Abstract
The mitochondrial genome is a significant target of exogenous and endogenous genotoxic agents; however, the determinants that govern this susceptibility and the pathways available to resist mitochondrial DNA (mtDNA) damage are not well characterized. Here we report that oxidative mtDNA damage is elevated in strains lacking Ntg1p, providing the first direct functional evidence that this mitochondrion-localized, base excision repair enzyme functions to protect mtDNA. However, ntg1 null strains did not exhibit a mitochondrial respiration-deficient (petite) phenotype, suggesting that mtDNA damage is negotiated by the cooperative actions of multiple damage resistance pathways. Null mutations in ABF2 or PIF1, two genes implicated in mtDNA maintenance and recombination, exhibit a synthetic-petite phenotype in combination with ntg1 null mutations that is accompanied by enhanced mtDNA point mutagenesis in the corresponding double-mutant strains. This phenotype was partially rescued by malonic acid, indicating that reactive oxygen species generated by the electron transport chain contribute to mitochondrial dysfunction in abf2Δ strains. In contrast, when two other genes involved in mtDNA recombination, CCE1 and NUC1, were inactivated a strong synthetic-petite phenotype was not observed, suggesting that the effects mediated by Abf2p and Pif1p are due to novel activities of these proteins other than recombination. These results document the existence of recombination-independent mechanisms in addition to base excision repair to cope with oxidative mtDNA damage in Saccharomyces cerevisiae. Such systems are likely relevant to those operating in human cells where mtDNA recombination is less prevalent, validating yeast as a model system in which to study these important issues.
Mitochondria are important cellular targets for spontaneous and induced DNA damage (3, 7, 36, 38, 43), and mutations in mitochondrial DNA (mtDNA) cause diseases and likely contribute to late-onset neurodegenerative disorders and the aging process in humans (25, 51). Oxidative damage persists longer in mtDNA than it does in nuclear DNA (52) and appears to be a major contributor to mtDNA mutagenesis in vivo. In addition, decreased mitochondrial oxidative phosphorylation capacity is linked to oxidative stress pathways that perturb other cellular components and functions (52). The susceptibility of mtDNA to oxidative damage has been hypothesized to arise from a combination of biological determinants (36), including the proximity of mtDNA to reactive oxygen species that are by-products of normal respiration, the lack of a compact nucleosome structure to protect mtDNA from damage, and a paucity of mtDNA damage-processing pathways relative to those known to exist in the nucleus (6). However, it is largely unknown if and how these factors ultimately contribute to the susceptibility of mtDNA to damage.
DNA damage caused by reactive oxygen species is removed by the base excision repair pathway (32), which is initiated by the action of glycosylases that excise specific damaged bases from DNA. The action of these enzymes results in an apurinic/apyrimidinic (AP) site, which is subsequently cleaved either by a separate AP endonuclease or by an AP lyase activity that is associated with the DNA glycosylase. Following an additional DNA end-processing step, the AP site is removed and converted into a substrate for DNA polymerase and ultimately DNA ligase, which fill and seal the DNA gap, respectively, thus repairing the damage. The discovery that an AP endonuclease and specific DNA glycosylases are localized to mitochondria has clearly confirmed the existence of a mitochondrial base excision repair pathway (3, 7, 38, 48), which was postulated earlier based on mtDNA repair studies in mammalian cells (26). As is the case for the nucleus, the budding yeast Saccharomyces cerevisiae has begun to serve as an informative model system for mtDNA repair and damage tolerance pathways. With regard to oxidative damage, several components of the base excision repair pathway have been localized to mitochondria in yeast. This includes the major AP endonuclease, Apn1p, (50) and mitochondrial forms of Ogg1p (46) and Ntg1p (1, 54), DNA glycosylases that initiate repair of oxidatively damaged purines and pyrimidines, respectively.
Based on the fact that multiple pathways exist for the repair and tolerance of DNA damage in bacteria and in the nuclei of eukaryotic cells (27), it seems likely that mitochondria also do not rely solely on base excision repair for this purpose. Consistent with this idea is that yeast mitochondria contain at least two proteins implicated in mtDNA recombination, Pif1p, a DNA helicase (24), and Mhr1p, a protein of unknown function, both of which appear to be also involved in mtDNA repair or damage tolerance (17, 18, 28). In addition, Msh1p, a homolog of the bacterial mismatch repair protein MutS, is localized to and functions in mitochondria (5). These observations indicate that base excision repair, recombination, and mismatch repair pathways exist in yeast mitochondria to help maintain the mitochondrial genome. To date, the existence of other DNA repair or damage tolerance pathways in mitochondria, such as nucleotide excision repair and translesion synthesis, has not been demonstrated in any organism. In addition, whether the mitochondrial repair and tolerance pathways functionally interact to form a compensatory network, as has been shown to occur in bacteria (32, 33) and in the yeast nucleus (12, 47), remains to be determined.
Recombination appears to be one pathway involved in mtDNA damage resistance in yeast. In addition to Mhr1p and Pif1p, there are three proteins that have documented functions in recombination in mitochondria, Cce1p, Nuc1p, and Abf2p. Cce1p (also called Mgt1p) is a cruciform cutting endonuclease that resolves Holliday junctions in mtDNA that occur during homologous recombination (15, 23). As a result, cce1 null mutations lead to an ∼10-fold increase in the number of recombination junctions in wild-type mtDNA (30) and also affect the segregation properties of certain mutated mtDNA molecules, a process that is influenced by the presence of recombination junctions (29). Nuc1p is a multifunctional nuclease that is required for the vast majority of DNase activity in mitochondria (9), including a DNA exonuclease activity implicated in DNA end-processing steps required to initiate homologous recombination. As a result, nuc1 null mutations result in an ∼50% reduction in mtDNA recombination frequency (56). Finally, Abf2p is an abundant DNA-binding component of the mtDNA nucleoid (31) that is a member of the high-mobility group (HMG) box family of proteins (10). This class of proteins binds unusual DNA structures and can bend and wrap DNA (11, 16). In mitochondria, Abf2p influences multiple processes involving mtDNA, including transcription (34), segregation during cell division (57), and the stabilization of recombination junctions (30).
The existence of substantial mtDNA recombination is a major difference between yeast and mammalian cells. While biochemical activities capable of catalyzing certain steps involved in recombination in vitro have been isolated from mammalian mitochondria (49), whether mtDNA recombination occurs in mammals to any appreciable extent is still debated (14). Thus, the involvement of recombination as an mtDNA damage tolerance mechanism in mammalian cells is likely to be significantly less than that in yeast.
In order to elucidate oxidative DNA damage resistance pathways in mitochondria, we analyzed mutant yeast strains that lack the mitochondrion-localized DNA glycosylase, Ntg1p, alone and in combination with null mutations in the ABF2, PIF1, NUC1, and CCE1 genes. Our results demonstrate that Ntg1p is involved in the repair of oxidative base damage in mtDNA and indicate that Pif1p and Abf2p reduce this type of mtDNA damage via novel functions of these proteins other than recombination.
MATERIALS AND METHODS
Yeast strains.
All yeast strains used in this study are derivatives of DBY2006 (α his3Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2) and were grown in standard synthetic dextrose (SD), yeast extract-peptone-dextrose (YPD), or yeast extract-peptone-glycerol (YPG) as described previously (44). To create an NTG1 plasmid-shuffle (45) strain, DBY2006 was transformed with a PCR product that contained the NTG1 locus with the coding region replaced with kanamycin resistance cassette (KanMX4) in order to disrupt the chromosomal NTG1 gene by homologous recombination. Antibiotic-resistant transformants were selected on YPD medium containing G418 (200 μg/ml) and screened by PCR and Southern analysis of genomic DNA for insertion of the KanMX4 marker into the chromosomal NTG1 locus. A G418-resistant strain that had the NTG1 gene disrupted was then transformed with the plasmid pRS316-NTG1 that contained a wild-type copy of the NTG1 gene expressed from its own promoter to create the strain TWO1 (α his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 ntg1Δ::KanMX4 pRS316-NTG1 [URA3 CEN/ARS]). This strain is functionally wild type due to the plasmid-borne copy of the NTG1 gene complementing the chromosomal disruption. Growth of this strain on medium containing 2 g of 5-fluoroorotic acid (5-FOA; United States Biological, Inc.) per liter as described previously (37) selects for growth of cells that have spontaneously lost the URA3/NTG1 plasmid (45) and are therefore ntg1 null.
The following strains are derivatives of TWO1 that were made by standard marker disruption as described above for NTG1, except that nutritional markers were used instead of the KanMX4 cassette: abf2Δ strain, TWO4 (α his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 ntg1Δ::KanMX4 abf2Δ::TRP1 pRS316-NTG1 [URA3 CEN/ARS]); pif1Δ strain, TWO5 (α his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 ntg1Δ::KanMX4 pif1Δ::HIS3 pRS316-NTG1 [URA3 CEN/ARS]); nuc1Δ cce1Δ strain, TWO6 (α his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 ntg1Δ::KanMX4 nuc1Δ::LEU2 cce1Δ::HIS3 pRS316-NTG1 [URA3 CEN/ARS]). Growth of these strains in liquid or on solid 5-FOA-containing medium selected for the corresponding combination null mutant strains: abf2Δ ntg1Δ, pif1Δ ntg1Δ, and nuc1Δ cce1Δ ntg1Δ. Lastly, by using the same marker disruption strategy described above, abf2Δ and pif1Δ single-mutant derivatives of DBY2006 were generated, which allowed analysis of these backgrounds without having to maintain the pRS316-NTG1 plasmid. All strains were maintained on YPG medium to sustain mitochondrial respiration competence.
To create a strain that moderately overexpressed Abf2p from its own promoter, DBY2006 was transformed with the CEN plasmid pRS313-ABF2 (34). The genomic fragment in this plasmid has been shown to increase expression of Abf2p two- to threefold (57). The corresponding isogenic wild-type strain was created by transforming DBY2006 with pRS313 without the ABF2 genomic insert.
Oxidative mtDNA damage assays.
Yeast mtDNA was isolated by a standard differential centrifugation/sucrose-gradient purification procedure as described elsewhere (35). Where indicated, mtDNA (8 μg) was heavily damaged by treatment with hydrogen peroxide (0.3 M) in the presence of copper sulfate (0.1 mM) for 20 min at 37°C in a total reaction volume of 50 μl, essentially as described earlier (21). After treatment, 250 μl of 0.3 M sodium acetate was added to the sample and mtDNA was ethanol precipitated, washed twice with 70% ethanol, and resuspended in Tris-EDTA buffer (pH 8.0). This control peroxide-damaged mtDNA and the mtDNA isolated from the indicated yeast strains were incubated for 30 min at 37°C with 2 μg of recombinant Ntg1p in a 25-μl reaction volume containing 15 mM potassium phosphate buffer (pH 6.8), 10 mM EDTA, 10 mM β-mercaptoethanol, and 40 mM KCl. The reaction was terminated by heating the sample at 60°C for 5 min, and products were resolved by electrophoresis through a 0.4% agarose gel. Recombinant Ntg1p used in these experiments was expressed in Escherichia coli as described previously (53), and the N-terminally tagged protein was purified by glutathione-agarose chromatography followed by Mono-S fast-performance liquid chromatography. The purity was approximately 80%, with the remaining 20% consisting mostly of glutathione S-transferase peptide.
Petite-mutant induction assays
The petite-mutant induction assay is based on the fact that growth of yeast cells on YPG (glycerol-containing) medium requires mitochondrial respiration, while growth on YPD (glucose-containing) medium is possible without it (42). To determine the rate of formation of spontaneous petite mutants, a culture of each yeast strain to be tested was grown to near saturation in 5 ml of YPG medium to maintain respiration competence. This starter culture was then diluted to an optical density at 600 nm of 0.05 into either 5 ml (see Fig. 3B) or 20 ml (see Fig. 2) of SD medium (with appropriate nutritional supplements required by each strain) with or without 5-FOA or 5 ml of YPD with or without 50 mM malonic acid (see Fig. 3A). Samples from the experimental cultures were removed immediately (zero point) and/or at specific time points after growth at 30°C with shaking (200 rpm) on a rotary shaker. Identical samples from each culture were then diluted and plated onto YPD and YPG plates and allowed to grow at 30°C until colonies formed (2 to 5 days). The number of colonies on each set of plates was determined (100 to 1,000 CFU/plate in all experiments), and the percentage of respiration-competent cells was calculated as follows: number of colonies on YPG/number of colonies on YPD × 100 (see Fig. 2). Alternatively, the percentage of respiration-incompetent (petite) colonies was also calculated from the ratio of white colonies to total colonies on YPD plates (see Fig. 3). Respiration-competent cells form red colonies on YPD medium due to the disruption of the ADE2 gene in our strain backgrounds, while petite cells form white colonies (42).
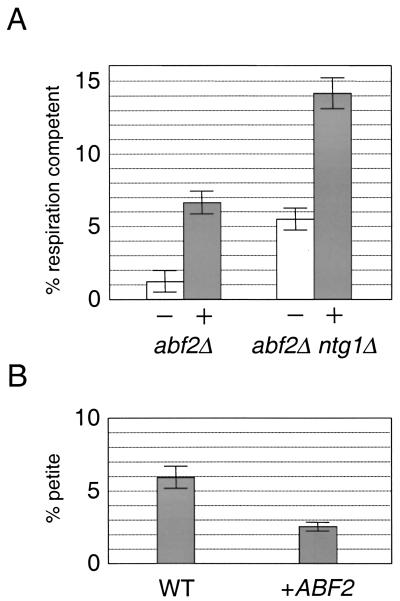
FIG. 3.
Malonic acid or overexpression of Abf2p reduces the rate of spontaneous petite-mutation formation. (A) Results of a petite-mutant induction assay on strains DBY2006 abf2Δ and TWO5 are shown. In the case of TWO5, the pRS316-NTG1 plasmid was chased from the strain with 5-FOA (making it abf2Δ ntg1Δ) prior to the experiment and maintained on YPG medium (see Materials and Methods). The ordinate represents the percentage of respiration-competent cells calculated after approximately six generations of growth in YPD medium in the absence (−, white bars) or presence (+, filled bars) of 50 mM malonic acid. (B) Results of a petite-mutant induction assay on DBY2006 containing a CEN plasmid expressing Abf2p (pRS313-ABF2 [34]), labeled +ABF2, compared to the isogenic control strain containing a pRS313 plasmid without an ABF2 genomic insert, labeled WT. Here the ordinate indicates the percentage of petite cells after approximately six generations of growth in SD medium that selected for maintenance of the indicated plasmids. In panels A and B, the top of each bar represents the mean of three independent measurements and each error bar indicates ± 1 standard deviation from the mean.
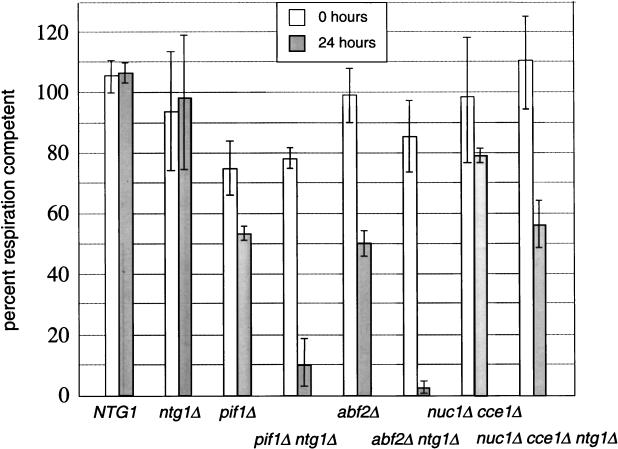
FIG. 2.
Null mutations in PIF1 and ABF2 each exhibit a synthetic-petite phenotype with an ntg1 null mutation. Shown are the results of petite-mutant induction assays performed on parallel cultures of strains TWO1, TWO4, TWO5, and TWO6 grown in SD medium and SD medium plus 5-FOA (to initiate plasmid shuffle; see Materials and Methods). The genotype of each strain during the experiment is indicated at the bottom of the figure. For example, the strain TWO1 is wild type when grown in SD medium (labeled NTG1) and NTG1 null when grown in SD plus 5-FOA (labeled ntg1Δ). The ordinate represents the percentage of respiration-competent cells calculated at the 0-h (open bars) and 24-h (filled bars) time points in the assay. The top of each bar represents the mean of three independent measurements, and each error bar indicates ± 1 standard deviation from the mean.
Erythromycin resistance assays.
The conditions of the erythromycin resistance assay were adapted from the protocol of Chi and Kolodner (5). Individual colonies from indicated strains were used to inoculate 5 ml of YPG medium, and the cultures were grown at 30°C to an optical density at 600 nm of 2 to 4. Cultures were diluted, and cells were plated onto solid YPG medium and grown at 30°C until single colonies formed. Next, 10 to 15 individual colonies were used to inoculate separate 5-ml cultures of YPG and grown at 30°C in a roller drum. After 48 h, a small sample was removed to determine the total number of respiration-competent cells by plating onto YPG. The remainder of each culture was plated onto solid YPG medium containing erythromycin (1 mg/ml) and grown at 30°C for 6 to 7 days until drug-resistant colonies formed. The mutation frequency reported in Table 1 was calculated as follows: number of erythromycin-resistant colonies/total number of colonies. Potential jackpot cultures were identified as outliers statistically by using the Grubbs test (19). These values were removed from the calculations of mutation frequency presented in Table 1.
TABLE 1.
Enhanced frequency of erythromycin-resistant mutants in abf2Δ ntg1Δ and pif1Δ ntg1Δ double-mutant strains
| Strain | Avg no. of cells plated (108) | Avg. no. of erythromycin- resistant mutants | Mutant frequencya | Fold increaseb |
|---|---|---|---|---|
| NTG1 | 7.71 | 0.7 | 1.13 × 10−9 | 1 |
| ntg1Δ | 9.13 | 1.9 | 2.59 × 10−9 | 2.3 |
| pif1Δ | 5.69 | 16 | 3.34 × 10−8 | 29 |
| pif1Δ ntg1Δ | 5.28 | 31 | 6.35 × 10−8 | 56 |
| abf2Δ | 6.19 | 1.2 | 2.09 × 10−9 | 1.8 |
| abf2Δ ntg1Δ | 5.22 | 25 | 5.68 × 10−8 | 50 |
Calculated from the results of plating 25 to 30 independent cultures.
Calculated relative to the wild-type (NTG1) strain.
RESULTS
Ntg1p functions in mitochondria to reduce oxidative mtDNA damage.
Ntg1p removes oxidized pyrimidines via N-glycosylase activity to create an abasic site that is subsequently processed by its AP lyase activity to cause DNA strand breakage. Utilizing these activities, we developed a simple electrophoresis assay to assess levels of oxidative damage in yeast mtDNA based upon the conversion of slowly migrating mtDNA species (∼80-kb monomeric units) into a population of smaller, faster-migrating mtDNA fragments following treatment with purified Ntg1p in vitro. To test the validity of this assay, total yeast mtDNA was prepared from a wild-type strain and treated with purified Ntg1p alone or after incubation with hydrogen peroxide to artificially introduce oxidative DNA damage to the sample in vitro. Treatment of purified mtDNA with hydrogen peroxide alone, which introduces an extensive amount of both oxidative base damage and DNA strand breaks under the conditions used (21), resulted in significant conversion of monomeric mtDNA into a smear of faster-migrating mtDNA fragments (Fig. 1, lane 3). Subsequent incubation of the peroxide-treated mtDNA with purified Ntg1p resulted in the complete conversion of the monomeric mtDNA species to a smear of even-faster-migrating mtDNA fragments (Fig. 1, lane 4). This effect was specific for oxidative damage introduced by peroxide treatment in vitro because incubation of purified yeast mtDNA with Ntg1p alone had no effect (Fig. 1, lane 2). Therefore, this assay provides a convenient method to detect oxidative base damage in yeast mtDNA.
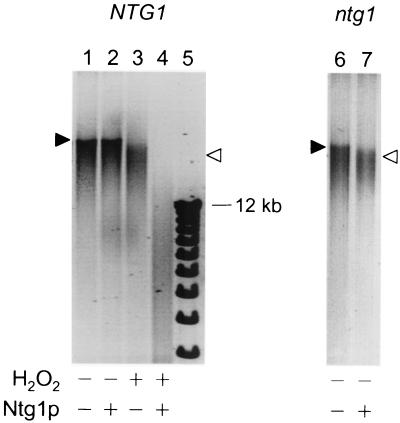
FIG. 1.
Increased oxidative mtDNA damage levels in an ntg1Δ strain. Negative images of ethidium bromide-stained agarose gels of the products of an Ntg1p-based assay for oxidative mtDNA damage are shown. mtDNA isolated from wild-type (NTG1, lane 1) and ntg1 null (ntg1, lane 6) yeast strains ran predominantly as a slowly migrating species (indicated by the solid arrowhead). Purified mtDNA left untreated (−) or treated (+) with hydrogen peroxide (H2O2), to artificially introduce an extensive level of oxidative mtDNA damage (21), or purified Ntg1p, to introduce strand breaks at sites of oxidative damage in vitro, is indicated. Lane 1, wild-type mtDNA; lane 2, wild-type mtDNA treated with Ntg1p; lane 3, hydrogen peroxide-damaged wild-type mtDNA; lane 4, hydrogen peroxide-damaged wild-type mtDNA treated with purified Ntg1p; lane 5, DNA size markers; lane 6, mtDNA from an ntg1Δ strain; lane 7, mtDNA from an ntg1Δ strain treated with purified Ntg1p. The increase in mobility of mtDNA species resulting from DNA strand breaks introduced by treatment with H2O2 and/or Ntg1p in vitro is indicated by the open arrowheads (which refer to lanes 3 and 7). The data shown are from a representative experiment of three independent trials.
By using this assay, the relative amount of spontaneous oxidative base damage in an ntg1 null yeast strain was assessed. Compared to the wild-type control strain (Fig. 1, lanes 1 and 2), mtDNA isolated from an ntg1Δ yeast strain exhibited increased degradation of mtDNA as a result of treatment with purified Ntg1p in vitro (Fig. 1, lanes 6 and 7). In fact, the amount of strand breakage introduced by treatment of mtDNA from the ntg1Δ strain with Ntg1p in vitro was comparable to that introduced into wild-type mtDNA by hydrogen peroxide treatment (Fig. 1, compare lanes 1 and 3 to lanes 6 and 7, respectively). These data indicate that the absence of Ntg1p in vivo results in an increased amount of oxidative damage remaining in the mitochondrial genome.
abf2 and pif1 null mutations exhibit a synthetic-petite phenotype with ntg1 null mutations.
Spontaneous formation of yeast petite mutants is a measure of mtDNA integrity (42), the loss of which causes reduction or absence of expression of mtDNA-encoded subunits of the mitochondrial oxidative phosphorylation system. We implemented a petite-mutant induction assay that employs a yeast strain (TWO1) that is engineered to allow plasmid-shuffle (45) of NTG1. Growth of TWO1 in 5-FOA-containing medium allowed the selection of ntg1Δ cells that have lost the NTG1 plasmid, which is the only source of the NTG1 gene in the strain. Despite the fact that increased oxidative mtDNA base damage persists (Fig. 1), no increase in spontaneous petite-mutant formation rate was observed for the ntg1Δ strain after 24 h of growth in glucose medium (Fig. 2). In fact, the ntg1Δ strain did not exhibit any increase in spontaneous petite-mutant formation compared to the isogenic wild-type control strain, even after extended growth periods (>30 generations; data not shown).
Recombination is a DNA damage tolerance pathway functioning in bacteria and in the nuclei of eukaryotes (4), and recent evidence suggests that it may also be involved in similar processes in yeast mitochondria (28). In addition, nuclear recombination rates are increased when base excision repair capacity is compromised in yeast (47), suggesting that under certain conditions recombination is induced to compensate for reduced DNA repair capacity. Therefore, we reasoned that if recombination can compensate for loss of Ntg1p function, then a cooperative effect on spontaneous petite-mutant formation rate will be observed in strains that are null for both NTG1 and genes involved in recombination in mitochondria. In yeast this type of genetic interaction is termed synthetic (20), and therefore, we refer to this phenotype a “synthetic-petite” phenotype. To test this hypothesis, we created yeast strains in which genes involved in mitochondrial recombination (ABF2, CCE1, NUC1, and PIF1) were inactivated in a TWO1 genetic background. These strains were then tested for synthetic-petite phenotypes with ntg1 null mutations using the petite-mutant induction assay. An increase in the spontaneous petite-mutant formation rate was observed when pif1Δ or abf2Δ single mutant strains were grown in glucose medium (Fig. 2). However, the petite-mutant induction rate was greatly enhanced in the corresponding pif1Δ ntg1Δ and abf2Δ ntg1Δ double-mutant strains (Fig. 2), indicating that abf2 and pif1 null mutations each exhibit a synthetic-petite phenotype with an ntg1 null mutation. In contrast, no increase in spontaneous petite mutant formation was observed in cce1Δ or nuc1Δ single null mutant strains or in the corresponding cce1Δ ntg1Δ or nuc1Δ ntg1Δ strains when analyzed in the same manner (data not shown). In fact, no significant increase in the spontaneous petite-mutant formation rate was observed even in a cce1Δ nuc1Δ double-null mutant strain (Fig. 2) and only a weak synthetic effect on petite-mutant induction was observed in the corresponding cce1Δ nuc1Δ ntg1Δ triple-null mutant strain (Fig. 2). These results indicate that both Pif1p and Abf2p function in the resistance to oxidative mtDNA damage in mitochondria in a manner largely independent of recombination.
Spontaneous petite-mutant formation in abf2Δ strains is mediated by activity of the mitochondrial electron transport chain.
Malonic acid is an analog of succinate that competitively inhibits the mitochondrial enzyme succinate dehydrogenase, a tricarboxylic-cycle enzyme and a component of the mitochondrial electron transport chain (complex II). In yeast, addition of malonic acid reduces, but does not completely inhibit, the electron transport chain and therefore causes a reduction in the amount of reactive oxygen species produced as by-products of respiration (28, 40). To determine if mitochondrial reactive oxygen species contribute to the observed petite-mutant induction phenotypes, we conducted a petite-mutant induction assay in the presence of malonic acid. In abf2Δ and the abf2Δ ntg1Δ strains, the presence of malonic acid significantly increased (2.5- to 5-fold) the number of respiration-competent cells recovered after 24 h of growth in YPD medium (Fig. 3A). Malonic acid had no effect on the spontaneous rate of petite-mutant formation in any of our wild-type genetic backgrounds and did not rescue the petite-mutant induction phenotype of a pif1Δ strain (data not shown). Partial rescue of the petite-mutant induction phenotype of the pif1Δ ntg1Δ strain by malonic acid was occasionally observed; however, this effect was highly variable (data not shown). These results support the notion that reactive oxygen species generated by the mitochondrial electron transport chain can contribute to the spontaneous petite phenotype.
Moderate overexpression of Abf2p reduces spontaneous petite formation.
Overexpression of Abf2p (>10-fold) causes dramatic instability of mtDNA and induction of petite mutations (57). However, moderate (two- to threefold) overexpression of Abf2p can be achieved by placing the ABF2 gene under the control of its own promoter on a CEN plasmid (57). To determine if moderate overexpression of Abf2p had any effect on the rate of spontaneous petite-mutant formation, we transformed the yeast strain DBY2006 with the plasmid pRS313-ABF2 (34) and performed a petite-mutant induction assay. Compared to the corresponding isogenic control, a strain overexpressing Abf2p in this manner exhibited a significant reduction (∼twofold) in the rate of formation of spontaneous petite mutants (Fig. 3B). These data are consistent with a role for Abf2p in protecting cells from spontaneous mtDNA damage.
abf2Δ and pif1Δ strains exhibit increased mtDNA mutagenesis that is enhanced in the absence of Ntg1p.
Specific point mutations in the large (21S) and small (15S) rRNA genes encoded by mtDNA result in resistance to erythromycin, an antibiotic that specifically inhibits mitochondrial translation (8). Thus, acquisition of erythromycin resistance provides a direct readout of mtDNA point mutagenesis in vivo (5). Using this assay, we measured mtDNA mutagenesis in ntg1Δ, abf2Δ, and pif1Δ single-mutant strains and in abf2Δ ntg1Δ and pif1Δ ntg1Δ double-mutant strains (Table 1). Only a small (∼twofold) increase in mtDNA mutagenesis was observed in the ntg1 null mutant strain. However, similar to the results of the petite-mutant induction assays (Fig. 2), a significant increase in mtDNA mutagenesis was observed in the pif1Δ strain (∼30-fold), and this increase was enhanced, in an apparently additive manner, in the pif1Δ ntg1Δ strain to ∼60-fold. In contrast to the results of the petite-mutant induction assay, little effect on mtDNA point mutagenesis was observed in the abf2Δ strain (∼twofold). However, the abf2Δ ntg1Δ strain exhibited a large increase (∼50-fold), indicating that the simultaneous loss of Abf2p and Ntg1p has a strong synergistic effect on mtDNA mutagenesis. Altogether, these data point to involvement of both Abf2p and Pif1p in preventing mtDNA mutagenesis but suggest that the mechanism through which each is acting is different.
DISCUSSION
To gain insight into how eukaryotic cells resist damage to the mitochondrial genome, we have investigated yeast strains that are sensitized toward spontaneous oxidative mtDNA damage because they lack a mitochondrion-localized base excision repair enzyme, Ntg1p. Cultures of strains null for NTG1 alone or in combination with null mutations in four distinct genes involved in mtDNA maintenance and recombination (ABF2, CCE1, NUC1, and PIF1) were analyzed using a petite-mutant induction assay, which measured the rate of loss of mitochondrial respiration capacity that results primarily from the corruption of mtDNA integrity (42). In addition, mtDNA point mutagenesis in these strains was ascertained directly by using an in vivo erythromycin-resistance assay.
An increased level of oxidative mtDNA damage (Fig. 1) and correspondingly a small increase (∼twofold) in spontaneous mtDNA mutagenesis (Table 1) were observed in ntg1 null mutant strains. This is the first functional evidence to support direct involvement of Ntg1p in mitochondrial base excision repair, a role predicted from its demonstrated activity in vitro (41, 53) and mitochondrial localization in vivo (54). Interestingly, ntg1Δ strains do not exhibit any significant increase in spontaneous petite-mutant formation rate (Fig. 1). This is likely a reflection of the fact that mtDNA is present in multiple copies and therefore can handle an increased oxidative damage and/or mutational load due to complementation of mutant alleles by wild-type alleles present on separate mtDNA molecules. This threshold effect is a well-documented phenomenon in mitochondrial disease patients who often harbor a mixture of wild-type and mutated mtDNA molecules, a situation known as heteroplasmy (51). The fact that ntg1Δ yeast strains do not exhibit a petite phenotype and have only modest increases in mtDNA mutagenesis also suggests that, in the absence of this base excision repair enzyme, mtDNA remains protected from excessive oxidative damage by other mechanisms. For example, there may be redundancy in Ntg1p function in mitochondria by other DNA glycosylases present in the organelle (46) or, perhaps more likely, other mtDNA damage resistance pathways might be able to compensate for the lack of Ntg1p function. The latter scenario is supported by the observation that DNA repair and tolerance pathways in bacteria and in the nuclei of eukaryotes have overlapping specificity and appear to form an operational network that ensures the efficient removal of spontaneous DNA damage (12, 32, 33, 47). The existence of such a network suggests that the absence or inactivity of one pathway can be compensated for to some degree by the presence of the others. This concept, in part, was the rationale for the present study, in which we hypothesized that inactivation of other pathways involved in mtDNA damage resistance would result in synthetic-petite-mutant induction phenotypes in conjunction with an ntg1 null mutation.
Recombination is an important DNA damage tolerance pathway in the yeast nucleus (4), and recently it has been demonstrated that when base excision repair is compromised, nuclear recombination rates are substantially increased as part of an apparently compensatory response (47). In addition, it appears that recombination is involved to some degree in tolerating oxidative damage in yeast mitochondria (28). The results of this study do not discount recombination as a DNA damage tolerance mechanism in yeast mitochondria; however, the fact that pif1Δ and abf2Δ mutations exhibit a strong synthetic-petite phenotype with ntg1 null mutations while cce1Δ and nuc1Δ mutations do not (Fig. 1) suggests that Pif1p and Abf2p function in mtDNA damage resistance via activities other than, or in addition to, recombination.
Two additional lines of evidence support the concept that the observed synthetic-petite phenotypes observed in our experiments are due, at least in part, to oxidative damage to mtDNA coupled with reduced repair or resistance capacity. First, the petite-mutant induction rates of abf2Δ and abf2Δ ntg1Δ strains were significantly reduced by the presence of malonic acid in the growth medium (Fig. 3). Malonic acid has been shown to reduce oxidative mtDNA damage in yeast (28, 40), suggesting that this type of damage is mediated by reactive oxygen species generated during normal mitochondrial respiration. Second, mtDNA point mutagenesis was substantially elevated in pif1Δ, pif1Δ ntg1Δ, and abf2Δ ntg1Δ strains (Table 1).
Our results also revealed interesting differences regarding the relative contributions of Pif1p and Abf2p to the observed phenotypes. For example, abf2Δ single mutants exhibited little increase in mtDNA mutagenesis while that of pif1Δ single mutants was substantially elevated (30-fold increase compared to the wild type [Table 1]). Despite these differences, these strains displayed similar rates of spontaneous petite-mutant formation (Fig. 1). This demonstrates that increased mtDNA point mutagenesis causes an increased rate of spontaneous petite-mutant formation but that the influence of other factors can contribute substantially (equally, in the case of abf2Δ strains) to this phenotype. One such factor is most likely large-scale rearrangement or complete loss of the mtDNA genome, a major contributor to the formation of spontaneous petite mutants in yeast (42). The fact that mtDNA point mutagenesis is elevated to similar levels in the pif1Δ ntg1Δ and abf2Δ ntg1Δ strains while the levels in the pif1Δ and abf2Δ single-mutant strains differ substantially again implicates a role for both Pif1p and Abf2p in preventing mtDNA mutagenesis but suggests that the mechanism by or the degree to which each protein is doing so is markedly different. This is supported further by the observation that the degree of synergism in mtDNA mutagenesis rates between an abf2 null mutation and an ntg1 null mutation is significantly greater than that observed between pif1 and ntg1 (Table 1) and by the fact that malonic acid partially rescues the petite-mutant induction phenotype of abf2Δ strains, but not pif1Δ strains.
Though Pif1p and Abf2p have been implicated to some degree in mtDNA recombination, these proteins also have functions unrelated to recombination. For example, Abf2p is an abundant mitochondrial DNA-binding protein of the HMG box family (10) that can bend and wrap DNA (11, 16), as well as influence mtDNA segregation (57), transcription (34), and nucleoid structure (31). Likewise, Pif1p is a 5′-to-3′ DNA helicase that has been implicated in mtDNA repair (18) and the nuclear form of the enzyme is involved in the inhibition of bidirectional rDNA replication (22) and telomere length regulation (39). While the mechanism underlying the synthetic interactions between Abf2p, Pif1p, and Ntg1p in mitochondria is not entirely clear, we speculate that recombination-independent functions of each of these proteins can influence the degree to which mtDNA is damaged or repaired. For example, it is possible that Pif1p and Abf2p together or independently influence the structure or accessibility of mtDNA in a manner that facilitates the binding or activity of Ntg1p or other repair proteins. This could involve a general function such as packaging of the mtDNA into nucleoids (31) or promoting a specific DNA or protein/DNA conformation at sites of DNA damage.
Alternatively, models that invoke novel functions of Pif1p and Abf2p can be envisioned. Abf2p, through its DNA-binding and bending activity (11, 16) and ability to influence overall nucleoid structure, may provide shielding that reduces mtDNA damage in much the same manner that nucleosome structure can protect nuclear DNA from damage (13, 36, 55). Alterations in Abf2p levels also result in changes in mtDNA copy number (57). Decreased mtDNA copy number in abf2Δ strains may lead to increased susceptibility to oxidative mtDNA damage. Likewise, increased mtDNA copy numbers in strains that moderately overexpress Abf2p (57) may provide a buffer against mtDNA damage. The fact that moderate overexpression of Abf2p results in a significant reduction in the spontaneous petite-mutant formation rate (Fig. 3B) is consistent with Abf2p providing resistance to mtDNA damage either through an mtDNA-shielding or mtDNA copy-number-mediated mechanism. In the case of Pif1p, because nuclear and mitochondrial forms of the protein are derived from the same gene, its known function in the nucleus likely provides insight into its function in mitochondria. For example, if Pif1p is a region-specific (ribosomal DNA and telomere) helicase that inhibits DNA replication in the nucleus, as has been postulated by others (2), it may likewise function to govern the rate of mtDNA replication. In this model, lack of Pif1p would result in a reduced time frame for mtDNA repair to occur. If this were the case, replication through unrepaired, damage-containing regions would occur, resulting in an increase in mtDNA mutations and perhaps aborted replication complexes. This would result in a synthetic-petite phenotype in ntg1 null strains in which mtDNA repair is compromised.
While our results do not implicate recombination as a major mechanism that can compensate for lack of Ntg1p in yeast mitochondria, they also do not eliminate the possibility that recombination is one mechanism for repair or tolerance of oxidative mtDNA damage in yeast (28). Our results do, however, strongly suggest that recombination-independent mechanisms are also functioning to reduce the susceptibility of mtDNA to oxidative damage. Any recombination-independent mechanism is likely to be significant for understanding how mtDNA integrity is maintained in vertebrate cells, for which mtDNA recombination has not been clearly demonstrated (14). This is particularly relevant with regard to the functions of Abf2p and Pif1p, because human homologs of these genes exist (2, 34). An increased understanding of mtDNA damage resistance mechanisms will provide new insights into the physiologic significance of mtDNA mutagenesis and perhaps into how to counteract the pathological consequences resulting in damage to mtDNA caused by endogenous and exogenous genotoxic agents. Genetic model systems, such as the S. cerevisiae system described here, will continue to be instrumental in elucidating these processes.
Acknowledgments
This work was supported by grant DAAD19-00-1-0560 (G.S.S.) from the Army Research Office and CA-78622 (P.W.D.) from the National Institutes of Health. N.D. is supported by an NIH Predoctoral Training Program in Genetics (GM-08490).
T.W.O. and N.A.D. contributed equally to this work.
REFERENCES
- 1.Alseth, I., L. Eide, M. Piravano, T. Rognes, E. Seeberg, and M. Bjoras. 1999. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 19:3779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessler, J. B., J. Z. Torres, and V. A. Zakian. 2001. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 11:60-65. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen, D. F., K. G. Pinz, and R. M. Perez-Jannotti. 2001. Enzymology of mitochondrial base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 68:257-271. [DOI] [PubMed] [Google Scholar]
- 4.Broomfield, S., T. Hryciu, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486:167-184. [DOI] [PubMed] [Google Scholar]
- 5.Chi, N. W., and R. D. Kolodner. 1994. Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J. Biol. Chem. 269:29984-29992. [PubMed] [Google Scholar]
- 6.Clayton, D. A., J. N. Doda, and E. C. Friedberg. 1974. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 71:2777-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croteau, D. L., R. H. Stierum, and V. A. Bohr. 1999. Mitochondrial DNA repair pathways. Mutat. Res. 434:137-148. [DOI] [PubMed] [Google Scholar]
- 8.Cui, Z., and T. L. Mason. 1989. A single nucleotide substitution at the rib2 locus of the yeast mitochondrial gene for 21S rRNA confers resistance to erythromycin and cold-sensitive ribosome assembly. Curr. Genet. 16:273-279. [DOI] [PubMed] [Google Scholar]
- 9.Dake, E., T. J. Hofmann, S. McIntire, A. Hudson, and H. P. Zassenhaus. 1988. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 263:7691-7702. [PubMed] [Google Scholar]
- 10.Diffley, J. F. X., and B. Stillman. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA 88:7864-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diffley, J. F., and B. Stillman. 1992. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 267:3368-3374. [PubMed] [Google Scholar]
- 12.Doetsch, P. W., N. J. Morey, R. L. Swanson, and S. Jinks-Robertson. 2001. Yeast base excision repair: interconnections and networks. Prog. Nucleic Acids Res. Mol. Biol. 68:29-39. [DOI] [PubMed] [Google Scholar]
- 13.Enright, H. U., W. J. Miller, and R. P. Hebbel. 1992. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 20:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyre-Walker, A., and P. Awadalla. 2001. Does human mtDNA recombine? J. Mol. Evol. 53:430-435. [DOI] [PubMed] [Google Scholar]
- 15.Ezekiel, U. R., and H. P. Zassenhaus. 1993. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 240:414-418. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, R. P., T. Lisowsky, M. A. Parisi, and D. A. Clayton. 1992. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267:3358-3367. [PubMed] [Google Scholar]
- 17.Foury, F., and J. Kolodynski. 1983. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foury, F., and A. Lahaye. 1987. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 6:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubbs, F. E. 1969. Procedure for detecting outlying observations in samples. Technometrics 11:1-21. [Google Scholar]
- 20.Guarente, L. 1993. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 9:362-366. [DOI] [PubMed] [Google Scholar]
- 21.Helland, D., P. Doetsch, and W. Haseltine. 1986. Substrate specificity of a mammalian DNA repair endonuclease that recognizes oxidative base damage. Mol. Cell. Biol. 6:1983-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 23.Kleff, S., B. Kemper, and R. Sternglanz. 1992. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahaye, A., H. Stahl, D. Thines-Sempoux, and F. Foury. 1991. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 10:997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson, N., and D. A. Clayton. 1995. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29:151-178. [DOI] [PubMed] [Google Scholar]
- 26.LeDoux, S. P., and G. L. Wilson. 2001. Base excision repair of mitochondrial DNA damage in mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 68:273-284. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 28.Ling, F., H. Morioka, E. Ohtsuka, and T. Shibata. 2000. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 28:4956-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockshon, D., S. G. Zweifel, L. L. Freeman-Cook, H. E. Lorimer, B. J. Brewer, and W. L. Fangman. 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947-955. [DOI] [PubMed] [Google Scholar]
- 30.MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. USA 95:6739-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman, S. M., O. Zelenaya-Troitskaya, P. S. Perlman, and R. A. Butow. 1996. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 24:386-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsen, H., and H. E. Krokan. 2001. Base excision repair in a network of defense and tolerance. Carcinogenesis 22:987-998. [DOI] [PubMed] [Google Scholar]
- 33.Otterlei, M., B. Kavli, R. Standal, C. Skjelbred, S. Bharati, and H. E. Krokan. 2000. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 19:5542-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisi, M. A., B. Xu, and D. A. Clayton. 1993. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 13:1951-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Querol, A., and E. Barrio. 1990. A rapid and simple method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res. 18:1657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter, C., J. W. Park, and B. N. Ames. 1988. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA 85:6465-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodeheffer, M. S., B. E. Boone, A. C. Bryan, and G. S. Shadel. 2001. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 276:8616-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer, D. E., and B. Van Houten. 1999. Repair of DNA damage in mitochondria. Mutat. Res. 434:161-176. [DOI] [PubMed] [Google Scholar]
- 39.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76:145-155. [DOI] [PubMed] [Google Scholar]
- 40.Senbongi, H., F. Ling, and T. Shibata. 1999. A mutation in a mitochondrial ABC transporter results in mitochondrial dysfunction through oxidative damage of mitochondrial DNA. Mol. Gen. Genet. 262:426-436. [DOI] [PubMed] [Google Scholar]
- 41.Senturker, S., P. Auffret van der Kemp, H. J. You, P. W. Doetsch, M. Dizdaroglu, and S. Boiteux. 1998. Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 26:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shadel, G. S. 1999. Yeast as a model for human mtDNA replication. Am. J. Hum. Genet. 65:1230-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 44.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 46.Singh, K. K., B. Singala, H. A. Sikder, and C. Schwimmer. 2001. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 29:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takao, M., H. Aburatani, K. Kobayashi, and A. Yasui. 1998. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 26:2917-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thyagarajan, B., R. A. Padua, and C. Campbell. 1996. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 271:27536-27543. [DOI] [PubMed] [Google Scholar]
- 50.Vongsamphanh, R., P. K. Fortier, and D. Ramotar. 2001. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol. 21:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 52.Yakes, F. M., and B. Van Houten. 1997. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA 94:514-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You, H. J., R. L. Swanson, and P. W. Doetsch. 1998. Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry 37:6033-6040. [DOI] [PubMed] [Google Scholar]
- 54.You, H. J., R. L. Swanson, C. Harrington, A. H. Corbett, S. Jinks-Robertson, S. Senturker, S. S. Wallace, S. Boiteux, M. Dizdaroglu, and P. W. Doetsch. 1999. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry 38:11298-11306. [DOI] [PubMed] [Google Scholar]
- 55.Yu, L., I. H. Goldberg, and P. C. Dedon. 1994. Enediyne-mediated DNA damage in nuclei is modulated at the level of the nucleosome. J. Biol. Chem. 269:4144-4151. [PubMed] [Google Scholar]
- 56.Zassenhaus, H. P., and G. Denniger. 1994. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr. Genet. 25:142-149. [DOI] [PubMed] [Google Scholar]
- 57.Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman, and R. A. Butow. 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics 148:1763-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]