Abstract
Immunoglobulin μ alternative RNA processing is regulated during B-cell maturation and requires balanced efficiencies of the competing splice (μm) and cleavage-polyadenylation (μs) reactions. When we deleted sequences 50 to 200 nucleotides beyond the μs poly(A) site, the μs/μm mRNA ratio decreased three- to eightfold in B, plasma, and nonlymphoid cells. The activity could not be localized to a smaller fragment but did function in heterologous contexts. Our data suggest that this region contains an RNA polymerase II pause site that enhances the use of the μs poly(A) site. First, known pause sites replaced the activity of the deleted fragment. Second, the μ fragment, when placed between tandem poly(A) sites, enhanced the use of the upstream poly(A) site. Finally, nuclear run-ons detected an increase in RNA polymerase loading just downstream from the μs poly(A) site, even when the poly(A) site was inactivated. When this μ fragment and another pause site were inserted 1 kb downstream from the μs poly(A) site, they no longer affected the mRNA expression ratio, suggesting that pause sites affect poly(A) site use over a limited distance. Fragments from the immunoglobulin A gene were also found to have RNA polymerase pause site activity.
Elements that modulate gene expression by altering RNA polymerase elongation (reviewed in references 9, 35, 43, and 49) have been identified near the 5′ end of genes (e.g., myc and human immunodeficiency virus) (18, 20, 26, 27), within the coding region of genes (e.g., histone H3.3 and apolipoprotein A-I) (22, 37, 46), and downstream from some (3, 14), but not all (51), cleavage-polyadenylation [poly(A)] sites. The elements found downstream from the α globin (14) and complement C2 (3) poly(A) sites were shown to contribute to transcriptional termination. The α globin element is contained within a 92-bp fragment that is located about 300 nucleotides (nt) downstream from the α globin poly(A) site (14). The C2 element is a 156-bp fragment that begins just downstream from the C2 poly(A) site; despite its close proximity, it is not required for efficient use of the C2 poly(A) site (3). These elements were called RNA polymerase pause sites because they appeared to have a kinetic affect on RNA polymerase II transcription (3, 14); when placed between two competing reactions, their effect on gene expression was consistent with these elements causing RNA polymerase to pause or slow its elongation rate. This was true when they were placed between tandem poly(A) sites (3, 14), between tandem promoters (13), and between a regulated exon and its intronic regulatory region (38). In addition, in nuclear run-on assays, which measure RNA polymerase loading along a transcription unit, an increase in RNA polymerase loading was seen over these elements, again suggesting that RNA polymerase was being slowed or temporarily paused, so there was a higher probability of finding RNA polymerase molecules over those regions (3, 14). More recently, the activity of these pause sites and their effect on upstream poly(A) sites was examined in an in vitro coupled transcription cleavage-polyadenylation assay (52, 53). These elements enhanced the use of an upstream poly(A) site in a transcription-dependent manner but did not enhance poly(A) site use in a simple cleavage-polyadenylation assay (52). RNA polymerase pausing by the C2 (MAZ4) pause site was detected in the absence of a functional poly(A) site and also when the template was transcribed by purified RNA polymerase, suggesting that this element may have intrinsic RNA polymerase pause activity (52). While the definitive in vitro experiments to specifically call these elements pause sites (for examples, see references 18 and 49) have not been performed, because this term has been used to describe these elements based on the kinetic effect they have on RNA polymerase (1, 3, 4, 13, 14, 52, 53), we will continue to refer to them in this paper as pause sites.
Sequences with similar RNA polymerase pausing activities are also found downstream of poly(A) sites between tandemly arranged genes in fission yeast (1). Interestingly, these elements in mammalian and yeast genes seem to contain multiple redundant sequences; no subfragments have been found to contain the activity of the whole (1, 3). Some of these pause sites have activity in both the sense and antisense orientations, depending on the specific assay used (1, 3, 13, 14, 38). To date, no sequence homology has been found among RNA polymerase pause sites that would allow them to be identified solely by sequence comparisons; pause sites have always been defined functionally. In addition, how different pause sites slow RNA polymerase transcription is not always clear. However, there seem to be multiple ways to slow an elongating RNA polymerase (reviewed in reference 49). A few, but not all, DNA binding proteins have been shown to pause RNA polymerase transcription (4, 10). Sequences in the nascent RNA that can form RNA secondary structure, newly synthesized U-rich or UC-rich sequences, and sequences that bend DNA have all been associated with RNA polymerase pausing (for examples, see references 18, 49, and 52).
Immunoglobulin (Ig) genes have received much attention as a model system for regulated alternative RNA processing (reviewed in reference 29). During B-lymphocyte maturation from a B cell to a plasma cell, there is a switch in the Ig mRNA content of the cells; B cells produce similar amounts of mRNA encoding membrane and secreted forms of Ig protein while plasma cells predominantly produce mRNA encoding the secreted Ig. The 3′ end of the IgM (μ) pre-mRNA contains competing RNA processing signals that have balanced efficiencies: a poly(A) site that creates the 3′ end of the secretory-specific (μs) mRNA is located within the intron (Cμ4-M1) that is spliced to form the membrane-specific mRNA (μm). The strengths of these two reactions are balanced and their relative use is regulated during B-lymphocyte maturation. Since non-Ig genes that contain a similar splice versus cleavage-polyadenylation competition are also regulated in lymphoid cell lines (28) and in transgenic mice (42), the factors that modulate the RNA processing choices are not Ig-gene-specific but rather must be general processing factors. In support of this, the μs/μm (pA/splice) mRNA ratio is modulated by changes in the CstF 64,000-molecular-weight protein (64K protein) levels (45) and/or by modulation of CstF 64K activity (12, 50).
The μs poly(A) site is central to the regulation of μs/μm mRNA, and it has been studied in a chimeric context (33, 34) and within the μ gene itself (M. L. Peterson, unpublished data) to understand better how its use is modulated during B-cell development. While making mutations to sequences within and surrounding the μs poly(A) site, we identified a fragment located 50 to 200 bp downstream of the μs poly(A) site that we show here contains an RNA polymerase II pause site similar to those found downstream of the α globin and C2 poly(A) sites (3, 14). When this region was deleted, a three- to eightfold decrease in the pA/splice mRNA ratio was seen in B cells, plasma cells, and nonlymphoid cells. Much of this effect was lost when this region was subdivided by smaller deletions. The effect of the deletion could be seen in a heterologous context, and this region, as a 158-bp fragment, could enhance the use of a heterologous poly(A) site. Several observations have led us to suggest that this region is an RNA polymerase II pause site that enhances use of the μs poly(A) site. First, the pause sites from the α globin and C2 (MAZ4) genes could replace the activity of the deleted fragment. Second, placing the 158-bp μ fragment between tandem poly(A) sites enhanced use of the upstream site, as seen with other pause sites. Finally, nuclear run-ons with fragments spanning the Ig μ gene detected an increase in RNA polymerase loading just downstream from the μs poly(A) site in plasma cells. This increased loading was still detected when the μs poly(A) site was inactivated but not when both the μs poly(A) site and the 158-bp region were deleted. When this 158-bp μ fragment and the α globin pause site were inserted 1 kb downstream from the μs poly(A) site, they no longer had an effect on the pA/splice mRNA expression ratio. This suggests that there is a limited distance over which RNA polymerase pause sites can affect the use of an upstream poly(A) site. The μ pause site appears to function similarly in B cells and plasma cells and therefore, likely modulates the overall usage of the μs poly(A) site rather than contributing directly to the μs/μm mRNA processing regulation. Interestingly, we find similar RNA polymerase II pause site activity within fragments downstream from the αs poly(A) site in the IgA gene.
MATERIALS AND METHODS
Plasmid construction.
All of the mouse Cμ gene modifications were constructed in the pSV2Cμ plasmid used previously (Fig. 1) (for examples, see references 30 to 32) by a two-step cloning procedure. First, the modification was made in a pUC9 subclone containing a 2,318-bp PstI fragment, the modification was confirmed by DNA sequencing, and the ApaI-KpnI fragment containing the modification was cloned into pSV2Cμ. The ΔU (made for us by Jeff Wilusz) and B mutations were made by following a megaprimer mutagenesis protocol (39) starting with the wild-type or ΔU plasmid, respectively. The sequence of the ΔU mutation is shown in Fig. 1A, and the B mutation changes TCCTCC to AAATGG at the positions shown in Fig. 1A and 2. The ΔNH, ΔBH, and ΔNB deletions (Fig. 2) were made by digesting the mutation-containing plasmids with NotI-HindIII, BstXI-HindIII, or NotI-BstXI, respectively, making the ends blunt with Klenow, and religating the plasmids. ΔNH-NH(+) and ΔNH-NH(−) were made by digesting ΔU with NotI-HindIII, making the ends blunt with Klenow, and then adding extra copies of the 158-bp NotI-HindIII fragment before ligating. The Cμ genes that contain known pause sites in place of the NH fragment (see Fig. 4), ΔNH-α(+), ΔNH-α(−), ΔNH-MAZ4(+), and ΔNH-MAZ4(−), were made by digesting ΔU with NotI-HindIII, making the ends blunt with Klenow, isolating the vector fragment, and inserting blunt-ended α globin or MAZ4 pause site fragments. The α globin fragment was a 110-bp HpaI-BglII fragment from SΔ3 (14), and the MAZ4 fragment was a 165-bp PstI-BamHI fragment from PAC-MAZ4 (also called C2x4) (4). The Cμ genes containing fragments from the IgA intron (see Fig. 7), ΔNH-IgA228(+), ΔNH-IgA228(−), ΔNH-IgA193(+), and ΔNH-IgA193(−) were made as described above, except that the fragments inserted in place of the NotI-HindIII fragment were the 228-bp BanI-ApaLI and 193-bp ApaLI-BanI fragments isolated from an IgA intron subclone (41).
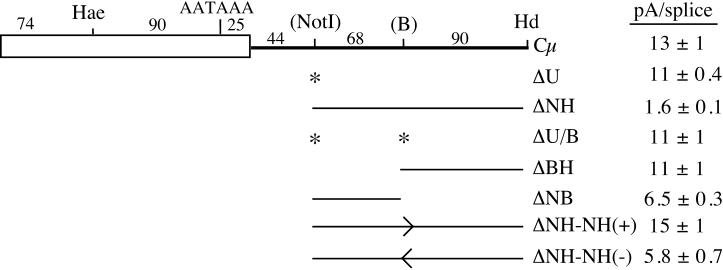
FIG. 1.
(A) Map of the chimeric SVneo-Cμ transcript in pSV2Cμ and its derivatives. The open box with dotted edges is the SV40-neo sequence, the filled boxes are mouse Cμ exons that are common to μs and μm mRNA, the open box is the μs-specific exon, and the hatched boxes are the μm-specific exons. The Cμ4-M1 splice reaction that directly competes with cleavage-polyadenylation at the μs poly(A) site is shown along with the size of this intron. The region between the Cμ4 5′ splice site and the HindIII (Hd) restriction site is expanded below: Hae, HaeII; AATAAA, part of the μs poly(A) signal; end of the open box, μs poly(A) cleavage site; NotI, new NotI restriction site created by the ΔU mutation; B, new BstXI restriction site created by the B mutation. The distances between the identified sequences, counting from the middle of the restriction enzyme recognition sequences, are shown. The sequence immediately downstream from the μs cleavage site is shown along with the sequence of the ΔU mutation; the G's and T's are shown in bold. The region removed by the NH deletion (ΔNH) is delineated. P (PstI), A (ApaI), and K (KpnI) respresent sites used in subcloning. (B) Deleting the 150-bp NH fragment decreases the pA/splice expression ratio in lymphoid and nonlymphoid cells. S1 nuclease analysis of RNA isolated from cells transiently transfected with the constructs shown above each lane is shown. Data with RNA from the nonlymphoid HepG2 cell line, the M12 B-cell line, and the S194 plasma cell line are shown. The S1 nuclease probe distinguishes RNA cleaved and polyadenylated at the μs poly(A) site (pA) from spliced μm mRNA (splice) (31). The pA/splice expression ratios of these representative gels were quantitated by PhosphorImager analysis and are shown below each lane.
FIG. 2.
The NH fragment cannot be subdivided without losing the effect of the full NH deletion. The μs poly(A) site region is as shown in Fig. 1A, and the mutations made in each construct are diagrammed. The asterisks indicate the presence of site-directed mutations, and the bars delineate the sequences deleted. The arrows indicate reinsertions of the NH fragment in the sense (+) or antisense (−) orientation. The pA/splice ratio of RNA from HepG2-transfected cells was quantitated by S1 analysis; the averages ± standard deviations of at least two analyses of multiple transfections are shown.
FIG. 4.
The activity of the NH fragment can be replaced with RNA polymerase pause sites from the α globin and C2 (MAZ4) genes but only when located close to the μs poly(A) site. The 3′ end of the μ gene is as shown in Fig. 1A. The mutations in each construct are diagrammed. The bars delineate deleted sequences, and the arrows indicate the insertion of the pause site fragments either in place of the NH fragment or at the KpnI site in the sense (+) or antisense (−) orientation. RNA from HepG2-transfected cells was quantitated by S1 analysis, and the pA/splice ratio averages ± standard deviations of at least two analyses of multiple transfections are shown.
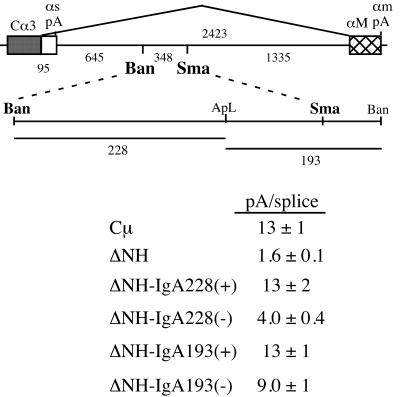
FIG. 7.
Fragments from the IgA gene can replace the activity of the NH fragment. The 3′ end of the mouse IgA gene, which is similar in stricture to the IgM gene, is shown with symbols as described for Fig. 1A; the size of the Cα3-M1 intron and the distances between identified restriction sites are shown. Sequences between the BanI (Ban) and SmaI (Sma) restriction sites were shown to be required for correct αs/αm mRNA processing (11). Two subfragments from this region, the 228-bp BanI-ApaLI (ApL) and the 193-bp ApaLI-BanI fragments, were put in place of the NH fragment in the Cμ gene (Fig. 1A) in both the sense (+) and antisense (−) orientations. RNA from HepG2-transfected cells was quantitated by S1 analysis; the pA/splice ratio averages ± standard deviations of at least two analyses of multiple transfections are shown.
The Cμ genes containing the pause sites inserted in the KpnI site in the Cμ4-M1 intron, ΔNH-KNH(+), ΔNH-KNH(−) (data not shown), ΔNH-Kα(+), and ΔNH-Kα(−) (see Fig. 4), were made by linearizing pSV2CμΔNH with KpnI, making the ends blunt with Klenow, and inserting the 158-bp NotI-HindIII fragment or the 110-bp HpaI-BglII α globin pause site fragment.
The plasmids containing poly(A) sites in the intron of the major histocompatibility complex (MHC) class I Dd gene (Fig. 3), Ddμs, DdμsΔNH, and Ddγs, are similar to simian virus 40 (SV40) Igk-Ddαs (28, 42), except that the poly(A) sites inserted at the blunt-ended NcoI site were a 280-bp HaeII-HinfI μs poly(A) site, a 160-bp HaeII-NotI μsΔNH poly(A) site, and a 185-bp EcoRI-HindIII γ2b secretory-specific site (provided by Chris Milcarek), respectively. To make Ddγs-NH(+) and Ddγs-NH(−), the 158-bp NotI-HindIII fragment was inserted at a blunt-ended SphI site at the 3′-most end of the γs poly(A) site fragment in Ddγs.
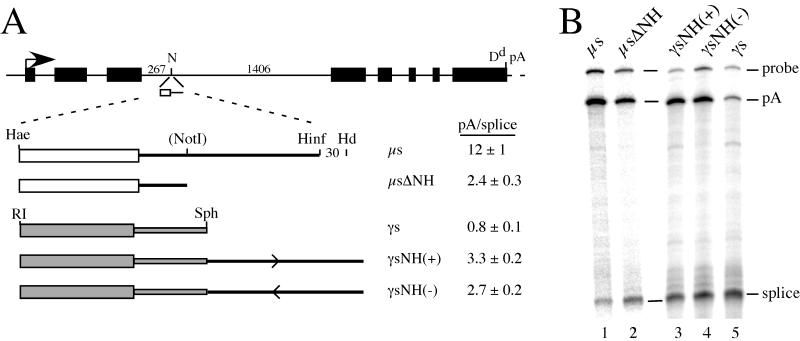
FIG. 3.
The effect of the NH fragment is seen in a heterologous context and can act on a heterologous poly(A) site. (A) The μs and γ2b secretory-specific (γs) poly(A) sites, with or without the NH fragment, were placed in the NcoI (N) site of the third intron of the MHC class I Dd gene. The μs fragments are represented as in Fig. 1A, except the μs fragment ends at the HinfI (Hinf) site. The EcoRI (RI)-SphI (Sph) γs fragments are shown as gray boxes. (B) RNA from HepG2 cells transfected with the constructs shown above each lane was analyzed by S1 nuclease mapping by using a probe that distinguishes spliced RNA (splice) from RNA cleaved and polyadenylated at the inserted poly(A) site (pA) (28); the protected bands are labeled. The pA/splice ratio was quantitated, and the averages ± standard deviations of at least two analyses of multiple transfections are shown in panel A.
The constructs that were used to make stable S194 plasma cell lines were derived from plasmid pR-SP6, which contains an intact Ig gene (31). The genes contained either the wild-type μs poly(A) site (Cμ), previously called μs-m (31), or a deletion of the μs poly(A) site (pA21 and μm-2). pA21 was made by using the megaprimer mutagenesis protocol (39), and it deletes 21 nt that include the AAUAAA of the μs poly(A) site and the surrounding AU-rich region (34). The μm-2 deletion removes the 313-bp HaeII-HindIII fragment that contains the entire μs poly(A) site and the downstream NH fragment (Fig. 1A).
The poly(A) competition (PAC) vector and positive controls containing the α globin, C2, and MAZ4 (C2x4) pause sites (3, 4) were provided by Nick Proudfoot. To make PAC-NH (see Fig. 5), the PAC vector was digested with XbaI and the ends were made blunt with Klenow and ligated with the blunt-ended 158-bp NotI-HindIII μ fragment.
FIG. 5.
The NH fragment has RNA polymerase pause site activity in a PAC assay. The NH fragment was inserted into the PAC vector between the α globin poly(A) (α glo pA) and SPA sites, and its activity was compared to the previously characterized α globin (α glo pause) and C2 (MAZ4) pause sites. The plasmids were transfected into HepG2 cells, and the RNA was analyzed by S1 nuclease mapping by using the probe diagrammed. The S1 probe contained the C2 pause site, so the SPA band results from the mismatch between the probe and the RNAs analyzed. The MAZ4 RNA is derived from C2 sequences, and thus, the protected fragment is slightly longer than for the other RNAs. The percent use of the α poly(A) site was quantitated by PhosphorImager analysis; a representative gel is shown.
Cell culture and DNA transfections.
The M12 B-cell line was grown in RPMI 1640 media supplemented with 10% fetal bovine serum and 50 μM 2-mercaptoethanol. The S194 plasmacytoma cell line was grown in Dulbecco modified Eagle medium supplemented with 10% horse serum. The HepG2 human hepatoma cell line was grown in Dulbecco modified Eagle-F12 medium (1:1) supplemented with 10% fetal bovine serum and 10 μg of insulin/ml.
The B cells and plasma cells were transiently transfected by following the DEAE-dextran protocol (17), and the HepG2 cells were transiently transfected by following a calcium phosphate procedure (44). Plasma cells were stably transfected with linear plasmids by electroporation followed by selection in 500 μg of G418/ml.
RNA preparation and analysis.
Cytoplasmic RNA was prepared from B cells and plasma cells 40 h after transfection (40). A hot phenol procedure was used to isolate total RNA from transfected HepG2 cells (44). S1 nuclease analysis was used to analyze the expression from the transfected genes. For the Cμ-derived (31) and Dd-derived (28) genes, probes that distinguish spliced from cleaved and polyadenylated mRNA were 3′ end-labeled with α [32P]dCTP and the Klenow fragment of DNA polymerase I, hybridized to RNA overnight at 50°C, and digested with 60 U of S1 nuclease for 30 min at 37°C. For the PAC vector RNA, a 3′ end-labeled probe derived from the PAC vector containing the C2 pause site (3) was used to compare the amount of RNA cleaved at the first and second poly(A) sites (see Fig. 6). The protected fragments were separated on 6% acrylamide-7 M urea gels which were dried and quantitated by PhosphorImager analysis. Multiple RNA samples were analyzed at least twice to obtain the pA/splice expression ratios reported.
FIG. 6.
RNA polymerase loading is increased just downstream from the NH fragment. (A) S1 analysis of S194 cells stably transfected with the construct shown above each lane. Cμ, the intact Ig gene; pA21, the Ig gene with a 21-nt deletion of the μs poly(A) site; μm-2, the Ig gene with a 313-bp deletion encompassing both the μs poly(A) site and the NH fragment. The position of the probe protected by the spliced (splice) and cleaved and polyadenylated (pA) RNA is shown. RNA that is not spliced in the pA21 and μm-2 constructs protects the S1 probe to the location of the deletion endpoints (mismatch). (B) Nuclear run-on analysis from intact IgM genes stably expressed in S194 plasma cells. RNA that was labeled in vitro in isolated nuclei from cells transfected with the IgM gene identified on the right was hybridized to M13 DNA containing the fragments shown above each lane and identified in panel C. (C) Diagram of the μ gene with the fragments subcloned into M13 for use as probes. Probe 1, 610-bp PstI-HindIII fragment; probe 2, 532-bp HindIII-NcoI fragment; probe 3, 456-bp HaeIII fragment; probe 4, 275-bp XbaI-MboII fragment; probe 5, 469-bp HindIII-DraI fragment; probe 6, 446-bp EcoRI-BglII fragment (note that probe 6 was not included in the experiment for which results are shown in panel B but was included in other independent experiments). The vector mp18 DNA was used as a negative control. H, HindIII site. (D) Quantitation of nuclear run-ons performed with S194 cell nuclei containing the intact Ig gene (Cμ), the Ig gene with a 21-nt deletion of the μs poly(A) site (pA21), and the Ig gene with a 313-bp deletion encompassing both the μs poly(A) site and the NH fragment (μm-2). The hybridized RNA was quantitated by PhosphorImager analysis, corrected for background hybridization and for the number of U's contained within each probe, and compared to the signal over probe 1 to obtain the relative polymerase loading for each probe. Each run-on was performed at least three times, although not all probes were used in each analysis; those bars shown without error bars are a result of a single hybridization.
Nuclear run-ons.
Nuclear run-ons were performed with S194 cells stably transfected with intact Ig genes containing either a wild-type μs poly(A) site (Cμ) or a μs poly(A) site inactivated by a 21-nt deletion (pA21) or a 313-bp HaeII-HindIII deletion (μm-2). Nuclei were isolated, and nascent transcripts were labeled for 10 min with [32P]UTP as described previously (19). Labeled RNA was isolated by using Trizol reagent (Life Technologies) according to the manufacturer's protocol. These conditions allow RNA polymerase molecules to resume transcription and incorporate labels over the next several hundred nucleotides. The labeled RNA was hybridized for 48 h with immobilized M13 subclones of the mouse Cμ gene: probe 1, 610-bp PstI-HindIII fragment; probe 2, 532-bp HindIII-NcoI fragment; probe 3, 456-bp HaeIII fragment; probe 4, 275-bp XbaI-MboII fragment; probe 5, 469-bp HindIII-DraI fragment; and probe 6, 446-bp EcoRI-BglII fragment. The hybridized filters were washed, treated with RNase A, and quantitated by PhosphorImager analysis. The relative RNA polymerase loading was calculated by first correcting each probe signal for background hybridization and the number of U's within the probe and then comparing the signal of each to that of probe 1. Since the pA21 and μm-2 deletions remove portions of probe 1, their signals were corrected for the number of U's remaining in the construct.
RESULTS
Sequences far downstream of the μs poly(A) site affect the μs/μm mRNA ratio.
Since the μs poly(A) site is central to the regulated competition between cleavage-polyadenylation and Cμ4-M1 splicing during B-cell development, the sequences in and around this site have been mutagenized (33, 34; Peterson, unpublished) to better understand the features that contribute to its regulation. The location of the GU-rich downstream sequences of the μs poly(A) site differs from many poly(A) sites. The μs poly(A) site contains two potential elements, one located close to the cleavage site and one located beginning about 40 nt downstream (Fig. 1A). We altered the far-downstream sequence by making a site-directed mutation that created a NotI restriction site (ΔU). This mutation had no effect on the pA/splice expression ratio in B cells, plasma cells, or nonlymphoid cells (Fig. 1B), suggesting that these sequences are not a part of the downstream elements of the μs poly(A) site. We used this unique NotI restriction site to determine whether there were any sequences located farther downstream that affected μs poly(A) site use by deleting 158 bp between the NotI and HindIII sites in construct ΔNH (Fig. 1A). When transiently expressed in M12 B cells, S194 plasma cells, and HepG2 liver hepatoma cells, ΔNH had a three- to eightfold-lower pA/splice mRNA expression ratio than did the wild-type or ΔU constructs (Fig. 1B). Since the effect of the NH deletion was detected in all cell types, we continued to use the HepG2 cell line to further analyze this fragment because it transfects more efficiently than the lymphoid cell lines.
Since most standard poly(A) site downstream sequence elements are found within 30 nt of the cleavage site (57) and the NH fragment extends from about 50 to 200 nt beyond the μs poly(A) site, this suggests that this fragment contains another type of element that affects the pA/splice expression ratio. When the sequence of the mouse and human genes in this region are compared, the most striking common feature is that the 5′ portion of both sequences is very pyrimidine rich; the first half of the mouse NH fragment is about 75% pyrimidine, while the second half is only about 50% pyrimidine. While the cleavage-polyadenylation enhancer identified downstream from the calcitonin poly(A) site in the calcitonin/CGRP gene also has a pyrimidine-rich sequence, its activity requires adjacent 3′ and 5′ splice site-like sequences (23, 24) and these are not found in the NH fragment. To separate the pyrimidine-rich and non-pyrimidine-rich sequences, we made a site-directed mutation in the ΔU gene to create a unique BstXI restriction site near the 3′ end of the pyrimidine-rich sequence (ΔU/B) (Fig. 2). This mutation itself had no effect on the pA/splice mRNA expression ratio (Fig. 2). We used the new restriction site to make two smaller deletions in this region, between the NotI and BstXI sites and between the BstXI and HindIII sites (ΔNB and ΔBH, respectively) (Fig. 2) and expressed these constructs in the HepG2 cell line. Surprisingly, neither of the smaller deletions had the same effect as the original NH deletion. Deleting the 3′ portion (ΔBH) had no effect on the pA/splice ratio, whereas deleting the 5′ pyrimidine-rich region (ΔNB) had only a partial effect. We also reinserted the NH fragment in the sense and antisense orientations and found that it had partial activity when in the antisense orientation (Fig. 2). Since a discrete element cannot be identified within the 158-bp NH fragment, it is unlikely that it contains a cleavage-polyadenylation enhancer similar to that in the calcitonin/CGRP gene (23-25). It is also unlikely that this region contains a G-rich auxiliary poly(A) element like the sequence near the core GU/U-rich downstream elements in the SV40 late (5) and other poly(A) sites (2) since the NH fragment contains no runs of G longer than 2 nt. Other less-definable sequences that stimulate in vitro cleavage-polyadenylation have been identified within 30-to-60-nt fragments located just downstream from the GU-rich elements (8). However, since the NH fragment is located farther from the functional upstream GU-rich element of the μs poly(A) site (34; Peterson, unpublished) than are these previously identified auxiliary elements, it is less likely that this is the activity contained within the NH fragment. The fact that the NH fragment cannot be reduced in size without losing part of its activity is more consistent with it acting as an RNA polymerase pause site, as previous studies showed that these elements lost their full activity when subdivided (1, 3).
The NH fragment affects the μs and γs poly(A) sites in a heterologous context.
To test whether the effect of deleting the NH fragment from the μs poly(A) site could occur in a heterologous context, we placed the μs poly(A) site, with or without the NH fragment, into the third intron of the MHC class I Dd gene (Fig. 3A). It has been shown previously that the αs poly(A) site, when inserted into this intron, is used in competition with intron 3 splicing and that the processing of this chimeric mRNA is regulated between B-cell and plasma cell lines and in transgenic mice (28, 42). Other poly(A) sites placed in this location have a pA/splice expression ratio that is related to the strength of the poly(A) site (unpublished data). In the Dd gene, we found that the pA/splice expression ratio of the μs site lacking the NH fragment was about fivefold lower than that of the intact μs poly(A) site (Fig. 3). This effect is similar to what was seen in the μ gene, demonstrating that the activity within the NH fragment can be detected in this heterologous context. These results also indicate that the effect of the NH deletion is due to the loss of these sequences and not to a dominant effect of the specific Cμ intronic sequences brought into the proximity of the μs poly(A) site by the deletion.
RNA polymerase pause sites have been shown to increase the use of linked heterologous poly(A) sites (for examples, see references 3 and 53). To test whether the NH fragment would act in this manner, we placed it downstream of the mouse γ2b secretory poly(A) site (γs) that had been inserted into the Dd intron 3. The γs poly(A) site fragment, which included 65 nt of sequence downstream from the cleavage site, was not used very efficiently in the context of the Dd gene (Fig. 3), consistent with previous experiments using this poly(A) site (for an example, see reference 21). However, when the NH fragment was added downstream, use of the γs poly(A) site was enhanced, resulting in a fourfold increase in the pA/splice expression ratio in γsNH(+) (Fig. 3). The NH fragment also had a partial effect in the antisense orientation in γsNH(−).
Intron size cannot explain the effect of the NH deletion.
The size of the intron that is in competition with a poly(A) site has been shown to affect the pA/splice ratio in the Cμ (16, 31, 32, 48), Cα (41), and Dd (unpublished data) genes; when the intron size was decreased, the pA/splice ratio also decreased. Thus, while previous deletions were larger (>350 nt), it was possible that some of the effect of the NH deletion was due to a change in the intron size. To address this, we inserted the NH fragment, in both orientations, into the KpnI site about 1 kb downstream of the intron of the ΔNH construct [ΔNH-KNH(+) and ΔNH-KNH(−)] (data not shown). When the intron size was restored to that of the wild type in this way, the pA/splice ratio was not similarly restored; the expression ratio was very similar to that of ΔNH (data not shown). Therefore, the change in pA/splice expression ratio induced by the NH deletion is not caused by a change in the size of the Cμ4-M1 intron but rather must be due to the loss of the specific sequences in this fragment. This experiment also indicates that the activity within the NH fragment does not affect the expression ratio when located 1 kb downstream from the μs poly(A).
Known RNA polymerase II pause sites can substitute for the NH fragment.
If the NH fragment contains an RNA polymerase II pause site, then other fragments with similar activity may compensate for the loss of the NH fragment. To test this, we inserted the pause site from the α globin gene (14) and a tetramer of the MAZ site (MAZ4) from the C2 gene (3) downstream of the μs poly(A) site, in place of the NH fragment, to produce ΔNH-α(+) and ΔNH-MAZ4(+), respectively. Both of these sites completely restored the pA/splice expression ratio to that of the wild-type gene (Fig. 4). In the opposite orientation, these fragments had partial activity. Like the NH fragment, when the α globin pause site was placed in the KpnI site farther downstream in the intron in both orientations [ΔNH-Kα(+) and ΔNH-Kα(−)], the pause site did not significantly affect the pA/splice expression ratio (Fig. 4). These results show that the NH fragment contains an activity that can be replaced by other fragments previously characterized as RNA polymerase pause sites.
The NH fragment has activity in a PAC assay for pause sites.
It has been shown that RNA polymerase II pause sites, when placed between two poly(A) sites, will increase the relative use of the upstream site; this PAC assay has been used as an independent assay for RNA polymerase pause site activity (1, 3, 4). When we placed the NH fragment in this PAC vector, downstream from the weaker α globin poly(A) site and upstream from the stronger synthetic poly(A) (SPA) site, we detected an increase in use of the weaker upstream site (Fig. 5). The magnitude of the increase was similar to the enhancement seen with the α globin pause site but not as strong as that seen with the MAZ4 pause site. These data are consistent with the idea that the NH fragment has RNA polymerase pausing activity since it behaves in this assay like other pause sites.
RNA polymerase II loading is increased downstream from the μs poly(A) site.
Nuclear run-on assays, which measure relative RNA polymerase II loading across a gene, detect transient RNA polymerase pausing as an increase in run-on signal over one region compared to the signal over other parts of the gene. Using nuclei from plasma cells stably transfected with an intact immunoglobulin μ gene (Cμ), we measured RNA polymerase loading at various positions along the transcription unit (Fig. 6B and C). Consistent with the presence of an RNA polymerase pause site downstream from the μs poly(A) site, we detected a signal that was 2.3-fold higher over the fragment downstream from the μs poly(A) site than over the body of the gene (Fig. 6D, compare solid bars for probes 1 and 2). This increased polymerase loading was followed by a gradual decline as transcription terminated further downstream. The increase in run-on signal suggests that there was a higher density of RNA polymerase molecules in this region and/or in the region directly upstream, which corresponds to the NH fragment, consistent with the idea that this region causes RNA polymerase II pausing.
To determine whether this RNA polymerase pausing was dependent on the sequences in this region rather than on the use of the μs poly(A) site itself, perhaps because factors recruited to the poly(A) site induced RNA polymerase to pause, we measured RNA polymerase loading from two Ig μ genes containing μs poly(A) site deletions. We made a 21-nt deletion (pA21) that removed the AAUAAA and surrounding AU-rich sequences (34) and we deleted the μs poly(A) site and the downstream pause region (μm-2) as a 313-bp HaeII-HindIII fragment (Fig. 1A). We analyzed the RNA expressed from these genes by S1 nuclease mapping; any RNA from these genes that remains unprocessed or is cleaved and polyadenylated at a cryptic poly(A) site will protect the S1 probe to the deletion endpoint where the sequence diverges from the probe (mismatch) (Fig. 6A). The RNA detected from pA21 and μm-2 was mostly the spliced μm mRNA; the mismatch/splice ratio for both pA21 and μm-2 was 0.07 (Fig. 6A). This is in contrast to the pA/splice ratio of 40 for Cμ; this is a >500-fold reduction in the expression ratio (Fig. 6A). Since pA21 inactivates use of the μs poly(A) site but retains the pause site, if the pause site acts independently from poly(A) site recognition, we should see an increase in polymerase loading over probe 2, as in Cμ. Since μm-2 doesn't have the pause site, loading over probe 2 should not be elevated in this gene. This is what we observed in the run-on analysis of these two genes; the polymerase loading over probe 2 was slightly diminished in pA21 compared to Cμ, but it was clearly elevated when compared to μm-2 (Fig. 6B and D). These experiments support our proposal that the NH fragment contains sequences that cause RNA polymerase pausing. But they also suggest that poly(A) site recognition may augment the pausing over probe 2 in Cμ compared to that in pA21. Consistent with previous data which showed that a functional poly(A) site is required for transcription termination (36), the decline in polymerase loading over downstream probes seen with Cμ was not detected with pA21 and μm-2. Rather, termination in pA21 and μm-2 was apparent only with probe 6, which is likely due to use of the downstream μm poly(A) site (Fig. 6D).
Sequences downstream from the αs poly(A) site can substitute for the NH fragment.
The structure and the regulated alternative processing pathways of the different immunoglobulin isotype genes are similar, and it has been proposed that they are governed by similar regulatory mechanisms (reviewed in reference 29). The mouse IgA gene has a longer Cα3-M1 intron than the μ gene (Fig. 7), but the αs poly(A) site is also weaker than the μs poly(A) site (31). Thus, the efficiencies of these two reactions seem to have remained balanced to respond to RNA processing changes during B-cell maturation (41). It was recently reported that sequences within the Cα3-M1 intron, about 650 nt downstream from the αs poly(A) site, affected the pA/splice expression ratio (11). In these experiments, intron deletions that left a 348-nt BanI-SmaI fragment in place did not affect the pA/splice ratio, but when this fragment was also deleted, the pA/splice ratio decreased dramatically. While these sequences in the Cα3-M1 intron are located farther from the poly(A) site than in the NH fragment, they seem to have a similar effect on the pA/splice ratio. To determine if the sequences in this region of the IgA gene contain RNA polymerase pause site activity, we cloned two subfragments of the IgA region in place of the NH fragment in the μ gene (Fig. 7). Interestingly, both fragments were able to restore the pA/splice expression ratio to the wild-type level when placed in the sense orientation [ΔNH-IgA228(+) and ΔNH-IgA193(+)] (Fig. 7). In the antisense orientation, the 228-bp BanI-ApaLI fragment had only partial activity while the 193-bp ApaLI-BanI fragment had substantial activity. These results suggest that the IgA gene also contains RNA polymerase pause site(s) that modulate the pA/splice expression ratio like those in the IgM gene.
DISCUSSION
We have identified and partially characterized a 158-bp fragment, located about 50 nt downstream from the μs poly(A) site, that appears to act like an RNA polymerase II pause site. Deletion of this NH fragment decreases the pA/splice mRNA ratio, most likely by decreasing the use of the μs poly(A) site that is in competition with Cμ4-M1 splicing. This fragment has a similar effect on poly(A) sites in a heterologous context; use of both the μs and γs poly(A) sites placed in the intron of the Dd gene was higher when they were followed by the NH sequences. The pA/splice ratio of ΔNH could be restored to wild-type levels by replacing the NH fragment with two different well-characterized RNA polymerase pause sites but not by reinserting the NH fragment or another pause site fragment 1 kb further downstream. The results of two independent experiments are also consistent with the conclusion that the NH fragment has RNA polymerase pause site activity. In a PAC assay, the NH fragment had activity similar to that seen with other previously characterized pause sites. By using nuclear run-on assays, increased RNA polymerase loading was detected in the region downstream from this fragment whether the μs poly(A) site was functional or not; a similar increase was not seen when both the poly(A) site and the NH fragment were deleted. While some of our data could be interpreted to suggest that the NH fragment contains an auxiliary poly(A) element that enhances use of the μs poly(A) site directly, this explanation is not consistent with its activity in the nuclear run-on assays, with its location relative to the GU-rich sequences in the μs and γs poly(A) sites, or with the fact that the α globin and C2 pause sites, which are clearly not auxiliary poly(A) elements, can functionally replace the NH fragment. Thus, we conclude that the NH fragment contains RNA polymerase pause site activity which is able to enhance the use of the upstream μs poly(A) site.
In a previous mutational analysis of the μs poly(A) site, its use was reduced when the downstream GU-rich sequence and all sequences further downstream were deleted (34). In these experiments, poly(A) site activity was measured indirectly by the amount of luciferase produced by different poly(A) site-containing constructs. Since we mutated the GU-rich element in the ΔU construct and found, in the context of the rest of the μ gene, that these sequences had no effect on the pA/splice mRNA ratio, it is likely that the reduced μs poly(A) site activity seen previously was due to loss of the pause site sequences. This would suggest that the pause site could enhance the use of the upstream poly(A) site, even in the absence of a competing splice site. In a subsequent set of experiments with the μs poly(A) site, a secondary structure encompassing the downstream GU-rich sequences and the 5′ end of the NH fragment was shown to affect the use of the μs poly(A) site (33). It is possible that this structure could contribute to the pause site activity, but the entire activity found in the fragment cannot be due to this secondary structure, since deleting the 5′ half of the NH fragment resulted in only partial loss of activity. Also, the NH fragment that was placed downstream from the γs poly(A) site did not have this secondary structure and yet it activated use of this poly(A) site. In addition, a secondary structure in this region is not required since the activity of the NH fragment can be restored by other known pause sites that are highly unlikely to form a similar structure.
How pause sites act to slow RNA polymerase II has not been fully established. However, it is known that sequences in the nascent RNA as well as sequences in the DNA can influence RNA polymerase elongation, either because they form unusual RNA or DNA structures, form a weak DNA-RNA hybrid (26), or bind proteins that can cause the RNA polymerase to pause (reviewed in reference 49). It is also not yet clear whether the pause sites that have been identified downstream from poly(A) sites act in a way similar to or different from RNA polymerase pause and/or arrest sites found near the 5′ ends of transcription units (e.g., myc and human immunodeficiency virus) (18, 20, 26, 27) or those that cause regulated blocks to intergenic elongation (e.g., histone H3.3 and apolipoprotein A-I) (22, 37, 46). Multiple factors that control the elongation rate of RNA polymerase in vivo have been identified, thus making this a highly regulatable step in gene expression (9, 35, 43, 49). The pause sites we have identified here and those that were previously characterized do not have obvious sequence homologies. As described above, the NH fragment is predominantly pyrimidine rich, whereas the MAZ4 element is purine rich; the α globin and IgA fragments are neither purine nor pyrimidine rich. Sequences from the IgA gene that had pause site activity have several copies of a CCCTG motif that is also found in the NH fragment, but similar motifs are not found in either the α globin or MAZ4 pause sites. Thus, even though we have identified several new RNA polymerase pause sites, the new sequence information does not provide new insight into how these sequences may function at the molecular level.
A novel observation from our experiments is that the pause site location determines whether it will affect the pA/splice mRNA ratio. When located near the μs poly(A) site, it does affect the expression ratio, but when it is 1 kb downstream, it does not. Very little work has been done to explore the spatial relationships between poly(A) sites and pause sites. In vitro, when the MAZ4 pause site was moved 118 nt downstream from its original location, its ability to activate the upstream poly(A) site was not affected (53). However, activity over a larger distance was not investigated. The mechanism for the distance effect we observed is not clear. It could be that recognition of the poly(A) site must occur within a limited time after its synthesis, and a pause site can influence this event only within this time frame. If so, it may be that by the time RNA polymerase reaches the pause site 1 kb downstream, the decision to use or not use the μs poly(A) site has been made already. Two previous papers have described experiments, using cis antisense sequences against either 5′ splice sites (15) or poly(A) sites (7) located at various distances, to estimate the time between when a processing site appears in the nascent transcript and when it is actually used or bound by factors that commit it to being used. While experimental details differed, both papers observed that as an inhibitory cis-acting sequence was moved farther downstream, it was less able to inhibit the use of the processing site, suggesting that there is a limited time and/or distance over which RNA processing sites can be used. In addition, it was shown that the strength of the poly(A) site affected this distance (7). Our observation that a pause site could influence an upstream poly(A) site when it is 50 bp but not 1 kb beyond the site is consistent with these observations.
Based on the location of the NH pause site, just downstream from the μs poly(A) site that is central to μ alternative RNA processing, it is tempting to postulate a role for this element in μ processing regulation. However, to date, we have no evidence that the NH pause site is differentially recognized in B cells and plasma cells, since deleting this element decreased the pA/splice RNA ratio in both cell types. Yet the magnitudes of the decrease were not identical, so it is still possible that the element could contribute to μ processing regulation. This element might also have an impact on transcriptional read-through into the Cδ exons, which are located downstream from the IgM constant region exons and are spliced into the Ig mRNA to produce IgD-encoding mRNA (6). IgD expression is down-regulated, both in early B cells and in plasma cells, because transcription terminates before reaching the Cδ exons (47, 54-56). A comparison of the transcriptional run-on profiles of B cells and plasma cells expressing the μs poly(A) site deletions shown here, as well as μ genes containing pause site mutations, may shed some light on these possibilities. Nevertheless, our data indicate that the pause site is an integral part of the overall architecture of the μ gene and is important for modulating the balance of the competing splice and cleavage-polyadenylation reactions. It is intriguing that fragments with a similar RNA polymerase pause activity are present in the intron downstream from the αs poly(A) site in the IgA gene. Thus, pause sites may modulate the balance of the competing RNA processing reactions in both of these evolutionarily related transcription units. The location of these elements in the IgA gene is further from the αs poly(A) site than the NH fragment is from the μs poly(A) site in the IgM gene. It will also be important to probe the basis for the spacing requirement between poly(A) sites and downstream pause sites to better understand how these elements interact functionally.
Acknowledgments
We thank Nick Proudfoot for the α globin and MAZ4 pause sites, the PAC plasmids for pause site analysis, and helpful discussions. We also thank Jeff Wilusz for the ΔU mutation, Chris Milcarek for the γ2b secretory poly(A) site, Cam Dingle and Clarissa Cowan for making and analyzing the stable cell lines, and Cam Dingle, Brett Spear, and Caroline Kane for helpful discussions and comments on the manuscript.
This work was supported by grants MCB-9507513 and MCB-9808637 from the National Science Foundation.
REFERENCES
- 1.Aranda, A., and N. Proudfoot. 1999. Definition of transcriptional pause elements in fission yeast. Mol. Cell. Biol. 19:1251-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin, G. K., M. Boots, P. S. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashfield, R., P. Enriquez-Harris, and N. J. Proudfoot. 1991. Transcriptional termination between the closely linked complement genes C2 and factor B: common termination factor for C2 and c-myc? EMBO J. 10:4197-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashfield, R., A. J. Patel, S. A. Bossone, H. Brown, R. D. Campbell, K. B. Marcu, and N. J. Proudfoot. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagga, P. S., L. P. Ford, F. Chen, and J. Wilusz. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 23:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., and P. W. Tucker. 1984. The molecular biology of immunoglobulin D. Nature 307:417-422. [DOI] [PubMed] [Google Scholar]
- 7.Chao, L. C., A. Jamal, S. J. Kim, L. Huang, and H. G. Martinson. 1999. Assembly of the cleavage and polyadenylation apparatus requires about 10 seconds in vivo and is faster for strong than weak poly(A) sites. Mol. Cell. Biol. 19:5588-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, F., and J. Wilusz. 1998. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 26:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 10.Connelly, S., and J. L. Manley. 1989. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell 57:561-571. [DOI] [PubMed] [Google Scholar]
- 11.Coyle, J. H., and D. A. Lebman. 2000. Correct immunoglobulin α mRNA processing depends on specific sequence in the Cα3-αM intron. J. Immunol. 164:3659-3665. [DOI] [PubMed] [Google Scholar]
- 12.Edwalds-Gilbert, G., and C. Milcarek. 1995. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 15:6420-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggermont, J., and N. J. Proudfoot. 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 12:2539-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enriquez-Harris, P., N. Levitt, D. Briggs, and N. J. Proudfoot. 1991. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 10:1833-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eperon, L. P., I. R. Graham, A. D. Griffiths, and I. C. Eperon. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54:393-401. [DOI] [PubMed] [Google Scholar]
- 16.Galli, G., J. W. Guise, M. A. McDevitt, P. W. Tucker, and J. R. Nevins. 1987. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted μ-chain expression during B-cell development. Genes Dev. 1:471-481. [DOI] [PubMed] [Google Scholar]
- 17.Grosschedl, R., and D. Baltimore. 1985. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell 41:885-897. [DOI] [PubMed] [Google Scholar]
- 18.Keene, R. G., A. Mueller, R. Landick, and L. London. 1999. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and C-myc genes. Nucleic Acids Res. 27:3173-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley, D. E., and R. P. Perry. 1986. Transcriptional and post-transcriptional control of immunoglobulin mRNA production during B lymphocyte development. Nucleic Acids Res. 14:5431-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6:2201-2213. [DOI] [PubMed] [Google Scholar]
- 21.Lassman, C. R., S. Matis, B. L. Hall, D. L. Toppmeyer, and C. Milcarek. 1992. Plasma cell-regulated polyadenylation at the Igγ2b secretion-specific poly(A) site. J. Immunol. 148:1251-1260. [PubMed] [Google Scholar]
- 22.Lin-Lee, Y. C., S. M. Soyal, A. Surguchov, S. Sanders, W. Strobl, and W. Patsch. 1995. Thyroid hormone influences conditional transcript elongation of the apolipoprotein A-I gene in rat liver. J. Lipid Res. 36:1586-1594. [PubMed] [Google Scholar]
- 23.Lou, H., G. J. Cote, S. M. Berget, and R. F. Gagel. 1995. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol. Cell. Biol. 15:7135-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lou, H., R. F. Gagel, and S. M. Berget. 1996. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 10:208-219. [DOI] [PubMed] [Google Scholar]
- 25.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palangat, M., and R. Landick. 2001. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase. J. Mol. Biol. 311:265-282. [DOI] [PubMed] [Google Scholar]
- 27.Palangat, M., T. I. Meier, R. G. Keene, and R. Landick. 1998. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell 1:1033-1042. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, M. L. 1994. Regulated immunoglobulin (Ig) RNA processing does not require specific cis-acting sequences: non-Ig genes can be alternatively processed in B cells and plasma cells. Mol. Cell. Biol. 14:7891-7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson, M. L. 1994. RNA processing and the expression of immunoglobulin genes, p. 321-342. In E. C. Snow (ed.), Handbook of B and T lymphocytes. Academic Press, San Diego, Calif.
- 30.Peterson, M. L., M. B. Bryman, M. Peiter, and C. Cowan. 1994. Exon size affects competition between splicing and cleavage-polyadenylation in the immunoglobulin μ gene. Mol. Cell. Biol. 14:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson, M. L., and R. P. Perry. 1989. The regulated production of μm and μs mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the Cμ4-to-M1 splice. Mol. Cell. Biol. 9:726-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, M. L., and R. P. Perry. 1986. Regulated production of μm and μs mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the μs-μm intron. Proc. Natl. Acad. Sci. USA 83:8883-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, C., C. B. Kyriakopoulou, and A. Virtanen. 1999. Identification of a stem-loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res. 27:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, C., and A. Virtanen. 1997. The murine secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 25:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot, N. J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14:105-110. [DOI] [PubMed] [Google Scholar]
- 37.Reines, D., M. J. Chamberlin, and C. M. Kane. 1989. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 264:10799-10809. [PubMed] [Google Scholar]
- 38.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. J. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 40.Schibler, U., K. B. Marcu, and R. P. Perry. 1978. The synthesis and processing of the messenger RNAs specifying heavy and light chain immunoglobulins in MPC-11 cells. Cell 15:1495-1509. [DOI] [PubMed] [Google Scholar]
- 41.Seipelt, R. L., and M. L. Peterson. 1995. Alternative RNA processing of IgA pre-mRNA responds like IgM to alterations in the efficiency of the competing splice and cleavage-polyadenylation reactions. Mol. Immunol. 32:277-285. [DOI] [PubMed] [Google Scholar]
- 42.Seipelt, R. L., B. T. Spear, E. C. Snow, and M. L. Peterson. 1998. A nonimmunoglobulin transgene and the endogenous immunoglobulin μ gene are coordinately regulated by alternative RNA processing during B-cell maturation. Mol. Cell. Biol. 18:1042-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilatifard, A. 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 12:1437-1446. [DOI] [PubMed] [Google Scholar]
- 44.Spear, B. T., and S. M. Tilghman. 1990. Role of α-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol. Cell. Biol. 10:5047-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, A., L. Zhang, J. Herrmann, B. Wu, L. Kedes, and D. Wells. 1997. Cell-cycle-specific transcription termination within the human histone H3.3 gene is correlated with specific protein-DNA interactions. Genet. Res. 69:101-110. [DOI] [PubMed] [Google Scholar]
- 47.Tisch, R., N. Kondo, and N. Hozumi. 1990. Parameters that govern the regulation of immunoglobulin δ heavy-chain gene expression. Mol. Cell. Biol. 10:5340-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsurushita, N., and L. J. Korn. 1987. Effects of intron length on differential processing of mouse μ heavy-chain mRNA. Mol. Cell. Biol. 7:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 50.Veraldi, K. L., G. Arhin, K. Martincic, L.-H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung, G., L. M. Choi, L. C. Chao, N. J. Park, D. Liu, A. Jamil, and H. G. Martinson. 1998. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol. 18:276-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3:593-600. [DOI] [PubMed] [Google Scholar]
- 53.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, D., and P. W. Tucker. 1984. Transcriptional regulation of μ-δ heavy chain locus in normal murine B lymphocytes. J. Exp. Med. 160:564-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan, D., and P. L. Witte. 1988. Transcriptional regulation of μ and δ gene expression in bone marrow pre-B and B lymphocytes. J. Immunol. 140:2808-2814. [PubMed] [Google Scholar]
- 56.Yuan, D., P. L. Witte, J. Tan, J. Hawley, and T. Dang. 1996. Regulation of IgM and IgD heavy chain gene expression. J. Immunol. 157:2073-2081. [PubMed] [Google Scholar]
- 57.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]