Abstract
We previously reported the cloning and characterization of a novel nuclear hormone receptor transcriptional coactivator, which we refer to as NRC. NRC is a 2,063-amino-acid nuclear protein which contains a potent N-terminal activation domain and several C-terminal modules which interact with CBP and ligand-bound nuclear hormone receptors as well as c-Fos and c-Jun. In this study we sought to clone and identify novel factors that interact with NRC to modulate its transcriptional activity. Here we describe the cloning and characterization of a novel protein we refer to as NIF-1 (NRC-interacting factor 1). NIF-1 was cloned from rat pituitary and human cell lines and was found to interact in vivo and in vitro with NRC. NIF-1 is a 1,342-amino-acid nuclear protein containing a number of conserved domains, including six Cys-2/His-2 zinc fingers, an N-terminal stretch of acidic amino acids, and a C-terminal leucine zipper-like motif. Zinc fingers 1 to 3 are potential DNA-binding BED finger domains recently proposed to play a role in altering local chromatin architecture. We mapped the interaction domains of NRC and NIF-1. Although NIF-1 does not directly interact with nuclear receptors, it markedly enhances ligand-dependent transcriptional activation by nuclear hormone receptors in vivo as well as activation by c-Fos and c-Jun. These results, and the finding that NIF-1 interacts with NRC in vivo, suggest that NIF-1 functions to regulate transcriptional activation through NRC. We suggest that NIF-1, and factors which associate with coactivators but not receptors, be referred to as cotransducers, which act in vivo either as part of a coactivator complex or downstream of a coactivator complex to modulate transcriptional activity. Our findings suggest that NIF-1 may be a functional component of an NRC complex and acts as a regulator or cotransducer of NRC function.
Nuclear hormone receptors comprise a family of ligand-dependent transcription factors that have a broad effect on gene expression, growth, and development (2, 38, 39). These include the thyroid hormone receptors (TRs) for thyroid hormone (T3), the retinoic acid (RA) receptors (RARs) for all trans RA, the RARs and the retinoid X receptors (RXRs) for 9-cis RA, vitamin D receptor (VDR) for 1,25-(OH)2 vitamin D3, glucocorticoid receptor (GR), progesterone receptor, estrogen receptors (ERs), and the peroxisome proliferator-activated receptors (PPARs), which are regulated by a variety of lipophilic compounds. These receptors share a similar modular structure consisting of an N-terminal A/B domain, a DNA-binding C domain, and a D, E, and F ligand binding domain (LBD) (7, 38). The LBDs of nuclear receptors are organized into 12 helical regions, and the binding of ligand to the LBD of a DNA-bound receptor mediates a conformational change which recruits coactivators or coregulators, leading to transcriptional activation (38, 49).
Coactivators which have been identified include members of the p160 family (SRC-1/NCoA-1 [27, 42], TIF-2/GRIP-1/NCoA-2 [23, 50, 52] and AIB1/p/CIP/ACTR/RAC3/TRAM-1 [1, 12, 32, 48, 50]), the CBP/p300 family (9, 20, 27), RIP140 (8), NRC/ASC-2/PRIP/RAP250/TRBP (6, 28, 30, 34, 58), PGC-1 (44), ARA70 (56), p/CAF (4, 55), and NRIF3, which exhibits specificity for only the TRs and the RXRs (31). In addition to mediating the effects of nuclear hormone receptors, certain coactivators also appear to enhance the activity of other transcription factors such as NF-κB, c-Fos, and c-Jun (28).
The DRIPs/TRAPs are another class of factors which are recruited to ligand-bound nuclear hormone receptors (e.g., VDR and TR) (15, 45). The DRIPs and TRAPs are multiprotein complexes which appear to be similar, if not identical, and interestingly, are devoid of the p160 type of coactivators. Some of the polypeptides of the DRIP/TRAP complex also appear to be a part of the SMCC, CRSP, and ARC complexes (24, 40, 46). The DRIP/TRAP complexes associate with ligand-bound TR or VDR via an ∼220-kDa component referred to as PBP/TRAP220/DRIP205 (15, 45, 60), and other components of the complex interact with other transcription factors (24, 36, 40, 45, 46).
The association of coactivators with receptors occurs through receptor-interacting LXXLL modules of the coactivator (13, 22, 34, 37), which bind to a hydrophobic cleft in the ligand-bound receptor formed by several regions of the LBD (13, 14, 41). The p160 family of coactivators, RIP140, and TRAP220/DRIP205 contain multiple LXXLL motifs (22), which is consistent with the idea that a single molecule of the coactivator can bind a nuclear receptor dimer in vivo (13, 37).
We previously reported the cloning and characterization of NRC (nuclear receptor coactivator) (34) (also referred to as ASC-2/PRIP/RAP250/TRBP) from rat and human cells. NRC acts as a potent coactivator for nuclear hormone receptors (34) and other transcription factors, such as c-Fos, c-Jun, and NF-κB (28). The importance of NRC as an essential coregulator is reflected by the finding that NRC null mice are embryonic lethal (M. Mahajan and H. Samuels, unpublished data). NRC is organized into several modular domains which appear to play an important role in its function as a coactivator and coregulator for nuclear hormone receptors. NRC contains one functional LXXLL motif (LXXLL-1) that binds all nuclear receptors with high affinity. This appears to occur through the formation of NRC dimers, thus contributing two LXXLL motifs to bind nuclear receptor dimers (34). A region containing a second LXXLL motif (LXXLL-2) appears to be highly selective for estrogen-bound ERs. NRC harbors a potent N-terminal activation domain (AD1), which is as active as the VP16 activation domain, and a second activation domain (AD2), which overlaps with the receptor-interacting LXXLL-1 region. Receptor binding mediates a conformational change in NRC, resulting in enhanced activity of the coactivator (34). The C-terminal region of NRC appears to function as a modulatory domain which influences the overall activity of NRC. NRC binds CBP/p300 with high affinity in vivo (34) and in vitro (28), suggesting that NRC may be an important functional component of CBP/p300 complexes in the cell.
CBP and p300, which exhibit intrinsic histone acetyltransferase activity, function as transcriptional integrators for multiple factors, including p/CAF (a histone acetyltransferase) (55), NF-κB (43), the STATs (57), nuclear hormone receptors (9, 20, 27), the p160 family (50, 51), E1A (10), p53 (33), and NRC (28, 34). Although NRC appears to associate with CBP in vivo (34), the identity of other factors that are part of this or other NRC complexes that play a role in the action of NRC are unknown. In this study, we report the cloning and characterization of NRC-interacting factor 1 (NIF-1), which associates with and enhances the activity of NRC in vivo. NIF-1 is a novel nuclear protein of the recently proposed BED finger domain family (3) containing six zinc fingers which directly interacts with NRC but not with nuclear hormone receptors. Although NIF-1 does not bind directly to nuclear hormone receptors, it markedly enhances their ligand-dependent transcriptional activity in vivo. In addition, like NRC, NIF-1 also enhances the activities of c-Fos and c-Jun in vivo. Our results indicate that NIF-1 can be considered an example of an emerging new class of coregulators (which we refer to as cotransducers) which act as part of a complex in vivo to modulate coactivator activity.
MATERIALS AND METHODS
Yeast two-hybrid cDNA library from GH4C1 cells.
Poly(A)+ RNA isolated from GH4C1 cells was used for the synthesis of cDNA with a Stratagene cDNA synthesis system. cDNA was size fractionated and ligated with EcoRI-XhoI-digested pJG4-5, which conditionally expresses the cDNA as a fusion with the B42 activation domain in Saccharomyces cerevisiae (17). The construction of the cDNA library has been described earlier (34).
Yeast two-hybrid screen.
NRC-b (amino acids 849 to 2,063) (see Fig. 6A) was cloned into pEG202ΔPL, a modified yeast LexA expression vector, and used as bait in a two-hybrid screen. pEG202ΔPL was derived from the parent vector pEG202, as described earlier (34). All methods and transformation procedures have been described previously (34). The yeast strain EGY48 harboring the LacZ reporter pSH18-34 (17) and pEG202ΔPL-NRC-b was transformed with the GH4C1 pJG4-5 cDNA library. Transformants were directly screened on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) synthetic defined minimal medium (SD)-galactose-raffinose plates lacking Trp, Ura, His, and Leu. Putative positive clones were further purified on Trp−, Ura−, His−, and Leu− SD-galactose-raffinose plates. The purified clones were plated on SD-dextrose Trp−, Ura−, and His− plates to repress the expression of cDNAs from pJG4-5. Galactose-inducible interactions were verified upon replica plating each clone on Trp−, Ura−, His−, and Leu− X-Gal SD-galactose-raffinose and Trp−, Ura−, and His− X-Gal SD-dextrose plates. Yeast clones exhibiting a positive LacZ response on galactose-raffinose plates, but not on dextrose plates, were considered to be potential NRC-interacting clones. The putative cDNAs from positive clones were further verified against several different baits. The positive interactors were then sequenced and subjected to restriction digestion, size determination, and further analysis.
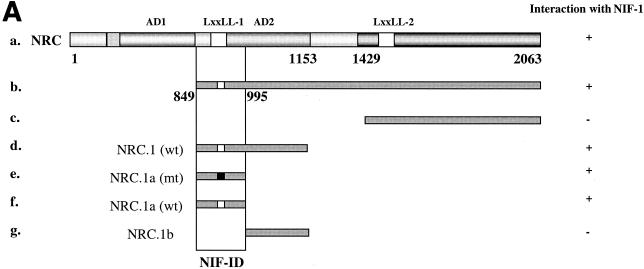
FIG. 6.
Identification of the NIF-ID of NRC. (A) Interaction of NIF-1 with NRC in yeast. Each of the LexA-NRC fusions was tested for interaction with various constructs of NIF-1 (Fig. 5) expressed as B42 fusions. All of the fragments of NRC containing the NIF-ID interact with NIF-1 clones containing the NRC-ID. MT fragments depicted are NRC clones containing mutations in the LXXLL-1 receptor interaction motif in which LVNLL was changed to AVNAA. +, positive; −, negative. (B) Binding of NIF-1 with NRC in vitro. NIF-1 was labeled with [35S]l-methionine by in vitro transcription-translation with reticulocyte lysates. Bacterially expressed and purified GST-NRC.1a (a 147-amino-acid region of NRC that contains the NIF-1-ID) bound to glutathione-agarose beads was incubated with 35S-labeled NIF-1. The samples were then electrophoresed in SDS gels and the 35S-NIF-1 bound to GST or GST-NRC.1a was visualized by autoradiography. One fifth of the amount of 35S-labeled NIF-1 used in the incubation was also electrophoresed in the same gel.
Plasmids.
Expression plasmids for nuclear receptors and various reporters have been described earlier (31, 34). A Flag-tagged sequence was introduced into the 5′ end of full-length NIF-1 cDNA by PCR and cloned into a pEX vector (24, 31). Other than the Flag tag, pEX-FlagNIF-1 is identical to pEX-NIF-1. Gal4-LBD MOR (mouse ERα [mERα]) was kindly provided by Malcom Parker (35). All plasmids described below were generated by either PCR or restriction enzyme digestion and verified by sequencing and expression studies. Human pEX-NRC and various NRC fragments in pJG4-5ΔPL (B42 fusions) and pEG202ΔPL (LexA fusions), such as human NRC-c (amino acids 1,429 to 2,063), rat NRC.1, wild-type (WT) and mutant (MT) and human NRC (amino acids 849 to 1,153), which is analogous to the residues found in rat NRC.1, have been previously described (34). B42 and LexA fusions of rat NRC.1a (amino acids 849 to 995, WT and MT) and NRC.1b (amino acids 995 to 1,153) were amplified by PCR with specific primers and cloned into both pEG202ΔPL and/or pJG4-5ΔPL yeast vectors, sequenced, and examined for protein expression. Rat NRC.1a was cloned as a glutathione S-transferase (GST) fusion in pGEX4T (GST-NRC.1a). LexA-human VDR (hVDR)-LBD was generously provided by David Moore, Baylor College of Medicine. hERα LBD in pJG4-5 was from Michael Garabedian, Department of Microbiology, New York University School of Medicine. LexA-hERα LBD was produced by releasing the LBD from pJG4-5 and cloning it into pEG202ΔPL.
Cloning of human NIF-1 and NIF-1 expression plasmids.
Human NIF-1 was cloned by screening a lambda gt10 phage library derived from a human cell line, NTera-2D1 (kindly provided by Naoko Tanese and Angus Wilson, Department of Microbiology, New York University School of Medicine). One of the positive phage clones (6B) containing a 4.5-kb cDNA insert was identified as a nearly full-length NIF-1 lacking 110 bp from the 5′ end. The 3′ end of the 6B phage includes a stop codon, about 315 bp of the 3′ untranslated region sequence, and a short poly(A) tail. Full-length NIF-1 was generated by ligating a 226-bp HindIII-XhoI PCR product of an expressed sequence tag (EST) (BE297231) (see below) containing a consensus Kozak homology at the 5′ end with an XhoI-EcoRI fragment of human NIF-1 (∼4.5 kb) released from the 6B clone. The full-length NIF-1 cDNA was cloned into the pEX (34) and pcDNA3 vectors (Invitrogen) and contains coding sequences of 4,029 bp, with HindIII at the 5′ end and EcoRI at the 3′ end. Green fluorescent protein (GFP)-NIF-1 was generated by releasing full-length NIF-1 cDNA from pEX-NIF-1 with HindIII-EcoRI and cloning it into pEGFP(C3) (Clontech). Human NIF-1 (6B) was also cloned into the EcoRI site of pJG4-5ΔPL. One of the EST clones (BE297231) (IMAGE Consortium) was sequenced completely, identified as an isoform of human NIF-1, and designated NIF-2. The NIF-2 cDNA lacking the first 222 nucleotides (N-terminal 74 amino acids) was released from pOTB7 with XhoI, end-filled, and cloned into the pJG4-5ΔPL and pEG202ΔPL yeast vectors at NcoI-filled ends. The following plasmids were cloned into the pJG4-5ΔPL and/or pEG202ΔPL yeast vectors (see Fig. 5). (i) Human NIF-1(f), the C-terminal region of human NIF-1 (representing amino acids 1043 to 1342), containing zinc finger 6, the leucine zipper-like motif, and the remainder of the C terminus was cloned into NcoI-filled XhoI sites. (ii) Human NIF-1(e), an SmaI-XhoI NIF-1 fragment representing amino acids 1138 to 1342, was ligated with NcoI-filled XhoI-cut vectors. This clone contains the leucine zipper-like motif and the remaining C terminus of NIF-1. (iii) Human NIF-1(b), a NotI-EcoRI-end-filled fragment representing amino acids 42 to 644 of the N-terminal region of NIF-1, was cloned into BamHI-end-filled yeast vectors. (iv) Human NIF-1(g), representing amino acids 1007 to 1150 and harboring zinc fingers 5 and 6, was generated by PCR with specific primers and cloned as an XhoI-EcoRI fragment.
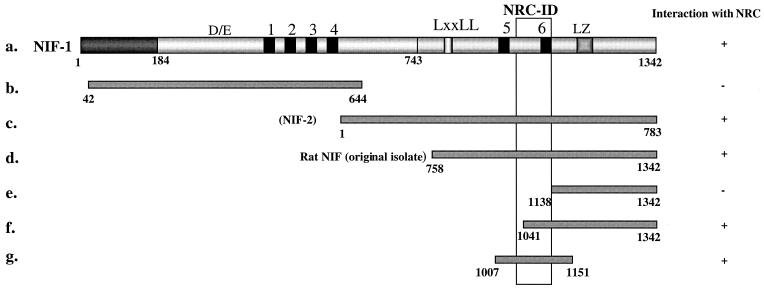
FIG. 5.
NIF-1 interacts with NRC in yeast through a region containing zinc finger 6. Various NIF constructs were generated as B42 fusions and tested against each of the LexA fusions of NRC shown in Fig. 6A, a to g, in two-hybrid interaction assays. Rat NIF is the original isolate from the GH4C1 library while NIF-2 is an isoform of human NIF-1 that lacks amino acids 184 to 743, which includes the DE region and zinc fingers 1 to 4. The numbers correspond to amino acids. All of the NIF fragments containing the NRC-ID interacted with NRC in two-hybrid assays. +, positive; −, negative.
Transfections into mammalian cells.
Transfections in HeLa cells were performed with the appropriate control vectors by using calcium-phosphate coprecipitation as described earlier (34). Various ligands, such as T3 for TR, 9-cis RA for RAR and RXR, and dexamethasone (Dex) for GR, were used at concentrations of 0.5 μM. TTNPB, which is selective for RAR, and LG100153, which is selective for RXR, were used at concentrations of 200 nM unless otherwise indicated. Typically, 1 μg of chloramphenicol acetyltransferase (CAT) reporter plasmid and 1 to 2 μg of the expression plasmids were used per sample unless otherwise indicated. All transfections were performed in duplicate or triplicate. The variation in CAT activity of the duplicate or triplicate samples was less than 10%, and each experiment was repeated at least two times. All CAT assays were performed as described earlier (34). All transfections in GH4C1 cells were performed by using the Lipofectamine-based reagent Geneporter 2 (GTS, San Diego, Calif.) according to the manufacturer's instructions. Reporter plasmids, −73 collagenase CAT (54) and ΔMTV-IR-CAT (16), were used at concentrations of 50 to 100 ng/sample, and other plasmids, pEX-NRC and pEX-NIF-1, were used at concentrations of 0.7 to 1.2 μg/sample. GFP-NIF-1 was transfected into Cos1 cells by using calcium-phosphate coprecipitation, and the cell distribution of GFP-NIF-1 was analyzed by fluorescent microscopy and Hoechst dye staining of the nucleus 48 h later.
Yeast and β-galactosidase assays.
All β-galactosidase assays were performed at least twice in duplicate or triplicate. Various ligands, such as T3 for the TRs, 9-cis RA for RXR and RAR, and estradiol (E2), were used at concentrations of 1 μM while deoxycorticosterone for GR was used at a concentration of 10 μM. Yeast colonies were first grown exponentially in Ura−, His−, and Trp− SD-dextrose medium, washed, diluted to the appropriate density, and incubated in Ura−, His−, and Trp− SD-galactose-raffinose medium followed by quantitation of β-galactosidase as described earlier (31, 34). β-galactosidase units are expressed as (OD420 × 1,000)/(minutes of incubation × OD600 of the yeast suspension), where OD420 and OD600 are optical density at 420 and 600 nm, respectively.
In vivo association of NIF-1 with NRC.
The mammalian GST expression vectors pEBG (expressing GST) and pEBG-NRC (expressing a GST fusion of full-length NRC) have been described earlier (34). pEX-FlagNIF-1 was cotransfected with pEBG or pEBG-NRC in 293T cells, and whole-cell extracts were prepared 36 h later, as described previously (34). Proteins remaining bound to the expressed GST proteins were purified with glutathione-agarose beads and processed for sodium dodecyl sulfate (SDS)-gel electrophoresis followed by Western blotting as described earlier (34). The Western blot was probed with M2 anti-Flag antibody to detect Flag-NIF-1.
In vitro binding of NIF-1 to GST-NRC.
GST-NRC.1a was expressed in SG1117 Escherichia coli by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) and then purified and immobilized to glutathione-agarose, as described previously (18, 19). NIF-1 was labeled by in vitro transcription-translation with [35S]l-methionine with rabbit reticulocyte lysates. Typically, 200 to 400 ng of GST protein bound to glutathione-agarose was used per assay. 35S-labeled proteins were mixed with GST or GST-NRC.1a beads. The samples were incubated at 4°C for 30 min in binding buffer (20 mM Tris-HCl [pH 7.7 at 25°C], 2 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol, 0.01% bovine serum albumin, 0.5 mM phenylmethylsulfonyl fluoride, 0.25% NP-40, and 0.25 μM zinc acetate). The samples were then washed with the same incubation buffer, and the bound 35S-labeled protein was analyzed by SDS-gel electrophoresis followed by autoradiography.
Nucleotide sequence accession number.
The nucleotide sequences of NIF-1 and NIF-2 have been deposited in GenBank and assigned the following accession numbers: NIF-1/NIF-2, accession number AF395833; rat NIF, accession numbers AF309071 and AY079168.
RESULTS
Identification of NIF-1, a novel zinc finger protein that interacts with NRC.
NRC interacts with CBP in vivo (34) and binds to and enhances transcriptional activation by ligand-bound nuclear hormone receptors as well as other factors such as NF-κB, c-Fos, and c-Jun (28, 34). Since the mechanism of transcriptional enhancement by NRC is not clearly understood, we sought to identify factors which may play a role in mediating these effects of NRC. In this study, we used a yeast two-hybrid screen to identify factors that functionally interact with NRC. A yeast LexA vector which expresses a fusion of the LexA DBD with NRC (amino acids 849 to 2063) was used as bait to screen the pJG4-5 GH4C1 cDNA library, which was used previously to identify NRC (34). pJG4-5 conditionally expresses cDNAs as B42 activation domain fusions. This screen identified a cDNA interactor (1.8 kb), which was found to be an ortholog of a putative transcript from a gene of unknown function identified in the human genome located on chromosome 20. The assembled transcript from this human genome sequence is predicted to encode a protein of 1,342 amino acids. We refer to this clone as NIF-1. Reverse transcription-PCR with mRNA from human T-47D and MCF-7 breast cancer cells, with primers from the predicted human sequence, identified an mRNA of the same size as that assembled from the NIF-1 genomic sequence. In addition, reverse transcription-PCR with GH4C1 mRNA, with primers from the predicted human cDNA sequence, indicated that an mRNA of similar size to the assembled NIF-1 sequence is expressed in GH4C1 cells.
Cloning, sequence, and predicted domain structure of NIF-1.
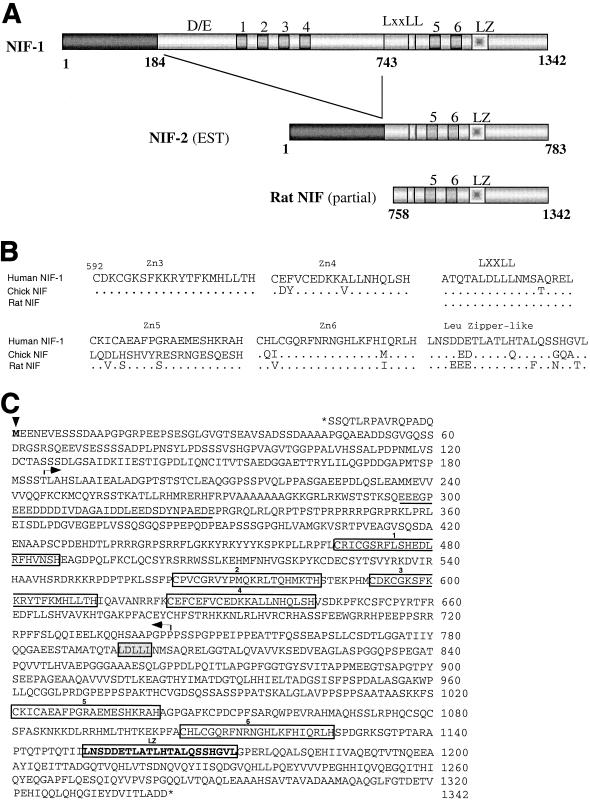
A human teratocarcinoma λgt10 cDNA library was screened with a 32P-NIF-1 cDNA probe generated from MCF-7 cells by PCR. Seven independent NIF-1 cDNA clones were identified. Upon comparison with the predicted transcript from the human genomic NIF-1 sequence, the longest clone isolated from the phage library was missing 110 nucleotides of coding sequence from the 5′ end, whereas the 3′ end extended beyond the stop codon and contains a poly(A) tail and a 3′ untranslated region sequence. A database search identified a number of ESTs, seven of which were sequenced completely. One of the ESTs (BE297231) was found to be a full-length alternatively spliced form of NIF-1 which we refer to as NIF-2. The 5′ end of this EST contained an authentic ATG and an in-frame stop codon upstream of the ATG, which is consistent with the predicted NIF-1 mRNA sequence. A PCR product containing the 110 nucleotides missing in NIF-1(6B) was generated from the EST DNA and ligated to NIF-1(6B) to generate a full-length NIF-1 clone. In addition to the human and rat NIFs, a GenBank search identified a NIF-related partial chicken cDNA clone (cFZF) (GenBank accession no. U27196) of unknown function.
Figure 1A compares the domain structure of the predicted amino acid sequence of NIF-1 with those of NIF-2 and the partial rat NIF cloned from GH4C1 cells with the yeast-two hybrid screen. NIF-1 contains 1,342 amino acids consisting of six predicted Cys-2/His-2-type zinc fingers, an LXXLL motif, a putative leucine zipper region near its C terminus, and a region of ∼35 amino acids rich in acidic amino acids towards the N terminus. Motif searches also indicate several putative protein kinase A and tyrosine kinase phosphorylation sites. In addition, a motif search identified that the region containing the first three Cys-2/His-2 zinc fingers of NIF-1 as a component of the recently described BED finger DNA-binding domain found in a number of transcriptional activators and repressors in Drosophila melanogaster (3, 21). Although the function of these BED finger domains is not understood, it has been suggested that these proteins may alter local chromatin architecture through association with insulator sequences in the DNA (3).
FIG. 1.
Sequence and predicted domain structure of NIF proteins. (A) Comparison of human and rat NIFs. Schematic representations of the functional domains identified in human NIF-1, NIF-2, and the partial rat NIF clone are shown. D/E represents an Asp- and Glu-rich acidic amino acid stretch of ∼35 amino acids. The LXXLL motif corresponds to the amino acids LDLLL. Zinc fingers of the Cys-2/His-2 type are dispersed and represented by numbers 1 through 6. LZ indicates the leucine zipper-like motif localized at the C terminus. NIF-2 was identified by sequencing an EST clone (BE297231) and appears to be an alternatively spliced isoform of human NIF-1. Rat NIF is a partial clone isolated from the GH4C1 pJG4-5 cDNA library deposited in GenBank under accession no. AF309071 and AY079168. (B) Similarity of the zinc fingers, LXXLL, and leucine zipper-like domains in human, rat, and chicken NIFs. The region of comparison includes amino acids 592 to 1172 and contains zinc fingers 5 and 6 and the LXXLL and leucine zipper regions. (C) Amino acid sequence and functional domains of human NIF-1. NIF-1 mRNA contains an open reading frame of 1,342 amino acids. The initiator Met indicated by the arrowhead is preceded by a short open reading frame and an in-frame stop codon. DE, an acidic region rich in Asp and Glu, is underlined. Zinc fingers 1 through 6 are boxed. The leucine zipper-like motif is indicated in bold and boxed. The LXXLL motif is boxed and lightly shaded. The amino acid sequence within the arrows (which includes the DE stretch and zinc fingers 1 through 4) is absent in NIF-2, an isoform of NIF-1. The nucleotide and amino acid sequences of NIF-1 and NIF-2 have been deposited in GenBank under accession number AF395833.
The zinc fingers, LXXLL, and putative leucine zipper regions of human NIF-1, rat NIF, and the chicken NIF clone are highly conserved, with some divergence of zinc finger 5 and the leucine zipper region. Interestingly, the LXXLL region is highly conserved in all three proteins (Fig. 1B). Overall, human NIF-1 and the partial rat NIF clone share 86% homology at the amino acid level while the chicken NIF clone exhibits less homology to NIF-1 (62%). The first 184 amino acids of NIF-1 are identical to those found in NIF-2. Interestingly, NIF-2 lacks the region of NIF-1 corresponding to amino acids 184 to 743, which harbors the DE region and zinc fingers 1 through 4 but is otherwise identical to NIF-1. Figure 1C illustrates the nucleotide and amino acid sequences of NIF-1 and NIF-2. These sequences have been deposed in GenBank.
Cell and tissue distribution of NIF-1.
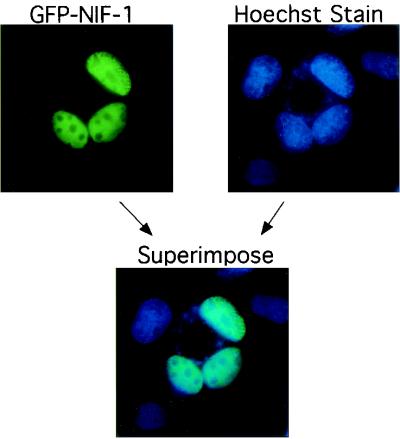
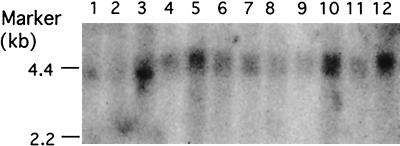
To study the subcellular localization of NIF-1, Cos1 cells were transfected with a pEGFP-NIF-1 expression vector and the cellular distribution of the GFP-NIF-1 was determined by fluorescent microscopy. As shown in Fig. 2, GFP-NIF-1 localizes exclusively to the cell nucleus, consistent with its possible function as a transcriptional regulator. A full-length 32P-labeled NIF-1 cDNA probe, predicted to identify both NIF-1 and NIF-2 mRNAs, was used to study the tissue distribution of human NIFs (Fig. 3). A multitissue Northern blot (Stratagene) was probed, and it detected a NIF-1 mRNA of ∼5 kb with relatively higher expression in the skeletal muscle, thymus, placenta, and blood. The colon, spleen, kidney, and lung showed moderate expression of NIF-1 mRNA, and the small intestine, heart, liver, and brain showed lower levels of expression of NIF-1 mRNA. Overexposure of the same blot detected an mRNA species of ∼2.5 kb, consistent with the size of NIF-2. This transcript was detected in the heart and skeletal muscle and to a lesser extent in the thymus, spleen, kidney, liver, placenta, and blood (not illustrated). NIF-2 was not detected in the small intestine and colon. The results of the Northern blot suggest that the NIF-1 mRNAs are of low abundance but are widely expressed.
FIG. 2.
NIF-1 is a nuclear protein. GFP-NIF-1 was transfected into Cos1 cells, and GFP fluorescence was detected in the nucleus (green). The nucleus was also stained with Hoechst stain (blue). GFP-NIF-1 fluorescence was also overlapped with the nuclear Hoechst stain as shown.
FIG. 3.
Northern blot of NIF-1 mRNAs in different tissues. NIF-1 mRNAs were detected with an MTN blot (Stratagene) containing poly(A)+ RNAs from the various tissues indicated. A NIF-1 mRNA of ∼5 kb was detected by probing the blot with 32P-labeled human NIF-1 cDNA. Lanes 1 through 12 contain RNAs from the brain, heart, skeletal muscle, colon, thymus, spleen, kidney, liver, small intestine, placenta, lung, and blood, respectively. A shorter mRNA of ∼2.5 kb, designated NIF-2, was detected upon longer exposure of the blot (not shown) and is described in the text.
NRC associates with NIF-1 in mammalian cells.
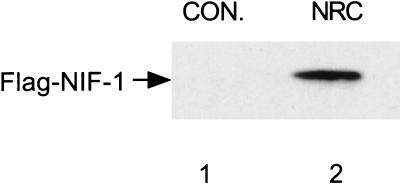
To document that NIF-1 can associate with NRC in vivo, we coexpressed a vector expressing Flag-tagged NIF-1 with mammalian GST vectors expressing GST (pEBG) or GST-NRC (pEBG-NRC) in 293T cells. Thirty-six hours later, the cells were lysed and the lysates were incubated with glutathione-agarose followed by SDS-gel electrophoresis and Western blotting with anti-Flag M2 antibody (Fig. 4). Flag-tagged NIF-1 was detected in cells expressing GST-NRC but not the GST control. These results indicate that NIF-1 can associate with NRC in mammalian cells.
FIG. 4.
NIF-1 associates with NRC in mammalian cells. The mammalian GST expression vectors pEBG (expressing GST) and pEBG-NRC (expressing a GST fusion of full-length NRC) were cotransfected with pEX-FlagNIF-1 in 293T cells. Whole-cell extracts were prepared 36 h later, and the proteins remaining bound to the expressed GST proteins were purified with glutathione-agarose beads and processed for SDS-gel electrophoresis followed by Western blotting as described earlier (34). The Western blot was probed with M2 anti-Flag antibody to detect Flag-NIF-1. Lane 1, pEBG control (CON.); lane 2, pEBG-NRC.
The C-terminal region of NIF-1 containing its sixth zinc finger interacts with NRC.
Although the original NIF isolate from GH4C1 cells lacks the N-terminal region of human NIF-1, it shares amino acid identity with the corresponding region of human NIF-1 (Fig. 1), suggesting that the C-terminal region of NIF-1 is likely involved in the interaction of NIF-1 with NRC. To map the region(s) of NIF-1 which interact with NRC, various domains of NIF-1 were conditionally expressed in yeast as B42 fusions from pJG4-5 and examined for interaction with a variety of LexA-NRC deletions, including the LXXLL-1 mutant of NRC which fails to bind nuclear hormone receptors (34) (Fig. 5). These studies indicated that the NRC interaction domain (NRC-ID) of NIF-1 maps to a 97-amino-acid C-terminal region of NIF-1 containing zinc finger 6. A much weaker interaction (10- to 20-fold less) was also found with the N-terminal region of the protein. The precise region mediating this weaker interaction was not mapped but may be mediated by zinc finger 1, which shares greater homology with zinc finger 6 than any of the other zinc finger motifs.
Identification of the NIF-ID of NRC.
A yeast two-hybrid assay was also used to identify the region of NRC which interacts with NIF-1 (Fig. 6A). Various regions of NRC were expressed as LexA fusions in yeast, and their interactions were compared with those of full-length NIF-1, NIF-2, and various deletions of NIF-1 conditionally expressed from pJG4-5. The NIF-1 interaction domain (NIF-ID) of NRC was localized to amino acids 849 to 995 of human NRC, which also contains the LXXLL-1 receptor interaction motif. To study the possible involvement or requirement of the NRC LXXLL-1 motif for direct interaction with NIF-1, we carried out yeast two-hybrid assays with LexA-NRC constructs containing either the WT (LVNLL) or MT (AVNAA) LXXLL-1 motif. The results indicate that LXXLL-1 is not required for the interaction of NRC with NIFs since the LXXLL-1 mutant forms of NRC interacted with NIF-1 as efficiently as the wild-type NRC forms.
The yeast two-hybrid data suggested that residues 849 to 995 of human NRC and the corresponding region of rat NRC are involved in the interaction with NIF-1. To document that this region of NRC binds to NIF-1 in vitro, we expressed this region of rat NRC as a GST fusion in E. coli and purified it with glutathione-agarose beads. 35S-labeled NIF-1, synthesized by in vitro transcription-translation in reticulocyte lysates, was incubated with ∼200 ng of purified GST or GST-NRC.1a at 4°C for 30 min in binding buffer with mild shaking. The GST-glutathione-agarose beads were washed, and the bound 35S-labeled proteins were analyzed by SDS-gel electrophoresis followed by autoradiography. As shown in Fig. 6B, 35S-labeled NIF-1 bound to GST-NRC.1a but not to GST, indicating that NIF-1 binds to the same region of NRC in vitro as determined in the yeast two-hybrid assay for which results are shown in Fig. 6A.
NIF-1 does not interact with nuclear hormone receptors but potentiates their ligand-dependent transcriptional activity.
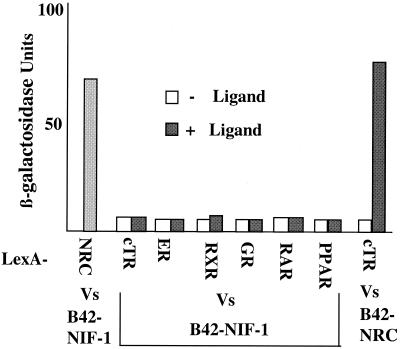
Since NIF-1 interacts with NRC, and NRC has been shown to be a potent coregulator of ligand-bound nuclear hormone receptors, we sought to determine whether NIF-1 could modulate nuclear receptor activity. As shown in Fig. 1, NIF-1 contains an LXXLL motif and might thus interact with nuclear hormone receptors directly, even though it was cloned by using NRC as bait. To examine for this possibility, we studied the interaction of full-length B42-NIF-1, conditionally expressed from pJG4-5, with LexA fusions of nuclear receptor LBDs (chicken TRα [cTRα], ERα, RXRα, GR, RARα, and PPARα) in yeast (Fig. 7). In addition, a LexA fusion of full-length cTRα was also tested against full-length B42-NIF-1 and gave results similar to those obtained with the cTRα LBD. NIF-1 did not interact with any of these receptors with or without a cognate ligand, but it interacted strongly with LexA-NRC. To document that the LexA LBD fusions were expressed and responded to ligand in yeast (as reported previously), similar studies were carried out with B42-NRC. As expected, B42-NRC interacted with the LexA-cTRα LBD in a T3-dependent manner (Fig. 7). As previously described (34), all other nuclear hormone receptors showed similar binding with B42-NRC in the presence of their cognate ligands (data not shown)
FIG. 7.
NIF-1 does not directly interact with nuclear receptor LBDs in yeast. NIF-1 was expressed as a B42 fusion and tested against LexA fusions of the following receptor LBDs: cTRα, hERα, hRXRα, hGR, hRARα, hPPARα, and NRC. T3-dependent interaction of LexA-cTRα was also verified against B42-NRC in the same assay as a positive control. Details of the β-galactosidase assay in yeast extracts are described in Materials and Methods. Vs, versus.
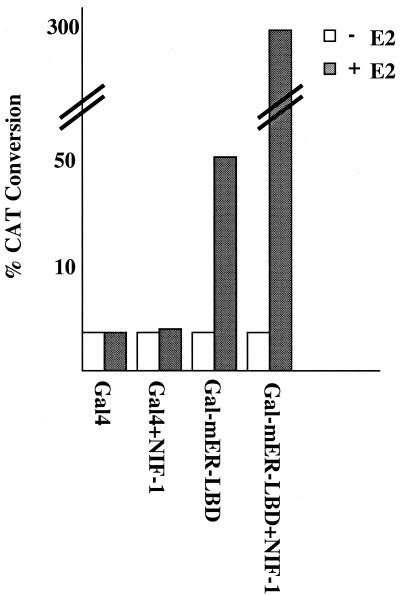
NIF-1 interacts with NRC but not with ligand-bound nuclear receptors, indicating that the LXXLL motif found in NIF-1 is not a functional interaction domain for the nuclear hormone receptors tested in the present study. The possibility remains, however, that the LXXLL in NIFs may display selective interaction with other receptors or orphans not tested. Given the fact that NRC is a potent coactivator in mammalian cells for ligand-bound nuclear receptors, and that NIF-1 binds NRC in yeast and in vitro, it is likely that NIF-1 might affect the coactivator function of NRC in vivo. We therefore carried out transfection studies to determine whether NIF-1 could enhance ligand-dependent receptor activity in mammalian cells. In our initial experiments, we examined whether NIF-1 could alter the estradiol-mediated transcriptional activation of the Gal4DBD fused to the mER-LBD (Gal4-mER-LBD) in HeLa cells (Fig. 8). Expression of NIF-1 did not alter transcriptional activity when expressed with the Gal4 DBD alone but enhanced the estradiol-mediated stimulation of Gal4-mER-LBD about sixfold, further indicating that receptor activity could be affected by NIF-1, albeit indirectly.
FIG. 8.
NIF-1 enhances ligand-dependent activation by Gal4-ER-LBD in HeLa cells. The Gal4 reporter pBL-G5-CAT2 was cotransfected in HeLa cells with vectors expressing the Gal4 LBD or the Gal4 LBD fusion of the mER-LBD with or without NIF-1. Cells were incubated with or without ligand, E2 (100 nM), for 40 h, and duplicate samples were then assayed for CAT activity. The experiment was repeated at least two times with similar results (refer to Materials and Methods for details about plasmids). +, with; −, without.
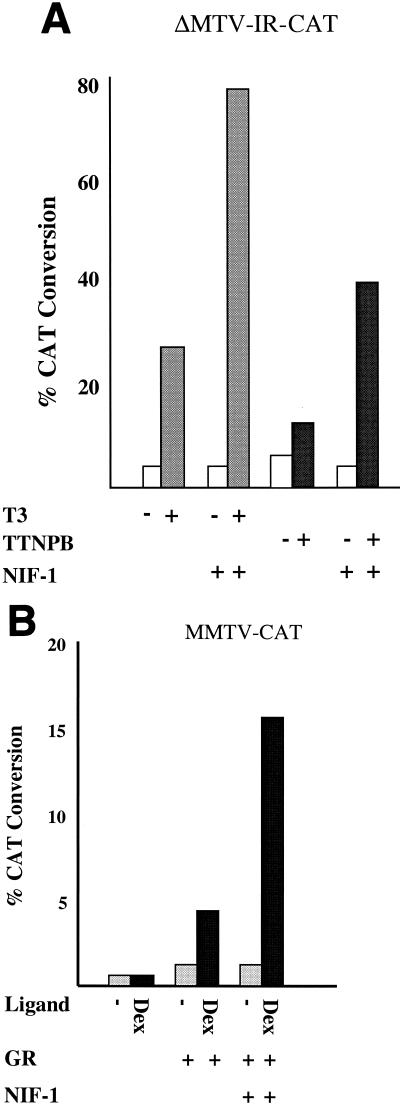
To study the effect of NIF-1 on the regulation of gene expression by wild-type receptors, we examined the effect of NIF-1 on the ligand-dependent activity of TR, RAR, and GR (Fig. 9). HeLa cells were transfected with the appropriate CAT reporter genes and with vectors expressing cTRα, hRARα, or hGR alone or with NIF-1. Ligand-dependent activation was studied by using T3 for TR, the RAR-selective ligand TTNPB for RAR, and Dex for GR. In each case, the expression of NIF-1 enhanced the extent of ligand-dependent activation by these receptors about threefold.
FIG. 9.
NIF-1 activates TR, RAR, and GR in HeLa cells. (A) HeLa cells were transfected with the ΔMTV-IR-CAT reporter and expression vectors for cTRα or hRARα and NIF-1 as indicated. The cells were incubated with T3 at a concentration of 1 μM and the RAR-specific ligand TTNPB at a concentration of 200 nM. All samples were analyzed in duplicate, and the experiment was repeated at least two times. Panel B is the same as panel A except that the mouse mammary tumor virus (MMTV)-long terminal repeat (LTR)-CAT reporter and an hGR expression vector were cotransfected with (+) or without (−) 500 nM Dex.
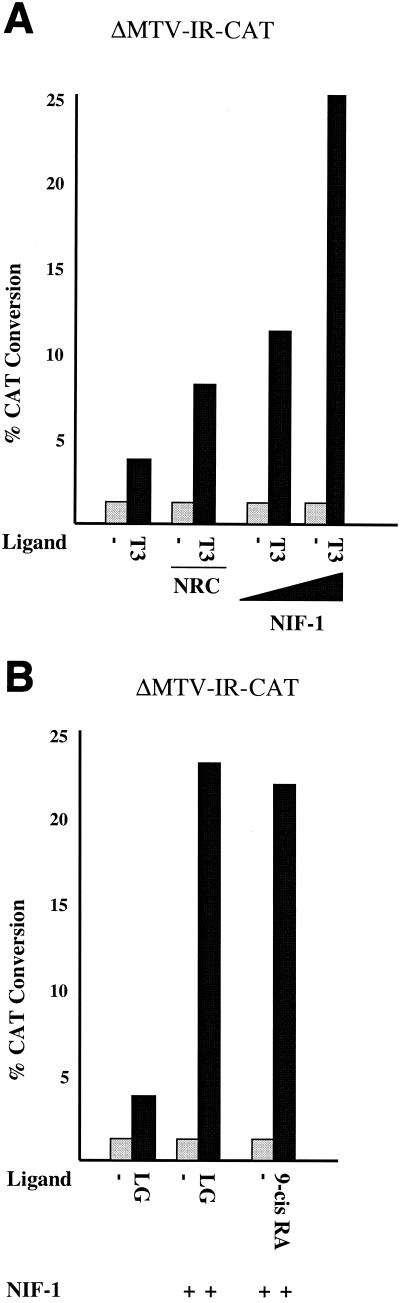
The effect of NIF-1 expression on transcriptional activation by endogenous TR and RXR was examined in GH4C1 cells (Fig. 10). NIF-1 enhanced T3 stimulation of endogenous TR activity about sixfold, and this effect of NIF-1 was greater than that found for NRC (about twofold), suggesting that NIF-1 may be more limiting for T3 stimulation in GH4C1 cells. NIF-1 also enhanced the activity of endogenous RXR about sixfold, as assessed with LG100153 (an RXR-specific ligand) and 9-cis RA.
FIG. 10.
Ligand-dependent activation of endogenous nuclear receptors by NIF-1 in GH4C1 cells. (A) Cells were cotransfected with the ΔMTV-IR-CAT reporter alone and with (+) or without (−) the NIF-1 or NRC expression plasmids at various concentrations. T3 ligand was used at a concentration of 1 μM. Each sample was analyzed in duplicate, and the experiment was repeated at least two times with similar results (refer to Materials and Methods for details about plasmids). Panel B is the same as panel A except that the RXR-specific ligand LG100153 and the RXR/RAR-specific ligand 9-cis RA were each used at concentrations of 200 nM.
NIF-1 potentiates the transcriptional activity of AP1.
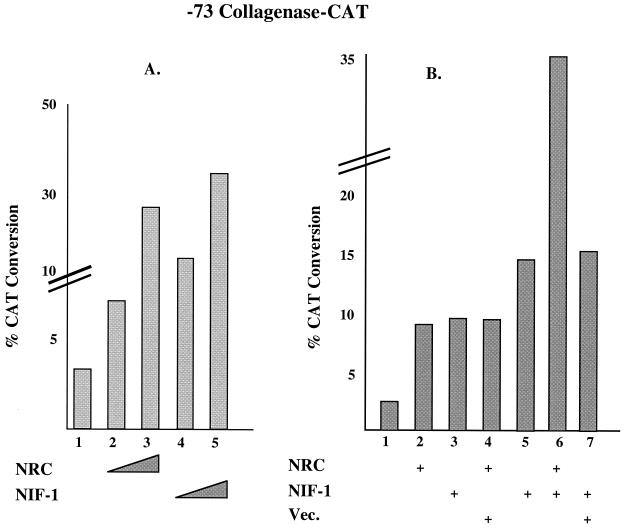
Since NIF-1 interacts with NRC, and NRC has been shown to be a potent coactivator of c-Fos and c-Jun (AP1) (28), we examined the effect of NIF-1 on the activity of endogenous AP1 in HeLa cells (Fig. 11). HeLa cells were transfected with a CAT reporter for AP1 activity, −73 collagenase-CAT (54), with and without vectors expressing NRC and/or NIF-1. NRC increased the activity of the −73 collagenase-CAT reporter about 9-fold while NIF-1 enhanced the activity about 10-fold (Fig. 11A). Expressing c-Fos and/or c-Jun in HeLa cells further enhanced the extent of activity of the −73 collagenase-CAT reporter, and the expression of NRC or NIF-1 further increased the extent of activation (not shown). Since the activity of the −73 collagenase-CAT reporter gene was similarly affected by NRC or NIF-1, we carried out cotransfection studies with smaller amounts of NRC or NIF-1 expression vectors to assess whether expression of both factors would lead to an effect greater than that found for each factor alone (Fig. 11B). In this setting, the expression of NRC resulted in a threefold stimulation while expression of NIF-1 led to a fivefold increase in the activity of the −73 collagenase-CAT reporter gene. Expression of both NRC and NIF-1 resulted in a 12-fold increase, further supporting the notion that NRC and NIF-1 functionally interact in the cell to enhance transcriptional activation.
FIG. 11.
NIF-1 and NRC activate AP1 activity in HeLa cells. (A) The −73 collagenase-CAT reporter plasmid driven by AP1 (c-Fos and/or c-Jun) was transfected with 1 and 3 μg of the expression plasmids for NRC or NIF-1. The samples were analyzed in duplicate, and the experiment was repeated at least two times with similar results. (B) The amount of expression vector for NRC used was 0.7 μg. The amount of NIF-1 expression plasmid used was 0.7 μg in lane 3 and 1.2 μg in lanes 5 to 7. The amount of vector control (Vec.) used was 0.7 μg. +, with.
DISCUSSION
Nuclear hormone receptors modulate a wide variety of developmental and physiological processes in vertebrates through the transcriptional regulation of target genes in specific tissues. A wide variety of studies indicate that the LBDs of these receptors play central roles in mediating transcriptional activation as a result of ligand binding, and this activity has been referred to as activation function-2, or AF-2. In certain nuclear receptors, the variable N-terminal A/B domain also plays an important role in mediating transcriptional activation (e.g., GR, ER, and progesterone receptor) and this activity has been referred to as activation function-1, or AF-1. Although AF-1 and AF-2 were defined functionally, an important question relates to defining the molecular determinants and protein-protein interactions that determine the activity of AF-1 and AF-2. Yeast two-hybrid screens and biochemical approaches have identified a number of factors which appear to function as coactivators or coregulators of AF-2 and/or AF-1 function. Although certain nuclear receptor A/B domains appear to contain an independent activation function, the integration of the activity of the N-terminal A/B domain with the LBD in the context of full-length receptors results in a mutually dependent function of AF-1 and AF-2.
A central question is how coactivator binding to a ligand-bound receptor leads to transcriptional activation. The finding that p160 coactivators can associate with CBP/p300 suggests that transcriptional enhancement of nuclear receptors by coactivators involves the recruitment of large coactivator-associated complexes to the promoter-bound ligand-bound receptor. In addition, coactivators may exist in dynamic association with different complexes, thereby leading to marked diversity in the extent of activation which may be dependent on cell type, the transcription factor, and possibly the promoter context. Thus, different DRIP/TRAP complexes have been reported to contain both common and unique components, which are thought to be involved in the modulation of different transcription factors. For example, DRIP/TRAP, ARC, CRSP, SRB-mouse mediator, and SMCC are related but distinct multiprotein complexes involved in activation of the nuclear hormone receptors SREBP-1a/Sp1, NF-κB (p65), Sp1, E1A/VP16, and p53 (5, 24, 40, 45). It is remarkable that most of the complexes share common polypeptides despite the fact that the transcription factors modulated by these protein complexes are structurally and functionally distinct. Interestingly, the NAT complex (47), which represses activated transcription, shares components with other complexes involved in the activation described above. Thus, it is becoming increasingly clear that these transcriptionally active complexes contain unique components and also share a number of common factors.
We recently described the cloning of a novel coactivator, which we refer to as NRC, which is part of a CBP complex in vivo that does not appear to include SRC-1. In addition, TRBP (NRC) has also been reported to associate with DRIP130, a common component of some activator complexes, including the DRIP/TRAP complex. Thus, as with other coactivators, NRC may exist as a component of distinct multiprotein complexes which may each mediate specific effects with a subset of transcriptional regulators. In this study, we described the cloning and characterization of a novel factor from rat and human cells which interacts in vitro and in vivo with NRC and modulates the function of NRC in cells. Based on its ability to interact with NRC, we refer to this factor as NIF.
Human NIF-1 is a 1,342-amino-acid nuclear protein containing six Cys-2/His-2 zinc finger domains, an N-terminal acidic sequence of ∼35 residues rich in Glu and Asp, and an LXXLL motif and a putative leucine zipper-like motif in the C-terminal region. NIF-1 contains several putative protein kinase A and tyrosine kinase phosphorylation sites. In addition, the first three Cys-2/His-2 zinc fingers appear to be part of the recently proposed BED finger DNA-binding domain (3). This domain is found in proteins thought to be involved in activation or repression through association with insulator sequences in the DNA (21) and may thus act to modulate local chromatin structure. Human NIF-1 and the partial rat NIF clone identified in the yeast two-hybrid screen share 86% homology at the amino acid level. In particular, the zinc finger domains, LXXLL region, and the leucine zipper-like motif are highly conserved. A GenBank search identified a NIF-related partial chicken cDNA clone (cFZF) of unknown function. cFZF shares 62% homology with the corresponding region of human NIF-1 with divergence at zinc finger 5 and the leucine zipper-like regions. Interestingly, the LXXLL region is highly conserved in all three proteins. Although this LXXLL motif does not mediate interaction with NRC or the ligand-bound nuclear hormone receptors that we examined, its conservation implies that it may serve an important function in mediating other protein-protein interactions.
An EST database search identified a number of human NIF ESTs. DNA sequencing indicated that one of the ESTs (BE297231) (∼2.2 kb) contained 5′ and 3′ coding sequences identical to those found in NIF-1. This cDNA appears to reflect an alternatively spliced form of NIF-1, which we refer to as NIF-2. NIF-2 lacks 559 residues between amino acids 184 and 743, containing zinc fingers 1 to 4. However, NIF-2 retains the NRC-ID, which includes zinc finger 6. In keeping with this, NIF-2 interacts with NRC in yeast two-hybrid assays. However, the role of NIF-2 with respect to NRC and its other functions remains to be elucidated. A multitissue Northern blot probed with full-length 32P-NIF-1 cDNA identified a widely expressed ∼5-kb transcript of low abundance and a less abundant ∼2.5-kb transcript which appears to be more restricted in its tissue expression. We assume that the ∼5-kb transcript is NIF-1 and the ∼2.5-kb transcript is NIF-2.
Full-length human NIF-1 binds NRC in vivo and in vitro, and extensive mapping with yeast two-hybrid assays indicates that the NRC-ID of NIF-1 occurs through a region containing zinc finger 6. Interestingly, a short region of 97 amino acids containing zinc finger 6, which is conserved in the rat and human NIFs and in chicken cFZF, appears to be sufficient for a strong interaction with NRC in yeast. A very weak interacting region containing zinc finger 1 was also detected (data not shown). Interestingly, zinc finger 1 shares a weak similarity with zinc finger 6. We also mapped a NIF-ID in NRC by using various regions of NRC in yeast two-hybrid assays. The domain was mapped to a 146-amino-acid region of NRC (amino acids 849 to 995) which also contains the LXXLL receptor-interacting domain of NRC. However, this LXXLL motif of NRC is not directly involved in the interaction of NRC with NIF-1 since mutation of the LXXLL motif (LVNLL to AVNAA), which eliminates NRC receptor interactions, did not alter the interaction of NRC with NIF-1. This suggests that NIF-1 and activated receptors could simultaneously interact with NRC. This finding is consistent with our observation that NIF-1 can enhance ligand-dependent transcriptional activation without directly interacting with nuclear hormone receptors.
We previously reported that NRC can enhance the activity of a large number of nuclear hormone receptors. In this study, we showed that NIF-1, which does not interact with receptors, also enhances the activity of expressed ER, TR, GR, and RAR in HeLa cells and endogenous TR and RXR in GH4C1 cells which contain NRC. In addition, the activity of c-Fos and c-Jun, which have been reported to be enhanced by NRC, are also enhanced by NIF-1. We presume that this modulation of ligand-bound nuclear hormone receptors by NIF-1 occurs through its interaction with NRC and not through the interaction of other factors. However, it is possible that NIF-1 could also be a component of other coactivator complexes not involving NRC. Further definition of whether NRC is required for the effect of NIF-1 on nuclear receptors, c-Fos, or c-Jun will require cells which do not express NRC.
Recently, in addition to NIF-1, three other factors, CAPER, PIMT, and CoAA (25, 26, 59), were reported to interact with NRC proteins (ASC-2/PRIP/TRBP). CAPER, PIMT, and CoAA are distinct proteins which each contain RNA binding motifs. In contrast, NIF-1 does not contain RNA binding motifs. CAPER was reported to interact directly with ERα and ERβ but not with TR, GR, RXR, or PPAR, and it was reported to enhance activation by ER about threefold. PIMT appears to contain a methyltransferase activity. However, enhancement of stimulation by RXR or PPAR (∼1.6-fold) did not require methyltransferase activity. Expression of CoAA enhanced the activity of GR, TR, and ER about threefold. Whether these changes reflect a direct or indirect interaction of PIMT or CoAA with nuclear receptors was not examined. Since CAPER, PIMT, and CoAA were each cloned as an interactor with NRC, further studies are needed to determine whether these factors, including NIF-1, are also integral components of other coactivator complexes in the cell.
Since NIF-1 does not directly associate with receptors but enhances their activities, it functions differently than previously described coactivators, which exert their effects through direct association with ligand-bound receptors. Thus, we suggest that NIF-1, and factors which behave similar to NIF-1, be referred to as cotransducers, which act in vivo either as part of a coactivator complex or downstream of a coactivator complex to modulate transcriptional activity. Examples of such factors include CARM1 and PRMT1 (11, 29, 53). How would a cotransducer such as NIF-1 enhance the activity of coactivators such as NRC? The mechanism(s) has not yet been defined, but it includes (i) the contribution of an activation surface, (ii) conformational alteration of a coactivator to expose an activation domain, (iii) interaction with other proteins to stabilize a multiprotein coactivator complex, (iv) direct association with the basal transcription machinery, or (v) modification of chromatin architecture as a BED domain protein. Since the Cys-2/His-2 class of zinc finger has been reported to be involved in DNA interactions, this raises the possibility that NIF-1 may directly bind DNA. Thus, in addition to being a component of a coactivator complex recruited to a transcription factor (e.g., nuclear receptors, c-Fos, and c-Jun) by a coactivator (e.g., NRC), NIF-1 might also act as a DNA-binding factor that modulates transcription by recruiting a coactivator complex to a specific target gene. Thus, NIF-1 may mediate its effects by acting through multiple mechanisms in the cell.
Acknowledgments
We thank Malcom Parker for the Gal4-mERα-LBD vector, David Moore for the LexA-hTRβ LBD, LexA-hGR LBD, LexA-hRXRα LBD, and LexA-hVDR LBD yeast vectors, Michael Garabedian for pJG4-5-hERα-LBD, Naoko Tanese for advice with phage screening, and Angus Wilson for the λgt10 human cDNA library. We also thank Shahana Mahajan for help.
This research was supported by NIH grant DK16636 (to H.H.S.). Sequence analysis and database searches were done through the New York University Medical Center Research Computing Resource, which receives support from the National Science Foundation.
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1269-1304. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L. 2000. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25:421-423. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, J. C. G., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, V. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 6.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. A. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 7.Carson-Jurica, M. A., W. T. Schrader, and B. W. O'Malley. 1990. Steroid receptor family: structure and functions. Endocr. Rev. 11:201-218. [DOI] [PubMed] [Google Scholar]
- 8.Cavailles, N., S. Dauvois, F. L'Horset, S. Lopez, S. Hoare, P. J. Kushner, and M. G. Parker. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti, D., V. J. LaMorte, M. C. Nelson, T. Nakajima, I. G. Schulman, H. Juguilon, M. Montminy, and R. M. Evans. 1996. Role of CBP/P300 in nuclear receptor signalling. Nature 383:99-103. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 13.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747-1749. [DOI] [PubMed] [Google Scholar]
- 15.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman, B. M., J. Casanova, B. M. Raaka, J. Ghysdael, and H. H. Samuels. 1992. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol. Endocrinol. 6:429-442. [DOI] [PubMed] [Google Scholar]
- 17.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 18.Hadzic, E., V. Desai-Yajnik, E. Helmer, S. Guo, S. Wu, N. Koudinova, J. Casanova, B. M. Raaka, and H. H. Samuels. 1995. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor α is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol. Cell. Biol. 15:4507-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadzic, E., I. Habeos, B. M. Raaka, and H. H. Samuels. 1998. A novel multifunctional motif in the N-terminal A/B domain of T3Rα modulates DNA-binding and receptor dimerization. J. Biol. Chem. 273:10270-10278. [DOI] [PubMed] [Google Scholar]
- 20.Hanstein, B., R. Eckner, J. DiRenzo, S. Halachmi, H. Liu, B. Searcy, R. Kurokawa, and M. Brown. 1996. p300 is a component of an estrogen receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:11540-11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart, C. M., O. Cuvier, and U. K. Laemmli. 1999. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma 108:375-383. [DOI] [PubMed] [Google Scholar]
- 22.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 23.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375-33383. [DOI] [PubMed] [Google Scholar]
- 26.Jung, D. J., S. Y. Na, D. S. Na, and J. W. Lee. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229-1234. [DOI] [PubMed] [Google Scholar]
- 27.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S.-C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 28.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, L. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, is a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 31.Li, D., V. Desai-Yajnik, E. Lo, M. Schapira, R. Abagyan, and H. H. Samuels. 1999. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol. Cell. Biol. 19:7191-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 34.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mak, H. Y., S. Hoare, P. M. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 37.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 39.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 40.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 41.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 42.Onate, S. A., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 44.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 45.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 46.Ryu, S., and R. Tjian. 1999. Purification of transcription cofactor complex CRSP. Proc. Natl. Acad. Sci. USA 96:7137-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 48.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 49.Toney, J. H., L. Wu, A. E. Summerfield, G. Sanyal, B. M. Forman, J. Zhu, and H. H. Samuels. 1993. Conformational changes in chicken thyroid hormone receptor α1 induced by binding to ligand or to DNA. Biochemistry 32:2-6. [DOI] [PubMed] [Google Scholar]
- 50.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 51.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voegel, J. J., M. J. S. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 54.Ways, D. K., W. Qin, P. Cook, P. J. Parker, J. B. Menke, E. Hao, A. M. Smith, C. Jones, J. M. Hershman, M. E. Geffner, F. Su, H. H. Samuels, and S. J. Usala. 1993. Dominant and non-dominant negative c-erbAβ1 receptors associated with thyroid hormone resistance syndromes augment TPA-induction of the collagenase promoter and exhibit defective T3-mediated repression. Mol. Endocrinol. 7:1112-1120. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 56.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. USA 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, J. J., U. Vinkemeier, W. Gu, D. Chakravarti, C. M. Horvath, and J. E. Darnell, Jr. 1996. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. USA 93:15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, Y., C. Qi, W. Q. Cao, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2001. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl. Acad. Sci. USA 98:10380-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]