Abstract
Unlike classical nuclear receptors that require ligand for transcriptional activity, the constitutive androstane receptor (CAR) is active in the absence of ligand. To determine the molecular contacts that underlie this constitutive activity, we created a three-dimensional model of CAR and verified critical structural features by mutational analysis. We found that the same motifs that facilitate ligand-dependent activity in classical receptors also mediated constitutive activity in CAR. This raises a critical question: how are these motifs maintained in an active conformation in unliganded CAR? The model identified several novel interactions that account for this activity. First, CAR possesses a short loop between helix 11 and the transactivation domain (helix 12), as well as a short carboxy-terminal helix. Together, these features favor ligand-independent docking of the transactivation domain in a position that is characteristic of ligand-activated receptors. Second, this active conformation is further stabilized by a charge-charge interaction that anchors the carboxy-terminal activation domain to helix 4. Mutational analysis of these interactions provides direct experimental support for this model. We also show that ligand-mediated repression of constitutive activity reflects both a displacement of coactivator and a recruitment of corepressor. Our data demonstrate that CAR utilizes the same conserved structural motifs and coregulator proteins as originally defined for classical nuclear receptors. Despite these remarkable similarities, our model demonstrates how a few critical changes in CAR can dramatically reverse the transcriptional activity of this protein.
Nuclear hormone receptors are transcription factors essential for virtually all aspects of physiology, including normal differentiation, development, and homeostasis. The transcriptional activity of these receptors is modulated by small lipophilic ligands, including the classical steroid hormones, thyroid hormone, retinoids, and other lipid metabolites (33). Upon binding ligand, classical nuclear receptors undergo a conformation change that results in the recruitment or displacement of a variety of coregulator proteins (15, 55). These coregulators include coactivators (PBP/DRIP205/TRAP220/TRIP2, SRC-1/NCoA-1, GRIP1/TIF2/NCoA-2, and ACTR/pCIP/AIB1/NCoA-3) (2, 4, 19, 30, 32, 38, 47, 56, 59) and corepressors (nuclear receptor corepressor [NCoR] and silencing mediator of retinoic acid and thyroid hormone receptors [SMRT]) (5, 21, 42). Both classes of coregulators utilize α-helical motifs (receptor interaction domains [RIDs]) to make direct contacts with a hydrophobic cleft (8, 11, 37, 43) on the surface of nuclear receptors (18, 22, 47). These coregulators form complexes with other proteins that function either by remodeling chromatin or by providing a bridge between the nuclear receptor and the basal transcription machinery. These interactions allow nuclear receptor ligands to activate or repress transcription of specific target genes (17).
Nuclear receptors have a common modular structure, including a highly conserved ligand binding domain (LBD) located at the C terminus of the protein. The LBDs of several nuclear receptors have been crystallized and found to possess a common overall structure, including 12 α-helices (H1 to H12) that enclose the ligand-binding pocket (53). A coregulator interaction surface, which includes helix 3 (H3), serves as a common interface that is utilized by both coactivators and corepressors.
The ability to discriminate between coactivators and corepressors is determined by the position of H12, which contains the ligand-dependent activation domain (AF2). In the absence of ligand or in the presence of certain antagonists, H12/AF2 is docked at the C-terminal end of the hydrophobic coregulator cleft, favoring corepressor interaction with certain receptors (3). In the presence of agonists, H12/AF2 switches to a position adjacent to the coregulator cleft, creating a distinct surface for coactivators (40). Thus, H12/AF2 functions as a ligand-dependent switch that toggles between the repressed, basal, and transcriptionally activated states. Depending on the receptor and type of ligand bound, H12/AF2 has been described in a continuum of positions that span the extremes of complete repression and complete activation (3, 37, 43, 50). Through these studies, it has become clear that fairly minor changes in the position of H12/AF2 can have a large impact on the transcriptional activity of nuclear receptors.
Unlike classical nuclear receptors that are activated by ligand, the constitutive androstane receptor (CAR) possesses strong transcriptional activity in the absence of ligand. This activity is observed in a variety of mammalian cells and in Saccharomyces cerevisiae (7) and represents an important new paradigm, as several orphan receptors are constitutively active. To date, two classes of ligands have been identified for CAR. The first class is exemplified by androstanol (5α-androstan-3α-ol), which acts as an inverse agonist that reverses the constitutive activity of CAR (14). The second class is defined by 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), a potent agonist that can both activate CAR and relieve androstanol-mediated repression (45, 48).
Experiments with knockout mice have shown that CAR controls hepatic xenobiotic metabolism (52) by regulating the expression of cytochrome P450 enzymes (20, 35, 45, 54), which metabolize a large number of pharmaceutical agents and xenotoxins (1, 10). CAR is thus an attractive drug target for the regulation of drug metabolism. However, an understanding of the molecular basis for the reversed transcriptional activity of CAR is required to fully exploit this receptor in screening and other drug discovery assays. We explored this question by creating a three-dimensional model of the CAR LBD based on the crystal structure of the closely related nuclear receptor human pregnane X receptor (PXR). The model was then tested experimentally by mutating key amino acid residues that appeared to account for the constitutive activity of CAR. With this approach, we show that the overall structure of CAR is remarkably similar to that of classical nuclear receptors. However, constitutive activity is mediated by a small number of structural features that have not been observed in other receptors. Our model demonstrates how a small set of unique interactions can dramatically alter the direction of transcriptional activity among nuclear receptors.
MATERIALS AND METHODS
Plasmids.
Mammalian expression vectors were derived from pCMX, which contains the cytomegalovirus promoter-enhancer followed by a bacteriophage T7 promoter for transcription in vitro (49). All CAR-related plasmids were derived from mouse CAR (GenBank accession no. AF009327) (7). CAR ΔAF2 spans amino acids 1 to 348 and is a deletion of the CAR H12/AF2 transcriptional activation domain. CAR ΔAF2 was constructed by replacing the BspEI/NheI fragment of CAR with synthetic oligonucleotides corresponding to amino acids 329 to 348 of CAR. CMX-CAR L352A and CAR E355A were constructed by replacing the BspEI/NheI fragment of CAR with synthetic oligonucleotides corresponding to amino acids 329 to 358 of CAR, encoding an alanine at either position 352 or 355 of the H12/AF2 domain.
All retinoid X receptor (RXR) plasmids were derived from human RXRα (GenBank accession no. NM_002957). RXR M454L/L455A was obtained from Ira Schulman. RXR K285A was created by site-directed mutagenesis (see below). RXR ΔAF2 is a deletion mutant spanning amino acids 1 to 443 and lacks the C-terminal H12/AF2 transactivation domain of RXR. RXR ΔAF2/K285A was constructed by replacing the SalI/NgoMIV fragment of RXR ΔAF2 with the corresponding fragment of RXR K285A.
Gal4 fusions contained the indicated sequences fused to the C-terminal end of the yeast Gal4 DNA-binding domain (amino acids 1 to 147; GenBank accession no. X85976). Gal-SRC-1 (human SRC-1, Asp 617 to Asp 769; GenBank accession no. U59302) contains the three canonical receptor interaction domains (RID1, RID2, and RID3). Gal-SMRT (human SMRT Arg 1109 to Gly 1330; GenBank accession no. U37146) contains RID1 and RID2. Gal-SMRT RID1 contains amino acids 1318 to 1330 of human SMRT, and Gal-SMRT RID2 contains amino acids 1110 to 1123.
VP16 fusions contained the 78 amino acids of the herpesvirus VP16 transactivation domain (Ala 413 to Gly 490; GenBank accession no. X03141) fused to the N termini of different proteins as follows: VP-CAR LBD (mouse CAR LBD, Leu 78 to Ser 358; GenBank accession no. AF009327), VP-VDR LBD (human vitamin D receptor [VDR] LBD, Glu 92 to Ser 428; GenBank accession no. XM_027197), VP-TRβ LBD (human thyroid hormone receptor beta [TRβ] LBD, Glu 173 to Asp 456; GenBank accession no. NM_000461), and VP-RARα LBD (human retinoic acid receptor alpha [RARα] LBD, Glu 156 to Pro 462; GenBank accession no. NM_000964).
The RXRα LBD (human RXRα LBD, Glu 203 to Thr 462; GenBank accession no. X52773) was cloned in a CMX-based vector containing a nuclear localization signal. CMX-βgal contains the Escherichia coli β-galactosidase coding sequences derived from pCH110 (GenBank accession no. U02445). Luciferase reporter constructs (TK-Luc) contain the herpesvirus thymidine kinase promoter (−105 to +51) downstream of the following response elements: UASGx4 for the Gal4 reporter, βRE2x2 (14) for the CAR reporter, and the acyl coenzyme A oxidase PPREx3 (13) for RXR (RXREx3).
Site-directed mutagenesis.
Site-directed mutagenesis was performed with the Stratagene Quick-Change PCR method according to the manufacturer's instructions. The primers used were as follows (only the oligonucleotides representing the upper DNA strand are shown): mouse CAR K187A, 5′-ATCATCAAGTTCACCGCGGATCTGCCGCTCTTC-3′; CAR K205E, 5′-ACCAGATCTCCCTTCTCGAGGGAGCGGCTGTGGAA-3′; CAR loop, an insertion of three alanine residues between E345 and L346, 5′-CTTCAGCGCTTGGAGGAAGCGGCCGCACTGTCTGCTATGACGCCG-3′; CAR C-terminal extension, an insertion of four alanines at the end of CAR, 5′-CTCGGGGAGATTTTGCAGTGCGGCCGCGGCATGAGGCCCAGGCTTGCAT-3′; RXR K285A, 5′-CTGGTGGAGTGGGCCGCGCGAATCCCACACTTCTCA-3′; SRC-1 RID1m (amino acids 633 to 637 [LVQLL], GenBank accession no. U59302, were changed to LAAAL), 5′-ACCAGTCACAAACTAGCGGCCGCTTTGACAACAACTGCC-3′; SRC-1 RID2m (amino acids 690 to 694 [LHRLL], GenBank accession no. U59302, were changed to LHAAA), 5′-CGGCATAAAATTCTACACGCGGCCGCACAGGAGGGTAGCCCCTCA-3′; and SRC-1 RID3m (amino acids 749 to 753 [LRYLL], GenBank accession no. U59302, were changed to LRYAA), 5′-CATCAGCTCCTACGCTATGCGGCCGCTAAAGATGAGAAAGAT-3′. All constructs were confirmed by sequencing. Western blots confirmed that all mutants were expressed at the same level in transfected CV-1 cells (data not shown).
Transient-transfection assays.
CV-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% resin-charcoal-stripped fetal bovine serum (FBS), 50 U of penicillin G per ml, and 50 μg of streptomycin sulfate per ml (DMEM-FBS) at 37°C in 5% CO2. One day prior to transfection, cells were plated to 50 to 80% confluence with phenol red-free DMEM-FBS. Cells were transiently transfected by lipofection as described before (12).
Reporter constructs (300 ng/105 cells) and cytomegalovirus-driven expression vectors (25 ng/105 cells) were added as indicated along with CMX-βgal (500 ng/105 cells) as an internal control. After 2 h, the liposomes were removed and replaced with fresh medium. Cells were treated for approximately 40 h with phenol red-free DMEM-FBS containing the indicated compounds at the following concentrations: 5 μM androstanol, 10 μM TCPOBOP, 100 nM LG268, 100 nM T3, 100 nM Am580, and 100 nM 1α,25-dihydroxy vitamin D3. After exposure to ligand, the cells were harvested and assayed for luciferase and β-galactosidase activity. Each value represents the average of triplicate data points from a single experiment; every experiment was repeated three or more times with similar results. No cytotoxicity was observed with any of the compounds when used at the indicated concentrations and treatment times.
Coactivator-corepressor recruitment assay.
Glutathione S-transferase (GST)-SRC-1 (RID1 to RID3) and GST-SMRT (RID1 and RID2) were expressed in E. coli and purified on glutathione-Sepharose columns. In vitro-translated CAR and RXR (0.6 to 1.2 μl) and GST-SRC-1 or GST-SMRT (5 μg) were incubated for 30 min at room temperature with 100,000 cpm of Klenow polymerase-labeled probes in 10 mM Tris (pH 8.0)-50 mM KCl-6% glycerol-0.05% NP-40-1 mM dithiothreitol-12.5 ng of poly(dI-dC) per ml and the indicated ligands. Complexes were electrophoresed through polyacrylamide gels in 0.5% TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). An optimized 32P-labeled DR-1 probe (5′-AGCTACCAGGTCAAAGGTCACGTAGCT-3′; underlining indi-cates the DR-1) was used for RXR homodimers, and the 32P-labeled βRE2 probe was used for CAR/RXR heterodimers.
Molecular modeling.
A molecular model of mouse CAR was built with the Homology module within InsightII 98.0, a molecular modeling package (Molecular Simulations Inc., San Diego, Calif.). Dynamics and energy minimization were carried out with Discover 2.9.7 within InsightII. The consistent valence force field was used throughout (9). The hypervariable loop connecting H1 and H3 was built with the ab initio loop builder within the Homology package and refined as described below. The synthetic ligand TCPOBOP was built with the fragment library within InsightII. Dynamics and minimization identified two stable conformations. The conformer that most closely resembled the PXR ligand, SR12813, was docked in the ligand binding cavity by superimposing homologous functional groups. The conformation of the hypervariable loop and the docking of TCPOBOP were simultaneously optimized with a hundred cycles of dynamics at 300 K (5 ps each), with each cycle followed by conjugate gradient minimization to a maximum derivative of 1.0 kcal/mol.
Of the 100 resulting minimized structures, the one with the lowest energy was selected for the final model. Docking of coactivator was achieved by superimposing the model of CAR onto the corresponding segments of the perxisome proliferator-activated receptor gamma (PPARγ) subunit of the recently solved PPARγ-RXRα-SRC-1 crystal structure (16). The PPARγ-RXRα coordinates were then deleted, leaving only CAR and the bound SRC-1 coactivator peptide. Interface interactions were optimized with conjugate gradient minimization to a maximum derivative of 1.0 kcal/mol. The quality of the resulting model was tested with Procheck (28), which reported that 82.6% of residues fell within most-favored regions of the Ramachandran plot, whereas the remainder fell in additionally allowed regions.
RESULTS
CAR H12/AF2 and H3 are essential for constitutive activity.
To determine the structural features that promote CAR activity, we asked whether the same motifs used for ligand-dependent activation in classical receptors are also used for constitutive activation by CAR. To address this question, we mutated conserved amino acid residues in H12/AF2 (L352A and E355A) and H3 (K187A) that contribute to the hydrophobic coregulator cleft on the surface of ligand-bound nuclear receptors (8, 11, 37, 43). These mutants were tested for their transcriptional activity and their ability to interact with SRC-1, a nuclear receptor coactivator that enhances CAR-mediated transactivation (36). For transfections, CV-1 cells were cotransfected with CAR expression vectors and a reporter construct containing a response element from the RARβ2 promoter (βRE2). As expected, CAR strongly activated transcription in the absence of ligands. This constitutive activity was repressed by androstanol, and TCPOBOP strongly activated the androstanol-repressed receptor (Fig. 1A). In contrast, mutations in either the H12/AF2 domain (L352A and E355A) or H3 (K187A) completely inhibited constitutive activity, and TCPOBOP failed to restore activity to wild-type levels (Fig. 1A).
FIG. 1.
Mouse CAR activity requires the same structural motifs as ligand-dependent activity by other nuclear receptors. (A) CV-1 cells were cotransfected with different CAR expression plasmids, a luciferase reporter construct containing two copies of the βRE2 response element, and CMX-βgal. Cells were treated with ligands (TCPOBOP at 10 μM and androstanol at 5 μM) for 40 h before luciferase and β-galactosidase activities were measured. (B) Coactivator recruitment assay was performed by mixing wild-type (WT) or mutant CAR, RXR, a 32P-labeled βRE probe, and the bacterially expressed RIDs of SRC-1. TCPOBOP (10 μM) was added to the mix where indicated. Heterodimer formation and migration were not changed by the various receptor mutants. (C) A mammalian two-hybrid experiment was performed by transfecting CV-1 cells with expression plasmids for Gal-SRC-1, VP-CAR LBD, the RXR LBD, and a luciferase reporter containing four copies of a Gal4 response element (UASGx4). Cells were treated for 40 h with ligands before luciferase and β-galactosidase activities were measured. Gal-SRC-1 refers to a fusion between the Gal4 DNA-binding domain and the three RIDs of SRC-1 (wild-type RIDs [WT] or mutations in either of the three LXXLL motifs [RID1m, RID2m, and RID3m]). Western blot analysis indicated that all mutants were expressed to levels equivalent to those of the wild-type proteins (data not shown). (D) Same as panel C except that VP-VDR LBD, VP-TRβ LBD, or VP-RARα LBD was used and the cells were treated with the corresponding ligands, as indicated. WT, wild type. Reporter activity refers to the luciferase value divided by the β-galactosidase value for each point. Fold activation represents the reporter activity of the receptor in the presence of ligand divided by the reporter activity of the same receptor in the absence of ligand.
To test the ability of the above mutants to recruit coactivator in vitro, we used a DNA-dependent electrophoretic mobility shift assay. This approach has an advantage over other assays in that it measures coactivator recruitment to native, DNA-bound complexes. In vitro-translated CAR was mixed with RXR, its obligate heterodimeric partner, a bacterially expressed SRC-1 coactivator fragment, and a 32P-labeled DNA probe (βRE). The resulting complexes were separated on nondenaturing polyacrylamide gels. As previously demonstrated, wild-type CAR interacts with SRC-1 in the absence of ligand, and the interaction is stabilized by the agonist TCPOBOP (Fig. 1B, lanes 2 and 3). Single point mutations in H12/AF2 (L352A or E355A) or H3 (K187A) specifically resulted in the complete loss of both constitutive and ligand-induced SRC-1 recruitment (Fig. 1B, lanes 4 to 9). The inability to recruit coactivator closely mirrors the inability of these mutants to activate transcription (Fig. 1A). These data demonstrate that constitutive and TCPOBOP-dependent activations by CAR require the same receptor motifs (H12/AF2 and H3) as ligand-dependent activation by classical receptors.
The above experiments identified receptor motifs required for CAR activity. We next sought to determine whether the coactivator surfaces recognized by CAR are similar to those used by classical nuclear receptors. The coactivator SRC-1 contains three canonical RIDs. The coactivator recruitment assay (Fig. 1B) clearly demonstrated that a fragment containing these three motifs was sufficient to interact with CAR.
Previous studies have demonstrated that a single RID makes a direct contact with the coregulator interaction surface of a single nuclear receptor. Moreover, each RID can preferentially interact with specific nuclear receptors (6, 8, 31, 34). We therefore asked which SRC-1 RIDs are required for the CAR-SRC-1 interaction. To address this question, mutations were individually introduced in each of the three RIDs, creating RID1m, RID2m, and RID3m, and the mutants were tested for their ability to interact with CAR in a mammalian two-hybrid assay. CV-1 cells were transfected with expression vectors for Gal-SRC-1, the LBD of CAR fused to the activation domain of VP16 (VP-CAR LBD), the RXR LBD, and a reporter construct containing four copies of a Gal4 response element (UASGx4).
Consistent with the in vitro assay, CAR interacted with GAL-SRC-1 in the absence of ligand (Fig. 1C). This interaction was stimulated by TCPOBOP and repressed by androstanol. When a mutation was introduced into RID1 of SRC-1, the CAR-SRC interaction was only partially lost, whereas mutation in RID2 or RID3 completely inhibited or dramatically reduced binding with or without ligand (Fig. 1C). In control experiments, no interactions were seen between CAR proteins and the Gal4 fragment (Fig. 1C). Western blot analysis demonstrated that all mutants were expressed at similar levels (data not shown). Although these SRC-1 mutants were inactive on CAR, they were functional in that they displayed the expected activity on other receptors, such as the VDR, TR, and RAR (Fig. 1D) (8, 34). These data demonstrate that RID2 is the most critical RID that mediates the interaction of SRC-1 with the CAR-RXR heterodimer.
CAR requires RXR for coactivator recruitment.
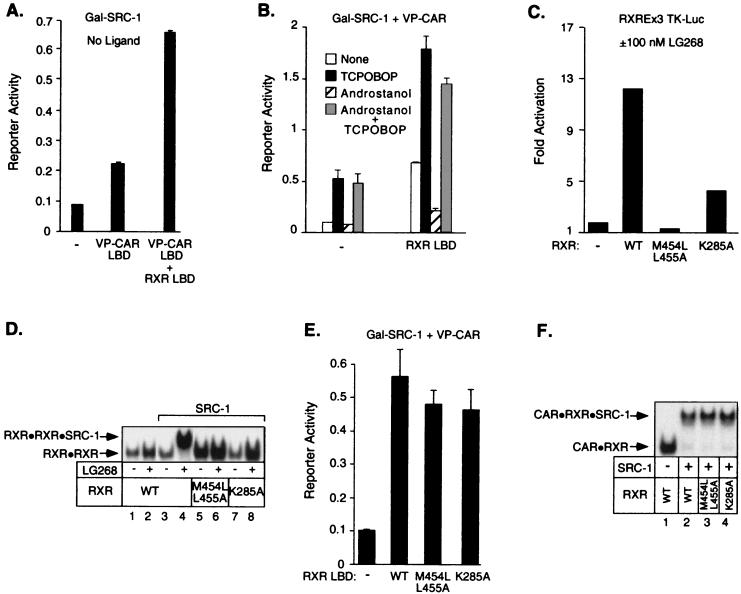
Since CAR forms a heterodimer with RXR, we wondered whether the exogenous RXR that was present in the above experiments was required for CAR activity. With the two-hybrid assay, only a weakly constitutive CAR-SRC-1 interaction was observed with endogenous RXR (Fig. 2A). Notably, this interaction was stimulated over threefold by the addition of exogenous RXR (Fig. 2A). We next tested the effect of CAR ligands on the RXR-dependent CAR-SRC-1 interaction. In the absence of added RXR, TCPOBOP recruited SRC-1 and androstanol had little effect (Fig. 2B). In contrast, exogenous RXR enhanced the constitutive CAR-SRC-1 interaction, and the ability of androstanol to displace SRC-1 became apparent (Fig. 2B).
FIG. 2.
Mouse CAR requires the RXR LBD, but not its coactivator interaction domains, for full activity. (A) A mammalian two-hybrid assay was performed as described for Fig. 1C except that no ligands were added. (B) A mammalian two-hybrid assay was performed as for Fig. 1C. (C) CV-1 cells were cotransfected with the indicated RXR expression plasmids, a luciferase reporter construct containing three copies of an RXRE, and CMX-βgal. Cells were treated for 40 h with 100 nM LG268 before luciferase and β-galactosidase activities were measured. (D) The coactivator recruitment assay was performed by mixing wild-type (WT) or mutant RXR, a 32P-labeled DR-1 probe, and the bacterially expressed RIDs of SRC-1. LG268 (100 nM) was added to the mix where indicated. Only the bound DNA complexes are shown. Heterodimer formation and migration were not changed by the various receptor mutants. (E) A mammalian two-hybrid experiment was performed by transfecting CV-1 cells with expression vectors for Gal-SRC-1, VP-CAR LBD, the indicated RXR LBD mutants, and a luciferase reporter containing four copies of a Gal4 response element (UASGx4). No ligands were added. (F) Coactivator recruitment assay was performed by mixing CAR, wild-type or mutant RXR, a 32P-labeled βRE probe, and the bacterially expressed RIDs of SRC-1. Only the bound DNA complexes are shown.
One possible explanation for these results is that the transactivation domain of RXR contributes to coactivator recruitment by CAR-RXR heterodimers. Alternatively, RXR may function allosterically to stabilize the active conformation of CAR. To distinguish between these possibilities, we studied the effects of point mutations in the RXR coactivator interaction domain. As expected (27), a double point mutation in RXR H12/AF2 (M454L/L455A) or mutation of K285A in the RXR H3 severely impaired RXR activity (Fig. 2C). Similarly, these RXR mutants were unable to recruit SRC-1 in vitro (Fig. 2D). We then asked whether the transcriptionally inactive RXR mutants could substitute for RXR in forming the ternary CAR-RXR-SRC-1 complex. With both the mammalian two-hybrid assay and the in vitro coactivator recruitment assay, we found that the RXR H12/AF2 and H3 mutants were as effective as wild-type RXR in recruiting coactivator (Fig. 2E and F). Thus, while RXR is required for full coactivator recruitment, the coactivator surface of RXR (H3 and H12/AF2) does not contribute to this interaction. These results suggest that RXR functions allosterically to stabilize the CAR-SRC-1 interaction.
Androstanol-dependent repression coincides with corepressor recruitment.
Corepressor proteins such as SMRT are thought to mediate basal repression by interacting with unliganded thyroid hormone and retinoic acid receptors. In these instances, ligand relieves basal repression by displacing corepressor from the receptor surface. Since CAR functions in a manner opposite that of these classical receptors, we wondered whether androstanol represses CAR activity by recruiting corepressor proteins. We first used mammalian two-hybrid assays to ask whether CAR can interact with the corepressor SMRT. We found that in the absence of the RXR LBD, androstanol promoted a weak but reproducible interaction between CAR and SMRT (Fig. 3A). However, the RXR LBD increased the interaction between CAR and SMRT in an androstanol-dependent manner (Fig. 3A). We then asked whether individual SMRT RIDs were sufficient for interaction with CAR-RXR heterodimers. In the mammalian two-hybrid assay, neither SMRT RID1 nor RID2 promoted an interaction with CAR (Fig. 3B). These data indicate that the androstanol-bound CAR-RXR heterodimer is the preferred target for SMRT and that two SMRT RIDs are required for this interaction. This is consistent with previous observations that two repression domains are required for corepressor-receptor interactions (25, 57).
FIG. 3.
Repression of CAR is mediated by an interaction with SMRT. (A) A mammalian two-hybrid assay was performed by transfecting CV-1 cells with expression plasmids for Gal-SMRT, VP-CAR LBD, the RXR LBD, and the UASGx4 luciferase reporter. Cells were treated for 40 h with ligands before luciferase and β-galactosidase activities were measured. Gal-SMRT refers to a fusion between the Gal4 DNA-binding domain and the two RIDs of SMRT. (B) A mammalian two-hybrid assay was performed as for panel A except that the Gal-SMRT constructs contained either the isolated RID1 or RID2. Western blot analysis indicated that all mutants were expressed to levels equivalent to those of the wild-type (WT) proteins (data not shown). (C) A mammalian two-hybrid assay was performed as for panel A with wild-type CAR or with a mutant lacking H12/AF2 (ΔAF2). (D) A coactivator recruitment assay was performed by mixing the indicated CAR and RXR constructs, a 32P-labeled βRE2 probe, and the bacterially expressed SMRT RIDs. Heterodimer formation and migration were not changed by the various receptor mutants.
Since it has been documented that the H12/AF2 domain can mask certain corepressor interactions (58), we deleted CAR AF2 and asked whether this favored the CAR-SMRT interaction. Indeed, removal of CAR AF2 enhanced the binding of SMRT in both the absence and presence of androstanol (Fig. 3C). To confirm the role of CAR AF2, we examined its effect on corepressor recruitment in vitro. CAR, RXR, a 32P-labeled βRE, and the bacterially expressed SMRT RID1 and RID2 fragment were mixed, and the resulting complexes were separated on nondenaturing polyacrylamide gels. Figure 3D shows that CAR, RXR, and SMRT form a complex when the H12/AF2 domain is deleted from both CAR and RXR (lane 2). Unlike with the mammalian two-hybrid assay, we did not detect an interaction between wild-type CAR, RXR, and SMRT (data not shown). This is likely due to the fact that the mammalian two-hybrid assay is more sensitive than the in vitro coactivator-corepressor recruitment assay. Nonetheless, these results confirm that H12/AF2 masks the corepressor interaction surfaces on the CAR-RXR heterodimer.
For classical receptors, coactivator and corepressor binding are mutually exclusive, as both types of coregulators have an overlapping hydrophobic recognition site in the vicinity of H3 (39). To determine whether CAR also utilizes a shared recognition site for both classes of coregulators, we examined the role of H3 in this interaction. We previously demonstrated that H3 is required for coactivator recruitment by both CAR (K187, Fig. 1A and B) and RXR (K285, Fig. 2C and D). We now examined the effect of these mutations on corepressor recruitment. Mutation of either K187A in CAR or K285A in RXR strongly reduced the association with SMRT (Fig. 3D, lanes 3 and 4), and the combination of both mutations completely abolished the interaction (Fig. 3D, lane 5). These data demonstrate that H3 is required for recruitment of both coactivator and corepressor. These data are also consistent with the results shown in Fig. 3A and B, which indicate that SMRT contacts both members of the CAR-RXR heterodimer. Together, these data further indicate that CAR utilizes motifs similar to those of classical receptors, though these motifs function in a reversed fashion: whereas H3 of classical receptor heterodimers bind SMRT in the absence of ligand, H3 of the CAR-RXR heterodimers binds SMRT in the presence of the inverse agonist, androstanol.
CAR structure reveals the basis for its constitutive activity.
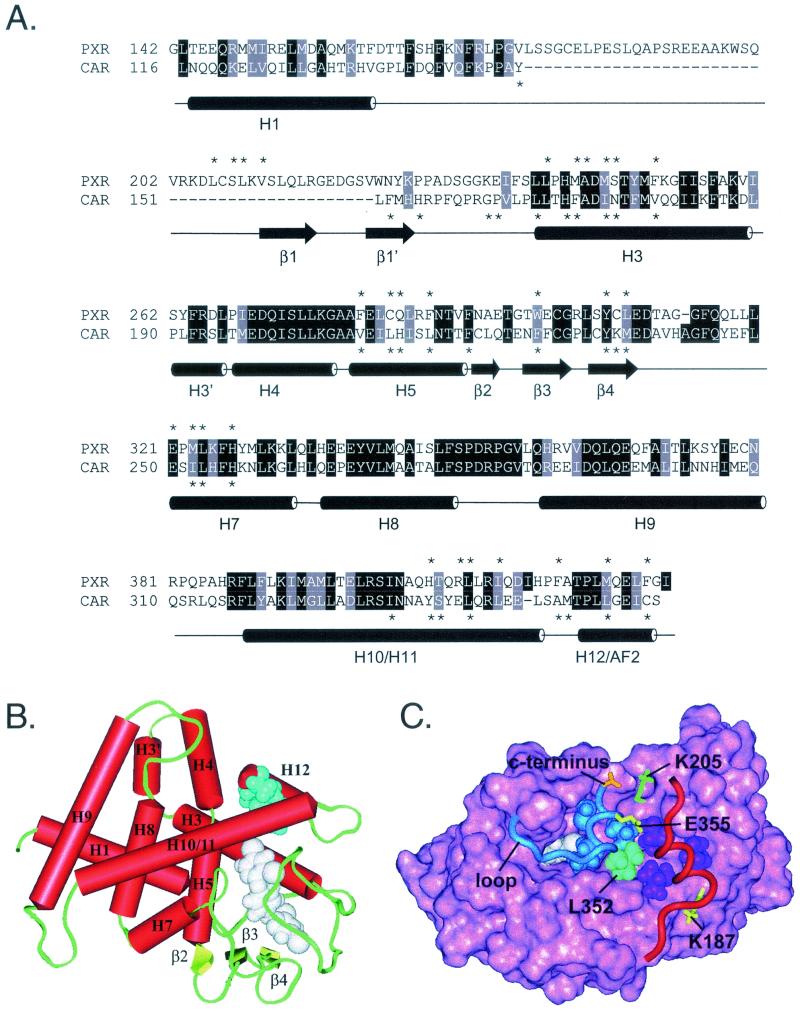
The above data demonstrate that the same motifs that facilitate transcriptional activity in classical receptors also mediate the reversed activity of CAR. This raises the fundamental question of how these motifs are maintained in an active conformation in unliganded CAR. To better understand this phenomenon, we established a homology model of the CAR LBD based on the structure of previously crystallized nuclear receptors. Initially, the CAR sequence was aligned with the sequences of other nuclear receptor LBDs of known structure (Protein Data Bank [PDB] codes 1LBD, 3ERD, 2PRG, 2LBD, 1A28, 1DB1, 1BSX, and 1ILH). The structure of human PXR (51) bound to the synthetic ligand SR12813 (PDB file 1ILH) was selected as the most appropriate structural template for three reasons: its sequence identity with CAR is the highest (35.1%), it has the potential to exhibit some constitutive activity (51), and PXR and CAR can bind to an overlapping array of ligands (35, 54), suggesting that the three-dimensional structures of their binding pockets are closely related.
A model based on the VDR was initially considered because the sequence identity between CAR and VDR (30.8%) is almost as high as that between CAR and PXR. However, the VDR-based model was abandoned for several reasons: the structure of the hypervariable loop connecting H1 and H3 in VDR was not compatible with the sequence of CAR (steric clashes and exposed hydrophobic residues were observed) and the ligand-binding cavity of the resulting model was largely filled with side chains, leaving insufficient room for TCPOBOP. As noted by others, cavity size is largely determined by the residues connecting β-strand 4 and H7 (51). VDR has a tightly packed helix in this region (H6) (41), whereas PXR has a flexible loop that projects away from the core (51), creating a much larger cavity (1,150 Å3 versus 697 Å3) (3). PXR and CAR recognize a similar array of xenobiotic ligands (35, 54), suggesting a corresponding similarity in the structures of their ligand-binding domains. Thus, unlike the VDR-based model, the structural features of the PXR-based model readily accommodate TCPOBOP binding.
The structure-based sequence alignment used for building the model is shown in Fig. 4A, and the predicted structural model is shown in Fig. 4B. Helices 10 and 11 in CAR are depicted as one continuous helix (H10/11), similar to that described for the crystal structure of PXR (51). Because of the relatively high sequence identity, the CAR structural model (Fig. 4B) closely resembles the crystal structure of PXR (root mean square deviation for aligned α carbons = 3.15 Å). Each has a relatively large internal cavity measuring ≈1,150 Å3. The 31 residues that line the cavity are indicated in Fig. 4A along with the cavity residues of PXR. As in PXR, nearly all the cavity residues are hydrophobic, but in most cases the actual amino acids differ. The size difference of these residues alters the shape of the cavity, exposing additional residues and masking others. To complete the model, the synthetic ligand TCPOBOP was docked in the ligand-binding cavity.
FIG. 4.
Homology model of the CAR ligand-binding domain. (A) Structure-based amino acid sequence alignment of human PXR and mouse CAR ligand-binding domains. Cylinders represent α-helices in PXR as defined in Protein Data Bank file 1ILH. Arrows represent β-strands. Specific α-helices (H) and β-strands (β) are identified by number. PXR lacks H2 and H6, which are found in most other LBDs. Identical residues are shown on a black background. Homologous residues are shaded gray. Asterisks indicate residues that line the internal ligand-binding cavity in the crystal structure of PXR or the model of CAR. Helices 10 and 11 are indicated as one continuous helix, as described in the PXR crystal structure (51). (B) Homology model of mouse CAR based on the crystal structure of human PXR. Specific structural elements are labeled. TCPOBOP (white) is shown in one of several possible conformations within the ligand-binding cavity, as described in Materials and Methods. Androstanol (blue) can mimic the hydrophobic side chains of H12 (L352, L353, I356, and C357). Thus, androstanol is shown superimposed on H12/AF2 and docked in the groove normally occupied by this helix when it assumes the active conformation (see Discussion). In this model, the residues that line the androstanol pocket are N175 (backbone oxygen only), T176, V179, Q180, K205, A208, V209, S337, Y338, L340, Q341, M349, and T350. (C) Top view of the mouse CAR homology model. Pink, solid rendering of mouse CAR LBD residues 116 to 344. Blue, ribbon rendering of residues 345 to 358, which constitute H12/AF2 and the loop that precedes it. L353, I356, and C357, residues that mimic the three leucines in the canonical LXXLL motif of CAR, are shown in blue space-filling representation. Cyan, L352, the residue that precedes the LXXLL motif and forms part of the hydrophobic cleft in which coactivator binds. Red, ribbon rendering of SRC-1 coactivator peptide. The three leucines of the SRC-1 RID2 LXXLL motif (690, 693, and 694) are shown in purple space-filling representation. Yellow, classical charge clamp residues. E355 neutralizes the positive dipole at the N terminus of the coactivator peptide, whereas K187 neutralizes the negative dipole at the C terminus. Orange, carboxylate group at C terminus of CAR. Green, K205, which forms a salt bridge with the C-terminal carboxylate to stabilize H12/AF2 in the active conformation. White, TCPOBOP docked in the ligand-binding cavity beneath H12. The loop between H11 and H12 is indicated and points to the junction between residues 345 and 346, the insertion site of three alanine residues.
Despite the overall structural similarities between the CAR model and the crystal structure of PXR, several significant differences are apparent. First, the extended loop between H1 and H3 in PXR is replaced by a very short loop in CAR. In PXR, this loop includes two β-strands that form one wall of the ligand-binding cavity and thus help dictate its specificity. Second, the loop connecting H11 to H12 is shorter in CAR than in other nuclear receptors. This limits the ability of H12/AF2 to assume the inactive conformation (along the C-terminal end of the coregulator cleft) and favors binding in the AF2 groove, which is characteristic of the activated receptors. Third, the three hydrophobic residues that anchor AF2 to its binding groove are smaller in CAR (A348, L353, and C357) than in PXR (F420, M425, and F429), allowing the CAR AF2 to sit deeper in its binding groove. Fourth, the C-terminal helix (H12/AF2) is shorter in CAR than in any other receptor. The shortness of H12 results in placement of the negatively charged carboxy terminus of the protein directly in the groove adjacent to K205 (Fig. 4B and C), which, acting like a charge clamp, neutralizes the free carboxy group as well as the negative dipole at the end of H12. In PXR, the C-terminal residue (I431) folds back onto the helix in order to occupy a hydrophobic patch. As a result, the carboxy terminus in PXR is 3 Å farther from the neutralizing lysine (K277), reducing its neutralizing effect.
Mutational analysis of the CAR structure.
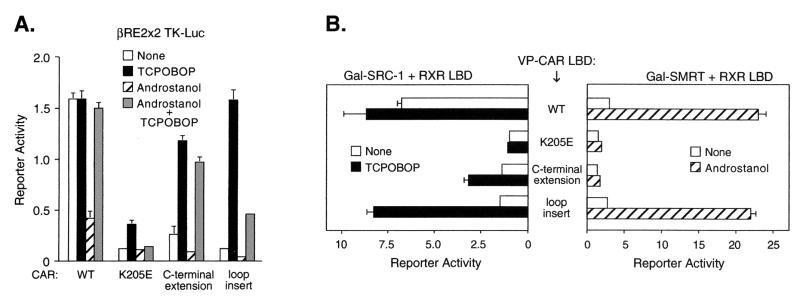
To test the importance of these structural differences, we introduced mutations in CAR that would be predicted to disrupt these critical interactions (Fig. 4C). First, we created a K205E mutation that replaces a positively charged lysine with a negatively charged glutamate residue, which is incapable of neutralizing the C terminus of H12. This should abolish constitutive activity by destroying the ability of this residue to anchor H12/AF2 in the active conformation. Indeed, the K205E mutant was not constitutively active in transfection experiments (Fig. 5A). This loss of activity did not result from a global defect in CAR activity because the K205E mutant retained the ability to bind DNA as an RXR heterodimer (data not shown). The impaired constitutive activity of this mutant was reflected by its inability to recruit the SRC-1 coactivator in the mammalian two-hybrid assay (Fig. 5B). Thus, these findings support the structural model which demonstrates that interaction of K205 with the C terminus of H12/AF2 is critical for maintaining CAR in a constitutively active conformation.
FIG. 5.
Structural features required for constitutive activity of CAR. (A) CV-1 cells were cotransfected with different CAR expression plasmids, a luciferase reporter construct containing two copies of βRE2, and CMX-βgal. Cells were treated with ligands for 40 h before luciferase and β-galactosidase activities were measured. (B) Mammalian two-hybrid assay was performed by transfecting CV-1 cells with different expression plasmids for VP-CAR LBD, RXR LBD, Gal-SRC-1 (left panel), or Gal-SMRT (right panel) and the luciferase reporter UASGx4. Cells were treated for 40 h with ligands before luciferase and β-galactosidase activities were measured.
In addition to losing constitutive activity, the K205E mutant exhibited only a weak response to TCPOBOP, and TCPOBOP failed to activate androstanol-repressed receptor, unlike in wild-type CAR (Fig. 5A). Moreover, the K205E mutant failed to recruit coactivator in response to TCPOBOP (Fig. 5B). These findings suggest that K205 may also contribute to activation of CAR by agonists such as TCPOBOP.
Next, we examined the role of the C-terminal helix, which is shorter in CAR than in classical receptors. The model indicates that the shortened C-terminal helix allows K205 to neutralize both the charged carboxy terminus and the negative dipole at the end of H12/AF2. To test this prediction, we added four alanine residues to the C-terminal end of CAR. Our model predicts that by extending the C terminus one helical turn, the C terminus would no longer be optimally aligned with K205, preventing the anchoring of H12/AF2 in the active conformation. Consistent with the model, the C-terminally extended receptor lost all constitutive activity (Fig. 5A) and failed to recruit the coactivator in the absence of ligand (Fig. 5B). Note that this effect was not due to a generalized defect, as this mutant retained the ability to respond to TCPOBOP (Fig. 5A) and to recruit coactivator in a TCPOBOP-dependent fashion (Fig. 5B). These findings further support our structural model. Moreover, the ability to selectively abolish constitutive activity in this mutant but not others (K205, L352A, and E355A) (Fig. 1A and B; Fig. 5A and B) implies that constitutive and agonist-inducible activity have both distinct and overlapping determinants.
Finally, we examined the role of the loop joining H11 and H12, which is shorter in CAR than in classical receptors. The structural model predicts that this shortened loop favors the folding of CAR H12/AF2 into the active conformation. A longer loop would extend the range of H12 beyond its active position, allowing it to reach the coregulator cleft and compete with SRC-1 for binding. We inserted three alanines in the H11-H12 loop, and as predicted, this mutation resulted in the loss of both constitutive activity (Fig. 5A) and ligand-independent coactivator recruitment (Fig. 5B). Again, we note that this effect was selective for constitutive activation, i.e., TCPOBOP-dependent activities were maintained (Fig. 5A and B). Taken together, these results validate our structural model and define unique structural features that are responsible for the constitutive activity of CAR.
Interestingly, the C-terminal extension mutant failed to interact with the corepressor SMRT in the presence of the inverse agonist (androstanol) (Fig. 5B). In contrast, the extended loop mutation did not alter the ability of CAR to interact with corepressor in the presence of androstanol (Fig. 5B). These findings are consistent with previous studies demonstrating that H12/AF2 masks the surface of interaction between corepressor and classical nuclear receptors (58). Overall, these findings further confirm the general similarities between CAR and classical receptors and demonstrate how a few critical changes can dramatically reverse the transcriptional activity of a protein.
DISCUSSION
CAR is different from classical nuclear receptors in that it has very strong constitutive activity that can be repressed by certain ligands. Similar constitutive activity is observed in several orphan nuclear receptors, indicating that this mode of regulation represents a new paradigm in hormone-regulated transcription. This stimulated us to determine the structural features that underlie the constitutive activity of CAR. It was unclear from the outset whether this activity would arise from a dramatic departure of CAR from the canonical nuclear receptor structure or whether more subtle changes could account for the reversed direction of CAR activity.
For classical nuclear receptors, it is well established that ligands function by promoting an exchange among coregulator proteins (reviewed in reference 17). For example, in the absence of ligands, the thyroid hormone and retinoic acid receptors repress basal transcription by interacting with corepressor proteins. These interactions are mediated via direct interactions between a single α-helical motif (RID, I/LXXI/VI) (22, 23) on the corepressor and a complementary hydrophobic coregulator cleft located in the vicinity of receptor helix 3. This receptor-corepressor interaction is partially masked by H12/AF2 in unliganded receptors (58). Upon binding ligand, the H12/AF2 helix, which contains a core LXXLL motif, is reoriented so that it competes for and displaces the corepressor RID. This reorientation simultaneously places H12/AF2 into an active conformation that can recruit coactivator proteins.
Nuclear receptor coactivators contain several α-helical RIDs that are also characterized by a consensus LXXLL core motif (18, 47). The ligand-activated conformation is characterized by a charge clamp that anchors a single coactivator RID in the coregulator cleft between a conserved glutamate in H12/AF2 and a conserved lysine in H3 (8, 11, 37, 43). Since binding of coactivator and corepressor to the coregulator cleft is mutually exclusive (39), the ligand acts as a switch that toggles H12/AF2 between the inactive (repressed) and active conformations.
An examination of the primary amino acid sequence of CAR (Fig. 4A) indicates that it retains the general characteristics of classical receptors. In particular, there is a high degree of conservation in critical motifs such as the H12/AF2 transactivation domain and the coregulator cleft. Although the activity of CAR is reversed compared to classical receptors, our mutation analysis is remarkable in that it demonstrates that motifs that are operative in classical receptors are also utilized by CAR (Fig. 1A and B). For example, the charge clamp residues that are required for ligand-activated transcription in classical receptors are essential for constitutive activity in CAR (K187 in H3 and E355 in H12/AF2). Furthermore, several residues immediately upstream of the H12/AF2 LXXLL motif are known to modulate transcriptional activity in classical receptors (8, 34). In CAR, the LXXLL core is defined by LGEIC, and there is an additional leucine (Fig. 4A, residue 352) immediately upstream of this motif. The equivalent residue in TRβ is essential for ligand activation and coregulator interaction (46). Similarly, we found that L352 in CAR is essential for constitutive activity (Fig. 1A) and coactivator recruitment. These findings indicate that CAR utilizes the same motifs for constitutive activity that were defined previously for ligand-dependent activation by classical receptors
We also examined the coactivator motifs that are required for constitutive recruitment of the coactivator SRC-1 (Fig. 1C and D). Of the three canonical RIDs identified previously, we found that RID2 is absolutely essential for activity. Similar results have been reported for classical nuclear receptors. Indeed, in control experiments we showed that the same RIDs are also critical for ligand-dependent interactions with the vitamin D, thyroid hormone, and retinoic acid receptors. Thus, not only does CAR utilize the same receptor motifs for transactivation, it also utilizes the same complementary coactivator interfaces as defined for classical receptors. Taken together, these findings suggest that the overall three-dimensional structure of CAR is similar to that of other members of the nuclear receptor family.
Although many structural features of CAR appear to be conserved, its constitutive activity represents a dramatic departure from the ligand-activated paradigm that has been established for classical receptors. This raises an important question: what structural features allow CAR to achieve ligand-independent transcriptional activation? While classical receptors utilize ligand to stabilize H12/AF2 in the transcriptionally active state, our data indicate that CAR must possess inherent structural features that mimic the function of ligand in nuclear receptors. We created a three-dimensional model of the CAR LBD to identify these critical features.
Examination of the predicted CAR structure immediately suggests several novel interactions that can account for its constitutive activity. CAR possesses a short loop between H11 and the H12/AF2 transactivation domain as well as a short carboxy-terminal helix. These structural features favor ligand-independent docking of the transactivation domain into a groove where it assumes a conformation that is characteristic of ligand-activated receptors. This active conformation is stabilized by a charge-charge interaction that anchors the carboxy-terminal activation domain in this active position via direct contacts with H4. Mutational analysis of these interactions provides direct experimental support for this model (Fig. 5).
These data indicate that the constitutive activity of CAR is mediated by a small number of novel interactions that are dramatic in that they result in constitutive activation of CAR. These intramolecular interactions result in an activated structure that is reminiscent of that achieved by classical ligand-receptor interactions. Further confirmation of this structure will await a detailed crystallographic or nuclear magnetic resonance analysis. Nonetheless, our studies demonstrate that the conserved three-dimensional scaffold of the nuclear receptor superfamily is remarkably efficient in that a few minor changes allow the evolution of both constitutive and ligand-activated receptors.
In addition to being constitutively active, CAR also departs from classical receptors in that one of its ligands, androstanol, acts as an inverse agonist that represses constitutive activity. We show that this inverse agonism reflects androstanol-dependent corepressor recruitment as well as coactivator displacement (14). These molecular events further highlight the reversed transcriptional properties of CAR, as ligand for classical receptors performs the exact opposite function, i.e., corepressor displacement and coactivator recruitment. It is interesting that corepressors were initially identified for their ability to repress basal transcription in unliganded or antagonist-occupied receptors (26). Subsequent studies demonstrated that certain estrogen receptor antagonists could recruit corepressors without significantly lowering basal transcriptional activity (24, 29, 44). Since coactivator binding and corepressor binding are mutually exclusive (39), these findings suggest that corepressors can act to maintain basal activity by preventing receptor-coactivator assembly.
Our findings suggest a third function for corepressors, i.e., to assist in the repression of constitutive activity in response to inverse agonists such as androstanol. This finding has important therapeutic implications, as CAR can activate the expression of a number of genes that promote the metabolism and clearance of large numbers of pharmaceutical agents (20, 35, 45, 54). When these pathways pose a therapeutic obstacle, it may be advantageous to repress the activity of these genes. Thus, the identification of drugs that strongly recruit corepressor to CAR might provide a means to specifically lower the expression of these genes below basal levels. In principle, this could be accomplished by screening for compounds that promote CAR-SMRT interactions. Indeed, similar synthetic ligands have already been identified for the retinoic acid receptor (26).
Although we demonstrate that androstanol can displace coactivator and recruit corepressor, the precise conformation changes that mediate this exchange remain to be determined. It is possible that androstanol occupies the TCPOBOP-binding cavity and promotes a switch in the location of H12/AF2 that would favor corepressor binding. This would represent a conformation change that is opposite that described for ligand-activated receptors. Interestingly, our model suggests that an alternative androstanol binding site may exist which is distinct from the traditional binding cavity that has been identified in classical receptors. We note that androstanol can mimic the hydrophobic side chains of H12/AF2, suggesting that androstanol may bind to the same groove that is occupied by H12/AF2 when it assumes the active conformation (Fig. 4B and its legend). According to this model, androstanol competes for and displaces H12/AF2 from its active site, suggesting how androstanol inhibits constitutive activity. Future structural studies will be required to verify this hypothesis.
Acknowledgments
We thank the Gonda family for support of facilities at the Gonda Diabetes & Genetic Research Center. We also thank Ira Schulman for RXR transactivation domain mutants.
The City of Hope Molecular Modeling Facility is supported by NCI grant CA33572.
REFERENCES
- 1.Anzenbacher, P., and E. Anzenbacherova. 2001. Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 58:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 3.Brzozowski, A. M., A. C. Pike, Z. Dauter, R. E. Hubbard, T. Bonn, O. Engstrom, L. Ohman, G. L. Greene, J. A. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753-758. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. D., and R. M. Evans. 1995. A transcriptional corepressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., B. A. Johnson, Y. Li, S. Aster, B. McKeever, R. Mosley, D. E. Moller, and G. Zhou. 2000. Both coactivator LXXLL motif-dependent and -independent interactions are required for peroxisome proliferator-activated receptor gamma (PPARγ) function. J. Biol. Chem. 275:3733-3736. [DOI] [PubMed] [Google Scholar]
- 7.Choi, H. S., M. Chung, I. Tzameli, D. Simha, Y. K. Lee, W. Seol, and D. D. Moore. 1997. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 272:23565-23571. [DOI] [PubMed] [Google Scholar]
- 8.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauber-Osguthorpe, P., V. A. Roberts, D. J. Osguthorpe, J. Wolff, M. Genest, and A. T. Hagler. 1988. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins 4:31-47. [DOI] [PubMed] [Google Scholar]
- 10.Ekins, S., and S. A. Wrighton. 1999. The role of CYP2B6 in human xenobiotic metabolism. Drug Metab. Rev. 31:719-754. [DOI] [PubMed] [Google Scholar]
- 11.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747-1749. [DOI] [PubMed] [Google Scholar]
- 12.Forman, B. M., E. Goode, J. Chen, A. E. Oro, D. J. Bradley, T. Perlmann, D. J. Noonan, L. T. Burka, T. McMorris, W. W. Lamph, et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687-693. [DOI] [PubMed] [Google Scholar]
- 13.Forman, B. M., P. Tontonoz, J. Chen, R. P. Brun, B. M. Spiegelman, and R. M. Evans. 1995. 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell E83:803-812. [DOI] [PubMed] [Google Scholar]
- 14.Forman, B. M., I. Tzameli, H. S. Choi, J. Chen, D. Simha, W. Seol, R. M. Evans, and D. D. Moore. 1998. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature 395:612-615. [DOI] [PubMed] [Google Scholar]
- 15.Freedman, L. P. 1999. Increasing the complexity of coactivation in nuclear receptor signaling. Cell 97:5-8. [DOI] [PubMed] [Google Scholar]
- 16.Gampe, R. T., Jr., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson, and H. E. Xu. 2000. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 5:545-555. [DOI] [PubMed] [Google Scholar]
- 17.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 18.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 19.Hong, H., K. Kohli, M. J. Garabedian, and M. R. Stallcup. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honkakoski, P., I. Zelko, T. Sueyoshi, and M. Negishi. 1998. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 18:5652-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 22.Hu, X., and M. A. Lazar. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93-96. [DOI] [PubMed] [Google Scholar]
- 23.Hu, X., Y. Li, and M. A. Lazar. 2001. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol. 21:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, T. A., J. K. Richer, D. L. Bain, G. S. Takimoto, L. Tung, and K. B. Horwitz. 1997. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol. Endocrinol. 11:693-705. [DOI] [PubMed] [Google Scholar]
- 25.Jeannin, E., D. Robyr, and B. Desvergne. 1998. Transcriptional regulatory patterns of the myelin basic protein and malic enzyme genes by the thyroid hormone receptors alpha1 and beta1. J. Biol. Chem. 273:24239-24248. [DOI] [PubMed] [Google Scholar]
- 26.Klein, E. S., J. W. Wang, B. Khalifa, S. A. Gavigan, and R. A. Chandraratna. 2000. Recruitment of nuclear receptor corepressor and coactivator to the retinoic acid receptor by retinoid ligands. Influence of DNA-heterodimer interactions. J. Biol. Chem. 275:19401-19408. [DOI] [PubMed] [Google Scholar]
- 27.Lala, D. S., R. Mukherjee, I. G. Schulman, S. S. Koch, L. J. Dardashti, A. M. Nadzan, G. E. Croston, R. M. Evans, and R. A. Heyman. 1996. Activation of specific RXR heterodimers by an antagonist of RXR homodimers. Nature 383:450-453. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 29.Lavinsky, R. M., K. Jepsen, T. Heinzel, J. Torchia, T. M. Mullen, R. Schiff, A. L. Del-Rio, M. Ricote, S. Ngo, J. Gemsch, S. G. Hilsenbeck, C. K. Osborne, C. K. Glass, M. G. Rosenfeld, and D. W. Rose. 1998. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. USA 95:2920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, J. W., F. Ryan, J. C. Swaffield, S. A. Johnston, and D. D. Moore. 1995. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374:91-94. [DOI] [PubMed] [Google Scholar]
- 31.Leers, J., E. Treuter, and J. A. Gustafsson. 1998. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol. Cell. Biol. 18:6001-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, L. B., D. J. Parks, S. A. Jones, R. K. Bledsoe, T. G. Consler, J. B. Stimmel, B. Goodwin, C. Liddle, S. G. Blanchard, T. M. Willson, J. L. Collins, and S. A. Kliewer. 2000. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275:15122-15127. [DOI] [PubMed] [Google Scholar]
- 36.Muangmoonchai, R., D. Smirlis, S. C. Wong, M. Edwards, I. R. Phillips, and E. A. Shephard. 2001. Xenobiotic induction of cytochrome P450 2B1 (CYP2B1) is mediated by the orphan nuclear receptor constitutive androstane receptor (CAR) and requires steroid coactivator 1 (SRC-1) and the transcription factor Sp1. Biochem. J. 355:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and coactivator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 38.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 39.Perissi, V., L. M. Staszewski, E. M. McInerney, R. Kurokawa, A. Krones, D. W. Rose, M. H. Lambert, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1999. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13:3198-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renaud, J. P., N. Rochel, M. Ruff, V. Vivat, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 378:681-689. [DOI] [PubMed] [Google Scholar]
- 41.Rochel, N., J. M. Wurtz, A. Mitschler, B. Klaholz, and D. Moras. 2000. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell 5:173-179. [DOI] [PubMed] [Google Scholar]
- 42.Seol, W., H. S. Choi, and D. D. Moore. 1995. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol. Endocrinol. 9:72-85. [DOI] [PubMed] [Google Scholar]
- 43.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 44.Smith, C. L., Z. Nawaz, and B. W. O'Malley. 1997. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol. Endocrinol. 11:657-666. [DOI] [PubMed] [Google Scholar]
- 45.Sueyoshi, T., T. Kawamoto, I. Zelko, P. Honkakoski, and M. Negishi. 1999. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. 274:6043-6046. [DOI] [PubMed] [Google Scholar]
- 46.Tagami, T., W. X. Gu, P. T. Peairs, B. L. West, and J. L. Jameson. 1998. A novel natural mutation in the thyroid hormone receptor defines a dual functional domain that exchanges nuclear receptor corepressors and coactivators. Mol. Endocrinol. 12:1888-1902. [DOI] [PubMed] [Google Scholar]
- 47.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional coactivator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 48.Tzameli, I., P. Pissios, E. G. Schuetz, and D. D. Moore. 2000. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell. Biol. 20:2951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umesono, K., K. K. Murakami, C. C. Thompson, and R. M. Evans. 1991. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uppenberg, J., C. Svensson, M. Jaki, G. Bertilsson, L. Jendeberg, and A. Berkenstam. 1998. Crystal structure of the ligand binding domain of the human nuclear receptor PPARγ. J. Biol. Chem. 273:31108-31112. [DOI] [PubMed] [Google Scholar]
- 51.Watkins, R. E., G. B. Wisely, L. B. Moore, J. L. Collins, M. H. Lambert, S. P. Williams, T. M. Willson, S. A. Kliewer, and M. R. Redinbo. 2001. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292:2329-2333. [DOI] [PubMed] [Google Scholar]
- 52.Wei, P., J. Zhang, M. Egan-Hafley, S. Liang, and D. D. Moore. 2000. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407:920-923. [DOI] [PubMed] [Google Scholar]
- 53.Wurtz, J. M., W. Bourguet, J. P. Renaud, V. Vivat, P. Chambon, D. Moras, and H. Gronemeyer. 1996. A canonical structure for the ligand-binding domain of nuclear receptors. Nat. Struct. Biol. 3:87-94. [DOI] [PubMed] [Google Scholar]
- 54.Xie, W., J. L. Barwick, C. M. Simon, A. M. Pierce, S. Safe, B. Blumberg, P. S. Guzelian, and R. M. Evans. 2000. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 14:3014-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamir, I., J. Dawson, R. M. Lavinsky, C. K. Glass, M. G. Rosenfeld, and M. A. Lazar. 1997. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc. Natl. Acad. Sci. USA 94:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, J., X. Hu, and M. A. Lazar. 1999. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol. Cell. Biol. 19:6448-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]