Abstract
Transcription factors of the interferon regulatory factor (IRF) family have been identified as critical mediators of early inflammatory gene transcription in infected cells. We recently determined that, besides IRF-3 and IRF-7, IRF-5 serves as a direct transducer of virus-mediated signaling. In contrast to that mediated by the other two IRFs, IRF-5-mediated activation is virus specific. We show that, in addition to Newcastle disease virus (NDV) infection, vesicular stomatitis virus (VSV) and herpes simplex virus type 1 (HSV-1) infection activates IRF-5, leading to the induction of IFNA gene subtypes that are distinct from subtypes induced by NDV. The IRF-5-mediated stimulation of inflammatory genes is not limited to IFNA since in BJAB/IRF-5-expressing cells IRF-5 stimulates transcription of RANTES, macrophage inflammatory protein 1β, monocyte chemotactic protein 1, interleukin-8, and I-309 genes in a virus-specific manner. By transient- transfection assay, we identified constitutive-activation (amino acids [aa] 410 to 489) and autoinhibitory (aa 490 to 539) domains in the IRF-5 polypeptide. We identified functional nuclear localization signals (NLS) in the amino and carboxyl termini of IRF-5 and showed that both of these NLS are sufficient for nuclear translocation and retention in infected cells. Furthermore, we demonstrated that serine residues 477 and 480 play critical roles in the response to NDV infection. Mutation of these residues from serine to alanine dramatically decreased phosphorylation and resulted in a substantial loss of IRF-5 transactivation in infected cells. Thus, this study defines the regulatory phosphorylation sites that control the activity of IRF-5 in NDV-infected cells and provides further insight into the structure and function of IRF-5. It also shows that the range of IRF-5 immunoregulatory target genes includes members of the cytokine and chemokine superfamilies.
As an initial response to viral infection, cells produce a spectrum of early inflammatory proteins that can activate cytolytic functions of T cells or directly inhibit viral replication. In vivo, these cytokines can control the growth of both lytic and nonlytic viruses and determine the outcome of viral infections (51). A prompt and regulated cellular response to viral infection is central to host defense. It is coordinated by a genetic regulatory network in which a given transcription factor controls the expression of a diverse set of target genes depending on the cell type and/or the nature of cellular stimuli. The functional diversity of such a transcription factor is dependent on its modification (e.g., phosphorylation) and/or interaction with other transcription factors that are coexpressed and/or activated in the infected cell (53, 54).
Interferon (IFN) regulatory factors (IRFs) are transcriptional mediators of virus- and IFN-induced signaling pathways and have been shown to be involved in antiviral defense, immune response, and cell growth regulation. IRF-1 was initially identified as a transcriptional activator of the IFNB gene in infected cells. Subsequently, nine cellular IRF genes (IRF-1, IRF-2, IRF-3, IRF-4/Pip/ICSAT, IRF-5, IRF-6, IRF-7, ICSBP/IRF-8, and ISGF3γ/p48/IRF-9 genes), as well as virus-encoded analogues of cellular IRFs, were identified (36, 42). Although members of the IRF family can function as transcriptional activators (e.g., IRF-1, IRF-3, IRF-5, and IRF-9), repressors (e.g., IRF-8), or both (e.g., IRF-2, IRF-4, and IRF-7), they have significant homology in the N-terminal 115 amino acids (aa), which comprise the DNA-binding domain (DBD). The unique function of a particular IRF can often be accounted for by cell type-specific expression, its intrinsic transactivation potential, and the ability to interact with IRF family members or other transcription factors and cofactors (3, 20, 59).
Three IRFs (IRF-3, IRF-5, and IRF-7) were found to function as direct transducers of virus-mediated signaling and play a crucial role in the expression of type I IFN genes (4, 7, 20, 36, 38, 45, 56, 59, 61) as well as chemokine gene expression. While IRF-3 is constitutively expressed in all cell types (2), constitutive expression of IRF-7 can be detected predominantly in cells of lymphoid origin (1). Nevertheless, in most cell types, expression of IRF-7 can be stimulated by type I IFN (1, 44, 46). Expression of IRF-5 seems to be restricted to B cells and dendritic cells; however, IRF-5 can be induced by type I IFN (7) in most cells of lymphoid origin, suggesting its role in innate immunity. All three of these IRFs were found to reside predominantly in the cytoplasm, and, upon the virus-induced phosphorylation of serine residues in the carboxyl terminus, these proteins translocate to the nucleus (1, 7, 31, 37, 41, 44, 57, 63). It was shown that carboxyl-terminal phosphorylation of IRF-3 facilitates its binding to transcriptional coactivator p300/CBP and to the PRDIII element in the IFNB promoter (21, 47, 57, 58, 63). IRF-7 also undergoes nuclear translocation after virus-induced serine phosphorylation in its carboxyl-terminal region, which is highly homologous to the corresponding region of IRF-3 (1, 44, 46). However, phosphorylated IRF-7 can also be detected in the nuclei of uninfected cells (3, 61). Both IRF-3 and IRF-7 were identified as components of the transcriptional complex-enhanceosome, which binds to the promoters of IFNA1 and IFNB genes in infected cells (3, 57). The importance of these two factors in the induction of early inflammatory genes was further strengthened by the generation of IRF-3−/− mice, which revealed only weak induction of type I IFN genes in response to viral infection (45). In addition, fibroblasts from IRF9−/− mice that were void of IRF-7 expression were unable to express type I IFN genes upon viral infection (45). These data indicate that IRF-7 plays a critical role in the induction of type I IFN genes. Similarly, in human cells that were not capable of expressing IRF-7, the virus-mediated induction of IFNA genes could be detected only after reconstitution of IRF-7 gene expression (61). These results demonstrate both the essential and distinct roles of IRF-3 and IRF-7, which together ensure the transcriptional regulation of diverse group of IFNA and B genes for an antiviral response.
Although IRF-3 and IRF-7 are necessary factors for virus-induced type I IFN gene expression, the observation that IRF-5 also induces expression of type I IFN genes in a virus-specific manner suggests that, in cells expressing IRF-5, IRF-7 may not be solely responsible for the activation (7). To understand the role of IRF-5 in innate immunity, the detailed assessment of IRF-5 function and identification of immunoregulatory genes controlled by IRF-5 are required. The goal of the present study was therefore to define elements in the IRF-5 polypeptide that regulate its transcriptional activation and nuclear translocation, as well as to identify the regulatory phosphorylation sites.
MATERIALS AND METHODS
Cell culture and virus.
HeLa and bovine tracheal cells (American Type Culture Collection [ATCC]) and 2fTGH cells, obtained from G. Stark (Cleveland Clinic Foundation, Cleveland, Ohio), were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS). BJAB cells were grown in RPMI 1640 with 10% FBS. Sendai virus was purchased from Specific Pathogen Free Avian Supply (Preston, Conn.), and Newcastle disease virus (NDV) was purchased from ATCC (VR-699). Infections were conducted with 640 hemagglutinin units/100-mm-diameter plate (80% confluence) for a given time period. Vesicular stomatitis virus (VSV) was obtained from Phil Marcus (University of Connecticut), and infections were conducted at a multiplicity of infection (MOI) of 2.0. Herpes simplex virus type 1 (HSV-1) was supplied by Gary Hayward (Johns Hopkins University, Baltimore, Md.), and infections were performed at an MOI of 1.0. Cells were treated with IFN for 8 h at 500 U/ml. 2fTGH IRF-5-overexpressing cells (2fTGH/IRF-5 cells) were generated as previously described (7). BJAB cells constitutively expressing IRF-5 (BJAB/IRF-5 cells) were generated by cotransfection of BJAB cells with N-terminal Flag-tagged IRF-5-expressing plasmid in which IRF-5 cDNA was under the control of the cytomegalovirus promoter (pCMV-Tag2B.IRF-5) and pSV2neo (ratio, 10:1, respectively). Transfected cells were selected by growth in G-418, and single clones were screened for the expression of IRF-5 by Western blot hybridization with a monoclonal anti-Flag antibody.
Plasmid construction and antibodies.
cDNAs encoding IRF-5 carboxyl-terminal deletion mutations were generated by 25 cycles of PCR amplification with Vent DNA polymerase. DNA oligonucleotide primers were synthesized and purified by Life Technologies CustomePrimers. The sequences for Gal4-IRF-5 carboxyl-terminal mutants were cloned into the pSG424 vector as previously described (7). The sequences for pCMV-Tag2B.IRF-5-encoded carboxyl-terminal deletion mutants were cloned into pCMV-Tag2B (Stratagene Inc.) at the SalI-XbaI site previously described (7). Green fluorescent protein (GFP)-IRF-5 N1, C1, and C2 were constructed by inserting the PCR-amplified SalI-BamHI coding sequence for the IRF-5 amino-terminal (N1) or IRF-5 carboxyl-terminal (C1 or C2) fragment into the pEGFP-C1 vector (Clontech). IRF-5 nuclear localization signal (NLS) mutants were generated from GFP-full-length IRF-5 (IRF-5fl) by overlap PCR mutagenesis with Vent DNA polymerase using the QuickChange site-directed mutagenesis kit (Stratagene). A full-length IRF-5 open reading frame was cloned at the SalI-NotI site in frame with the glutathione S-transferase (GST) gene in the pGEX4T-3 vector (Amersham Pharmacia Biotech). GST-IRF-5 N1 and C1 were constructed in the same manner. The point mutations of IRF-5 at serines 475 (S475A), 477 (S477A), and 480 (S480A) and the triple point mutation (3SA) were generated from pCMV-Tag2B.IRF-5 by using the QuickChange site-directed mutagenesis kit. Mutations were confirmed by sequencing and expression in 2fTGH cells by Western blot analysis with an anti-Flag antibody. IRF-3, IRF-5, Gal4-IRF-5, and GFP-IRF-5 expression plasmids, Gal4TKCAT, RANTES luciferase, HuIFNA1 and -A2, and HuIFNB soluble alkaline phosphatase (SAP) reporter plasmids, and anti-IRF-3 polyclonal antibodies were described previously (2, 7, 15, 21). The M2 anti-Flag monoclonal antibody was obtained from Sigma, anti-Gal4(DBD) polyclonal antibodies were obtained from Santa Cruz Biotechnology and Living Colors A.V., and the peptide polyclonal antibody (anti-GFP antibody) was from Clontech.
Reverse transcription-PCR (RT-PCR) analysis and antiviral assay.
One microgram of total RNA isolated by the cesium chloride method was reverse transcribed to cDNA with oligo(dT) primers in a 30-μl reaction mixture. From this mixture of cDNAs, IFNA, IRF-3, IRF-5, IRF-7, and β-actin cDNAs were amplified by PCR as previously described (4, 7, 61). Levels of biologically active IFN-α in the medium were determined by a cytopathic bioassay using bovine tracheal cells, which recognize IFN-α but not IFN-β, with VSV as the challenging virus (12). Individual IFNA subtypes were identified by cloning and subsequent sequencing of the PCR-amplified fragments as described recently (7, 61). A one-way analysis of variance (http://faculty.vassar.edu/∼lowry/VassarStats.html) was used to analyze the significance of observed differences in IFNA subtype expression. F values were calculated, and significance was determined by the null hypothesis: no significant difference between the two groups is considered if the F value is less than or equal to 1.
RPA.
To analyze the effect of IRF-5 on cytokine and chemokine gene transcription in uninfected and virus-infected cells, the stable BJAB/IRF-5 cell lines were used. For the RNase protection assay (RPA), total RNA was isolated from BJAB/IRF-5 cells by the cesium chloride method. Total RNA (5 to 10 μg) was subjected to RPA by using the hCK-5 chemokine template of the RiboQuant multiprobe RPA kit, in accordance with the manufacturer's instructions (BD Pharmingen, San Diego, Calif.). Briefly, high-specific-activity [α-32P]UTP-labeled antisense hCK-5 RNA probes were synthesized with T7 polymerase and purified by chloroform-isoamyl alcohol (50:1) extraction. The probe set was then hybridized in solution in excess to the target RNA (temperature gradient from 90 down to 56°C for 12 to 16 h); the free probe and other single-stranded RNA were then removed by digestion with RNases. The RNase-protected probes were purified by ethanol precipitation, resolved on denaturing polyacrylamide gels, and quantified by phosphorimaging. Values for chemokine mRNA levels were normalized to those for glyceraldehyde-3-phosphate dehydrogenase mRNA levels for each experiment.
Transient transfection and CAT, SAP, and luciferase assays.
In the transient- transfection assay, 2 × 106 2fTGH or HeLa cells were transfected with a constant amount of DNA (5 μg/60-mm-diameter plate, 8 μg/100-mm-diameter plate) by using Superfect transfection reagent (Qiagen). Equal amounts (2.5 μg) of chloramphenicol acetyltransferase (CAT), SAP, or luciferase reporter plasmid and IRF-5-expressing plasmid were transfected with the β-galactosidase expression plasmid (200 ng). The transfected cells were split 14 h later, incubated for another 6 h, and infected with Sendai virus or NDV for 16 h or treated with 75 μg of double-stranded RNA (dsRNA)/ml and 50 μg of cycloheximide/ml for 6 h. The CAT and SAP assays were performed as previously described (7). Luciferase activity was measured by the method of Ye et al. (60). Each experiment was repeated three times. The β-galactosidase expression levels were used to normalize the difference in transfection efficiency in all transient-transfection assays.
Subcellular localization of GFP-IRF-5 proteins.
2fTGH cells were transfected with GFP-IRF-5fl, GFP-IRF-5 N1, GFP-ΔmC1-IRF-5, GFP-IRF-5 C1, GFP-IRF-5 C2, GFP-ΔmN1-IRF-5, or GFP-ΔmNLS-IRF-5 expression plasmids (5 μg). Fourteen hours after transfection, cells were divided and seeded in chambered coverglass slides (Nunc, Naperville, Ill.), incubated for another 4 to 6 h, and infected with Sendai virus or NDV or left uninfected for 3 and 6 h. Cells were then examined under a fluorescence microscope at a wavelength of 507 nM. All pictures presented were recorded at a total magnification of ×200.
Metabolic labeling of IRF-5 serine mutant proteins with 32Pi.
2fTGH cells (3 × 106) were transfected with pCMV-Tag2B.IRF-5 or its mutants and incubated for 16 h. Cells were washed with saline and incubated for an additional 2 h in phosphate-free Dulbecco’s modified Eagle medium supplemented with 2% dialyzed FBS. Cells were then left uninfected or were infected with Sendai virus or NDV and labeled with 0.5 mCi of 32Pi (Amersham Inc.)/ml for 6 h. Samples were collected in lysis buffer (10 mM Nonidet P-40, 0.15 M NaCl, 0.01 M sodium phosphate [pH 7.2], 2 mM EDTA, 50 mM sodium fluoride, 0.2 mM sodium vanadate, protease inhibitor cocktail) and left on ice for 30 min. The whole-cell extracts (250 μg) were used for immunoprecipitation with an anti-Flag antibody, and precipitated proteins were resolved by sodium dodecyl sulfate-7% polyacrylamide gel electrophoresis (SDS-PAGE).
Expression of GST-IRF-5 fusion proteins and GST pull-down assay.
GST-IRF-5, GST-IRF-5 N1, and GST-IRF-5 C1 were transformed into BL21 bacteria and induced with 0.5 mM IPTG for 3 h. The GST fusion proteins were purified from bacterial lysates by affinity chromatography on a glutathione-agarose column (Sigma). Coomassie blue staining of the GST fusion proteins was used to quantify the amount of protein on the beads. Equal amounts of GST fusion proteins and beads were then applied in each pull-down assay. The binding proteins were isolated as a whole-cell extract in lysis buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 200 mM NaCl, 0.5% NP-40, 0.2 mM protease inhibitor cocktail) from uninfected or virus-infected 2fTGH/IRF-5 cells. GST fusion proteins immobilized on beads and whole-cell extracts were incubated in 300 μl of incubation buffer (10 mM Tris-Cl [pH 7.6], 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40, 500 μg of bovine serum albumin, and 8% glycerol) at 4°C for 2 h. The beads were then washed three times with incubation buffer, and the bound proteins were eluted and resolved by SDS- PAGE, transferred to Hybond transfer membranes (Amersham), and incubated with an anti-Flag antibody or an anti-IRF-3 monoclonal antibody (3). Immunocomplexes were detected by using the ECL system.
Immunoprecipitation and Western blot analysis of IRF-5 interactions.
2fTGH/IRF-5 cells were cotransfected with or without the GFP-IRF-5 expression plasmid to examine interactions between IRF-5/IRF-5 and IRF-5/IRF-3. Cells were uninfected or infected with Sendai virus or NDV for 6 h and then lysed in immunoprecipitation lysis buffer (20 mM HEPES [pH 7.9], 50 mM NaCl, 10 mM EDTA, 2 mM EGTA, 0.1% NP-40, 10% glycerol, 0.2 mM protease inhibitor cocktail). Extracts (250 μg) were incubated with 1 μg of anti-Flag antibody or 1 μg of anti-IRF-3 polyclonal antibody cross-linked to protein G- or A-Sepharose beads, respectively. Precipitates were washed four times with immunoprecipitation lysis buffer and eluted by boiling the beads for 3 min in 1× SDS loading buffer. Eluted proteins were separated by SDS-10% PAGE and transferred to membranes, and IRF-5 and IRF-3 were detected by anti-GFP polyclonal antibodies or anti-Flag monoclonal antibodies and anti-IRF-3 antibodies, respectively.
Chromatin immunoprecipitation assay.
2fTGH/IRF-5 cells (6 × 107) were infected with Sendai virus or NDV for 6 h or left uninfected. The proteins bound to DNA were cross-linked by addition of 11% formaldehyde in aqueous buffer (0.1 M NaCl, 1 mM EDTA, 50 mM HEPES, pH 8.0) to a final concentration of 1% for 30 min at 37°C (7, 57, 64). The reaction was stopped by addition of 0.125 M glycine. The cell pellets were washed with 5 ml of wash buffer (5 mM PIPES [piperazine-N,N′-bis{2-ethanesulfonic acid}] [pH 8.0], 85 mM KCl, 0.5% Nonidet P-40, 0.2 mM phenylmethylsulfonyl fluoride), resuspended in sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0) on ice, and lysed by sonication for 10 s. Samples were diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl) and cleared with protein A-Sepharose. Equal amounts of proteins were immunoprecipitated with 1 μg of anti-IRF-3 polyclonal or anti-Flag monoclonal antibody for 4 h at 4°C. Immunocomplexes were extensively washed and treated with RNase A (50 μg/ml), 0.5% SDS, and proteinase K (500 μg/ml). The cross-linked DNA-protein complexes were reverted by heating at 65°C for 6 h, and the DNA was recovered by phenol-chloroform extraction. DNA purified by precipitation with 2 M ammonium acetate-ethanol was used as a template for PCR amplification with universal primers corresponding to the regions of human endogenous IFNA genes that are conserved in all subtypes (61).
RESULTS
VSV and HSV-1 activate IRF-5.
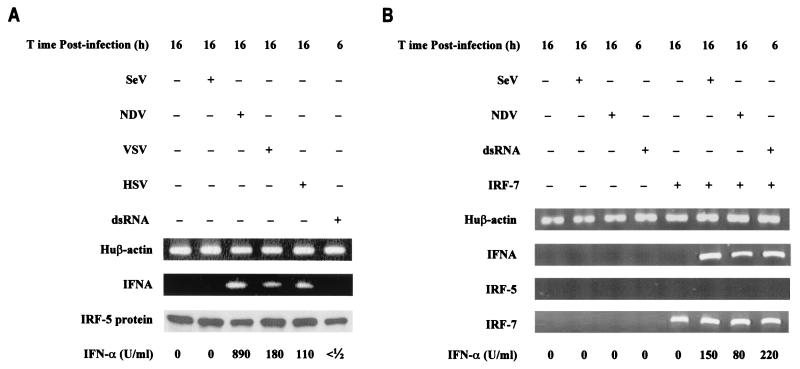
It was previously shown that overexpression of IRF-5 stimulated expression of IFNA genes in cells infected with NDV but not Sendai virus, suggesting virus-specific activation of IRF-5. This is in contrast to what was found for IRF-3 and IRF-7, where multiple viruses and inducers can activate each IRF (7, 23, 24, 49). An important question remaining is whether virus families and inducers other than NDV can activate IRF-5. We have therefore examined the activation of IRF-5 by VSV (Rhabdoviridae), HSV-1 (Herpesviridae), and dsRNA. The human fibrosarcoma (2fTGH) cell line, which expresses IRF-3 but not IRF-5 or IRF-7, is unable to induce IFNA gene transcription upon viral infection. Yet, it has been shown that expression of IFNA genes was rescued in the presence of IRF-5 or IRF-7 (Fig. 1) (7, 61). To determine which other inducers can activate IRF-5, the 2fTGH/IRF-5 cells were infected with Sendai virus, NDV, VSV, or HSV-1 or were treated with dsRNA and levels of synthetized IFN-α were measured by a cytopathic assay using bovine tracheal cells. As shown previously, NDV, but not Sendai virus, induced transcription of IFNA genes and expression of biologically active IFN-α in 2fTGH/IRF-5 cells (Fig. 1A). Both VSV- and HSV-1-infected 2fTGH/IRF-5 cells produced low levels of IFN-α, which could be detected by an antiviral assay. Furthermore, VSV infection of 2fTGH cells transfected with a plasmid expressing GFP-IRF-5 fusion protein resulted in the accumulation of GFP-IRF-5 in the nucleus (data not shown), suggesting posttranslational modification of IRF-5 by VSV that resembled phosphorylation and translocation of IRF-5 to the nucleus in NDV-infected cells (7). Combined, these results indicate that IRF-5 can be activated by Paramyxoviridae, Rhabdoviridae, and Herpesviridae to induce various amounts of biologically active IFN-α. Treatment of 2fTGH/IRF-5 cells with dsRNA did not lead to the stimulation of IFNA gene transcription, nor did it induce synthesis of biologically active IFN-α (Fig. 1A).
FIG. 1.
Activation of IRF-5 and endogenous IFNA genes by different inducers. (A) Reconstitution of endogenous IFNA gene expression in 2fTGH/IRF-5 cells infected with NDV, VSV, and HSV but not Sendai virus or dsRNA. IFNA cDNAs were amplified using primers corresponding to the conserved regions of all human IFNA genes (61). Human (Hu) β-actin-amplified fragments are shown at the bottom as a control for RNA levels. Levels of IRF-5 in cell lysates were determined by immunoblotting with an anti-Flag antibody, and biologically active IFNA was detected by an antiviral assay using bovine tracheal cells and VSV as the challenging virus (12). (B) 2fTGH cells expressing ectopic IRF-7 induce IFNA gene expression after infection with Sendai virus or NDV and after treatment with dsRNA. 2fTGH cells were left untransfected (left four lanes) or were transfected with IRF-7 expression plasmid (right four lanes) and were uninfected or were infected with Sendai virus or NDV for 16 h or were coincubated with dsRNA and cycloheximide for 6 h. RT-PCR for β-actin, IFNA, IRF-5, and IRF-7 amplification was performed as described in Materials and Methods.
Virus and IRF-dependent IFN-α subtype induction.
Since we have shown that IRF-5 can stimulate the induction of IFNA genes in VSV- and HSV-1-infected cells, we had an opportunity to closely examine whether the induced profile of IFNA gene subtypes is determined only by the preferential IRFs expressed or also by the specific viral infection. Previously we have shown that IFNA8 was the predominant subtype (45%) expressed in NDV-infected 2fTGH/IRF-5 cells and IFNA1/A13, -A4, and -A14 subtypes were expressed at significantly lower levels (12%) (Table 1) (7). In comparison, NDV-infected 2fTGH/IRF-7 cells preferentially induced IFNA1/A13 (40%) and IFNA4, -A10, and -A17 were expressed in the range of 12 to 19%. These data suggest that the profile of IFNA subtypes expressed may be determined by a particular IRF and not by the virus since, in 2fTGH/IRF-7 cells, infection with Sendai virus or NDV led to nearly identical levels of subtype expression (7, 61).
TABLE 1.
IFNA subtypes induced by VSV and NDV infection in 2fTGH cells expressing IRF-5c
| IFNA subtype | No. of positive clones/40 clones in VSV-infected cells | % Positive clones in cells infected with
|
|
|---|---|---|---|
| VSV | NDVa | ||
| A1/A13 | 2 | 5.0 | 13.3b |
| A2 | 2 | 5.0 | 3.3b |
| A4 | 2 | 5.0 | 11.7b |
| A5 | 7 | 17.5 | 3.3 |
| A7 | 0 | 0 | 1.7b |
| A8 | 10 | 25.0 | 45.0 |
| A10 | 8 | 20.0 | 5.0 |
| A14 | 0 | 0 | 11.7 |
| A17 | 4 | 10.0 | 0 |
| A21 | 5 | 12.5 | 5.0b |
Values taken from Barnes et al. (7).
Value not significantly different from value for VSV induction of IFNA genes based on a one-way analysis of variance analysis as described in Materials and Methods.
Data for the predominant subtype are in boldface.
To determine how the infecting virus affects the profile of expressed IFNA genes, we analyzed IFNA gene expression in 2fTGH/IRF-5 cells infected with VSV for 16 h. The RT-PCR amplification and cloning of the individual IFNA subtypes were described recently, and randomly selected clones were identified by sequence analysis (7, 61). Results from sequencing, displayed in Table 1, reveal that, in both VSV- and NDV-infected cells, IFNA8 was the predominant subtype expressed. However, under the described conditions, the IFNA5 (17.5%), -A10 (20%), and -A17 (10%) subtypes were detected at higher frequencies in VSV-infected cells than in NDV-infected cells (3, 5, and 0%, respectively). In contrast, IFNA subtypes A1/A13, A4, and A14 were detected more frequently in NDV-infected cells than in VSV-infected cells. IFNA17 was the only new subtype that was expressed after VSV infection but not after NDV infection. These results indicate that the levels of IFNA subtypes induced by VSV and NDV, not the subtypes themselves, were distinct. In contrast, the expression of IRF-7 and IRF-5 in NDV-infected 2fTGH cells resulted in expression of unique IFNA subtypes (7).
IRF-5 induces multiple chemokines in a virus-specific manner.
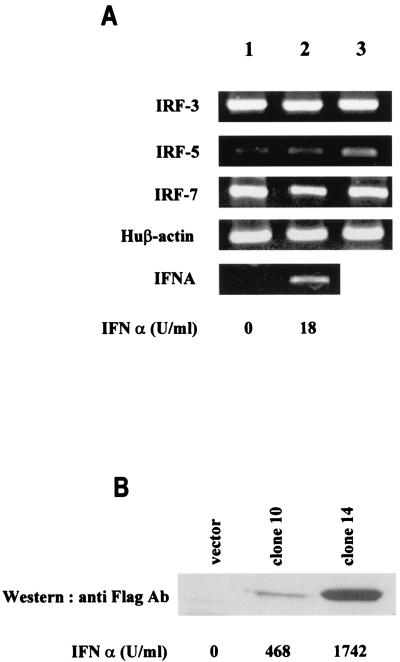
We have shown that the IRF-5 gene is expressed constitutively in dendritic cells (7), where high levels of IRF-5 were detected specifically in precursor dendritic cells (pDC2) that have been identified as high IFN-α producers (50; B. J. Barnes, unpublished data). Since constitutive expression of IRF-5 was also detected in some B-cell lines, such as Namalwa and Daudi, that are capable of producing high levels of IFN-α upon viral stimulation (Namalwa), we examined whether overexpression of IRF-5 in B cells would result in an increase in the virus-mediated induction of type I IFNs and possibly other cytokine and chemokine genes. To this end, we generated a stable B-cell line (BJAB) expressing different levels of IRF-5. In normal BJAB cells, expression of endogenous IRF-5 is nearly undetectable by RT-PCR, yet low levels of IRF-5 mRNA could be detected in NDV-infected cells and cells treated with type I IFN (Fig. 2A). Both IRF-3 and IRF-7 are well expressed in these cells, and NDV infection or type I IFN treatment did not greatly affect the levels of their expression. BJAB cells produce only low levels of endogenous IFN-α upon Sendai virus or NDV infection (∼18 U/ml).
FIG. 2.
Activation of endogenous IRFs and IFNA genes in infected BJAB cells. (A) The relative levels of IRF-3, IRF-5, IRF-7, and IFNA mRNA were determined by RT-PCR analysis of BJAB cells (lane 1), NDV-infected BJAB cells at 16 h postinfection (lane 2), and BJAB cells treated with IFN for 8 h (lane 3). Individual cDNAs were amplified by using primers specific for the IRFs or primers corresponding to the conserved regions of all human IFNA genes (4, 7, 61). Levels of biologically active IFN-α synthesized in these cells were determined by antiviral assay and are shown at the bottom. Huβ-actin, human β-actin. (B) The levels of Flag-tagged IRF-5 expressed in stably transfected, NDV-infected BJAB clones 10 and 14 were determined 16 h postinfection by immunoblotting with an anti-Flag antibody. Left lane, empty vector control, pCMVsport; middle lane, low-expressing IRF-5 clone 10; right lane, high-expressing IRF-5 clone 14. The levels of biologically active IFN-α in the medium of infected cells, shown at the bottom, were determined by an antiviral assay (12).
To determine the correlation between levels of IRF-5 expression and induction of endogenous IFN-α upon infection with Sendai virus or NDV, two stably transfected BJAB/IRF-5 clones, clone 10, a low expresser of IRF-5, and clone 14, a high expresser, were examined (Fig. 2B). While infection with Sendai virus induced only low levels of biologically active IFN-α (10 U/ml), the levels of IFN-α induced in NDV-infected BJAB/IRF-5 cells (clone 10, 468 U/ml; clone 14, 1,742 U/ml) directly correlated with the levels of expression of IRF-5 in these cells.
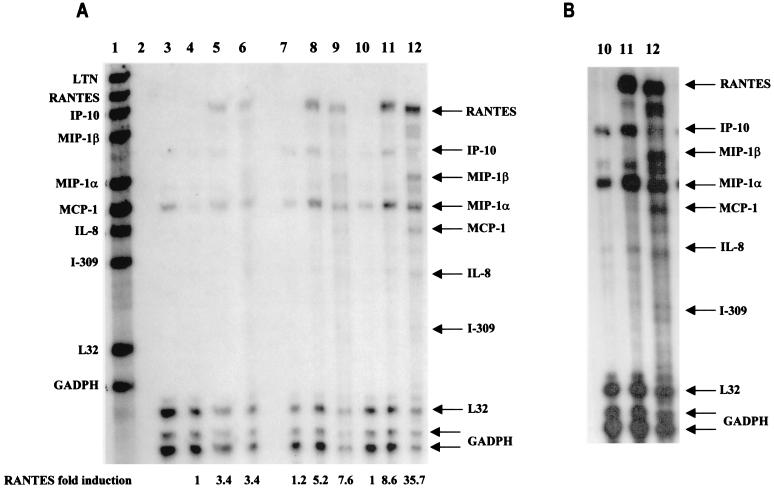
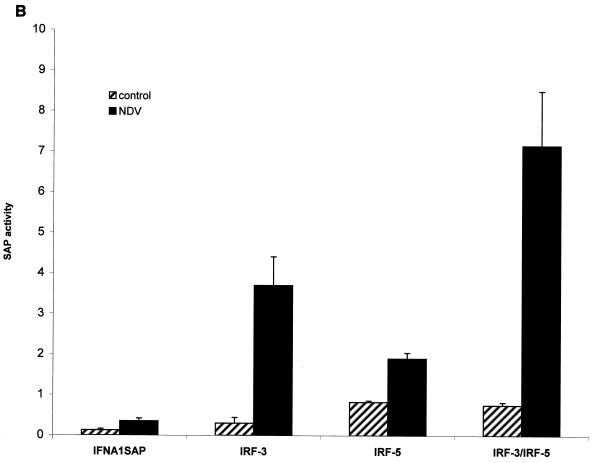
These two cell lines were also used to examine the IRF-5-induced expression of chemokine genes. Results from the RPA are shown in Fig. 3A. BJAB control cells (cells transfected with an empty vector) infected with Sendai virus or NDV (lanes 5 and 6, respectively) expressed only low levels of RANTES and macrophage inflammatory protein 1α (MIP-1α) transcripts. The relative levels of RANTES, MIP-1α, and IFN-α-inducible protein 10 (IP-10) transcripts were higher in BJAB/IRF-5 clone 10 cells infected with Sendai virus (lane 8) or NDV (lane 9) than in BJAB control cells. The stimulating effect of IRF-5 was more visible in the infected high expresser, clone 14 (Fig. 3A and B, lanes 10 to 12). In this cell line, a clear difference between Sendai virus and NDV stimulation could also be observed. For instance, while infection with Sendai virus led to an 8.6-fold increase in RANTES mRNA levels by clone 14 cells compared to levels produced by BJAB control cells, NDV infection of clone 14 cells gave a 35.7-fold increase in RANTES mRNA levels. In addition, the induction of other chemokine genes (e.g., MIP-1α, monocyte chemotactic protein 1 [MCP-1], MIP-1β, interleukin-8 [IL-8] genes) by viral infection was found to be consistently four- to fivefold greater in IRF-5-expressing cells than in control cells not expressing IRF-5. While RANTES, IP-10, and MIP-1α transcripts were detected in IRF-5-expressing cells infected with Sendai virus or NDV, MIP-1β, MCP-1, IL-8, and I-309 genes were induced only by infection with NDV. Thus, NDV infection of IRF-5-expressing cell lines induced the expression of a large number of chemokine genes. Altogether, these data indicate that IRF-5 targets expression of both common and distinct inflammatory genes in Sendai virus- and NDV-infected cells.
FIG. 3.
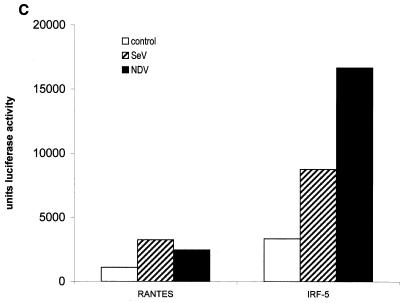
IRF-5 induces multiple cytokines and chemokines in infected BJAB cells. (A) IRF-5 induces expression of multiple chemokines in infected BJAB cell lines expressing either low levels of Flag-tagged IRF-5 (clone 10) or high levels of Flag-tagged IRF-5 (clone 14). Total RNA was isolated from BJAB/IRF-5 cells at 6 h postinfection and analyzed by RPA using the hCK-5 kit (Pharmingen) as described in Materials and Methods. Lane 1, hCK-5 probe template; lane 2, tRNA control; lane 3, human RNA control; lane 4, uninfected BJAB control cells; lane 5, Sendai virus-infected BJAB control cells; lane 6, NDV-infected BJAB control cells; lane 7, uninfected BJAB/IRF-5 clone 10; lane 8, Sendai virus-infected BJAB/IRF-5 clone 10; lane 9, NDV-infected BJAB/IRF-5 clone 10; lane 10, uninfected BJAB/IRF-5 clone 14; lane 11, Sendai virus-infected BJAB/IRF-5 clone 14; lane 12, NDV-infected BJAB/IRF-5 clone 14. Control cells are BJAB cells transfected with an empty vector. Fold induction of RANTES mRNA expression was normalized to the level of glyceraldehyde-3-phosphate dehydrogenase mRNA expression. The data are representative of two independent experiments. (B) Long exposure of RNA samples analyzed in lanes 10 to 12. (C) Activation of the RANTES luciferase reporter in infected cells expressing IRF-5. HeLa cells were left uninfected or were infected with Sendai virus or NDV for 16 h, and luciferase activity was measured as described in Materials and Methods.
It has been previously shown that IRF-3 is a direct activator of the RANTES gene promoter (30). Since we now have shown that IRF-5 also significantly increases the relative levels of RANTES transcripts in BJAB cells, we next examined whether IRF-5 transactivates the RANTES promoter using the RANTES luciferase reporter assay. HeLa cells were cotransfected with an IRF-5-expressing plasmid and a reporter plasmid containing the RANTES promoter in front of the luciferase gene and then were infected with either Sendai virus or NDV. As shown in Fig. 3C, the constitutive activity of this promoter was low but Sendai virus and NDV infection stimulated the RANTES promoter to similar levels. The transcription activity of this promoter was further enhanced (twofold) by cotransfected IRF-5. However, the IRF-5-mediated enhancement was higher in NDV-infected cells (five- to sixfold) than Sendai virus-infected cells (threefold). These data indicate that IRF-5 stimulates transcription of the RANTES promoter more efficiently in NDV-infected cells than in Sendai virus-infected cells.
Localization of a constitutive-activation domain.
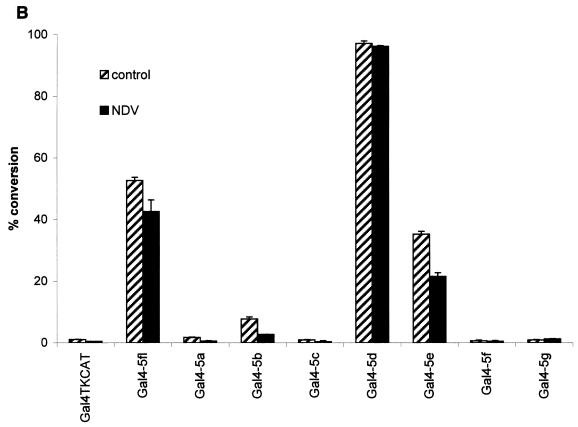
The IRF-5 transcription factor shares many structural features with IRF-3 and IRF-7, including the N-terminal DBD common to all family members, a putative IRF interaction domain in the C terminus, and a serine-rich domain located at the C terminus between aa 471 and 486. Because of the involvement of IRF-5 in the activation of type I IFNA genes and in the induction of chemokine genes involved with T-lymphocyte trafficking (Fig. 3), we analyzed the structural and functional properties of the IRF-5 protein as they related to transactivation. The transactivation potential of IRF-5 was shown previously by using the Gal4 TKCAT reporter assay (7), yet in these experiments the activation domain was not identified. To localize the transactivation domain, we cloned IRF-5 cDNA and different segments of IRF-5 cDNA lacking the DBD into the pSG424 vector in frame with the coding sequence for the Gal4 DBD (Fig. 4A) and tested the ability of the chimeric Gal4-IRF-5 proteins to activate transcription of the Gal4 TKCAT reporter plasmid. Figure 4B shows that the chimeric proteins containing IRF-5 aa 136 to 240 (IRF-5a), 136 to 345 (IRF-5b), and 136 to 410 (IRF-5c) did not activate transcription, whereas chimeric proteins containing aa 136 to 489 (IRF-5d) and 136 to 539 (IRF-5e) and IRF-5 aa 1 to 539 (IRF-5fl) stimulated transcription of the Gal4 TKCAT reporter. Immunoblot analysis of cell extracts revealed that all of the transfected plasmids tested were expressed at about the same level (Fig. 4C). These results indicated that aa 410 through 489 were necessary for transactivation of the Gal4 TKCAT reporter plasmid. To confirm whether this region alone could confer transactivation potential, we examined the ability of IRF-5 fusion constructs Gal4-IRF-5f (aa 410 to 489) and Gal4-IRF-5g (aa 410 to 539) to activate the Gal4 TKCAT reporter plasmid. Results indicate that, while aa 410 to 489 are required for transactivation, this region alone is not sufficient for activation.
FIG. 4.
Analysis of intrinsic transactivation potentials of various IRF-5 regions fused to the Gal4 DBD. (A) Schematic representation of seven IRF-5 deletion mutants. Straight lines and angled lines indicate included and excluded sequences, respectively. The Gal4 DBD and IRF-5 DBD are indicated. (B) Gal4 TKCAT reporter transient transfection assay. HeLa cells were cotransfected with the indicated plasmid expressing Gal4-IRF-5 fusion proteins (2.5 μg), Gal4 TKCAT reporter plasmid (2.5 μg), and the β-galactosidase plasmid (0.2 μg). Cells were uninfected or infected with NDV, and CAT activity was measured 48 h after transfection. Percent conversion after normalizing for β-galactosidase activity is expressed. (C) Levels of transfected IRF-5 protein isolated from uninfected HeLa cells or HeLa cells infected with NDV for 16 h were detected by Western blot analysis with the anti-Gal4 (DBD) polyclonal antibody.
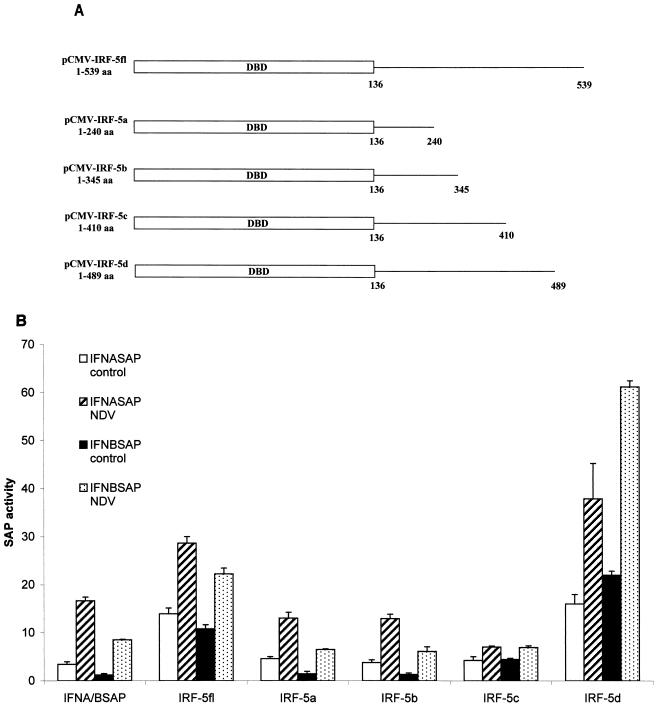
Furthermore, it is worth noting the large difference between the transactivation potentials of Gal4-IRF-5fl and Gal4-IRF-5d and that of Gal4-IRF-5e, indicating the presence of an autoinhibitory domain in the region between aa 489 and 539. Autoinhibitory domains in the C-terminal regions of IRF-3 and IRF-7 have been identified previously (4, 32, 33, 38). Interestingly, data from this reporter assay have not shown any substantial difference between the transactivation potential of IRF-5 in virus-infected cells and that in uninfected cells. However, when IRF-5fl cDNA and that of its C-terminal deletion mutants were cloned into a eukaryotic expression vector (Fig. 5A) and tested for their ability to activate transcription of the IFNA1 or IFNB SAP reporter plasmids, all constructs containing the transactivation domain (aa 410 to 489; IRF-5fl and IRF-5d), which also included a region (aa 471 to 486) rich in serine residues, were virus responsive (Fig. 5B). It is important that, while IRF-5 carboxyl-terminal deletion mutants IRF-5a, -5b, and -5c activate the IFNA1 and IFNB reporters, the observed levels of activation were lower than the levels of activation observed when the reporter plasmids were transfected alone (Fig. 5A; IFNA/BSAP). The IRF-5 mutant that was missing 50 aa at the carboxyl terminus (IRF-5d) was a more effective inducer of both IFNA1 and IFNB virus-responsive elements (VRE) than the full-length IRF-5 (two- to threefold). Taken together, these data indicate that the autoinhibitory domain affects the transcriptional activity of both the phosphorylated and unphosphorylated IRF-5 protein.
FIG. 5.
IRF-5 contains an autoinhibitory domain located between aa 489 and 539 as determined by an IFNA SAP reporter assay. (A) Schematic representation of four IRF-5 carboxyl-terminal deletion mutants. (B) Differential activation of IFNA and IFNB SAP promoters by IRF-5 deletion mutants in NDV-infected cells. 2fTGH cells were cotransfected with IRF-5 deletion plasmids (2.5 μg) and either IFNA1 or IFNB SAP reporter plasmids (2.5 μg) together with the β-galactosidase-expressing plasmid (0.2 μg). At 16 h posttransfection, cells were left uninfected or were infected for 16 h with NDV, and SAP activity was measured as previously described (7).
IRF-5 contains two functional NLSs.
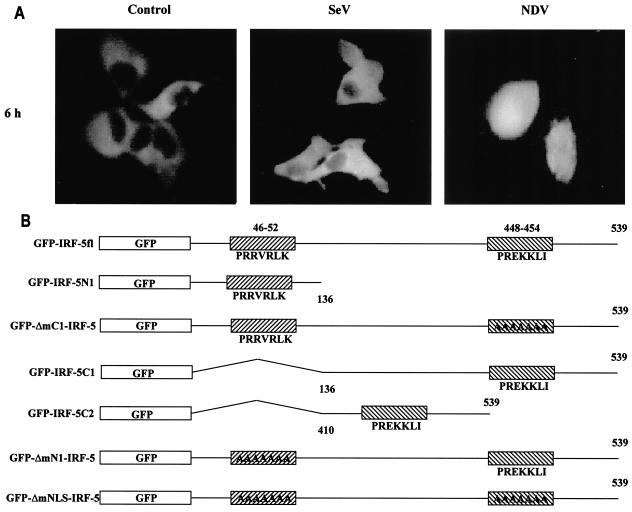
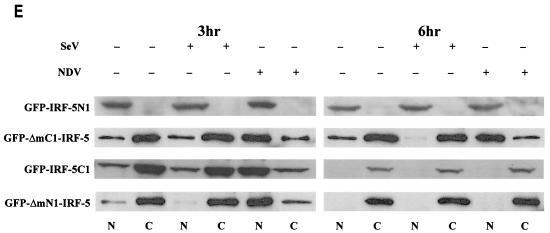
It has been previously shown that members of the IRF family of transcription factors, such as IRF-1, IRF-3, IRF-4, and IRF-9 (26, 27, 48), contain functional NLSs that are not strictly conserved between each family member but that reside in the amino terminus. Examination of the IRF-5 primary amino acid sequence did not reveal any regions significantly homologous to these known NLSs (Table 2). However, results from a search of the IRF-5 protein sequence and similarity (using PSORT II; Human Genome Center, Tokyo, Japan) indicated the presence of two putative NLSs, one residing in the amino-terminal domain, aa 46 to 52, and the other in the carboxyl terminus, aa 448 to 454 (Fig. 6B and Table 2). Recently we have shown by Western blot analysis and fluorescence microscopy that, in both uninfected and Sendai virus-infected 2fTGH cells, the ectopically expressed GFP-IRF-5 fusion protein resided primarily in cytoplasm but could also be detected in the nucleus; however, 2 to 3 h after NDV infection, GFP-IRF-5 was translocated to the nucleus, where it could be detected for 6 to 8 h after infection (Fig. 6A) (7). To examine whether each of the proposed individual NLSs was functional, deletion mutants consisting of either amino- or carboxyl-terminal regions of IRF-5 fused to GFP were constructed (Fig. 6B), along with NLS mutants, and the importance of each individual NLS on the translocation ability of IRF-5 was determined. When the carboxyl terminus deletion mutant, GFP-IRF-5 N1 (aa 1 to 136), was transfected into 2fTGH cells, the fusion protein accumulated in the nuclei of both Sendai virus- and NDV-infected cells as well as in the uninfected cells (Fig. 6C).
TABLE 2.
| IRFa | NLSb |
|---|---|
| IRF-5 | 46PRRVRLK448PREKKLI |
| IRF-1 | 120RKERKSKn5KSKTKRK |
| IRF-2 | 120KKGKKPKn5KVKnnKQ |
| IRF-3 | 77KR |
| IRF-4 | 66KGKFRn10KTRLR |
| IRF-8 (ICSBP) | 66KGKFKn10KTRLR |
| IFR-9 (p48) | 66KGKYKn10KTRLR |
Boldface indicates protein of interest.
n, number of amino acids localized between NLSs.
FIG. 6.
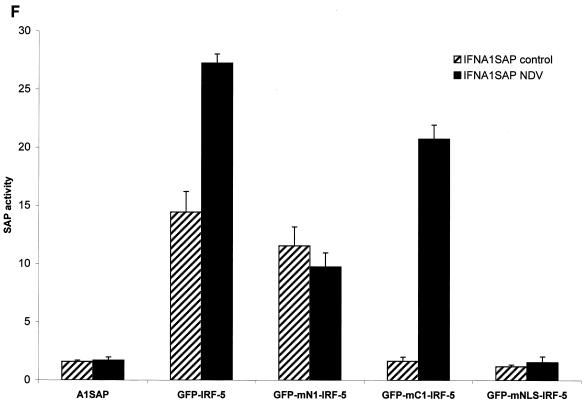
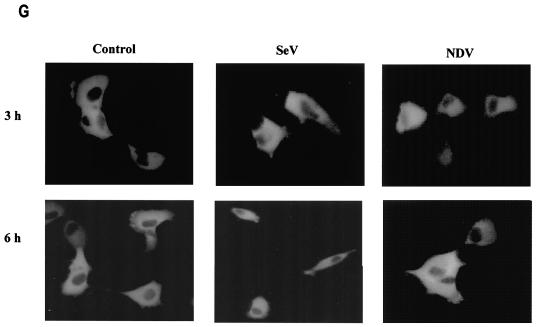
IRF-5 contains two functional NLSs. (A) NDV infection but not Sendai virus infection leads to the nuclear accumulation of full-length GFP-IRF-5 6 h postinfection. (B) Schematic representation of GFP-IRF-5 fusion proteins containing both amino- and carboxyl-terminal NLSs (GFP-IRF-5fl) and proteins containing either the amino-terminal NLS (GFP-IRF-5 N1 and GFP-ΔmC1-IRF-5), the carboxyl-terminal NLS (GFP-IRF-5 C1, GFP-IRF-5 C2, and GFP-ΔmN1-IRF-5), or both NLSs mutated (GFP-ΔmNLS-IRF-5). The individual NLS sequences areindicated. (C) Nuclear translocation of GFP-IRF-5 N1 is independent of virus infection. 2fTGH cells were transfected with GFP-IRF-5 N1, and subcellular localization was determined 3 and 6 h after virus infection by fluorescence microscopy as described in Materials and Methods. (D) GFP-IRF-5 C1 is responsive to viral infection yet is not retained in the nucleus. Subcellular localization was determined as described above. (E) The relative levels of GFP-IRF-5 N1, GFP-ΔmC1-IRF-5, GFP-IRF-5 C1, and GFP-ΔmN1-IRF-5 in cytoplasmic (C) and nuclear (N) extracts of uninfected and virus-infected cells were determined by immunoblotting with anti-Flag antibodies to detect IRF-5 localization. (F) Differential activation of the IFNA1 SAP reporter by IRF-5 NLS mutants. 2fTGH cells were cotransfected with IRF-5 expression plasmids (2.5 μg) and an IFNA1 SAP reporter plasmid (2.5 μg) together with the β-galactosidase-expressing plasmid (0.2 μg). At 16 h posttransfection, cells were left uninfected or were infected for 16 h with NDV, and SAP activity was measured as previously described (7). (G) Subcellular localization of GFP-ΔmNLS-IRF-5. The IRF-5 amino- and carboxyl-terminal NLSs are responsible for IRF-5 translocation to the nucleus.
To determine whether the carboxyl-terminal NLS also contributed to the nuclear localization of IRF-5, the amino terminus deletion mutant, GFP-IRF-5 C1 (aa 136 to 539), was transfected into 2fTGH cells and the cells were infected with virus (Fig. 6D). In uninfected cells, the GFP-IRF-5 C1 protein was detected only in the cytoplasm, yet in NDV-infected cells, but not in Sendai virus-infected cells, the distribution was altered and IRF-5 was detected primarily in the nucleus at 3 h postinfection. These results indicate that the IRF-5 C1 peptide containing the carboxyl-terminal NLS also contains residues that are responsive to the NDV-mediated activation and translocation of IRF-5. Results identical to those obtained for the GFP-IRF-5 C1 peptide were observed while examining the translocation ability of the GFP-IRF-5 C2 peptide (aa 410 to 539) (and a fusion construct containing aa 410 to 489; data not shown), indicating that the C-terminal NLS alone is functionally active and responsive to virus. At 6 h after NDV infection, GFP-IRF-5 C1 and C2 were no longer detected in the nucleus, but rather in the cytoplasm, suggesting that the carboxyl-terminal NLS of IRF-5 was not sufficient for nuclear retention.
Western blot analysis of IRF-5 N1 and C1 proteins present in cytoplasmic and nuclear extracts from uninfected and virus-infected cells confirmed the GFP data (Fig. 6E). The IRF-5 N1 peptide was detected primarily in the nuclei of both infected and uninfected cells at 3 and 6 h postinfection; however, at longer exposure times, IRF-5 N1 could be detected in the cytoplasm of these cells (data not shown). Whereas the IRF-5 C1 peptide was present predominantly in the cytoplasm of uninfected or Sendai virus-infected cells, IRF-5 C1 could be detected in the nuclei of NDV-infected cells at 3 h postinfection yet translocated to the cytoplasm at 6 h postinfection. The levels of IRF-5 C1 detected in the cytoplasm at 6 h postinfection were significantly lower than levels detected at 3 h postinfection, suggesting that cytoplasmic IRF-5 C1 is subject to degradation (Fig. 6E). The observed degradation appeared to be independent of virus infection, since the decreased protein levels were also detected in lysates from uninfected cells, and may instead be a result of three-dimensional structural changes due to the exposure of amino acid residues targeted for degradation by the amino-terminal deletion. These data show that both of the IRF-5 NLSs are functional and can mediate the nuclear transport of IRF-5 but that the 5′ NLS is sufficient and necessary for the nuclear retention of wild-type (wt) IRF-5 in infected cells.
The full-length GFP-IRF-5 constructs in which either the amino- or carboxyl-terminal NLS had been altered by a nonfunctional mutation (170 mN1-IRF-5 or 170 mC1-IRF-5, respectively) and which therefore contained only a single functional NLS show distinct properties. The IRF-5 peptide containing only the 3′ NLS (170 mN1-IRF-5) translocated to the nucleus at 3 h after NDV infection but was detected in the cytoplasm at 6 h postinfection (Fig. 6E). In contrast, the IRF-5 fusion peptide, containing only the 5′ NLS (GFP-170 mC1-IRF-5), was translocated and was retained in the nuclei of NDV-infected cells, thus behaving like wt IRF-5. The difference in the transactivation abilities of these two mutants was also interesting (Fig. 6F). While GFP-170 mC1-IRF-5 was able to activate the IFNA1 promoter in cells infected with NDV and not in uninfected cells, GFP-170 mN1-IRF-5 showed the same transactivating potential in infected and uninfected cells. These data indicate that the 5′ NLS is functional only when IRF-5 is phosphorylated while the 3′ NLS is recognized both in the phosphorylated and unphosphorylated IRF-5 polypeptide (Fig. 6F). To further delineate whether these two NLSs of IRF-5 are the only NLSs responsible for its nuclear translocation, we constructed a mutant IRF-5 fusion protein where both NLSs were made functionally inactive by alanine mutation (Fig. 6B). However, in both infected and uninfected cells, this peptide was retained in the cytoplasm (Fig. 6G) and was not transcriptionally active (Fig. 6F). Taken together, these results demonstrate that both NLSs are critical for the nuclear translocation of IRF-5 and its transcriptional activity and that translocation of IRF-5 to the nucleus is not mediated by IRF-3 heterodimerization or dimerization with other proteins that localize to the nucleus.
Mutational analysis of putative IRF-5 phosphorylation sites.
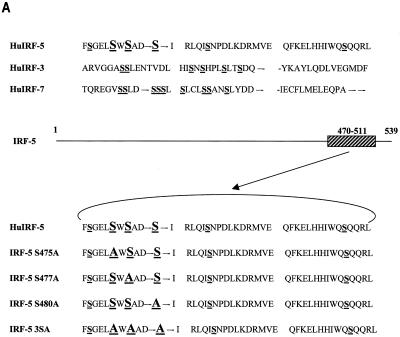
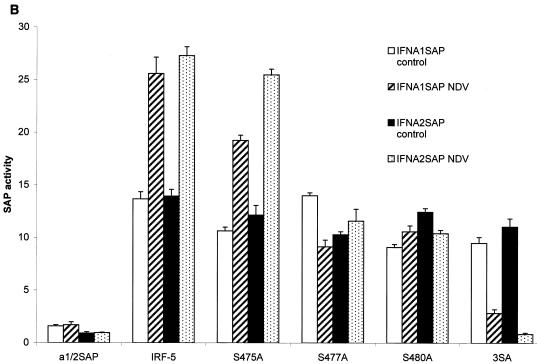
The virus-responsive transactivation domain of IRF-5, consisting of aa 410 to 489, contains a serine-rich cluster of amino acids (aa 471 to 486) similar to the regions in IRF-3 and IRF-7 containing serine residues targeted by virus-induced phosphorylation. On the basis of the homology to the IRF-3 and IRF-7 phosphorylation sites, we examined the role of serine residues 475, 477, and 480 in the carboxyl terminus of IRF-5 (Fig. 7A) by generating point mutations at these three serine residues in the full-length Flag-tagged IRF-5 protein. In these IRF-5 mutants, we either individually replaced Ser-475, Ser-477, or Ser-480 with alanine (IRF-5 S475A, IRF-5 S477A, or IRF-5 S480A) or mutated all three serines to alanines (IRF-5 3SA). To examine the functional activity of these mutants in infected cells, individual IRF-5 expression plasmids were cotransfected with IFNA1 or IFNA2 SAP reporter plasmids into 2fTGH cells. IRF-5 and IRF-5 S475A activated the reporters to similar extents in NDV-infected cells, suggesting that Ser-475 was not critical for virus-induced activation (Fig. 7B). On the other hand, activation by IRF-5 S477A and IRF-5 S480A mutants was reduced by two- to threefold compared to that of wt IRF-5. More importantly, the triple mutant, IRF-5 3SA, was unresponsive to virus-mediated activation, yet the ability of the IRF-5 3SA mutant to activate the IFNA1 and -A2 promoter was not significantly diminished in uninfected cells. It should be noted that all of the IRF-5 mutants were able to activate the IFNA1 and -A2 VRE in uninfected cells to nearly the same extent as wt IRF-5. These results indicate that, while Ser residues 477 and 480 are targets for the virus-induced phosphorylation and activation of IRF-5, they are not essential for IRF-5-mediated activation in uninfected cells.
FIG. 7.
Critical role of IRF-5 serine residues 477 and 480 in NDV-induced transactivation. (A) Carboxyl-terminal amino acid homology regions of IRF-5, IRF-3, and IRF-7. Potential serine phosphorylation sites of IRF-5 and serines phosphorylated in IRF-3 and IRF-7 are underlined. The amino acids targeted for alanine substitution are shown in large letters. Hu, human. (B) Expression plasmids (2.5 μg) encoding wt IRF-5 and IRF-5 with point mutations (S475A, S477A, S480A, and 3SA) were cotransfected with IFNA1 or IFNA2 SAP promoters (2.5 μg) into 2fTGH cells. Cells were left uninfected or were infected with NDV for 16 h, and SAP activity was measured as previously described (7).
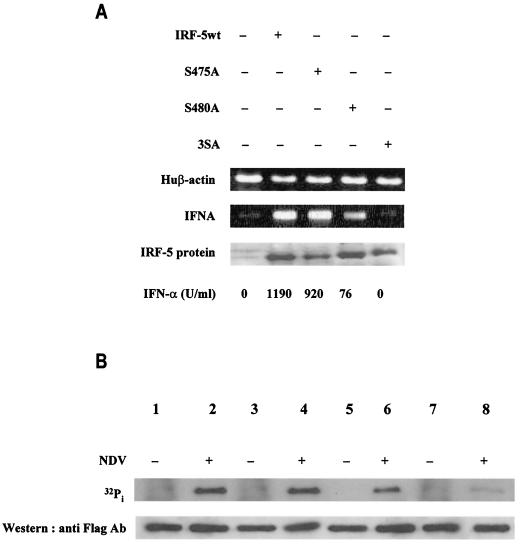
We next examined whether these mutants could also induce expression of the endogenous IFNA genes in NDV-infected 2fTGH cells. Since IRF-5 S477A and IRF-5 S480A were found to have the same transcriptional activity in the transient transfection assay, we compared the activities of the IRF-5 S475A, S480A, and 3SA mutants with the activity of wt IRF-5. Total RNA from 2fTGH cells transfected with wt IRF-5 and mutants IRF-5 S475A, S480A, and 3SA was analyzed 16 h after NDV infection by RT-PCR. The results revealed that wt IRF-5 and IRF-5 S475A induced expression of IFNA genes and synthesis of biologically active IFN-α to similar levels (Fig. 8A). In cells transfected with mutant IRF-5 S480A, levels of biologically active IFN-α induced by NDV were significantly lower and the relative levels of IFNA transcripts were also lower. Levels of biologically active IFN-α could not be detected after NDV infection of 2fTGH cells transfected with the triple serine mutant, IRF-5 3SA. Altogether, these data indicate that the NDV-mediated functional activation of IRF-5 in infected cells requires Ser-477 and -480. However, since the elimination of these two serines did not completely abolish the activation of IRF-5 (Fig. 7B), serines other than these two may be phosphorylated in NDV-infected cells.
FIG. 8.
Serine-477 and -480 are phosphorylated in NDV-infected cells and are essential for the IRF-5-mediated activation of IFNA genes. (A) Analysis of IFNA transcripts in 2fTGH cells transfected with wt IRF-5 or point mutant IRF-5 S475A, S480A, or 3SA and infected with NDV. IFNA cDNAs were amplified by PCR with primers corresponding to the regions of IFNA genes that are conserved in all IFNA subtypes (61). The nuclear levels of IRF-5 and mutants were determined by immunoblotting with an anti-Flag antibody. The levels of biologically active IFN-α synthesized in these cells as determined by an antiviral assay are shown at the bottom. (B) Specific phosphorylation of IRF-5 point mutants by NDV infection. 2fTGH cells were transfected with wt IRF-5 (lanes 1 and 2) or IRF-5 point mutants (lanes 3 and 4, S475A; lanes 5 and 6, S480A; lanes 7 and 8, 3SA) and left uninfected (−) or infected (+) with NDV. Cells were incubated for 6 h in media containing [32P]orthophosphate, and then the 32P-labeled IRF-5 was precipitated from cell lysates with an anti-Flag antibody and proteins were separated by electrophoresis in SDS-7% polyacrylamide gel, dried, and then exposed to a PhosphorImager screen (Molecular Dynamics, Inc., Sunnyvale, Calif.). Top, levels of radiolabeled IRF-5; bottom, levels of IRF-5 protein in cell lysates detected by immunoblotting with an anti-Flag antibody.
While determining the functional role of these serine residues, we also examined whether replacement with alanine affects the levels of phosphorylated IRF-5 detected by metabolic labeling. Accordingly, wt IRF-5 and the three Flag-tagged serine mutants, IRF-5 S475A, S480A, and 3SA, were transfected into 2fTGH cells and cells were infected with NDV and incubated with [32P]orthophosphate. 32P-labeled IRF-5 was detected by immunoprecipitation with anti-Flag antibodies, and levels of phosphorylation were determined by autoradiography. As shown in Fig. 8B, both the wt IRF-5 (lanes 1 and 2) and the S475A mutant (lanes 3 and 4) were phosphorylated to similar levels in NDV-infected cells while no phosphorylation of IRF-5 could be observed in uninfected cells at the same exposure. In comparison, the NDV-induced phosphorylation of the IRF-5 S480A mutant (lane 6) was about fourfold less than that of wt IRF-5. While phosphorylation of the 3SA mutant was nearly undetectable at the same exposure time (lanes 7 and 8), low levels of IRF-5 3SA phosphorylation could be detected with prolonged exposure. These data reveal that IRF-5 phosphorylation is not limited to just S475, S477, and S480 but that phosphorylation of the three serines is functional and necessary for the NDV-induced activation of IRF-5. In addition, it is noteworthy that very low levels of wt IRF-5, IRF-5 S475A, and IRF-5 S480A phosphorylation could be detected in uninfected cells after prolonged exposure. Last, the relative expression levels of all transfected plasmids in uninfected and NDV-infected cells were similar (Fig. 8B).
Dimerization of IRF-5 in virus-infected cells.
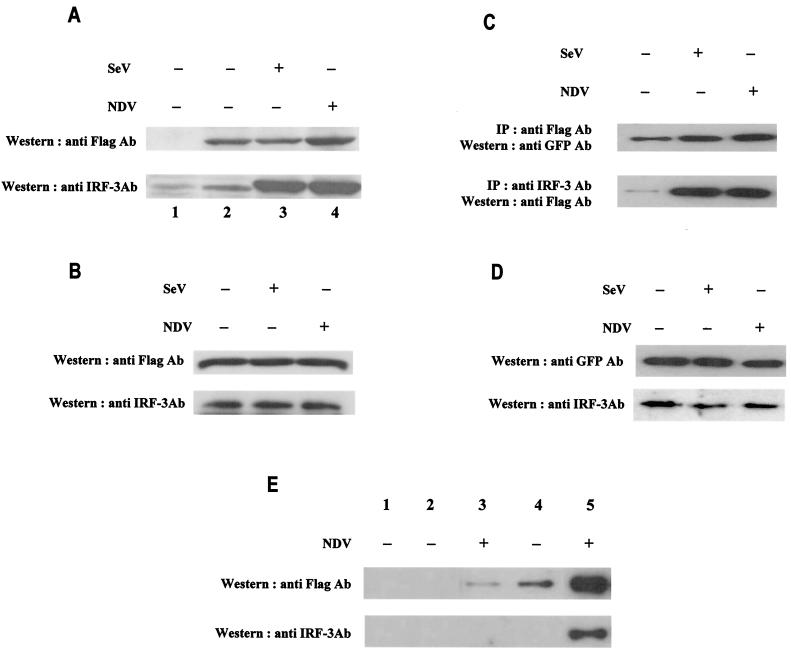
It was previously demonstrated that both IRF-3 and IRF-7 could form homo- and heterodimers in infected cells (31, 32, 38). To determine whether IRF-5 can also form dimers in uninfected or virus-infected cells, we performed GST pull-down and coimmunoprecipitation assays. In the GST pull-down assay, we measured the binding of Flag-tagged IRF-5 from cell lysates of uninfected and NDV-infected 2fTGH/IRF-5 cells to the immobilized GST-IRF-5 fusion protein. As shown in Fig. 9A, while IRF-5 did not bind GST alone (top, lane 1), the binding of IRF-5 from cell lysates of both uninfected and Sendai virus-infected cell lysates to GST-IRF-5 could be detected (top, lanes 2 and 3). However, the binding of IRF-5 from cell lysates of NDV-infected cells to GST-IRF-5 was enhanced about three- to fourfold (top, lane 4). Since the 2fTGH cells contain relatively high levels of endogenous IRF-3, we also measured the binding of IRF-3 to GST-IRF-5. The results show a strong binding of IRF-3 from the lysates of virus-infected cells to GST-IRF-5 while the binding of IRF-3 from the uninfected-cell lysates was much lower (Fig. 9A, bottom, lanes 1 to 4). The relative levels of IRF-3 and IRF-5 in 2fTGH/IRF-5 cells, as determined by Western blot analysis, were about the same (Fig. 9B)
FIG. 9.
Enhanced dimerization of phosphorylated IRF-5 in infected cells. (A) 2fTGH/IRF-5 cells were left uninfected or were infected with Sendai virus or NDV for 6 h. Dimerization of IRF-5 was analyzed by GST pull-down assay (10). Whole-cell extract (250 μg) was applied to GST-agarose beads (lane 1) or GST-tagged IRF-5-agarose beads (lanes 2 to 4), and specifically bound proteins were detected by Western blotting with an anti-Flag antibody (Ab) (top) or an anti-IRF-3 polyclonal antibody (bottom) as described in Materials and Methods. (B) Expression of Flag-tagged IRF-5 and endogenous IRF-3 in the cell lysates analyzed in panel A. Proteins detected represent 8% of input onto GST columns. (C) Detection of IRF-5 dimerization in infected cells. 2fTGH/IRF-5 cells were transfected with GFP-IRF-5 and then infected with Sendai virus or NDV for 6 h. Whole-cell extracts (250 μg) were immunoprecipitated (IP) with either an anti-Flag antibody (top) or an anti-IRF-3 polyclonal antibody (bottom) as described in Materials and Methods. The immunoprecipitated complexes were separated on SDS-7% PAGE gels and subsequently probed with an anti-GFP polyclonal antibody or an anti-Flag antibody to detect IRF-5. (D) Expression levels of endogenous IRF-5 and IRF-3 in the cell lysates of 2fTGH/IRF-5 cells analyzed in panel C. IRF-5 and IRF-3 were detected with an anti-GFP antibody or an anti-IRF-3 antibody, respectively. (E) Formation of the IRF-3/IRF-5 heterodimer is mediated through the IRF-5 carboxyl terminus. Whole-cell lysate (250 μg) from 2fTGH/IRF-5 uninfected or NDV-infected cells, as shown in panel B, was used for mapping the IRF-5 interaction domain. Cell lysates were applied to GST-agarose beads (lane 1) or IRF-5-agarose beads where IRF-5 was tagged with GST at the amino terminus (lanes 2 and 3) or the carboxyl terminus (lanes 4 and 5). Specifically bound proteins were detected by Western blotting with anti-Flag antibodies (top) or anti-IRF-3 antibodies (bottom), as described in Materials and Methods.
To determine whether we could also detect homo- and heterodimerization of IRF-5 in vivo, 2fTGH/IRF-5 cells were transfected with GFP-IRF-5 and left uninfected or infected with Sendai virus or NDV. Cell lysates were immunoprecipitated with an anti-Flag antibody (detecting the endogenous Flag-tagged IRF-5), and the coprecipitated transfected IRF-5 was detected by Western blotting with an anti-GFP antibody. Immunoblot analysis indicated that IRF-5 homodimerization was stronger in infected cells than in uninfected cells (Fig. 9C). A surprising finding was that formation of the IRF-5 homodimer was not specific and was observed in both Sendai virus- and NDV-infected cells, while we could detect phosphorylation and activation of IRF-5 only in NDV-infected cells, not Sendai virus-infected cells. The relative levels of IRF-3 and transfected GFP-IRF-5 in cell lysates were not affected by viral infection (Fig. 9D).
Experiments with an IRF-3-targeted ribozyme (62) revealed that both IRF-3 and IRF-5 were important for the effective induction of IFNA gene transcription in 2fTGH/IRF-5-infected cells (7). As a result, we next examined whether IRF-5 associated with IRF-3 in vivo. To this end, lysates from 2fTGH/IRF-5 cells were immunoprecipitated with an anti-IRF-3 antibody and the immunoprecipitated proteins were then resolved by SDS-PAGE and IRF-5 was detected by Western blot analysis with an anti-Flag antibody. Coprecipitation of IRF-5 with IRF-3 could be detected (Fig. 9C) only in lysates of infected cells, indicating that these two IRFs heterodimerize after virus infection. While Western blot analysis of the cell lysates used for coimmunoprecipitation revealed higher levels of GFP-tagged IRF-5 than of endogenous IRF-3 (Fig. 9D), IRF-3/IRF-5 heterodimerization appeared to dominate over IRF-5 homodimer formation after virus infection. However, we are unable to rule out the possibility that the differences in binding are due to differences in the affinities of the antibodies used, yet these data correlate with observations made from the GST-pull down assay in Fig. 9A and B. When 2fTGH/IRF-5 cells were immunoprecipitated with an anti-Flag antibody and IRF-3 was detected by Western blot analysis with the anti-IRF-3 antibody, formation of the IRF-3/IRF-5 heterodimer appeared to dominate over IRF-5 homodimer formation (data not shown).
Additionally, we mapped the region of IRF-5 that was important for homodimer formation by using amino- and carboxyl-terminal GST-IRF-5 fusion proteins (Fig. 9E). While IRF-5 did not bind to GST alone (top, lane 1), it weakly associated with the amino-terminal domain of IRF-5 (aa 1 to 136) in NDV-infected cells (lane 3). Moreover, IRF-5 associated with the carboxyl-terminal region of IRF-5 (aa 137 to 539) in uninfected cells, yet the interaction was greatly enhanced by NDV infection (Fig. 9E, lanes 4 and 5). Next, we mapped the domain of IRF-5 that interacted with IRF-3 by examining the binding of endogenous IRF-3 from uninfected and NDV-infected 2fTGH/IRF-5 cells to the GST-IRF-5 fusion proteins. IRF-3 did not bind the amino-terminal half (aa 1 to 136) of IRF-5 from uninfected or NDV-infected cells (bottom, lanes 2 and 3) yet bound specifically to the carboxyl-terminal region of IRF-5 in NDV-infected cells (bottom, lane 5). These data indicate that IRF-5 homodimerizes by its carboxyl-terminal region (aa 137 and 539), with possible overlap into the adjacent amino-terminal region, whereas the binding of IRF-3 to IRF-5 occurred only in infected cells and was limited to the carboxyl-terminal domain of IRF-5. Further experiments are needed to determine whether the domains by which IRF-5 homodimerizes or heterodimerizes with IRF-3 are overlapping or distinct.
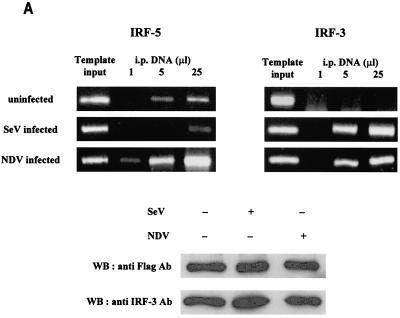
IRF-3 and IRF-5 bind to the endogenous IFNA promoter in infected cells.
Since our results indicated a strong association between IRF-5 and IRF-3 in infected cells, we sought to determine whether both of these factors bind to the endogenous IFN-α promoters in infected cells by using the chromatin immunoprecipitation assay. To this end, 2fTGH/IRF-5 cells were infected either with Sendai virus or NDV and, 6 h postinfection, proteins were cross-linked to DNA and the protein-DNA complexes were precipitated with either anti-Flag antibodies (detecting the Flag-tagged IRF-5) or anti-IRF-3 antibodies (3, 7). The DNA present in the precipitates was then amplified by PCR with universal primers recognizing all endogenous IFNA subtype promoters (61). As shown in Fig. 10A, the fragment corresponding to the IFNA promoters was amplified from the DNA used for immunoprecipitation (lane 1, template input). To show that the PCR amplification of the immunoprecipitated cross-linked DNA was quantitative, serial dilutions of immunoprecipitated DNA were used for PCR to indicate linear amplification. The endogenous IFNA promoters were amplified from the DNA of uninfected cells immunoprecipitated with anti-Flag antibodies but not from the DNA precipitated with anti-IRF-3 antibodies (Fig. 10A). These data suggest that IRF-5 associates with the endogenous IFNA promoters in uninfected cells and correlate well with the observation that IRF-5 activates expression of IFNA promoters in a SAP assay of uninfected cells. Very low levels of amplification from DNA precipitated with anti-Flag antibodies in Sendai virus-infected cells were observed, but IRF-5 binding to the IFNA promoters was greatly enhanced in NDV-infected cells. These results indicate that NDV-specific phosphorylation strongly enhances the binding of nuclear IRF-5 to the endogenous IFNA promoters. When the precipitation was done with anti-IRF-3 antibodies, bands of great intensity only from NDV- and Sendai virus-infected cells were detected, indicating that IRF-3 primarily associates with the IFNA promoters after virus infection. Thus, the binding of unmodified IRF-5 to the IFNA promoter was detected in the absence of IRF-3 binding (Fig. 10A, uninfected), yet, in NDV-infected cells where both IRF-3 and IRF-5 were phosphorylated, both IRF-3 and IRF-5 associated with the endogenous promoter. Western blot analysis has shown that the levels of IRF-3 and IRF-5 in infected and uninfected 2fTGH/IRF-5 cells were comparable.
FIG. 10.
Binding of IRF-5 and IRF-3 to IFNA promoters. (A) In vivo binding of IRF-5 and IRF-3 to the endogenous IFNA promoter as analyzed by the chromatin immunoprecipitation assay. 2fTGH/IRF-5 cells were infected with Sendai virus or NDV or were left uninfected. Cellular DNA and proteins were cross-linked and subjected to the chromatin immunoprecipitation assay as described in Materials and Methods. The immunoprecipitations were performed with either anti-Flag antibody (Ab) or anti-IRF-3 polyclonal antibody to detect IRF-5 and IRF-3 binding, respectively. DNA recovered from chromatin immunoprecipitation by heating was amplified by using universal primers specific for endogenous IFNA genes (61). Template input, amplification of the endogenous IFNA promoter region from DNA-protein complexes before immunoprecipitation. Immunoprecipitated (i.p.) DNA was resuspended in 60 μl of Tris-EDTA. Serial dilutions (1, 5, or 25 μl) were used as templates for PCR amplification to show that the response was in the linear range. Levels of IRF-5 (anti-Flag Ab) and IRF-3 protein in cell lysates as detected by Western blotting are shown. (B) Cooperation between IRF-3 and IRF-5 binding to IFNA1 VRE enhances IRF-5-induced IFNA1 expression. 2fTGH cells were cotransfected with IFNA1 SAP (1 μg) and IRF-3 (2 μg) or IRF-5 (2 μg) or IRF-3 (2 μg) and IRF-5 (2 μg). All transfections were performed with equal amounts of total DNA (5 μg); pUC19 was used as filler DNA. At 16 h posttransfection, cells were left uninfected or were infected with NDV for an additional 16 h, and SAP activity was measured as described in Materials and Methods. SAP activity is expressed after normalizing for β-galactosidase expression.
Since we have shown that formation of the IRF-3/IRF-5 heterodimer occurred primarily after virus infection, we next examined the consequence of heterodimerization and functional activation by a cotransfection assay. Using the IFNA1 SAP reporter plasmid, we determined that cotransfection of both IRF-3 and IRF-5 with the reporter yielded a synergistic effect, where activation was greatly enhanced over the activity observed with either IRF alone (Fig. 10B). Taken together, these results indicate that the IRF-3/IRF-5 heterodimer is a significantly stronger transactivator of the IFNA promoter in infected cells than either homodimer.
DISCUSSION
Most cell types produce IFN in response to virus infection, and there has been significant progress in understanding the cellular pathways activated upon infection with RNA viruses. For RNA viruses, many lines of evidence suggest that dsRNA is the trigger for the activation of IRF-3 (14, 49, 52, 58). It has been previously shown that both IRF-3 and IRF-7 are activated by viral infection and dsRNA, leading to the induction of type I IFN genes (1, 2, 21, 31, 41, 44, 46, 47, 57, 63). Recently we have demonstrated that another IRF, IRF-5, is specifically activated by NDV but not Sendai virus infection and could reconstitute the stimulation of IFNA genes in NDV-infected cells but not in Sendai virus-infected cells (7). In the present study, we show that two other viruses, VSV and HSV-1, can activate IRF-5, stimulate nuclear translocation, and induce expression of IFNA genes and synthesis of biologically active IFN-α proteins. These data indicate that both RNA and DNA viruses can activate IRF-5. However, it was unexpected to find that neither Sendai virus nor dsRNA treatment could activate IRF-5, suggesting that activation of IRF-5 may employ pathways different from those for activation of IRF-3 and IRF-7. The mechanism by which IRF-5 is activated in a virus-specific manner is not understood, yet results indicate that individual and distinct kinases may play an important role (B. J. Barnes, unpublished data).
IRF-7 was shown previously to be a critical factor for the induction of IFNA genes in both human and mouse fibroblast cells as well as in infected mice (37, 44-46, 61). Yet we have found little redundancy in the functions of IRF-5 and IRF-7. Not only is the response of IRF-5 to viral infection more restricted, the IFNA gene subtypes induced by IRF-5 in NDV-infected cells are distinct from the IFNA subtypes induced in NDV-infected IRF-7-expressing cells. Thus, while in NDV-infected human fibrosarcoma cells (2fTGH) IRF-7 primarily induced expression of the IFNA1 gene, in IRF-5-overexpressing cells, IFNA8 was the major IFNA subtype induced (7). These data suggest that the expression profile of IFNA subtypes is determined by distinct IRFs expressed in infected cells as well as by the ratio between the relative levels of expressed IRFs. For instance, modulation of the relative levels of IRF-3 in 2fTGH cells expressing IRF-7 resulted in an altered pattern of IFNA gene expression (62). The observation that IRF-5 can also be activated by VSV and HSV-1 infection allowed us to determine the role of the infecting virus in the induction of individual IFNA subtypes. NDV and VSV infection of IRF-5-expressing 2fTGH cells resulted in about the same profile of expressed IFNA genes, but with altered subtype levels. These results suggest that the profile of individual genes induced is determined by the IRFs and that the levels of expression are determined by the virus. We have previously observed that, in IRF-7-expressing cells, NDV and Sendai virus induced the same profile of IFNA subtypes, with IFNA1 as the major subtype (7, 61). Although it is not yet clear whether the individual IFN-α subtypes have distinct functions, it was shown that the IFN-α8 subtype has much higher antiviral activity than IFN-α1, thus indicating that the functions of IRF-5 and IRF-7 in immune response may be distinct.
Although the expression of IRF-5 is restricted to only few cell types, we have detected high constitutive expression in Namalwa B cells, monocytes, and dendritic cells, as well as precursor dendritic cells (pDC2) that express high levels of IFN-α upon viral infection (P. A. Fitzgerald-Bocarsly, W.-S. Yeow, A. Izaguirre, B. J. Barnes, and P. M. Pitha, 3rd Joint Meeting of ICS and ISCIR, abstr. 04013, 2000; B. J. Barnes unpublished data). The IRF-5-mediated activation of inflammatory genes in infected cells is, however, not limited to the type I IFN genes. In B cells expressing ectopic IRF-5, viral infection resulted in the induction of several CC chemokines including RANTES, MIP-1α and -β, MCP-1, I-309 (potent monocyte chemoattractant and inhibitor of apoptosis in thymic cells), and CXC chemokines IL-8 (neutrophil-activating factor) and IP-10. Most of the chemokines induced by virus in IRF-5-overexpressing cells have lymphocyte-chemotactic activity (5, 6) and are thus important for T-lymphocyte recruitment, suggesting a possible role for IRF-5 in lymphocyte trafficking. Association of IRF-3 and IRF-9 in the induction of RANTES and IP-10, respectively, was previously shown (30, 35). The disregulated activation of some of these chemokines has been observed in disorders associated with leukocyte infiltration and inflammatory disease. For example, patients diagnosed with multiple sclerosis, a chronic neuroinflammatory disease, show high levels of MIP-1α, IP-10, and RANTES in their cerebrospinal fluid (18, 39, 40). In rheumatoid arthritis, an inflammatory disease characterized by the infiltration of inflammatory cells into the synovium-lined joints, chemokines RANTES, MCP-1, MIP-1α, IL-8, and IP-10 were detected in the synovial fluid from actively involved joints (18). Thus, under certain conditions, IRF-5 may function not only as an inducer of the host defense but also as a mediator of pathogenic inflammation.
Interestingly, some of the chemokine genes, such as RANTES, IP-10, and MIP-1α genes, were induced in BJAB/IRF-5 cells by both NDV and Sendai virus while others were induced only by NDV (MIP-1β, MCP-1, IL-8, and I-309 genes). These data indicate that, in B cells expressing high levels of IRF-5, infection with NDV targets the expression of a larger set of inflammatory genes than Sendai virus, suggesting that the signaling pathways induced by NDV are more complex than those induced by Sendai virus. However, the possibility that a Sendai virus-encoded protein, such as Sendai virus C proteins, specifically interferes with the activation and function of individual IRFs and possibly other transcription factors required for the induction of the early inflammatory genes also has to be considered (17, 19, 25). Several viruses were shown to inhibit the function of IRFs as a part of viral mimicry (10, 13, 28, 29, 34, 43, 52). For instance, influenza virus-encoded protein NS1 (52) and human papillomavirus type 16 (HPV-16) (43) target the function of IRF-3, the leader peptide of Theiler virus (55) and Ebola virus VP35 (8, 11) may also target IRF-3, and kaposi sarcoma herpesvirus-encoded vIRFs target the function of IRF-1, IRF-3, and IRF-7 (10, 28, 29, 34, 43). Altogether, these data emphasize the overall complexity of the innate immune response to viral infection and its regulation.
Previously, distinct spliced variants of both IRF-3 and IRF-7 were identified, and some of these variants were shown to modulate the function of full-length IRF-3 and IRF-7 proteins (22, 65). Similarly, three different variants of IRF-5 have been recently identified. The IRF-5 cDNA we have cloned from dendritic cells, B cells, and HB2 breast endothelial cells (7; B. J. Barnes unpublished data) contains a 48-nucleotide (nt) deletion compared with the original IRF-5 sequence deposited in GenBank (U51127). Thus far, we have been unable to isolate the full-size IRF-5 mRNA (variant 1, NM 002200) in any of the cell lines analyzed. Another IRF-5 variant mRNA (variant 2, NM 032643), identified in a renal cell adenocarcinoma, also contains a 48-nt deletion, but adjacent to it is a 30-nt insertion. Sequence analysis of the IRF-5 gene indicated that variants 1 and 2 use an alternative exon for the 5′ untranslated region (UTR). Interestingly, both the deletion and insertion are localized in the PEST domain of IRF-5, and thus the function and stability of these IRF-5 variants may not be identical.
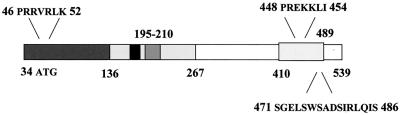
A structure-function analysis of our IRF-5 variant indicates the presence of multiple regulatory domains that control IRF-5 activity (Fig. 11). The serine-rich region located within the transactivation domain of IRF-5 (aa 410 to 489) confers both the constitutive and inducible expression of IRF-5. Furthermore, the C-terminal 50 aa of IRF-5 contains an autoinhibitory domain that reduces IRF-5 activity in both infected and uninfected cells. Both transactivation and autoinhibitory domains were also identified in IRF-3 and IRF-7 polypeptides (1, 32, 33, 38). While in IRF-3 and IRF-7 the transactivation domain is masked by an autoinhibitory domain in the absence of phosphorylation, the autoinhibitory domain of IRF-5 does not completely inhibit IRF-5 transactivation prior to phosphorylation since IRF-5 activates a variety of reporters in uninfected cells, including IFNA and IFNB promoters (7). However, a deletion of the autoinhibitory domain significantly increased the virus-induced activation of IFNA and -B SAP promoters (Fig. 5B).
FIG. 11.
Schematic representation of IRF-5. Structural and functional domains involved in posttranscriptional modification, subcellular translocation, and interaction with other IRF proteins are shown. aa 1 through 728 constitute the full-length IRF-5 protein. aa 35 through 136 represent the DBD (boxed), containing the tryptophan pentad repeats homologous in all IRF family members. The PEST domain is located between aa 152 and 267, where the internal deletion occurs (aa 195 through 210 [grey box]). A glutamate stretch unique to IRF-5 is located from aa 176 through 183 (black box), and a proline-rich region was identified between aa 189 and 340. IRF-5 also contains a C-terminal protein-interacting domain between aa 268 and 470, which contains the transactivation domain localized at aa 410 through 489 (boxed). Amino acids of the amino- and carboxyl-terminal NLSs are indicated at the top, along with the amino acids of the serine-rich cluster, labeled at the bottom.
The IRF-1 and IRF-2 proteins contain functional NLSs located immediately C-terminal of the DBD, between aa 120 and 140 (48). These are represented by a fairly traditional bipartite consensus NLS. The IRF-3 protein contains only two basic residues in its functional NLS (aa 77 and 78), which does not resemble a traditional or nontraditional monopartite or bipartite consensus NLS (26). IRF-4 and IRF-9 also contain a bipartite NLS within basic aa 62 to 90 that reveals very little homology with the NLS regions in IRF-1, IRF-2, and IRF-3 (27). In contrast, the IRF-5 protein contains two traditional monopartite NLSs not identified in the other characterized IRF family members (Table 2). Although both of these NLSs can translocate IRF-5 to the nucleus, our results show that their functions are not identical. The amino-terminal NLS (aa 46 to 52), localized in the DBD of IRF-5, contributes to the nuclear localization of IRF-5 and to its retention in the nucleus. The IRF-5 peptide containing only the 5′ NLS behaves like wt IRF-5 since it is localized in the nuclei of NDV-infected cells, while nuclear localization of the carboxyl-terminal NLS (aa 448 to 454) peptide was transient. These two NLSs are solely responsible for nuclear localization of the IRF-5 polypeptide since the IRF-5 mutant lacking both of these NLSs was localized in the cytoplasm in both infected and uninfected cells and lost its ability to transactivate the IFNA1 promoter. Interestingly, the transactivating abilities of these two NLSs are distinct. While the 5′ NLS was able to transactivate the IFNA1 promoter only in NDV-infected cells, the mutant containing the 3′ NLS activated this promoter in both infected and uninfected cells. These data indicate that, in uninfected cells, the 3′ NLS is exposed and responsible for the observed transactivating potential of IRF-5, while the 5′ NLS is masked by either an intramolecular interaction or association with another protein. Phosphorylation of serines in the carboxyl terminus of IRF-5 results in the exposure of the 5′ NLS and retention of IRF-5 in the nucleus and further enhancement of its transactivating potential. It remains to be determined whether retention of IRF-5 in the cytoplasm of uninfected cells is further facilitated by binding with other cytoplasmic proteins. It was shown that IRF-9, which contains a bipartite NLS, is retained in the cytoplasm by association with STAT2 (27) and intramolecular association between the interaction domain (IAD) and DBD of IRF-4 was previously demonstrated (9, 16). Recently it was also shown that a specific subset of importin-α receptors recognize the IRF-3 NLS and are involved with shuttling IRF-3 to the nucleus (26). Whether these proteins are also involved with the shuttling of IRF-5 into the nucleus has yet to be determined.
The IRF-3 and IRF-7 proteins contain a serine-rich domain localized in the C-terminal region between aa 382 and 410 and aa 471 and 487, respectively, that is targeted for virus-induced phosphorylation. It was shown that the nuclear localization of IRF-3 and IRF-7 and their transactivation potential are dependent on phosphorylation of Ser-385 and Ser-386 of IRF-3 and Ser-477 and Ser-479 of IRF-7 (32, 63). However, it has been shown that, in the IRF-3 polypeptide, these are not the only phosphorylation sites and phosphorylation may also occur at serines in the amino terminus of IRF-3 (49; T. Alce and P. M. Pitha, unpublished data). Phosphorylation of Thr-135 of IRF-3, resulting in nuclear accumulation, was recently identified (22). IRF-5 shows the presence of potential serine phosphorylation sites between aa 471 and 486, and mutational analysis indicated that Ser-477 and Ser-480 of IRF-5 are phosphorylated in cells infected with NDV. These residues also play a critical role in the IRF-5-mediated activation of IFNA promoters in infected cells. It should be noted, however, that these serine residues are not the only phosphorylation targets in the IRF-5 polypeptide since low levels of phosphorylation could be detected in IRF-5 3SA, which lacks serine residues 475, 477, and 480. Interestingly, while the transactivation activity of this mutant in infected cells was negligible, this promoter could still transactivate IFNA promoters in uninfected cells, indicating that the phosphorylation of these serines (475, 477, and 480) is not critical for IRF-5-mediated transactivation in uninfected cells. These data are in agreement with our finding that IRF-5 can also activate IFNA promoters in uninfected cells. Phosphorylation of IRF-5 also facilitates self-association and homodimer formation. However, the fact that some homodimer formation could be detected also in uninfected cells suggests that phosphorylation is not an absolute requirement for IRF-5 self-association. Thus, while IRF-3 can homodimerize only after it is phosphorylated (33, 63), both IRF-5 and IRF-7 can form homodimers, even in uninfected cells (32).
Unlike IRF-5 homodimerization, formation of the IRF-3/IRF-5 heterodimer could be detected only in infected cells, suggesting that phosphorylation of both IRF-5 and IRF-3 (33) leads to a consequential conformational change allowing the interaction between these two proteins to occur. Our results further suggest that the IRF-3/IRF-5 heterodimer is functionally active and plays an important role in the IRF-5-mediated activation of cellular genes. Both of these factors were found to bind the endogenous IFNA promoter in virus-infected cells and are part of the IFNA enhanceosome assembled on the promoters of IFNA genes in NDV-infected cells but not in Sendai virus-infected cells. Decreases in the levels of endogenous IRF-3 due to the presence of an IRF-3-specific ribozyme (62) decreased the in vivo binding of both IRF-5 and IRF-3 to this promoter (7). Interestingly, while in uninfected cells we could detect low levels of IRF-5, but no IRF-3, binding to the endogenous IFNA promoter, in cells infected with Sendai virus, we could detect only the binding of IRF-3. These data suggest that (i) both IRF-3 and IRF-5 need to be phosphorylated to heterodimerize, (ii) the binding affinity of the IRF-5 homodimer to the endogenous IFNA promoter is lower than the binding affinity of the IRF-3/IRF-5 heterodimer, and (iii) the IRF-3/IRF-5 heterodimer is a more effective transactivator than the IRF-5 homodimer.
In conclusion, our data demonstrate that, although there is structural similarity between IRF-5 and virus-activated IRF-3 and IRF-7, IRF-5 exhibits some unique features and functional characteristics that are not shared by the other two IRFs. These include, but are not limited to, (i) IRF-5-mediated activation of a unique subset of IFNA genes, which extends as well to a large number of chemokine genes; (ii) the presence of two functional NLSs in the IRF-5 polypeptide that are absent in the other IRFs; (iii) nuclear translocation and transactivation in uninfected cells, and (iv) virus-specific phosphorylation and activation. These data, together with the previously published studies on the characterization of IRF-3 and IRF-7, indicate that the expression and function of IRF-5 are distinct from those of IRF-3 and IRF-7 and that therefore the mechanisms by which they elicit an immune response may be complementary rather than redundant.
Acknowledgments
We express deep gratitude to Leslie Meszler from the Cell Imaging Core Facility at Johns Hopkins Oncology Center for excellent technical assistance. We also thank Moon Shin for the RANTES luciferase reporter plasmid and Kenneth M. Izumi for the BJAB B-lymphoma cell line.
This work was supported by NIH grants AI-19737-19 and AI-26123-12 to P.M.P. and an Anticancer Drug Development Pharmacology-Oncology training grant to B.J.B.
REFERENCES
- 1.Au, W. C., P. A. Moore, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 2.Au, W.-C., P. A. Moore, W. Lowther, Y.-T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au, W. C., and P. M. Pitha. 2001. Recruitment of multiple interferon regulatory factors and histone acetyltransferase to the transcriptionally active interferon a promoters. J. Biol. Chem. 276:41629-41637. [DOI] [PubMed] [Google Scholar]
- 4.Au, W. C., W. S. Yeow, and P. M. Pitha. 2001. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology 280:273-282. [DOI] [PubMed] [Google Scholar]
- 5.Baggiolini, M., B. Dewald, and B. Moser. 1997. Human chemokines: an update. Annu. Rev. Immunol. 15:675-705. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini, M., B. Dewald, and B. Moser. 1994. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv. Immunol. 55:97-179. [PubMed] [Google Scholar]
- 7.Barnes, B. J., P. A. Moore, and P. M. Pitha. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276:23382-23390. [DOI] [PubMed] [Google Scholar]
- 8.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass, A. L., E. Kehrli, C. F. Eisenbeis, U. Storb, and H. Singh. 1996. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10:2335-2347. [DOI] [PubMed] [Google Scholar]
- 10.Burysek, L., W. S. Yeow, B. Lubyova, M. Kellum, S. L. Schafer, Y. Q. Huang, and P. M. Pitha. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callahan, R., C. S. Cropp, G. R. Merlo, D. S. Liscia, A. P. Cappa, and R. Lidereau. 1992. Somatic mutations and human breast cancer. A status report. Cancer 69:1582-1588. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, S. C., S. K. Chattopadhyay, J. W. Hartley, H. C. I. Morse, and P. M. Pitha. 1991. Aberrant expression of cytokine genes in peritoneal macrophages from mice infected with LP-BM5 MuLV, a murine model of AIDS. J. Immunol. 146:121-127. [PubMed] [Google Scholar]
- 13.Choi, J., R. E. Means, B. Damania, and J. U. Jung. 2001. Molecular piracy of Kaposi's sarcoma associated herpesvirus. Cytokine Growth Factor Rev. 12:245-257. [DOI] [PubMed] [Google Scholar]
- 14.Daly, C., and N. C. Reich. 1993. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol. Cell. Biol. 13:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent, C. L., S. J. Macbride, N. A. Sharp, and D. R. Gewert. 1996. Relative transcriptional inducibility of the human interferon-alpha subtypes conferred by the virus-responsive enhancer sequence. J. Interferon Cytokine Res. 16:99-107. [DOI] [PubMed] [Google Scholar]
- 16.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 17.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerard, C., and B. J. Rollins. 2001. Chemokines and disease. Nat. Immunol. 2:108-115. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 20.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 21.Juang, Y., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99:2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T., T. Y. Kim, W. G. Lee, J. Yim, and T. K. Kim. 2000. Signaling pathways to the assembly of an interferon-beta enhanceosome. Chemical genetic studies with a small molecule. J. Biol. Chem. 275:16910-16917. [DOI] [PubMed] [Google Scholar]
- 24.Kim, T., T. Y. Kim, Y. H. Song, I. M. Min, J. Yim, and T. K. Kim. 1999. Activation of interferon regulatory factor 3 in response to DNA-damaging agents. J. Biol. Chem. 274:30686-30689. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau, J. F., J. P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M., B. Damania, X. Alvarez, V. Ogryzko, K. Ozato, and J. U. Jung. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 20:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 30.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320-34327. [DOI] [PubMed] [Google Scholar]
- 33.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majumder, S., L. Z. Zhou, P. Chaturvedi, G. Babcock, S. Aras, and R. M. Ransohoff. 1998. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J. Immunol 161:4736-4744. [PubMed] [Google Scholar]
- 36.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 37.Marie, I., J. E. Durbin, and D. E. S. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marie, I., E. Smith, A. Prakash, and D. E. Levy. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McManus, C., J. W. Berman, F. M. Brett, H. Staunton, M. Farrell, and C. F. Brosnan. 1998. MCP-1, MCP-2 and MCP-3 expression in multiple sclerosis lesions: an immunohistochemical and in situ hybridization study. J. Neuroimmunol. 86:20-29. [DOI] [PubMed] [Google Scholar]
- 40.Miyagishi, R., S. Kikuchi, T. Fukazawa, and K. Tashiro. 1995. Macrophage inflammatory protein-1 alpha in the cerebrospinal fluid of patients with multiple sclerosis and other inflammatory neurological diseases. J. Neurol. Sci. 129:223-227. [DOI] [PubMed] [Google Scholar]
- 41.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 43.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 45.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 46.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 47.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 48.Schaper, F., S. Kirchhoff, G. Posern, M. Koster, A. Oumard, R. Sharf, B. Z. Levi, and H. Hauser. 1998. Functional domains of interferon regulatory factor I (IRF-1). Biochem. J. 335:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 50.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 284:1835-1837. [DOI] [PubMed] [Google Scholar]
- 51.Slifka, M. K., and J. L. Whitton. 2000. Antigen-specific regulation of T cell-mediated cytokine production. Immunity 12:451-457. [DOI] [PubMed] [Google Scholar]
- 52.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 54.Tjian, R., and T. Maniatis. 1994. Transcriptional activation: a complex puzzle with few easy pieces. Cell 77:5-8. [DOI] [PubMed] [Google Scholar]
- 55.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaughan, P. S., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1997. Interferon regulatory factors: growth control and histone gene regulation—it's not just interferon anymore. J. Mol. Med. 75:348-359. [DOI] [PubMed] [Google Scholar]
- 57.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 58.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, R., A. J. van Wijnen, C. van Der Meijden, M. X. Luong, J. L. Stein, and G. S. Stein. 2001. The cell cycle control element of histone H4 gene transcription is maximally responsive to interferon regulatory factor pairs IRF-1/IRF-3 and IRF-1/IRF-7. J. Biol. Chem. 276:18624-18632. [DOI] [PubMed] [Google Scholar]
- 60.Ye, J., X. Zhang, and Z. Dong. 1996. Characterization of the human granulocyte-macrophage colony-stimulation factor gene promoter: an AP1 complex and an Sp1-related complex transactivate the promoter activity that is suppressed by a YY1 complex. Mol. Cell. Biol. 16:157-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeow, W. S., W. C. Au, Y. T. Juang, C. D. Fields, C. L. Dent, D. R. Gewert, and P. M. Pitha. 2000. Reconstitution of virus-mediated expression of interferon alpha genes in human fibroblast cells by ectopic interferon regulatory factor-7. J. Biol. Chem. 275:6313-6320. [DOI] [PubMed] [Google Scholar]
- 62.Yeow, W.-S., W.-C. Au, W. J. Lowther, and P. M. Pitha. 2001. Downregulation of IRF-3 levels by ribozyme modulates the profile of IFNA subtypes expressed in infected human cells. J. Virol. 75:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, J., C. Angelin-Duclos, J. Greenwood, J. Liao, and K. Calame. 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20:2592-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]