Abstract
The nonhistone chromosomal protein high-mobility group 1 protein (HMG-1/HMGB1) can serve as an activator of p53 sequence-specific DNA binding (L. Jayaraman, N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives, Genes Dev. 12:462-472, 1998). HMGB1 is capable of interacting with DNA in a non-sequence-specific manner and causes a significant bend in the DNA helix. Since p53 requires a significant bend in the target site, we examined whether DNA bending by HMGB1 may be involved in its enhancement of p53 sequence-specific binding. Accordingly, a 66-bp oligonucleonucleotide containing a p53 binding site was locked in a bent conformation by ligating its ends to form a microcircle. Indeed, p53 had a dramatically greater affinity for the microcircle than for the linear 66-bp DNA. Moreover, HMGB1 augmented binding to the linear DNA but not to the microcircle, suggesting that HMGB1 works by providing prebent DNA to p53. p53 contains a central core sequence-specific DNA binding region and a C-terminal region that recognizes various forms of DNA non-sequence specifically. The p53 C terminus has also been shown to serve as an autoinhibitor of core-DNA interactions. Remarkably, although the p53 C terminus inhibited p53 binding to the linear DNA, it was required for the increased affinity of p53 for the microcircle. Thus, depending on the DNA structure, the p53 C terminus can serve as a negative or a positive regulator of p53 binding to the same sequence and length of DNA. We propose that both DNA binding domains of p53 cooperate to recognize sequence and structure in genomic DNA and that HMGB1 can help to provide the optimal DNA structure for p53.
Mutation of the p53 gene is one of the most frequent events in the process of oncogenesis, and it is estimated that approximately 50% of human malignancies contain mutations in this pivotal gene (59). p53 is a transcription factor activated by diverse signals resulting from a variety of cellular stresses, including hypoxia, DNA damage, oncogene overexpression, and viral infection (38, 42, 59). Upon activation, p53 induces a number of downstream targets that play important roles in cell cycle arrest and apoptosis, including p21 (19), mdm2 (7, 51, 62), GADD45 (35), bax (43), cyclin G (49), and many other candidates (17, 64, 66). The in vivo importance of p53 transactivation is underscored by the fact that 93% of all known tumor-derived mutations occur in the DNA binding domain of the protein (23). Mice expressing p53 with a mutated transactivation domain (L25Q/W26S) are incapable of inducing cell cycle arrest in response to various forms of stress and are tumor prone (33), arguing that the ability of p53 to transactivate its target genes is important for its tumor suppressor function.
p53 contains features of a classical transcriptional activator, including an activation domain within its N terminus (codons 20 to 60), a central “core” sequence-specific DNA binding domain (codons 100 to 300), and an oligomerization domain within its C terminus (codons 326 to 355). The central DNA binding region has been shown to bind to sites containing two copies of a rather loose (RRRCA/T T/AGYYY) consensus sequence (18) that is found in the vicinity of many of its target gene promoters. What makes p53 somewhat unique among transcription factors is the fact that its C terminus contains another DNA binding domain that recognizes various forms of DNA in a structure-specific rather than a sequence-specific manner. Three- and four-way junctions (39) and stem-loops are recognized by the C terminus (36). In addition, the C terminus binds single-stranded ends (4, 5, 29), insertion or deletion mismatches (40), recombination intermediates (16), gamma-irradiated DNA (53), gapped DNA (67), and DNA aggregates (63). C-terminal interactions with different forms of DNA have different regulatory effects on the central DNA binding domain. For instance, binding of the C terminus to single-stranded DNA or p53 C-terminal peptide-induced DNA aggregates results in a increase in sequence-specific binding by the central DNA binding domain (29, 53, 63, 67), while binding of the C terminus to gapped DNA inhibits sequence-specific binding by the core (67).
The centrality of p53 as a factor that responds to many forms of stress with diverse cellular outputs suggests that it must be extensively regulated itself. Many modifications and interactions have been shown to take place for both the N-terminal and the C-terminal domains of p53. Some of these have been reported to alter its affinity for DNA (32). N-terminal antibodies can stabilize heat-sensitive DNA binding (21, 25) and cause a decrease in the rate of dissociation of p53 from DNA (13). The following have all been reported to activate sequence-specific DNA binding by p53: deletion of C-terminal residues 363 to 393 (26); addition of C-terminally derived peptides in trans (27); binding by antibody 421 (26), DNA-K (26), or c-Abl (48); phosphorylation by casein kinase II (26), protein kinase C (57), or cyclin-dependent kinases (60); and acetylation by p300/CBP (22, 54). The precise mechanisms by which the above events alter the affinity of p53 for DNA have been the subject of much debate (1). Regardless, as a result of the above studies, which were largely based on experiments with electrophoretic mobility shift assays (EMSAs), which measure binding to small oligonucleotides containing p53 binding sites, the C terminus has long been viewed as a negative regulator of sequence-specific DNA binding by the core domain; however, recent results have suggested that this is not the case (20).
High-mobility group (HMG) 1 protein (HMG-1/HMGB1) was previously identified as a novel enhancer of p53 DNA binding in vitro and transactivation in vivo (31). HMGB1 is a member of the HMG superfamily of proteins and is one of the canonical members (with HMG-2/HMGB2) of the HMG domain subfamily. HMGB1 is composed of two homologous HMG boxes followed by an extremely acidic C-terminal tail. HMGB1 is capable of binding various forms of DNA non-sequence specifically through its HMG boxes but has a particularly high affinity for underwound, bent, or distorted forms of DNA, including cisplatinated DNA. The HMG domain is an L-shaped bundle of alpha helices that use their concave face to bind DNA in the minor groove, resulting in a widened minor groove and a significant bend in the helix. Although the structures of HMGB1 or its derivatives complexed to unmodified DNA have yet to be determined, the crystal structure of the Drosophila melanogaster homolog HMGD, which is composed of a single HMG box most similar to box B of HMGB1, has been published, and the bending angle of the bound DNA has been measured at 111° (44). HMGD is somewhat more efficient in bending DNA than HMGB1, and so the bending angle of the latter may be somewhat smaller (44, 50, 52). Increasing evidence indicates a role for these very abundant proteins (about 1 copy per 10 to 15 nucleosomes) as facilitators of large nucleoprotein complexes (reviewed in reference 58). In addition to its effect on p53, HMGB1 has been shown to increase the DNA binding and either the transactivation or the repression of a growing number of transcription factors, including Oct-1/2 (68), steroid hormone receptors (10), adenovirus major late transcription factor (61), Hox proteins (65), and Rel proteins (11, 15, 41).
Here, in the process of dissecting the molecular mechanism by which HMGB1 augments p53 DNA binding, we have gained insight into the roles of p53 domains in regulating its interactions with DNA.
MATERIALS AND METHODS
Construction, expression, and purification of recombinant HMGB1.
HMGB1 full-length and mutant proteins were cloned by PCR into the NheI and BglII sites of the pRSETC vector and verified by sequencing. The recombinant proteins were then expressed in the BL21(DE3)/pLysS strain of Escherichia coli. Cultures were grown at room temperature to an optical density at 600 nm of 0.5 to 0.8, induced with 0.75 mM isopropyl-β-d-thiogalactoside (IPTG) for 4 h, and then harvested. The proteins were purified in a single step with a nickel-agarose column, except for the full-length protein, which required subsequent purification with a Q-Sepharose column.
Purification of recombinant p53 proteins.
p53 proteins were purified from insect cells that had been infected with recombinant baculoviruses expressing hemagglutinin (HA)-tagged or untagged versions of p53. In either case, cells were infected and extracts that were bound to either anti-p53 polyclonal antibody 421 or anti-HA monoclonal antibody 12CA5 columns were prepared as described previously (30). To elute p53 from the antibody 421 column, a buffer containing 50% ethylene glycol, 0.5 M NaCl, 20 mM Tris-HCl (pH 8.5), 1 mM EDTA (pH 8), 1 mM dithiothreitol, and 10% glycerol was used; for the anti-HA antibody column, the corresponding antigenic peptide (YPYDVPDYA) purchased from SynPep (Dublin, Calif.) was used.
EMSAs.
EMSAs were performed with 20-μl reaction mixtures containing 20 mM HEPES (pH 7.9), 25 mM KCl, 0.5 mM EDTA (pH 8.0), 10% glycerol, 2 mM MgCl2, 2 mM spermidine, 0.025% NP-40, 75 ng of salmon sperm DNA, and 0.15 nmol (or 3 ng) of 32P-labeled oligonucleotide. For some experiments (see Fig. 1C and D), 32-bp blunt-ended oligonucleotides containing the GADD45 site (Invitrogen/Gibco) had the following sequences: 5′-TAGAGCGAACATGTCTAAGCATGCTGGCGTCG-3′ and 5′-CGACGCCAGCATGCTTAGACATGTTCGCTCTA-3′. For all other experiments, the 66-bp oligonucleotides described below were used. Probes were labeled with [γ-32P]ATP and T4 polynucleotide kinase. After 30 min of incubation at room temperature, the reaction mixtures were subjected to electrophoresis on a 4% native polyacrylamide gel (30:1 acrylamide/bisacrylamide) containing 0.5× Tris-borate-EDTA buffer at 165 V for 1.5 h at room temperature. DNA-protein complexes were quantified by phosphorimaging with ImageQuant software.
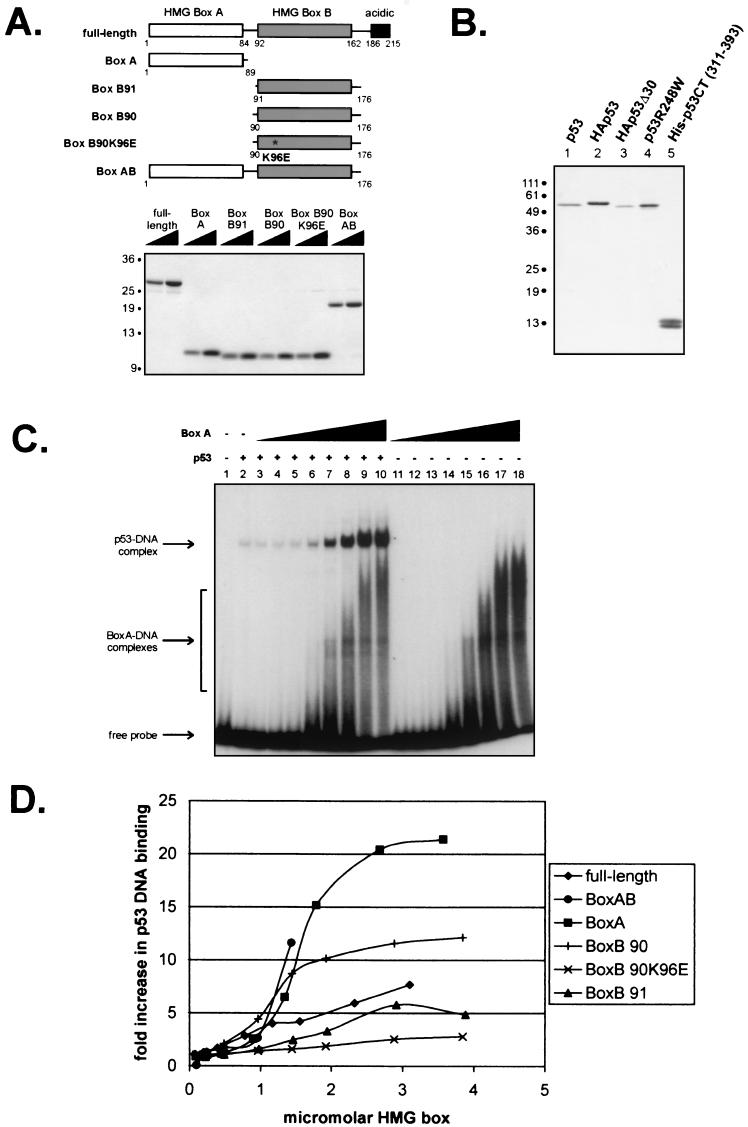
FIG.1.
Isolated HMG boxes efficiently increase p53 DNA binding. (A) Schematic diagram and Coomassie blue-stained gel of purified six-histidine-tagged full-length and truncated mutant forms of HMGB1. (B) Silver-stained gel of immunopurified wild-type and mutant p53 proteins. (C) Representative autoradiogram of an EMSA showing the effects of increasing concentrations of HMGB1 box A (0.09, 0.22, 0.45, 0.89, 1.34, 1.79, 2.68, and 3.57 μM [lanes 3 to 10 and lanes 11 to 18, respectively]) on the binding of p53 (8.5 nM) to a 32-bp oligonucleotide containing the GADD45 p53 binding site. (D) Graphic summary of the effects of the different forms of HMG1B shown in panel A on p53 DNA binding, as determined by EMSA. The units on the x axis take into account the fact that the full-length and box AB constructs have two DNA binding motifs per molecule. The curve for HMG1B box AB terminates at 1.4 μM HMG box because at higher concentrations, this construct bound the labeled DNA probe so well that it obscured the region containing the p53-DNA complex on gels.
Ligation-mediated circularization assay and purification of 66-bp circles.
Sixty-six-base-pair oligonucleotides with NheI overhangs and containing the p53 binding site from the GADD45 promoter were synthesized by Invitrogen/Gibco and had the following sequences: wild-type, 5′-CTAGCTGATATCGAATTCTCGAGCAGAACATGTCTAAGCATGCTGGGCTCGAGAATTCCTGCAGCG-3′ and 5′-CTAGCGCTGCAGGAATTCTCGAGCCCAGCATGCTTAGACATGTTCTGCTCGAGAATTCGATATCAG-3′; and mutant, 5′-TAGCTGATATCGAATTCTCGAGCAGAAAATTTCTAAGAATTCTGGGCTCGAGAATTCCTGCAGCG-3′ and 5′-CTAGCGCTGCAGGAATTCTCGAGCCCAGAATTCTTAGAAATTTTCTGCTCGAGAATTCGATATCAG-3′.
Assays were performed essentially as previously described (50). Briefly, 10-μl reaction mixtures contained 12 nM 32P-labeled 66-bp oligonucleotides, 50 mM HEPES (pH 7.5), 50 mM potassium glutamate, 10 mM magnesium acetate, 1 mM ATP, and HMGB1. The reaction mixtures were incubated at room temperature for 10 min. T4 DNA ligase (0.33 Weiss U; New England Biolabs) was then added, and incubation was continued at 16°C for 30 min and was followed by 15 min of heat inactivation at 65°C. Exonuclease III (6.25 U; New England Biolabs) was then added to the mixtures, followed first by incubation at 37°C for 45 min and then by heat inactivation at 70°C for 20 min. Finally, sodium dodecyl sulfate was added to a final concentration of 0.5% with 200 μg of proteinase K/ml, and the mixtures were subjected to a final incubation at 37°C for 30 min prior to resolution on an 8% polyacrylamide gel (60:1 acrylamide/bisacrylamide) containing 10% glycerol and 1× Tris-borate-EDTA buffer at 30 V for 14 h.
To purify the circular products, ligation reactions were performed as described above, except that, prior to gel electrophoresis, samples were phenol extracted two times and then precipitated in ethanol. After electrophoresis, the linear and circular species were located in the gel by autoradiography, excised, and eluted. The samples were subsequently precipitated in ethanol, quantitated by scintillation, and then used for EMSAs.
RESULTS
Isolated HMG boxes are more efficient in augmenting p53 DNA binding than full-length HMGB1.
HMGB1 consists of three functional domains: two homologous HMG boxes termed box A and box B as well as an acidic C-terminal domain (Fig. 1A). We generated constructs containing both HMG box A and B domains (box AB, amino acids 1 to 176), the isolated HMG box A domain (box A, amino acids 1 to 89), two different versions of the HMG box B domain (box B90, amino acids 90 to 176, and box B91, amino acids 91 to 176), and a mutant of box B90 in which lysine 96 is changed to aspartic acid (box B90 K96E). Box B90 K96E was previously shown to have a markedly reduced affinity for supercoiled DNA (56). In addition, box B proteins that differ by one amino acid at the N terminus were shown to have significantly different abilities to bend DNA (55). DNAs encoding these deletion forms of HMGB1 were cloned into a six-histidine-encoding vector and purified from E. coli (Fig. 1A).
The abilities of the HMGB1 mutants to increase DNA binding by baculovirus-expressed immunopurified p53 were tested by using EMSAs. A silver-stained gel of the p53 proteins used in this study is shown in Fig. 1B. Results from a typical assay with HMGB1 box A are shown in Fig. 1C, and the graphed data obtained from testing all of the above-mentioned mutant forms of HMG1B are shown in Fig. 1D. It was somewhat surprising that each of the isolated HMG boxes was capable of stimulating p53 DNA binding to a greater extent than the full-length protein. Box A enhanced DNA binding by about 20-fold, and box B90 enhanced p53 DNA binding by about 10-fold. The box AB mutant was the most efficient and, at the highest protein concentrations, could not be tested because box AB bound the entire amount of probe and obscured the region containing the p53-DNA complex band on the autoradiogram (data not shown). The box B90 K96E mutant showed a markedly reduced ability to increase DNA binding by p53 relative to the box B90 construct, performing even more poorly than full-length HMGB1 in this assay. In addition, box B91, which differs by only one amino acid from box B90, increased p53 DNA binding to about the same extent as the full-length protein, or only about half as well as box B90.
DNA bending is sufficient for HMGB1 enhancement of sequence-specific DNA binding.
We noted a correlation between the appearance of HMGB1-DNA complexes and the increase in p53 DNA binding, as shown in Fig. 1C. How might DNA binding by HMGB1 work to enhance p53 DNA binding? It has been shown that p53 induces a bend toward the major groove in the target DNA upon binding that has been estimated at 51 to 57° (6, 46, 47) and 40 to 48° (14). Furthermore, there is a positive correlation between the intrinsic flexibilities of different p53 response elements and the affinity of p53 for the site (45). HMGB1 itself has a well-documented ability to bind to DNA in a non-sequence-specific manner (58). HMGB1 interacts with DNA in the minor groove, causing a concomitant distortion in the DNA helix (58). Therefore, it was possible that the ability of HMGB1 to augment p53 DNA binding was related to its ability to interact with DNA and induce a conformational change in the target DNA. Because the ability of HMGB1 to bind to DNA is an intrinsic requirement for its DNA-bending property, it is possible that HMGB1 increases p53 DNA binding by providing a prebent substrate.
To explore this possibility, we took advantage of a ligation-mediated circularization assay that is based upon the empirical observation that DNA shorter than 125 bp will not circularize upon the addition of ligase because of inherent limitations in DNA flexibility (50). However, if a DNA-bending protein is added together with ligase, oligonucleotides as small as 59 bp can be circularized. Circular DNAs that arise from reaction mixtures containing both T4 DNA ligase and a DNA-bending protein such as HMGB1 can be confirmed by treatment with exonuclease III, which will digest linear but not circular DNA molecules (Fig. 2A ). Using this assay, we found that constructs that were able to bind DNA in a filter binding assay were also able to circularize the 66-bp probe (data not shown). One mutant (the box B90 K96E mutant) that was extremely defective in binding to DNA was completely unable to bend DNA in this assay. Unfortunately, however, with respect to the other mutants, the assay was not sufficiently quantitative to allow us to correlate the strengths of DNA binding and DNA bending. As a result, we were unable to separate the abilities of HMGB1 to bind and bend DNA from its abilities to augment p53 DNA binding.
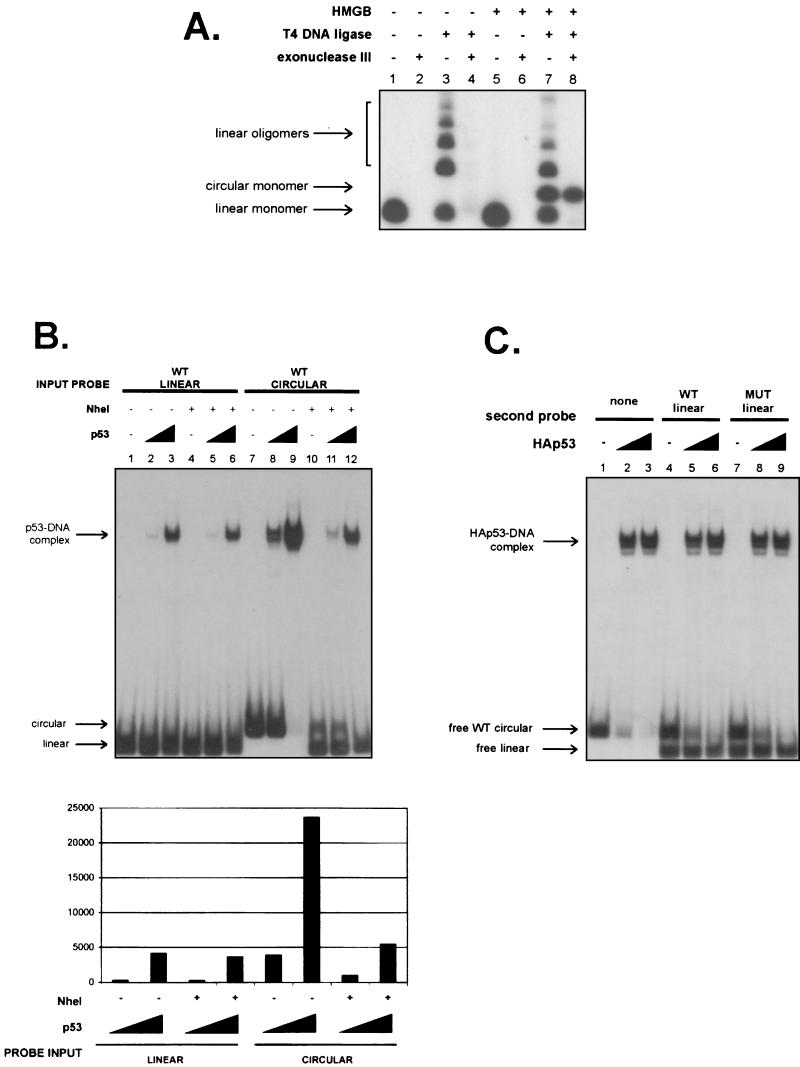
FIG.2.
p53 has a higher affinity for DNA that has been prebent by HMGB1. (A) Representative example of a ligation-mediated circularization assay with 58 nM full-length HMGB1. Reaction mixtures contained HMGB1, T4 DNA ligase, and exonuclease III. Arrows indicate relative migration positions of linear monomers (lanes 1, 3, 5, and 7), linear oligomers (lanes 3 and 7), and circular monomers (lanes 7 and 8). (B) DNA binding reaction mixtures contained either linear or circular 32P-labeled 66-bp DNA segments containing the GADD45 p53 binding site that had been either treated or not treated with restriction endonuclease NheI at 37°C for 14 h in the presence or absence of wild-type (WT) p53 (1.1 and 14 nM [lanes 2 and 3, lanes 5 and 6, lanes 8 and 9, and lanes 11 and 12, respectively]). Quantitation by phosphorimaging is shown below the gel. (C) Reaction mixtures with HA-p53 (15 and 30 nM [lanes 2 and 3, lanes 5 and 6, and lanes 8 and 9, respectively]) contained 32P-labeled wild-type circular 66-bp DNA (3.5 nM) and 32P-labeled wild-type (WT) or mutant (MUT) linear DNA (3.5 nM). Migration positions of free circular and linear probes are indicated.
Nevertheless, the ability to generate circular 66-bp DNA molecules containing a p53 binding site allowed us to test the hypothesis that p53 binds better to HMGB1-bent DNA. A 66-bp oligonucleotide containing the p53 response element from the GADD45 promoter was therefore used to generate and purify exonuclease-resistant microcircles that are dependent on the presence of HMGB1 in ligation mixtures (Fig. 2A). Indeed, when we compared equivalent quantities of linear and circular GADD45 site-containing molecules, there was a striking increase in the affinity of p53 for the latter form of DNA (Fig. 2B, compare lanes 2 and 3 to lanes 8 and 9). We confirmed that the increased number of p53-DNA complexes was in fact the result of the change in the DNA structure of the probe (and not copurifying HMGB1) by showing that linearization of the probe reverted the p53 DNA binding levels to those of the linear probe (Fig. 2B). We considered the alternate possibility that DNA ends were somehow inhibitory to p53 DNA binding and the relevant characteristic of the circular DNA was that it no longer had such ends. This scenario was excluded by mixing the linear and circular probes and verifying that the addition of the linear probe could not inhibit binding to the microcircle (Fig. 2C). In fact, the linear DNA was not capable of competing with the circular DNA for binding to p53, as evidenced by the near disappearance of the circular DNA probe while the free linear probe persisted at the higher level of p53 (Fig. 2C, lanes 6 and 9). Additionally, ligation of the ends of the linear 66-bp DNA creates a sequence resembling a p53 consensus half-site that is 18 bp from the GADD45 site. To ensure that this sequence did not contribute to the increased binding by p53, a second microcircle that contains only the GADD45 consensus site and lacks this extra p53 half-site was generated. p53 bound with virtually the same affinities to both microcircles, thus ruling out the possibility that the presence of an extra half-site contributes to the increased binding to microcircular DNA (data not shown).
It was important to assess whether and to what extent p53 bound site specifically to the microcircle. These questions were addressed by comparing the abilities of p53 to bind to purified circles containing wild-type and mutated GADD45 p53 binding sites (Fig. 3A). p53 had a 30-fold higher affinity for the circular form of the 66-bp DNA fragment than the linear form containing the wild-type GADD45 response element sequence. Surprisingly, p53 also had a higher affinity for the mutated binding site-containing circular DNA than the wild-type linear DNA. The level of p53 binding to the wild-type GADD45 site circle, however, was still four- to sixfold higher than that to the mutant GADD45 site circle. These results thus suggest a role for both sequence-specific and structure-specific affinities of p53 for the binding site-containing circular DNA. Note that there are two p53-DNA complexes with very similar migration profiles. We believe that these represent two slightly different conformations of either p53 or DNA or both. We do not believe that the difference between the two migratory forms is pertinent to the mechanisms that we are studying here, but they are interesting.
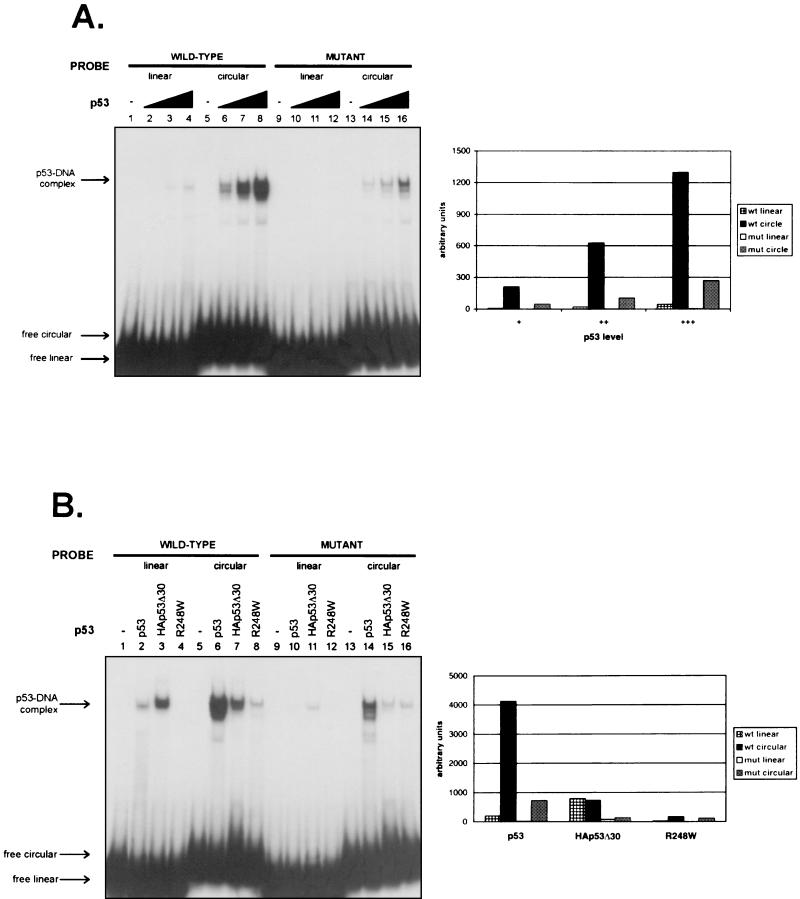
FIG. 3.
Prebending of DNA by HMGB1 is sufficient for enhancement of p53 sequence-specific DNA binding in vitro and requires both the core DNA binding domain and the C terminus of p53. (A) p53 (0.23, 0.46, 0.91, and 1.8 nM [lanes 2 to 4, lanes 6 to 8, lanes 10 to 12, and lanes 14 to 16, respectively]) was incubated with either wild-type (wt) or mutant (mut) 32P-labeled linear or circular DNA, run on a native gel, and autoradiographed. On the right is a graphic representation of the quantitation of the p53-DNA complexes by phosphorimaging. (B) p53, HA-p53Δ30, and R248W (each at 1.8 nM) were compared for their relative sequence- and structure-specific affinities for DNA by EMSAs. Gels were autoradiographed and then quantitated by phosphorimaging, the results of which are shown in the adjacent graph.
Both the central and the C-terminal DNA binding domains of p53 are required for the increased sequence-specific affinity of p53 for the microcircle.
Based on the somewhat unexpected observation that p53 had a higher affinity for the mutant circular probe than the wild-type GADD45 linear probe (Fig. 3A), it was of interest to determine the region(s) of p53 responsible for its increased binding to the microcircle. To address this question, two different mutant versions of p53 purified from baculoviruses were tested by using EMSAs. One mutant lacks the most C-terminal 30 amino acids (p53Δ30) and has been shown to have a higher affinity than full-length p53 for small oligonucleotides containing consensus sites (26). The other variant is a tumor-derived core domain mutant form of p53 (R248W) that is incapable of binding specifically to target DNA sequences but should still retain at least some C-terminal non-sequence-specific DNA binding ability. Figure 1B shows the silver-stained gel of the proteins used.
These different forms of p53 were compared for their relative levels of binding to wild-type and mutated binding site-containing linear and circular DNAs. Figure 3B shows a typical gel and graphic quantitation of the results. The data shown are the results of the determination of carefully precalibrated concentration curves for each of these proteins such that binding by equivalent amounts of p53 proteins is shown. The results of this experiment are summarized as follows. First, full-length p53 again bound far better to the circular form than to the linear form of either wild-type or mutated 66-bp DNA. Second, although p53Δ30 had, as expected, a higher affinity than full-length p53 for the wild-type linear probe, it did not discriminate between linear and circular forms. Third, and perhaps most strikingly, p53Δ30 bound less efficiently (by a factor of 2 to 5) than full-length p53 to the circular form of wild-type DNA. Note that because the p53Δ30 form of p53 was HA tagged at its N terminus, we also compared it to HA-tagged wild-type p53. Although binding by the untagged full-length protein was somewhat higher than that by the tagged form, both untagged and HA-tagged versions of full-length p53 showed the same general trends in binding to the different probes (data not shown). Fourth, p53Δ30 bound poorly to either of the mutant probes. Finally, the only detectable binding by R248W was to the circular DNA; this binding was without any sequence specificity.
The interesting implication of these results is that the C terminus is required for the increased affinity of p53 for the circle. In addition, it is important that the amount of circular DNA bound by full-length p53 was larger than the sum of DNAs bound by p53Δ30 and R248W, suggesting that the central and C-terminal domains bind the circle more efficiently in cis than in trans. We conclude that the C terminus of p53 is required for structure-specific recognition and that the central DNA binding domain imparts sequence specificity to the interaction with a circularized binding site.
Amino acids 311 to 393 of p53 are sufficient for high-affinity recognition of microcircle structure and are required in cis.
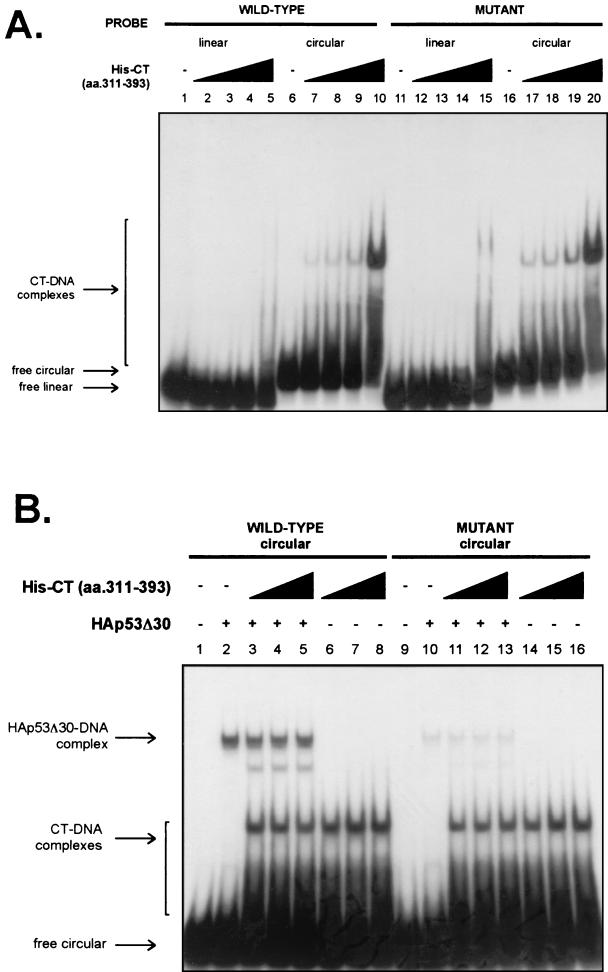
The results shown above prompted us to ask whether the C terminus of p53 can by itself bind with a higher affinity for circular DNA than for linear DNA. A fragment of p53 containing amino acids 311 to 393 tagged with six histidines at the N terminus and purified over a nickel-agarose column (Fig. 1B) was used in DNA binding assays containing both linear and circular probes. As shown in Fig.4A, the C terminus showed a significantly higher affinity for the microcircle than for the linear DNA in a non-sequence-specific manner remarkably reminiscent of the behavior of the R248W mutant, which has a mutant core domain and an intact C terminus. The binding of the C terminus to the circular probe was observed with only 58 nM protein, while an input more than 10-fold higher (706 nM) was necessary to detect binding to the linear probe (Fig. 4A). Moreover, titration of the C terminus in trans to a reaction mixture containing p53Δ30 failed to increase its binding to the circular probe and even competitively reduced its binding at higher concentrations (Fig. 4B). The failure to enhance binding in trans rules out the possibility that the requirement of the C terminus is to confer or stabilize some alternate structure of the target DNA, as has been suggested for the increase in p53 binding to small oligonucleotides in the presence of various basic proteins and the C terminus in trans (63). A novel complex was seen when the C terminus was added to p53Δ30 (Fig. 4B, lanes 3, 4, and 5); the identity of this complex is unknown at present. Taken together, our data show that a high-affinity interaction of p53 with a circularized binding site requires both DNA binding domains of p53 to be in the cis configuration.
FIG. 4.
The C terminus of p53 binds efficiently to bent DNA. (A) The purified six-histidine-tagged human p53 C terminus (amino acids [aa.] 311 to 393; 58, 175, 350, and 706 nM [lanes 2 to 5, lanes 7 to 10, lanes 12 to 15, and lanes 17 to 20, respectively]) was incubated with wild-type and mutant linear and circular DNAs and analyzed by EMSAs. (B) HA-p53Δ30 (1.1 nM) was incubated with the six-histidine-tagged C terminus of p53 (98, 196, and 392 nM [lanes 3 to 5, lanes 6 to 8, lanes 11 to 13, and lanes 14 to 16, respectively]) in the presence of wild-type and mutant circular probes and analyzed by EMSAs.
HMGB1 cannot increase the binding of p53 to microcircles.
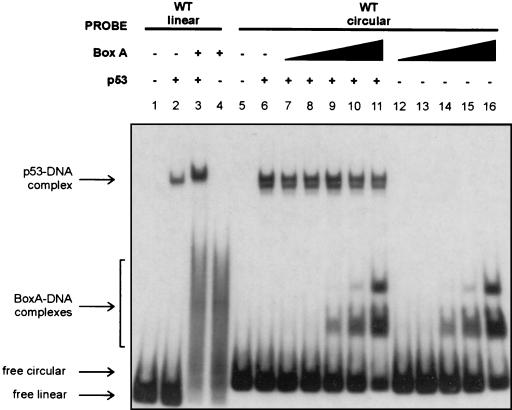
If the structure of the DNA imparted by HMGB1 is responsible for its ability to enhance p53 DNA binding, the addition of HMGB1 to reaction mixtures containing a circular probe should not result in further augmentation of p53 DNA binding. In Fig.5 we show that the addition of one of the strongest enhancers of p53 DNA binding, the box A fragment of HMGB1, did not increase p53 DNA binding to the circular probe. In this experiment, the ability of HMGB1 to enhance p53 DNA binding to the linear form of the 66-bp probe was also confirmed (Fig. 5, lanes 2 and 3). Note that a larger amount of HMGB1 was used in the p53 experiment with linear DNA than with circular DNA. This difference was necessary because HMGB1 has a much higher affinity for circular DNA than for linear DNA (12) (here it was on the order of ∼200- to 500-fold) and larger amounts of HMGB1 essentially outcompete p53 for circular DNA. Given that we found a strong correlation between the ability of HMG1B to bind to linear DNA and to increase p53 DNA binding (Fig. 1C), conclusions from this experiment are valid because at the amount required for detectable HMGB1 box A binding to the linear probe, it enhanced p53 DNA binding, while with the circular probe (Fig. 5, lanes 9 to 11), it actually inhibited p53 DNA binding, possibly through a competitive mechanism. These data argue that the conformational change imparted to DNA by HMGB1 constitutes the mechanism by which it is able to increase p53 DNA binding. The implications of these findings are discussed below.
FIG. 5.
HMGB1 cannot augment the binding of p53 to circular DNA. Increasing amounts of purified HMGB1 box A (0.22, 0.45, 2.2, 4.4, and 11 nM [lanes 7 to 11 and lanes 12 to 16, respectively]) were added to mixtures either containing or lacking p53 (0.9 nM) and containing a purified circular probe with the wild-type (WT) GADD45 binding site. As a positive control, p53 (6.9 nM) was incubated with HMGB1 box A (2.2 μM) in the presence of a linear probe containing the wild-type binding site.
DISCUSSION
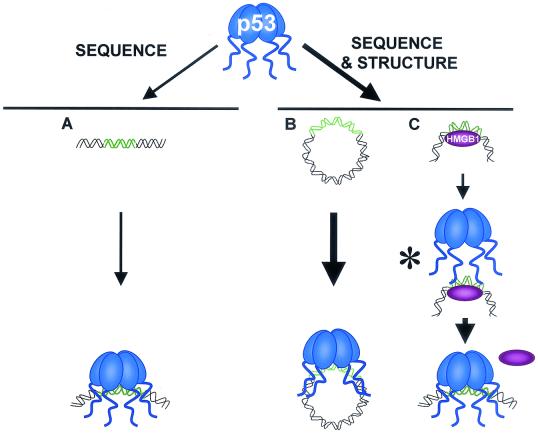
We have discovered that the structural context of the binding site plays a crucial role in its recognition by p53. Our observation that p53 binds far less efficiently to linear DNA than to circular DNA provides an explanation for the ability of HMGB1 to increase the interaction of p53 with DNA. Our data are summarized in Fig.6A and B, and a model depicting the mechanism by which HMGB1 enhances p53 DNA binding is depicted in Fig. 6C. We propose that HMGB1 binds and bends DNA, allowing it to form a configuration that is recognized by the C terminus of p53. This step facilitates the interaction of the core domain with its cognate sequence. The combined effect of the two p53 DNA binding domains results in an increase in the affinity of the full-length protein for the circular DNA that is greater than the added affinity of each domain alone. We suggest that under these conditions, HMGB1 is then rapidly displaced from the p53-DNA complex, as we have not been able to detect a ternary complex.
FIG. 6.
Efficient binding by p53 to DNA requires both sequence-specific core-DNA interactions and structure-specific C terminus-DNA interactions. (A) p53 has only sequence-specific affinity for its binding site in the context of short, linear DNA through its central DNA binding domain. (B) When the binding site is within bent DNA, p53 has both sequence-specific affinity for its binding site through its central DNA binding domain and structure-specific affinity through its C terminus. The combined affinities result in an increase in p53 binding to target sites. (C) HMGB1 provides prebent DNA to p53. The asterisk indicates a potentially unstable or nonexistent p53-HMGB1-DNA complex. Although pairwise interactions between p53 and DNA, HMGB1 and DNA, and p53 and HMGB1 have been demonstrated, there is currently no evidence for a ternary complex.
Our conclusion that the main mode by which HMGB1 increases p53 DNA binding is by providing prebent DNA is supported by at least three lines of evidence. First, HMGB1 augments the binding of p53 to a linear segment of DNA but is not capable of stimulating the binding of p53 to the same sequence in the form of a microcircle locked into a bent conformation. Not even the box A fragment, which is one of the most efficient versions of HMGB1 in bending DNA and in enhancing p53 DNA binding to linear DNA, is capable of enhancing p53 binding to the microcircle. Second, the increase in p53 DNA binding caused by HMGB1 box A is comparable to the relative increase in p53 binding to circular DNA versus linear DNA (30- and 20-fold, respectively). Third, we repeatedly tried to generate 66-bp microcircles in the DNA bending assay with p53 instead of HMGB1 but were not successful (data not shown), consistent with the conclusion that p53 does not bend DNA as efficiently as HMGB1.
Although these data imply that DNA bending by HMGB1 and the subsequent recognition of the bent structure by the p53 C terminus are the main mechanisms by which HMGB1 augments p53 DNA binding, the scenario is complicated by other considerations. It was previously shown that HMGB1 is capable of increasing binding by p53Δ30 to short oligonucleotides (31), while here we show that p53Δ30 did not bind with a higher affinity to the microcircle than to linear DNA. Although a full explanation for this potential contradiction is not yet available, the following should be considered. HMGB1 increases DNA binding by full-length p53 to a significantly greater extent than it does that by p53Δ30 (31; unpublished data), supporting our observation that the C terminus is a component of HMGB1 enhancement of p53 DNA binding. Additionally, the 66-bp circle may not be optimally bent for the core of p53, which could prefer a more moderately bent DNA structure (the core is the only DNA binding domain in p53Δ30). While it has been reported that, like HMGB1, the p53 core domain induces a bend toward the major groove, the bend angle induced by p53 is much smaller. Moreover, even though we constrained the motion of the ends of the DNA somewhat with ligation when we created the microcircles with HMGB1, it is not clear how different the structure of a circle is after the removal of HMGB1. Thus, the bent conformation of the microcircle may not precisely resemble the bend induced by HMGB1. That HMGB1 cannot increase p53 binding to purified circles, though, argues against this possibility. Whatever the explanation, in our experimental model, the major contribution of HMGB1 is to provide DNA that is sufficiently distorted such that recognition by the C terminus is the dominant effect.
It also remains possible that HMGB1 enhancement of p53 DNA binding involves another mechanism instead of or in addition to DNA bending. Protein-protein interactions between p53 and HMGB1 have been documented. It was previously shown that p53 and HMGB1 can interact in vitro (31), and it was subsequently reported by Imamura et al. that amino acids 1 to 91 of HMGB1 and amino acids 363 to 376 of p53 are required for their protein-protein interactions (28). It is difficult to extrapolate the relevance of the interactions mapped by Imamura et al. to our study because the deletion mutants used in their experiments differ in potentially significant ways from the mutant proteins that we have used. Therefore, it is unclear at present whether or not protein-protein interactions play a role in the ability of HMGB1 to increase p53 DNA binding. Furthermore, in the context of cellular chromatin, HMGB1 may act to augment p53 function by modes other than the mechanisms that we suggest here. Many lines of evidence suggest that in cells, HMGB1 serves as an architectural factor, stabilizing large nucleoprotein structures that require an increase in DNA flexibility or a particular stereospecific conformation of the promoter DNA (12, 58). In fact, the topology of DNA bound by p53 was reported to affect its binding. Supercoiling was shown to reduce p53 binding to the mdm2 (but not p21) promoter (37).
The role of the p53 C terminus has long been a subject of interest. While this region of p53 can recognize various forms of DNA, it has also been shown to be able to regulate the sequence-specific interactions of the central core domain with DNA. Until recently, all evidence suggested that the main function of the C terminus is to negatively regulate these interactions. Noncovalent interactions with proteins such as antibodies or covalent modifications by phosphorylation or acetylation increase the ability of p53 to bind to short oligonucleotides, and a C-terminal deletion version of p53 binds more effectively to DNA than does the full-length version of the protein. Hypotheses to explain these observations have ranged from modification-induced allosteric changes in p53 (24, 26, 60) to negative interference by the C terminus resulting from its own nonspecific interactions with DNA (2, 3, 9) to aggregation of target DNA (63). It has become clear that the very assay by which p53 binding is measured can influence whether or not a regulatory effect of the C terminus can be discerned. For example, Cain et al. (13) found that while the C-terminus-specific antibody can augment p53 binding to a 400-bp p21 promoter fragment in an EMSA, it does not do so when binding to the same fragment is measured by DNase I footprinting. Our present study extends these observations by showing that in experiments in which both the same length and sequence of DNA (66-bp GADD45 site-containing DNA) and the same assay (EMSA) were used, the C terminus of p53 could either negatively or positively influence the affinity of p53 for such DNA, depending exclusively on its conformation.
We speculate that a positive role in providing structure recognition is likely to be the dominant effect of the C terminus in cells. The DNA structure in chromatin is prominent, and even its interaction with nucleosomes has a significant impact on its structure. In fact, while the microcircles used here contain 66 bp, only 80 bp form a 360° turn around the nucleosome core particle (approximately 140 bp wrap around each core particle about 1.75 times). Additionally, recent studies have suggested that as the structure predominates, the negative effect of the C terminus is reduced. The acetylation of sites within the p53 C terminus strongly increases the binding of p53 to short oligonucleotides (20, 22, 54) but less so to longer DNA molecules or a chromatinized plasmid template (20). Actually, we found that while the CDK phosphorylation of p53 within the linker of the C terminus increased p53 binding to the linear DNA fragment, it did not enhance binding to the circle (data not shown). Recent studies assessing DNA binding to promoter sites in vivo by using chromatin immunoprecipitation assays also support this conclusion (8, 34). These reports argue that the amount of p53 that is bound to a promoter is largely a function of the level of p53 in the cell rather than a reflection of the extent of its posttranslational modifications, such as acetylation, in clear contrast to data showing sharp increases in the binding of p53 in vitro to short oligonucleotides upon acetylation or phosphorylation of the C terminus. Our results and recent results from other laboratories urge careful consideration of the nature, context, and secondary structure of the binding site used in p53 DNA binding assays.
Acknowledgments
We thank Ella Freulich for expert technical assistance as well as Vanesa Gottifredi, Masha Poyurovsky, Charles Di Como, Jinwoo Ahn, Nicole Baptiste, and members of the Prives laboratory for helpful advice and guidance.
This work was supported by NCI grant CA58316.
REFERENCES
- 1.Ahn, J., and C. Prives. 2001. The C-terminus of p53: the more you learn the less you know. Nat. Struct. Biol. 8: 730-732. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. E., B. Woelker, M. Reed, P. Wang, and P. Tegtmeyer. 1997. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol. Cell. Biol. 17:6255-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayed, A., F. A. Mulder, G. S. Yi, Y. Lu, L. E. Kay, and C. H. Arrowsmith. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8: 756-760. [DOI] [PubMed] [Google Scholar]
- 4.Bakalkin, G., G. Selivanova, T. Yakovleva, E. Kiseleva, E. Kashuba, K. P. Magnusson, L. Szekely, G. Klein, L. Terenius, and K. G. Wiman. 1995. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 23:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakalkin, G., T. Yakovleva, G. Selivanova, K. P. Magnusson, L. Szekely, E. Kiseleva, G. Klein, L. Terenius, and K. G. Wiman. 1994. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc. Natl. Acad. Sci. USA 91:413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balagurumoorthy, P., H. Sakamoto, M. S. Lewis, N. Zambrano, G. M. Clore, A. M. Gronenborn, E. Appella, and R. E. Harrington. 1995. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc. Natl. Acad. Sci. USA 92:8591-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 9.Bayle, J. H., B. Elenbaas, and A. J. Levine. 1995. The carboxyl-terminal domain of the p53 protein regulates sequence-specific DNA binding through its nonspecific nucleic acid-binding activity. Proc. Natl. Acad. Sci. USA 92:5729-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickman, J. M., M. Adam, and M. Ptashne. 1999. Interactions between an HMG-1 protein and members of the Rel family. Proc. Natl. Acad. Sci. USA 96:10679-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 13.Cain, C., S. Miller, J. Ahn, and C. Prives. 2000. The N terminus of p53 regulates its dissociation from DNA. J. Biol. Chem. 275:39944-39953. [DOI] [PubMed] [Google Scholar]
- 14.Cherny, D. I., G. Striker, V. Subramaniam, S. D. Jett, E. Palecek, and T. M. Jovin. 1999. DNA bending due to specific p53 and p53 core domain-DNA interactions visualized by electron microscopy. J. Mol. Biol. 294:1015-1026. [DOI] [PubMed] [Google Scholar]
- 15.Decoville, M., M. J. Giraud-Panis, C. Mosrin-Huaman, M. Leng, and D. Locker. 2000. HMG boxes of DSP1 protein interact with the rel homology domain of transcription factors. Nucleic Acids Res. 28:454-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudenhoffer, C., G. Rohaly, K. Will, W. Deppert, and L. Wiesmuller. 1998. Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol. Cell. Biol. 18:5332-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 19.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 20.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 21.Friedlander, P., Y. Legros, T. Soussi, and C. Prives. 1996. Regulation of mutant p53 temperature-sensitive DNA binding. J. Biol. Chem. 271:25468-25478. [DOI] [PubMed] [Google Scholar]
- 22.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 23.Hainaut, P., and M. Hollstein. 2000. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 77:81-137. [DOI] [PubMed] [Google Scholar]
- 24.Halazonetis, T. D., and A. N. Kandil. 1993. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 12:5057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, S., T. R. Hupp, and D. P. Lane. 1996. Allosteric regulation of the thermostability and DNA binding activity of human p53 by specific interacting proteins. CRC Cell Transformation Group. J. Biol. Chem. 271:3917-3924. [DOI] [PubMed] [Google Scholar]
- 26.Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 27.Hupp, T. R., A. Sparks, and D. P. Lane. 1995. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell 83:237-245. [DOI] [PubMed] [Google Scholar]
- 28.Imamura, T., H. Izumi, G. Nagatani, T. Ise, M. Nomoto, Y. Iwamoto, and K. Kohno. 2001. Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J. Biol. Chem. 276:7534-7540. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman, L., and C. Prives. 1995. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81:1021-1029. [DOI] [PubMed] [Google Scholar]
- 30.Jayaraman, L., E. Freulich, and C. Prives. 1997. Functional dissection of p53 tumor suppressor protein. Methods Enzymol. 283:245-256. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman, L., N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman, L., and C. Prives. 1999. Covalent and noncovalent modifiers of the p53 protein. Cell Mol. Life Sci. 55:76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jimenez, G. S., M. Nister, J. M. Stommel, M. Beeche, E. A. Barcarse, X. Q. Zhang, S. O'Gorman, and G. M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37-43. [DOI] [PubMed] [Google Scholar]
- 34.Kaeser, M. D., and R. D. Iggo. 2002. From the cover: chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity invivo. Proc. Natl. Acad. Sci. USA 99:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 36.Kim, E., N. Albrechtsen, and W. Deppert. 1997. DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene 15:857-869. [DOI] [PubMed] [Google Scholar]
- 37.Kim, E., G. Rohaly, S. Heinrichs, D. Gimnopoulos, H. Meissner, and W. Deppert. 1999. Influence of promoter DNA topology on sequence-specific DNA binding and transactivation by tumor suppressor p53. Oncogene 18:7310-7318. [DOI] [PubMed] [Google Scholar]
- 38.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 39.Lee, S., L. Cavallo, and J. Griffith. 1997. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem. 272:7532-7539. [DOI] [PubMed] [Google Scholar]
- 40.Lee, S., B. Elenbaas, A. Levine, and J. Griffith. 1995. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell 81:1013-1020. [DOI] [PubMed] [Google Scholar]
- 41.Lehming, N., A. Le Saux, J. Schuller, and M. Ptashne. 1998. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 43.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 44.Murphy, F. V. T., R. M. Sweet, and M. E. Churchill. 1999. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 18:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaich, A. K., E. Appella, and R. E. Harrington. 1997. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J. Biol. Chem. 272:14842-14849. [DOI] [PubMed] [Google Scholar]
- 46.Nagaich, A. K., V. B. Zhurkin, S. R. Durell, R. L. Jernigan, E. Appella, and R. E. Harrington. 1999. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc. Natl. Acad. Sci. USA 96:1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagaich, A. K., V. B. Zhurkin, H. Sakamoto, A. A. Gorin, G. M. Clore, A. M. Gronenborn, E. Appella, and R. E. Harrington. 1997. Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J. Biol. Chem. 272:14830-14841. [DOI] [PubMed] [Google Scholar]
- 48.Nie, Y., H. H. Li, C. M. Bula, and X. Liu. 2000. Stimulation of p53 DNA binding by c-Abl requires the p53 C terminus and tetramerization. Mol. Cell. Biol. 20:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto, K., and D. Beach. 1994. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 13:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paull, T. T., M. J. Haykinson, and R. C. Johnson. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 7:1521-1534. [DOI] [PubMed] [Google Scholar]
- 51.Perry, M. E., J. Piette, J. A. Zawadzki, D. Harvey, and A. J. Levine. 1993. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc. Natl. Acad. Sci. USA 90:11623-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pil, P. M., C. S. Chow, and S. J. Lippard. 1993. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl. Acad. Sci. USA 90:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed, M., B. Woelker, P. Wang, Y. Wang, M. E. Anderson, and P. Tegtmeyer. 1995. The C-terminal domain of p53 recognizes DNA damaged by ionizing radiation. Proc. Natl. Acad. Sci. USA 92:9455-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stros, M. 1998. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 273:10355-10361. [PubMed] [Google Scholar]
- 56.Stros, M., and J. Reich. 1998. Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA. Eur. J. Biochem. 251:427-434. [DOI] [PubMed] [Google Scholar]
- 57.Takenaka, I., F. Morin, B. R. Seizinger, and N. Kley. 1995. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J. Biol. Chem. 270:5405-5411. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ′architectural' DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 59.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., and C. Prives. 1995. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature 376:88-91. [DOI] [PubMed] [Google Scholar]
- 61.Watt, F., and P. L. Molloy. 1988. High mobility group proteins 1 and 2 stimulate binding of a specific transcription factor to the adenovirus major late promoter. Nucleic Acids Res. 16:1471-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 63.Yakovleva, T., A. Pramanik, T. Kawasaki, K. Tan-No, I. Gileva, H. Lindegren, U. Langel, T. J. Ekstrom, R. Rigler, L. Terenius, and G. Bakalkin. 2001. p53 latency. C-terminal domain prevents binding of p53 core to target but not to nonspecific DNA sequences. J. Biol. Chem. 276:15650-15658. [DOI] [PubMed] [Google Scholar]
- 64.Yu, J., L. Zhang, P. M. Hwang, C. Rago, K. W. Kinzler, and B. Vogelstein. 1999. Identification and classification of p53-regulated genes. Proc. Natl. Acad. Sci. USA 96:14517-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zappavigna, V., L. Falciola, M. Helmer-Citterich, F. Mavilio, and M. E. Bianchi. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981-4991. [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
- 67.Zotchev, S. B., M. Protopopova, and G. Selivanova. 2000. p53 C-terminal interaction with DNA ends and gaps has opposing effect on specific DNA binding by the core. Nucleic Acids Res. 28:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwilling, S., H. Konig, and T. Wirth. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]