell cycle length within two cell divisions, indicating that, as for the wild type, aging is asymmetric. Mutant dna2 mothers also became sterile as they progressed through their life span (Table 1), demonstrating a disruption of transcriptional silencing, another age-specific phenotype of wild-type cells (82).
TABLE 1.
Sterility test for dna2-1 MATa, W303 MATa, and sgs1Δ MATaa
| Strain | Generation | Shmoo | Total | % Sterile |
|---|---|---|---|---|
| W303 | 2-3 | 34 | 44 | 16 |
| 7-9 | 20 | 20 | 0 | |
| 9-15 | 13 | 24 | 46 | |
| dna2-1 | 2-3 | 43 | 50 | 14 |
| 2-3 | 10 | 20 | 50 | |
| 5-8 | 2 | 13 | 85 | |
| 5-9 | 2 | 13 | 85 | |
| sgs1Δ | 6-10 | 4 | 10 | 60 |
| 13-19 | 1 | 12 | 92 |
The ability of a dna2-1 MATa strain to respond to 10 μg of alpha factor per ml very early in its life and at near the halfway point of its life span, which is 8 generations, was examined. The ability of dna2-1 MATa, sgs1Δ MATa, and W303 MATa strains to respond to alpha factor was examined as described previously (82) by using sterile filter paper rectangles soaked with 10 μg of alpha factor per ml. Responses were recorded as the number of cells with shmoo morphology after 3 and 5 h of exposure. After removal of the filter paper, cells were moved to 0.5 cm beyond the site of the test and were examined to make sure they recovered and resumed division. Cells that failed to resume division were not counted in the data set on the assumption that they could be on the road to death and, thus, were beyond the possibility of response.
dna2-2 mutants also showed, like the wild type (78), age-related nucleolar enlargement and fragmentation. Early in the life of wild-type mother cells, the nucleolus is unitary and resembles a cap on the nuclear periphery but becomes enlarged and fragmented at around the average life span (80). Figure 2A shows dna2-2 cells stained with both DAPI, to reveal the entire nucleus (dark blue), and with monoclonal antibody to the abundant nucleolar protein Nop1p (bright green), a protein involved in ribosome biogenesis (1). The young cells all have typical nucleolar morphologies, as shown in the examples (Fig. 2A, Young). In contrast, greater than 90% of the dna2-2 cells isolated at 7 to 9 generations, about half their maximum life span, have enlarged and/or fragmented nucleoli (pale green or aqua Nop-1 staining) (Fig. 2A, Old). The change in Nop-1 color between young and old cells is probably due to the large amount of ribosomal DNA (see below) and/or relative dilution of Nop-1. The striking difference between the populations of young and old cells confirms that the nucleolar phenotype is not a feature of the general dna2 mutant population but that it is, as for the wild type, directly correlated with age.
Since genomic instability forms the basis of our model for premature aging, we next evaluated genomic stability in young and old dna2 cells by using the rDNA as an assay. Wild-type mother cells amplify rDNA, both intrachromosomally and in the form of ERCs, as they progress through their life span (79). We therefore compared the rDNA in young dna2-2 cells and in cells at about half their maximum life span. As shown in Fig. 2B, equal amounts of undigested DNA from the young and old cells were separated on an agarose gel by conventional electrophoresis (Fig. 2B, left) and analyzed by Southern blotting with an rDNA probe (Fig. 2B, right). When the gel was blotted and probed with labeled rDNA, the amount of rDNA-specific hybridization increased dramatically (apparently 10-fold in this experiment) in the old cells versus that in the young cells, representing amplification of this region. This large increase in the apparent amount of rDNA is likely an overestimate which might be explained if the altered nucleoli result in greater relative recovery of rDNA in the old than in the young cells. In addition, there seemed to be significant rDNA-specific degradation in old cells compared to that for young cells. This degradation was not noted on the ethidium bromide-stained gel and thus did not represent general DNA degradation or a problem with the method used to prepare DNA. Nor was it observed in DNA isolated from eighth-generation wild-type cells (data not shown). Identification of ERCs (5 kb) by one-dimension gel analysis was hampered by the extensive degradation of the rDNA in the dna2-2 mutant. Circular DNA can be separated from linear degradation products by two-dimension gel analysis in the presence of intercalating agents (Fig. 2C) (79). As a positive control for ERCs we used the isogenic sgs1Δ strain, which accumulates ERCs at the same rate as the wild type (29, 62, 79). ERCs are visible as the dark arc of topoisomers appearing below the diagonal of linear DNA in the sgs1Δ DNA shown in Fig. 2C and represent about 12.5% of total DNA. In the dna2-2 strain at about 8 generations (Fig. 2C, left), ERCs are observed as a faint arc of much lower intensity than that of the sgs1Δ strain, representing about 1.6% of the total DNA. These results were repeated in at least three separate experiments (data not shown). We conclude that while accumulation of ERCs may be important for the aging of sgs1Δ (79), high copy numbers of ERCs probably do not account for the cessation of cell division in old dna2 mothers. Since rDNA recombination is elevated (see also below), the lack of accumulation of ERCs may be due to failure of ERCs to replicate efficiently in the dna2 mutant. Nevertheless, we do find evidence (amplification of the rDNA, breakage of the rDNA, and formation of a small number of ERCs) that late-generation dna2 mutants show genomic instability in the rDNA, as do old wild-type or sgs1Δ mutants. Since DNA2 is a replication gene, we propose that the increased rDNA in old dna2 mutants is a symptom of an underlying defect in DNA replication, leading to recombinogenic structures in the rDNA. The difference between young and old dna2 populations shows that this damage accumulates with age. The observed increase in fragmentation of the rDNA with increasing age is evidence of DSB formation. We have previously shown that strains carrying dna2 mutations require RAD52 for optimal growth, suggesting that DNA damage resulting from the failure of Dna2p to perform its function requires recombinational repair (8, 9).
While some DNA replication mutants have been shown to be generally hyperrecombinogenic (27), this phenotype has not been related to cellular aging in any previous study. Furthermore, both our lab and others have failed to observe general increased recombination in dna2 mutants by assays similar to those used for other replication mutants (19, 27). To verify that dna2 mutants show hyperrecombination in the rDNA, loss of an ADE2 marker gene from the rDNA was assayed as described previously (79, 80). As shown in Table 2, recombination in the rDNA was increased in dna2-2 strains compared to that in the wild-type.
TABLE 2.
rDNA recombinationaa
| Strain | Loss rate (SEM) of rDNA::ADE2 marker (103) |
|---|---|
| WT | 2 (0.19) |
| dna2-2 | 6.1 (1) |
| fobΔ | <0.03 |
| dna2-2fob1Δ | <0.03 |
| sgs1Δ | 12.3 (2.6) |
| rad27Δ | 19.5 (3.5) |
An ADE2 marker was inserted into the rDNA region of the indicated strains, all of which are isogenic to W303, using the plasmid and method described by others (80). The loss rate of the rDNA::ADE2 marker was determined as described by Kaeberlein et al. (37). Approximately 30,000 colonies were examined for each strain. Average values are given from at least three experiments. WT, wild type.
Only one difference from old wild-type arrested mother cells (in addition to lack of ERCs) was noted. Over 90% of dna2 cells ceased dividing as budded cells, often multiply budded, whereas wild-type mothers stop growing largely as unbudded cells, as reported by others (60% unbudded, 20% small buds, and 20% large buds [62]). In a recent report that used a different method to isolate old cells, however, 35% of wild-type cells were shown to arrest with large buds and fail to enter the next cell division, so there is variation in the terminal arrest (52). The abnormal morphology of the old dna2 mutants might be expected for a DNA replication mutant and may reflect that the balance between lesions and their repair is different between the dna2 mutants and the wild type.
Extension of dna2 life span under conditions that also extend wild-type life span.
The shortened life span of sgs1Δ cells has been reinvestigated recently; the latter cells arrest as large budded cells, leading to the suggestion that stochastic cell death, as well as normal aging processes, contributes to the life span of sgs1 cells (62). Two tests were applied to show that the sgs1Δ mother cells in the second half of their life span, in contrast to those arresting early, were arresting due to acceleration of the normal aging process. The most widely accepted model for yeast aging suggests that reduced transcriptional silencing is responsible, since overexpression of SIR2 extends life span, both in yeast and in Caenorhabditis elegans, and since deletion of SIR2 shortens yeast life span (37, 82, 90). To assess if transcriptional silencing was related to the end of life span in dna2 mutants, an extra copy of SIR2 was introduced into the replication-proficient dna2-2 mutant. As shown in Fig. 3, maximum life span was extended, although the average life span was not (Table 3), just as has also been shown for sgs1Δ mutants (62). This suggests that while there may be stochastic death of cells early in the dna2 life span, the second half of the life span is similar to that of normal aging in that it is extended by increasing silencing or decreasing recombination, two outcomes of an increase in SIR2 gene dosage (24).
FIG. 3.
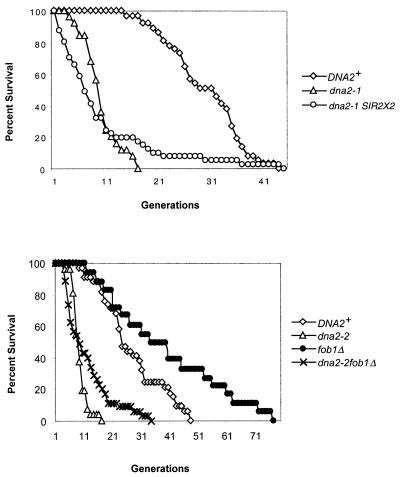
Life span is extended in dna2 strains with an extra copy of SIR2 or fob1Δ. (A) An extra copy of SIR2 extends dna2 life span. All strains were isogenic derivatives of A364a for the SIR2 study. Wild-type or dna2-1 cells carrying an extra copy of SIR2 were created with a plasmid from David Shore. The Xho-PstI fragment from pRS415 was cloned into the pRS404 plasmid. The resulting pRS304SIR2 plasmid was cut with SnaBI and transformed into the dna2-1 or wild-type strain. The constructs were verified by Southern blotting. (B) Deletion of FOB1 extends the maximum life span of dna2 mutants. For the fob1Δ study, strains were from the A364a background (20). Strain 7585-2-3 MATα trp1 leu2 ura3 his3 dna2-2 sol3Δ::LEU2 fob1Δ::HIS3 and the isogenic DNA2sol3Δ::LEU2 fob1Δ::HIS3 strain were constructed with a fob1Δ::HIS3-containing plasmid from M. Kobayashi (46). Similar results were obtained with respect to maximum life span in W303 DNA2, W303 dna2-2, W303 DNA2fob1Δ::HIS3, and W303 dna2-2fob1Δ::HIS3 strains, except that the fob1Δ strains had a smaller effect on the average life span of both wild-type and dna2-2 strains. Maximum life span extension by fob1Δ was similar. Strains and average and maximum life spans are further described in the text and in Table 3.
TABLE 3.
Summary of life span data determined in this study
| Straina | Avg life span (generations) |
|---|---|
| WT (7585-2-2) | 26.55 ± 13.91 |
| dna2-1 | 8.1 ± 2.5 |
| dna2-2 | 8.2 ± 2.5 |
| WT (SS111) | 30.4 ± 5.4 |
| dna2-20 | 4.9 ± 1.9 |
| WT | 22.9 ± 5.9 |
| DNA2/pGal-CEN-DNA2 | 18.9 ± 8.5 |
| WT (W303) | 24.3 ± 10.2 |
| dna2-2 (W303) | 7.3 ± 3.8 |
| sgs1Δ | 14.7 ± 10 |
| dna2-2sgs1Δ | 3.2 ± 1 |
| WT (7585-2-2) | 26.55 ± |
| fob1Δ | 38.22 ± 13.91 |
| dna2-2fob1Δ | 13.08 ± 7.69 (compare with row 3, dna2-2) |
| dna2-1::2XSIR2′ | 8.2 ± 2.5 |
| WT (SS111) | 26.55 ± 13.91 |
| pol1-14 | 11.5 ± 8.6 |
| pol1-17 | 21.2 ± 7.6 |
| rth1/rad27Δ | 11.6 ± 7.0 |
| ctf4Δ (WT 7585-2-2) | 6.4 ± 4.4 |
WT, wild type.
The second test applied in the case of sgs1Δ was to measure the effect of deletion of FOB1 on the life span of the older sgs1Δ mothers, since elimination of Fob1p increases normal life span (13, 62). Fob1p is required for the expansion and contraction of the rDNA repeats in yeast (46). Fob1 appears to play two related roles. Fob1 protein blocks replication forks arising from the ribosomal autonomously replicating sequence at a specific sequence called the RFB (see Fig. 4). It has been proposed that stalling of replication forks at the RFB gives rise to DSBs, promoting recombination within the rDNA and, thus, expansion and contraction of the rDNA (22, 46). Fob1 is also required for association of different rDNA repeats via the E element, which contains both the polI enhancer and adjacent RFB, however (92). Thus, Fob1p might stimulate recombination either through causing DSBs or by increasing pairing of the rDNA repeats (92). Whether increased rDNA recombination and amplification in dna2 mutants is due to increased pausing and DSBs in the rDNA or to increased pairing of breaks for repair (Fig. 2 and Table 2), we would expect a dna2-2fob1Δ double mutant to exhibit reduced rDNA recombination and extended life span. As shown in Fig. 3B, the double mutant dna2-2fob1Δ does have a longer maximum life span than the dna2-2 strain without the fob1Δ mutation. In addition, the average life span of dna2-2fob1Δ (13.08 ± 7.69 generations) is increased compared to that of the dna2-2 single mutant (8.2 ± 2.47 generations), just as the average life span of the wild type (26.55 ± 13.91 generations) is extended in the fob1Δ strain (38.22 ± 13.91 generations). In addition to extending life span, introduction of the fob1Δ mutation into the dna2-2 strain decreased the rate of loss of ADE2 from the rDNA (Table 2). The extension of life span and reduction in rDNA recombination in dna2-2fob1Δ again suggests that dna2 mutants undergo premature aging by the same processes involved in wild-type aging and that it involves hyperrecombination as either cause or effect (Fig. 2). The experiment does not distinguish between rDNA events and non-rDNA events, however, since life span is not restored to wild-type levels. We propose that the dna2 aging is due to accelerated normal aging, though there may be additional cell death due to replication errors, in analogy to the proposition that the life span of sgs1Δ mutants is a composite of normal aging and cell death due to defects in recombination (62).
FIG. 4.
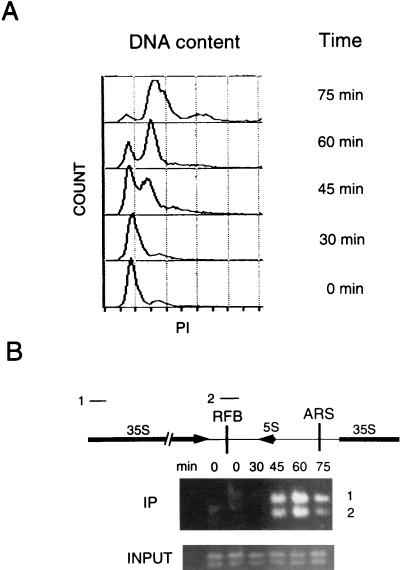
ChIP assays with synchronized cells demonstrate that Dna2p associates with rDNA maximally during S phase. (A) Cells were synchronized with mating pheromone and released as described in Materials and Methods. Flow cytometry was performed as described previously (12). Propidium iodide (PI) measures DNA content. (B) ChIP assays were performed with a strain in which the DNA2 open reading frame was precisely replaced by DNA2 fused to 9 myc tags in strain W303 pep4bar1, such that the fusion protein is expressed under the control of its endogenous promoter (12). The strain shows the same growth rates as the parental strain and is designated DNA2TMTH. Cross-linking, cell lysis, shearing, immunoprecipitation, and PCR amplification were carried out as described in Materials and Methods. The DNAs amplified from the anti-myc 9E10 monoclonal antibody immunoprecipitates are labeled IP and correspond to DNAs associated with Dna2p. The PCR products 1 and 2 correspond to the rDNA regions labeled 1 and 2 in the diagram. Control PCRs using as template the total genomic DNA in the extracts showed that each fragment amplified equally at each time point (labeled INPUT). The single-copy ARS1 probe showed no signal, and there is no internal control for rDNA, which has a very high copy number. The data shown are from a 25-cycle amplification (see Materials and Methods). The same procedure, carried out on a strain in which DNA2 was not fused to myc, yielded no PCR product (data not shown).
ChIP studies were carried out as a function of cell cycle phase position to confirm that Dna2p was associated with rDNA, that it was associated during DNA replication, and that, therefore, the hyperrecombination was likely a direct effect of the dna2 mutation (Fig. 4). As shown in Fig. 4, Dna2p is absent from the rDNA during G1, arrives at the rDNA at the beginning of S phase, associates with the rDNA maximally in S phase, and shows reduced association in G2 phase. Probes from two different rDNA regions, one spanning the RFB and one within the 35S-coding region, showed similar timing of association (Fig. 4 and see below). The same dramatic difference between G1 and S/G2 was seen for many different numbers of cycles of amplification (data not shown), suggesting the robustness of the result. The temporal pattern of association of DNA2 with the rDNA suggests that its primary role in the nucleolus is associated with the replication of the rDNA (S phase) or with repair of rDNA damaged during replication or of damage remaining after passage of the replication fork (S and G2 phases, but not G1). (The absence of a band in G1 is not due to the absence of the protein, which Western blotting shows is present throughout the cell cycle [data not shown]). To prevent confusion, we point out that Dna2 is not solely associated with the rDNA in S phase. It is also associated with other replicating sequences (12).
The parsimonious interpretation is that the rDNA amplification in the dna2 mutant arises due to replication fork stress, resulting in an increase in recombination in an attempt to repair replication fork damage. Zou and Rothstein (100) have used the rDNA as an assay to demonstrate directly that Holliday recombination intermediates accumulate in the rDNA in every cell cycle, appearing maximally during S phase, i.e., with the same temporal pattern as Dna2p association with rDNA. They also showed that at least six DNA replication mutations, including cdc9, which is synthetically lethal with dna2 (32), increase the frequency of Holliday structures in the rDNA. They proposed, as we do for dna2, that spontaneous damage during DNA replication gives rise to these structures throughout the genome. What is new here is that we find that this genetic instability is correlated in dna2 mutants with shortened life span.
Mutations in replication genes whose products interact with DNA2 lead to short life spans.
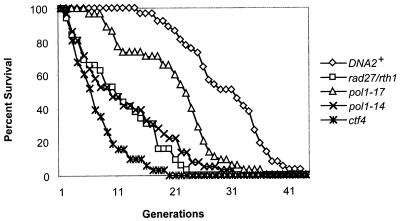
If the defect that underlies the genomic instability observed in the yeast rDNA reflects global replication fork stress, and if this stress contributes to aging, then other DNA replication mutants might be expected to exhibit reduced life spans. In Fig. 5, life spans of cells with mutations affecting polα (pol1-14 and pol1-17), FEN-1 (rad27Δ), and CTF4 (ctf4Δ) are shown. All of these genes interact genetically with dna2 (9, 20). polα is essential for initiation of Okazaki fragments; Fen1 is involved in maturation of Okazaki fragments; and CTF4 prevents chromosome loss and is required for sister chromatid cohesion (49) and encodes a protein that binds to the catalytic subunit of polα, Pol1p (64, 65). As anticipated, three of the four mutations tested that affected DNA replication also reduced life span (rad27Δ, average life span of 11.6 ± 7.0 generations; ctf4Δ, average life span of 6.4 ± 4.4 generations; pol1-14, average life span of 11.5 ± 8.6 generations). One DNA polymerase mutant, pol1-17, had an average life span comparable to its isogenic parent (21.2 ± 7.6 generations) at the permissive temperature. Interestingly, this mutant has a normal growth profile at the permissive temperature and serves as an important comparison for pol1-14, which shows serious growth defects and abnormally high DNA content by fluorescence-activated cell sorter analysis at both permissive and restrictive temperatures (11). Extensive nucleolar fragmentation and enlargement occurs in the aged cells of pol1-14 (data not shown) as in those of dna2-2 (Fig. 2A). As mentioned, cdc9 and rad27 show hyperrecombination in the rDNA (100). We verified this for the rad27Δ strain used in these studies by showing an increase in loss of the ADE2 marker from the rDNA (Table 2). These results support the idea that a replication defect, either in fork propagation or in repair of damaged replicating DNA, shortens life span. We note that others have shown that mutants in a replication initiation protein (encoded by CDC6) that functions at origins of replication only at G1/S, in contrast to the elongation mutants, extend life span (79), while the CDC7 gene, which is required for firing of origins throughout S phase and not just at G1/S and is also required for postreplication repair, is required for maximum life span (35).
FIG. 5.
Many, but not all, DNA replication mutants show reduced life span. Average life spans are the following (the numbers dissected are in parentheses): pol1-14, 11.5 ± 8.6 generations (n = 36); pol1-17, 21.2 ± 7.6 generations (n = 35); rth1/rad27Δ, 11.6 ± 7.0 generations (n = 32); ctf4Δ, 6.4 ± 4.4 generations (n = 31); DNA2, which is strain SS111, 30.4 ± 5.4 generations (n = 37). The isogenic parental strain for the pol1 and rad27 mutants is SS111 (see the legend to Fig. 1). ctf4 is isogenic to the dna2-2 strain shown in Fig. 1A that has an average life span of 8.2 ± 2.5 generations (n = 33) and to the wild-type strain 7585-2-2 (Tim Formosa), 26.55 ± 13.91 generations (n = 31).
DISCUSSION
The central questions in the study of yeast aging are what kind of damage accumulates in mother cells leading to cessation of cell division and what explains the asymmetric accumulation in mothers versus daughters. We propose, on the basis of the premature replicative senescence of DNA replication mutants, that damage accumulates in mother cells at least in part due to replication fork collapse. It is difficult to distinguish in our experiments whether incomplete replication in itself or failure to stabilize stalled replication forks or to efficiently repair them is the proximal cause of the shortened life span. Furthermore, we suggest that the results obtained with replication mutants represent an exaggerated case of spontaneous replication errors that occur in wild-type cells in every generation and, thus, allow us to comment on the processes occurring during normal aging. We find the asymmetry between the life expectancy of old dna2 mothers and the daughters of these mothers to be a strong argument that normal aging is being measured. The nucleolar disruption, rDNA amplification, and sterility, found only in late-generation dna2 mothers, also argue that aging is occurring.
Our studies do not explain the asymmetry of aging. They may provide a clue, however. One surprising finding in our studies is that ERCs do not accumulate in the dna2 mutants, as they do in the wild type and in the prematurely aging sgs1Δ mutants. Nevertheless, there does seem to be a strong connection between aging of dna2 mutants and events involving the rDNA, supporting previous hypotheses that the rDNA constitutes an AGE locus (40). First, the nucleoli of dna2 and other replication mutants are enlarged and fragmented and do not appear to be properly localized in the older cells. Second, there is amplification of the rDNA itself during the life span of dna2 mutants, and there is hyperrecombination in the rDNA but not general hyperrecombination. Third, deletion of FOB1 extends both the average and the maximum life span of dna2 mutants. All of the effects of the fob1Δ mutation documented to date are related to the rDNA. The extension of wild-type life span by the fob1Δ mutation had previously been attributed to the absence of ERCs, but that cannot be the case for dna2 mutants, since dna2 mutants do not accumulate ERCs. Fourth, the extension of maximum life span in dna2 strains with an extra copy of SIR2 also points to the involvement of the rDNA locus in the aging process of dna2 strains (62). SIR2 overexpression causes both increased silencing and decreased recombination in the rDNA (24, 83). SIR2 overexpression also leads to reduced frequency of initiation of DNA replication in the rDNA repeats and accordingly to a reduced number of forks paused at the RFBs in each repeat (P. Basero and E. Schwob, personal communication). One hypothesis is that inheritance of the rDNA may be asymmetric with respect to some aspect of its transcription, replication, and nucleolar morphology. One might propose that after DNA replication, undamaged chromosomes remain silenced and are selected for transfer into the daughter cells. However, there is no evidence for such a mechanism. The recent appreciation that the nucleolus is not just the site of ribosome biogenesis, as previously supposed, but also plays significant roles in transcriptional silencing and in exit from mitosis also suggest that further investigation of nucleolar and rDNA inheritance may illuminate studies of the mechanisms of yeast aging (77, 84, 91). The dna2 mutants may offer a useful genetic background for studying the potential contribution of these factors to the asymmetry of aging, since the mutants do not accumulate ERCs.
The replication fork lesion model for yeast aging, or replicative senescence, is consistent with two specific hypotheses for causes of yeast aging based on previous work but adds an additional dimension and suggests how the pathways may intersect. First, the recombinational aging model specifically hypothesized that aging results from inappropriate recombination in the rDNA repeats, resulting in extrachromosomal rDNA circles which are asymmetrically inherited. This hypothesis was originally supported by the ability of ectopic generation of an ERC to accelerate aging (79). Recently a sufficient number of cases have been documented in which there is either no correlation or an anticorrelation between aging and ERCs, such that ERCs no longer provide an explanation of the mechanism of yeast aging; instead, they appear to be another symptom (62, 79). The replication model we propose simply suggests that recombination is a sequel to a more primary defect in replication fork propagation/postreplication repair of replication lesions. It nicely accommodates premature aging defects in recombination mutants like rad52 (71), because recombinational repair may be necessary for repair of replicative damage. It is supported by the observed spontaneous increase in recombination specifically during S phase and in DNA replication mutants (100).
Second, a defective silencing model has been proposed in which aging results from a loss of the ability to silence inappropriate gene expression (31, 40, 55). This mechanism is supported by life span extension from deleting a histone deacetylase gene that is required for silencing at some loci (41), age-related losses in telomeric gene silencing (40, 43), relocation of the Sir proteins to the rDNA during aging (40), life span reduction in a sir2 deletion mutant, and life span extension by overproduction of SIR2 (37). It has been pointed out that the yeast cell is most vulnerable to changes in silencing during S phase as chromatin reassembles (36). In the replication stress model of aging, the relocation of the Sir proteins could be directed to aid in remodeling of chromatin during repair of damage to the replication fork. An increasing number of observations implicate replication genes in silencing (3, 15, 59, 66, 81, 99). Another link between silencing, recombination, and replication could be the fact that rDNA recombination is increased in sir2 mutants (24). In addition, old dna2 cells show early sterility (Table 1), suggesting defects in silencing in the silent mating type loci and reorganization of the Sir complex. During normal aging, a likely source of replication errors is endogenous oxidative DNA damage, which is likely to increase at high metabolic rates. The recent demonstration that Sir2 histone deacetylase requires NAD+ as a cofactor and/or that Sir2 may mediate the breakdown of NAD+ links the extension of life span by overproduction of SIR2 to the metabolic state of the cells (31, 55, 88, 89). Reformation of chromatin after recombinational repair of replication blocks due to oxidative damage in rapidly growing cells might require SIR2. Our finding that introduction of an extra copy of SIR2 into dna2 mutants increases the maximum life span significantly is consistent with the latter proposal.
The premature aging of sgs1Δ is consistent with the replicative damage hypothesis we propose. sgs1Δ mutants are viable but show increased recombination, sister chromatid exchange, and chromosome instability (22, 69, 94, 95). The double mutant sgs1Δsrs2Δ is inviable, and sgs1ts srs2Δ strains are defective in DNA synthesis and rDNA transcription, suggesting that SGS1 may be at the replication fork (53). This lethality can be overcome by a rad51 mutation (23, 62). One way to explain the suppression is that a putative intermediate in damaged replication fork processing accumulates in the double mutant but is not lethal if it is prevented from entering the recombination pathway (48). Others have found that Sgs1p is an integral component of the S-phase checkpoint response in yeast, binding to Rad53p and in the same epistasis group with polɛ (21). They suggest the role of Sgs1p is to monitor replication fork progress; for instance, to detect stalled forks. This suggestion would make the results with sgs1 consistent with the replication stress hypothesis for yeast life span determination. The increased severity of the defect in sgs1Δdna2 double mutants suggests divergence of function between the two helicases at some point in the complicated process. One possible scenario is that Sgs1p is required to resolve fork damage (39), while Dna2p is required to prevent damage. It is also important that the Sgs1 homolog in Xenopus laevis xBLM is absolutely required for replication in vitro (54).
Even if studies with yeast replication mutants using life span assays do not relate to the mechanisms limiting normal life span in yeast, which we think is unlikely, further studies with yeast using mutants like dna2 and sgs1 may shed light on human helicase diseases, which may also be diseases of DNA replication. Both BLM and WRN cell lines have replication defects (26, 57, 73, 85). Recently, BLM helicase has been proposed to be an antirecombinase which, like bacterial RuvAB, can promote branch migration of Holliday structures and thus might resolve reversed replication forks without entry into the recombination pathway (39). WRN, on the other hand, carries a helicase/nuclease, similar to Dna2p in that the helicase and nuclease seem to act in concert biochemically, though perhaps differing in substrate specificity (6, 76). WRN helicase interacts with replication proteins and proteins involved in DSB repair (for a review see reference 6). The mouse BLM−/BLM− knockout is embryonic lethal, and Blm−/Blm− chicken cells show increased sister chromatid exchange which is dependent on Rad54 and therefore on homologous recombination (for a review see reference 18). RecQ4 is a related helicase and is affected in Rothmund-Thomson syndrome, but its enzymatic properties are not well characterized (45). It will be interesting to test complementation of yeast dna2 mutant phenotypes by vertebrate WRN, BLM, and RecQ4 genes, as has already been done for yeast sgs1Δ strains (29, 97).
In conclusion, since aging is likely due to multiple factors, we mention that enhanced response to stress is implicated in lengthening life span. In yeast, overproduction of Lag1p or Ras2p leads to an extension of the life span, possibly by controlling antistress mechanisms (34). These mechanisms are not addressed in the present study but may interact with the mechanisms we have discussed if they lead to replicative damage. One such connection was suggested by a recent study profiling gene expression during yeast aging. In a sip2Δ strain that has accelerated aging and is a regulator of the Snf1p glucose-sensing pathway, which regulates shifts between energy expenditure and energy storage, DNA2 and SGS1 are significantly (more than twofold) overexpressed (56).
Acknowledgments
We thank S. M. Jazwinski for invaluable help in setting up the life span determinations and G. M. Martin and Masayasu Nomura for reading an earlier form of the manuscript. We thank John Aris for the Nop1 antibody and the Caltech-ERATO center for use of the Nikon microscope for image processing, Jessica Brown for characterization of nucleoli in pol1-14, and Meghan McFarlane for dissection of some life spans.
This research was supported by National Science Foundation POWRE grant MCB9805943 and by the Pomona College Research Committee (L.L.M.H). It was also supported by National Institutes of Health grant GM25508, National Science Foundation grant MCB9985527 to J.L.C., and National Science Foundation RUI grant MCB113937 to L.L.M.H.
REFERENCES
- 1.Aris, J. P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashrafi, K., D. Sinclair, J. Gordon, and L. Guarente. 1999. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:9100-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelrod, A., and J. Rine. 1991. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, S., E. Choi, K. Lee, J. Park, S. Lee, and Y. Seo. 1998. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273:26880-26890. [DOI] [PubMed] [Google Scholar]
- 5.Bae, S. H., K.-H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. [DOI] [PubMed] [Google Scholar]
- 6.Bohr, V. A., M. Cooper, D. Orren, A. Machwe, J. Piotrowski, J. Sommers, P. Karmakar, and R. Brosh. 2000. Werner syndrome protein: biochemical properties and functional interaction. Exp. Gerontol. 35:695-702. [DOI] [PubMed] [Google Scholar]
- 7.Budd, M. E., and J. L. Campbell. 1995. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budd, M. E., and J. L. Campbell. 2000. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459:173-186. [DOI] [PubMed] [Google Scholar]
- 9.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budd, M. E., W.-C. Choe, and J. L. Campbell. 1995. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270:26766-26769. [DOI] [PubMed] [Google Scholar]
- 11.Budd, M. E., K. D. Wittrup, J. E. Bailey, and J. L. Campbell. 1989. DNA polymerase I is required for DNA replication but not for repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe, W., M. Budd, O. Imamura, L. Hoopes, and J. L. Campbell. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 13.Defossez, P.-A., R. Prusty, M. Kaeberlein, S.-J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 3:447-455. [DOI] [PubMed] [Google Scholar]
- 14.Dua, R., D. L. Levy, and J. L. Campbell. 1998. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 273:30046-30055. [DOI] [PubMed] [Google Scholar]
- 15.Ehrenhofer-Murray, A. E., R. T. Kamakaka, and J. Rine. 1999. A role for the replication proteins PCNA, RF-C, polymerase ɛ and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis, N. A. 1997. DNA helicases in inherited human disorders. Curr. Opin. Gen. Dev. 7:354-363. [DOI] [PubMed] [Google Scholar]
- 17.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 18.Enomoto, T. 2001. Function of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication. J. Biochem. 129:501-507. [DOI] [PubMed] [Google Scholar]
- 19.Fiorentino, D. F., and G. R. Crabtree. 1997. Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S Phase. Mol. Biol. Cell. 8:2519-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Formosa, T., and T. Nitiss. 1999. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full DNA2 helicase activity is not essential for growth. Genetics 151:1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frei, C., and S. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 22.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase TOP3 interacts with SGS1, a DNA helicase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771-776. [DOI] [PubMed] [Google Scholar]
- 25.Guarente, L., and C. Kenyon. 2000. Genetic pathways that regulate ageing in model organisms. Nature 408:255-262. [DOI] [PubMed] [Google Scholar]
- 26.Hand, R., and J. German. 1975. A retarded rate of DNA chain growth in Bloom's syndrome. Proc. Natl. Acad. Sci. USA 72:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartwell, L. H., and D. Smith. 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayflick, L. 1965. The limited in vitro lifespan of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 29.Heo, S.-J., K. Tatebayashi, I. Ohsugi, A. Shimamoto, Y. Furuichi, and H. Ikeda. 1999. Bloom's syndrome gene suppresses premature aging caused by Sgs1 deficiency in yeast. Genes Cells 4:619-625. [DOI] [PubMed] [Google Scholar]
- 30.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 31.Imai, S.-I., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nat. Genet. 403:795-800. [DOI] [PubMed] [Google Scholar]
- 32.Ireland, M. J., S. S. Reinke, and D. M. Livingston. 2000. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jazwinski, S. M. 1990. An experimental system for the molecular analysis of the aging process: the budding yeast Saccharomyces cerevisiae. J. Gerontol. 45:B68-B74. [DOI] [PubMed]
- 34.Jazwinski, S. M. 1999. Molecular mechanisms of yeast longevity. Trends Microbiol. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 35.Jazwinski, S. M., N. K. Egimlez, and J. B. Chen. 1989. Replication control and cellular life span. Exp. Gerontol. 24:423-436. [DOI] [PubMed] [Google Scholar]
- 36.Jazwinski, S. M., S. Kim, C.-Y. Lai, and A. Benguria. 1998. Epigenetic stratification: the role of individual change in the biological aging process. Exp. Gerontol. 6:571-580. [DOI] [PubMed] [Google Scholar]
- 37.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:257-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang, J.-Y., E. Choi, S.-H. Bae, K.-H. Lee, B.-S. Gim, H.-D. Kim, C. Park, S. A. MacNeill, and Y.-S. Seo. 2000. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics 155:1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Nat. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy, B. K., M. Gotta, D. Sinclair, K. Mills, D. McNabb, M. Murthy, S. Pak, T. Laroche, S. M. Gasser, and L. Guarente. 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89:381-391. [DOI] [PubMed] [Google Scholar]
- 41.Kim, S., A. L. Benguria, and S. M. Jazwinski. 1999. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 10:3125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, S., P. A. Karchman, A. Benguria, and S. M. Jazwinski. 1999. Experimentation with the yeast model. CRC Press, Boca Raton, Fla.
- 43.Kim, S., B. Villeponteau, and S. M. Jazwinski. 1996. Effect of replicative age on transcriptional silencing near telomeres in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 219:370-376. [DOI] [PubMed] [Google Scholar]
- 44.Kirchman, P., S. Kim, C.-Y. Lai, and S. M. Jazwinski. 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Gen. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi, T., D. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi, T., M. Hidaka, M. Nishizawa, and T. Horiuchi. 1992. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233:355-362. [DOI] [PubMed] [Google Scholar]
- 48.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouprina, N., K. E., V. Bammolpv, V. Bliskovsky, R. Gizatullin, A. Kirillov, V. Zakharyev, P. Hieter, and F. Spencer. 1992. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5736-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuo, C.-L., C.-H. Huang, and J. L. Campbell. 1983. Isolation of yeast DNA replication mutants using permeabilized cells. Proc. Natl. Acad. Sci. USA 80:6465-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzminov, A. 1999. Recombination repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Bio. Rev. 63:751-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laun, P., A. M. Pichova, F., J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K.-U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 53.Lee, S.-K., R. E. Johnson, S.-L. Yu, L. Prakash, and S. Prakash. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339-2342. [DOI] [PubMed] [Google Scholar]
- 54.Lian, S., J. Graham, and H. Yan. 2000. The function of Xenopus Bloom's syndrome protein homolog (xBLM) in DNA replication. Genes Dev. 14:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin, S.-J., P.-A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life -span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 56.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276:36000-36007. [DOI] [PubMed] [Google Scholar]
- 57.Lonn, U., S. N. Lonn, U. G. Winblad, and J. German. 1990. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 50:3141-3145. [PubMed] [Google Scholar]
- 58.Lopes, M., C. Cotta-Ramusino, A. Pellicioloi, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 59.Martin, A. A., I. Donne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGlynn, P., R. G. Lloyd, and K. J. Marians. 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 98:8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McVey, M., M. Kaeberlein, H. A. Tissenbaum, and L. Guarente. 2001. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 157:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michel, B. S., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miles, J., and T. Formosa. 1992. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase α, acts in DNA metabolism in vivo. Mol. Cell. Biol. 12:5274-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miles, J., and T. Formosa. 1992. Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc. Natl. Acad. Sci. USA 89:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller, A. M., and K. A. Nasmyth. 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312:247-251. [DOI] [PubMed] [Google Scholar]
- 67.Mortimer, R. K., and J. R. Johnston. 1959. Life span of individual yeast cells. Nature 183:1751-1752. [DOI] [PubMed] [Google Scholar]
- 68.Murray, A., and J. Szostak. 1983. Pedigree analysts of plasmid segregation in yeast. Cell 34:961-970. [DOI] [PubMed] [Google Scholar]
- 69.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nat. Gen. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 70.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park, P. U., P.-A. Defossez, and L. Guarente. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3848-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson, C. R. D., J. R. Cryar, and J. W. Gaubatz. 1984. Constancy of ribosomal RNA genes during aging of mouse heart cells and during serial passage of WI-38 cells. Arch. Gerontol. Geriatr. 3:115-125. [DOI] [PubMed] [Google Scholar]
- 73.Poot, M., H. Hoehn, T. M. Runger, and G. M. Martin. 1992. Impaired S phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 202:267-273. [DOI] [PubMed] [Google Scholar]
- 74.Pringle, J. R., A. Adams, D. Drubin, and B. Hararer. 1991. Immunofluorescence methods for yeast. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 75.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 76.Shen, J. C., and L. A. Loeb. 2000. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 28:3260-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. S. Chen, J. Jang, A. Shechenko, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 78.Sinclair, D., K. Mills, and L. Guarente. 1998. Aging in Saccharomyces cerevisiae. Annu. Rev. Microbiol. 53:533-560. [DOI] [PubMed] [Google Scholar]
- 79.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles-a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 80.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutations. Science 177:1313-1316. [DOI] [PubMed] [Google Scholar]
- 81.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Nahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smeal, T., J. Claus, B. Kennedy, F. Cole, and L. Guarente. 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84:633-642. [DOI] [PubMed] [Google Scholar]
- 83.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 84.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 85.Takeuchi, F., F. Hanaoka, M. Goto, I. Akaoka, T. Hon, M. Yamada, and T. Miyamoto. 1982. Altered frequency of initiation sites of DNA replication in Werner's syndrome cells. Hum. Genet. 60:365-368. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka, T., and K. Nasmyth. 1998. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17:5182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanner, K. G., J. Landry, R. Sternglanz, and J. M. Denu. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97:14178-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a SIR2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 91.Visintin, R., R. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 92.Wai, H., K. Johzuka, L. Vu, K. Eliason, T. Kobayashi, T. Horiuchi, and M. Nomura. 2001. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21:5541-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang, T.-C. V., and K. C. Smith. 1986. Postreplication repair in ultraviolet-irradiated human fibroblasts: formation and repair of DNA double-strand breaks. Carcinogenesis 7:389-392. [DOI] [PubMed] [Google Scholar]
- 94.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 96.Wolff, S., J. Bodycote, and R. B. Painter. 1974. Sister chromatid exchanges induced in Chinese hamster cells by UV irradiation of different stages of the cell cycle: the necessity for cells to pass through S. Mutat. Res. 25:73-81. [DOI] [PubMed] [Google Scholar]
- 97.Yamagata, K., J. Kato, A. Shimamoto, et al. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Nat. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu, C.-E., J. Oshima, Y.-H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]
- 99.Zhang, H., K. Shibahara, and B. Stillman. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408:221-225. [DOI] [PubMed] [Google Scholar]
- 100.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 90:87-96. [DOI] [PubMed] [Google Scholar]