Abstract
Luman is a human basic leucine zipper transcription factor that, like the herpes simplex virus transcription factor VP16, requires the host cell factor, HCF, for activity. Although both HCF and Luman have been implicated in cell growth, their biological roles have not been clearly defined. Luman conforms to a type II membrane-associated glycoprotein with its carboxyl terminus embedded in cellular membranes and its amino terminus, which contains all its identified functional domains, in the cytoplasm. Here we show that Luman is processed by regulated intramembrane proteolysis (RIP). The site 1 protease (S1P), a Golgi apparatus-resident enzyme responsible for catalyzing the first step in the RIP pathway of the sterol regulatory element binding proteins (SREBPs) and ATF6, may also be involved in the processing of Luman. Thus, processing of Luman was highly stimulated by brefeldin A, a compound that causes the reflux of Golgi apparatus enzymes to the endoplasmic reticulum (ER). In addition, coexpression of Luman with S1P containing a KDEL ER retrieval signal resulted in virtually quantitative cleavage of Luman in the absence of any treatment. Finally, Luman contains a sequence, RQLR, immediately downstream from the transmembrane domain which bears similarity to the consensus S1P cleavage site identified by others. Substitution of arginine residues within this motif abolished S1P cleavage, providing robust evidence that S1P is involved in Luman processing. We observed that following S1P cleavage, the majority of the cleaved Luman was retained in cytoplasmic membranes, indicating that an additional step or enzymes yet to be identified are involved in complete cleavage and release to yield the product which ultimately enters the nuclei of cells.
Luman (also known as LZIP and CREB3) is a basic leucine zipper transcription factor of the CREB/ATF gene family. It possesses a potent N-terminal acidic activation domain and a basic-leucine zipper motif (bZIP) (15, 23-26). The primary structure of Luman appears to be strongly conserved, and Luman homologues in mice (LZIP [8]), cattle (our unpublished results), and fruit flies (dCREB-A/BBF-2 [1, 37]) have been identified. We and others (15, 24) originally identified Luman when screening for cellular ligands of the human host cell factor (HCF, also known as C1 factor), a protein required by the herpes simplex virus (HSV) transactivator VP16. Luman interacts with HCF through the tetrapeptide DHTY (15, 24, 25), which is homologous to the EHAY HCF binding sequence of VP16. This motif, as (D/E)HXY, is conserved in the VP16 homologues of other alphaherpesviruses as well as in the homologues of Luman in mice, cattle, and fruit flies. Luman can bind and activate genes containing cyclic AMP response elements (CREs), although its natural target has not been identified. Similarly, although Luman has been implicated in the regulation of cell growth (18), its biological role in this process has not been clearly defined. Luman mRNA is present in a wide range of adult and fetal tissues (24), although it is not clear if the protein is as ubiquitous.
Luman contains a transmembrane domain that allows it to associate with the endoplasmic reticulum (ER), and Luman retained in the ER sequesters most of the cellular HCF at this location (23). HCF is expressed in most tissues and is located in the nuclei of cells. However, in the neurons of dorsal root ganglia, HCF appears to be sequestered in the cytoplasm, and its translocation to the nucleus correlates with events that lead to the reactivation of latent HSV (20). Although the mechanism for the retention of HCF in the cytoplasm has not been identified, our preliminary results suggest that Luman may play a role in this process (23). Our observations suggest that the movement of Luman and HCF from the ER to the nuclei of neurons may influence the reactivation of HSV from latency. This hypothesis is supported further by the observation that Luman can activate promoters of HSV genes thought to be required for reactivation from latency (23). The mechanism by which Luman and HCF are released from a cytoplasmic location is not known, but we proposed previously that Luman may be processed by a specific pathway known as regulated intramembrane proteolysis (RIP).
RIP is a mechanism that allows for a rapid response to regulatory signals that mediate a variety of cellular processes such as differentiation, lipid metabolism, and response to unfolded proteins (reviewed by Brown et al. [7]). To date, five proteins of the RIP family have been identified: sterol regulatory element binding protein (SREBP), amyloid precursor protein (APP), Notch, Ire1, and ATF6 (7). RIP proteins have some common features. All RIP proteins span the membrane bilayer at least once. They can be classified as either type I or type II membrane-spanning proteins based on the orientation of the amino terminus in the membrane. In addition, intramembrane cleavage leading to the release of the proteins from membranes occurs only after a portion of the extracytosolic sequence is removed by a primary cleavage (3, 5, 6, 9, 28). Reduction of the extracytosolic segment to fewer than 30 amino acids seems to be a prerequisite for secondary intramembrane cleavage (7).
The best-characterized RIP protein is SREBP-2, a member of the SREBP family of transcription factors, which regulate genes involved in cholesterol and fatty acid metabolism (6). SREBP-2 is bound to membranes of the ER and nuclear envelope in a hairpin orientation, with both its amino and carboxyl termini projecting into the cytosol. The SREBP-2 amino-terminal domain contains the acidic transcriptional activation sequence and basic helix-loop-helix and leucine zipper domains needed to mediate DNA binding, nuclear entry, and dimerization (33, 34). The carboxyl-terminal domain has a regulatory role. In response to sterol depletion, the carboxyl terminus complexes with the ER protein SCAP (SREBP cleavage-activating protein), which moves SREBP into the post-Golgi compartment (11, 30, 34). The first cleavage in the Golgi lumen is catalyzed by site-1 protease (S1P), a membrane-associated serine protease of the subtilisin family (35). S1P cleaves after the leucine residue of the consensus sequence RXX(L/K) (14) (RSVL in SREBP-2) and creates two protein segments (12). S1P cleavage depends on the prior transport of SREBP by SCAP to the Golgi apparatus. However, this requirement can be bypassed experimentally if activated S1P is delivered to the ER either by treatment of cells with the drug brefeldin A or by transfection of cells lacking SCAP with plasmids expressing an active form of S1P linked to the ER retrieval sequence KDEL (11). Following cleavage of SREBP by S1P, the amino-terminal portion of SREBP is cleaved 3 amino acids within the membrane-spanning region by site-2 protease (S2P) (13, 31). The released amino-terminal product then translocates into the nucleus, where it activates transcription by binding promoters containing sterol-regulating elements (6, 13). S2P cleavage requires the sequence DRSR outside the membrane and residues NP near the middle of the membrane-spanning helix (13, 41, 42).
ATF6 is a type II leucine zipper transmembrane protein and, like SREBP, is located in the ER. Accumulation of improperly folded proteins in the ER, calcium depletion, or inhibition of glycosylation leads to induction of ER chaperones, a phenomenon termed the unfolded protein response (19, 27, 32, 43). This response is mediated by the proteolytic cleavage of the 90,000-molecular-weight (90K) type II transmembrane form of ATF6. The resulting 50K amino-terminal portion (p50ATF6), which contains the transcription activation region and DNA binding and dimerization motifs, moves into the nucleus and activates the genes for the ER chaperone GRP78 and other proteins involved in assisting in the folding of ER proteins (16, 22, 40). Like SREBP, ATF-6 also contains an S1P consensus cleavage site in its extracytosolic domain and is processed by S1P and S2P enzymes (40, 42).
Structurally, Luman has many of the characteristics of a RIP protein. It contains an amino-terminal region with an activation domain, a DNA binding domain, and a leucine zipper (23, 24, 26) (see Fig. 5). This amino-terminal portion is followed by a transmembrane domain and a carboxyl terminus of undefined function (23). In addition, in cells transiently expressing Luman, most of the protein is located in the ER (23). In this study, we wanted to determine if Luman is processed by proteolysis, leading to the translocation of its amino terminus to the nucleus. We show that, like SREBP and ATF6, Luman is a glycoprotein and that its amino terminus is released from the ER by intramembrane proteolysis to enter the nucleus.
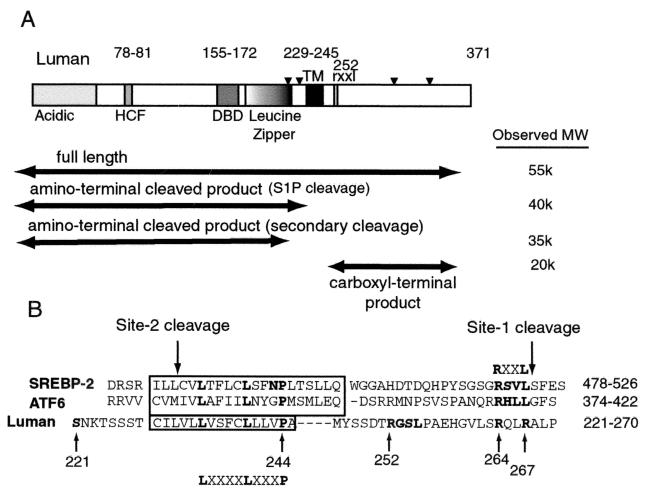
FIG. 5.
(A) Schematic representation of Luman, with observed molecular weights of products. Arrowheads, possible glycosylation sites; HCF, HCF binding domain; DBD, DNA binding domain; TM, transmembrane domain, rxxl, putative S1P recognition motif; Acidic, acidic activation domain. The molecular weights of Luman and its fragments were determined by comparing their electrophoretic mobilities with those of protein molecular weight markers electrophoresed on the same gel. (B) Comparison of amino acid sequences of the transmembrane regions of human SREBP-2, ATF6, and Luman. Potentially important amino acids in Luman and the sites for the S221Op, R252A, R264G, and R267G mutations are boldfaced and numbered. Downward arrows point to the S1P and S2P cleavage sites for SREBP-2, and the LXXXXLXXXP sequence in the transmembrane domains of the three proteins is indicated.
MATERIALS AND METHODS
Cell culture and materials.
Vero cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% newborn bovine serum and penicillin-streptomycin. All restriction endonucleases, DNA-modifying agents, and molecular reagents were obtained from Life Technologies unless stated otherwise. Cultures of neurons from rat dorsal root ganglia were prepared and maintained as described by Andersen et al. (2). To determine if Luman was glycosylated, either transfected cells were treated with tunicamycin (Sigma) or cell lysates were treated with endoglycosidase H (Endo H) or p-N-linked deglycosidation enzyme (PNGase F) (New England Biolabs) as directed by the manufacturer. Lysates representing equivalent numbers of cells were then separated by electrophoresis, and Luman polypeptides were detected by immunoblotting.
Plasmids.
The construction of pcLuman, which comprises the coding sequences for Luman linked at its amino terminus to oligonucleotides coding for the FLAG epitope, has been described previously (24). In this article this plasmid will be called pcFL-Lu. An influenza hemagglutinin (HA) epitope was attached to the carboxyl terminus of the coding sequence of full-length Luman by site-directed mutagenesis (see below) using oligonucleotide primers coding for the amino acid sequence YPYDVPDYA. This plasmid was called pcFL-Lu-HA. Plasmid TK3 (pTK3) was constructed by replacing a BglII-HindIII fragment, which contains the cytomegalovirus (CMV) promoter in pcDNA3 (Invitrogen), with a BglII-HindIII fragment containing the HSV thymidine kinase (TK) promoter from pRL-TK (Promega). Coding sequences for FL-Lu-HA and mutants of Luman were transferred to pTK3 from pcDNA3 as HindIII-XhoI fragments.
An additional construct (pJS6) for the expression of Luman, containing a different eptiope tag (SV5) at its N-terminal end together with the HA tag at its C terminus, was constructed by first inserting the SV5 epitope sequence between the HindIII and BamHI sites of pcDNA3 (Invitrogen) and then inserting the sequence for the HA epitope between the XhoI and XbaI sites. The coding sequence for Luman was amplified from pcLuman by using primers AAAAAGCTTATGGAGCTGGAATTGGATGC and GATCCTCGAGGCCTGAGTATCTGTCCTG. This amplified product was cloned between the BamHI and XhoI sites. An identical construct (pJS7) for the expression of doubly tagged LumanΔTM was constructed by using the same primers to amplify the coding sequence from pcLumanΔTM.
Plasmid pBB15 contains 143 bp of HSV DNA upstream of the start site of the latency-associated transcript (LAT) linked to coding sequences for chloramphenicol acetyltransferase (CAT) (4). Plasmids expressing S1P, pCMV-S1P(1-997)Myc-KDEL, and pCMV-S1P(1-997)Myc-KDAS, were kindly provided by Joseph Goldstein (University of Texas Southwestern Medical Center).
Site-directed mutagenesis.
Mutants were constructed by using the Quickchange site-directed mutagenesis kit (Stratagene) with modifications described by Wang and Malcolm (38). To replace serine 221 in the Luman coding sequence with the Opal stop codon TGA (S221Op), we used oligonucleotides GTGATTGAGATATGAAACAAAACCAGC and GCTGGTTTTGTTTCATATCTCAATCAC. The mutation removed a unique EcoRV site. To replace arginine 252 in the putative S1P cleavage site with alanine (R252A), we used oligonucleotides TACTCCTCTGACACAGCCGGGAGCCTGCCAGCT and AGCTGGCAGGCTCCCGGCTGTGTCAGAGGAGTA. The mutation also introduced a unique NciI site into the Luman coding sequences. To replace arginine 264 with glycine, the oligonucleotide pair GGAGTGTTGTCCGGGCAGCTTCGTGCC and GGCACGAAGCTGCCCGGACAACACTCC was used. The mutation introduced a unique NciI site. To replace arginine 267 with glycine, the oligonucleotide pair TCCCGCCAGCTTGGGGCCCTCCCCAGT and ACTGGGGAGGGCCCCAAGCTGGCGGGA was used. The mutation introduced a unique ApaI site. Mutants were first screened for the presence of the restriction sites introduced and then confirmed by sequencing using the Sequenase sequencing kit (Amersham).
Antibodies.
The preparation of antiserum against glutathione S-transferase (GST)-Luman has been described elsewhere (23). To remove antibodies against GST, the serum was diluted 1/10 in phosphate-buffered saline (PBS) containing 10% newborn calf serum and absorbed for 1 h at 4oC with glutathione Sepharose beads (Pharmacia) to which GST had been bound. Sodium azide was added to the supernatant (0.02%). To prepare negative serum, antibodies against both GST and Luman were removed by absorption with GST-Luman trapped on glutathione Sepharose beads. This absorption reduced the level of anti-Luman antibodies in the serum to the extent that we were not able to use it to detect Luman in transfected-cell lysates by immunofluorescence, immunohistochemistry, or immunoblotting. Monoclonal antibodies against influenza virus HA and against FLAG were purchased from Sigma, monoclonal antibodies against Myc were purchased from Invitrogen, and polyclonal antibodies against the amino terminus of calnexin were purchased from Stressgen.
Transfections, immunofluorescence, and assays for transcription activation.
Plasmid DNA was introduced into cells by the calcium phosphate method as previously described (10, 23). Vero cells or COS cells in six-well plates were plated at densities of 2.5 × 105 cells per well for immunofluorescence and 1 × 106 cells per well for CAT assays. One hundred-millimeter-diameter plates were seeded at a density of 4 × 106 cells per plate. Five micrograms of DNA was used to transfect cells in six-well plates, and 20 μg of DNA was used for the 100-mm-diameter plates. For CAT assays, 250 ng of pCMVBGal, a plasmid specifying beta-galactosidase (β-Gal), was added to each transfection. Lysates were assayed for β-Gal (36) and for CAT by using an enzyme-linked immunosorbent assay kit (Roche). CAT values were adjusted for transfection efficiency by using β-Gal values.
During the addition of proteosome inhibitors, cells were incubated at 37°C for 20 h after transient transfection. The medium was removed and replaced with fresh medium containing 25 μg of N-acetyl-leucinal-leucinal-norleucinal (ALLN)/ml or 5 μM MG132 (Calbiochem). ALLN and MG132 were dissolved in dimethyl sulfoxide. Nontreated cells received equivalent amounts of dimethyl sulfoxide. Endo H and PNGase F experiments were performed according to the supplier's instructions (New England Biolabs). For treatment with brefeldin A (Sigma), transfected cells were treated with 1 μg of brefeldin per ml of medium. Cells were harvested 7 h after addition of brefeldin A. Stock solutions (5 mg/ml) of the drug were prepared in absolute alcohol. Control cultures received an equivalent amount of alcohol.
Cells were processed for immunofluorescence as outlined previously (23) and were observed with a Zeiss Axioskop microscope with an epifluorescence attachment. Images were captured by using the Northern Eclipse image analysis system and software (Empix Imaging, Inc.). Final images and figures for this report were prepared by using Adobe Illustrator 7.0 and Adobe Photoshop 4.0.
In additional experiments (see Fig. 3), transfected COS cells (grown on coverslips) were fixed for 15 min in methanol at −20°C. Coverslips were blocked with 10% newborn calf serum in PBS and then incubated with the primary antibody (an anti-SV5 monoclonal antibody at a dilution of 1/1,000) in blocking solution. After incubation at 37°C for 30 min, coverslips were washed three times in PBS. The secondary antibody was an anti-mouse antibody tagged with Alexa488 (Molecular Probes) which was applied at a dilution of 1/200. Dual-channel fluorescence images were acquired on a Zeiss LSM410 laser scanning confocal microscope.
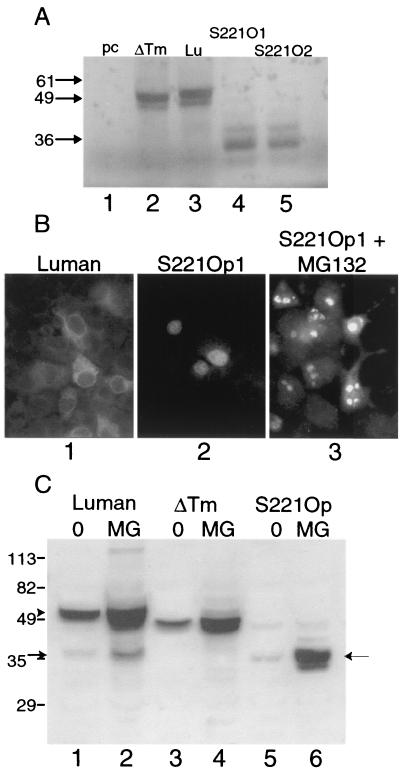
FIG. 3.
A construct expressing the amino-terminal portion of Luman. (A) Plasmids specifying Lu or ΔTm, and two independent clones in which the codon for serine 221, which lies just amino-terminal to the transmembrane domain, was replaced with an Opal codon (S221O1 and S221O2), were expressed in the TnT in vitro transcription-translation system and analyzed on an SDS-10% polyacrylamide gel. (B) Detection of Luman and Luman S221Op in transfected cells by immunofluorescence. Note the cytoplasmic location of Luman and the nuclear location of S221Op as well as the relatively few cells that show detectable S221Op. Panel 3 shows S221Op-expressing cells treated with the proteosomal inhibitor MG132. (C) Immunoblots of Luman, Luman ΔTm, and Luman S221Op expressed in Vero cells with or without MG132, separated on an SDS-8% polyacrylamide gel. The arrowhead and arrow on the left indicate full-length Luman and the potential amino-terminal cleaved product of Luman, respectively. The arrow on the right indicates S221Op. Note that relatively little S221Op was detected unless proteosomal digestion was suppressed by MG132.
Immunoblotting.
Proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels with 8, 10, or 12% polyacrylamide and were blotted to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad). Protein bands were detected by standard immunoblotting procedures with various antibodies and chemiluminescence by use of a Western blotting Detection System kit (Amersham Pharmacia Biotech). The molecular weights of bands were calculated from the electrophoretic mobilities of BenchMark prestained protein markers (Life Technologies).
Cellular fractionation.
Nuclear and membrane fractions were prepared by a modification of a procedure described by Wang et al. (39). Vero cells in 100-mm-diameter dishes were transfected with 20 μg of plasmid DNA per dish. Two days later, cells were scraped into PBS and collected by centrifugation. They were then resuspended in 10 ml of hypotonic buffer B (10 mM HEPES-KOH [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, and 0.5 mM EGTA) and kept on ice for 10 min. Cells were then collected by centrifugation and again resuspended in buffer B (5 times the packed cell volume) containing the proteolytic inhibitors pepstatin, aprotinin, and E64. After being kept on ice for 10 min, the cells were passed through a 1-in., 22-gauge needle 15 times and then through a 1-in., 23-gauge needle 5 times. Removal of the cell membrane was monitored by observing 10 μl of the cells under a microscope after staining with trypan blue. The procedure resulted in about 80% of the cells being reduced to blue-staining nuclei. Nuclei were collected by centrifugation at 10,000 × g for 5 min and were extracted in hypertonic buffer C (20 mM HEPES-KOH [pH 7.4], 25% glycerol, 0.5 M NaCl, 1.5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA) containing protease inhibitors. The 10,000 × g supernatant was centrifuged at 100,000 × g for 1 h to pellet cell membranes.
RESULTS
Luman is modified posttranslationally.
The molecular weight of Luman, calculated from its derived amino acid sequence, is about 44K. Luman produced in vitro by use of the TnT transcription-translation system (Promega), which is limited in its ability to posttranslationally modify proteins, had an electrophoretic mobility consistent with a protein of about 50K (Fig. 1A, lane 3). In contrast, Luman detected in lysates of transfected cells by using anti-Luman serum was larger, with a molecular weight of 55K to 60K (Fig. 1A, lane 1). At 55K Luman appeared to be considerably larger than its predicted molecular weight of 44K or the apparent molecular weight of the protein produced in vitro, suggesting that it was modified posttranslationally.
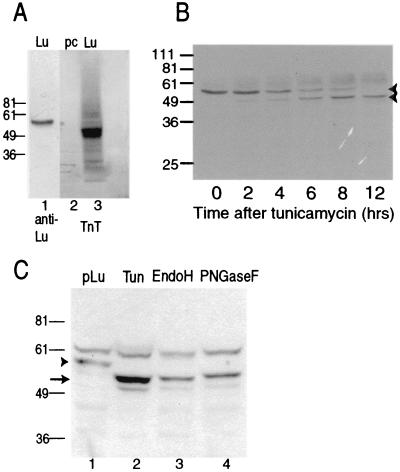
FIG. 1.
Luman is posttranslationally modified by glycosylation. (A) Immunoblot of Luman from transfected Vero cells (lane 1) and autoradiograph of the in vitro TnT system (Promega) charged with pcDNA3 (lane 2) or pcFL-Lu (lane 3). Cell and TnT lysates were separated on an SDS-8% polyacrylamide gel and blotted. Strips were then analyzed by immunoblotting or by autoradiography. (B) Immunoblot analysis of Luman-expressing cells after incubation in the presence of tunicamycin. Vero cells in six-well plates were transfected with pcFL-Lu. After 24 h, the medium was replaced with fresh medium containing tunicamycin at a concentration of 2 μg/ml. At the indicated times, cells were lysed, separated on an SDS-10% polyacrylamide gel, and analyzed by immunoblotting with an anti-Luman antibody. (C) Luman is glycosylated. Lysates of Luman-expressing cells treated with tunicamycin (Tun) or membranes from fractionated Luman-expressing cells that were either left untreated (pLu) or digested with Endo H or PNGase F were separated on an SDS-10% polyacrylamide gel and analyzed by immunoblotting. Lanes represent lysates from equivalent numbers of cells. The positions of protein molecular weight markers (in thousands) electrophoresed on the same gels are indicated on the left. The arrowhead points to Luman, while the arrow indicates unglycosylated (lane 2) or deglycosylated (lanes 3 and 4) Luman. The 61K band present in all lanes is a cellular protein that reacts nonspecifically to the anti-Luman serum.
We next investigated whether Luman contained N-linked carbohydrate. Tunicamycin, which blocks N-linked glycosylation, was added to cells 24 h after transfection with pcFL-Lu, and samples were collected every 2 h (Fig. 1B). Before treatment with tunicamycin and as long as 6 h after treatment, Luman was predominantly a 55K protein. Two hours after treatment, a smaller band at 50K appeared, and after 12 h in the presence of tunicamycin, the 50K band predominated.
Sequence analysis showed that Luman contains four potential sites for N-linked glycosylation (see Fig. 5A). To confirm that the 50K protein, made in the presence of tunicamycin, represented an unglycosylated form of Luman, lysates of transfected cells were incubated with either Endo H or PNGase F to remove carbohydrate side chains. As shown in Fig. 1C, addition of Endo H (lane 3) or PNGase F (lane 4) converted the 55K Luman polypeptide to a band with an apparent molecular weight of 50K. These results indicate that Luman, like ATF6 (16), contains N-linked carbohydrate chains that account, at least partially, for its larger apparent molecular weight. All detectable Luman in the cells was reduced to a lower-molecular-weight band by Endo H or PNGase F, suggesting that it had not been moved into the Golgi apparatus for oligosaccharide modification. These results support our previous observations (23) that most of the Luman in the cells is associated with the ER.
The amino-terminal portion of Luman.
In addition to the 55K-to-60K glycosylated Luman, immunoblots of cells expressing the protein occasionally revealed a 40K polypeptide (Fig. 2A, lane 1) that reacted both to a polyclonal anti-Luman serum and to monoclonal antibodies directed against an artificial amino-terminal epitope. To determine if this polypeptide represented the amino-terminal product of RIP, we treated Luman-expressing cells for increasing amounts of time with brefeldin A. As a control, cells expressing Luman ΔTm, a mutant of Luman that lacks its transmembrane domain and is therefore not retained in the ER (23), were treated in a similar manner. For SREBP and ATF6, brefeldin A promotes cleavage of the ER-resident proteins by S1P. The active form of the protease is resident in the Golgi apparatus but refluxes to the ER in the presence of brefeldin A. Over a period of 8 h, brefeldin A induced the specific accumulation of the 40K protein (Fig. 2A, lanes 2 to 7). In contrast, brefeldin A had no effect on Luman ΔTm (Fig. 2A, lanes 9 to 14). These results indicate that a Golgi apparatus-resident enzyme was responsible for cleavage of ER-associated Luman. Note that while brefeldin A treatment caused the accumulation of the 40K protein, the relative amounts of the intact and cleaved products varied. On certain occasions cleavage was quantitative (see Fig. 4 and 6), while on others substantial amounts of the intact protein remained (Fig. 2).
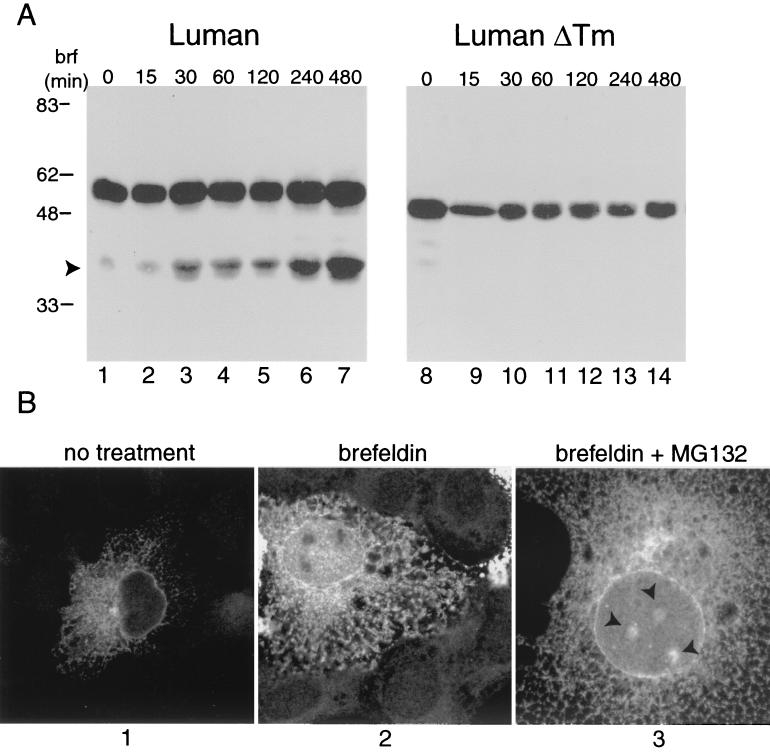
FIG. 2.
Effect of brefeldin A on Luman. (A) COS cells transfected with 2 μg of expression vectors for Luman (lanes 1 to 7) (pJS6) or Luman ΔTM (lanes 8 to 14) (pJS7). At 24 h posttransfection, the medium was replaced with Dulbecco's modified Eagle's medium containing 1 μg of brefeldin A(brf)/ml, and cells were incubated for the times indicated. Total-cell extracts were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis using an SV5 monoclonal antibody (dilution, 1/10,000) that recognized the extreme amino termini of the Luman proteins. Arrowhead indicates the 40K polypeptide. (B) COS cells grown on coverslips were transfected with pJS6. At 24 h posttransfection, cells were treated with 1 μg of brefeldin A/ml alone or with 10 μM MG132, as indicated. Cells were fixed in methanol and examined by confocal microscopy. Arrowheads indicate sites of Luman accumulation in the nucleus.
FIG. 4.
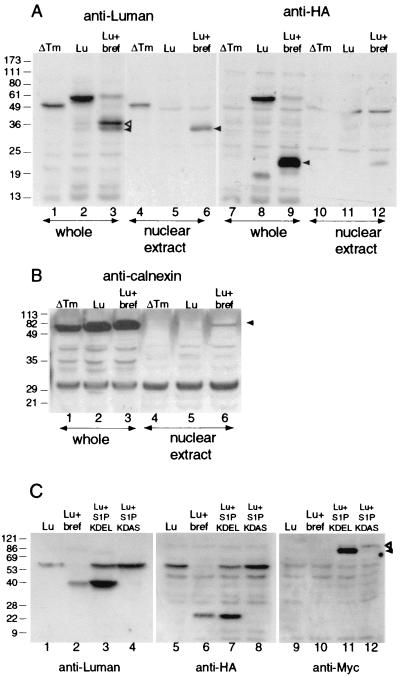
Effect of brefeldin A on Luman. (A) Detection of brefeldin A-generated fragments in the nucleus. Cells were transfected with pcFL-Lu[ΔTM] (ΔTM) or pcFL-Lu-HA (Lu). All cultures were treated with MG132 to suppress degradation of the activated amino-terminal Luman fragment. One culture of cells transfected with pcFL-Lu-HA was also treated with brefeldin A (Lu + bref) from 12 h after transfection. Forty hours after transfection, cells were harvested and fractionated to collect the nuclear fraction, and whole and nuclear fractions were separated on an SDS-12% polyacrylamide gel and immunoblotted with antibodies against Luman or HA. Open arrowhead in lane 3, 40K polypeptide; solid arrowhead in lanes 3 and 6, 35K polypeptide; solid arrowhead in lane 9, ∼20K fragment. (B) Cells were treated as described in the legend to panel A and immunoblotted with antibodies against calnexin (arrowhead). (C) Cultures were transfected with pcFL-Lu-HA either alone or with pS1P-KDEL or pS1P-KDAS. Cultures transfected with pcFL-Lu-HA were either left untreated or treated with brefeldin A. Lysates were electrophoresed on SDS-10% polyacrylamide gels and immunoblotted with an anti-Luman serum or an anti-HA or anti-Myc monoclonal antibody. Open arrowhead, S1PA; solid arrowhead, S1PB and S1PC (14).
FIG. 6.
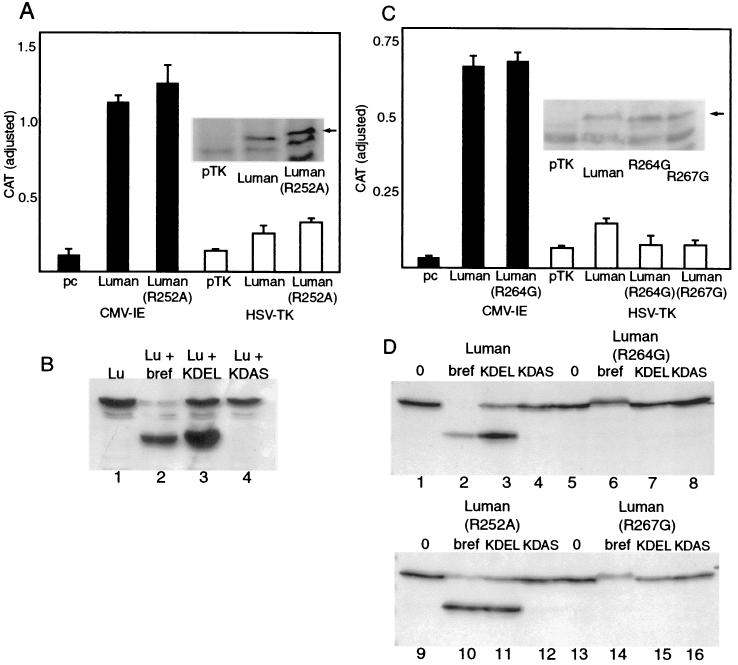
Effects of mutation of arginines 252, 264, and 267 at the putative S1P cleavage sites on generation of the amino-terminal cleaved fragment of Luman. (A) Vero cells were transfected with plasmids expressing either FL-Lu-HA (Luman) or FL-Lu-HA(R252A) expressed from the CMV IE promoter or an HSV TK promoter. The CMV IE and HSV TK promoters containing blank plasmids, pcDNA3 and pTK3, were used as controls. Cells were cotransfected with pBB15, a reporter plasmid with coding sequences for CAT linked to the basal HSV LAT and pCMV-β-Gal. CAT activity is adjusted for transfection efficiency. (Inset) Western blot of lysates of cells transfected with the pTK constructs. Arrow indicates the location of bands representing Luman and Luman(R252A). The lower-molecular-weight band in all lanes is due to nonspecific binding of the anti-Luman antibody. (B) Lysates of cells expressing LuFL-Lu-HA(R252A), either left untreated (Lu), treated with brefeldin A (Lu + bref), or cotransfected with either pS1P-KDEL or pS1P-KDAS, were separated on SDS-10% polyacrylamide gels and probed with anti Luman serum. (C) Cells transfected with pBB15 and (from left to right) either pcDNA, pcFL-Lu-HA (Luman), pcFL-Lu-HA(R264G), pTK3, pTKFL-Lu-HA (Luman), pTKFL-Lu-HA(R264G), or pTKFL-Lu-HA(R267G) were assayed for CAT activity. (Inset) Western blot of lysates of cells transfected with the pTK constructs. (D) Cells transfected with plasmids expressing Luman or its mutants (R252A, R264G, or R267G), either alone or in conjunction with plasmids expressing either S1PKDEL or S1PKDAS, were analyzed by immunoblotting. Cells expressing Luman or mutant proteins alone were either left untreated (lanes 1, 5, 9, and 13) or treated with brefeldin A (lanes 2, 6, 11, and 14).
Consistent with the accumulation of the 40K cleavage product, brefeldin A treatment also led to an increase in accumulation of nuclear Luman as detected with the N-terminal tag (Fig. 2B; compare panels 1 and 2). Again, though, it should be noted that substantial amounts of Luman still remained associated with the ER in the presence of the drug. Addition of the proteosomal inhibitor MG132 as well as brefeldin A caused accumulation of Luman in nuclear structures (Fig. 2B, panel 3).
To help us characterize the amino-terminal portion of Luman, we replaced a serine residue (S221) that lies just upstream of the transmembrane region with an Opal termination codon (S221Op). Since the products of RIP are generally short-lived, we examined two independent S221Op clones in the TnT in vitro transcription-translation system. Presumably, proteins produced in this system are less likely to be targeted for proteolytic degradation. Both clones produced proteins of about 35K (Fig. 3A, lanes 4 and 5). Plasmids expressing full-length Luman or Luman ΔTm specified larger proteins of about 50K or 49K, respectively. In contrast to the situation in cells expressing full-length Luman, Luman S221Op was located mainly in the nuclei of cells (Fig. 3B), although considerably fewer cells contained S221Op than full-length Luman when comparable amounts of plasmid DNA were used for transfection. The low number of cells expressing Luman S221Op was reflected in the small amount of protein detected in immunoblots (Fig. 3C, lane 5). This reduction in the level of Luman S221Op protein in cells was probably due to rapid proteolytic degradation of the protein, since transfected cells treated with the proteosome inhibitor MG132 contained levels of protein comparable to those in cells expressing full-length Luman (Fig. 3C, lane 6). In cultures expressing Luman S221Op and treated with MG132, this protein was detected in as many cells as full-length Luman (Fig. 3B, panel 3). In these cells Luman S221Op was concentrated in what appeared to be nuclear or juxtanuclear structures.
Together our results show that treatment with brefeldin A causes the accumulation of a specific cleavage product of Luman (Fig. 2) and that this protein (40K) has an electrophoretic mobility consistent with a molecule slightly larger than the unstable mutant of Luman (S221Op) which would resemble the putative product of RIP (Fig. 3). We next determined whether the 40K Luman product induced in brefeldin A-treated cells was selectively located in the nuclei of cells. We treated Luman-expressing cells with brefeldin A and analyzed whole-cell and nuclear extracts. As a control we examined untreated cells expressing Luman ΔTm. Since the products of RIP are usually degraded rapidly, we also treated cells with the proteosomal inhibitor MG132. Following 7 h of treatment with brefeldin A, most of the Luman in the cell lysate was a doublet of about 40K and 35K (Fig. 4A, lane 3). Much of the faster-migrating protein in the doublet could be eluted from the nuclei of cells (Fig. 4A, lane 6). In parallel experiments, Luman ΔTm could also be eluted from nuclei (Fig. 4A, lane 4), while full-length Luman was absent from nuclear extracts (lane 5). The 40K and 35K proteins in brefeldin A-treated cells did not contain the carboxyl-terminal HA epitope (Fig. 4A, lane 9), suggesting that they represented the amino-terminal portion of Luman. In brefeldin A-treated cells, most of the HA epitope was associated with a fragment of about 20K (Fig. 4A, lane 9) and only very minute amounts of this polypeptide were associated with nuclear extracts. To confirm that the nuclear extracts were uncontaminated by ER proteins, we screened all samples for calnexin, which resides in the ER. This protein, while abundant in whole-cell lysates, was largely absent from the nuclear fractions (Fig. 4B; compare lanes 1, 2, and 3 with lanes 4, 5, and 6). The relatively small level of calnexin contamination of the nuclear fraction of brefeldin A-treated cells (Fig. 4B, lane 6) does not account for the almost quantitative recovery of the 35K but not the 40K band in this fraction. Incidentally, the 35K band in brefeldin A-treated cells has the same electrophoretic mobility as the Luman S221Op product (data not shown).
Cleavage of Luman can be promoted by location of active S1P in the ER.
The initial cleavage of SREBP by S1P requires the SCAP-mediated transport of SREBP to the Golgi apparatus. S1P itself is initially present in an inactive state in the ER as S1PA. It is transported to the Golgi apparatus and activated by self-catalyzed cleavage (S1PC) (14). Reflux of active S1P to the ER by brefeldin A or expression of an active form of S1P, linked to the ER retrieval signal, KDEL, obviates the need for SCAP, leading to the processing of ER-associated SREBP.
Since our experiments with brefeldin A suggested that a protease resident in the Golgi apparatus might catalyze the cleavage of ER-associated Luman, we determined whether S1P could cleave Luman. We either treated Luman-expressing cells with brefeldin A or cotransfected them with a plasmid expressing either S1P-KDEL, which is retained in the ER, or S1P-KDAS, which is not. In S1P-KDEL-expressing cells, most of the Luman was reduced to a 40K band (Fig. 4C, lane 3) that comigrated with the 40K Luman band in brefeldin A-treated cells (lane 2). Like the brefeldin A-generated band, the 40K band in cells expressing S1P-KDEL also did not contain the carboxyl-terminal HA epitope (Fig. 4C, lanes 6 and 7). Both brefeldin A-treated and S1P-KDEL-expressing cells contained a comigrating HA-linked fragment of about 20K (Fig. 4C, lanes 6 and 7). A probe of the immunoblots with antibodies against a Myc epitope, which is linked to S1P-KDEL and KDAS (Fig. 4C, lanes 9 to 12), confirmed that while S1P-KDEL-expressing cells contained the faster-migrating, active form of S1P (lane 11), cells expressing S1P-KDAS contained only the slower-migrating, inactive form (lane 12). These results strongly suggest that the cleavage of Luman in brefeldin A-treated cells is catalyzed by S1P. Surprisingly, while treatment of cells with brefeldin A and expression of S1P-KDEL had profound effects on the integrity of full-length Luman as seen on immunoblots (Fig. 4C, lanes 2 and 3), the treatments caused only a small, though consistent, increase in nuclear fluorescence. Most of the protein remained in the ER (data not shown). This is probably because while S1P separates the amino and carboxyl portions of Luman, the amino portion remains associated with the ER, and as with SREBP and ATF6, its release from membranes requires a second cleavage catalyzed by an as yet unidentified protease.
Site of S1P cleavage.
Seven amino acids downstream from the transmembrane domain of Luman is the sequence RGSL (beginning at residue 252), which resembles the RXXL motif identified as the recognition site for S1P in SREBP and ATF6 (Fig. 5B). While the protein is cleaved after the leucine residue in the motif, replacement of the arginine in the motif with any amino acid except lysine blocks cleavage of SREBP (12) and ATF6 (42) by S1P. To determine if the RGSL sequence in Luman represents its S1P cleavage site, we mutated the arginine residue in the motif to alanine (R252A). Figure 6A and B show that the mutation had no effect on the ability of Luman, expressed from either the CMV immediate-early (IE) promoter or the HSV TK promoter, to activate the HSV LAT promoter (pBB15). The mutation also failed to block the processing of the protein in brefeldin A-treated or S1P-KDEL-expressing cells (compare Fig. 4C, lanes 1 to 4, with Fig. 6D, lanes 9 to 12). However, there are two additional arginine residues located just downstream of R252, at positions 264 and 267, within the RQLR sequence, which retains close similarity to the RXXL S1P cleavage sites in SREBP and AFT6. To determine if R264 or R267 plays a role in S1P cleavage, we changed either residue to glycine. Figure 6D shows that in contrast to wild-type Luman and Luman R252A, mutation of either R264 or R267 eliminated cleavage in brefeldin A-treated cells (compare lanes 2 and 10 with lanes 6 and 14) and in cells expressing S1P-KDEL (compare lanes 3 and 11 with lanes 7 and 15). These results suggested that both R264 and R267 contribute to the S1P cleavage site in Luman. However, like R252A, neither R264G nor R267G had an effect on the ability of Luman, expressed from the CMV IE promoter, to activate the HSV LAT promoter (Fig. 6C, first three bars; also data not shown). Since cells expressing large amounts of Luman from the CMV IE promoter might process the protein by an aberrant pathway, we examined the mutants cloned into pTK3. Comparable levels of the mutant proteins and Luman were expressed from the less-active HSV TK promoter (inset). However, the mutants were unable to activate transcription from the HSV LAT promoter (Fig. 6C). This is in contrast to the R252A mutation, which, even when the mutant was expressed from the HSV TK promoter, failed to suppress the ability of Luman to activate transcription (compare open bars in Fig. 6A and C). Our results suggest that while S1P activates Luman by cleavage at the RQLR sequence, in cells expressing large amounts of Luman the transcriptional factor is delivered to the nucleus by an alternative pathway. While brefeldin A in most cases led to the almost-quantitative conversion of the 60K full-length Luman to the 40K cleaved product (Fig. 4C, lane 2, and 6D, lane 2), it did not improve the ability of Luman, expressed either from the CMV IE or from the HSV TK promoter, to activate the HSV LAT promoter (data not shown). This suggests that a second cleavage, required for release of Luman from the ER membrane, may be a rate-limiting step in the proteolytic activation of Luman.
DISCUSSION
Luman/LZIP/CREB3 is a basic leucine zipper-containing human transcription factor that, like the HSV transactivator VP16, requires HCF for activity. It is present in neurons of mammalian dorsal root ganglia, the primary site of latency for alphaherpesviruses, and it can activate promoters of HSV genes that may be responsible for the establishment of, and reactivation from, latency (23). These observations led us to hypothesize that the normal cellular role of Luman is to activate gene expression in response to stress and that herpesviruses may have evolved to use it as a trigger for reactivation. Recently, we observed that Luman is retained in the ER, and we proposed that it may be processed in a manner similar to that of certain other regulatory proteins that are sequestered in cellular membranes and are activated by RIP. The RIP process allows cells to respond rapidly to physiological crises by activating premade transcription factors. Here we report further characterization of Luman and provide results which indicate that Luman is indeed a member of the subclass of membrane-anchored transcription factors which are activated by RIP and that S1P, the Golgi apparatus-resident protease that processes SREBP and ATF6, is involved.
We determined that Luman is a glycoprotein (Fig. 1). On SDS-polyacrylamide gels Luman had an electrophoretic mobility consistent with a molecular weight of 55K to 60K. Based on amino acid composition, the predicted molecular weight of Luman is about 44K (24). The increased electrophoretic mobility of Luman is at least partly due to glycosylation, since treatment with tunicamycin or removal of N-linked carbohydrate side chains by Endo H or PNGase F yielded a 50K protein (Fig. 2). Luman contains four possible N-linked glycosylation sites at amino acid residues 203, 222, 307, and 348. SREBP and ATF6 are also modified posttranslationally by glycosylation (16, 43, 44), although other forms of modification that might alter electrophoretic mobility, such as phosphorylation, ubiquitination, methylation, and acetylation, cannot be ruled out.
Our results are consistent with the following model for the arrangement of Luman in the ER and its release by RIP. Luman is embedded in ER membranes, with its amino terminus directed toward the cytoplasm and its carboxyl terminus in the lumen of the ER. The process of activation begins with the transport of the protein to the Golgi apparatus by an as yet unknown mechanism. Here the first proteolytic cleavage of the protein by S1P at the sequence RQLR generates a membrane-associated 40K amino-terminal fragment and a 20K luminal carboxyl-terminal fragment. A second cleavage, possibly by S2P, leads to release of the amino-terminal fragment, which is transported to the nucleus to activate downstream genes. All cleavage products of Luman, particularly the transcriptionally active nuclear fragment, are degraded rapidly.
The transport of Luman from the ER to the Golgi apparatus in cultured cells and its subsequent cleavage are inefficient, and only trace amounts of the cleavage products are observed in the absence of brefeldin A or S1P-KDEL (Fig. 2A, lane 1). For SREBP, the protein SCAP is key to the activation of SREBP by sterols (17, 29-31). SCAP escorts SREBP to the Golgi apparatus in preparation for S1P cleavage. A portion of the membrane attachment domain of SCAP contains a sterol-sensing region (29, 30), and in the presence of sterols the ability of SCAP to escort SREBP to the Golgi apparatus is suppressed. No similar accessory protein with a regulatory role has been identified for ATF6, and we have not identified such a partner for Luman. HCF, the only protein known to associate with Luman, probably does not fulfill this role, in view of the fact that a mutant of Luman in which all three conserved amino acids in the HCF binding domain were altered (DHTY78AGTA) was still processed to 40K and 20K polypeptides in the presence of brefeldin A or S1P-KDEL (data not shown).
The primary cleavage of Luman likely occurs in the Golgi apparatus and is catalyzed by S1P, since brefeldin A, which causes a reflux of Golgi apparatus enzymes to the ER, increased the efficiency of the cleavage (Fig. 2A and 4A). and S1P-KDEL, targeted to the ER, also resulted in the 40K and 20K amino- and carboxyl-terminal fragments (Fig. 4C; compare lane 2 with lane 3 and lane 6 with lane 7). In SREBP and ATF6 the recognition site for S1P has been identified as the sequence RXXL. S1P cleaves the bond connecting leucine to the next amino acid (12, 40, 42). For these proteins the arginine at position 4 in the motif can be replaced by lysine with some decrease in the cleavability of the protein; however, replacement with alanine prevents cleavage of the protein. The leucine at position 1 appears to be more flexible, as replacement with arginine or phenylalanine leads only to some decrease in cleavability, although replacement with valine abolishes it (12). Luman also contains an RXXL (RGSL) motif downstream of the transmembrane domain. This motif is conserved in the bovine homologue of Luman (unpublished data), while in the mouse homologue, LZIP, the terminal leucine in the motif is replaced with a valine. Replacement of leucine with valine at position 1 abolishes cleavage of SREBP (12) and greatly reduces cleavage of ATF6 (42). Our results show that instead of the RGSL sequence, the S1P recognition sequence in Luman is RQLR. Mutation of either of the arginine residues in the motif (Fig. 6D) blocked cleavage of Luman in either brefeldin A-treated or S1P-KDEL-expressing cells. Both mutations also eliminated the ability of Luman, expressed from the HSV TK promoter, to activate transcription. Interestingly, blocking of cleavage by S1P did not affect the ability of Luman R264G or Luman R267G to activate transcription if the proteins were expressed from the more powerful CMV IE promoter (Fig. 6C). This suggests that, as with ATF6 (42), expression of large amounts of Luman leads to aberrant processing of the molecule, overriding the S1P-catalyzed process.
As seen with SREBP and ATF6 the amino-terminal fragment of Luman, generated as a result of S1P cleavage, remained associated with cellular membranes. While in both brefeldin A-treated and S1P-KDEL-expressing cells, most of Luman was converted to the 40K product (Fig. 4C, lanes 2 and 3), most of the protein remained associated with the ER. In addition, brefeldin A did not improve the ability of Luman to activate transcription. This suggests that a second cleavage is required for release of Luman from the ER and translocation to the nucleus. For SREBP and ATF6 this enzyme has been identified as S2P. No stringent requirements for particular amino acids at the S2P cleavage have been identified (31), although there is a requirement for asparagine and proline residues in the transmembrane domains of SREBP. The residues are adjacent in SREBP and are separated by 2 amino acids in ATF6. Mutation of either residue in SREBP has no effect, but simultaneous alteration of both residues prevents S2P cleavage (41), suggesting that the requirement for both residues is not absolute. In addition to the asparagine residue, the transmembrane regions of SREBP and ATF6 contain the sequence LXXXXLXXXP. Although the transmembrane domain of Luman does not contain an asparagine residue, it does contain a proline as part of the LXXXXLXXXP motif (Fig. 5B). This similarity among the transmembrane domains of the three proteins suggests that, although we have not yet identified an enzyme responsible for the secondary cleavage of Luman, the three proteins may be released from the membrane by similar mechanisms.
The released amino-terminal products of both SREBP and ATF6 are extremely unstable and are seen in the nucleus only in the presence of inhibitors of proteosomal degradation. In agreement with these observations, when we treated Luman-expressing cells with brefeldin A as well as the proteosomal inhibitor MG132, whole-cell lysates contained a doublet of about 40K and 35K (Fig. 4A, lane 3). Only the lower band (Fig. 4A, lane 7) was associated with nuclear extracts, suggesting that this might represent the active amino-terminal form of Luman, which is normally rapidly destroyed by proteosomal degradation. The instability of the amino-terminal product could be demonstrated by the extreme instability of the Luman S221Op mutant, which should structurally resemble the released amino-terminal portion of Luman. While almost comparable amounts of Luman and Luman S221Op were made in vitro (Fig. 3A; compare lane 3 with lanes 4 and 5), the same plasmids yielded vastly disparate amounts of protein when transfected into cells (Fig. 3C; compare lanes 1 and 5). The S221Op protein was barely visible in immunoblots unless proteosomal degradation was suppressed with MG132 (Fig. 3C, lane 6). Surprisingly, while both S221Op and ΔTm mutants accumulated in the nuclei of cells, they were considerably less efficient at activating transcription than wild-type Luman (data not shown). The instability of the mutant proteins cannot entirely explain this decrease in transcriptional efficiency, since the released form of Luman would also be as unstable. Perhaps Luman must be processed correctly through a membrane-linked pathway for its amino-terminal portion to function as an efficient activator.
In summary, our results show that Luman is a member of small class of transcription factors whose main mechanism of control is RIP. We are currently attempting to identify compounds or treatments that would induce different forms of cellular stress and stimulate Luman cleavage. Of various compounds tested to date, including tunicamycin, the calcium ionophore A23187, the phorbol ester PMA (phorbol 12-myristate-13-acetate) (19, 21), and inducers known to activate herpesvirus latency (dibutyl cAMP, sodium butyrate, and dexamethasone), none have induced significant increases in the amino-terminal cleavage product of Luman. However, it is possible that, as with other proteins in this class, overproduction of Luman from the CMV promoter may mask the effects of these inducers. Further analyses, including, e.g., the use of weaker promoters for expression of more physiological levels of Luman, are currently under way.
Acknowledgments
This work was funded by an operating grant to V.M. from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
We thank Michael Brown and Joseph Goldstein for providing S1P-KDEL- and S1P-KDAS-expressing plasmids.
REFERENCES
- 1.Abel, T., R. Bhatt, and T. Maniatis. 1992. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 6:466-480. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, P. L., C. A. Webber, K. A. Kimura, and D. J. Schreyer. 2000. Cyclic AMP prevents an increase in GAP-43 but promotes neurite growth in cultured adult rat dorsal root ganglion neurons. Exp. Neurol. 166:153-165. [DOI] [PubMed] [Google Scholar]
- 3.Annaert, W., and B. De Strooper. 1999. Presenilins: molecular switches between proteolysis and signal transduction. Trends Neurosci. 22:439-443. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, A. H., and P. O'Hare. 1990. Regulation and cell type-specific activity of a promoter located upstream of the latency-associated transcript of herpes simplex virus type 1. J. Virol. 64:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. [DOI] [PubMed] [Google Scholar]
- 6.Brown, M. S., and J. L. Goldstein. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96:11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 8.Burbelo, P. D., G. C. Gabriel, M. C. Kibbey, Y. Yamada, H. K. Kleinman, and B. S. Weeks. 1994. LZIP-1 and LZIP-2: two novel members of the bZIP family. Gene 139:241-245. [DOI] [PubMed] [Google Scholar]
- 9.Chan, Y. M., and Y. N. Jan. 1999. Presenilins, processing of beta-amyloid precursor protein, and Notch signaling. Neuron 23:201-204. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. A., and H. Okayama. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques 6:632-638. [PubMed] [Google Scholar]
- 11.DeBose-Boyd, R. A., M. S. Brown, W. P. Li, A. Nohturfft, J. L. Goldstein, and P. J. Espenshade. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99:703-712. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, E. A., M. S. Brown, J. L. Goldstein, and J. Sakai. 1997. Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the luminal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272:12778-12785. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, E. A., U. P. Dave, J. Sakai, J. L. Goldstein, and M. S. Brown. 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273:17801-17809. [DOI] [PubMed] [Google Scholar]
- 14.Espenshade, P. J., D. Cheng, J. L. Goldstein, and M. S. Brown. 1999. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274:22795-22804. [DOI] [PubMed] [Google Scholar]
- 15.Freiman, R. N., and W. Herr. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua, X., J. Sakai, Y. K. Ho, J. L. Goldstein, and M. S. Brown. 1995. Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J. Biol. Chem. 270:29422-29427. [DOI] [PubMed] [Google Scholar]
- 18.Jin, D. Y., H. L. Wang, Y. Zhou, A. C. Chun, K. V. Kibler, Y. D. Hou, H. Kung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozutsumi, Y., M. Segal, K. Normington, M. J. Gething, and J. Sambrook. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462-464. [DOI] [PubMed] [Google Scholar]
- 20.Kristie, T. M., J. L. Vogel, and A. E. Sears. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. USA 96:1229-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, A. S. 1992. Mammalian stress response: induction of the glucose-regulated protein family. Curr. Opin. Cell Biol. 4:267-273. [DOI] [PubMed] [Google Scholar]
- 22.Li, M., P. Baumeister, B. Roy, T. Phan, D. Foti, S. Luo, and A. S. Lee. 2000. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, R., and V. Misra. 2000. Potential role for Luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J. Virol. 74:934-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, R., P. Yang, P. O'Hare, and V. Misra. 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, R., P. Yang, S. Padmakumar, and V. Misra. 1998. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. J. Virol. 72:6291-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciano, R. L., and A. C. Wilson. 2000. N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc. Natl. Acad. Sci. USA 97:10757-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan, D. R., M. J. Gething, and J. Sambrook. 1994. The cellular response to unfolded proteins: intercompartmental signaling. Curr. Opin. Biotechnol. 5:540-545. [DOI] [PubMed] [Google Scholar]
- 28.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 29.Nohturfft, A., R. A. DeBose-Boyd, S. Scheek, J. L. Goldstein, and M. S. Brown. 1999. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. USA 96:11235-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nohturfft, A., D. Yabe, J. L. Goldstein, M. S. Brown, and P. J. Espenshade. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102:315-323. [DOI] [PubMed] [Google Scholar]
- 31.Rawson, R. B., N. G. Zelenski, D. Nijhawan, J. Ye, J. Sakai, M. T. Hasan, T. Y. Chang, M. S. Brown, and J. L. Goldstein. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1:47-57. [DOI] [PubMed] [Google Scholar]
- 32.Roy, B., and A. S. Lee. 1999. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 27:1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai, J., A. Nohturfft, D. Cheng, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272:20213-20221. [DOI] [PubMed] [Google Scholar]
- 34.Sakai, J., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1998. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 273:5785-5793. [DOI] [PubMed] [Google Scholar]
- 35.Sakai, J., R. B. Rawson, P. J. Espenshade, D. Cheng, A. C. Seegmiller, J. L. Goldstein, and M. S. Brown. 1998. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2:505-514. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Smolik, S. M., R. E. Rose, and R. H. Goodman. 1992. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol. Cell. Biol. 12:4123-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using Quickchange site directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 39.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 41.Ye, J., U. P. Dave, N. V. Grishin, J. L. Goldstein, and M. S. Brown. 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. USA 97:5123-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, C., F. E. Johansen, and R. Prywes. 1997. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 17:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]