Abstract
RSC is an essential chromatin remodeling complex in Saccharomyces cerevisiae that performs central roles in transcriptional regulation and cell cycle progression. Here we identify Htl1 as a novel factor that associates with the RSC complex both physically and functionally. We isolated HTL1 through a genetic screen for mutants that displayed additive growth defects with a conditional mutation in the protein kinase C gene (PKC1), which has been suggested through genetic connections to interact functionally with RSC. Several lines of evidence connect HTL1 to RSC function. First, an htl1Δ mutant displayed temperature-sensitive growth and a G2/M cell cycle arrest at restrictive temperatures, a phenotype similar to that of strains with conditional mutations in essential RSC components. Second, we isolated RSC3, which encodes a component of the RSC complex, as a dosage suppressor of the htl1Δ growth arrest. Third, an htl1Δ mutant displayed additive growth defects with conditional rsc3 alleles. Fourth, overexpression of HTL1 suppressed the growth defect of a strain with a conditional mutation in another RSC component, RSC8. Finally, we demonstrate that Htl1 is a nuclear protein that can associate in vivo with a fraction of the RSC complex. We propose that an RSC-Htl1 complex acts coordinately with protein kinase C to regulate the G2/M transition.
Changes in chromatin structure play an important and dynamic role in gene regulation (19, 23). Chromatin remodelers are multiprotein complexes comprising an ATPase “engine” required for nucleosome repositioning (4, 24, 48) and various additional components thought to be important for the regulation and targeting of such complexes to genomic loci (38). Chromatin remodeling complexes are thought to activate or repress gene expression by exposing or concealing regulatory sequences. Two such complexes in Saccharomyces cerevisiae include the SWI/SNF complex and the RSC complex (named RSC for remodels the structure of chromatin). RSC is an abundant complex of at least 16 subunits, which was purified on the basis of homology of several of its subunits to those of the SWI/SNF complex (5). Of the two, only RSC is essential for viability, serving an important function in cell cycle progression from G2 to M (1, 5, 7, 45).
The RSC complex exists in distinct forms, containing either Rsc1 or Rsc2, and with or without Rsc3 and Rsc30 (1, 5, 6). Moreover, the RSC subunit Sfh1 is phosphorylated during the G1 phase of the cell cycle (7), suggesting that chromatin remodeling by RSC is regulated at multiple levels. Additionally, rsc mutations affect expression of genes involved in cell wall biogenesis, ribosome biogenesis, the nitrogen discrimination pathway (NDP), carbon source utilization, and the TOR pathway. Recently, whole-genome occupancy studies have revealed the occupancy of RSC at hundreds of yeast genes, including those involved in NDP, carbon source utilization, the TOR pathway, histone genes, and tRNA genes (9, 34).
Genetic observations have revealed a functional link between RSC components and the cell wall integrity signaling pathway controlled by the Rho1 GTPase and Pkc1 (1, 7a, 14). This pathway monitors and regulates cell wall biogenesis during vegetative growth and in response to pheromone-induced morphogenesis (31). The master switch for cell wall signaling is Rho1, which is activated by several members of a family of cell surface sensors (12, 17, 22, 36, 39, 46).
Active Rho-GTP binds to and activates protein kinase C (21, 35), which is encoded by PKC1 (32). Loss of PKC1 function, or any of the components of the mitogen-activated protein (MAP) kinase cascade under its control (31), results in a cell lysis defect that is attributable to a deficiency in cell wall construction (29, 30, 37). The MAP kinase cascade is a linear pathway comprised of a MEKK (Bck1 [8, 27]), a pair of redundant MEKs (Mkk1/2 [15]), and a MAP kinase (Mpk1/Slt2 [26, 33]). One consequence of signaling through the MAP kinase cascade is the activation of the Rlm1 transcription factor (10, 47). Signaling through Rlm1 regulates the expression of at least 25 genes, most of which have been implicated in cell wall biogenesis (18).
Because the growth defect of a pkc1 null mutant is more severe than that of any of the pathway components that function downstream of this protein kinase, we have proposed that Pkc1 regulates a bifurcated pathway (30). To elucidate the nature of the second pathway branch and to identify novel targets of Pkc1, we conducted a screen for mutations that displayed additive growth defects with a pkc1ts mutation. Here we report the isolation of HTL1, a gene previously reported to be required for genomic stability and for growth at elevated temperatures (25). We demonstrate that HTL1 is important for cell cycle progression. Loss of function of HTL1 results in a G2/M arrest at restrictive temperatures that is similar to that observed in strains with conditional mutations of essential RSC subunits. Finally, we show that Htl1 interacts with the RSC complex both physically and functionally.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YEPD (1% Bacto yeast extract, 2% Bacto Peptone, 2% glucose) with or without 10% sorbitol. Synthetic minimal (SD) medium (40) supplemented with the appropriate nutrients was used to select for plasmid maintenance and gene replacement. Synthetic complete (SC) medium with or without 5-fluoroorotic acid (5-FOA) (0.1% [3]) was used to assess the viability of rsc3 htl1Δ mutants. Yeast cells were transformed by the lithium acetate method (16). Escherichia coli DH5α was used to propagate all plasmids. E. coli cells were cultured in Luria broth medium (1% Bacto Tryptone, 0.5% Bacto yeast extract, 1% NaCl) and transformed by standard methods.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 1783 | MATaa | I. Herskowitz |
| 1788 | MATa/MATαa | I. Herskowitz |
| DL376 | MATapkc1Δ::LEU2a | 29 |
| DL1248 | MATα stt1-1 pRS315[PKC1 ADE3]b | This study |
| DL2312 | MATabck1Δ::HIS4a | This study |
| DL2315 | MATa/MATα bck1Δ::HIS4/BCK1a | This study |
| DL2745 | MATabck1Δ::HIS4 htl1Δ::TRP1a | This study |
| DL2749 | MATahtl1Δ::TRP1a | This study |
| DL2751 | MATα htl1Δ::TRP1a | This study |
| DL2754 | MATa/MATα htl1Δ::TRP1/htl1Δ::TRP1a | This study |
| DL2822 | MATα “pet18” isolate of DL1248#1b | This study |
| DL2823 | MATα “pet18” isolate of DL1248#2b | This study |
| MCY3890 | MATaswh3-ts16 (rsc8ts)c | 44 |
| YBC628 | MATarsc3Δ::HIS3 pRS316[RSC3]d | 1 |
| YBC840 | MATarsc3Δ::HIS3 pRS315[rsc3-1]d | 1 |
| YBC842 | MATarsc3Δ::HIS3 pRS315[rsc3-2]d | 1 |
| YBC843 | MATarsc3Δ::HIS3 pRS315[RSC3]d | 1 |
| YBC906 | MATarsc3Δ::HIS3 pRS315[rsc3-3]d | 1 |
| YBC928 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trp1Δ63 pep4Δ::Kanmx | This study |
| YBC1334 | MATarsc3Δ::HIS3 htl1Δ::TRP1 pRS316[HTL1] pRS315[RSC3]d | This study |
| YBC1336 | MATarsc3Δ::HIS3 htl1Δ::TRP1 pRS316[HTL1] pRS315[rsc3-1]d | This study |
| YBC1338 | MATarsc3Δ::HIS3 htl1Δ::TRP1 pRS316[HTL1] pRS315[rsc3-2]d | This study |
| YBC1340 | MATarsc3Δ::HIS3 htl1Δ::TRP1 pRS316[HTL1] pRS315[rsc3-3]d | This study |
EG123 background (leu2-3,112 trp1-1 ura3-52 his4 can1).
ade2 ade3 ura3 leu2 trp1 his3.
W303-1A background (trp1-1 leu2-3,112 his3-11,15 ura3-1 ade2-1 can1-100).
his3Δ200 leu2Δ1 lys2-128Δ trp1Δ63 ura3-52.
Synthetic lethal screen and isolation of HTL1.
S. cerevisiae strain DL1248 was grown in SD medium to an A600 of 0.5, washed, and resuspended in 0.9% KCl to a density of 3 × 107 cells/ml. The cells were then irradiated for 60 s with a 254-nm-wavelength UV lamp (0.5 J m−2 s−1), which resulted in an approximately 25% loss in plating efficiency. The irradiated cells were harvested, resuspended in YEPD, and grown overnight in the dark at 23°C. They were then plated onto YEPD (approximately 150 colonies/plate) and allowed to grow at 23°C for 3 to 4 days. Colonies that appeared entirely red with no white sectors were picked, patched onto YEPD, and grown at 37°C for 2 to 3 days. Five temperature-sensitive isolates were transformed with a centromeric library (in pRS314), plated on SD medium lacking tryptophan, and incubated at 23°C overnight. Plates were then shifted to 37°C for two additional days to select for complementing clones. Plasmids were rescued from colonies arising at 37°C. Of the five original temperature-sensitive synthetic lethal isolates, two (DL2822 and DL2823) were complemented by single plasmids with overlapping sequences from the right arm of chromosome III. The plasmid (designated A3) rescued from DL2822 possessed a 6.6-kb insert that contained five complete open reading frames (ORFs) (PET18, MAK31, HTL1, HSP30, and YCR022C), 1,092 bp of the 3′ end of MAK32, and 924 bp of the 3′ end of YCR023C. The plasmid rescued from DL2823 (designated A4) possessed an insert of approximately 5.4 kb that contained five complete ORFs (MAK32, PET18, MAK31, HTL1, and HSP30), a partial Ty element, and 296 bp of the 3′ end of YCR022C. A 527-bp EcoRV-HindIII fragment containing only HTL1/YCR020W-B was the smallest segment that allowed sectoring of strains DL2822 and DL2823 at 23°C and growth at 37°C. This fragment was subcloned into a 2μm plasmid, YEp352 (13), and a centromeric plasmid, pRS316 (42).
Southern blotting for HTL1.
The 666-bp HindIII fragment containing HTL1 was 32P labeled using the Multiprime random priming kit (Invitrogen Life Technologies). This labeled fragment was used to probe HindIII-digested genomic DNA (5 μg) from strains DL1248 or DL2822 separated on an agarose gel and transferred to a nitrocellulose membrane.
Genomic deletion of HTL1.
To disrupt the genomic copy of HTL1, 795 bp of sequence 5′ to the HTL1 start codon and 1,030 bp of sequence 3′ of the HTL1 stop codon were amplified in separate PCRs from genomic DNA of strain 1783. The 5′ fragment was amplified with primers that placed a NotI site at the end adjacent to the HTL1 coding sequence and a BamHI site at the opposite end. The 3′ fragment was amplified with primers that placed a KpnI site adjacent to the HTL1 coding sequence and a BamHI site at the opposite end. These fragments were ligated in a three-molecule reaction to the NotI and KpnI sites of the integrative plasmid pRS304 (42) to create a unique BamHI site between the fragments. The resulting plasmid, pRS304[htl1Δ::TRP1], was linearized with BamHI and used to transform the yeast strain to tryptophan prototrophy. Deletion of HTL1 in Trp+ transformants was confirmed by PCR. All primers were obtained from Invitrogen Life Technologies.
Isolation of RSC3 as a dosage suppressor of the htl1Δ growth defect.
A diploid yeast strain (DL2754 [htl1Δ/htl1Δ]) was used to avoid isolation of recessive suppressor mutations. This strain was cultured at 23°C in 50 ml of YEPD to an A600 of 0.8. Cells were transformed with a 2μm URA3-marked library (in pRS202; gift of F. Spencer) and plated on SD medium lacking uracil. Plates were incubated at 23°C for 1 day and shifted to 34°C for 2 to 3 days to select directly for suppressors. The smallest insert identified that was capable of suppressing the growth defect was a 4.3-kb fragment with one complete ORF (RSC3), 264 bp of the 3′ end of CPR5, and 551 bp of the 3′ end of GPI11. A 3.4-kb PvuII/MscI fragment containing RSC3 was subcloned into the SmaI site of the 2μm vector pRS426. This clone was capable of suppressing the temperature sensitivity of strain DL2754.
FACS analysis.
Cultures of strains 1788 and DL2754, grown in YEPD plus 10% sorbitol at 23°C, were diluted to A600 of 0.2 with an equal volume of YEPD plus 10% sorbitol prewarmed to 54°C and cultured at 38°C for either 5 or 10 h (DL2754 loses viability upon longer maintenance at restrictive temperatures). The wild-type culture (strain 1788) was periodically diluted with prewarmed medium to maintain the cells in the logarithmic growth phase. Aliquots of 1 ml were drawn after 5 and 10 h at restrictive temperatures and prepared for fluorescence-activated cell sorting (FACS) analysis by a wash with phosphate-buffered saline (PBS) followed by overnight fixation in 70% ethanol at 4°C. Cells were washed with PBS, treated extensively with RNase A (1 mg/ml for 5 h at 37°C), washed again, and stained with 50 μg of propidium iodide per ml. Cells were sonicated briefly and diluted to a final concentration of 106 cells/ml for FACS analysis, which was performed with a Becton Dickinson FACSCalibur cytometer at the Johns Hopkins Flow Cytometry Core Facility. Another 1-ml aliquot of each culture was drawn at each time point for microscopy, washed with PBS, and resuspended in VectaShield with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) for microscopic analysis.
Construction of HA- and FLAG-tagged HTL1.
To create HTL1-3xHA, HTL1 was PCR amplified from an HTL1 plasmid using primers that introduced a SacI site 794 bp 5′ to the HTL1 translational start site and omitted the stop codon from the HTL1 coding sequence. This fragment was introduced into YEp352[3xHA] (39), creating an in-frame fusion at the 3′ end of HTL1, with a glycyl codon between the HTL1 and hemagglutinin (HA)-coding sequences. This epitope-tagged form of Htl1 (Htl13xHA) was fully functional, as judged by its ability to complement the htl1Δ growth defect when expressed from a centromeric plasmid. This construction was used for indirect immunofluorescence microscopy with a Zeiss Axioskop microscope, as described previously (20). To create FlagHTL1, HTL1 was PCR amplified with a forward primer designed to contain a BamHI site immediately upstream of the Flag coding sequence (8 amino acids preceded by an ATG) followed by 24 bp of HTL1 sequence (omitting the endogenous start codon). The reverse primer was designed to place an EcoRI site downstream of the HTL1 stop codon. The PCR product was introduced into pRS426 containing the methionine-repressible MET25 promoter and the CYC1 transcriptional terminator (p426[MET25]) to yield pMET-Flag-HTL1. This plasmid complemented the htl1Δ growth defect even when expression of FlagHtl1 was repressed with methionine.
Association of Flag-tagged Htl1 with RSC complex.
Yeast strain YBC928 was transformed to uracil prototrophy with either pMET-Flag-HTL1 or empty vector. Transformants were cultured at 28°C in YEPD; extracts were prepared as described previously (6). For immunoprecipitation of FlagHtl1, anti-Flag M2 affinity gel or control beads (Sigma) were incubated for 2 h with 300 μg of protein extract in immunoprecipitation (IP) buffer A (50 mM Tris-Cl [pH 7.5], 10% glycerol, 100 mM NaCl, 2 mM EDTA, β-mercaptoethanol). Precipitates were recovered and washed in IP buffer A containing 250 mM NaCl and 0.05% Tween 20 and eluted with 0.2 mg of Flag peptide (Sigma) per ml in IP buffer A. Proteins from crude extracts, Flag-Htl1-depleted supernatant fractions, and eluates were separated on a gradient sodium dodecyl sulfate (SDS)-10 to 20% (acrylamide) polyacrylamide gel (Bio-Rad), and transferred to polyvinylidene difluoride membranes for immunoblot detection of FlagHtl1 with anti-Flag antibodies (Sigma) and of Sth1 with anti-Sth1 antibodies (1). The extracts described above were also used for immunoprecipitation of Sth1. Anti-Sth1 antibodies were conjugated to protein A-Sepharose beads (Sigma) at a concentration of 0.5 mg/ml in IP buffer A; the beads and antibodies h were then incubated for 2 h with 300 μg of protein extract and treated as described above, except that immunoprecipitates were eluted with SDS sample buffer.
RESULTS
Mutation of HTL1is synthetically lethal with pkc1.
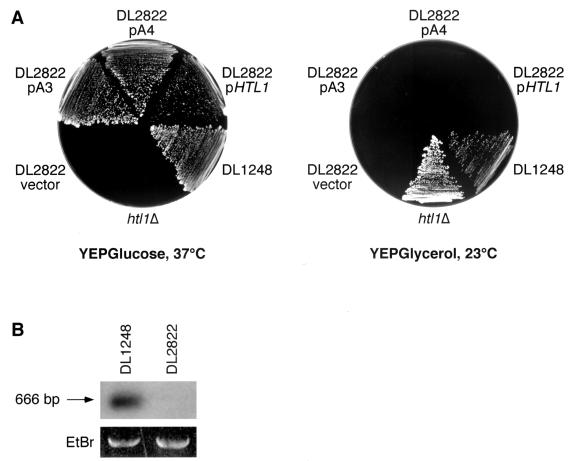
To identify genes whose loss of function results in an additive growth defect with pkc1, we constructed a strain (DL1248) that harbors a genomic pkc1ts allele (stt1-1 [49]), ade2 and ade3 mutations, and a plasmid bearing wild-type copies of both PKC1 and ADE3. This strain can survive loss of the PKC1-ADE3 plasmid at 23°C (but not at 37°C), yielding white sectors within red colonies. Mutants of this strain that cannot survive loss of the plasmid-borne PKC1, and therefore fail to form sectors, were isolated after UV mutagenesis. Of 150 nonsectoring colonies isolated, five were identified as temperature sensitive for growth at 37°C, indicating the presence of a new temperature-sensitive mutation. This secondary phenotype facilitated isolation of the affected gene. These mutants were transformed with a centromeric library of genomic yeast DNA (27) to isolate genes capable of complementing their temperature-sensitive growth defects. Of these mutants, one was complemented by MPK1, one was complemented by PKC1, and one was complemented by a previously uncharacterized nonannotated ORF (designated RIN1 [A. K. Sobering, M. J. Romeo, H. A. Vay, and D. E. Levin, submitted for publication]). The remaining two mutants (DL2822 and DL2823) were both rescued by plasmids (designated A3 and A4, respectively) containing overlapping fragments from the right arm of chromosome 3, with PET18, MAK31, HTL1, and HSP30 in common. Deletion analysis of these plasmids revealed that the smallest segment allowing sectoring of strains DL2822 and DL2823 at 23°C and growth at 37°C was a 527-bp EcoRV-HindIII fragment containing only HTL1/YCR020W-B (named HTL for high temperature lethal [25]) (Fig. 1A). The HTL1 gene encodes a recently identified 78-amino-acid protein with no known homologs.
FIG. 1.
A deletion through HTL1 confers an additive growth defect with pkc1ts. (A) The mutation in strain DL2822 results in separable growth defects at 34°C and on medium with glycerol as the sole carbon source. Strain DL2822, transformed with centromeric plasmids bearing HTL1 alone, plasmid A3 (PET18, MAK31, HTL1, HSP30, and YCR022C), plasmid A4 (MAK32, PET18, MAK31, HTL1, and HSP30), or vector (pRS314), was streaked onto YEP plus glucose or YEP plus glycerol and incubated at the indicated temperatures for 3 days. The parental strain, DL1248, and an htl1Δ strain (DL2751) are included. (B) HTL1 has been lost from the genome of DL2822. Genomic DNA (5 μg) from strain DL2822 and parental strain DL1248 was digested with HindIII, separated on an agarose gel, and subjected to Southern blot analysis using the 666-bp HindIII fragment bearing HTL1 as a probe (top blot). The bottom blot shows an ethidium bromide (EtBr)-stained ribosomal repeat fragment from the same gel as a quantitation control.
To determine if strains DL2822 and DL2823 bear mutations in HTL1, we attempted to PCR amplify from these strains the genomic regions containing HTL1. However, we were not able to isolate PCR products from either mutant even when using primers up to 3 kb flanking either side of HTL1, suggesting that HTL1 had been deleted from their genomes. Previously described PET18 mutants (28) resulted from spontaneous deletions of greater than 10 kb from the right arm of chromosome 3 (43). Such mutants are temperature sensitive due to loss of HTL1 (25) and unable to grow on nonfermentable carbon sources. The latter phenotype is known to result from loss of function of another, as yet unidentified gene within the deleted region (25). We found that like the previously described mutants, strains DL2822 and DL2823 were unable to grow on rich medium containing glycerol as the sole carbon source (Fig. 1A). This defect was not complemented by either plasmid A3 (bearing PET18, MAK31, HTL1, HSP30, and YCR022C), or plasmid A4 (bearing MAK32, PET18, MAK31, HTL1, and HSP30), indicating that this strain had suffered a genomic deletion that extends beyond the region from MAK32-YCR022C. A Southern blot of genomic DNA verified the absence of HTL1 in DL2822 (Fig. 1B). Taken together, these results suggest that loss of HTL1 function is synthetically lethal with stt1-1.
To examine the behavior of a null mutation in HTL1 alone, we constructed a genomic deletion of HTL1 in wild-type strain 1788. Heterozygous htl1Δ::TRP1 diploids were induced to sporulate, and haploid segregants were dissected. Tryptophan prototrophy cosegregated with temperature sensitivity for growth at or above 34°C, consistent with the previous report by Lanzuolo et al. (25). As anticipated, the htl1Δ mutants were capable of growth on glycerol-containing medium (Fig. 1A).
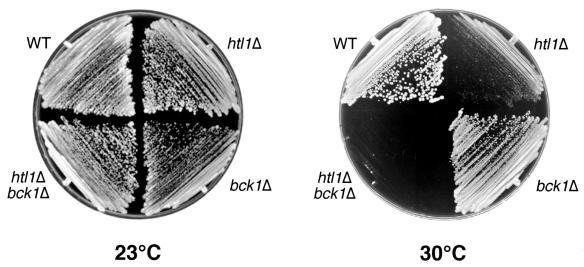
We next tested for additive growth defects caused by combining null mutations in HTL1 and either PKC1 or BCK1. Doubly heterozygous diploids were generated by crossing an htl1Δ::TRP1 strain (DL2751) with a pkc1Δ::LEU2 strain (DL376). A total of 56 tetrads were dissected and allowed to germinate at 23°C on medium containing 10% sorbitol for osmotic support. Although both single mutants and wild-type segregants were recovered at the expected frequencies, no viable Leu+ Trp+ spores were recovered, indicating that the combined loss of PKC1 and HTL1 functions is lethal. HTL1 was also deleted in a heterozygous bck1Δ/BCK1 diploid (DL2315). The resultant transformants were induced to sporulate, and haploid segregants germinated at 23°C. Although the bck1Δ htl1Δ segregants were able to grow on YEPD at 23°C, they failed to grow at or above 30°C (Fig. 2). This restrictive temperature was 4°C lower than that of the htl1Δ mutant, which grew poorly at 30°C. This result indicates only mild additivity of the htl1Δ and bck1Δ growth defects. Additionally, the double mutant displayed a cell lysis defect, as judged by the presence of nonrefractile ghosts (not shown) that was similar to that of bck1Δ mutants at restrictive temperatures. However, we were not able to detect a cell wall deficiency in htl1Δ mutants using tests for hypersensitivity to the cell wall lytic enzyme zymolyase or the cell wall antagonist caffeine (data not shown).
FIG. 2.
Null mutations in BCK1 and HTL1 result in mildly additive growth defects. Wild-type (WT) (1783), bck1Δ (DL2312), htl1Δ (DL2749), and bck1Δ htl1Δ (DL2745) yeast strains were streaked onto YEPD, plated, and incubated at the indicated temperatures for 3 days.
A null HTL1 mutant undergoes a cell cycle-specific growth arrest at nonpermissive temperatures.
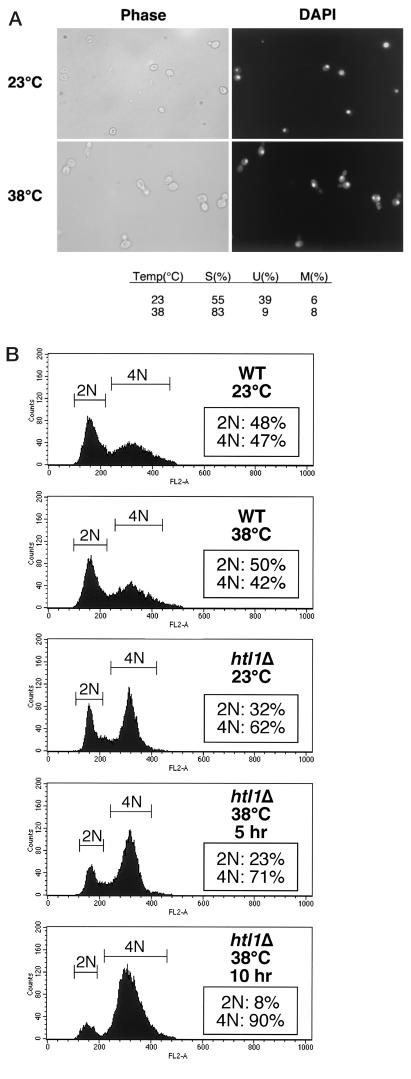
Microscopic examination of htl1Δ cells at restrictive temperatures revealed that this mutant arrested growth with a nearly uniform morphology. For all mating types, 80 to 90% of cells arrested with single, medium- to large-sized buds, single nuclei, and no evidence of cell lysis (Fig. 3A). To determine whether htl1Δ cells arrest in the cell division cycle before or after DNA replication, we assessed the DNA content of this mutant cultivated at either low or high temperatures. Log-phase cultures of diploid wild-type (1788) or htl1Δ cells (DL2754) were shifted from 23 to 38°C for 5 or 10 h to measure DNA content by FACS analysis. The htl1Δ mutant displayed an enriched population of 4N cells at low temperatures compared to the wild-type population, suggesting a delay at the G2/M boundary (Fig. 3B). Moreover, at restrictive temperatures, 90% of the htl1Δ cells accumulated 4N nuclei, indicating a postreplicative arrest at G2/M. Similar results were obtained with haploid htl1Δ cells (data not shown).
FIG.3.
Deletion of HTL1 results in a cell cycle-specific growth arrest at restrictive temperatures. (A) Phase-contrast microscopy and fluorescence microscopy (DAPI staining) of log-phase cultures of DL2754 (htl1Δ/htl1Δ) growing at 23°C or arrested for 10 h at 38°C. The fractions of cells that appear to have single buds (S), no buds (unbudded [U]), or multiple buds (MB) were determined from counts of at least 100 cells. (B) Flow cytometric analysis of cultures of either strain 1788 (wild type [WT]) or DL2754 incubated at 23°C or shifted to 38°C for the indicated times.
RSC3 is a dosage suppressor of htl1Δ.
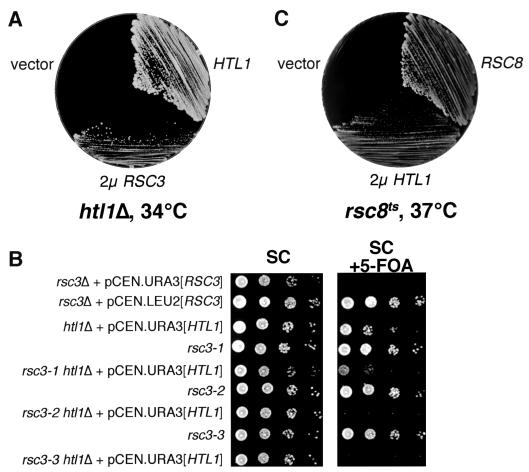
To gain an understanding of the htl1Δ growth defect, we isolated multicopy suppressors of an htl1Δ/htl1Δ mutant (DL2754). Suppressors were selected directly at 34°C on SD plates lacking uracil after transformation with a 2μm URA3-marked genomic library. Of ∼80,000 transformants selected, 8 colonies arose at the restrictive temperature. Among the plasmids rescued from these colonies, four contained wild-type HTL1, and four possessed overlapping DNA fragments from the right arm of chromosome IV. The only complete gene common to all four plasmids was RSC3 (named RSC for remodels the structure of chromatin) (data not shown). Deletion analysis revealed that RSC3 alone, when expressed from a 2μm plasmid, was sufficient to allow an htl1Δ/htl1Δ mutant to grow at 34°C (Fig. 4A). The RSC3 gene encodes an essential component of the RSC complex, which alters gene expression by remodeling of chromatin (1). Rsc3 is highly similar to well-characterized fungal DNA-binding proteins (such as Gal4), as it contains an essential binuclear zinc cluster followed by a leucine zipper. Some genes involved in the maintenance of cell wall integrity are among those regulated by RSC (1). Interestingly, conditional mutations in RSC3 and other essential genes encoding RSC members arrest growth at the G2/M boundary (1, 7, 9, 45), with a terminal morphology that is very similar to that observed for htl1Δ cells. However, overexpression of HTL1 failed to suppress the growth defects of any of the three conditional rsc3 alleles.
FIG. 4.
Genetic connections between HTL1 and RSC. (A) Overexpression of RSC3 suppresses the growth defect of an htl1Δ mutant. Strain DL2754 (htl1Δ/htl1Δ) harboring 2μm plasmid pRS426 [RSC3], centromeric plasmid pRS316 [HTL1], or vector (pRS426) were streaked onto YEPD and incubated at 34°C for 3 days. (B) Deletion of HTL1 results in additive growth defects with conditional rsc3 mutations. Yeast strains (from top to bottom, YBC628, YBC843, YBC1334, YBC840, YBC1336, YBC842, YBC1338, YBC906, and YBC1340) were grown in selective medium at room temperature overnight. The saturated cultures and a series of 10-fold dilutions were spotted onto SC plates or SC plates containing 5-FOA and incubated at 28°C for 3 days. (C) Overexpression of HTL1 suppresses the growth defect of an rsc8ts mutant. Yeast strain MCY3890 (swh3-ts16), transformed with the 2μm plasmid YEp352 [HTL1], or YEp352 alone, was streaked with the isogenic wild-type strain W303-1A (RSC8) onto YEPD and incubated at 37°C for 3 days.
Rsc30 is an RSC subunit that is almost identical to Rsc3 in the binuclear zinc cluster region. Several lines of evidence suggest that Rsc3/30 operate as a heterodimer within the RSC complex but perform different functions (1). In contrast to RSC3, overexpression of RSC30 from a 2μm plasmid did not suppress the growth defect of htl1Δ/htl1Δ mutants (data not shown), consistent with the different roles these components serve in RSC function.
Deletion of HTL1 results in additive growth defects with rsc3 mutations.
Because RSC3 is an essential gene, we tested a set of conditional rsc3 mutations for additive growth defects with htl1Δ. The htl1Δ mutation was introduced into strains bearing one of three temperature-sensitive alleles of RSC3 (rsc3-1, -2, and -3 [1]) and a centromeric URA3-based plasmid with HTL1. These strains were tested for the ability to survive loss of the plasmid-borne HTL1 at 23°C by plating on 5-FOA-containing medium to evict the plasmid. Figure 4B shows that both the rsc3-2 htl1Δ and rsc3-3 htl1Δ mutants were unable to grow on 5-FOA-containing medium, indicating that loss of HTL1 is lethal in these mutants. Together with the observation that RSC3 is a dosage suppressor of htl1Δ, these results suggest that HTL1 and RSC3 function within the same pathway.
HTL1 is a dosage suppressor of an rsc8tsmutant.
The RSC8 gene encodes a component of the RSC complex, and homologs of Rsc8 are present in all SWI/SNF family remodeling complexes (5, 44). Though the precise role of Rsc8 is not known, studies on its human homolog, BAF155/170, suggest that this subunit interacts with the ATPase subunit Brg1 and also with gene-specific transcriptional activators such as EKLF (2). Treich et al. (44) reported previously that a DNA fragment bearing the MAK31 and HTL1 loci could suppress the growth defect of an rsc8ts mutant (swh3-ts16) when maintained at high copy number. To test whether this suppression was the result of HTL1 overexpression, a 2μm plasmid bearing only HTL1 was introduced to the swh3-ts16 mutant (MCY3890 [44]). This clone was able to suppress the growth defect of swh3-ts16 (Fig. 4C), demonstrating that HTL1, not MAK31, is the multicopy suppressor of swh3-ts16 obtained by Treich et al. (44), and further supporting the notion that Htl1 functions in the same pathway as the RSC complex.
Htl13xHA resides in the nucleus.
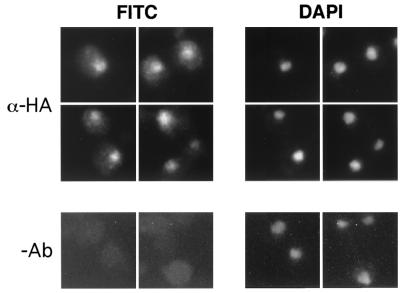
The RSC complex resides in the nucleus (9, 45). As an initial step toward establishing a physical connection between Htl1 and RSC, we determined the intracellular location of Htl1. Htl1 was fused at its C terminus to a triple-HA epitope (3xHA). Htl13xHA expressed from a 2μm plasmid colocalized with DAPI, indicating nuclear localization (Fig. 5). Similar results were obtained when Htl13xHA was expressed from a centromeric plasmid but with a less intense signal (data not shown).
FIG. 5.
Htl13xHA resides in the nucleus. Wild-type diploid cells (strain 1788), harboring HTL13xHA on a 2μm plasmid were grown to mid-log phase in YEPD, fixed, and subjected to indirect immunofluorescence microscopy with mouse monoclonal anti-HA antibody (12CA5) (α-HA) and fluorescein isothiocyanate (FITC)-conjugated secondary antibody. Nuclear DNA was visualized with DAPI. The no-antibody (−Ab) control cells (bottom) were treated with secondary antibody only.
Htl1 can associate with the RSC complex.
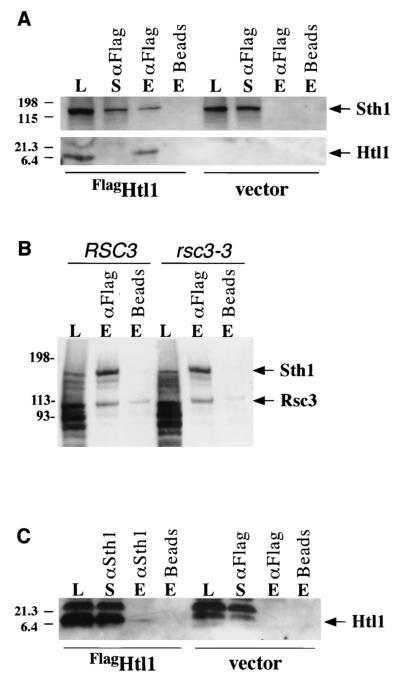
The genetic connections between HTL1 and components of the RSC complex established above, combined with the nuclear localization of Htl1, suggested that this protein might physically associate with the RSC complex. To explore this possibility, we tested for in vivo association of Htl1 with the Sth1 ATPase subunit of RSC (11). For this purpose, Htl1 was fused at its N terminus to a Flag epitope (FlagHTL1) and overexpressed in yeast cells (YBC928) from the MET25 promoter. Immunoprecipitation of FlagHtl1 from extracts coprecipitated a small proportion of the Sth1 (Fig. 6A, top blot). The observed coprecipitation reflects a true association of FlagHtl1 with RSC, because Sth1 was not detected in the absence of either FlagHtl1 or anti-Flag antibodies. Interestingly, Sth1 was also detected in the supernatant fraction (Fig. 6A, top blot) that had been immunodepleted of overexpressed FlagHtl1 (Fig. 6A, bottom blot), suggesting that Htl1 is associated with only a subset of RSC complexes. In a similar immunoprecipitation of FlagHtl1, either Rsc3 or its mutant form Rsc3-3 was detected with the coprecipitated Sth1 (Fig. 6B), indicating that Htl1 can associate with the form of RSC that contains Rsc3. In reciprocal immunoprecipitations, overexpressed FlagHtl1 was weakly detected in Sth1 immune complexes (Fig. 6C), supporting the conclusion that Htl1 can associate with a fraction of the RSC complex in vivo. We could not detect Htl13xHA expressed at endogenous levels from its own promoter in complex with Sth1, suggesting that the RSC-associated fraction may be small.
FIG. 6.
FlagHtl1 associates with the RSC complex in vivo. (A and C) Extracts were prepared from yeast strain YBC928 bearing either pMET-Flag-HTL1 or vector. Immunoprecipitations were conducted by incubating 300 μg of extract with either anti-Flag M2 affinity gel (αFlag) (A) or anti-Sth1 (αSth1) conjugated to protein A beads (C). Control beads with no antibodies were included for both. Samples were eluted either with Flag peptide (A) or with SDS sample buffer (C), separated by SDS-polyacrylamide gel electrophoresis, and subjected to immunoblot analysis with either anti-Sth1 or anti-Flag M2 antisera. Load (L) (60 μg), immunodepleted supernatant (S) (18%), and eluate (E) (50%) samples are shown. (B) Extracts were prepared from yeast strains YBC843 (RSC3) and YBC906 (rsc3-3) bearing pMET-Flag-HTL1, and FlagHtl1 was immunoprecipitated and eluted as described above for panel A. Samples were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot analysis with both anti-Sth1 and anti-Rsc3 antisera. Load (L) (60 μg) and eluate (E) (50%) samples are shown. The positions of molecular size standards (in kilodaltons) are shown to the left of the blots.
DISCUSSION
The RSC chromatin remodeling complex has emerged as an important transcriptional regulator of many genes, including those important for cell wall biogenesis, NDP, ribosome biogenesis, and carbohydrate metabolism (1, 9, 34). Additionally, RSC is essential for progression of the cell cycle through G2/M. Here, we report the association of Htl1 with RSC complex and functional links between RSC/Htl1 and Pkc1 signaling. HTL1 is a 78-amino-acid protein required for growth at 37°C and for maintenance of fertility and ploidy at permissive temperatures (25). Here, we isolated HTL1 in a genetic screen for additive growth defects with a conditional mutation in PKC1 (stt1-1). We now demonstrate that HTL1 is required for cell cycle progression through the G2/M transition at temperatures above 33°C and that the Htl1 protein can associate with the RSC complex. We propose that Pkc1 and RSC are coregulators of the G2/M transition.
Htl1 can associate with the RSC complex.
We present four lines of evidence supporting the conclusion that Htl1 interacts physically and functionally with the RSC complex. First, at restrictive temperatures, htl1 null mutants exhibit a uniform cell cycle arrest in G2/M that is similar to that of strains with conditional mutations in some essential components of the RSC complex (e.g., alleles of STH1, SFH1, RSC3, and RSC9 [1, 7, 9, 45]). For reasons that are not clear, the cell cycle-specific arrest of an htl1 null mutant was not observed in a previous study (25). Strain background differences may explain this disparity, as may the possible acquisition of suppressor mutations. Second, conditional rsc3 haploid strains accumulate cells with 4N DNA content (1), as was shown for htl1Δ haploids (25). Third, multicopy suppression experiments revealed a genetic interaction between HTL1 and RSC components. Specifically, overexpression of RSC3, an essential DNA-binding component of the RSC complex, suppressed the growth defect of an htl1 null mutant. Additionally, overexpression of HTL1 suppressed the growth defect of a conditional rsc8 mutant. The observation that an increased dose of HTL1 could not suppress the growth defect of any of three conditional rsc3 mutations suggests that its overexpression can compensate for some RSC defects, but not others. Fourth, Htl1 is a nuclear protein that when overexpressed was found to associate in vivo with known RSC complex components Rsc3 and Sth1.
A recent quantitative study of proteins associated with the general transcription factor TFIID (41), which is a multisubunit complex comprised of the TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs), revealed that all known RSC components were among the constellation of proteins that displayed preferential association with TBP (compared with TAFs). Htl1 was among the other, non-RSC proteins identified in this population. Our results, which indicate that Htl1 can associate with RSC, provide an explanation for the reported association of Htl1 with TBP.
Why was Htl1 not identified in previous characterizations of RSC components? The most likely explanation is that Htl1 appears to exist in substoichiometric amounts within the RSC complex. We found that cell extracts that were immunodepleted of overexpressed FlagHtl1 still possessed some of the Sth1 subunit. Moreover, it was possible to detect FlagHtl1 associated with immunoprecipitates of Sth1 only when the former was greatly overproduced. This may be significant in light of the observation that RSC exists in forms that either possess or are devoid of Rsc3/30 (5). The genetic interaction we observed between HTL1 and RSC3 may reflect a specific association of Htl1 with the form of RSC that contains Rsc3/30. The Rsc3/30 proteins exist as a heterodimeric complex that can form outside of the RSC complex (1). Interestingly, although these closely related components serve overlapping roles, they may act antagonistically for certain functions. Our finding that the htl1 null growth defect was suppressed by overexpression of RSC3, but not RSC30, underscores the functional differences between these components.
Interactions between the RSC complex and Pkc1.
Recent studies have raised the possibility that RSC function is regulated by Pkc1. First, a genetic screen for multicopy suppressors of the growth defect of a temperature-sensitive mutation in STH1/NPS1, which encodes the DNA-dependent ATPase of the RSC complex, yielded upstream components of the cell wall integrity pathway, including PKC1 (14). Overexpression or mutational activation of elements of the MAP kinase cascade controlled by Pkc1 failed to suppress sth1, prompting these investigators to suggest that the observed effect of Pkc1 is not mediated by the MAP kinase pathway. Additive growth defects between conditional alleles of STH1 (nps1-105) and PKC1 (stt1-1) also support a functional connection to RSC (14). A recent study by Chai et al. (7a) confirmed these findings using another allele of STH1 (sth1-3) and also demonstrated additive growth defects between sth1-3 and a null mutation in the cell surface sensor for Pkc1 activation encoded by WSC1. Second, mutation of either RSC3 or RSC30 alters the expression of several genes implicated in cell wall biogenesis, and the sensitivity to caffeine and formamide displayed by conditional RSC3 mutants is suppressed by overexpression of PKC1 (1). Moreover, rsc3 mutations are lethal in combination with a pkc1 null allele (even in the presence of osmotic support), but not a bck1 null allele. Collectively, these observations strongly suggest a functional link between Pkc1 and RSC.
Loss of Pkc1 function results in cell lysis resulting from a deficit in cell wall construction (29, 30). However, loss of function in the MAP kinase cascade regulated by Pkc1 results in cell lysis only at elevated temperatures (30), indicating that Pkc1 regulates at least one other pathway that contributes to cell wall integrity. We found that loss of HTL1 function was lethal in the presence of pkc1 mutations but resulted in only mild defect additivity with a bck1 null allele, similar to the behavior of other rsc mutants. Although the observed defect additivity suggests that htl1 mutants may suffer a deficiency in cell wall biogenesis, we observed no evidence of cell lysis at restrictive temperatures and were unable to detect a cell wall defect through tests for hypersensitivity to cell wall stresses.
An interesting alternative explanation for the observed defect additivity between pkc1 and htl1 invokes a role for Pkc1 in passage through G2/M. Depletion of Pkc1 results in cell lysis at the bud tip during a distinct period in the cell cycle, previously interpreted to reflect the point at which the cell is most sensitive to cell wall stress (30). Such cells die uniformly with single, postreplication nuclei and small buds (32). This is also the case for conditional alleles of pkc1 (29). Hosotani et al. (14) recently extended these observations, finding that a conditional pkc1 mutant (stt1-1) undergoes a G2/M delay at permissive temperatures. These investigators suggested that Pkc1 might have a cell cycle function that is independent of cell wall metabolism. Because we isolated an htl1 mutant in a synthetic lethal screen with stt1-1, our finding that an htl1 null mutant also displays a G2/M delay at permissive temperatures suggests that a double pkc1 htl1 mutant may succumb to a G2/M block. A cell cycle block could explain our inability to rescue this double mutant by osmotic support.
Models for RSC/Htl1 function and Pkc1 cooperativity in cell wall biogenesis and cell cycle progression.
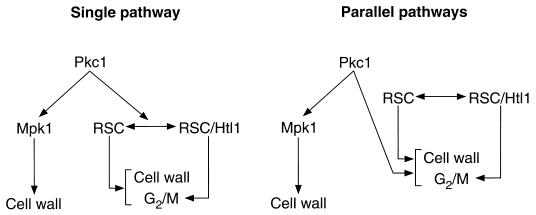
The striking defect additivity observed between pkc1Δ and either rsc3 alleles (1) or htl1Δ suggests that cell viability requires either intact Pkc1 function or intact RSC function. However, this relationship (and other data) does not establish whether these factors function together in a single pathway or coordinately in parallel pathways (Fig. 7). In considering a single pathway, Pkc1 could phosphorylate RSC components and alter RSC function at loci important for cell wall biogenesis and G2/M progression. Clearly, Pkc1 regulation of RSC would have to be limited to a subset of RSC functions, because null mutations in most RSC components are lethal even in the presence of osmotic support (pkc1Δ mutants are viable under these conditions). Moreover, RSC occupies hundreds of genes not related to Pkc1 function. However, to date, we have been unable to detect phosphorylation of purified RSC by Pkc1 in vitro (unpublished observations), though we have not ruled out the possibility that RSC is a Pkc1 target in vivo. A second model equally consistent with our data are that Pkc1 and RSC act coordinately in parallel pathways to regulate cell wall biogenesis and passage through G2/M. In this case, RSC components are not direct targets of Pkc1. Instead, the functions of both of these factors are required for proper expression of genes involved in cell wall biogenesis and passage through the G2/M transition. In either case, our data strongly suggest that Htl1 assists RSC primarily for proper passage through G2/M phase (Fig. 7), as htl1 null mutants phenotypically copy the G2/M cell cycle block observed in certain rsc mutants but lack the cell wall phenotypes associated with rsc3 mutations.
FIG. 7.
Models for the interaction of Pkc1 with RSC in the regulation of cell wall biogenesis and G2/M.
Acknowledgments
We thank Takahiro Negishi for assistance with htl1 suppressor screening and Southern blotting, Forrest Spencer for providing the genomic library, and Marian Carlson for providing yeast strains.
This work was supported in part by NIH grants GM48533 to D.E.L. and GM60415 to B.R.C. Training grant 5T32CA09110 supported M.J.R. and A.K.S. Training grant T32GM07464 supported M.L.A.-H. B.R.C. is an Assistant Investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, B. R. 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23:20-25. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Y., B. R. Cairns, R. D. Kornberg, and B. C. Laurent. 1997. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 17:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Chai, B., J. Hsu, J. Du, and B. C. Laurent. 2002. Yeast RSC function is required for organization of the cellular cytoskeleton via an alternative PKC1 pathway. Genetics 161:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costigan, C., S. Gehrung, and M. Snyder. 1992. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol. Cell. Biol. 12:1162-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 10.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, J., I. Nasir, B. K. Benton, M. P. Kladde, and B. C. Laurent. 1998. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 150:987-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, J. E., A. M. Muers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 14.Hosotani, T., H. Koyama, M. Uchino, T. Miyakawa, and E. Tsuchiya. 2001. PKC1, a protein kinase C homologue of Saccharomyces cerevisiae, participates in microtubule function through the yeast EB1 homologue, BIM1. Genes Cells 6:775-788. [DOI] [PubMed] [Google Scholar]
- 15.Irie, K., M. Takase, K. S. Lee, D. E. Levin, H. Araki, K. Matsumoto, and Y. Oshima. 1993. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 13:3076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, J. J., S. M. Nilius, and J. J. Heinisch. 1998. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 258:148-155. [DOI] [PubMed] [Google Scholar]
- 18.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 19.Kadonaga, J. T. 1998. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307-313. [DOI] [PubMed] [Google Scholar]
- 20.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 21.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 271:9193-9195. [DOI] [PubMed] [Google Scholar]
- 22.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 25.Lanzuolo, C., S. Ederle, A. Pollice, F. Russo, A. Storlazzi, and J. F. Pulitzer. 2001. The HTL1 gene (YCR020W-b) of Saccharomyces cerevisiae is necessary for growth at 37°C, and for the conservation of chromosome stability and fertility. Yeast 18:1317-1330. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1) mediates signaling by protein kinase C. Mol. Cell. Biol. 13:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, K. S., and D. E. Levin. 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibowitz, M. J., and R. B. Wickner. 1978. Pet18: a chromosomal gene required for cell growth and for the maintenance of mitochondrial DNA and the killer plasmid of yeast. Mol. Gen. Genet. 165:115-121. [DOI] [PubMed] [Google Scholar]
- 29.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin, D. E., B. Bowers, C. Chen, Y. Kamada, and M. Watanabe. 1994. Dissecting the protein kinase C/MAP kinase signaling pathway of Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 40:229-239. [PubMed] [Google Scholar]
- 31.Levin, D. E., and B. Errede. 1995. The proliferation of MAP kinase signaling pathways in yeast. Curr. Opin. Cell Biol. 7:197-202. [DOI] [PubMed] [Google Scholar]
- 32.Levin, D. E., F. O. Fields, R. Kunisawa, J. M. Bishop, and J. Thorner. 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62:213-224. [DOI] [PubMed] [Google Scholar]
- 33.Martin, H., J. Arroyo, M. Sanchez, M. Molina, and C. Nombela. 1993. Activity of the yeast MAP kinase homolog Slt2 is critically required for cell integrity at 37°C. Mol. Gen. Genet. 241:177-184. [DOI] [PubMed] [Google Scholar]
- 34.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai. 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5931-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozaki, K., K. Tanaka, H. Imamura, T. Hihara, T. Kamayema, H. Nonaka, H. Hirano, Y. Matsuura, and Y. Takai. 1996. Rom1p and Rom2p are small GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP-binding protein in Saccharomyces cerevisiae. EMBO J. 15:2196-2207. [PMC free article] [PubMed] [Google Scholar]
- 37.Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J.-L. Carpentier, L. S. Klig, and M. A. Payton. 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12:4896-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 39.Rajavel, M., B. Philip, B. M. Buehrer, B. Errede, and D. E. Levin. 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toh-e, A., and Y. Sahashi. 1985. The PET18 locus of Saccharomyces cerevisiae: a complex locus containing multiple genes. Yeast 1:159-171. [DOI] [PubMed] [Google Scholar]
- 44.Treich, I., L. Ho, and M. Carlson. 1998. Direct interaction between Rsc6 and Rsc8/Swh3, two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 26:3739-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchiya, E., M. Uno, A. Kiguchi, K. Masuoka, Y. Kanemori, S. Okabe, and T. Mikayawa. 1992. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2 phase control. EMBO J. 11:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for the maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, C., T. Tsukiyama, D. Gdula, P. Georgel, M. Martinez-Balbas, G. Mizuguchi, V. Ossipow, R. Sanaltzopoulos, and H. M. Wang. 1998. ATP-remodeling of chromatin. Cold Spring Harbor Symp. Quant. Biol. 63:525-534. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, S., E. Ikeda, I. Uno, and H. Mitsuzawa. 1992. Characterization of a staurosporine- and temperature-sensitive mutant, stt1, of Saccharomyces cerevisiae: STT1 is allelic to PKC1. Mol. Gen. Genet. 231:337-344. [DOI] [PubMed] [Google Scholar]