Abstract
[PSI+] strains of the yeast Saccharomyces cerevisiae replicate and transmit the prion form of the Sup35p protein but can be permanently cured of this property when grown in millimolar concentrations of guanidine hydrochloride (GdnHCl). GdnHCl treatment leads to the inhibition of the replication of the [PSI+] seeds necessary for continued [PSI+] propagation. Here we demonstrate that the rate of incorporation of newly synthesized Sup35p into the high-molecular-weight aggregates, diagnostic of [PSI+] strains, is proportional to the number of seeds in the cell, with seed number declining (and the levels of soluble Sup35p increasing) in the presence of GdnHCl. GdnHCl does not cause breakdown of preexisting Sup35p aggregates in [PSI+] cells. Transfer of GdnHCl-treated cells to GdnHCl-free medium reverses GdnHCl inhibition of [PSI+] seed replication and allows new prion seeds to be generated exponentially in the absence of ongoing protein synthesis. Following such release the [PSI+] seed numbers double every 20 to 22 min. Recent evidence (P. C. Ferreira, F. Ness, S. R. Edwards, B. S. Cox, and M. F. Tuite, Mol. Microbiol. 40:1357-1369, 2001; G. Jung and D. C. Masison, Curr. Microbiol. 43:7-10, 2001), together with data presented here, suggests that curing yeast prions by GdnHCl is a consequence of GdnHCl inhibition of the activity of molecular chaperone Hsp104, which in turn is essential for [PSI+] propagation. The kinetics of elimination of [PSI+] by coexpression of a dominant, ATPase-negative allele of HSP104 were similar to those observed for GdnHCl-induced elimination. Based on these and other data, we propose a two-cycle model for “prionization” of Sup35p in [PSI+] cells: cycle A is the GdnHCl-sensitive (Hsp104-dependent) replication of the prion seeds, while cycle B is a GdnHCl-insensitive (Hsp104-independent) process that converts these seeds to pelletable aggregates.
[PSI+] is a non-Mendelian epigenetic element originally identified in a genetic screen by its ability to increase the efficiency of tRNA-mediated nonsense suppression in Saccharomyces cerevisiae (6, 7). Based on genetic arguments, Wickner (47) was the first to suggest that the [PSI+] determinant was a self-replicating prion form of translation termination factor Sup35p (also called eRF3 [43, 50]). In accordance with Prusiner's original prion hypothesis (33), Sup35p is able to exist in one of two different states: the normal soluble form, required for translation termination, and an “infectious” (i.e., transmissible) aggregated form, i.e., as a prion. In [PSI+] cells, the altered prion form of Sup35p directs newly synthesized Sup35p molecules to adopt the same conformation. This autocatalytic conversion generates new [PSI+] prion “seeds,” which are stably transmitted from mother to daughter cell, where the self-propagation of the altered protein state continues.
Direct evidence for a link between Sup35p and the [PSI+] determinant came from the demonstration that [PSI+] is inducible de novo by overexpression of the SUP35 gene in a non-prion-containing (i.e., [psi−]) cell and requires continued synthesis of Sup35p for its maintenance (3, 4, 9, 12, 44). Subsequently, a series of elegant experiments, both in vivo and in vitro (see references 37 and 38 for reviews), confirmed the prion-like properties associated with Sup35p in [PSI+] strains. For example, in [PSI+] cells, Sup35p is found largely in a proteinase K-resistant form while, in prion-free [psi−] cells, Sup35p remains soluble and sensitive to proteinase K digestion (30, 31). The aggregation of Sup35p can therefore explain the [PSI+] phenotype; in [PSI+] cells, Sup35p is largely inactivated by its sequestration into high-molecular-weight complexes, thereby precluding it from efficiently performing its role as a translation termination factor.
Studies on the conversion of mammalian prion protein PrP to transmissible prion PrPSc form have generated a number of different models to explain how a prion protein determinant can be propagated and transmitted (see reference 2 for a review). A detailed in vitro analysis of yeast Sup35p self-directed aggregation led Serio et al. (39) to suggest that a process of nucleated conformational conversion of Sup35p most probably occurs in the establishment of the [PSI+] determinant. This mechanism involves both the nucleated-polymerization and the template assembly processes proposed for PrP (2), and Serio et al. (39) proposed that conformational conversion of the native Sup35p occurs by the seeding activity of a nucleated complex. These nuclei (or intermediate oligomers) would be formed by the conformational modification of the partially unfolded Sup35p protein. Once the intermediate polymers reach a critical concentration, assembly of large polymers can occur through a templating mechanism.
Physicochemical analyses of Sup35p have shown that the N-terminal portion of the protein (the so-called N domain or prion-forming domain) contains the minimal region responsible for [PSI+] propagation in vivo and directs amyloid fiber formation in vitro (16, 23, 32). The Sup35p amyloid fibers have features in common with the amyloid proteins implicated in several human diseases (for reviews, see references 1, 11, 34, and 48). However, it remains to be established whether the high-molecular-weight aggregates seen in vivo are amyloid fibers of Sup35p and whether or not they are the seeds necessary for the propagation of the [PSI+] determinant.
Studies on the maintenance and transmission of [PSI+] and other yeast prion determinants have shown an absolute requirement for molecular chaperones Hsp70 and Hsp104 (5, 10, 21, 27, 28, 41). However, in vitro Sup35p amyloid formation does not require Hsp104 (16, 23, 32). Two hypotheses have been proposed to explain the role of Hsp104 in yeast prion maintenance. In the first model Hsp104 is required for the native protein to reach an intermediate conformational state necessary either for efficient conformational conversion (5) or for forming oligomeric complexes (30, 39). In the second model, Hsp104 is not implicated in the conversion mechanism but rather acts as a “disaggregase,” breaking down the Sup35p prion aggregate into smaller polymeric complexes in order to facilitate prion partitioning and segregation to the nascent daughter cells during cell division (24, 31).
One potential route to studying the role of Hsp104 in yeast prion propagation, i.e., replication of the prion determinant and its transmission to daughter cells, is through inhibition studies using the chaotropic salt guanidine hydrochloride (GdnHCl). Growth of [PSI+] cells in the presence of 1 to 5 mM GdnHCl results in rapid elimination of [PSI+] cells from the population (14, 45). Based on the kinetics of elimination we have proposed that GdnHCl inhibits the replication of prion seeds (14). The demonstration that GdnHCl inhibits Hsp104-dependent acquisition of thermotolerance and the refolding of heat-damaged proteins suggests that GdnHCl inhibits Hsp104 activity, probably by inhibiting the ATPase function of the protein (15) since this activity is essential for [PSI+] maintenance (5). In this paper we explore both the mechanism by which GdnHCl cures cells of [PSI+] seeds and the relationship between prion seed formation and Sup35p aggregation in [PSI+] propagation. Furthermore, we argue that GdnHCl inhibition studies can be used to explore the role of Hsp104 in yeast prion propagation in vivo.
MATERIALS AND METHODS
Strains and plasmids.
The two strains of S. cerevisiae used in this study were a “strong” [PSI+] [PIN+] derivative of strain 74-D694 (5) (generously provided by Susan Liebman, University of Illinois at Chicago) and a [psi−] [pin−] derivative obtained after curing with 3 mM GdnHCl treatment. For the analysis of the phenotype conferred by the Sup35Hp version, strain MT700/9d containing plasmid pYK810 was used (29). This strain contains a sup35::kanMX disruption cassette, and viability is obtained by the presence of the SUP35 gene on a centromeric URA3 plasmid.
To generate a galactose-regulated histidine-tagged version of Sup35p, the following was done. Plasmid pUKC639 (K. M. Jones, personal communication), a multicopy LEU2 plasmid, was digested with BamHI and HindIII, and the polylinker of plasmid pET-21a (Novagen), containing a sequence coding for a C-terminal hexahistidine tag, was inserted between GAL1 promoter and HSP26 terminator sequences, generating pUKC1800. The coding sequence of SUP35 was then amplified by PCR using the following primer pair: primer 1 (5′ CGAGTTGCATATGTCGGATTCAAACCAAG 3′) and primer 4 (5′ GAGGCTCTCGAGCTCGGCAATTTTAACAATTTTAC 3′). This reaction generates flanking NdeI and XhoI sites, respectively. Following digestion the PCR fragment was cloned into pUKC1800, similarly digested with NdeI and XhoI, to generate plasmid pUKC1809, coding for Sup35p in frame with a hexahistidine tail at the C terminus of the molecule. This construct has two additional amino acids (Leu and Glu) at the end of the Sup35p polypeptide before the hexahistidine tag.
To compare the phenotypes generated by Sup35Hp and wild-type Sup35p, pUKC1512SUP (29), containing the SUP35 gene on a centromeric HIS3 plasmid, was digested with EcoRV and NotI. This removed the Sup35pMC-encoding domain, which was replaced by an EcoRV-NotI fragment from pUKC1809 containing the coding sequence for the equivalent MC domain fused to the hexahistidine-encoding tail. This generated plasmid pUKC1602.
Plasmid pUKC815 is a single-copy URA3 vector containing the Escherichia coli lacZ gene under the control of the constitutive PGK1 promoter (42). The related plasmid used to monitor the level of termination codon readthrough (i.e., pUKC819; see below) contains an in-frame premature UGA termination codon (42).
Culture conditions.
Standard yeast growth media and growth temperature (30°C) were used (40). The cells were grown on a glucose-based synthetic medium (YNBD) for plasmid maintenance (40). GdnHCl treatment was performed on YPD medium (10 g of yeast extract, 15 g of Bacto Peptone, and 20 g of glucose/liter) by using a GdnHCl concentration of 3 mM. For slow growth a glucose-depleted YPD containing glucose at 2 g/liter instead of 20 g/liter was used. The GAL1 promoter was induced by incubating cells on YPGal (YPD containing 20 g of galactose/liter instead of glucose as the carbon source). To repress the GAL1 promoter, the cells were transferred back to YPD. The ade1-14 suppression phenotype in strain 74-D694 and the ade2-1 suppression phenotype in strain MT700/9d were assessed by colony color on 1/4YPD medium (13).
Coexpression of an ATPase-negative mutant (Hsp104KT).
Strain 74-D694 [PSI+] was transformed with plasmid pUKC1831, which contains the ATPase-negative mutant allele of HSP104, namely, hsp104 K218T,K620T (5), under the control of the GAL1 promoter (15). Mid-exponential-phase cultures in YNBD were used to inoculate complete synthetic medium containing 20 g of raffinose and 20 g of galactose/liter. Growth was monitored regularly by measuring the optical density of the culture at 600 nm (OD600). Cultures were maintained in the exponential phase of growth by reinoculation into fresh complete medium with raffinose and galactose when the OD reached approximately 0.4. Aliquots were removed from the culture both at the time of inoculation and during the course of the experiment and plated onto 1/4YPD agar plates to score colony color (red and white) phenotypes.
Analysis of Sup35p aggregates by subcellular fractionation.
Crude yeast extracts were prepared by lysis of yeast cells with glass beads in buffer P (10 mM phosphate buffer [pH 7.5], 250 mM NaCl, 2 mM phenylmethylsulfonyl fluoride, and one tablet of a protease inhibitor cocktail [Boehringer]) at a cell suspension of approximately 3.3 × 108 cells/ml. To test the sensitivity of Sup35p to detergents, crude extracts were also prepared by lysis of yeast cells in buffer ST (buffer P containing 2% sodium dodecyl sulfate [SDS] and 1% Triton X-100). The resulting total crude extract was fractionated by ultracentrifugation at 100,000 × g for 15 min at 4°C with a TL100 centrifuge (Beckman). The soluble fraction was recovered, and an equal volume of buffer P or buffer ST was added to the pellet fraction. An aliquot of the total crude extract was also kept. After incubation for 10 min at 95 to 100°C in 2× loading buffer, identical volumes of each fraction (in a range from 10 to 40 μl) were subjected to SDS-10% polyacrylamide gel electrophoresis (PAGE) and Western blot analysis as previously described (13). No preliminary low-speed centrifugation was used to remove cell debris in order to allow us to analyze the entire amount of Sup35p present in the cells. Because cell debris was also pelleted after the ultracentrifugation step, special care was taken to solubilize the pellet in SDS loading buffer at 95°C prior to SDS-PAGE.
Similar results were obtained when fractionation was performed with buffer P or buffer ST except that [psi−] cells showed a slightly higher level of Sup35Hp in the pellet fraction when buffer P was used (10 to 5%) than when buffer ST was used (5 to 0%). Therefore the presence of detergent did not destabilize the Sup35p aggregate or the newly synthesized Sup35Hp-containing [PSI+] aggregate at 4 or 25°C. Only when a temperature higher than 90°C was applied did the aggregate dissolve in the presence of 1.5 to 2% SDS. However, it should be noted that the presence of detergents may destabilize the interaction between soluble Sup35p and the translation machinery (or the cells debris) such that Sup35p may be pelleted during the centrifugation procedure used. We observed also that, for [psi−] cells, a high cell density (>15 OD units/ml) resulted in the partial precipitation of soluble Sup35p. For these reasons all fractionation procedures were performed at the same cellular concentration of 15 OD units/ml using buffer ST.
Western blot analysis.
After resolution of the protein samples by SDS-10% PAGE, the fractionated proteins were transferred to a nitrocellulose membrane (Sartorius) by using a semidry blotting system (Bio-Rad) under the conditions recommended by the manufacturer. Sup35p and Sup35Hp were detected by using affinity-purified polyclonal antibodies raised against full-length S. cerevisiae Sup35p and by using an anti-penta-His polyclonal antibody (Qiagen), respectively. Antirabbit or antimouse polyclonal antibodies, linked with peroxidase, were used as secondary antibodies. The antibody-antigen interaction was revealed with the ECL reagent (Amersham-Pharmacia).
Monitoring and quantifying the [PSI+] and [psi−] phenotypes.
Suppression of the ade1-14 (UGA) allele (for strain 74-D694) or the ade2-1 (UAA) allele (for strain MT700/9d) was assessed as previously described (13, 29) by determining colony phenotype by both color (red, nonsuppressed; pink, partially suppressed; white, suppressed) and the extent of growth in the absence of adenine. Conversion of [PSI+] to [psi−] during 3 mM GdnHCl treatment was assessed by determining colony color.
A quantitative method for measuring translation termination codon readthrough was performed as previously described by Stansfield et al. (42) with plasmids pUKC815 (PGK1-lacZ) and pUKC819 (PGK1-UGA-lacZ). Strains transformed with either pUKC819 or pUKC815 were grown in appropriate synthetic medium to an OD600 of 0.4 to 0.6 nm with or without GdnHCl (3 mM). Aliquots of the culture were harvested by centrifugation and put immediately on ice. Appropriate amounts of cells were resuspended in 800 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0). Cell suspensions were vortexed after the addition of 50 μl of chloroform and 20 μl of 0.1% SDS and preincubated at 30°C for 15 min. The reactions were started by the addition of 200 μl of 4-mg/ml ONPG (o-nitrophenyl-β-d-galactopyranoside). Reaction mixtures were incubated at 30°C, and reactions were terminated by the addition of 500 μl of 1 M Na2CO3. Following a brief centrifugation to pellet the cell debris, the A420 and A550 of the supernatant were measured and β-galactosidase activity was calculated as previously described (42).
Assaying the recovery of heat-inactivated luciferase.
Strain 74-D694 and a corresponding isogenic strain with an hsp104::LEU2 disruption were each transformed with plasmid pGPDluxAB(HIS), which codes for the constitutive expression of a temperature-sensitive luciferase fusion (15). Transformants were grown at 30oC in a YNBD-based selective medium and inoculated in YPD for one generation until the culture reached an OD600 of 0.3 to 0.4. The culture was then incubated at 37oC for 1 h to induce the heat shock response or alternatively kept at 30oC for the same period of time. GdnHCl (3 mM) was then added to the culture, and the culture was further incubated at 30oC for either 1 h or five generations of growth. The luciferase reactivation assay was then carried out essentially as described previously (15). Luciferase activity was measured by the addition of 5 ml of n-decylaldehyde (Sigma) to 300 ml of the cell culture, and luminescence was immediately quantified in a BioOrbit 1253 luminometer.
RESULTS
GdnHCl affects Hsp104-dependent processes within one cell generation.
Growth of [PSI+] cells in the presence of 3 mM GdnHCl leads to the appearance of [psi−] cells after a lag of some four or five generations of growth, with a further seven or eight generations being needed to generate an essentially [psi−] population (14). If GdnHCl rapidly blocks the replication of the [PSI+] seeds, the appearance of [psi−] cells would be the consequence of the dilution out of the preexisting [PSI+] seeds by cell division (14). This conclusion was based on the kinetics of appearance of [psi−] colonies on rich, glucose-based medium after removal of GdnHCl from the culture. Thus, any cell carrying at least one [PSI+] seed at the time of plating in the absence of GdnHCl would give rise to a [PSI+] colony if seed replication was sufficiently rapid to regenerate enough seeds to ensure continued transmission by cell division.
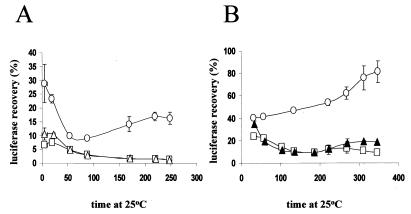
A key component of this model is an assumption that GdnHCl rapidly inhibits [PSI+] seed replication, i.e., within the first cell generation. Although our kinetic data are entirely consistent with this assumption (14), the data shown in Fig. 1 provide direct evidence that GdnHCl has a rapid effect on cellular functions important for [PSI+] propagation. We (15) and others (22) have provided in vivo evidence that GdnHCl inhibits Hsp104-dependent processes (e.g., the refolding of heat-inactivated luciferase). The data in Fig. 1 demonstrate that cells grown for only 1 h in 3 mM GdnHCl show the same level of inhibition of Hsp104-dependent luciferase reactivation as do cells grown for five generations in GdnHCl (Fig. 1A) or cells with an hsp104::LEU2 disruption (Fig. 1B).
FIG. 1.
Recovery of heat-inactivated luciferase in the presence of 3 mM GdnHCl and absence of Hsp104. (A) Strain 74-D694 [PSI+] grown at 30°C for 1 h in the presence (□) or absence (○) of GdnHCl (3 mM) or grown in the presence of 3 mM GdnHCl for five generations (▵) before assaying for luciferase reactivation as described in Materials and Methods. Luciferase activity was determined before heat treatment and at several subsequent time points and expressed as percentages of the activity before the heat treatment. (B) Cells were grown at 30oC and then shifted to 37oC for 1 h before being placed back at 30oC, when 3 mM GdnHCl was added. Luciferase reactivation was carried out as for panel A. The strains used were 74-D694 [PSI+], either untreated (○) or grown in the presence of 3 mM GdnHCl for 1 h (□) and [psi−] strain 74-D694 hsp104::LEU2 (▴).
GdnHCl induces antisuppression in [PSI+] cells.
Our model for GdnHCl-induced curing of [PSI+] predicts that, at the point at which seed-free [psi−] cells begin to appear in a GdnHCl-treated culture, the majority of cells in the population would have a significantly reduced number of seeds and a concomitantly higher proportion of soluble Sup35p (14). A higher concentration of Sup35p available for translation termination would lead to a reduction in the nonsense suppression phenotype (i.e., antisuppression [8]) in such treated cells.
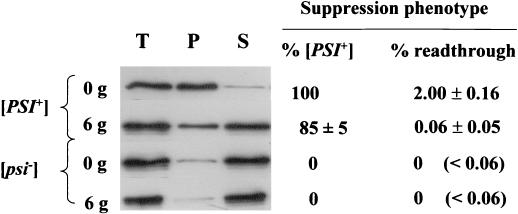
Detection of [PSI+] cells by the nonsense suppression phenotype of colonies is delayed from the event that triggers curing by the time required for single cells to form colonies on the agar surface. To explore the validity of our model, we therefore determined the physical state of and the defect in translation termination in a GdnHCl-treated [PSI+] culture. [PSI+] strain 74-D694 was grown for six generations in the presence or absence of GdnHCl (3 mM). After six generations, approximately 85% of the cells plated still contained at least one [PSI+] seed and formed pure mitotically stable [PSI+] colonies whereas most (i.e., >80%) of the Sup35p was found in the soluble fraction (Fig. 2). Using a lacZ-based reporter assay (42) we also determined the efficiency of translation termination at the UGA codon in the GdnHCl-treated cells. These data show that the nonsense suppression phenotype of the GdnHCl-treated culture was essentially the same as that for a [psi−] culture, even though approximately 85% of the cells in that culture were able to produce [PSI+] colonies when plated onto YPD.
FIG. 2.
Subcellular distribution of Sup35p in [PSI+] and [psi−] cells grown in 3 mM GdnHCl. [PSI+] and [psi−] derivatives of strain 74-D694 were grown in YNBD to stationary phase and then transferred to YPD medium. After six generations of growth (6g) in the presence of 3 mM GdnHCl a subcellular fractionation analysis of Sup35p was performed as described in Materials and Methods by using an anti-Sup35p polyclonal antibody. Samples from the 0g time point were similarly studied. [PSI+]-to-[psi−] conversion after GdnHCl treatment was analyzed by plating cells on 1/4YPD medium and visually determining the percentage of white ([PSI+]) colonies at each time point. Approximately 500 colonies were counted for each time point. The translation termination readthrough assays were performed as described in Materials and Methods. The levels of β-galactosidase in the pUKC819 transformants were expressed as percentages of the level seen in the control pUKC815 transformants. The percentages of readthrough shown (with standard deviations) were calculated from the averages of two repetitions for each of two independent transformants. T, total crude extract; S, soluble fraction; P, pellet fraction.
These data confirm that Sup35p is largely in a soluble (i.e., nonpelletable) form in GdnHCl-treated [PSI+] cells prior to the appearance of [psi−] cells. However, the low number of remaining [PSI+] seeds must rapidly replicate upon removal of the GdnHCl, presumably in less than one cell division, to ensure that a sufficient number of seeds are present prior to the next cell division to allow for efficient inheritance of the [PSI+] phenotype (14). Therefore GdnHCl treatment induces an antisuppressor phenotype in [PSI+] cells before finally eliminating the [PSI+] determinant. This finding raises the question of whether GdnHCl blocks the polymerization of newly synthesized Sup35p into aggregates or simply dissolves preexisting aggregates to generate high levels of soluble Sup35p.
Assaying the fate of newly synthesized Sup35p in GdnHCl-treated [PSI+] cells.
To monitor the fate of newly synthesized Sup35p in [PSI+] cells following exposure to GdnHCl, we constructed a Sup35p derivative carrying a C-terminal hexa-His tag. The coding sequence of the associated gene (which we designate SUP35H) was placed under the control of the galactose-inducible GAL1 promoter to generate plasmid pUKC1809. The hexa-His-tagged Sup35p (Sup35Hp) could therefore be distinguished from endogenous wild-type Sup35p by Western blot analysis using an anti-penta-His antibody.
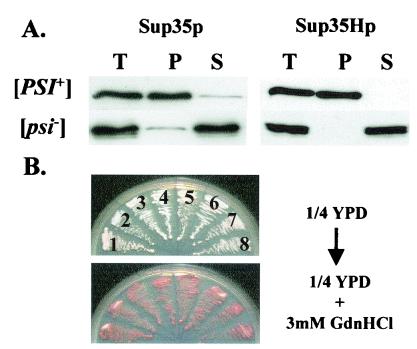
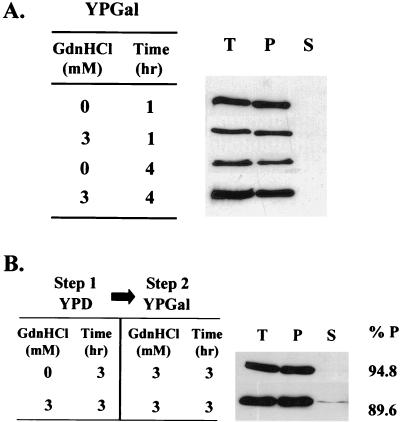
We confirmed that the hexa-His tag at the C terminus of Sup35p did not alter the prion-like behavior of this protein as follows. [PSI+] and [psi−] derivatives of strain 74-D694 were transformed with plasmid pUKC1809 by using glucose-based selective medium in order to repress transcription of the SUP35H gene. Three independent transformants of each strain were then grown to mid-exponential phase in the same medium and then transferred to a galactose-based rich medium (YPGal) to induce transcription of the SUP35H gene. Cells were removed at 30-min intervals posttransfer, and the steady-state levels and subcellular distribution of Sup35Hp were determined. After 60 min Sup35Hp was detected in both [PSI+] and [psi−] cells, with maximal steady-state levels of Sup35Hp being reached after 6 h (data not shown). The newly synthesized Sup35Hp molecules behaved identically to the endogenous Sup35p in these strains, i.e., they were aggregated in [PSI+] cells and soluble in [psi−] cells (Fig. 3A). Thus, seeded polymerization of Sup35Hp was not prevented by the C-terminal hexa-His tag, nor did it trigger nonspecific aggregation in the [psi−] cell.
FIG. 3.
The addition of a hexahistidine tag on the C terminus of Sup35p does not alter its prion-like behavior in [PSI+] cells. (A) Subcellular distribution of Sup35p and Sup35Hp in the [PSI+] and [psi−] derivatives of the 74-D694 strain transformed with plasmid pUKC1809. After growth to log phase on YPD medium (A600 = 0.5) the cultures were transferred to YPGal medium to induce Sup35Hp synthesis. Cell samples were removed every 30 min over a 6-h incubation period and analyzed as described in the legend to Fig. 1 by using either an anti-Sup35p (data not shown) or an antipentahistidine polyclonal antibody (right) to detect Sup35p or Sup35Hp, respectively. For Sup35Hp identical results were obtained at the different time points and the results shown are those obtained after 2 h of growth in YPGal. The analysis of wild-type Sup35p (left) was undertaken before transferring the cells to YPGal medium. (B) The [PSI+]-associated suppression phenotype is not altered by the presence of the C-terminal hexahistidine tag. Plasmids pUKC1512SUP (encoding wild-type Sup35p) and pUKC1602 (encoding Sup35Hp) were separately introduced into [PSI+] strain MT700/9d sup35::kanMX in place of plasmid pYK810 (encoding the wild-type Sup35p). After growth on 5-fluoroorotic acid, the [PSI+]-related suppression phenotypes of both transformants were analyzed by determining colony color on 1/4YPD (top) and after growth in the presence of 3 mM GdnHCl (bottom). Four independent clones were used for each plasmid: pUKC1512SUP, clones 1 to 4; pUKC1602, clones 5 to 8. T, S, and P are as defined for Fig. 2.
To confirm that the [PSI+] seeds containing only Sup35Hp were stably inherited, the SUP35H gene was introduced into the haploid [PSI+] strain (MT700/9d) which carries a sup35::kanMX allele, with viability being maintained by a plasmid-borne wild-type allele of SUP35 (29). The SUP35H coding sequence was placed under the control of the natural SUP35 promoter (generating single-copy, HIS3-based plasmid pUKC1602) to ensure the Sup35Hp was not overexpressed. The wild-type SUP35 gene was then replaced by the SUP35H gene by plasmid shuffling using 5-fluoroorotic acid selection. pUKC1602 transformants showed the characteristic [PSI+] phenotype, i.e., white or pink colonies indicative of adenine prototrophy (strains 5 to 8, Fig. 3B) seen with a nontagged SUP35 control (carried on plasmid pUK1512SUP; strains 1 to 4, Fig. 3B). Both the pUKC1602 and the pUKC1512SUP [PSI+] transformants were equally efficiently cured of [PSI+] by growth in the presence of 3 mM GdnHCl (Fig. 3B, bottom). Western blot analysis confirmed that both Sup35p and Sup35Hp fractionated identically between the soluble and pellet fractions in both the [PSI+] and [psi−] strains (data not shown). Therefore the addition of a hexa-His tag to the C terminus of Sup35p does not modify its prion-like behavior or the replication and transmission of [PSI+] seeds.
Growth in the presence of GdnHCl does not promote disaggregation or proteolysis of preexisting Sup35p aggregates.
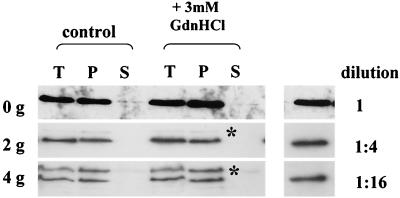
GdnHCl is a chaotropic salt, well known for its properties as a protein denaturant. One possible explanation for its [PSI+]-curing properties is that it dissociates Sup35p aggregates, thereby destroying their continued seeding potential. This would result in the apparent solubilization of Sup35p, as seen in Fig. 2. To test this possibility, we grew [PSI+] strain 74-D694 carrying plasmid pUKC1809 (GAL1-SUP35H) on YPGal medium for 4 to 5 h (approximately one generation) to generate a strain with a high level of Sup35Hp-containing aggregates. The cells were then returned to fresh YPD medium for 2 h to repress transcription of the SUP35H gene. Finally, the cells were transferred to fresh YPD medium, either with or without GdnHCl (3 mM), and cell samples were removed before growth and after two and four generations of growth and the aggregation state of Sup35Hp was determined (Fig. 4). Even after four generations of growth, no soluble Sup35Hp was evident in the GdnHCl-treated cells and the same amount of Sup35Hp as that in cells grown without GdnHCl was present. The experiment could not be continued beyond four generations because of the further dilution of the aggregates and concomitant loss of the signal.
FIG. 4.
GdnHCl does not dissociate preexisting Sup35p aggregates in a [PSI+] strain. [PSI+] strain 74-D694 was transformed with plasmid pUKC1809 (carrying SUP35H under the control of the GAL1 promoter) and transferred to YPGal for 4 to 5 h to induce synthesis of Sup35Hp. Synthesis was then stopped by transferring the cells to YPD medium for 2 h, after which time the cell cultures were split, one-half being diluted into fresh YPD medium and the other being diluted into identical medium containing GdnHCl (3 mM final concentration). The cells were then incubated further to allow them to go through four generations of growth with cell samples being taken at zero (0g), two (2g), and four generations (4g) for subcellular fractionation analysis as described in the legend to Fig. 1. The preexisting Sup35Hp was detected with antipentahistidine antibodies. To assess the relative levels of Sup35Hp in the pellet (P) fraction after two and four generations, the control (0g) P sample was also diluted either 1:4 or 1:16 prior to SDS-PAGE and Sup35Hp was detected with the same antipentahistidine antibody (right). An additional band detected by the antipentahistidine antibody (∗) was variably detected in samples analyzed but was not reproducibly present in any one particular sample. The anti-Sup35p antibody did not detect this protein (data not shown). T and S are as defined for Fig. 2.
The reduction in the level of detectable Sup35Hp in the aggregates in the presence of GdnHCl could be accounted for by simple dilution out of the preexisting aggregates. This was shown by comparing the Western blot signals for second- and fourth-generation cells with the signal generated by 1:4 and 1:16 dilutions, respectively, of the time zero sample (Fig. 4). These data therefore demonstrate that GdnHCl does not lead to resolubilization by deaggregation of Sup35p aggregates that exist in a [PSI+] cell prior to the addition of GdnHCl and are not consistent with extensive GdnHCl-induced proteolysis of aggregates.
GdnHCl does not prevent newly synthesized Sup35p from entering Sup35p aggregates in a [PSI+] strain.
One possible explanation for the GdnHCl-induced inhibition of [PSI+] seed replication is that it alters the conformation of preexisting seeds or aggregates so that they cannot catalyze the conformational change of newly synthesized Sup35p to the prion form. To determine whether newly synthesized Sup35p molecules can continue to enter high-molecular-weight Sup35p aggregates in GdnHCl-treated [PSI+] cells, [PSI+] strain 74-D694 expressing both the wild-type Sup35p and the Sup35Hp protein was first grown for three generations on YNBD (glucose-based) selective medium to ensure retention of the plasmid and repression of the GAL1-SUP35H gene. Cells were then transferred to YPGal medium containing 3 mM GdnHCl and incubated at 30°C for either 1 or 4 h. After 1 h, approximately 95% of newly synthesized Sup35Hp was present in the pellet fraction centrifuged at 100,000 × g, a proportion that did not change on incubation for a further 3 h (Fig. 5A). This indicates that sufficient seeds remained in the GdnHCl-treated cells even after approximately one generation of growth to ensure conversion of the majority of the newly synthesized Sup35Hp molecules into an aggregated form. The data shown in Fig. 3A show that aggregation is not due simply to overproduction of Sup35Hp since the Sup35Hp is only found in a soluble form when overproduced in the [psi−] control. It should be noted that the [PSI+] determinant was eliminated by GdnHCl at similar rates in YPD and YPGal media (15) (data not shown).
FIG. 5.
The influence of growth in the presence of GdnHCl on the physical state of newly synthesized Sup35p in [PSI+] cells. (A) [PSI+] strain 74-D694 carrying plasmid pUKC1809 (expressing GAL1-SUP35H) was grown overnight in YNBD to ensure plasmid retention and to repress Sup35Hp synthesis. Cells were then transferred to YPGal medium either with or without GdnHCl (3 mM) and grown for either 1 or 4 h. The subcellular distribution of Sup35Hp at these two time points was then determined as described in the legend to Fig. 2 with an antipentahistidine antibody. T, S, and P are as defined for Fig. 2. (B) The 74-D694 [PSI+] strain carrying plasmid pUKC1809 was grown overnight in YNBD medium. Cells were then transferred to YPD medium, either with or without GdnHCl (3 mM), and incubated for a further 3 h. Finally, both cultures were transferred to YPGal medium containing 3 mM GdnHCl. After a further incubation for 3 h the subcellular location of the Sup35Hp was assessed as described for panel A with an antipentahistidine antibody. The proportion of Sup35Hp in the pellet fraction (%P) was determined by densitometry.
To test whether the four- to five-generation lag period observed prior to the appearance of [psi−] cells in GdnHCl-treated cultures reflected the time required by GdnHCl to prevent polymerization of the newly synthesized Sup35p into pelletable aggregates, we determined whether incubation in the presence of 3 mM GdnHCl prior to shifting cells to YPGal medium altered the aggregation behavior of Sup35Hp. 74-D694 [PSI+] pUKC1809 cells were initially grown for 3 h in YPD containing 3 mM GdnHCl (approximately 0.6 generations) before being transferred to a galactose-based medium containing 3 mM GdnHCl. After a 3-h incubation in the YPGal containing 3 mM GdnHCl, Sup35Hp was still able to form aggregates (Fig. 5B) although a slight increase in the level of soluble Sup35Hp was observed. These data show that GdnHCl does not prevent newly synthesized Sup35Hp from being seeded by preexisting Sup35p seeds and that GdnHCl does not alter the conformational state of preexisting seeds such that they are unable to catalyze polymerization of newly synthesized Sup35Hp molecules.
Reducing the number of [PSI+] seeds reduces the ability of newly synthesized Sup35p to enter aggregates.
Most [PSI+] cells have to undergo at least four or five generations of growth in the presence of GdnHCl before [psi−] cells begin to appear in the culture (14). By monitoring the kinetics of [PSI+] loss from such cells, Eaglestone et al. (14) concluded that the data were consistent with approximately 60 [PSI+] seeds per cell; seed replication was blocked by GdnHCl, leading to a cell division-dependent reduction in seed number by dilution assuming random segregation of seeds to daughter cells. Our data (Fig. 5) show that incubation of [PSI+] cells in 3 mM GdnHCl for less than one generation did not prevent Sup35Hp from entering aggregates, although under such conditions the predicted seed number per cell would be approximately cut in half assuming random segregation (14). We therefore next determined whether incubation beyond one generation in the presence of 3 mM GdnHCl, to reduce the seed number still further, influenced the ability of newly synthesized Sup35p to form aggregates, i.e., does a decline in the seed number influence the rate of polymerization of newly synthesized Sup35p?
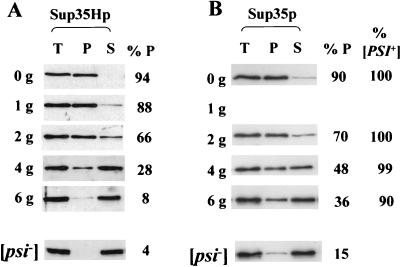
Strain 74-D694 [PSI+] pUKC1809 was grown in YPD-3 mM GdnHCl for between one and six generations, with cell samples being transferred at various times to a fresh YPGal-3 mM GdnHCl and incubated for a further 2 h to induce Sup35Hp synthesis. As shown in Fig. 6A, the greater the number of generations of growth in the presence of GdnHCl, the less newly synthesized Sup35Hp was able to enter aggregates when induced in the presence of GdnHCl, even though the total amounts of newly synthesized Sup35Hp were similar at all time points examined (compare T tracks in Fig. 6A). Increasing the amount of newly synthesized Sup35Hp molecules did not restore efficient polymerization since incubating the six-generation sample for 4 h rather than 2 h on YPGal containing 3 mM GdnHCl gave the same relative levels of soluble and pelletable Sup35Hp (data not shown).
FIG. 6.
Reducing the number of [PSI+] seeds by GdnHCl reduces the ability of newly synthesized Sup35p to enter high-molecular-weight aggregates. (A) [PSI+] strain 74-D694, transformed with plasmid pUKC1809, was grown in YPD medium containing 3 mM GdnHCl for up to six generations (0g to 6g), and then, at various points, the cells were transferred to YPGal medium containing 3 mM GdnHCl for a further 2 h to allow for induction of synthesis of the Sup35Hp. For each of the samples taken, the subcellular distribution of the Sup35Hp was assessed as described above. T, S, and P are as defined for Fig. 2. (B) The cell samples were also analyzed for the degree of aggregation of the endogenous Sup35p prior to the transfer to YPGal by using a polyclonal anti-Sup35p antibody. The proportions of Sup35p and Sup35Hp proteins in the pellet fraction (%P) were determined by densitometry. Cells were also plated on 1/4YPD before the 2-h YPGal incubation to assess the percentage of [PSI+] cells in the population being analyzed. The subsequent incubation for 2 h in YPGal did not significantly alter the relative numbers of [PSI+] cells (data not shown).
These same samples were also analyzed for the aggregation state of the endogenous Sup35p prior to the transfer to the YPGal-3 mM GdnHCl medium to ascertain the relative distribution of Sup35p between the aggregated and soluble forms (Fig. 6B). The levels of soluble Sup35p increased markedly over the six generations of growth, although the level of pelletable endogenous Sup35p after six generations (36% of total) was significantly higher than that seen for the Sup35Hp (8% of the total). This difference is presumably due to the stability of the preexisting endogenous Sup35p aggregates even when grown in the presence of GdnHCl (Fig. 4).
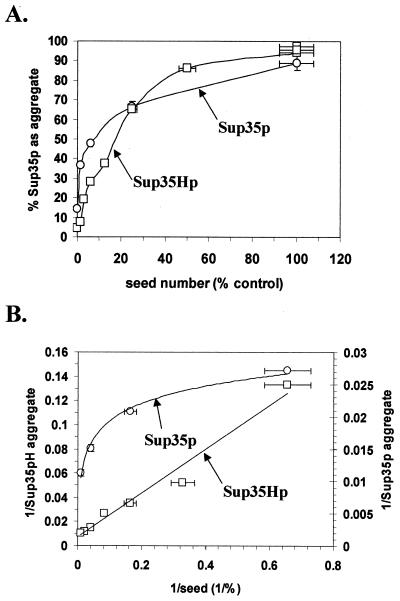
The data in Fig. 6 suggest that, as the number of [PSI+] seeds in a cell is reduced by a GdnHCl-induced block in their replication, the ability of newly synthesized Sup35p to aggregate is similarly reduced. In other words, the rate of seeded polymerization of Sup35p is dependent on the number of seeds in the cell, as is demonstrated by the plots in Fig. 7 using the data shown in Fig. 6. For example, after six generations of growth in 3 mM GdnHCl, approximately 90% of the cells still contained on average one or two [PSI+] seeds, with 36% of the endogenous Sup35p in an aggregated form. Yet greater than 90% of the newly synthesized Sup35Hp remained soluble (Fig. 6). These findings suggest that GdnHCl does not affect the polymerization of Sup35p molecules per se but rather blocks the generation of the [PSI+] seeds necessary to drive the polymerization process. Furthermore, the [PSI+] seeds need not necessarily be the aggregated forms of Sup35p pelleted at 100,000 × g. Compare the six-generation time point in Fig. 6, where, prior to induction of Sup35Hp synthesis, 36% of the endogenous Sup35p is in the pellet fraction and 90% of the cells are [PSI+] (i.e., contain [PSI+] seeds) yet less than 8% of the newly synthesized Sup35Hp is able to form aggregates.
FIG. 7.
The kinetics of Sup35p aggregation in vivo. Quantification of the proportion of Sup35p and Sup35Hp was performed by densitometric scanning of the Western blots shown in Fig. 6. For each data point the sum of the area corresponding to the pellet and the soluble fractions equaled the total fraction with no greater than 10% variation. The seed numbers were deduced from the kinetics of elimination of [PSI+] from cells over six generations of growth in YPD containing 3 mM GdnHCl (14). The intermediate seed numbers for each generation were calculated as a simple exponential function to reach the total seed number. (A) Plot of the percentage of Sup35p present in the pellet fraction (i.e., the fraction that corresponded to the aggregated Sup35p) against the estimated seed number. (B) Plot of the inverse of each data point from panel A.
GdnHCl blocks [PSI+] seed replication in slow-growing, glucose-starved cells.
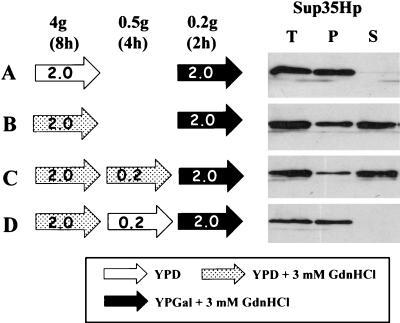
To determine whether prolonged exposure to GdnHCl in the absence of rapid cell division (and hence diluting out of seeds) would affect the rate of aggregation, we transferred [PSI+] cells to YPD with 1/10 the normal level of glucose (0.2% rather than 2%) and determined the ability of the [PSI+] cells to promote Sup35p aggregation after GdnHCl was removed. Specifically, 74-D694 [PSI+] pUKC1809 was grown for four generations (approximately 8 h) in YPD without or with GdnHCl (3 mM), the latter in order to reduce the number of [PSI+] seeds. Under such conditions, the culture would be expected to contain less than 1% [psi−] cells (14). A portion of the cells were then transferred to a fresh YPD medium with 1/10 the level of glucose and incubated for a further 4 h with or without GdnHCl (3 mM). In the low-glucose medium, the growth rate was significantly reduced, leading to only a 50% increase in cell density over the 4-h period. Finally, the cells were transferred to YPGal-3 mM GdnHCl for a further 2 h to induce the expression of the SUP35H gene and to assess the ability of the newly synthesized Sup35Hp to enter aggregates. Control experiments, in which the ability of the newly synthesized Sup35Hp to enter aggregates without the 4-h glucose starvation was examined, were also carried out in parallel. Figure 8 summarizes both the protocols used and the results obtained.
FIG. 8.
Influence of GdnHCl on the aggregation of newly synthesized Sup35p in slowly dividing [PSI+] cells growing on glucose-limited medium. [PSI+] strain 74-D694 transformed with plasmid pUKC1809 was grown for 2 h in YPGal containing 3 mM GdnHCl following four different preincubation regimens (A to D) as indicated. For cultures C and D this included growth in glucose-depleted YPD containing 0.2% glucose rather than the standard 2% glucose. The fate of the newly synthesized Sup35Hp following the 2-h growth on YPGal was then assessed as described in the legend to Fig. 1 by using an antipentahistidine antibody. T, S, and P are as defined for Fig. 2.
Incubation without GdnHCl allowed >95% of the newly synthesized Sup35Hp to appear in aggregates (Fig. 8A), whereas incubation in the presence of GdnHCl (Fig. 8B), even without substantial cell division (Fig. 8C), allowed only partial (i.e., 20 to 30%) aggregation. In contrast, in the GdnHCl-treated cells, subsequently incubated without substantial cell division in the absence of GdnHCl, >95% of Sup35p was found in the aggregated form (Fig. 8D). Therefore, slowly dividing glucose-starved [PSI+] cells can fully regain their ability to generate de novo new [PSI+] seeds in the absence of GdnHCl (Fig. 8D) while those incubated in slow-growth conditions in the presence of GdnHCl retain this ability (Fig. 8C). These data further support the notion that aggregation is promoted by the number of seeds present. The slight reduction in aggregated Sup35Hp in the glucose-starved cells versus the nonstarved cells treated with GdnHCl (compare Fig. 8C and B) is most likely a consequence of the low levels of growth (0.5 generation) seen during the 4-h incubation period, which would result in some dilution out of seeds.
[PSI+] seed replication is an exponential process that does not require ongoing protein synthesis.
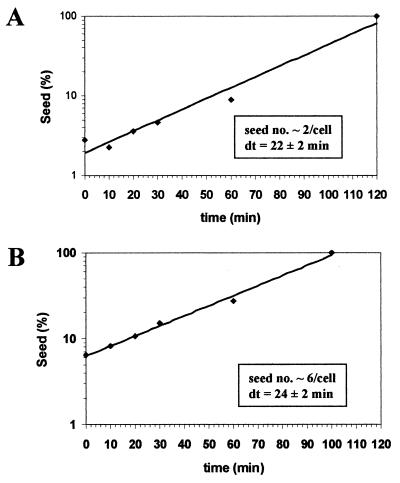
The experiments described above suggest that GdnHCl reduces the rate of aggregation of newly synthesized Sup35p by reducing the effective seed number in a growth-dependent manner, an effect that is rapidly reversed when the GdnHCl is removed. We therefore exploited this finding to analyze the kinetics of seed replication following removal of the GdnHCl. Strain 74-D694 [PSI+] pUKC1809 was grown for six generations in YPD-3 mM GdnHCl to produce a population largely comprising [PSI+] cells but with seed numbers reduced on average to one or two per cell. The cells were then resuspended in fresh low-glucose (0.2%) YPD medium without GdnHCl to allow seed replication in the absence of a high rate of cell division. At various subsequent time intervals, cell samples were removed, diluted into YPD-3 mM GdnHCl, and incubated at 30°C. From this culture, cells were removed at various times and plated onto YPD to score the proportion of [psi−] cells in the population. By the method described by Eaglestone et al. (14), we were able to use the kinetics of [PSI+] elimination to estimate the approximate number of [PSI+] seeds remaining following release from GdnHCl treatment. The resulting estimated seed numbers were then plotted against time of removal from the GdnHCl, and the data from two independent experiments are shown in Fig. 9. The increase in [PSI+] seed number following removal of GdnHCl was found to be exponential, with seed number doubling approximately every 22 to 24 min over the 2-h period measured. In the experiment starting with a slightly lower seed number (Fig. 9A), no replication was evident in the first 20 min although no such lag was seen when starting with a slightly higher seed number (Fig. 9B). These data are consistent with the rate of prion seed replication not being directly linked to the rate of cell division.
FIG. 9.
Rate of prion seed replication in a [PSI+] strain following a release from a GdnHCl-induced block. [PSI+] strain 74-D694 was grown on YPD containing 3 mM GdnHCl for either 6 (A) or 4.5 (B) generations to generate a population of cells with only a few seeds per cell (14). The cells were then resuspended in fresh YPD (low-glucose [0.2%]) medium without GdnHCl, and cell samples were taken every 10 min for the first 30 min and then at 60 and 120 min. By the 2-h time point the cells had not gone through one generation. The average number of [PSI+] seeds per cell in the cell populations at each of the time points was determined by establishing the rate of [PSI+] curing by 3 mM GdnHCl essentially as described by Eaglestone et al. (14).
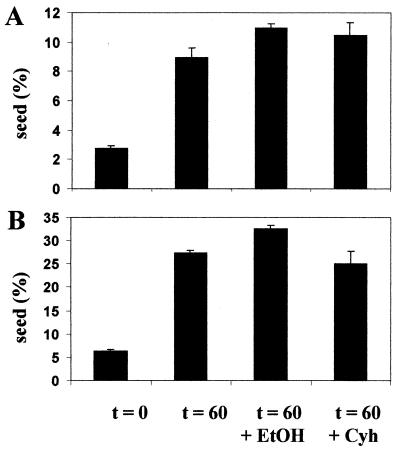
To determine whether ongoing protein synthesis was important for [PSI+] seed replication, [PSI+] cells were grown for six generations in YPD-3 mM GdnHCl and then were resuspended in low-glucose (0.2%) medium containing 10 μg of cycloheximide/ml to block protein synthesis. This concentration of cycloheximide was the same as that used to block protein synthesis in the luciferase reactivation assay (15) (Fig. 1). Since the cycloheximide was dissolved in 0.1% ethanol, the effect of the ethanol alone was also examined in parallel. The seed number after 60 min was determined by determining the kinetics of GdnHCl-induced [PSI+] elimination as described above. In two independent experiments (Fig. 10) no significant reduction in seed number in the cycloheximide- or ethanol-treated cells was observed; indeed there appeared to be a slightly higher seed number in the ethanol-treated cells. Therefore, we conclude that in vivo the [PSI+] seed generation process does not require either rapid cell division or ongoing protein synthesis. As the Sup35p present in cells is largely soluble after six generations of growth in YPD-3 mM GdnHCl (Fig. 6), this suggests that seed regeneration requires only the preexisting Sup35p and Hsp104 molecules, with replication occurring via an exponential process that is not coupled to cell division.
FIG. 10.
[PSI+] seed replication does not require ongoing cell division or protein synthesis. [PSI+] strain 74-D694 was grown in YPD with 3 mM GdnHCl for six generations to generate a cell population containing a low average seed number per cell. The cells were then resuspended in fresh low-glucose (0.2%) YPD (YPD0.2) without GdnHCl to allow seeds to begin to replicate (see Fig. 8). In parallel, cells from the same culture were also transferred to fresh YPD0.2 with either 10 μg of cycloheximide (Cyh)/ml in 0.1% ethanol (EtOH) or 0.1% ethanol alone. Cells were removed from these cultures at the beginning (t = 0) and after 60 min of incubation (t = 60), and the average seed number in the cell population was determined essentially as described by Eaglestone et al. (14). The results of two independent experiments (A and B) using cultures with slightly different starting seed numbers are shown.
These data are in agreement with our proposal (14) that GdnHCl induces a reversible block in [PSI+] seed replication. Furthermore, we show that the decrease in seed number in the presence of GdnHCl occurs in a growth-dependent manner. This argues against GdnHCl causing physical inactivation of its cellular target (for example, Hsp104) or some kind of posttranslational, covalent modification to Sup35p that would inhibit its seeded polymerization.
Curing of [PSI+] by inhibition of Hsp104 ATPase activity.
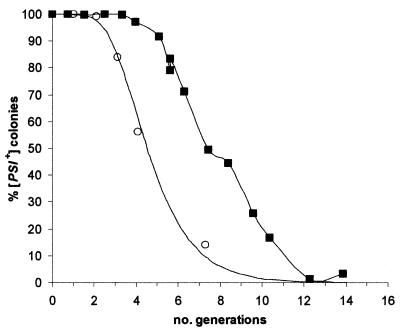
Wegrzyn et al. (46) recently suggested that [PSI+] curing by elimination of Hsp104 activity through deletion of the HSP104 gene is too rapid to be by the same mechanism as the curing induced by growth in GdnHCl. This raises the question of whether [PSI+] curing by GdnHCl and that by Hsp104 inactivation occur by different mechanisms. The curing kinetics that Wegrzyn et al. (46) show for the loss of [PSI+] during outgrowth of a Δhsp104 haploid feature a lag distinctive of curing by normal (not defective) segregation after inhibition of replication of [PSI+] seeds by GdnHCl. The lag is shorter than the one that we commonly observe (14) but would be characteristic of cells in which the number of seeds at the start is lower than we estimated for our standard haploid [PSI+] cells (14). Their data could simply reflect the fact that the Δhsp104 ascospores contain only approximately one-fourth of the cytoplasm of the original diploid cell. In Fig. 11 we show a plot of the [PSI+] elimination data of Wegrzyn et al. (from Table 2 in reference 46) during the outgrowth of the Δhsp104 haploid spores and overlay this with the elimination curve for a haploid cell with 16 seeds (i.e., one-fourth of the number we predict for a haploid Hsp104+ strain [14]) predicted by our GdnHCl curing model (14). The two overlap.
FIG. 11.
The kinetics of [PSI+] elimination by Hsp104 depletion or inactivation. The data of Wegrzyn et al. (46) for the elimination of [PSI+] in haploid ascospores containing an hsp104 gene deletion are plotted (○) together with a continuous line indicating the predicted kinetics of [PSI+] elimination from a cell containing 16 [PSI+] seeds calculated by using the model of Eaglestone et al. (14). Also shown for comparison are [PSI+] elimination data obtained by inactivation of endogenous Hsp104 by coexpression of an ATPase-negative mutant HSP104 K218T,K620T (▪).
To further support our supposition that the kinetics of elimination of [PSI+] by GdnHCl mimic what happens when the ATPase activity of Hsp104 is inactivated, we examined in detail the kinetics of [PSI+] elimination in [PSI+] cells engineered to overexpress a dominant ATPase-negative allele of HSP104, namely, the HSP104 K218T,K620T allele (5). Unlike Wegrzyn et al. (46) we scored any colony that contained at least one [PSI+] (white, Ade+) sector as [PSI+] since at the time of plating the progenitor cell must have contained at least one [PSI+] seed. The kinetics of [PSI+] elimination observed (Fig. 11) exactly matched what we observed with cells incubated in the presence of 3 mM GdnHCl, namely, a four-generation lag with complete elimination after a further seven or eight generations of growth (14). When we plotted our data using the criterion applied by Wegrzyn et al. (46), i.e., only counting clonally pure white Ade+ colonies as [PSI+], we saw essentially the same kinetics of elimination as they observed using a similar construct (data not shown).
DISCUSSION
One of the major physical properties defining both yeast and mammalian prions is aggregation-prone conformational changes. In [PSI+] yeast cells, Sup35p proteins are found as partially proteinase K-resistant high-molecular-weight aggregates (30, 31). This property is efficiently transmitted to daughter cells during cell division by, it is assumed, conformational modification of newly synthesized Sup35p seeded by the prion form of Sup35p in the [PSI+] mother cell. Neither the molecular nature of the transmitted seed nor the mechanism of transmission of such seeds has been established for any yeast prion. Continued propagation of the [PSI+] determinant therefore requires both seed replication and efficient mother-to-daughter transmission of seeds during cell division. Several lines of evidence have indicated that molecular chaperone Hsp104 is required for the continued propagation of yeast prion seeds (5, 10, 27, 41) and that Hsp104 is inactivated by growth in the presence of protein denaturant GdnHCl (15, 22). In this paper we have used GdnHCl to explore the mechanism by which the [PSI+] determinant is propagated in vivo and its implications for the role of Hsp104 in this process.
The state of Sup35p in GdnHCl-grown cells.
During growth in GdnHCl, cells are cured of [PSI+] such that when the cells are transferred to a medium without GdnHCl, they grow into stable [psi−] colonies (45). When this curing is monitored by plating cells to form colonies on solid medium, a lag of several generations before [psi−] colonies start to appear is observed (14). Late in this lag, however, we find that cells become phenotypically [psi−] while remaining genetically [PSI+], in that (i) soluble Sup35p accumulates and, because of this, (ii) the efficiency of translation termination increases (Fig. 2). These characteristics are observed at a time during growth in GdnHCl when 85% or more of the cells are still capable of growing into [PSI+] colonies and thus presumably retain at least one prion seed. In other words, cells become phenotypically [psi−] before losing the ability to propagate [PSI+].
The discrepancy between phenotype and genotype in GdnHCl-treated [PSI+] cells can be explained in one of three ways: (i) sequestration of soluble Sup35p into aggregates is proportional to the numbers of [PSI+] seeds in cells, which in turn are reduced by segregation during growth in GdnHCl, (ii) GdnHCl blocks aggregation of newly synthesized Sup35p, or (iii) GdnHCl causes the breakdown of preexisting aggregates to the soluble form of Sup35p. By studying the fate of both newly synthesized Sup35p and preexisting Sup35p in GdnHCl-treated cells, we show that growth in GdnHCl does not inhibit aggregation of newly synthesized Sup35p nor does it cause preformed aggregates to break down and release soluble Sup35p. Rather, we show that the rate of incorporation of newly synthesized Sup35p into high-molecular-weight aggregates is proportional to the number of seeds, i.e., is inversely proportional to the number of generations of growth in GdnHCl.
As shown in Fig. 7, the rate of incorporation of Sup35Hp into aggregates shows first-order kinetics in relation to estimated seed number. Seeds are regenerated exponentially when cultures which are phenotypically [psi−] but genetically [PSI+] are transferred to GdnHCl-free medium. Remarkably this regeneration of seeding capacity is independent of a need for ongoing protein synthesis, and the measurable seed number doubles approximately every 20 min. Thus a cell containing only 1 seed would, when released from the GdnHCl-induced block, generate 64 seeds within 2 h, i.e., prior to the next cell division. This regeneration may reflect the recovery of Hsp104 activity upon removal of GdnHCl (15, 22).
In attempting to understand the significance of our results we must first clarify what it is we are trying to explain. Most studies, particularly those involving in vitro experiments, focus on the means by which recombinant forms of the native Sup35p or the Sup35p NM domain form amyloid fibers (16, 32, 39). In addition, how the prion form comes to be propagated has to be considered; in other words, what is the molecular form of Sup35p actually passed from cell to cell. All the genetic evidence suggests that inheritance depends on a limited number of Sup35p “particles” (14, 26) whose numbers double in synchrony with cell division and which are partitioned more or less randomly at cell division. The reason we need to be concerned with this extra dimension of prion biology is that here we demonstrate a separation between Sup35p protein aggregation and propagation, i.e., replication and transmission, of the [PSI+] determinant.
Curing of yeast prions by GdnHCl or by the inactivation of Hsp104: is the same mechanism involved?
The evidence provided by Ferreira et al. (15) and Jung and Masison (22) is consistent with GdnHCl inhibiting the activity of Hsp104 required for yeast prion propagation. This is most likely to be via direct inhibition of the Hsp104 ATPase activity, and Glover and Lindquist (17) have reported that trace amounts of GdnHCl do indeed inhibit Hsp104 ATPase activity. However, Wegrzyn et al. (46) have reported that the kinetics of [PSI+] elimination by GdnHCl are not the same as those seen when the HSP104 gene is genetically inactivated. However, in Fig. 11 we show that their data can be explained through their use of haploid Δhsp104 ascospores, which would be expected to contain only approximately one-fourth of the number of seeds. This would result in a reduced lag period of approximately two generations, as is observed for GdnHCl curing (14). Furthermore, our detailed kinetic analysis of [PSI+] elimination by coexpression in a [PSI+] strain of a dominant ATPase-negative allele of the HSP104 gene (Fig. 11) indicates that the kinetics are essentially identical to what one would see for GdnHCl curing if one scored as [psi−] only those colonies containing no white [PSI+] sectors. It is this use of different scoring criteria that may explain the differences between our data (15) (Fig. 11) and those of Wegrzyn et al. (46).
While we cannot rule out the additional possibility that GdnHCl has pleiotropic effects that may effect the inactivation of Hsp104 through a yet to be identified primary cellular target, the circumstantial evidence strongly suggests that Hsp104 inactivation is an entirely plausible mechanism. Demonstration that there is no other GdnHCl-sensitive determinant necessary for seed replication would require the isolation of a GdnHCl-incurable Hsp104 mutant although, given that GdnHCl probably binds competitively to one or both of the ATPase domains of Hsp104, such a mutation is likely to also inactivate the ATPase activity of Hsp104, which in turn would generate a [psi−] strain.
Aggregation of Sup35p in [PSI+] cells is seed dependent but not Hsp104 dependent.
An important conclusion from our studies is that, unlike [PSI+] propagation, conversion of soluble Sup35p to the diagnostic prion form (i.e., high-molecular-weight aggregates) does not require Hsp104 activity. Although Hsp104 is required for the in vivo replication of the prion, it is not apparently required for the conformational change of newly synthesized Sup35p to the prion form. This conclusion is based on the observation that newly synthesized Sup35p continues to become part of aggregates in cells grown in the presence of GdnHCl, i.e., in conditions that fully inhibit Hsp104 activity. Furthermore, Zhou et al. (49) and we (F. Ness and M. F. Tuite, unpublished data) have shown that de novo formation of [PSI+] by overexpression of the NM domain of Sup35p is not blocked in the presence of GdnHCl. These indications of the irrelevance of Hsp104 to Sup35p aggregation are reinforced by the fact that, in in vitro studies, seeded conversion of soluble Sup35p into amyloid fibers occurs in the absence of Hsp104 and depends only on the presence of preexisting fibers (32, 39).
The presence of GdnHCl causes, during growth, a marked reduction in the rate of incorporation of newly synthesized Sup35p into high-molecular-weight aggregates before cells become genetically [psi−]. Although we observe that this loss is gradual during the first five or six generations of growth in GdnHCl, at the concentration of GdnHCl used we get almost immediate (in less than one generation) inhibition of Hsp104 activity (Fig. 1). The gradual loss of prion-forming activity cannot therefore be explained by a gradual inactivation of Hsp104. Instead, we show that the rate of incorporation of newly synthesized Sup35p into aggregates is related in a simple way to the numbers of some preexisting structures. We conclude that it is prion seed replication alone that is sensitive to GdnHCl, while the conversion of native Sup35p to its highly aggregated form, which is dependent on such seeds, is not. We take this to indicate that seed replication is dependent on Hsp104 activity while seed-dependent aggregation is not.
The nature of [PSI+] seeds.
What are [PSI+] seeds? They might be the high-molecular-weight aggregated form of Sup35p found in [PSI+] cells. Alternatively, they may be relatively low-molecular-weight prion-synthesizing complexes and, as such, would perhaps include a templating prion molecule. [PSI+] seeds would be expected to have two properties; one is the ability to convert native Sup35p to a prion form, which would then aggregate with other Sup35p molecules in either a native or prion form, and the other is self-replication. Of these two properties, only the second would be Hsp104 dependent. This conclusion embraces both the monomer-directed conformational-change model (18, 33) and the nucleated-polymerization mechanism (20, 24, 25). However, one hypothesized role for Hsp104, namely, that it is needed for newly synthesized native Sup35p to reach an altered physical state or to break an energy barrier before being converted into the aggregated form (5, 30, 36, 39), is ruled out. Rather, this function is wholly fulfilled by preexisting seeds, as we have shown that a large part (more than 50%) of newly synthesized Sup35p continues to form aggregates even after three generations of growth in GdnHCl-containing medium, although Hsp104 is inactivated in such conditions. Our data also suggest that Hsp104 activity is not required for segregation of seeds since that continues randomly in cells dividing in 3 mM GdnHCl.
Wegrzyn et al. (46) interpret their results under a single unifying theory that the role of Hsp104 is to break up prion aggregates, with successful heritable stability being the consequence of a balance between the total loss of aggregates (and presumably prions) and the generation of sufficient aggregates that can be partitioned successfully to daughter cells. In other words aggregates are seeds and Hsp104 is necessary for replication This neatly explains both the reciprocity between Hsp104 activity and the appearance of cells with large aggregates of Sup35p-green fluorescent protein (GFP) fusion proteins (46) and the correlation between the number of cells with atypically large aggregates and the number of [psi−] cells in populations underexpressing Hsp104.
Do our data rule out the possibility that the high-molecular-weight Sup35p aggregates are the replicating seed structures? The rate of incorporation of newly synthesized Sup35p in the presence of GdnHCl is not related to the proportion of Sup35p present as such aggregates but rather to the estimated number of seeds in the culture. Incorporation into such aggregates decreases proportionately to seed number, while the amount of aggregate found remains high over much of the time of the curing experiment (Fig. 6). Neither numbers of seeds nor incorporation is related to the amount of aggregate, nor is the amount of aggregate present related to the declining numbers of seeds. Seeds and aggregate seem to be different entities, with the pelletable high-molecular-weight aggregates most likely representing “dead-end” products with no seeding activity per se.
It is, however, possible to reconcile replicating seeds with aggregates. No one has yet reported accurate data on the sizes of Sup35p aggregates in a [PSI+] cell, and, until we know the numbers of pelletable aggregates present (not merely the amount of Sup35p present in such aggregates) and how these numbers relate to Hsp104 activity, we cannot say whether aggregates decline in numbers during growth in the presence of GdnHCl. If such aggregates consist of amyloid fibers and if one or both ends of a fiber provided the template for prionization of native Sup35p molecules (19, 35), then the function of Hsp104 might be to cut fibers and create new ends (24), an activity consistent with its known role as a protein “remodeling factor” (17). Whether one or both ends were templates, a single cleavage event would double their numbers and the replication would thus proceed exponentially. The templating step itself would be independent of Hsp104, so newly synthesized Sup35p would continue to be incorporated into aggregates and the amount of aggregate would remain high. If Hsp104 activity was simply confined to cleaving the amyloid fibers, we would not expect to see the release of soluble Sup35p from preexisting fibers. However, what would happen during curing by GdnHCl is that no new ends would be created by this mechanism and the existing ends would be diluted out until cells started to segregate with no fibers. The relationship between the numbers of fiber ends per cell and the rate of incorporation of newly synthesized Sup35p would also show first-order kinetics (Fig. 7).
A two-cycle model for prionization of Sup35p in [PSI+] cells.
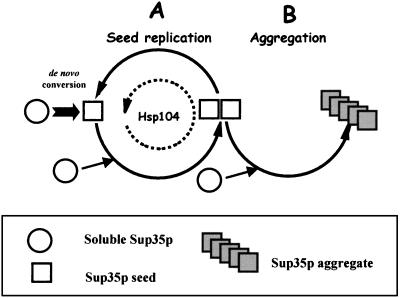
Our data indicate that replication of prion seeds is sensitive to GdnHCl (i.e., Hsp104 dependent) whereas the de novo formation of prion seeds (49) and formation of aggregates (this paper) are not (i.e., are Hsp104 independent). However, Sup35p aggregates do not break down in the presence of GdnHCl once they are formed, nor do they appear to break down in the control cells, at least after four generations of growth in the absence of GdnHCl (Fig. 4). Taking full account of the new data that we present here, we propose the model illustrated in Fig. 12. In [PSI+] cells there are two cycles involving prionization of soluble Sup35p: cycle A replicates seeds, whereas cycle B converts the seeds to high-molecular-weight aggregates. Hsp104 carries out cycle A, whereas cycle B operates independently of Hsp104. Both cycles draw on the pool of soluble Sup35p in the cell. If Hsp104 activity is lowered or inactivated either by GdnHCl treatment or by HSP104 gene repression, the balance is tipped toward aggregate formation in cycle B. There may be fewer aggregates than seeds, but they can get steadily larger since aggregation is independent of Hsp104 and less soluble Sup35p is being used to generate new seeds. When Sup35p-GFP fusions are being overexpressed, large aggregates of this material would form. Also, in those conditions where the levels of Hsp104 are perhaps insufficient to maintain seed number, not only does [PSI+] become more unstable but also less soluble Sup35p is sequestered and the deficiency in translation termination is alleviated.
FIG. 12.
A two-cycle model for prionization of Sup35p in [PSI+] cells. Cycle A replicates the [PSI+] seed (squares), which can either be a conformationally altered monomer or a multimeric form of Sup35p and which is dependent on Hsp104. Cycle B converts these seeds to aggregates, which themselves do not have seeding activity. Cycle B is independent of Hsp104 as is the de novo conversion of the soluble nonprion form of Sup35p (circles) to the seed form in the absence of a preexisting seed.
If Hsp104 activity is high enough, the competition for soluble Sup35p is tipped toward cycle A. The amount of aggregate formed is proportional to the number of seeds, and thus seed number is correlated with depletion of soluble Sup35p and with the level of stop codon readthrough. Wegrzyn et al. (46) observed that Sup35p-GFP aggregates are smaller in strains expressing normal levels of Hsp104 than in strains with lower levels of Hsp104. It may be that many more aggregates are formed because of the larger numbers of seeds but that they do not grow as big as they do when the same amount of fusion protein is present but there are fewer aggregates for it to condense with during cycle B. This interpretation is supported by our recent finding (C. Lawrence and M. F. Tuite, unpublished data) that the degree of formation of Sup35p-GFP prion aggregates in [PSI+] cells is strain specific, which in turn reflects the level of endogenous expression of Hsp104 in the different strains
Acknowledgments
The work described in this paper was supported by research grants from the Biotechnology and Biological Sciences Research Council (BBSRC), The Wellcome Trust, and the European Commission (contract no. BI104-98-6045).
REFERENCES
- 1.Caughey, B. 2000. Transmissible spongiform encephalopathies, amyloidoses and yeast prions: common threads? Nat. Med. 6:751-754. [DOI] [PubMed] [Google Scholar]
- 2.Caughey, B. 2001. Interactions between prion protein isoforms: the kiss of death. Trends Biochem. Sci. 26:235-242. [DOI] [PubMed] [Google Scholar]
- 3.Chernoff, Y. O., I. L. Derkach, A. Dagkesamanskaya, V. Tikhomironva, M. D. Ter-Avanesyan, and S. Inge-Vechtomov. 1988. Nonsense-suppression by amplification of translational protein factor gene. Dokl. Akad. Nauk SSSR (Biol. Sci.) 301:1227-1229. [PubMed] [Google Scholar]
- 4.Chernoff, Y. O., I. L. Derkach, and S. G. Inge-Vechtomov. 1993. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24:268-270. [DOI] [PubMed] [Google Scholar]
- 5.Chernoff, Y. O., S. L. Lindquist, B.-I. Ono, S. G. Inge-Vechtomov, and S. W. Liebman. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880-884. [DOI] [PubMed] [Google Scholar]
- 6.Cox, B. S. 1965. ψ, a cytoplasmic suppressor of super-suppression in yeast. Heredity 20:505-521. [Google Scholar]
- 7.Cox, B. S., M. F. Tuite, and C. S. McLaughlin. 1988. The psi factor of yeast: a problem in inheritance. Yeast 4:159-178. [DOI] [PubMed] [Google Scholar]
- 8.DePace, A. H., A. Santoso, P. Hillner, and J. S. Weissman. 1998. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93:1241-1252. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch, I. L., Y. O. Chernoff, V. V. Kushnirov, S. G. Inge-Vechtomov, and S. W. Liebman. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derkatch, I. L., M. E. Bradley, P. Zhou, Y. O. Chernoff, and S. W. Liebman. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobson, C. M. 1999. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24:329-332. [DOI] [PubMed] [Google Scholar]
- 12.Doel, S. M., S. J. McCready, C. R. Nierras, and B. S. Cox. 1994. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137:659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaglestone, S. S., B. S. Cox, and M. F. Tuite. 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 18:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaglestone, S. S., L. W. Ruddock, B. S. Cox, and M. F. Tuite. 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira, P. C., F. Ness, S. R. Edwards, B. S. Cox, and M. F. Tuite. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40:1357-1369. [DOI] [PubMed] [Google Scholar]
- 16.Glover, J. R., A. S. Kowal, E. C. Schirmer, M. M. Patino, J. J. Liu, and S. Lindquist. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89:811-819. [DOI] [PubMed] [Google Scholar]
- 17.Glover, J. R., and S. Lindquist. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73-82. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, P. M., P. Bamborough, V. Daggett, S. B. Prusiner, and F. E. Cohen. 1997. The prion folding problem. Curr. Opin. Struct. Biol. 7:53-59. [DOI] [PubMed] [Google Scholar]
- 19.Inoue, Y., A. Kishimoto, J. Hirao, M. Yoshida, and H. Taguchi. 2001. Strong growth polarity of yeast prion fiber revealed by single fiber imaging. J. Biol. Chem. 276:35227-35230. [DOI] [PubMed] [Google Scholar]
- 20.Jarrett. J. T., and P. T. Lansbury. 1993. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 73:1055-1058. [DOI] [PubMed] [Google Scholar]
- 21.Jung, G., G. Jones, R. D. Wegryzn, and D. C. Masison. 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, G., and D. C. Masison. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43:7-10. [DOI] [PubMed] [Google Scholar]
- 23.King, C. Y., P. Tittmann, H. Gross, R. Gebert, M. Aebi, and K. Wuthrich. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushnirov, V. V., and M. D. Ter-Avanesyan. 1998. Structure and replication of yeast prions. Cell 94:13-16. [DOI] [PubMed] [Google Scholar]
- 25.Lansbury, P. T., and B. Caughey. 1995. The chemistry of the scrapie reaction: the “ice 9” metaphor. Chem. Biol. 2:1-5. [DOI] [PubMed] [Google Scholar]
- 26.McCready, S. J., B. S. Cox, and C. S. McLaughlin. 1977. The extrachromosomal control of nonsense suppression in yeast: an analysis of the elimination of [psi+] in the presence of a nuclear gene PNM. Mol. Gen. Genet. 150:265-270. [DOI] [PubMed] [Google Scholar]
- 27.Moriyama, H., H. K. Edskes, and R. B. Wickner. 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperoneYdj1p. Mol. Cell. Biol. 20:8916-8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newnam, G. P., R. D. Wegrzyn, S. L. Lindquist, and Y. O. Chernoff. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19:1325-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parham, S. N., C. G. Resende, and M. F. Tuite. 2001. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 20:2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patino, M. M., J. J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622-626. [DOI] [PubMed] [Google Scholar]
- 31.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 32.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1997. In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277:381-383. [DOI] [PubMed] [Google Scholar]
- 33.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136-144. [DOI] [PubMed] [Google Scholar]
- 34.Rochet, J. C., and P. T. Lansbury. 2000. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 10:60-68. [DOI] [PubMed] [Google Scholar]
- 35.Scheibel, T., A. S. Kowal, J. D. Bloom, and S. L. Lindquist. 2001. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr. Biol. 11:366-369. [DOI] [PubMed] [Google Scholar]
- 36.Schirmer, E. C., and S. Lindquist. 1997. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc. Natl. Acad. Sci. USA 94:13932-13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serio, T. R., and S. L. Lindquist. 1999. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu. Rev. Cell Dev. Biol. 15:661-703. [DOI] [PubMed] [Google Scholar]
- 38.Serio, T. R., and S. L. Lindquist. 2000. Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol. 10:98-105. [DOI] [PubMed] [Google Scholar]
- 39.Serio, T. R., A. G. Cashikar, A. S. Kowal, G. J. Sawicki, J. J. Moslehi, L. Serpell, M. F. Arnsdorf, and S. L. Lindquist. 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289:1317-1321. [DOI] [PubMed] [Google Scholar]
- 40.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sondheimer, N., and S. Lindquist. 2000. An epigenetic modifier of protein function in yeast. Mol. Cell 5:163-172. [DOI] [PubMed] [Google Scholar]
- 42.Stansfield, I., A. Akhmaloka, and M. F. Tuite. 1995. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 27:417-426. [DOI] [PubMed] [Google Scholar]
- 43.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. L. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ter-Avanesyan, M. D., A. R. Dagkesamanskaya, V. V. Kushnirov, and V. N. Smirnov. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuite, M. F., C. R. Mundy, and B. S. Cox. 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98:691-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegrzyn, R. D., K. Bapat, G. P. Newnam, A. D. Zink, and Y. O. Chernoff. 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21:4656-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickner, R. B. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264:566-569. [DOI] [PubMed] [Google Scholar]
- 48.Wickner, R. B., K. L. Taylor, H. K. Edskes, M. L. Maddelein, H. Moriyama, and R. T. Roberts. 2000. Prions of yeast as heritable amyloidoses. J. Struct. Biol. 130:310-322. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, P., I. L. Derkatch, and S. W. Liebman. 2001. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol. Microbiol. 39:37-46. [DOI] [PubMed] [Google Scholar]
- 50.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. G. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]