Abstract
A large number of cytoplasmic tRNAs are imported into the kinetoplast-mitochondrion of Leishmania by a receptor-mediated process. To identify the sequences recognized by import receptors, mitochondria were incubated with a combinatorial RNA library. Repeated cycles of amplification of the imported sequences (SELEX) resulted in rapid selection of several import aptamers containing sequence motifs present in the anticodon arm, the D arm, the V-T region, and acceptor stem of known tRNAs, confirming or suggesting the presence of import signals in these domains. As predicted, truncated derivatives of tRNAIle(UAU) containing the D arm or the V-T region were imported in vitro. Four aptamers were studied in detail. All were imported in vitro as well as in transiently transfected cells, using the same pathway as tRNA, but their individual import efficiencies were different. Two types of aptamers were discernible: the A arm and D arm homologues (type I), which were efficiently transferred across the inner mitochondrial membrane, and the V-T homologues (type II), which were not. Remarkably, subnanomolar concentrations of type I RNAs stimulated the entry of type II RNAs into the matrix, whereas type II RNAs inhibited inner membrane transfer of type I RNAs. Moreover, tRNATyr(GUA) and tRNAIle(UAU) interacted with one another as type I and type II, respectively. Such cooperative and antagonistic interactions may allow the use of a limited number of receptors to recognize a large number of tRNAs of variable affinity and enable the maintenance of a properly balanced tRNA pool for mitochondrial translation.
The import of nuclear-encoded, cytoplasmic tRNAs into mitochondria has been documented for a growing number of protist, plant, and other species which lack the full complement of tRNA genes in their mitochondrial genomes (reviewed in reference 21). The complexity of this process is underlined by the observation that the numbers and identities of imported species vary widely: from one in yeast (16), to 11 in potato (15), to at least 24 in kinetoplastid protozoa and apicomplexans, which lack all mitochondrial tRNA genes (8, 23).
How mitochondria select only the required tRNA species from the cytoplasmic multitude is still a mystery. According to one school of thought, the structure of the tRNA as a whole is recognized (9). Alternatively, specific sequence motifs or short structures present on individual tRNAs could serve as the basis for discrimination. In Tetrahymena cells, the anticodon of tRNAGln apparently acts as an import determinant (20). In yeast, both the anticodon domain and the acceptor stem of tRNALys appear to play a role in import (7). In contrast, the D arm of Leishmania tRNAIle (10) and tRNATyr (13) contain import signals, but replacement of this region of tRNAIle by the D arm of tRNAGln(CUG) (which, of itself, does not contain any signal [11, 18]) produces a hybrid molecule which is still imported (10); whether tRNAIle contains more than one import signal is unresolved at present. Studies with mutant forms of the D arm of tRNATyr showed further that the RNA structural requirements are different for transfer through the outer, as opposed to the inner, membrane (2). This suggests the presence of distinct receptors at the two membranes. TAB, an outer membrane receptor (1), is immunochemically distinct from the inner membrane receptor (2), but the identity of the latter is unknown.
In this paper, we have asked the following question: are there multiple sequence motifs recognized by the import receptors, and if so, are the corresponding RNAs imported by independent pathways using distinct receptors? To identify the range of import signals recognized by mitochondrial receptors, the SELEX procedure (5) was used to isolate, from a combinatorial RNA library, those sequences (aptamers) that are imported with high efficiency. A subset of these aptamers was studied in more detail for their import characteristics alone and in combination. We show that conserved motifs in different tRNA domains can potentially act as import signals, that two types of RNAs are discernible, differing in their ability to cross the inner membrane, and that these two types interact with each other at the inner membrane in a manner suggestive of allosteric regulation at the inner membrane receptor complex.
MATERIALS AND METHODS
Cell culture and preparation of mitochondria.
Promastigotes of Leishmania tropica strain UR6 were cultured on solid blood agar medium (6) supplemented with 150 μg of biopterin/ml and 50 μg of adenine/ml. Mitochondria were purified from DNase I-treated lysates by Percoll gradient centrifugation and stored in 50% glycerol storage buffer, as described previously (14). The final protein concentration of gradient-purified fractions was 8 to 10 mg/ml.
Preparation of substrate for SELEX.
DNA templates were prepared by PCR amplification of the oligonucleotide O-58 (GGAATTCTAATACGACTCACTATAGGGACGCAGGGACTGTAN16ATGTGACTGTAGCGG), which contains a 16-base random sequence in the middle (estimated complexity, 4.29 × 109), flanked by a T7 RNA polymerase promoter sequence at the 5′ end followed by a 5′ anchor sequence and a 3′ anchor sequence. The 5′ and 3′ anchors for the amplification of selected RNAs included sequences flanking the D domain of Leishmania tRNATyr(GUA) (11). The two primers used for the PCR were O-59(GGAATTCTAATACGACGACTCACTATAGGGACGCAGGGACTGTA,containing an EcoRI linker, the T7 RNA polymerase promoter, and five 3′-terminal bases identical to the D arm flanking sequence) and O-60 (GCCCAAGCTTCCGCTACAGTCACAT, complementary to the 3′ end sequence of O-58 and containing a HindIII linker). For PCR, 0.1 pmol of O-58 and 25 pmol each of primers O-59 and O-60 were added to a 100-μl reaction mixture containing 10 mM Tris-HCl, pH 8, 50 mM KCl, 1.5 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, and 5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer). After heat activation of the enzyme at 90°C for 10 min, amplifications were carried out for 30 cycles of 90°C for 30 s, 60°C for 30 s, and 72°C for 30 s. PCR products were gel eluted after nondenaturing 6% polyacrylamide gel electrophoresis and used for the preparation of substrate RNA. Transcription was performed with 10-μl reaction mixtures containing 40 mM Tris-HCl, pH 7.5, 6 mM MgCl2, 1 mM spermidine, 1 mM dithiothreitol (DTT), 0.5 mM concentrations (each) of CTP, ATP, and GTP, 0.25 mM UTP, 1 μCi of [α-32P]UTP, and 10 U of T7 RNA polymerase at 37°C for 90 min. Then, 1 U of RQ1 DNase (Promega) was added and the reaction mixture was further incubated at 37°C for 15 min. RNA was recovered by phenol-chloroform extraction and ethanol precipitation.
In vitro selection protocol.
Random-sequence RNA (pool 0; 1 pmol), prepared as described above, was incubated with 1 mg of mitochondria in a 200-μl reaction mixture containing 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 4 mM ATP at 37°C for 15 min, and then RNases A and T1 were added to final concentrations of 2.5 μg/ml and 50 U/ml, respectively, and RNase digestion continued at 37°C for 15 min. The mitochondria were washed with isotonic sucrose-Tris-EDTA buffer (250 mM sucrose, 10 mM Tris-HCl, pH 8, 1 mM EDTA) by centrifugation, and imported RNA was isolated by guanidinium isothiocyanate extraction and ethanol precipitation (13). Selected RNA was annealed with 25 pmol of primer O-60 in Superscript II (BRL) reverse transcription buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2) at 65°C for 5 min, followed by slow cooling to room temperature. Then, DTT (2 mM), deoxynucleoside triphosphates (0.4 mM [each]), and 50 U of Superscript II reverse transcriptase were added and the reaction mixtures (10 μl) were incubated at 42°C for 50 min. The cDNA was amplified by PCR using primers O-59 and O-60, as described above. The selected pool (pool 1 DNA) was subjected to a second round of transcription, import, reverse transcription, and PCR. By repetition of this cycle, pools 2 to 5 were generated. At each step, the PCR product was gel purified and precautions were taken to avoid contamination of imported sequences with unimported RNA.
Complexity analysis.
The complexities of selected pools were compared by partial RNase digestion, as previously described (5). 32P-labeled RNA (105 cpm) and yeast tRNA (2 μg) in 10 μl of RNase T1 digestion buffer (20 mM sodium citrate, pH 5, 1 mM EDTA, 7 M urea, 0.02% xylene cyanol, 0.02% bromophenol blue) was denatured by heating at 92°C for 5 min, followed by rapid chilling on ice. RNase T1 (1 U) was added, and the reaction mixtures were incubated at 55°C for 10 min and then placed on ice. The reaction mixtures were directly loaded on a 12% polyacrylamide sequencing gel for electrophoresis and autoradiography.
Cloning and sequencing of aptamers.
Pool 4 DNA (obtained after the fourth cycle) was digested with EcoRI and HindIII and cloned into pUC19 vector. Escherichia coli XL1-Blue cells were transformed and 36 positive colonies were obtained. Twenty-one of these were sequenced manually by the dideoxy chain termination method using Klenow fragment and both forward and reverse sequencing primers.
Preparation of import assay substrates.
RNA from pools 0 to 5 was prepared by transcription from PCR-amplified DNA templates using T7 RNA polymerase, as described above, except that the UTP concentration was reduced to 10 μM and 10 to 20 μCi of [α-32P]UTP was used. Aptamer RNAs were prepared by runoff transcription of the corresponding HindIII-linearized clones. tRNATyr and tRNAGln(CUG) were synthesized from linearized plasmids pSKB1 and pSKB-2, respectively (1). tRNATyr D-arm oligoribonucleotide (26-mer) was prepared by oligonucleotide transcription, as described earlier (13). Effector RNAs were similarly synthesized, except that the amount of [α-32P]UTP in the transcription reaction mixture was reduced to one-tenth and the UTP concentration was raised from 10 to 250 μM. Thus, the specific activity of effector RNAs is 1/250 that of substrates. tRNAIle(UAU) was obtained using DNA template generated by PCR amplification of the Leishmania tarentolae tRNAIle gene cloned in plasmid pGCN8. PCR amplification was carried out with the primers O-65 (GGAATTCTAATACGACTCACTATAG) and O- 66 (TGGTGCTCCCGGCGGGGC), corresponding to the 5′ and 3′ regions of the tRNA, respectively (22). tRNAIle(1-41) was synthesized by runoff transcription of the PCR-amplified gene restricted with HpaII, which cuts at positions 41 and 66 within the gene (22). To isolate tRNAIle(42-66), the intact tRNAIle transcript was annealed to the 25-nucleotide (nt) HpaII fragment spanning positions 42 to 66, in 80% formamide-40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid] (pH 6.4)-0.4 M NaCl-1 mM EDTA at 55°C for 18 h, the nonhybridized RNA digested with RNase A plus RNase T1 as previously described (3) and the protected RNA recovered by proteinase K treatment, phenol extraction, and ethanol precipitation. Substrates and effectors were quantified on the basis of the specific activity of the label, the counts per minute incorporated in the transcription reaction, and the number of U residues in the RNA; in the case of complex mixtures of unknown composition (pools P0 to P5), the U content was assumed to be 25%.
In vitro import assays.
Total uptake into mitochondria was assayed as described above for in vitro selection, except that 100 μg of mitochondria was incubated with high-specific-activity 32P-labeled RNA in 20-μl reaction mixtures. Separation of submitochondrial compartments was performed as previously described (17). Briefly, the outer membrane was solubilized with digitonin and the soluble outer membrane plus intermembrane space fraction was separated from mitoplasts by centrifugation. The mitoplasts were then subjected to freeze-thaw lysis to separate the matrix (soluble) and inner membrane (insoluble) fractions. Marker enzyme assays were performed as previously described (17). To study the effect of uncouplers, mitoplasts were preincubated with 50 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 10 min on ice before the import reaction. The recovered RNA was analyzed by urea-6% polyacrylamide gel electrophoresis and quantified by autoradiography and scintillation counting of dried gel bands.
Binding assays and determination of Kd values.
Binding reactions were carried out by incubating 1.56 to 100 fmol of 32P-labeled RNA in 10-μl reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 100 mM KCl, and 50 μg of mitochondria at 0°C for 30 min; under these conditions, binding is sequence specific (2, 12). Unbound RNA was removed by washing with isotonic sucrose, and the bound RNA was recovered and quantified as described above. Scatchard plots of b/f versus b, where b = bound RNA concentration and f = free RNA concentration, yielded the following equation: Kd = −1/slope.
Import assay in transiently transfected cells.
Details of this procedure have been previously described (2). Briefly, promastigotes of L. tropica strain UR6 were electroporated with 32P-labeled RNA and incubated at 22°C for 10 min. Electroporated cells were lysed by hypotonic syringe passage, the particulate fraction containing mitochondria was treated with DNase I, RNase A, and RNase T1, and RNA was isolated as described above.
RESULTS
Selection of import aptamers from a random pool.
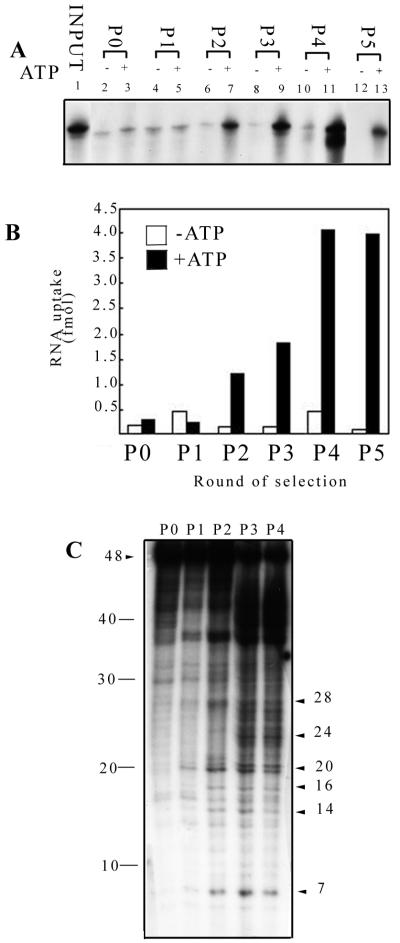
A pool of random 16-mer sequences was incubated with isolated mitochondria in the presence of ATP. Excess, nonimported RNA was digested with RNase, the mitochondria were washed, and mitochondrial RNA was recovered and amplified by reverse transcription-PCR (RT-PCR) using anchoring 5′ and 3′ primers. RNA was synthesized from the amplified DNA by using T7 RNA polymerase, and the import-amplification cycle was repeated. RNA from each round (designated Pn, where n = number of cycles, the nonselected initial pool being P0) was tested for in vitro import. ATP-dependent uptake increased after two cycles of selection-amplification and attained saturation by the fourth cycle, indicating positive selection for importable sequences (Fig. 1A and B).
FIG. 1.
In vitro selection for RNA import aptamers in isolated Leishmania mitochondria. Mitochondria were incubated with a combinatorial library of RNA sequences under optimized conditions for import. Imported RNAs were isolated after RNase treatment and amplified by RT-PCR to generate double-stranded DNA templates for the next round of selection. (A) Import of RNA isolated after increasing numbers of selection cycles. 32P-labeled RNAs (100 fmol), synthesized to high specific activity from templates generated at each round, were incubated with mitochondria (100 μg of protein) in the absence (lanes 2, 4, 6, 8, 10, and 12) or presence (lanes 3, 5, 7, 9, 11, and 13) of 4 mM ATP for 15 min at 37°C, and the RNase-resistant RNA was analyzed. P0, starting pool; P1 to P5, RNAs after one to five rounds of selection, respectively. Lane 1, input pool 0 RNA, 2 fmol. Lanes 12 and 13 are from a separate experiment carried out under identical conditions. (B) Amount of RNA imported as a function of pool number in the absence or presence of ATP. (C) Complexity analysis by RNase fingerprinting. Shown (from left to right) are partial RNase T1 digests of P0 to P4 RNA, respectively. Arrowheads on the right indicate sizes of positively selected bands. Marker sizes are shown at the left.
To assess the relative sequence complexities of the selected pools, the one-dimensional RNase T1 fingerprints of RNAs of P0 to P4 were compared (Fig. 1C). P0 RNA yielded an essentially uniform ladder of bands, as expected from a completely random sequence. In vitro selection resulted in the appearance of a number of specific bands, and a decline in the background ladder, indicating the increasing abundance of specific RNA sequences. Notably, the appearance of these bands coincided with the increase in import efficiency at cycle 2 (compare Fig. 1B and C). Reamplification of P0, P1, or P2 RNA in the absence of mitochondrial selection did not alter the import efficiency of the pool (data not shown), ruling out any significant bias at the transcription and amplification steps.
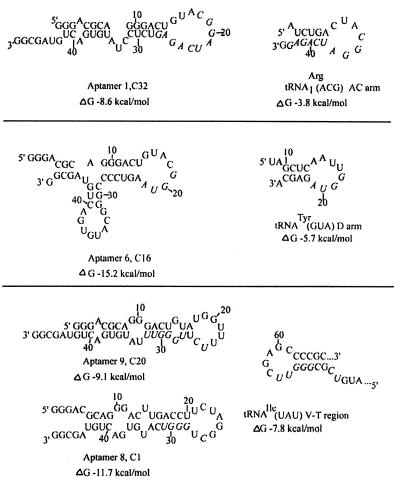
Pool 4 DNA was cloned and several clones were sequenced (Table 1). About one-quarter of the clones contained the sequence CGGAUCAGAGU (aptamers 1 to 3). Aptamers 1 to 3 have identical sequences in the first 12 positions and also share the motif YAGAGY with aptamers 4 and 4A, and this motif is present in the D domain of a number of kinetoplastid tRNAs (Table 1). The D domain motif GUAGAGC, which is known to be part of an import signal (2, 13), was found in 21% of available Leishmania and Trypanosoma tRNA sequences but in only 6% of yeast tRNAs. Moreover, aptamers 1 and 2 are identical in 12 contiguous positions to the anticodon domain of Leishmania tRNAArg(ACG); this region includes the anticodon itself (Table 1; Fig. 2). Aptamers 5 and 6 constitute a second major group; they are identical to each other except for a G→A transition at the sixth position of the selected sequence. They share the motif YGGUARR with the conserved D arm motif on tRNAs (Table 1). More extensive homology with available kinetoplastid sequences was not observed, possibly due to the incomplete nature of the current database (containing only 14 tRNA sequences from Leishmania and 14 from trypanosomes; http://www.uni-bayreuth.da/departments/biochemie/tRNA). However, these aptamers potentially form a hairpin structure which closely resembles the D domain hairpin, especially in the positioning of the shared motif in the loop and stem regions (Fig. 2); in the absence of more tRNA sequence information, these aptamers were therefore tentatively grouped with the D domain homologues. The motif UAGGAC in aptamer 7 was identified in the D domain of two Leishmania tRNAs. Aptamers 8 and 9 constitute another group with the motif CUG3-4U and form similar secondary structures with pyrimidine-rich eight-base loops, a base-paired GGG motif in the stem flanked on the 5′ side by a UC or CU dinucleotide. A similar GU-rich motif, UC(G/A)(U/C/G)GGGUU, was present in the V-T region of 5 out of 14 (36%) of Leishmania or Trypanosoma tRNAs (Table 1) but was relatively rare in yeast tRNA (2 out of 51, or 4%). The above secondary structural features were also present in the V-T regions of the corresponding tRNAs, but the alignment of the loop was different (Fig. 2). Aptamer 10 is homologous to the V-T region of the two tRNAArg isoacceptors. Aptamers 11 and 12 contained runs of GY; similar but shorter runs were present in the acceptor stems of a number of tRNAs (Table 1).
TABLE 1.
Homology of aptamers with tRNAs
| Aptamer no. | Aptamer sequencea | Clone(s) with sequence (%)b | Homologous tRNAc
|
||
|---|---|---|---|---|---|
| Domain | Species | Sequence | |||
| 1 | CGGAUCAGAGUCUCCU | 25, 32, 39, 40, 57 (24) | A | Arg1(ACG) | ACGGAUCAGAG |
| Arg2(UCG) | UCGGAUCUGAG | ||||
| 2 | CGGAUCAGAGUCUCCC | 8 (5) | |||
| 3 | CGGAUCAGAGUCGGCU | 4 (5) | |||
| 4 | CGGGCUGAUGUAGAGC | 21 (5) | D | Tyr(GUA) | UGGUAGAGC |
| Lys(UUU) | UGGUAGAGC | ||||
| 4A | GGGUUAGAGCUGUUCG | 5 (5) | Trp(CCA) | UGGUAGAGC | |
| Arg1(ACG) | UGGAAGAGC | ||||
| 5 | CGGUAGGUCCCCGGGC | 36, 45, 47 (14) | Gln2(UUG) | UGGUAGAGC | |
| Phe(AAG) | UGGGAGAGC | ||||
| 6 | CGGUAAGUCCCCGGGC | 16 (5) | |||
| 7 | CCUUACUAGGACUGUA | 9 (5) | D | Ile(UAU) | UUAGGAC |
| Val(CAC) | UUAGGAC | ||||
| 8 | CCUUCUAGGCUGGGUC | 1 (5) | V-T | Lys(UUU) | UCGUGGGUU |
| 9 | UGGUUUUCUGGGGUUU | 20, 29, 41 (14) | Val(UAC) | UCGCGGGUU | |
| Val(CAC) | UCGCGGGUU | ||||
| Ile(UAU) | UCGCGGGUU | ||||
| Thr(AGU) | UCAGGGGUU | ||||
| 10 | AUUGUUGCAUGGUUGA | 43 (5) | V-T | Arg1(ACG) | GUUGCAGGUU |
| Arg2(UCG) | GUUGCAGUU | ||||
| 11 | ACGUAGUCCGCGUGCG | 24 (5) | AS | Lys(UUU) | GAGUGCG |
| Val(UAC) | CCGCGU | ||||
| 12 | CCCUCUGCCGGUGUGG | 26 (5) | AS | Gly(UCC) | UGUGG |
| Arg1(ACG) | GUGUGG | ||||
Only the selected 16-nt segment of each 48-nt aptamer is shown; the anchor sequences are shown in Fig. 2. Aptamers were matched against L. tarentolae and Trypanosoma brucei cytoplamic tRNA sequences in the Sprinzl tRNA database (http://www.unibayreuth.da/departments/biochemic/tRNA) except for the L. tarentolae tRNAVal(CAC) and tRNAPhe(AAG) sequences, which were previously published (24).
Numbers in parentheses are percentages of listed clone(s) with a particular sequence.
D, D arm; A, anticodon arm; V, variable loop; T, T arm; AS, acceptor stem. The amino acid specificity and anticodon sequence of each tRNA are shown. All tRNAs were from L. tarentolae except for tRNAGln2(UUG), which was from T. brucei. All sequences contain contiguous runs of at least five matching bases. Anticodons are underlined.
FIG. 2.
Secondary structures of selected aptamers and. homologous tRNA domains. Structures were derived by energy minimization using the MFOLD program (http://bioinfo.math.rpi.edu/∼mfold/rna/). Calculated stabilities (ΔG values) at 37°C and 1 M Na+ are shown. In each aptamer, the selected region (positions 18 to 33) is flanked by the 5′ anchor (positions 1 to 17) and 3′ anchor (positions 34 to 48). Aptamers are placed with the homologous tRNA domain shown at the right; conserved bases are shown in italics.
Significance test for aptamer-tRNA homologies.
The observed structural similarities between individual aptamers and different domains of kinetoplastid tRNAs suggest that, in addition to the well-characterized D arm import signal, other tRNA domains may play an important role. Moreover, the same tRNA may contain signals in different domains. For example, the D domain of tRNAIle(UAU) was homologous to aptamer 7 (Table 1) while the V-T region was homologous to aptamers 8 and 9 (Table 1; Fig. 2). We therefore tested the prediction that both the D arm and V-T region contain import signals.
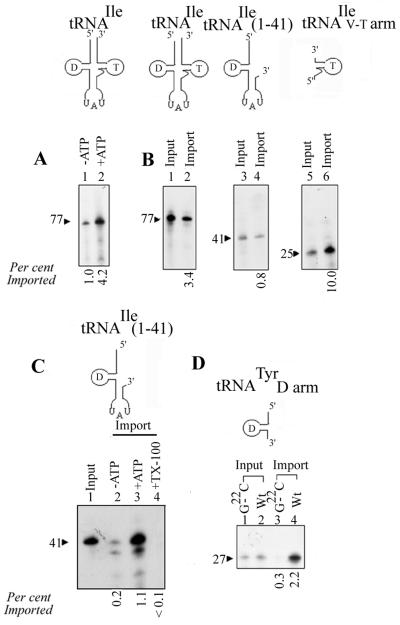
Truncated derivatives of tRNAIle were prepared (see Materials and Methods) and tested for import into isolated mitochondria. About 3.4% of the 77-nt tRNAIle was imported in an ATP-dependent manner (Fig. 3). In contrast, the import efficiency of tRNAIle(1-41), which contains the D and anticodon domains, was 0.8%, i.e., one-quarter of that of the intact molecule. This low level of RNase protection was ATP stimulated and completely sensitive to treatment of the mitochondria with Triton X-100 (Fig. 3D). Conversely, the 25-nt tRNAIle(42-66), containing the variable and T domains, was imported three times more efficiently than the intact molecule. This enhanced import is not due simply to nonspecific uptake of short oligonucleotides since the system is sequence specific, as shown by the sensitivity of import to a single point mutation in the 27-nt tRNATyr D arm oligonucleotide (Fig. 3E; see also reference 2). Moreover, similar truncation experiments with tRNATyr showed the lack of importability of the 3′ half of the molecule, including the V and T domains (13). These results therefore indicate the presence of a previously unknown, strong import signal in the V-T region of tRNAIle, as predicted by the homology analysis.
FIG. 3.
Import of derivatives of tRNAIle(UAU). (A) Import of intact tRNAIle(UAU) into mitochondria in absence (lane 1) or presence (lane 2) of 4 mM ATP. (B) The uptakes of full-length and truncated versions of tRNAIle(UAU) by intact mitochondria (100 μg) were compared. Left, full-length tRNAIle; lane 1, input, 2 fmol; lane 2, 50 fmol of RNA incubated with mitochondria and assayed for import. Middle, RNAIle(1-41); lane 3, input, 1 fmol; lane 4, 50 fmol of RNA assayed for import. Right, tRNAIle(42-66); lane 5, input, 2 fmol; lane 6, 50 fmol of RNA assayed for import. (C) Effect of ATP on import of RNAIle(1-41) (100 fmol). Import reactions were carried out in the absence (lane 2) or presence (lanes 3 and 4) of 4 mM ATP. After the incubation, Triton X-100 (0.5%) was added to reaction mixture 4, followed by RNase in all reaction mixtures. Lane 1, input (1 fmol). (D) Sequence-specific import of the tRNATyr D arm. Wild-type or G22→C mutant form of the D arm oligonucleotide (50 fmol) was assayed for import (lanes 4 and 3, respectively). Lanes 1 and 2, input RNA, 1 fmol. The fraction of each RNA imported is shown below the lanes.
Import characteristics of individual aptamers.
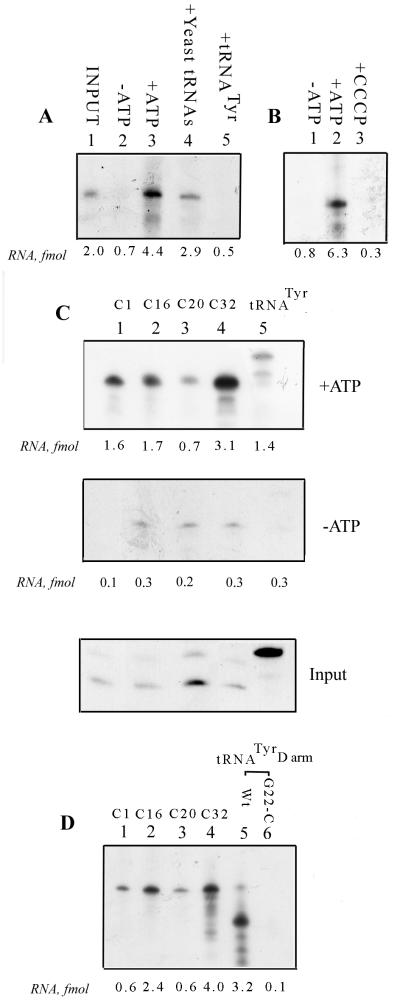
Four RNAs from the P4 selection pool were analyzed in this study: clone 1 (C1), C16, C20, and C32 (aptamers 8, 6, 9, and 1, respectively; Fig. 2). In vitro import of C32 was ATP dependent and competed out by an excess of tRNATyr, whereas yeast tRNA was a much less effective competitor (Fig. 4A ), indicating that the aptamer is specifically recognized by the import receptor for tRNATyr. ATP-dependent transfer of C32 into mitoplasts, obtained by digitonin permeabilization of mitochondria, was observed (Fig. 4B). Inner membrane transfer of C32 required a proton-motive force, as shown by its sensitivity to the protonophore uncoupler CCCP (Fig. 4B). The import of C1, C16, and C20 was stimulated by ATP, but the relative overall import efficiencies were different, C32 being the most efficient (Fig. 4C).
FIG. 4.
In vitro import of individual aptamers. (A) Import characteristics of C32. 32P-labeled C32 RNA (100 fmol) was incubated with 100 μg of mitochondria in the absence (lane 2) or presence (lanes 3 to 5) of 4 mM ATP for 15 min at 37°C. Lanes 4 and 5, yeast tRNA or low-specific-activity tRNATyr (1 pmol each) was included as competitor. (B) Mitoplast import assay. 32P-labeled C32 RNA (100 fmol) was incubated with mitoplasts prepared from 100 μg of mitochondria in either the absence of ATP (lane 1) or presence of 4 mM ATP (lanes 2 and 3). Lane 3, 50 μm CCCP was included. (C) Comparison of C1, C16, C20, and C32 (lanes 1 to 4, respectively) and tRNATyr (lane 5) for import into intact mitochondria. RNA (50 fmol) and mitochondria (50 μg) were used in each case. Top, ATP added; middle, no ATP; bottom, input (1 fmol each). (D) Binding of aptamers to intact mitochondria. Mitochondria (50 μg) were incubated at 0°C for 30 min with 32P-labeled RNAs (20 fmol) from C1, C16, C20, C32 (lanes 1 to 4), tRNATyr D arm wild-type (lane 5), or tRNATyr D arm G22→C mutant (lane 6). Bound RNA was recovered after washing the mitochondria.
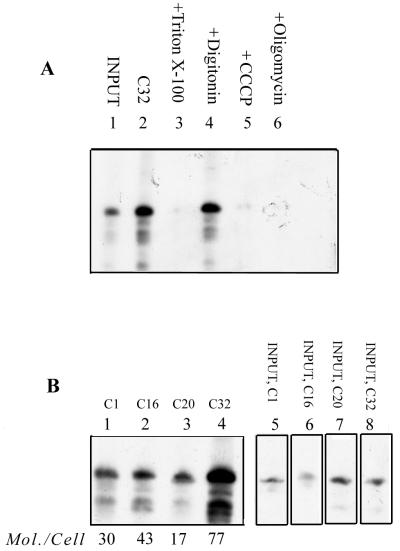
To examine the import of these aptamers in transiently transfected cells, Leishmania promastigotes were electroporated with 32P-labeled RNAs, and after a brief incubation, the mitochondrial fraction was isolated and treated with RNase and the internalized RNA was analyzed by gel electrophoresis. Approximately 77 molecules of C32 RNA per transfected cell were recovered (Fig. 5A). Treatment of the mitochondrial fraction with Triton X-100 rendered the RNA sensitive to RNase, indicating entrapment in a membrane vesicle. Importantly, RNA uptake was sensitive to the mitochondrial inhibitors CCCP and oligomycin, which dissipate the proton gradient and inhibit F1F0 ATPase, respectively. Transfer of tRNA to the mitochondrial matrix in vitro and in transfected cells was previously shown to be inhibited by both of these reagents (2, 17). Moreover, the RNA remained completely RNase resistant in the presence of digitonin, which selectively permeabilizes the outer membrane (Fig. 5A) (17). This shows that the observed signal represents RNA completely localized in the matrix. As is the case in vitro, the efficiencies of transfer of the other aptamers were less than that of C32 (Fig. 5B). The variation in mitochondrial uptake of the different aptamers was not due to different intracellular concentrations of the transfected RNAs, since the total amount of labeled RNA recovered from the transfected cells was about the same in all cases (data not shown).
FIG. 5.
Transfer of aptamers into the mitochondrial matrix in transfected cells. (A) Transfection of C32 into Leishmania promastigotes. Cells (5 × 108 in 0.4 ml) were transfected with 1 pmol of 32P-labeled RNA in the absence (lanes 2 to 4) or presence (lane 5) of 10 μM CCCP or of 50 μM oligomycin (lane 6). Aliquots of the transfected cells were lysed, and the mitochondrial fraction was treated with RNase and DNase in the absence (lanes 2, 5, and 6) or presence of 0.5% Triton X-100 (lane 3) or 320 μM digitonin (lane 4). Lane 1, input C32 RNA, 1 fmol. (B) Transfection of promastigotes with 1 pmol each of C1, C16, C20, and C32 (lanes 1 to 4, respectively). Lanes 5 to 8, input C1, C16, C20, and C32, respectively (1 fmol each). The number of molecules of each aptamer recovered per viable cell (viability posttransfection, 30%) is shown below the lanes.
To assess the interaction of each aptamer with mitochondrial surface receptors, mitochondria were incubated with 32P-labeled RNAs under conditions which minimize nonspecific binding (Fig. 4D). Specific, but quantitatively different, binding of all aptamers to mitochondria was observed. About 30% of the bound C32 RNA was internalized under the import conditions used (data not shown). Moreover, correlation of the observed binding to import was shown by the lack of binding of a mutant tRNATyr D arm oligonucleotide that is not imported (Fig. 4D) (2). From titration data and Scatchard analysis, the Kd's of binding of C1, C16, C20, and C32 to the outer membrane were determined to be 1.0, 0.46, 1.19, and 0.1 nM respectively, compared with the value of 0.9 nM for the D arm of tRNATyr.
Selection of sorted RNAs.
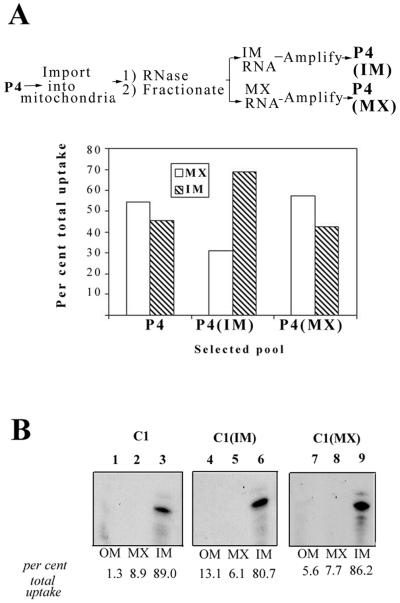
The foregoing SELEX experiment was performed with total RNA internalized into intact mitochondria. It was shown previously that the import process can be kinetically and biochemically subdivided into outer and inner membrane transfer steps (2, 17). Moreover, structural variants of the D arm import signal have defined intrinsic inner membrane transfer efficiencies; some are transferred with high efficiency, and others remain stuck to the inner membrane (2). It was therefore expected that in a SELEX-type situation involving a mixture of sequences, the inner membrane would filter out those that have a low inner membrane transfer efficiency, resulting in purification of the matrix-associated pools to an efficiency approaching 100%. To test this, SELEX pool P4, a mixture of aptamers (Table 1), was incubated with intact mitochondria, and after RNase treatment, the mitochondria were fractionated into inner membrane and matrix compartments and the RNA contained in each fraction was amplified separately and tested for intramitochondrial localization (Fig. 6A). The parental pool had an inner membrane transfer efficiency of about 50%. The inner membrane-associated pool had a lower efficiency (30%), but surprisingly, the efficiency of the matrix-associated pool was about the same as that of the parental pool (Fig. 6A). Repetition of the cycle of fractionation and amplification of the matrix-associated fraction did not result in further enhancement of inner membrane transfer efficiency (data not shown). One possible explanation of this result is that the inner membrane is leaky, leading to failure of the fractionation; this leakiness could be intrinsic to the membrane preparation, or somehow induced by the fractionation-amplification procedure. To test for leakiness, a control experiment was performed with a single RNA (aptamer 8, C1), which was predominantly retained at the inner membrane (see below). If the membrane is leaky, the matrix localization of C1 should increase upon repeated fractionation-amplification. In fact, the inner membrane transfer efficiency of C1 continued to be low after repeated cycles, arguing against any obvious leakiness of the inner membrane (Fig. 6B).
FIG. 6.
Intramitochondrial sorting of pool P4 RNA. (A) P4 RNA (100 fmol) was incubated with intact mitochondria (100 μg), and after RNase treatment, the outer membrane was solubilized with digitonin and the mitoplasts were separated into inner membrane (IM) and matrix (MX) fractions by freeze-thaw lysis. RNA associated with each fraction was recovered, reverse transcribed, and amplified by PCR. Each DNA pool was then transcribed with T7 RNA polymerase in the presence of radiolabeled UTP to yield pools P4 (IM) and P4 (MX), respectively. These pools were individually incubated with mitochondria, and their intramitochondrial distributions were quantified by fractionation as described above. The percentages of total internalized RNA associated with the inner membrane or matrix for the parental pool P4 and the sorted pools P4 (IM) and P4 (MX) are shown. (B) Fractionation-amplification of C1. C1 RNA was incubated with mitochondria, the inner membrane and matrix fractions were isolated, and RNA from each fraction was amplified by RT-PCR to yield inner membrane and matrix DNA, respectively. RNA was prepared from each PCR product and incubated with mitochondria, and the intramitochondrial location determined. The panels show fractionation experiments with the parental RNA (left) and the two sorted RNAs (middle and right).
We therefore hypothesized that the transfer efficiency of an individual RNA, when present alone, is different from that present in a mixture with other RNAs. Allosteric interactions between aptamers using the same pathway could lead to modulation of efficiency. In this scenario, one RNA (the effector) would bind to a multi-site or multimeric receptor, inducing an allosteric conformational change resulting in increasing or decreasing the affinity for a second RNA (the substrate). Alternatively, the effector may induce the opening or closing of import channels (gating), thereby stimulating or inhibiting transfer of substrate.
Inter-RNA interactions.
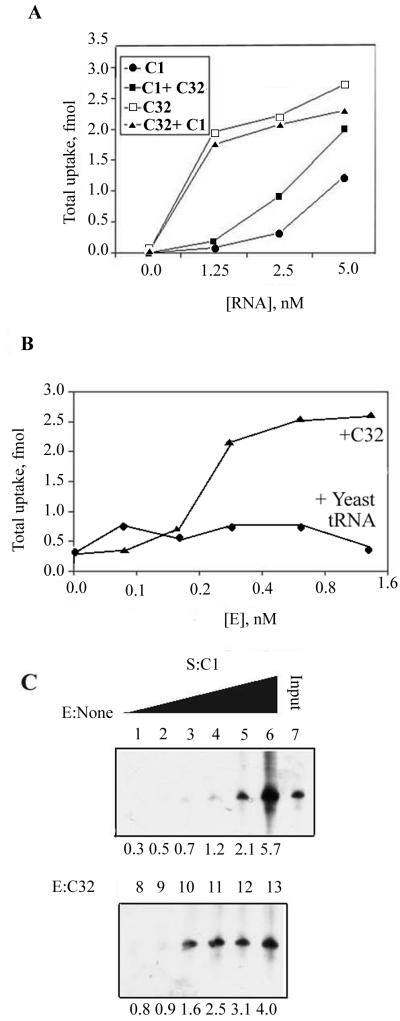
To test this possibility, mitochondria were incubated with high-specific-activity, 32P-labeled aptamer (substrate) in the presence or absence of one-tenth the concentration of a second aptamer (effector) of low-specific activity and total uptake was assayed. In the presence of C32 effector, the uptake of C1 was stimulated; the effect was more pronounced at lower concentrations of substrate (Fig. 7A ). By contrast, C1 effector had no significant effect on import of C32 substrate (Fig. 7A). No signal was detected in presence of effector alone, a result of the combination of low concentration and low specific activity of the effector. Moreover, treatment of effector with micrococcal nuclease resulted in the loss of stimulatory activity, indicating that the effector is RNA in nature (data not shown). Titration with effector showed that there was a sharp rise in C1 uptake between 0.15 and 0.3 nM C32; comparable concentrations of nonspecific RNAs such as yeast tRNA had no significant effect (Fig. 7B). Binding of C1 to intact mitochondria was also stimulated by C32 (Fig. 7C); the Kd value for C1 was reduced from 1.0 to 0.25 nM in presence of 1 nM C32 (data not shown). Thus, C32 was able to stimulate binding as well as uptake of C1 through the outer membrane. However, the magnitude of stimulation was low (about twofold in most experiments) and the total uptakes of C16 and C20 were not affected by C32 (Fig. 8). Thus, with these particular combinations of RNAs, allosteric effects at the outer membrane are marginal or insignificant.
FIG. 7.
Interactions between aptamers at the outer membrane. (A) Effect of C32 on total uptake of C1 and vice versa. The indicated concentrations of high-specific-activity substrate were incubated with mitochondria in the absence or presence of a 0.5 nM concentration of the other RNA as low-specific-activity effector (E), and the total uptake was measured. (B) Specificity of C32 effect. The total uptake of C1 substrate (2.5 nM) in the presence of indicated concentrations of C32 effector or total yeast tRNA was assayed. (C) Effect of C32 on binding of C1 to intact mitochondria. Binding assays were performed with 0.312, 0.625, 1.25, 2.5, 5.0, or 10.0 nM C1 in the absence (lanes 1 to 6, respectively) or presence (lanes 8 to 13) of 1 nM C32. The amounts of bound C1 (in femtomoles) are indicated at the bottom of each panel. Lane 7, input C1 (2 fmol).
FIG. 8.
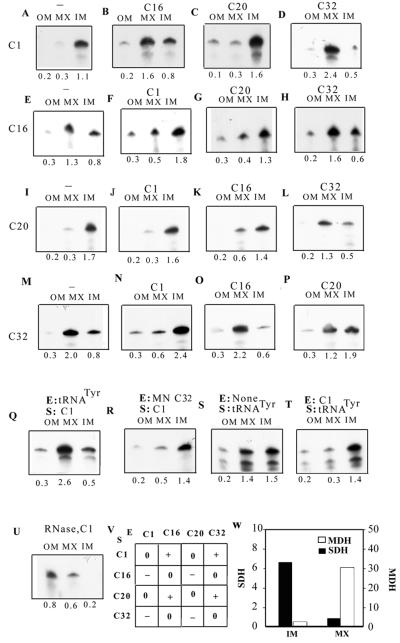
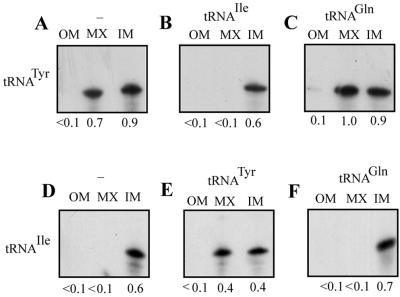
Interactions between aptamers at inner membrane. Intramitochondrial distribution assays were performed with high-specific-activity substrate (S; 100 fmol) and low-specific-activity effector (E; 10 fmol) in the following combinations, and the RNA associated with the outer membrane plus intermembrane space (OM), inner membrane (IM), and matrix (MX) fractions was analyzed. (A) Substrate, C1, no effector; (B) substrate, C1; effector, C16; (C) substrate, C1; effector, C20; (D) substrate, C1; effector, C32; (E) substrate, C16; no effector; (F) substrate, C16; effector, C1; (G) substrate, C16; effector, C20; (H) substrate, C16; effector, C32; (I) substrate, C20; no effector; (J) substrate, C20; effector, C1; (K) substrate, C20; effector, C16; (L) substrate, C20; effector, C32; (M) substrate, C32; no effector; (N) substrate, C32; effector, C1; (O) substrate, C32; effector, C16; (P) substrate, C32; effector, C20; (Q) substrate, C1; effector, tRNATyr; (R) substrate, C1; effector, micrococcal-nuclease-treated C32; (S) substrate, tRNATyr; no effector; (T) substrate, tRNATyr; effector, C1; (U) substrate, C1; no effector, mitoplast treated with RNase before separation of inner membrane and matrix fractions. (V) Summary of interaptamer interactions at the inner membrane. +, stimulation of inner membrane transfer; −, inhibition; 0, no significant effect. The absolute amount of RNA in each band was obtained by liquid scintillation counting; relative scan densities varied, depending on specific activities of the RNAs, exposure times of the autoradiograms, etc. (W) Specific activities (nanomoles of substrate converted/minute/milligram of protein) of succinate dehydrogenase (SDH) and malate dehydrogenase (MDH) in inner membrane and matrix fractions.
We next examined RNA-RNA interactions at the inner membrane. By fractionation of the mitochondria into outer membrane plus intermembrane space, inner membrane and matrix compartments, the role of effectors in the intramitochondrial distribution of substrate RNAs was studied (Fig. 8). Enzyme assays confirmed the separation of inner membrane (succinate dehydrogenase) and matrix (malate dehydrogenase) markers by this procedure (Fig. 8W). The results can be summarized as follows. (i) In the absence of effector, both C1 and C20 were primarily confined to the inner membrane (Fig. 8A and I); this membrane-bound RNA was sensitive to RNase following permeabilization of the outer membrane with digitonin (compare Fig. 8A and U, inner membrane fractions), indicating association with the outer surface of the inner membrane. (ii) By contrast, C16 and C32 were primarily localized to the matrix in the absence of effector (Fig. 8E and M). (iii) In the presence of C32 effector, the “address” of C1 dramatically changed from inner membrane to matrix (compare Fig. 8A and D). Matrix entry of C20 was similarly stimulated by C32 (Fig. 8I and L). Pretreatment of C32 with micrococcal nuclease resulted in loss of activity (Fig. 8R), indicating that the effector is RNA in nature. (iv) Low levels of C16 effector similarly stimulated matrix entry of C1 (Fig. 8B), and to a lesser extent, of C20 (Fig. 8K). C16 had no significant effect on the distribution of C32 (Fig. 8O) and vice versa (Fig. 8H). (v) C1 and C20 had no effect on each other with respect to intramitochondrial distribution (Fig. 8C and J), but each inhibited the entry of either C16 or C32 into the matrix (Fig. 8F, G, N, and P). (vi) These effects were not confined to the aptamers. Thus, tRNATyr could effect the relocation of C1 (compare Fig. 8A and Q) and, conversely, C1 inhibited matrix transfer of tRNATyr (Fig. 8S and T).
These results indicate the occurrence of both positive and negative interactions between RNAs at the inner membrane. Those RNAs that can, by themselves, be transferred to the matrix may stimulate others, while the ones that are intrinsically poor matrix localizers have an inhibitory effect.
Since the interacting aptamers are homologous to different domains of tRNAs (Table 1), we tested the possibility that the corresponding tRNAs themselves interact in this manner. Two tRNAs were chosen: tRNATyr, containing the D domain homology, and tRNAIle, with the conserved V-T sequence (Table 1). As shown in Fig. 9, translocation of tRNATyr to the matrix was inhibited by low concentrations of tRNAIle but not by tRNAGln(CUG), which is not imported (11). Conversely, tRNAIle by itself or in presence of tRNAGln(CUG) was restricted to the inner membrane, whereas about 50% was relocated to the matrix in presence of tRNATyr (Fig. 9). These results show that, like the aptamers, intact tRNAs interact specifically at the inner membrane.
FIG. 9.
Interactions between tRNATyr and tRNAIle. Intramitochondrial distribution assays were performed with substrate (50 fmol) and effector (5 fmol) in the following combinations. (A) Substrate, tRNATyr; no effector; (B) substrate, tRNATyr; effector, tRNAIle; (C) substrate, tRNATyr; effector, tRNAGln(CUG); (D) substrate, tRNAIle; no effector; (E) substrate, tRNAIle; effector, tRNATyr; (F) substrate, tRNAIle; effector, tRNAGln(CUG).
DISCUSSION
In this report, we demonstrate the use of SELEX to select a variety of high-efficiency import aptamers from a random-sequence library. The aptamers were then referred to the tRNA sequence database to identify candidate import signals in different tRNA domains. This approach led to the identification of a new putative import signal in tRNAIle. The aptamers have variable intrinsic efficiencies of transfer across the outer and inner mitochondrial membranes, and moreover, interact with each other at the inner membrane by an allosteric mechanism. The results have important implications for the regulation of RNA import.
Previous studies have shown that mitochondria from kinetoplastid protozoa (Leishmania and trypanosomes) do not require cytoplasmic fractions to import tRNAs (12, 19, 25), implying that selectivity resides at the mitochondrial surface and not in soluble factors. This was borne out in the present instance by the ease with which a large number of import aptamers were selected: only four cycles of selection-amplification were necessary to achieve saturation of import efficiency by using a 16-mer library (Fig. 1). The aptamers had nanomolar affinities for mitochondria, in the same range as that of natural sequences, and were imported in vitro as well as in transfected cells (Fig. 4, 5, 7, and 8). The ease of selection may be attributed to the presence of high-affinity mitochondrial receptors, recognition of small rather than large RNA structures, and efficient removal of background sequences by RNase treatment. We also attempted to separate a preselected pool into distinct populations based on their intrinsic inner membrane transfer efficiencies (Fig. 6) but were unsuccessful because of the apparent leakiness of the inner membrane caused by allosteric interactions (see below).
Examination of the nucleotide sequences (Table 1) and putative secondary structures (Fig. 2) of the aptamers showed the occurrence of a number of groups, each composed of molecules that are related to each other and to specific domains in kinetoplastid tRNAs. The significance of these homologies with regard to the physiological tRNA import process is indicated by the following findings. (i) The motif YAGAGY, present in the D domain of several kinetoplastid tRNAs and previously shown to be important for import (2, 13), was independently recovered in the major group of aptamers (numbers 1 to 4). A second D domain motif, shared by tRNAIle and tRNAVal(CAC), was found in aptamer 7. An import signal in the D arm of tRNAIle has been previously observed (10, 19). We note that the 5′ half of tRNAIle, containing the D domain, is significantly less efficient than the intact molecule (Fig. 3), whereas the 5′ half of tRNATyr, with a similar secondary structure, is more efficient than intact tRNATyr (13). The lower efficiency of the tRNAIle D domain may be due to the presence of a different sequence motif (UAGGAC) from that in tRNATyr (UAGAGC; Table 1). (ii) Aptamer C32 is almost identical in sequence to the anticodon arm of tRNAArg(ACG) (Fig. 2). This is an indication of the possible involvement of the A domain as a trypanosomatid import signal. The anticodon is known to be an import determinant in Tetrahymena (20). (iii) The motif CUG3-4U, present in aptamers 8 and 9, pointed to the possible role of the V-T region of some tRNAs, and this was verified for the case of tRNAIle (Fig. 3). The presence of a signal in the V-T region offers an explanation for the observation that a hybrid tRNAIle molecule containing the D arm of tRNAGln(CUG)—which, by itself, does not contain a signal—is nonetheless imported in vivo (10). A role for the variable region of tRNAThr in mitochondrial targeting in Leishmania was also indicated by mutagenesis experiments (4); this tRNA contains the motif UCAG4UU in the V-T region (Table 1). (iv) Finally, no aptamers were obtained which share motifs with tRNAGln(CUG) or tRNAHis. These species, which are poorly or not all imported in vivo (11, 24), have D and V-T sequences that lack the conserved motifs in Table 1. With the limited information currently available, it is becoming evident that different tRNAs may present different domains to the import receptor(s) and, furthermore, that some tRNAs may contain multiple signals.
Using aptamers and intact tRNAs, we show for the first time the occurrence of regulation of mitochondrial RNA import through positive and negative interactions between different RNAs (Fig. 8 to 10). Based on their effector properties, the four aptamers studied could be classified into two types. Type I aptamers (C16 and C32) have high intrinsic efficiencies of inner membrane transfer, while type II aptamers (C1 and C20) have low inner membrane transfer efficiencies. Type I RNAs stimulate matrix transfer of type II but are themselves inhibited by type II. The relevance of these findings to tRNA import is suggested by the finding that tRNATyr interacts with the aptamers as a type I molecule (Fig. 8). Moreover, similar interactions between tRNATyr and tRNAIle were observed (Fig. 9). The results confirm the type I character of tRNATyr and show further that tRNATle is a type II molecule with respect to inner membrane translocation. All three type I RNAs, i.e., C16, C32, and tRNATyr, contain the motif YGG followed by YAGAGY or YAAGY, which is present in the D or A domain of many tRNAs (Table 1). On the other hand, type II RNAs (C1, C20, and tRNAIle) contain the conserved V-T motif UG3-4U (Table 1).
By modulating the affinities of different RNAs for the inner membrane receptor, cooperative and antagonistic interactions enable a balanced pool of RNAs to be obtained in the matrix for mitochondrial translation. Thus, the import efficiency of an individual RNA when present alone (i.e., the intrinsic value) and the efficiency of the same RNA present in a mixture (the in vivo situation) may be different; furthermore, since type I RNAs are inhibited by others whereas type II RNAs are stimulated, the net result of these interactions would be to bring the actual transfer efficiencies of different RNAs closer to one another, i.e., to reduce the variation observed with intrinsic transfer efficiencies. To examine whether this is the case for the four aptamers, their intrinsic outer membrane (Fig. 4) and inner membrane (Fig. 8) transfer efficiencies were used to compare the ratio of the aptamers expected to be transferred to the matrix in the absence of allosteric interactions (the noninteractive model) or in the presence of such interactions (the interactive model), with the corresponding ratios observed in vivo (Fig. 5). Two points may be noted from the analysis presented in Table 2. First, the noninteractive ratios of pairs of type I and type II RNAs, e.g., C16-C20 or C32-C20, were significantly higher than those observed in vivo. This is consistent with the occurrence of positive interactions on type II and/or negative interactions on type I in vivo, thereby reducing the ratio. Second, the large variations in noninteractive efficiencies could be reduced by considering allosteric effects. Thus, the expected noninteractive ratio of C1:C16:C20:C32:tRNATyr in the matrix was estimated to be 3.4:8.6:1:19.8:6.3, with a large statistical variance of more than 40 (Table 2). If the positive and negative interactions observed in Fig. 8 were factored in, the corresponding ratio was altered to 2.8:0.7:1:1.2:0.5 and the variance was dramatically reduced to 0.8. In vivo, the observed ratio was 1.8:2.5:1:4.5:1.8:2.2 and the variance was only 1.4. Thus, the low variance observed in vivo is in accord with the interactive model. However, the in vivo ratio is different from the interactive ratio; this is probably due to the fact that the in vitro value was calculated on the basis of interaptamer interactions at a substrate-to-effector ratio of 10:1 (Fig. 8), whereas in vivo, the effector would be a complex mixture of tRNAs of indeterminate composition. Preliminary experiments also support the existence of allosteric regulation in vivo (our unpublished data).
TABLE 2.
Comparison of relative inner membrane transfer efficiencies according to noninteractive and interactive models with in vivo values
| Clone | In vitro OM transfer (fmol/50 μg)a | In vitro IM transferb
|
In vivo IM transferf
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Noninteractive
|
Interactive
|
No. of molecules/cell | Ratio | ||||||
| Efficiencyc | Transfer (fmol)d | Ratioe | Efficiency | Transfer (fmol) | Ratio | ||||
| C1 | 1.7 | 0.20 | 0.34 | 3.4 | 0.75 | 1.28 | 2.8 | 30 | 1.8 |
| C16 | 1.6 | 0.54 | 0.86 | 8.6 | 0.19 | 0.30 | 0.7 | 43 | 2.5 |
| C20 | 0.7 | 0.14 | 0.10 | 1 | 0.65 | 0.46 | 1 | 17 | 1 |
| C32 | 3.1 | 0.64 | 1.98 | 19.8 | 0.18 | 0.56 | 1.2 | 77 | 4.5 |
| tRNA (Tyr) D arm | 1.1 | 0.61 | 0.67 | 6.7 | NDg | ND | ND | 30 | 1.8 |
| tRNA (Tyr) | 1.4 | 0.45 | 0.63 | 6.3 | 0.16 | 0.22 | 0.5 | 37 | 2.2 |
In vitro outer membrane (OM) transfer per 50 μg of intact mitochondria was determined using 50 fmol of RNA. Data are from Fig. 3C and 4C.
IM, inner membrane; noninteractive, assuming independent transfer; interactive, assuming stimulation of C1 and C20 or inhibition of C32 and C16. Maximum values observed are shown in Fig. 8.
Efficiency, fraction of total internalized RNA in the matrix fraction; determined by intramitochondrial distribution assay (data from Fig. 8 and reference 2).
Transfer was calculated as follows: outer membrane transfer × intrinsic efficiency.
Ratio, relative proportion compared to the lowest value (i.e., that of C20). Statistical variance = σx2, where σ is the standard deviation of x values. The variances of the ratios of the noninteractive and interactive inner membrane transfers are 42.7 and 0.8, respectively.
The number of molecules of RNA in the mitochondrial fraction per viable transfected cell was determined. Data for C1 to C32 are from Fig. 5, data for the tRNATyr D arm are from reference 2, and data for tRNATyr are our unpublished data. The variance of the ratios is 1.42.
ND, not determined.
At present, one can only speculate on the mechanism of allosteric control. The effector RNA may influence the binding of the substrate to an inner membrane receptor through conformational changes in the protein (classical positive or negative cooperativity). Alternatively, or additionally, the receptor: RNA interaction could regulate the opening or closing of import channels (gating). In support of the first mechanism, we have recently observed the presence of an allosterically regulated RNA binding activity in inner membrane extracts (our unpublished data). It is noteworthy that both type I and type II sequences contain oligopurine cores flanked by pyrimidines: YAGAGY and UG3-4U, respectively (Table 1). It is not inconceivable that the conformation of the same RNA binding site can be altered to accommodate distinct but related oligopurine motifs. Such flexibility would enable one or a limited number of receptors to recognize a large number of tRNAs, some with type I and others with type II import signals.
Acknowledgments
This work was supported by a grant from the Department of Science and Technology, Government of India. S.N.B. and S.C. were supported by Fellowships from the Council of Scientific and Industrial Research and the University Grants Commission, respectively.
We thank Andre Schneider for the gift of the pGCN8 plasmid.
REFERENCES
- 1.Adhya, S., T. Ghosh, A. Das, S. K. Bera, and S. Mahapatra. 1997. Role of an RNA binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem. 272:1396-21402. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya, S. N., S. Mukherjee, and S. Adhya. 2000. Mutations in a tRNA import signal define distinct receptors at the two membrane of Leishmania mitochondria. Mol. Cell. Biol. 20:7410-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik, M., S. Das, and S. Adhya. 1991. Evidence for translation control of β-tubulin synthesis during differentiation of Leishmania donovani. Parasitology 103:197-205. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D.-H. T., X. Shi, and Y. Suyama. 1994. In vivo expression and mitochondrial import of normal and mutated tRNAthr in Leishmania. Mol. Biochem. Parasitol. 64:121-133. [DOI] [PubMed] [Google Scholar]
- 5.Ciesiolka, J., M. Illangasekare, I. Majerfeld, T. Nickles, M. Welch, M Yarus, and S. Zinnen. 1996. Affinity selection-amplification from randomized ribooligonucleotide pools. Methods Enzymol. 267:315-335. [DOI] [PubMed] [Google Scholar]
- 6.Das, S., and S. Adhya. 1990. Organization and chromosomal localization of β-tubulin genes in Leishmania donovani. J. Biosci. 15:239-248. [Google Scholar]
- 7.Entelis, N., S. Kieffer, O. A. Kolesnikova, R. P. Martin, and I. A. Tarassov. 1998. Structural requirements of tRNALys for its import into yeast mitochondria. Proc. Natl. Acad. Sci. USA 95:2838-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, K., and S. L. Hajduk. 1990. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 265:19203-19215. [PubMed] [Google Scholar]
- 9.Hauser, R., and A. Schneider. 1995. tRNAs are imported into the mitochondria of Trypanosoma brucei independent of their genomic context and of their genetic origin. EMBO J. 14:4212-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima, B. D., and L. Simpson. 1996. Sequence dependent in vivo importation of tRNAs into the mitochondrion of Leishmania tarentolae. RNA 2:429-440. [PMC free article] [PubMed] [Google Scholar]
- 11.Lye, L. F., D. H. T. Chen, and Y. Suyama. 1993. Selective import of nuclear encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol. Biochem. Parasitol. 58:233-246. [DOI] [PubMed] [Google Scholar]
- 12.Mahapatra, S., and S. Adhya. 1996. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane bound receptors. J. Biol. Chem. 271:20432-20437. [DOI] [PubMed] [Google Scholar]
- 13.Mahapatra, S., S. Ghosh, S. K. Bera, T. Ghosh, A. Das, and S. Adhya. 1998. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 26:2037-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahapatra, S., T. Ghosh, and S. Adhya. 1994. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 22:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marechal-Drouard, L., P. Guillemaut, A. Cosset, M. Arbogast, F. Weiber, J. H. Weil, and A. Dietrich. 1990. Transfer RNA of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic Acids Res. 18:3689-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, R. P., J. M. Schneller, A. J. Stahl, and G. Dirheimer. 1979. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 18:4600-4605. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee, S., S. N. Bhattacharyya, and S. Adhya. 1999. Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J. Biol. Chem. 274:31249-31255. [DOI] [PubMed] [Google Scholar]
- 18.Nabhoiz, C. E., R. Hauser, and A. Schneider. 1997. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl tRNA synthetase activities. Proc. Natl. Acad. Sci. USA 94:7903-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio, M. A., X. Liu, H. Yuzawa, J. D. Alfonzo, and L. Simpson. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA 6:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusconi, C. P., and T. R. Cech. 1996. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 10:2870-2880. [DOI] [PubMed] [Google Scholar]
- 21.Scheinder, A., and L. Marechal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell. Biol. 10:509-513. [DOI] [PubMed] [Google Scholar]
- 22.Shi, X., D. H.-T. Chen, and Y. Suyama. 1994. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear encoded tRNAs between the cytosol and mitochondria. Mol. Biochem. Parasitol. 65:23-37. [DOI] [PubMed] [Google Scholar]
- 23.Simpson, A. M., Y. Suyama, H. Dewes, D. Cambell, and L. Simpson. 1989. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 17:5427-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suyama, Y., S. Wong, and D. A. Campbell. 1998. Regulated tRNA import in Leishmania mitochondria. Biochim. Biophys. Acta 1396:138-142. [DOI] [PubMed] [Google Scholar]
- 25.Yermovsky-Kammerer, A. E., and S. L. Hajduk. 1999. In vitro import of a nuclear encoded tRNA into the mitochondria of Trypanosoma brucei. Mol. Cell. Biol. 9:6253-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]