Abstract
Progression through mitosis is associated with reversible phosphorylation of many nuclear proteins including that of the high-mobility group N (HMGN) nucleosomal binding protein family. Here we use immunofluorescence and in vitro nuclear import studies to demonstrate that mitotic phosphorylation of the nucleosomal binding domain (NBD) of the HMGN1 protein prevents its reentry into the newly formed nucleus in late telophase. By microinjecting wild-type and mutant proteins into the cytoplasm of HeLa cells and expressing these proteins in HmgN1−/− cells, we demonstrate that the inability to enter the nucleus is a consequence of phosphorylation and is not due to the presence of negative charges. Using affinity chromatography with recombinant proteins and nuclear extracts prepared from logarithmically growing or mitotically arrested cells, we demonstrate that phosphorylation of the NBD of HMGN1 promotes interaction with specific 14.3.3 isotypes. We conclude that mitotic phosphorylation of HMGN1 protein promotes interaction with 14.3.3 proteins and suggest that this interaction impedes the reentry of the proteins into the nucleus during telophase. Taken together with the results of previous studies, our results suggest a dual role for mitotic phosphorylation of HMGN1: abolishment of chromatin binding and inhibition of nuclear import.
Progression through mitosis is associated with reversible phosphorylation of many cellular constituents, including components of the nuclear membrane, transcription factors, and chromatin binding proteins. For DNA and chromatin binding proteins, it has been suggested that the global phosphorylation at the onset of mitosis displaces the transcriptional machinery from chromatin and prevents the association of the proteins with their cognate binding sites, thereby facilitating the condensation of the chromatin fiber (12, 21, 23, 27, 29, 35, 36). We recently demonstrated that high-mobility group N (HMGN) nonhistone proteins are highly phosphorylated during mitosis, that this modification abolished their ability to bind to nucleosomes (33), and that the proteins do not colocalize with DNA or histones during mitosis (15, 33). HMGN are nucleosome binding proteins present in the nuclei of most growing vertebrate cells. The binding of HMGN to the nucleosome core reduces the compaction of the chromatin fiber and enhances the transcriptional potential of chromatin templates (reviewed in references 4 and 5). Thus, their displacement from chromatin is compatible with the general concept that mitotic condensation of the chromatin fiber is associated with the inactivation of nuclear proteins that are involved in chromatin decondensation and transcriptional activation.
The end of mitosis is associated with the dephosphorylation of lamins, re-formation of the nuclear membrane, and reestablishment of nuclear import activity. The exact fate of the phosphorylated regulatory factors and chromatin binding proteins at this stage has not been investigated in detail. Phosphorylation plays an essential role in the interaction of proteins with nuclear import and export factors (17) and with 14.3.3 proteins (10). As such, phosphorylation plays an important role in regulating nuclear transport and determining the intracellular compartmentalization of proteins (17, 30). We have reported that HMGN proteins relocate with chromatin only in late telophase after the formation of the nuclear membrane (15, 33); however, the relationship between the phosphorylation state of HMGN proteins and their reentry into the newly formed nucleus has not yet been studied.
In this report we demonstrate that phosphorylation of the nucleosomal binding domain (NBD) of HMGN1 proteins impairs their ability to enter the nucleus. Using specific point mutations, we demonstrate that the inability to enter the nucleus is a specific consequence of phosphorylation rather than inactivation of the bipartite nuclear localization signal (NLS) by the presence of negative charges in the proteins. We demonstrate that phosphorylation promotes the binding of HMGN1 to specific isotypes of 14.3.3 proteins both in vivo and in vitro and suggest that the mitotic phosphorylation of the NBDs of HMGN proteins serves not only to abolish their binding to nucleosomes but also to recruit specific 14.3.3 proteins. Recruitment of 14.3.3 may serve as another mechanism to ensure that HMGN proteins do not bind to mitotic chromatin and may also retard the nuclear reentry of HMGNs in late telophase. Our results provide a direct link between mitotic phosphorylation, nuclear import, and chromatin binding and may be of general relevance to understanding the consequences of this modification on the function of chromatin binding proteins.
MATERIALS AND METHODS
HMGN1 proteins and antibodies.
Recombinant wild-type and mutant HMGN1 proteins were generated and purified as previously described (6, 33). Antibodies to the HMGN proteins and to their phosphorylated forms were prepared and characterized as previously described (33). The anti-B23 antibody was previously described (9).
In vitro phosphorylation of the recombinant HMGN1 proteins.
Recombinant HMGN1 and various mutants were phosphorylated with protein kinase C (PKC) (Upstate Biotechnology) in buffer A supplemented with a lipid coactivator solution at a final concentration of 50 μg of phosphatidylserine per ml and 50 μg of diacylglycerol per ml (33).
Nuclear import assay.
The NLS-allophycocyanin conjugate and HMGN1 specifically labeled with 5-iodoacetamide fluorescein at position 88 (HMGN1-88c-5IAF) were prepared as described previously (15). HMGN1-88c-5IAF (300 ng) was phosphorylated with 0.5 μl of PKC (Calbiochem) in a total volume of 10 μl in the phosphorylation buffer recommended by the manufacturer for 1 h at 37°C. The labeling reaction mixture contained trace amounts of γ-32P-labeled ATP. The phosphorylated proteins were stored frozen at −20oC. The levels of phosphorylation were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. The import assays were performed essentially as described previously, using digitonin-permeabilized HEp-2 cells and Xenopus laevis egg extracts (15). The egg extracts were preincubated for 30 min at room temperature with an ATP-regenerating system either with or without 10 μM okadaic acid (ICN). The transport mixture contained phosphorylated protein and 15 μl of preincubated Xenopus egg extract to give a final concentration of 10 mg of extract per ml, 20 mM HEPES (pH 7), 11 mM potassium acetate, 2 mM dithiothreitol (DTT), 1 mM EGTA, 2 mM ATP, 20 mM creatine phosphate, 100 μg of creatine phosphokinase (Sigma) per ml, 0.5 mM GTP, and 12 μg of import substrate per ml. After import for 1 h at 22°C, the cells were fixed with 2% formaldehyde for 10 min and immediately examined by confocal laser scanning microscopy, using identical settings for all the proteins. The stability of the phosphorylation in the egg extracts was tested in parallel, using 150 ng of phosphorylated protein incubated with egg extract that either did or did not contain okadaic acid. The reaction mixture was analyzed by SDS-PAGE and autoradiography.
For nuclear export inhibition assays, mouse fibroblasts were cultured for 5 h in the presence of 20 ng of leptomycin B (Sigma) per ml as previously described (3). Cells were immunostained as described below.
Microinjection of proteins into cells.
Solutions used for microinjection were prepared immediately before injection and centrifuged at 10,000 × g for 30 min at 4°C to remove insoluble particles. These solutions contained purified HMGN1 or HMGN1S20,24E mutant proteins, both labeled at position 88 with 5IAF (15), at a final concentration of 1 mg/ml and Texas red-labeled dextran of 70 kDa (Molecular Probes) at 2 mg/ml (as a marker for injected cells). Forty-eight hours before injection, 3 × 105 mouse embryonic fibroblasts were plated in CoverGlass chambers (LAB TEK-Nalge Nunc International) and cultured in Dulbecco modified Eagle medium (DMEM) (Invitrogen). Before injection, the medium was replaced with DMEM supplemented with 5 mM HEPES (pH 7.4). Injections were performed with a Micromanipulator 5171 and a Transjector 5246 (Eppendorf) installed on an Axiovert 25 inverted microscope (Zeiss). Microinjections were performed under air pressure using sterile glass capillaries (Femtotips; Eppendorf). Fluorescent proteins and Texas red-labeled dextran were visualized by epifluorescence using an Axiovert S100 microscope (Zeiss). Digital images were captured using a charge-coupled device camera (SenSys).
Transfection into HmgN1−/− fibroblasts and HeLa cells.
HmgN1−/− fibroblasts were generated from 13-day-old embryos of mice bearing a targeted disruption of the HmgN1 gene. Northern, Western, and immunocytochemical analyses demonstrated that the mice and cells derived from these mice do not express HMGN1 mRNA and do not contain HMGN1 protein (Y. Birger, unpublished data). Plasmids expressing either native or mutant human HMGN1 or their green fluorescent protein (GFP) fusion forms were transfected into the fibroblast by the Lipofectamine 2000 method (Gibco-BRL). Cells expressing the GFP fusion proteins were detected by confocal immunofluorescence microscopy.
FRAP.
Fluorescence recovery after photobleaching (FRAP) was performed as described previously (28). A spot approximately 3 μm in diameter in HeLa cells expressing GFP-labeled proteins was bleached for 152 ms using the 488-nm-wavelength laser line of an argon laser with a nominal output of 40 mW set at 75% intensity. The fluorescence intensity in the photobleached area was measured at 152-ms intervals.
GST pull down assay.
Glutathione S-transferase (GST)-14.3.3 bacterial expression vectors were a gift from H. Piwnica-Worms, Washington University. The proteins were expressed in BL-21 cells and purified using glutathione beads as recommended by the manufacturer (Sigma). Glutathione agarose beads (25 μl) coated with 5 μg (each) of GST-14.3.3η, -σ, and -ζ isoforms were mixed with 50 μg of recombinant HMGN1, PKC-phosphorylated HMGN1, or HMGN1S20,24E mutant, in 1 ml of TBSN buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% Igepal Ca-630 (Sigma-Aldrich), 5 mM EGTA, 1.5 mM EDTA, 1 mM DTT) complemented with Complete protease inhibitor (Roche Diagnostic). The mixture was incubated with end-over-end shaking for 2 h at 4°C. Beads were recovered by centrifugation and washed four times in TBSN buffer. Washed beads were mixed with 60 μl of SDS-PAGE sample buffer, boiled, and centrifuged, and the proteins in the supernatants were analyzed by SDS-PAGE and by immunoblotting with either anti-HMGN1 or anti-NBD2P antibody which specifically recognizes phosphorylated HMGN1.
Immunoprecipitation of HMGN1-14.3.3 complexes from HMGN1-FLAG-expressing HeLa cells.
HeLa cells stably expressing HMGN1-FLAG protein were established by a retroviral transduction method (31). Cells were cultured in DMEM with 10% (vol/vol) fetal bovine serum (FBS) (Gibco-BRL). To enrich in mitotic cells, confluent cultures were treated with nocodazole (500 ng/ml) and harvested after 20 h. The cells were resuspended in BC100 buffer (20 mM Tris [pH 8.0], 10% glycerol, 100 mM KCl, 0.2 mM EDTA, 2 mM DTT, 50 mM NaF, 100 μM Na3VO4, 100 μM okadaic acid, protease inhibitor cocktail). To prepare cell extracts, freeze-thawed cells were passed 10 times through a 25-gauge needle. These extracts were mixed with anti-FLAG-agarose (Sigma-Aldrich) that was preequilibrated with BC100 buffer, and the mixture was incubated for 12 h at 4°C with rotatory shaking. Agarose beads were recovered by centrifugation and washed two times with BCT buffer adjusted to 500 mM KCl and two times with BCT buffer. The bound proteins were released from the anti-FLAG-agarose by adding 100 μM FLAG peptide and incubating for at least 4 h at 4°C. The released proteins were fractionated by SDS-PAGE (25) and analyzed by Western blotting with specific antibodies against 14.3.3 isoforms (Santa Cruz Biotechnology) with the ECL plus kit (Amersham).
Confocal microscopy.
Cells were grown on coverslips in DMEM (Life Technologies, Inc.) supplemented with 10% FBS (GIBCO-BRL) at 37°C in a 5% CO2 incubator, washed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde in PBS for 10 min at room temperature, washed with PBS, and incubated for 20 min in TNBS buffer (PBS, 0.1% Triton X-100, 1% FBS, 0.1% NaN3). The coverslips were incubated with primary antibody at 1 μg/ml in TNBS buffer overnight at room temperature in a moist chamber, washed with PBS, and incubated with secondary antibody labeled with either Texas red or fluorescein for 2 h at room temperature in a moist chamber. DNA was stained with TO-PRO3 (Molecular Probes). Following final wash steps, coverslips were inverted onto glass slides using the ProLong antifade reagent (Molecular Probes) as the mounting medium. Fluorescent cells were examined with an epifluorescence microscope (Optiphot; Nikon, Tokyo, Japan) equipped with a confocal system (MRC-1024; Bio-Rad Laboratories, Hercules, Calif.). Sequential excitation at wavelengths of 568, 488, and 647 nm was provided by a 15 mW krypton/argon laser (American Laser), and sequential images were collected using LaserSharp software (Bio-Rad).
RESULTS
HMGN but not phospho-HMGN colocalizes with nuclear DNA during telophase.
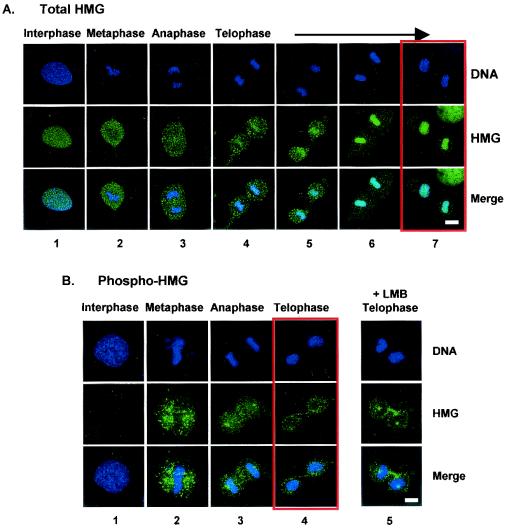
Previous biochemical studies indicated that the NBDs of HMGN proteins are highly phosphorylated only during mitosis (33). Confocal immunofluorescence microscopy with antibodies specific for HMGN1 or their phosphorylated NBDs reveals that phosphorylation affects the intracellular distribution of the proteins (Fig. 1). In interphase cells, antibodies to HMGN1 produce a strong nuclear signal, while antibodies to phosphorylated HMGN1 do not produce a signal at all (Fig. 1, panels 1). In metaphase and anaphase, both antibodies produce strong signals, indicating that phosphorylated HMGN proteins are present throughout the entire intracellular space (Fig. 1, panels 2 and 3). The protein does not colocalize with DNA, an observation that is in full agreement with our previous findings that the mitotic phosphorylation of the NBD abolishes the ability of HMGN proteins to bind to chromatin (33). In early telophase, the HMGN protein starts to accumulate in the newly formed nucleus and colocalizes with the DNA (Fig. 1A, panels 4). As the cells pass through telophase, an increasing fraction of the HMGN protein moves from the cytoplasm into the nucleus, and in late telophase, the great majority of the protein colocalizes with the DNA, and only traces of protein are found in the cytoplasm (Fig. 1A, panels 6 and 7). In contrast, the phosphorylated protein never colocalizes with DNA, and even in late telophase, the great majority of the immunofluorescent signal remains in the cytoplasm (Fig. 1B, panels 4). Our finding that both the nuclear entry and DNA colocalization of HMGN proteins are associated with dephosphorylation of their NBDs raises the possibility that the phosphorylated species is not efficiently imported into the nucleus. To exclude the possibility that the phosphorylated protein enters the nucleus but is then rapidly exported, we also examined cells grown in the presence of leptomycin B, an inhibitor of nuclear export. The confocal immunofluorescence images with antibody to phosphorylated HMGN in the leptomycin B-treated cells were indistinguishable from those obtained with cells not treated with leptomycin B. In agreement with previous findings that HMGN proteins are phosphorylated only during mitosis (33), interphase cells did not produce a fluorescent signal (not shown). More significantly, in late telophase, the distribution of the phosphorylated HMGN proteins in the leptomycin B-treated cells was not altered, and the great majority of the immunofluorescent signal remained in the cytoplasm, clearly distinct from the DNA signal (Fig. 1B, panels 5). The data suggest that phosphorylated HMGN proteins do not enter the nucleus.
FIG. 1.
Relocalization of HMGN1, but not of phosphorylated HMGN1, with nuclear DNA during telophase. Confocal immunofluorescence microscopy of mouse embryonic fibroblasts with antibodies elicited against either the C-terminal domain of mouse HMGN1 (A) or against the phosphorylated NBD of HMGN1 (B). The locations of the antibodies were determined by indirect immunofluorescence with fluorescein-labeled goat anti-rabbit immunoglobulin G. Note the increased colocalization of HMGN1 with DNA and the gradual decrease in extrachromosomal HMGN1 as the cell progresses through telophase. The panels boxed in red depict cells in late telophase. Note the differences in color of the nucleus in the merged panels in panels A and B. In panel B, panels 5 are confocal images from cells grown in the presence of leptomycin B (LMB). Bars, 10 μm.
Nuclear import of HMGN is abolished by phosphorylation but not by negative charges.
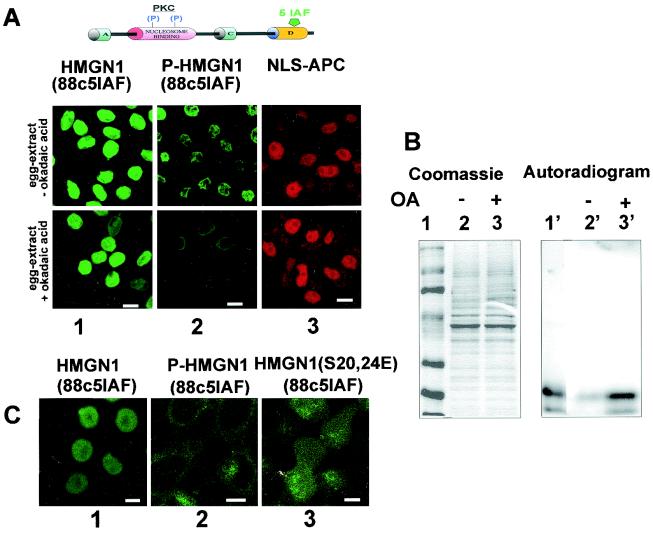
To test directly whether the state of phosphorylation affects the nuclear entry of HMGN1 proteins, we first produced an intrinsically fluorescent HMGN1 protein by reacting an Ala88Cys point mutant of HMGN1 (15, 38) with 5IAF and then phosphorylated the NBD of the protein with PKC using trace amounts of γ-32P-labeled ATP. Next, we added equal amounts of either phosphorylated or nonphosphorylated 5IAF-labeled HMGN1 to digitonin-permeabilized HEp-2 cells, used an X. laevis egg extract as a source of import factors, and visualized the locations of the fluorescent HMGN1 proteins (Fig. 2A). Based on the fluorescence intensity, the import of the phosphorylated protein was significantly less efficient than that of the nonphosphorylated proteins (Fig. 2A, top row, compare panels 1 and 2). Since the X. laevis extracts contain phosphatases that dephosphorylate the PKC-phosphorylated HMGN1 (Fig. 2B), the protein that entered the nucleus could be the unphosphorylated species. We therefore inhibited the phosphatases by treating the extracts with okadaic acid (Fig. 2B) and used the okadaic acid-treated extracts as a source of import factors that would facilitate the entry of the fluorescently labeled HMGN1 into the permeabilized HEp-2 cells. Fluorescence microscopy indicated that the okadaic acid treatment did not interfere with the import of either wild-type HMGN1 (Fig. 2A, bottom row, panel 1) or the NLS-conjugated autofluorescent protein allophycocyanin (Fig. 2A, bottom row, panel 3) but strongly inhibited the nuclear entry of the phosphorylated HMGN1 protein (Fig. 2A, bottom row, panel 2). Therefore, we conclude that phosphorylation of the NBD prevents the nuclear entry of HMGN1.
FIG. 2.
Phosphorylation of HMGN blocks nuclear import. (A) Inhibition of phosphatase activity blocks nuclear import of phosphorylated HMGN1. Fluorescein-labeled HMGN1 is diagrammed at the top of panel A; 5IAF is the fluorescent acetamide-fluorescein attached to the C-terminal domain of the protein, P denotes PKC-phosphorylated serines in the NBD of HMGN1, and NLC-APC is the NLS-allophycocyanin conjugate. Confocal images visualizing the import of fluorescein-labeled HMGN1 into permeabilized HEp-2 cells, mediated by a X. laevis egg extract that contained or did not contain okadaic acid. In the absence of okadaic acid, the phosphorylated HMGN1 protein (P-HMGN1) was imported into the nucleus less efficiently than the nonphosphorylated HMGN1, as evidenced by the intensity of the fluorescent signal (compare panels 1 and 2 in top row). In the presence of okadaic acid, the import of the phosphorylated protein but not that of the nonphosphorylated protein was totally blocked (compare panels 1 and 2 in bottom row). (B) Stability of HMGN1 phosphorylation in Xenopus egg extracts treated with okadaic acid and analyzed by SDS-PAGE. 5IAF-labeled HMGN1, phosphorylated with 32P ATP by PKC, was incubated in Xenopus egg extract that contained (+) or did not contain (−) okadaic acid (OA). An aliquot of the phosphorylated protein was added to the molecular size markers (lane 1). Note that okadaic acid inhibited the phosphatase activity in the egg extract (compare lane 2′ to lane 3′). (C) Negative charges in the NBD do not inhibit nuclear import. Import of fluorescently labeled HMGN1, phosphorylated HMGN1 (P-HMGN1), or the negatively charged HMGN1S20,24E mutant with extracts containing okadaic acid. Note that the phosphorylated HMGN1 protein does not enter the nucleus, but the double point mutant does (compare panel 2 to panel 3). Bars, 20 μm.
The NLS of the HMGN proteins consists of two elements separated by a stretch of about 30 amino acids, which include the two serine residues in the highly conserved, positively charged NBD (15). Phosphorylation of these two serine residues significantly changes the net charge of the most conserved region of the protein and potentially could induce conformational changes that would disrupt the function of the bipartite NLS. To critically examine whether the introduction of two negative charges is sufficient to impair nuclear import, we generated the double point mutant HMGN1S20,24E, in which the two serine residues in the HMGN NBD are mutated to glutamic acid. We then compared the nuclear entry of this mutant to that of the wild type or phosphorylated wild-type HMGN1. Okadaic acid-treated Xenopus egg extracts facilitated the nuclear entry of the HMGN1S20,24E double mutant protein; however, a significant portion of the protein was also found in the cytoplasm (Fig. 2C, panel 3). In contrast, all of the wild-type protein concentrated into the nucleus (Fig. 2C, panel 1). We previously demonstrated that the HMGN1S20,24E double mutant does not bind to nucleosomes (33). Our findings suggest that the glutamic acid double mutant enters the nucleus, but since it does not bind to nucleosomes, it is not efficiently retained and could be exported into the cytoplasm.
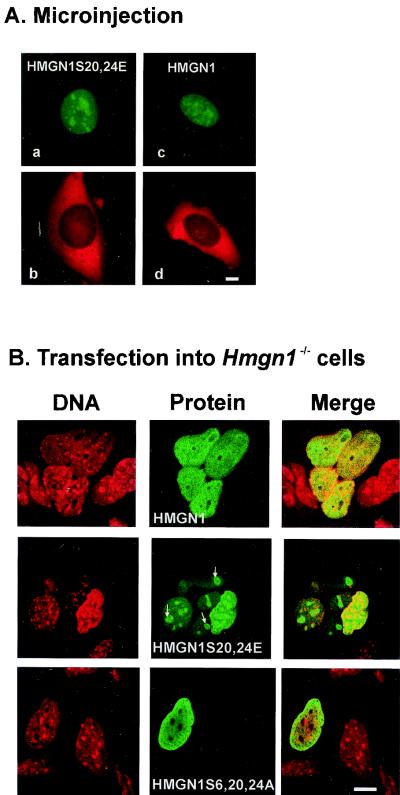
Microinjection of the 5IAF-labeled wild-type HMGN1 protein or of the negatively charged, double mutant HMGN1S20,24E protein into the cytoplasm of intact mouse embryonic fibroblasts indicates that both proteins enter into and are retained in the nucleus. High-molecular-weight dextran tagged with Texas red which was coinjected with the proteins remains in the cytoplasm, an indication that the nuclear membrane of the injected cells is intact and prevents the nuclear entry of large molecules lacking NLSs (Fig. 3A). We found that the nuclear entry of phosphorylated proteins cannot be studied by this approach, because the microinjected proteins are rapidly dephosphorylated by the cytoplasmic phosphatases. We conclude, however, that negative charges do not prevent the entry of the protein into the nucleus.
FIG. 3.
Negative charges in the HMGN NBD do not inhibit nuclear import. (A) Fluorescence microscopy of mouse embryonic fibroblasts injected with solutions containing fluorescently labeled (green) wild-type or mutant HMGN1 and high-molecular-weight dextran labeled with Texas red. The dextran remains in the cytoplasm, while both proteins enter the nucleus. (B) Confocal laser scanning fluorescence microscopy visualizing the location of either the DNA or the transfected proteins in HmgN1−/− mouse fibroblasts. The proteins expressed in the various cells are indicated in the figure. Since the endogenous protein is not expressed in these cells, the transfected protein was visualized by indirect immunofluorescence with antibodies specific to HMGN1. Note that the wild-type protein is evenly distributed throughout the interphase nucleus, while the double mutant with a negatively charged NBD forms aggregates (white arrows). Bars, 20 μm.
To avoid any possible effects due to the chemical modifications of the proteins or possible artifacts due to microinjection, we expressed wild-type HMGN1, the negatively charged HMGN1S20,24E double mutant, and a triple mutant of HMGN1 in which serines 6, 20, and 24 were mutated to alanine (mutant HMGN1S6,20,24A) in mouse fibroblasts lacking endogenous HMGN1 protein, i.e., HmgN1−/− cells. The expressed proteins are not tagged by either GFP or 5IAF. The HmgN1−/− cells, which will be described in detail elsewhere (Birger, unpublished), were derived from knockout mice in which the endogenous Hmgn1 gene was disrupted by targeted insertion. Detailed Northern and Western analyses verified that cells derived from these mice do not contain HMGN1 transcripts and do not express the endogenous HMGN1 protein (not shown). Since these cells do not contain endogenous HMGN1, the locations of the proteins expressed from the transfected vectors can be visualized with antibodies specific for human HMGN1.
Immunofluorescence analysis of HmgN1−/− fibroblast cultures transfected with plasmids expressing either wild-type human HMGN1 (Fig. 3B), the HMGN1S20,24E double mutant (Fig. 3B), or the HMGN1S6,20,24A triple mutant indicate that all the proteins enter the nucleus. As expected, the plasmid did not transfect all of the cells; therefore, part of the HmgN1−/− fibroblasts did not produce a green fluorescent signal (red nuclei in the merge column). In transfected interphase cells, all the proteins were found exclusively in the nucleus. However, whereas the wild-type HMGN1 mutant protein was evenly dispersed throughout the nucleus (Fig. 3A), the HMGN1S20,24E mutant was unevenly dispersed and formed aggregates (Fig. 3B, middle row), suggesting that the intranuclear organization of this negatively charged double mutant is different from that of the wild-type HMGN1. As expected, in mitosis, the negatively charged double mutant does not bind to chromosomes (33).
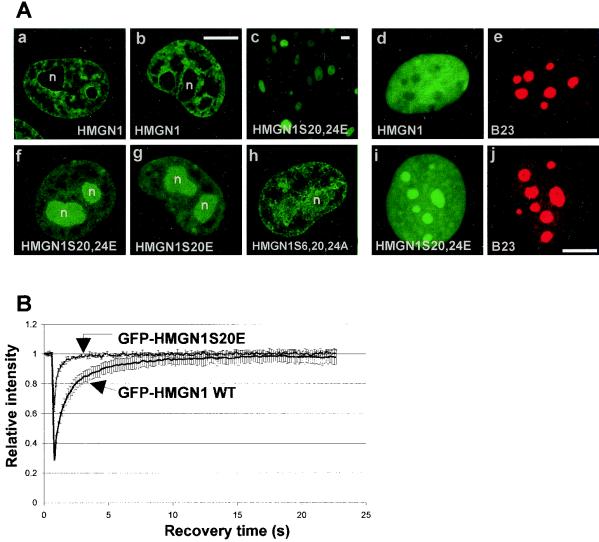
The major site of interaction of HMGN with chromatin is their NBD (8). To test whether mutations in the NBD of HMGN1 alters the intranuclear organization of the protein in living cells, we visualized the locations of HMGN1-GFP fusion proteins expressed in growing HeLa cells. Whereas wild-type HMGN-GFP is unevenly dispersed throughout the entire nonnucleolar space in the nucleus, the negatively charged double mutant HMGN1S20,24E-GFP and the negatively charged single mutant HMGN1S20E-GFP localized mainly to the nucleolus (Fig 4A). Immunofluorescence analysis of fixed cells with antibodies to B23 which specifically stain the nucleoli verified that the double point mutant HMGN was localized in the nucleoli. The triple mutant HMGN1S6,20,24A-GFP, whose nucleosome binding ability is partially impaired (not shown), was also altered, and the protein was found throughout the entire nucleus, including the nucleoli. These studies suggest a direct link between the chromatin binding ability and the intranuclear location of HMGN proteins and provide additional support for the conclusion that interphase nuclei do not contain significant levels of phosphorylated proteins.
FIG. 4.
Mislocalization of negatively charged HMGN1 mutants. Wild-type HMGN1 and GFP-labeled HMGN1 mutant proteins expressed in HeLa cells are shown. The type of GFP-labeled protein is indicated in the various panels. Note that wild-type HMGN1 is distributed throughout the entire nonnucleolar space in the nucleus (a and b). The negatively charged HMGN1S20,24E mutant is efficiently expressed in and enters the nucleus in all the HeLa cells (c and f). This double point mutant mislocalizes to the nucleolus (f), as does the single point mutant (g). The triple mutant HMGN1S6,20,24A, which is not negatively charged, is distributed throughout the entire nucleus (h). The nucleolar localization in living cells was independently verified by immunofluorescence with anti-B23 using fixed cells (cells expressing wild-type protein [d and e] and cells expressing mutant HMGN1 [i and j]). n, nucleolus. Bars, 20 μm (b and j) and 50 μm (c). (B) FRAP analysis of the intranuclear mobility of the wild-type (WT) HMGN1 and single point mutant HMGN1S20E. Note that the fluorescence recovery of the mutant is faster than that of the wild type, indicating that the mutant is more mobile.
We demonstrated previously that HMGN1S20,24E does not bind to nucleosomes and that in living cells this mutant is more mobile than the wild-type protein, an indication that its residence time on chromatin is shortened (33). To test whether a single negative charge in the NBD disrupts the interaction of the protein with chromatin, we used FRAP to compare the intranuclear mobility of the HMGN1S20E mutant to that of the wild-type HMGN1 protein. In FRAP experiments, a small area in a living cell expressing a fluorescently tagged protein is bleached with a high-intensity laser, and the rate of recovery of the fluorescence in the photobleached area is recorded. The rate of fluorescence recovery is an indication of the rate at which freely mobile molecules migrate into the photobleached spot. The rate of mobility is inversely related to their residence time at their target binding sites. HMGN1 is a highly mobile protein (32), and 80% of the prebleach intensity was reached in approximately 3 s (Fig. 4B). However, the mobility of the HMGN1S20E mutant was even faster, and 80% of the prebleach fluorescence intensity was reached within 1 s, i.e., at approximately the same time as free GFP (32).
The negative charge in the NBD affects the chromatin interaction and the intranuclear distribution, but not the nuclear entry of HMGN proteins. Taken together, the data suggest that inhibition of nuclear import is due to the presence of phosphorylated serine residues and not to the functional disruption of the bipartite NLS by the negative charges. We also note the absence of fluorescent signal in the cytoplasm of cells expressing the HMGN-GFP fusion proteins (Fig. 3B and 4A). Thus, although the mutated proteins cannot bind to nucleosomes and mislocalize to the nucleolus, they are retained within the nucleus and are not exported into the cytoplasm.
Phosphorylated HMGN1 binds to 14.3.3ζ protein.
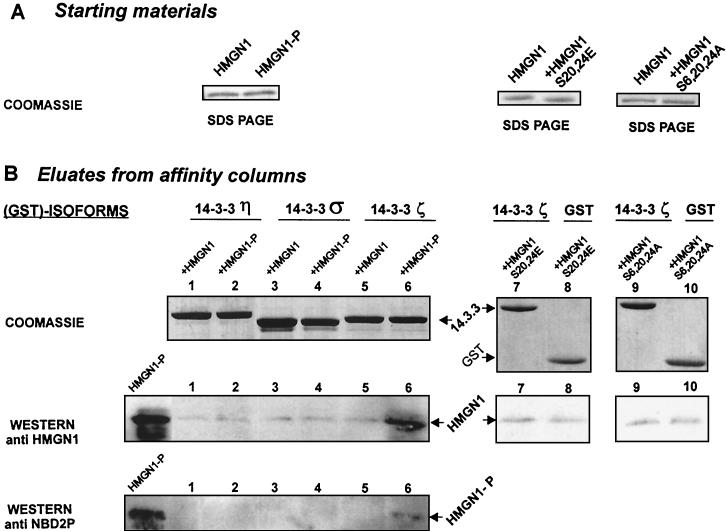
The sequence surrounding the two phosphorylated serines in the NBDs of HMGN proteins, RRSARLS, corresponds to the canonical Rx1-2Sx2-3S recognition motifs of 14.3.3 proteins (10). 14.3.3 proteins are known to be involved in cellular targeting processes, including nuclear import (10, 30). Therefore, it is possible that the inability of phosphorylated HMGN1 to enter the nucleus is linked to 14.3.3 binding. To test whether phosphorylated HMGN1 specifically interacts with 14.3.3 proteins, we incubated equal amounts of either nonphosphorylated or PKC-phosphorylated HMGN1 (Fig. 5A) with glutathione agarose tagged with GST or with the GST-fused 14.3.3η, 14.3.3σ, or 14.3.3ζ isoform. After extensive washing, the beads were treated with a buffer containing SDS and β-mercaptoethanol, thereby releasing the 14.3.3 proteins and other bound molecules from the immobilizing support. The released proteins were fractionated by electrophoresis in SDS-containing polyacrylamide gels. Coomassie blue staining of the gels indicates that the various slurries contained equal amounts of the14.3.3 isoforms, while the results of Western analysis indicate that a significant amount of HMGN1 protein was retained only when the phosphorylated protein was specifically bound by the 14.3.3ζ isoform (Fig. 5B). The trace amount of HMGN1 present in all the lanes is additional proof that equal amounts of proteins were loaded on the gels and that 14.3.3η and 14.3.3σ do not preferentially bind phosphorylated HMGN1. The GST control columns also bound only traces of HMGN1 (not shown). Western analysis with antibodies to phosphorylated proteins (anti-NBD2P) verified that the HMGN1 protein eluted from the 14.3.3ζ beads was phosphorylated (Fig. 5B, lanes 6).
FIG. 5.
Specific binding of phosphorylated HMGN1 (HMGN1-P) to 14.3.3ζ. (A) Polyacrylamide gel analysis of the starting materials, applied to the affinity columns. The SDS-polyacrylamide gels demonstrate that the solutions contained equal amounts of protein. (B) Analysis of the eluates from the 14.3.3 affinity columns. Either nonphosphorylated or PKC-phosphorylated HMGN1 was added to the GST-14.3.3 isoforms indicated. Coomassie blue staining of SDS-polyacrylamide gels of the eluates indicates that each fraction contained equal amounts of 14.3.3 protein. The results of Western analysis indicate a specific interaction between phosphorylated HMGN1 (HMGN1-P) and 14.3.3ζ. Equal trace amounts of HMGN1S20,24E bound to agarose beads containing 14.3.3ζ or GST (lanes 7 and 8). The HMGN1S6,20,24A mutant also does not bind to 14.3.3ζ (lanes 9 and 10).
Next, we tested whether the introduction of negative charges in the NBD of HMGN1 was sufficient to facilitate binding to 14.3.3ζ and incubated the double point mutant HMGN1S20,24E with either control gluthatione agarose beads or with the same beads containing the immobilized 14.3.3ζ isoform. The HMGN1S20,24E mutant did not bind to either of the beads (Fig. 5B, lanes 7 and 8), indicating that the negative charge by itself is not sufficient to facilitate specific interaction between HMGN1 and 14.3.3ζ proteins. Therefore, we conclude that the interaction between HMGN1 and the 14.3.3ζ isoform is specific to the phosphorylated form HMGN1 and is not due to a nonspecific negative charge effect. Thus, phosphorylation of the NBD of HMGN1 leads to inhibition of nuclear import and promotes interactions with 14.3.3ζ protein.
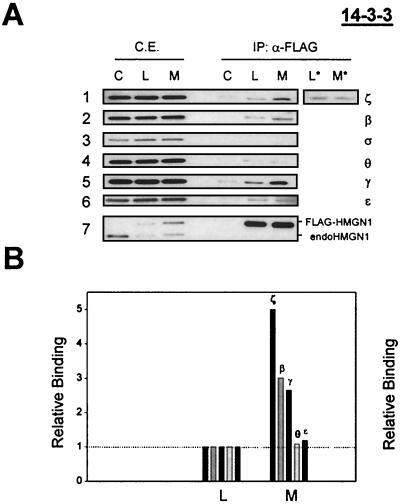
HMGN1 is associated with 14.3.3 in mitotic HeLa cells. The NBD of HMGN protein is phosphorylated only during mitosis (33). Therefore, to test whether HMGN1 proteins are indeed associated with 14.3.3 proteins in vivo, we isolated HMGN1-associated proteins from cells that were growing logarithmically or were arrested in mitosis. To facilitate the isolation of HMGN1-associated proteins, we first generated HeLa cells stably expressing FLAG-tagged HMGN1 and used affinity chromatography on columns containing FLAG antibodies to separate the tagged HMGN1 proteins from the cell extracts. The results of Western analysis with antibodies specific to HMGN1 indicate that the levels of the FLAG-tagged HMGN1 protein were approximately equal to those of the endogenous protein. The affinity columns efficiently purified the FLAG-tagged HMGN1 protein from both logarithmically growing and mitotic cells (Fig. 6A, blot 7). The equal intensity of the signal obtained from the Western analyses verified that equal amounts of HMGN1 proteins were loaded on and recovered from the affinity columns.
FIG. 6.
Coimmunoprecipitation reveals preferential associations of HMGN1 with specific isoforms of 14.3.3 proteins in mitotic cells. (A) Total cell extracts (C.E.) prepared from control cells (C), from logarithmically growing cells expressing FLAG-tagged HMGN1 (L), or from mitotic cells expressing the FLAG-tagged HMGN1 (M) were treated with immobilized anti-FLAG antibodies (α-FLAG). Lanes L* and M* contain extracts not treated with phosphatase inhibitors and the mitotic HMGNs are not phosphorylated. The proteins in the extracts (C.E.) and the materials bound to the affinity column (IP: α-FLAG) were fractionated by SDS-PAGE, and the contents of the various 14.3.3 isoforms were visualized by immunoblotting with specific antibodies. The antibodies used to develop the blots are indicated on the right under the 14.3.3 heading. Each of the three C.E. lanes, lanes C, L, and M, contained 5 μg of cell extract. In the IP: α-FLAG lanes, each lane contained 1/40th of the material recovered from the affinity columns. Each strip is a separate gel. The lowest strip shows a Western blot with anti-HMGN1 antibody, demonstrating the relative levels of HMGN1 and FLAG-HMGN1 in the cells. IP, immunoprecipitation. (B) Quantification of the relative enrichment of the signal obtained with the various anti-14.3.3 antibodies. In each case, the signal obtained with the FLAG-bound proteins from the logarithmically growing cells (L) was set at 1.0 (dotted line). In the mitotic cell preparation (M), about 65% of the cells were in metaphase.
The results of Western analysis with antibodies specific for the various 14.3.3 isoforms confirms that equal amounts of extracts (Fig. 6A) prepared from either nontransformed, control cells (Fig. 6A, lanes C) or from logarithmically growing cells (Fig. 6A, lanes L) and mitotic cells (Fig. 6A, lanes M) expressing HMGN-Flag were loaded on the columns. Quantitative analysis of the Western blots obtained with the proteins recovered from the affinity columns (Fig. 6A) indicate that the relative amounts of 14.3.3 proteins bound to HMGN in mitotic extracts were significantly higher than those in extracts prepared from growing cells (Fig. 6B). In these experiments, it is necessary to include phosphatase inhibitors in the solutions used to prepare the proteins. In the absence of phosphatase inhibitors, the mitotic HMGN1 is rapidly dephosphorylated and only trace amounts of 14.3.3ζ are bound to the protein extracted from either logarithmically growing or mitotic cells (Fig. 6A, lanes L* and M*). Compared to logarithmically growing cells, the amount of 14.3.3ζ associated with HMGN1 during mitosis increased fivefold, that of 14.3.3β increased threefold, and that of 14.3.3γ increased 2.7-fold, a total increase of 10.7-fold. Our finding that 14.3.3ζ is the major isotype that interacts with HMGN1 is in full agreement with the in vitro binding assay results (Fig. 6B). Considering that under our synchronization conditions, only approximately 70% of the cells were in mitosis and that in the logarithmically growing cell culture approximately 5% of the cell were in mitosis (33), we estimate that the total increase in 14.3.3 proteins associated with HMGN1 during mitosis is more than 16-fold over that in logarithmically growing cells. Thus, mitotic phosphorylation of the NBD of HMGN promotes binding of 14.3.3 proteins.
DISCUSSION
We report here that phosphorylation of serine residues in the NBD of chromosomal protein HMGN1 inhibits its entry into the nucleus and facilitates interaction with 14.3.3 proteins. Since the NBDs of the HMGN proteins are phosphorylated only during mitosis (33), we suggest that the interaction of HMGN with 14.3.3 proteins is a direct consequence of mitotic phosphorylation. Because 14.3.3 proteins are known to affect the subcellular localization of their binding partners (30), we link these two findings and suggest that the interaction of HMGN with 14.3.3 proteins retards their entry into the nucleus.
Interaction of HMGN1 with 14.3.3 proteins.
The results from several types of experiments support the conclusion that HMGN1 specifically associates with 14.3.3 proteins. First, in vitro binding assay results indicate that only the phosphorylated form of HMGN1 interacts strongly with the ζ isoform of 14.3.3 proteins. Second, the HMGN1S20,24E mutant that contains the same negative charges as the phosphorylated proteins does not bind to the 14.3.3ζ proteins, an indication that the HMGN1-14.3.3ζ interaction requires the presence of a phosphorylated NBD. Third, immunoprecipitation experiments with cell extracts reveal specific interactions between HMGN and 14.3.3 proteins in extracts from cells in mitosis, the phase in the cell cycle in which the HMGN1 proteins are highly phosphorylated (33). Significantly, 14.3.3ζ is the major isoform that interacts with HMGN1 in vivo, providing further support for the in vitro binding assays and for the specificity of the interaction.
It is well documented that the various isoforms of 14.3.3 proteins interact with and regulate the cellular locations of numerous proteins, thereby affecting many cellular processes, including cell division, cell signaling, and apoptosis (10, 39). Our studies are the first to report that 14.3.3 proteins interact with structural proteins that are known to directly affect the structure of the chromatin fiber. A role for 14.3.3 proteins in regulating the transcriptional activity of chromatin has already been suggested by the finding that the interaction of 14.3.3 with histone deacetylases (HDACs) sequesters the proteins in the cytoplasm, while loss of this interaction translocates the HDACs into the nucleus and enhances transcriptional repression, presumably due to enhanced histone deacetylation (10, 13, 40). Similarly, by sequestering the transcriptional regulator RSG in the cytoplasm, 14.3.3 proteins affect the endogenous amounts of gibberellins and regulate many aspects of plant development (18). In all these cases, the interaction of 14.3.3 with their protein partners is mediated by the recognition of specific phosphorylated serines. In this respect, the interaction of HMGN1 protein with 14.3.3 is similar in that it occurs only when the HMGN1 NBD is phosphorylated. Indeed, the sequence surrounding the phosphorylated serines in this NBD corresponds to a known 14.3.3 binding element (10). A significant difference between the HMGN proteins and the chromatin-modifying activities, such as HDAC, is that the HMGN proteins are always found in the nucleus, while the other activities are known to shuttle between the nucleus and cytoplasm.
During mitosis, the NBD of HMGN is phosphorylated, and the negative charge abolishes their ability to bind to nucleosomes (33). Since the 14.3.3 binding module of HMGN proteins is their NBD, this interaction could serve as an additional mechanism for preventing the interaction of HMGN with chromosomes, a novel role for 14.3.3 in regulating cellular activities. In addition, 14.3.3 may also retard the reentry of HMGN into the newly formed nucleus in late telophase.
Regulating the nuclear entry of HMGN.
HMGN proteins are the smallest nuclear proteins known to be actively transported into the nucleus (15). The proteins are displaced from mitotic chromatin but colocalize with the nuclear DNA at the end of telophase, as soon as the nuclear membrane is re-formed (15) (Fig. 1). Formation of the nuclear membrane precedes the onset of nuclear import (14). Since phosphorylated proteins fail to enter the nucleus, our results suggest that the proteins are rapidly dephosphorylated in the time interval between the formation of the nuclear membrane and the onset of nuclear import. Throughout interphase, the conserved serines in the NBD remain dephosphorylated (33), and the proteins are found only in the nucleus (Fig. 1). The results of our immunofluorescence (Fig. 1 and 3), microinjection (Fig. 3A), and HMGN-GFP expression (Fig 4A) analyses, as well as the results of cell fractionation experiments of Louie et al. (26), clearly demonstrate that 90% or more of the proteins are located in the nucleus throughout the nonmitotic stages of the cell cycle. These findings and our observations that HMGN1 mutants that do not bind to chromatin do not accumulate in the cytoplasm suggest that these proteins do not shuttle constantly between the nucleus and cytoplasm. The dephosphorylated stage could be maintained by the action of phosphatases, since treatment of K562 cells with okadaic acid leads to HMGN phosphorylation (26). The phosphorylated species were found in the cytoplasm, providing additional support that phosphorylation of the serines in the conserved NBD is incompatible with nuclear localization. Indeed, the intranuclear location of HMGN proteins bearing negative charges in the NBD is different from that of the native proteins (Fig. 4A). This observation together with the results of FRAP analysis (Fig. 4B) indicates that the negative charges impair the interaction of HMGN with chromatin.
Functional consequences of HMGN mitotic phosphorylation.
Mitotic phosphorylation introduces negatively charged phosphates into the NBDs of HMGN proteins. This process abolishes their ability to bind to nucleosomes (33), promotes interactions with 14.3.3 proteins, and impairs their nuclear import. Our studies uncouple the general effect of negative charge from those specific to the phosphate ion (Table 1). The native, unmodified protein enters the nucleus and binds to nucleosomes but does not bind to 14.3.3. The phosphorylated protein binds to 14.3.3 but does not enter the nucleus and does not bind to chromatin. The negatively charged HMGN1S20,24E double mutant does not bind to 14.3.3 and enters the nucleus but does not bind to nucleosomes. Taken together, the data indicate that the negative charge abolishes the interaction with chromatin, while the phosphate promotes binding to 14.3.3 and impairs nuclear import.
TABLE 1.
Uncoupling the effect of negative charge from that of phosphorylation
| Protein | Nucleosome binding | Nuclear import | 14.3.3 binding |
|---|---|---|---|
| Wild-type HMGN1 | Yes | Yes | No |
| Phosphorylated HMGN1 | No | No | Yes |
| HMGN1-S20,24E mutant | No | Yes | No |
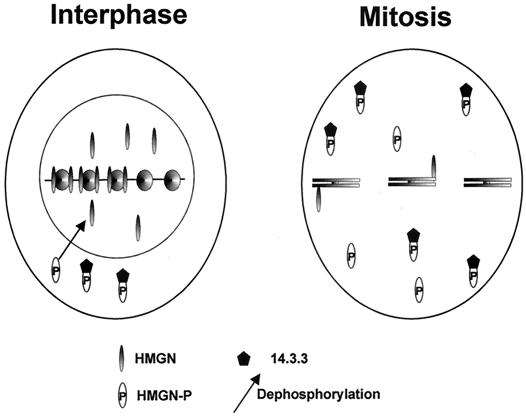
The model presented in Fig. 7 provides a conceptual framework for understanding the role of mitotic phosphorylation in regulating the cellular function of HMGN proteins. Throughout interphase, the HMGN proteins are located in the nucleus in a dynamic state (32), and their organization is related to transcriptional activity (16). Taken together with numerous results indicating that the proteins unfold the chromatin fiber and enhance transcription from chromatin templates (5, 7), the data suggest that the interaction of HMGN with chromatin facilitates access to the nucleosomal DNA.
FIG. 7.
Model of the effect of mitotic phosphorylation on the chromatin interaction and intracellular trafficking of HMGN proteins. In the interphase nucleus, nonphosphorylated proteins are found either associated with nucleosomes or in the nucleoplasm. In mitotic cells, the NBDs of most of the HMGN proteins are phosphorylated. The phosphorylated protein is not bound to chromosomes and can form a complex with 14.3.3. At the end of mitosis, the phosphorylated protein is temporarily sequestered in the cytoplasm by its association with 14.3.3 protein. Nuclear entry is associated with dephosphorylation of the NBD. HMGN-P, phosphorylated HMGN.
Mitosis is associated with a global inhibition of transcription and with compaction of the chromatin; however, at the nucleosomal level, the organization of the chromatin fiber is not grossly altered, and features characteristic of active chromatin are present in mitotic chromosomes (11, 20, 24, 27). Furthermore, recent results suggest that during mitosis, a set of chromatin-remodeling enzymes remain active and a subset of genes are highly transcribed (22, 42). Therefore, even during this stage of the cell cycle, unmodified HMGN proteins may access nucleosomes and interfere with the orderly progression of mitosis. Mitotic phosphorylation of the HMGN NBD and interactions with 14.3.3 proteins prevent the binding of HMGN proteins to nucleosomes in mitotic chromatin.
The interaction of HMGN with 14.3.3 also sequesters the proteins in the cytoplasm until the nuclear membrane is fully formed, prior to the onset of nuclear import activity (14). It may be significant that the serine residues affecting nuclear import are the same as those affecting chromatin interaction. By preventing nuclear entry until the chromatin is fully remodeled for the next round of transcription, phosphorylation of the conserved serine residues in the NBD of HMGN provides a tight link between nuclear import and transcription from chromatin templates.
Since numerous regulatory proteins such as SWI/SNF (29, 37), TFIID (36) Oct I (35), cJun (1), Sp1 (27) and HMGs (34) are phosphorylated and displaced from chromatin during mitosis (2, 12) and since phosphorylation affects nuclear import (2, 17, 19, 41), it is likely that the results presented here are of general relevance to understanding the functional effects of mitotic phosphorylation on the intracellular trafficking of nuclear proteins.
Acknowledgments
Marta Prymakowska-Bosak and Robert Hock contributed equally to this work.
We thank H Piwnica-Worms (Washington University) for a gift of GST-14.3.3 bacterial expression vectors and Yaffa Rubinstein for constructive criticisms of the manuscript.
Part of this work was supported by grant HO 1804/2 from the Deutsche Forschungsgemeinschaft to R.H.
REFERENCES
- 1.Bakiri, L., D. Lallemand, E. Bossy-Wetzel, and M. Yaniv. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19:2056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulikas, T. 1995. Phosphorylation of transcription factors and control of the cell cycle. Crit. Rev. Eukaryot. Gene Expr. 5:1-77. [PubMed] [Google Scholar]
- 3.Brunet, A., F. Kanai, J. Stehn, J. Xu, D. Sarbassova, J. V. Frangioni, S. N. Dalal, J. A. DeCaprio, M. E. Greenberg, and M. B. Yaffe. 2002. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156:817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin, M., P. S. Becerra, M. P. Crippa, D. A. Lehn, J. M. Pash, and J. Shiloach. 1991. Recombinant human chromosomal proteins HMG-14 and HMG-17. Nucleic Acids Res. 19:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin, M., L. Trieschmann, and Y. V. Postnikov. 1995. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin. Cell Biol. 6:247-255. [DOI] [PubMed] [Google Scholar]
- 8.Cook, G. R., M. Minch, G. P. Schroth, and E. M. Bradbury. 1989. Analysis of the binding of high mobility group protein 17 to the nucleosome core particle by 1H NMR spectroscopy. J. Biol. Chem. 264:1799-1803. [PubMed] [Google Scholar]
- 9.Dundr, M., T. Misteli, and M. O. Olson. 2000. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150:433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 11.Gazit, B., H. Cedar, I. Lerer, and R. Voss. 1982. Active genes are sensitive to deoxyribonuclease I during metaphase. Science 217:648-650. [DOI] [PubMed] [Google Scholar]
- 12.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 13.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraguchi, T., T. Koujin, T. Hayakawa, T. Kaneda, C. Tsutsumi, N. Imamoto, C. Akazawa, J. Sukegawa, Y. Yoneda, and Y. Hiraoka. 2000. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113:779-794. [DOI] [PubMed] [Google Scholar]
- 15.Hock, R., U. Scheer, and M. Bustin. 1998. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J. Cell Biol. 143:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hock, R., F. Wilde, U. Scheer, and M. Bustin. 1998. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 17:6992-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood, J. K., and P. A. Silver. 1999. In or out? Regulating nuclear transport. Curr. Opin. Cell Biol. 11:241-247. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi, D., S. Ishida, J. Fukazawa, and Y. Takahashi. 2001. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13:2483-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 20.Kerem, B., R. Goitein, G. Diamond, H. Cedar, and M. Marcus. 1984. Mapping of DNAse I sensitive regions on mitotic chromosomes. Cell 38:493-499. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, K., V. V. Rybenkov, N. J. Crisona, T. Hirano, and N. R. Cozzarelli. 1999. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98:239-248. [DOI] [PubMed] [Google Scholar]
- 22.Krebs, J., C. Fry, M. Samuels, and C. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn, A., A. Vente, M. Doree, and I. Grummt. 1998. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 284:1-5. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, M. T., B. Iyer, and R. J. Schwarz. 1982. Condensation of chromatin into chromosomes preserves an open configuration but alters the DNase I hypersensitive cleavage sites of the transcribed gene. Nucleic Acids Res. 10:4565-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Louie, D. F., K. K. Gloor, S. C. Galasinski, K. A. Resing, and N. G. Ahn. 2000. Phosphorylation and subcellular redistribution of high mobility group proteins 14 and 17, analyzed by mass spectrometry. Protein Sci. 9:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Balbas, M. A., A. Dey, S. K. Rabindran, K. Ozato, and C. Wu. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29-38. [DOI] [PubMed] [Google Scholar]
- 28.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 29.Muchardt, C., J. C. Reyes, B. Bourachot, E. Leguoy, and M. Yaniv. 1996. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 15:3394-3402. [PMC free article] [PubMed] [Google Scholar]
- 30.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 32.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 33.Prymakowska-Bosak, M., T. Misteli, J. E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves, R. 1992. Chromatin changes during the cell cycle. Curr. Opin. Cell Biol. 4:413-423. [DOI] [PubMed] [Google Scholar]
- 35.Segil, N., M. Geurmah, A. Hoffman, R. Roeder, and N. Heintz. 1996. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10:2389-2400. [DOI] [PubMed] [Google Scholar]
- 36.Segil, N., S. B. Roberts, and N. Heintz. 1991. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814-1816. [DOI] [PubMed] [Google Scholar]
- 37.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trieschmann, L., B. Martin, and M. Bustin. 1998. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc. Natl. Acad. Sci. USA 95:5468-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hemert, M. J., H. Y. Steensma, and G. P. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23:936-946. [DOI] [PubMed] [Google Scholar]
- 40.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wisniewski, J., E. Schultze, and B. Sapetto. 1994. DNA binding and nuclear translocation of insect HMG-1 proteins are inhibited by phosphorylation. Eur. J. Biochem. 225:687-693. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]