Abstract
Several aspects of eukaryotic mRNA processing are linked to transcription. In Saccharomyces cerevisiae, overexpression of the mRNA export factor Sub2p suppresses the growth defect of hpr1 null cells, yet the protein Hpr1p and the associated THO protein complex are implicated in transcriptional elongation. Indeed, we find that a pool of heat shock HSP104 transcripts are 3′-end truncated in THO complex mutant as well as sub2 mutant backgrounds. Surprisingly, however, this defect can be suppressed by deletion of the 3′-5′ exonuclease Rrp6p. This indicates that incomplete RNAs result from nuclear degradation rather than from a failure to efficiently elongate transcription. RNAs that are not degraded are retained at the transcription site in a Rrp6p-dependent manner. Interestingly, the addition of a RRP6 deletion to sub2 or to THO complex mutants shows a strong synthetic growth phenotype, suggesting that the failure to retain and/or degrade defective mRNAs is deleterious. mRNAs produced in the 3′-end processing mutants rna14-3 and rna15-2, as well as an RNA harboring a 3′ end generated by a self-cleaving hammerhead ribozyme, are also retained in Rrp6p-dependent transcription site foci. Taken together, our results show that several classes of defective RNPs are subject to a quality control step that impedes release from transcription site foci and suggest that suboptimal messenger ribonucleoprotein assembly leads to RNA degradation by Rrp6p.
The physical separation of mRNA biogenesis and bulk protein synthesis in eukaryotic cells necessitates molecular transport between the nuclear and cytoplasmic compartments. To form a mature translatable mRNA, several processing steps must be completed accurately in the nucleus. In successful formation of mature translatable mRNAs, the 5′ end of the transcript acquires a methylated guanosine cap structure, introns are excised, and the 3′ end is cleaved and polyadenylated (for a review, see reference 31). These pre-mRNA modifications serve to protect the transcript from the cellular degradation systems as well as prepare it for downstream events. Over the past few years, it has been established that pre-mRNA processing is carefully coordinated with transcription, and processing events are thought to occur mostly while the pre-mRNA is nascent, i.e., still emerging from the transcription machinery (29). Nuclear mRNA export is tightly linked to pre-mRNA processing and, consequently, probably also to transcription. In metazoans, although not strictly required for export, pre-mRNA splicing has been found to make loading of export factors onto the mRNA more efficient: the exon junction complex associates with the mRNA as a consequence of splicing, and the exon junction complex-associated proteins Aly/REF and UAP56 are important for productive mRNA export (14, 22, 24, 25, 28, 32). The Saccharomyces cerevisiae orthologues of these two proteins (the RNA binding protein Yra1p and the DECD-box RNA helicase Sub2p, respectively) are also involved in mRNA export in this organism (18, 37, 38, 40). Aly/Yra1p is believed to recruit the Mex67p/Mtr2p heterodimer (TAP/p15 in metazoans), which in turn mediates contact to the nuclear pore complex (2, 20, 21, 22, 24, 36, 37, 40, 43). Mex67p/Mtr2p recruitment to the messenger ribonucleoprotein (mRNP) might lead to Sub2p displacement, as in vitro studies have shown that Mex67p/Mtr2p binding and Sub2p binding to Yra1p are mutually exclusive (38).
Only a small fraction of S. cerevisiae transcripts contain introns, making pre-mRNA splicing insufficient to link the general mRNA population to nuclear export. Instead, 3′-end formation seems to play a major role in this organism. In temperature-sensitive (ts) mutants of the 3′-end processing factors rna14, rna15, and poly(A) polymerase (pap1), mRNA nuclear export is blocked (4, 16). Furthermore, uncapped T7 polymerase-directed transcripts are efficient substrates for nuclear export only if they are cleaved and polyadenylated, whereas unadenylated transcripts terminated due to a T7 terminator sequence are sequestered within the nucleus (11). These results indicate that, in contrast to the 5′-end cap, a proper 3′ end is required for nuclear export.
We recently discovered that a block to nuclear export leads to rapid and dramatic effects on the polyadenylation and subnuclear localization of two heat shock mRNAs, SSA4 and HSP104 (19). In a number of export mutants, these transcripts were sequestered at or near their sites of transcription and the mRNAs had poly(A) tails approximately twice as long as those found in a wild-type strain. Interestingly, transcripts synthesized without a poly(A) tail in the pap1-1 mutant were also retained in transcription site foci. This retention required Rrp6p as well as other components of the nuclear exosome, which is a large complex of exonucleolytic enzymes; i.e., in exosome mutants, unadenylated as well as hyperadenylated mRNAs are released from transcription sites and proceed in their metabolism (16). Although the molecular basis for exosome-mediated transcription site retention is not understood, these observations suggest that events closely linked to transcription play important roles in defining an export-competent mRNA. As a consequence, it is highly likely that early mRNA export factors will be found associated with sites of active transcription. Indeed, chromatin immunoprecipitation assays have shown that the RNA binding proteins Yra1p and Npl3p associate with mRNA while it is still in close proximity to chromatin (26). Sub2p/UAP56 is also thought to act at an early step in mRNA metabolism. Yeast SUB2 interacts genetically with YRA1, and the products of the two genes interact physically (38). Furthermore, mutations of SUB2 lead to a general defect in mRNA export and cause Rrp6p-dependent stalling of SSA4 and HSP104 RNAs at or near transcription sites (18, 38). It has recently been shown that both Yra1p and Sub2p are associated with the heterotetrameric THO-protein complex, which is genetically and physically linked to the transcription machinery (7, 8, 39). This observation suggested a role for members of the THO complex in mRNA export, which has indeed previously been determined to exist (39). Although the mechanistic rationale underlying these transport defects at the molecular level is presently unclear, they could be due to inefficient loading of export factor(s) onto the nascent transcript. Alternatively, the lesion could be indirect: for example, through more subtle effects on 3′-end formation.
Inspired by evidence linking SUB2 genetically and physically to HPR1 (17, 35), we have analyzed the status of nascent HSP104 transcripts in sub2 and hpr1 mutant strains as well as in strains with deletions of other components of the Hpr1p-containing THO-protein complex (8, 9). We find that a substantial pool of nuclear HSP104 RNA is 3′-end truncated in these mutants. Moreover, a share of transcripts are rapidly sequestered at or near transcription sites. Deletion of the RRP6 exonuclease restores a quasinormal level of full-length transcripts and reverses the transcript sequestration phenotype. This indicates that HSP104 transcripts produced in these mutants are subject to detection and partial degradation by Rrp6p. We have also analyzed transcripts produced in RNA 3′-end cleavage mutants as well as transcripts terminated by a self-cleaving ribozyme. In both cases, transcripts are retained within transcription site foci. All of the results show that several classes of defective RNPs are subject to a quality control step at or near the site of transcription.
MATERIALS AND METHODS
Yeast strains and plasmids.
Strains used in this study are listed in Table 1. The mtr2-26 and rat7-1 strains were kindly provided by Ed Hurt and Chuck Cole, respectively, and have previously been described (15, 34). In these strains, by using standard procedures, the RRP6 gene was replaced with the Escherichia coli kanamycin resistance (KANr) gene to create mtr2-26/Δrrp6 and rat7-1/Δrrp6. All other strains used are derived from W303 or have been backcrossed to W303 multiple times. Double mutants were created by standard genetic crosses. Original strains were kindly provided by H. Klein (Δhpr1), A. Aguilera (Δmft1 and Δtho2), and P. Thuriaux (Δgcn5, Δppr2, and Δrbp9). rna14-3, rna15-2, and Δxrn1 strains were a gift from F. Lacroute and M. Minet.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 | ura3-1 ade2-1 his3-11,5 trp1-1 leu2-3,112 can1-100 | F. Lacroute |
| DLY23 | W303 but mata sub2::HIS pCM188-SUB2 (URA3) | 27 |
| DLY33 | W303 but mata sub2::HIS pCM185-sub2-201 (TRP1) | 27 |
| DLY157 | W303 but matα hpr1::HIS | This studya |
| DLY223 | W303 but mata tho2::KANr | This studyb |
| DLY224 | W303 but mata mft1::KANr | This studyb |
| DLY200 | W303 but sub2::KANrhpr1::HIS pCM188-SUB2 (URA3) | This study |
| DLY232 | W303 but sub2::KANrtho2::KAN pCM188-SUB2 (URA3) | This study |
| DLY233 | W303 but sub2::KANrmft1::KAN pCM188-SUB2 (URA3) | This study |
| DLY123 | W303 but matα rrp6::KIURA3 | This study |
| DLY124 | W303 but sub2::HIS rrp6::KIURA3 pCM185-sub2-201 (TRP1) | 18 |
| DLY190 | W303 but rrp6::KANrhpr1::HIS | This study |
| DLY241 | W303 but mft1::KANrrrp6::KIURA | This study |
| DLY207 | W303 but mata ski2::HIS | This study |
| DLY208 | W303 but sub2::HIS ski2::HIS pCM185-sub2-201 (TRP1) | This study |
| DLY234 | W303 but hpr1::HIS ski2::HIS | This study |
| DLY199 | W303 but matα xrn1::TRP1 | M. Minet |
| DLY202 | W303 but sub2::HIS xrn1::TRP1 pCM188-sub2-201(URA) | This study |
| DLY210 | W303 but hpr1::HIS xrn1::TRP1 | This study |
| DLY142 | W303 but matα rna14-3 | F. Lacroute |
| DLY128 | W303 but matα rna15-2 | F. Lacroute |
| DLY171 | W303 but rna14-3 rrp6::KANr | This study |
| DLY172 | W303 but rna15-2 rrp6::KANr | This study |
| DLY237 | W303 but matα gcn5::HIS | This studyc |
| DLY238 | W303 but mata ppr2::URA | This studyc |
| DLY236 | W303 but matα rpb9::HIS | This studyc |
| DLY235 | W303 but gcn5::HIS rrp6::KANr | This study |
| DLY242 | W303 but ppr2::URA rrp6::KANr | This study |
| DLY243 | W303 but rpb9::HIS rrp6::KANr | This study |
| rat7-1 | mata trp1Δ63 ura3-52 leu2Δ1 rat7-1 | 15 |
| rat7-1/Δrrp6 | rat7-1 but rrp6::KANr | S. Denome |
| mtr2-26 | mata leu2 ura3 trp1 his3 MTR2::HIS3 pRS315-LEU2-mtr2-26 | 34 |
| mtr2-26/Δrrp6 | mtr2-26 but rrp6::KANr | This study |
Original strains provided by H. Klein.
Original strain provided by A. Aguilera.
Original strains provided by P. Thurianx.
Plasmids expressing green fluorescent protein (GFP) RNAs terminated by a cis-cleaving hammerhead ribozyme element (pRS304-2μm-polII-GFP-RZ) and the glyceraldehyde-3-phosphate dehydrogenase 3 (TDH3) 3′-end processing signal (pRS304-2μm-polII-GFP) were constructed as follows. The 2μm region of Yep24 was PCR amplified and cloned into the AatII site of pRS304 to yield pRS304-2μm. The TDH3 locus, from 670 bp upstream of the start codon to 500 bp downstream of the stop codon, was cloned into pRS304-2μm to yield pRS304-2μm-GPD. BamHI and SalI cloning sites replaced the TDH3 open reading frame (ORF). The GFP ORF was PCR amplified from pJK19-1 (kindly provided by P. A. Silver) and cloned into pRS304-2μm-GPD to yield pRS304-2μm-polII-GFP. pRS304-2μm-polII-GFP-RZ was created similarly, except that the 3′-flanking sequence was replaced with a cis-cleaving hammerhead ribozyme element (5′-CCT GTC ACC GGA TGT GTT TTC CGG TCT GAT GAG TCC GTG AGG ACG AAA CAG G).
Culture growth, RNA isolation, and in vivo protein labeling.
For experiments involving temperature shifts, yeast cultures were grown in yeast extract-peptone-dextrose medium at 25°C. Temperature shifts were performed by mixing a relevant volume of 25°C culture with an equal volume of 49°C medium, after which incubation was continued at 37°C for the specified times. For experiments involving GFP RNA expression, W303 wild-type cells transformed with pRS304-2μm-polII-GFP-RZ or pRS304-2μm-polII-GFP and Δrrp6 cells transformed with pRS304-2μm-polII-GFP-RZ were grown at 30°C in tryptophan dropout medium to an optical density at 600 nm of 0.3. For isolation of total RNA, cells were generally collected by centrifugation, washed once with ice-cold H2O, and stored at −80°C. Total RNA was extracted using the hot acid phenol method (1).
RNA analysis.
For Northern blot analysis of HSP104 RNA, 20 μg of total RNA was used. In DNA oligonucleotide-RNase H digestion reactions, DNA oligonucleotide DL163 (5′-ACATTTTCATCACGAGATTTACCC, directed against nucleotides [nt] 2583 to 2606 of HSP104 RNA) and DNA oligonucleotide DL195 (5′-TGAAGGCAGCTAGAATATGTATAGG, directed against nt 94 to 118 of HSP104 RNA) were used in experiments measuring levels of 3′ and 5′ ends, respectively. To facilitate quantitation of the 3′ ends, an oligo(dT)18 oligonucleotide was included for these RNase H reactions. To detect signals from the HSP104 5′ and 3′ ends, membranes were probed sequentially with radiolabeled oligonucleotides DL190 (5′-TTGAGCCAACGTCAAAATCGTTAGAGCCCTTTCTGTAAATTGCGTTTGGTCGTTCAT, directed against nt 1 to 57 of HSP104 RNA) and DL164 (5′-TTATCGTCATCACCTAACGTGTCAGCCCCTATAGTAGCTTCGTGATTTGGTAGAACTTCC, directed against nt 2631 to 2690 of HSP104 RNA), respectively. Radioactive signals of individual samples were quantitated on a PhosphorImager and normalized to the amount of U4 RNA. The integrity of HSP104 RNA in mutant strains, at a given time after temperature shift, was calculated as the level of HSP104 RNA 5′ or 3′ end in the mutant relative to the wild-type strain. RNA was resolved by using 6% denaturing polyacrylamide gel electrophoresis.
RNAs that failed to terminate correctly in the rna14-3 and rna14-3/Δrrp6 strains were analyzed using RNase H and oligonucleotides D1 and D2 directed against sequences 221 and 300 nt downstream of the stop codon, respectively.
For Northern blot analysis of GFP RNAs, 2 μg of total RNA was cleaved with RNase H by using oligonucleotide KD290 (5′-GAA CGC TTC CAT CTT CAA TGT TGT) and, where applicable, oligo(dT)18. Oligonucleotide KD290 is complementary to the GFP ORF, ∼200 nt upstream of the stop codon. Membranes were probed with radiolabeled oligonucleotide KD282 (5′-GCA GCC AGA TCC TTT GTA TAG TTC ATC CAT GCC ATG) and washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 15 min, twice at 25°C and twice at 42°C. Bands were visualized by autoradiography.
FISH analysis.
Fixation and preparation of cells for fluorescent in situ hybridization (FISH) analysis were done as previously described (19). Detection of HSP104 mRNA was done using Cy3-labeled THJ203, THJ204, THJ205, and THJ206 probes (19), poly(A)+ RNA was localized using a Cy3-labeled oligo(dT)70 probe, and GFP RNA was localized using CY3-labeled KD209, KD210, KD211, and KD212 probes, as described previously (11).
RESULTS
Genetic interactions between SUB2 and members of the THO protein complex.
It has been recently shown that the severe growth phenotype associated with deletion of SUB2 is partially suppressed by mutations in the RAD3 gene, a helicase and a component of transcription factor II H (18). SUB2 overexpression also suppresses the thermosensitive growth defect conferred by deletion of the HPR1 gene (12). These data inspired a closer investigation of the interaction between SUB2 and HPR1. To this end, we analyzed whether a SUB2 thermosensitive allele, sub2-201, was growth impaired in the absence of the inessential HPR1 gene. Indeed, the double mutant is inviable (Fig. 1). The sub2-201 mutation was also synthetically lethal with a yra1-8 (GFP) allele (kindly provided by F. Stutz), and yra1-8 (GFP) was synthetically lethal with the Δhpr1 mutation (reference 41 and data not shown).
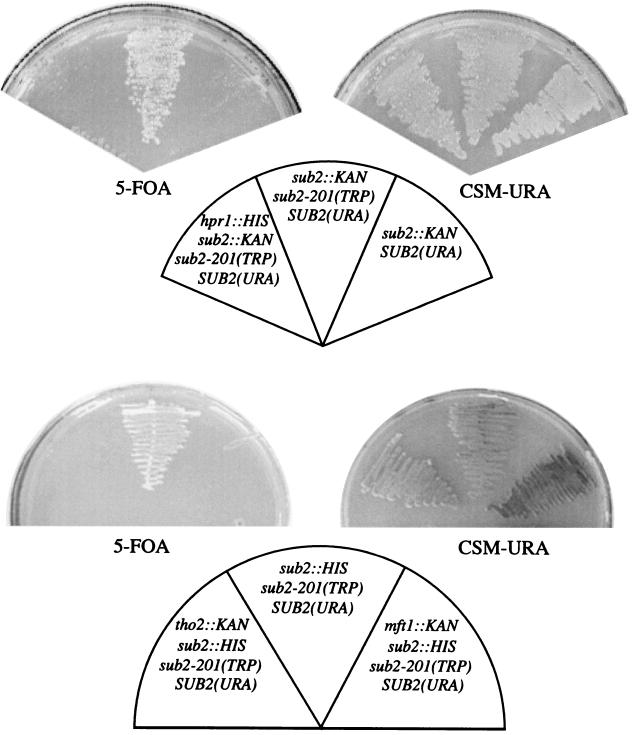
FIG. 1.
The sub2-201 mutation is synthetically lethal with deletion of any one of the HPR1, THO2, and MFT1 genes of the THO complex. Δsub2, Δsub2/Δhpr1, Δsub2/Δtho2, and Δsub2/Δmft1 cells, expressing Sub2-201p from a TRP1-marked plasmid and Sub2p from a URA3-marked plasmid, were grown at 24°C on medium containing 5-FOA (to rid the cells of Sub2p) or on URA dropout plates (CSM-URA), as indicated.
The Sub2p, Yra1p, and Hpr1p proteins have recently been purified, together with THO protein complex members (9, 17, 39). The core THO complex consists of the Tho2p, Hpr1p, Mft1p, and Thp2p proteins, and deletion of any of these four factors leads to accumulation of 3′-end-truncated transcripts (9). To assess whether the genetic interactions linking SUB2, YRA1, and HPR1 extend to other members of the THO complex, we attemped the construction of sub2-201/Δtho2 and sub2-201/Δmft1 double mutants. As shown in Fig. 1, neither double mutant was viable. This result is consistent with that of a recent report from the Hurt laboratory of work performed with the sub2-85 mutant allele (39).
HSP104 transcripts are sequestered in Rrp6p-dependent transcription site foci in strains from which individual THO components are deleted.
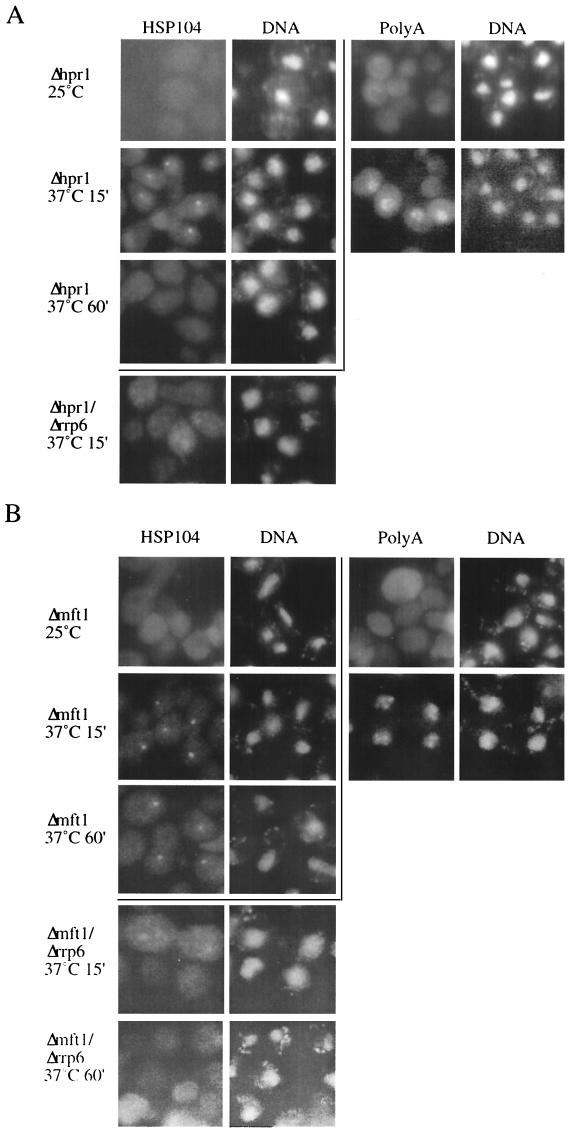
Thermosensitive mutants of Sub2p show nuclear accumulation of mRNA at the restrictive temperature (18, 38). Because of the genetic and physical association between SUB2 and HPR1, we analyzed the fate of transcripts produced in an hpr1 null environment. The Δhpr1 mutant was shifted to 37°C, and HSP104 RNA localization was assayed by FISH analysis utilizing sequence-specific probes. Under these experimental conditions, heat shock gene expression is transiently induced and HSP104 expression follows a pseudo-pulse-chase profile. At the 15-min time point, a single nuclear dot appeared in the majority of the cells (Fig. 2A, second row), which was a result identical to that previously observed with a number of ts mutant strains that inhibit mRNA export (16, 18, 19). The dot reflects retention of the transcript at or near its site of transcription (reference 19 and data not shown). However, this nuclear sequestration phenotype was more transient than previously observed for other mRNA export mutants; i.e., the HSP104 dot disappeared after a 60-min incubation at 37°C (Fig. 2A, third row). Since HSP104 RNA levels, as measured by Northern blotting, do not decrease over time (Fig. 3A), this result indicates an incomplete block to transcription site release, leading to the disappearance of the HSP104 RNA in situ signal after the transcriptional shutoff. The mRNA nuclear export phenotype of the Δhpr1 strain was also evident when poly(A)+ RNA localization was assayed using an oligo(dT) in situ probe (Fig. 2A, right panel).
FIG.2.
Deletion of HPR1 or MFT1 leads to Rrp6p-dependent transcription site focus accumulation of HSP104 RNA. (A) HSP104 and poly(A)+ RNA FISH analysis of Δhpr1 cultures grown at 25°C or temperature shifted to 37°C for 15 or 60 min and HSP104 RNA FISH analysis of Δhpr1/Δrrp6 cultures temperature shifted to 37°C for 15 min, as indicated. (B) RNA FISH analysis of Δmft1 and Δmft1/Δrrp6 cultures, as described for panel A. For the Δhpr1/Δrrp6 strain, HSP104 RNA FISH analysis was also performed on a culture grown for 60 min at 37°C. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole).
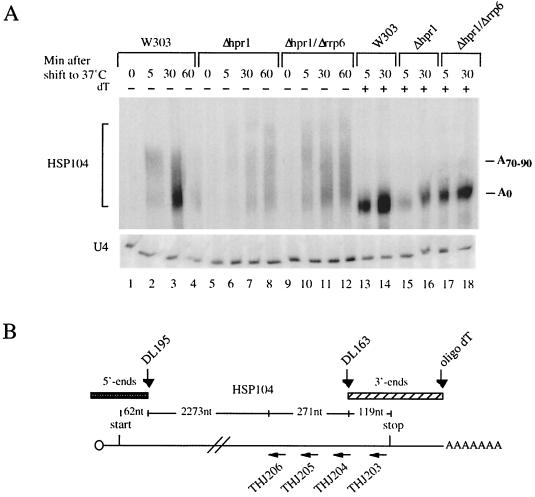
FIG. 3.
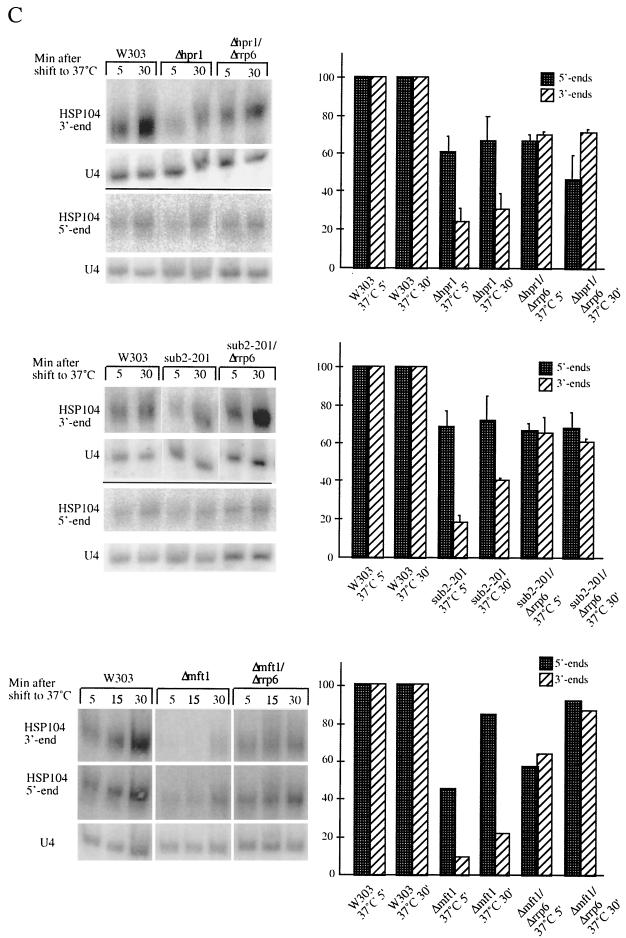
Rrp6p-dependent generation of 3′-end-truncated transcripts in Δhpr1, sub2-201, and Δmft1 mutant backgrounds. (A) Northern analysis of HSP104 RNA (or U4 snRNA as a control) isolated from W303, Δhpr1, and Δhpr1/Δrrp6 cultures harvested at the indicated time points after a 37°C temperature shift. Prior to gel electrophoresis, all samples were treated with RNase H and the DL163 oligonucleotide complementary to nt 2583 to 2606 of the HSP104 gene. The row above the upper gel (dT) indicates whether the RNA sample was also treated with oligo(dT) to remove the poly(A) tail. HSP104 RNA was visualized using a radiolabeled DNA oligonucleotide (DL164) complementary to nt 2631 to 2690 of the HSP104 gene. (B) Schematic representation of the HSP104 RNA, indicating positions cleaved by RNase H and the DNA oligonucleotides DL195, DL163, and oligo(dT). Positions recognized by the HSP104 RNA FISH probes (THJ203 to THJ206) are also shown. (C) Top row, Northern analysis of HSP104 RNA isolated from W303, Δhpr1, and Δhpr1/Δrrp6; middle row, Northern analysis of HSP104 RNA isolated from W303, sub2-201, and sub2-201/Δrrp6; bottom row, Northern analysis of HSP104 RNA isolated from W303, Δmft1, and Δmft1/Δrrp6. All cultures were harvested at the indicated time points after a 37°C temperature shift. To measure 5′-end levels, oligonucleotide DL195 complementary to nt 94 to 118 of the HSP104 gene was used in the RNase H cleavage reaction and a radiolabeled DNA oligonucleotide spanning the first 57 nt of HSP104 was used for hybridization. HSP104 3′-end abundance was measured as described for panel A. All samples were treated with oligo(dT) in the RNase H reaction to facilitate quantitation analysis. To quantitate relative HSP104 5′- and 3′-end levels, signals were normalized to the amount of U4 RNA and expressed in comparison to the percentage of RNA produced in a wild-type strain (set to a value of 100) at the same time point.
Retention of aberrant transcripts in transcription site foci requires components of the nuclear exosome (16). The 3′-5′ exonuclease Rrp6p likely defines the nuclear exosome, as it localizes to the nucleus and does not purify with the cytoplasmic exosome (6, 30). Deletion of RRP6 from the Δhpr1 strain resulted in almost the full disappearance of the HSP104 RNA nuclear dot at the 15-min time point. Instead, a diffuse HSP104 signal could be detected, which in some cells showed a mild nuclear accumulation (Fig. 2A, lower row). Disappearance of the nuclear dot is not due to lack of HSP104 RNA, since transcript levels were actually increased in the Δhpr1/Δrrp6 double-mutant strain compared to the Δhpr1 single-mutant strain (Fig. 3A). Furthermore, deletion of cytoplasmic degradation components (i.e., XRN1 or SKI2) did not result in dot relief in the Δhpr1 context (data not shown). Finally, Hsp104p protein synthesis was diminished in the Δhpr1 strain after 15 min compared to a wild-type strain and normal levels were restored in the Δhpr1/Δrrp6 double-mutant strain (data not shown). We conclude that Rrp6p, and possibly the nuclear exosome, are involved in HSP104 RNA transcription site retention in the Δhpr1 strain.
We next wanted to evaluate the involvement of other components of the THO complex in nuclear mRNA export and their relationships with RRP6. To this end, we analyzed the status of HSP104 transcripts in strains from which the inessential THO complex components MFT1 and THO2 (kindly provided by A. Aguilera) were deleted. As in Δhpr1 cells, in Δmft1 HSP104 RNA localized to a nuclear dot after a 15-min incubation at 37°C (Fig. 2B, second row). The signal intensity decreased slightly over time, although not as much as in the Δhpr1 strain (compare Fig. 2A and B). Under similar conditions, nuclear accumulation of poly(A)+ RNA was also detectable in this strain (Fig. 2B, right panel). Similar results were obtained for the Δtho2 strain (data not shown).
The contribution of Rrp6p to transcription site retention was evaluated by constructing a Δmft1/Δrrp6 strain (a Δtho2/Δrrp6 strain was not viable; see below). HSP104 RNA localization in Δmft1/Δrrp6 resembled that in Δhpr1/Δrrp6 (Fig. 2B, fourth and fifth rows). Northern blot analysis confirmed the presence of HSP104 RNA in the Δmft1/Δrrp6 strain (data not shown).
What is the nature of the transcription site-sequestered HSP104 transcripts in these strains? HSP104 RNA was detected with FISH probes directed towards the 3′ end of the ORF (Fig. 3B). When we utilized a single probe (THJ203) that hybridizes to a HSP104 region immediately upstream of the stop codon, nuclear dots could also be detected (data not shown). We conclude that HSP104 transcripts with sequences at or very close to the stop codon are retained in transcription site foci.
Rrp6p-mediated degradation of HSP104 transcripts in the Δhpr1, Δmft1, and sub2-201 mutant strains.
Using our transcriptional pseudo-pulse-chase heat shock assay, we analyzed HSP104 transcripts biochemically by Northern blotting. In wild-type cells, HSP104 transcription is transiently induced at 37°C and a polyadenylated species first appears that is exported from the nucleus, deadenylated, and degraded in the cytoplasm (a representative gel is shown in Fig. 3A). In contrast, HSP104 RNA expressed in hpr1 null cells did not seem to proceed in its metabolism but accumulated with slightly longer poly(A) tails (Fig. 3A; compare lanes 1 to 4 with lanes 5 to 8). The overall amount of HSP104 RNA was increased in the Δhpr1/Δrrp6 double mutant compared to the Δhpr1 single mutant (Fig. 3A; compare lanes 15 and 16 with lanes 17 and 18; see below for quantitation). Removal of HSP104 RNA poly(A) tails by oligo(dT) and RNase H revealed that the clear majority of transcripts had been polyadenylated.
Members of the THO complex have previously been implicated in transcription elongation (10). To determine Hpr1p, Mft1p, and Sub2p contributions to HSP104 RNA quality, we analyzed the status of HSP104 RNA 5′ ends and 3′ ends in W303, Δhpr1, Δmft1, and sub2-201 cells. Total RNA, harvested after a temperature shift, was subjected to oligonucleotide-directed RNase H cleavage and subsequent Northern analysis. Use of specific cleaving oligonucleotides and hybridization probes allowed independent assessment of 5′- and 3′-end abundance (Fig. 3B). In the Δhpr1 and sub2-201 background, the amount of 3′-end fragments derived from the HSP104 RNA was significantly reduced at the 5- and 30-min time points (to approximately 20% of wild-type levels) (Fig. 3C [see right panels for quantitation]). On the other hand, the 5′ ends were much less affected, which indicates the existence of 3′-end-truncated HSP104 transcripts in these two mutant strains. The presence of incomplete transcripts was even more pronounced in the Δmft1 strain, with the 3′ ends almost undetectable at the 5- and 15-min time points (Fig. 3C, lower part).
Given the proposed role for the THO protein complex, incomplete RNAs could be due to defective transcriptional elongation. However, deletion of the RRP6 gene from the Δhpr1, sub2-201, and Δmft1 backgrounds resulted in the almost complete restoration of HSP104 3′-end levels relative to HSP104 5′-end levels (Fig. 3C). This was not due to a general Rrp6p-mediated increase in mRNA abundance, since the levels of HSP104 3′ ends (and 5′ ends) were unaffected in rrp6 null cells compared to those of a wild-type strain (data not shown).
mRNA degradation in the cytoplasm occurs by two principal mechanisms: a major cap-dependent 5′-3′ pathway and a minor cap-independent 3′-5′ pathway (6). The exonuclease Xrn1p and the helicase Ski2p are nonessential, cytoplasmic factors involved in the 5′-3′ and 3′-5′ degradation pathways, respectively. Deletion of XRN1 (and to a minor extent, SKI2) in a wild-type strain resulted in stabilization of HSP104 RNA at the 30-min time point, as would be expected when cytoplasmic degradation of mRNA is inhibited (data not shown). However, deletion of XRN1 or SKI2 in a sub2-201 or Δhpr1 background did not result in general HSP104 RNA stabilization or restoration of a proper 5′-end:3′-end ratio, in contrast to what was observed in the Δhpr1/Δrrp6 and sub2-201/Δrrp6 double-mutant strains (data not shown). This strongly indicates that significant proportions of RNAs produced in Δhpr1, sub2-201, and Δmft1 environments are specifically degraded in the nucleus and suggests that Rrp6, and possibly the nuclear exosome, plays a major role in this process.
Specific genetic interactions between the THO complex and components of the nuclear exosome.
Rrp6p modulation of HSP104 localization and 3′-end abundance suggests a functional relationship between the THO protein complex and the nuclear exosome. Indeed, Δhpr1/Δrrp6 and Δmft1/Δrrp6 double-mutant strains are severely growth impaired at 24 and 30°C and inviable at all temperatures above 30°C (Table 2). Furthermore, a Δtho2/Δrrp6 strain is inviable at all temperatures tested (data not shown). We have previously reported an interaction between the sub2-201 mutation and deletion of RRP6 (18), and this observation extends to mutant YRA1 alleles, which are growth impaired or lethal in combination with exosome component mutations (41). Thus, SUB2, YRA1, HPR1, MFT1, and THO2 all show a genetic interaction with RRP6. Growth impairment of the sub2-201/Δrrp6 and Δhpr1/Δrrp6 mutants could be linked to general aspects of mRNA degradation. However, deletion of SKI2 or XRN1 does not affect growth of the sub2-201 mutant at any temperature. Similarly, Δhpr1/Δxrn1 and Δhpr1/Δski2 double-mutant strains show growth rates identical to that of a Δhpr1 single mutant (Table 2). Thus, the link between HPR1 and SUB2 on the one hand and the RNA degradation machinery on the other appears to be restricted to the nucleus.
TABLE 2.
Temperature sensitivity of strains
| Strain | Growth rate at a temp (°C) of:
|
|||
|---|---|---|---|---|
| 24 | 30 | 34 | 37 | |
| W303 | +++ | +++ | +++ | +++ |
| Δrrp6 | +++ | +++ | +++ | + |
| sub2-201 | +++ | +++ | +++ | − |
| Δxm1 | ++ | ++ | ++ | ++ |
| Δski2 | +++ | +++ | +++ | +++ |
| sub2-201/Δ rrp6 | +++ | + | − | − |
| sub2-201/Δ xrn1 | +++ | +++ | +++ | − |
| sub2-201/Δ ski2 | +++ | +++ | +++ | − |
| Δhpr1 | +++ | +++ | +++ | + |
| Δhpr1/Δrrp6 | ++ | + | − | − |
| Δhpr1/Δxrn1 | +++ | +++ | +++ | + |
| Δhpr1/Δski2 | +++ | +++ | +++ | + |
| Δmft1 | +++ | +++ | +++ | + |
| Δmft1/Δrrp6 | ++ | + | − | − |
| Δtho2 | ++ | ++ | + | − |
| Δtho2/Δrrp6 | − | − | − | − |
| rna14-3 | +++ | ++ | + | − |
| rna14-3/Δrrp6 | +++ | ++ | ++ | − |
| rna15-2 | +++ | ++ | ++ | − |
| rna15-2/Δrrp6 | +++ | ++ | ++ | − |
| mtr2-26 | +++ | ++ | − | − |
| mtr2-26/Δrrp6 | +++ | ++ | − | − |
| rat7-1 | +++ | +++ | +++ | − |
| rat7-1/Δrrp6 | +++ | +++ | +++ | − |
| Δrpb9 | ++ | +++ | +++ | ++ |
| Δrpb9/Δrrp6 | ++ | +++ | +++ | + |
| Δgcn5 | +++ | +++ | +++ | +++ |
| Δgcn5/Δrrp6 | +++ | +++ | +++ | + |
| Δppr2 | +++ | +++ | +++ | ++ |
| Δppr2/Δrrp6 | +++ | +++ | +++ | + |
Sub2p and Yra1p have been shown to be involved in mRNA export (18, 37, 38). Therefore, we sought to determine whether the synthetic phenotype of nuclear exosome mutations with lesions in SUB2, HPR1, MFT1, THO2, and YRA1 would extend to other nuclear mRNA export factors. Growth of strains harboring two known mRNA export factor mutations combined with an RRP6 deletion was tested at various temperatures. None of these double mutants (rat7-1/Δrrp6 or mtr2-26/Δrrp6) grew slower than their single-mutant counterparts (Table 2). This genetic interaction pattern might reflect the fact that Yra1p, Sub2p, and the THO protein complex function at early transcription-associated steps in mRNA export, whereas the roles of Rat7p and Mtr2p are linked to later steps associated with nuclear pore complexes.
We also tested for a genetic interaction between RRP6 and bona fide transcription elongation factors. As shown in Table 2, growth of cells from strains with a deletion of the PPR2 gene (transcription elongation factor TFIIS), RPB9 (RNA polII component), or GCN5 (SAGA- and ADA-complex-associated histone acetyltransferase) was unaffected by deletion of the RRP6 gene. Together, the genetic results strongly suggest that the interaction of THO complex components and the SUB2 helicase with RRP6 is related to a function that is specific for this set of proteins.
Read-through transcripts are retained and partially degraded in an Rrp6p-dependent manner.
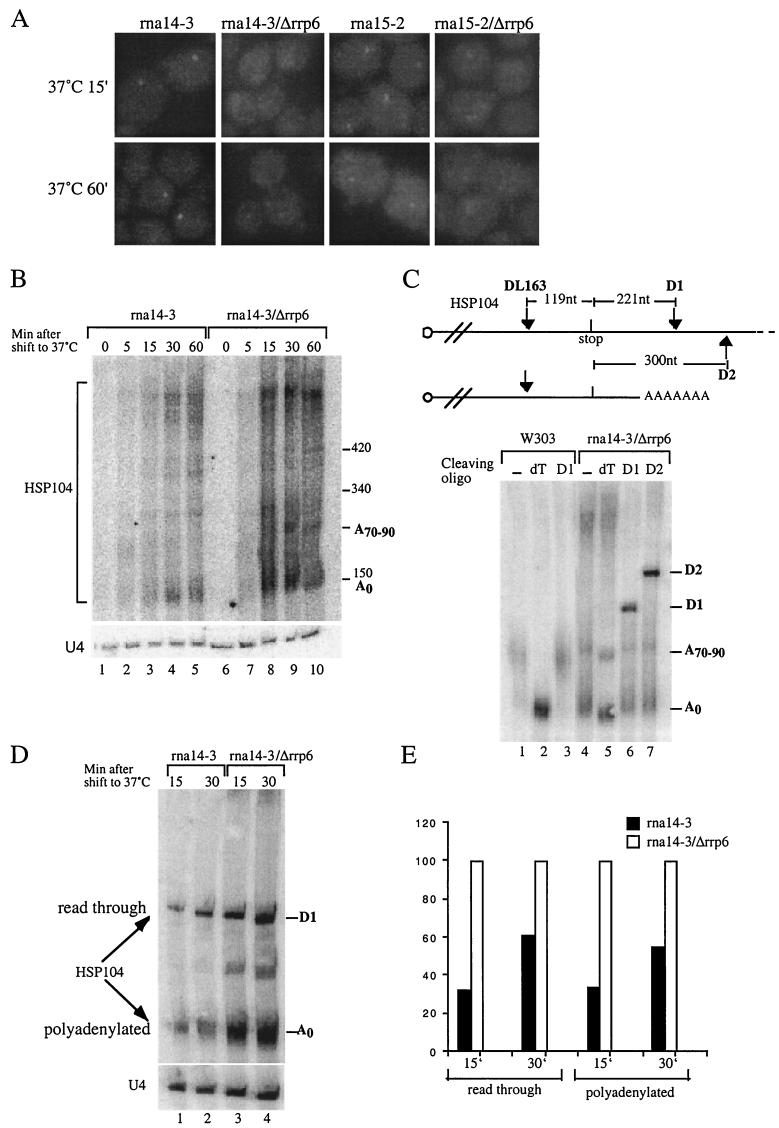
It has been suggested that nonadenylated, 3′-OH ends trigger the degradation activity of Rrp6p (5). Data shown in the previous sections show that HSP104 RNA produced in sub2- or THO complex-defective mutants are retained and degraded in the nucleus. It is not entirely clear whether these transcripts are adenylated (see Discussion). With polyadenylation and free 3′-OH ends in mind, we turned to the 3′-end cleavage and transcription termination mutants rna14-3 and rna15-2 (31). As shown in Fig. 4A, HSP104 transcripts accumulate in an intranuclear dot at the nonpermissive temperature in these strains. Deletion of RRP6 significantly reduces this signal in both mutant backgrounds, despite a general increase in HSP104 RNA levels (Fig. 4B to E). A fraction of the HSP104 transcripts produced in the rna14-3 and rna15-2 strains appear much longer and heterogeneous than in a wild-type strain, which results in a diffuse Northern blot signal (Fig. 4B [also Fig. 4C; compare lanes 1 and 4] and data not shown). These longer transcripts derive from defective 3′-end cleavage and/or transcriptional termination in these strains, as read-through HSP104 transcripts can be cleaved by RNase H and DNA oligonucleotides D1 and D2, positioned downstream of the canonical poly(A) addition site (Fig. 4C and data not shown). HSP104 transcripts polyadenylated at the correct site could also be detected (Fig. 4C to D). Both read-through and correctly polyadenylated transcripts appear to be substrates for Rrp6p, since the amount of both species increases approximately threefold at the 15-min time point in the rna14-3/Δrrp6 double mutant compared to the rna14-3 single mutant (Fig. 4D and E).
FIG.4.
HSP104 transcripts harboring extended 3′ untranslated regions accumulate in Rrp6p-dependent transcription site foci. (A) HSP104 mRNA FISH analysis of rna14-3, rna14-3/Δrrp6, rna15-2, and rna15-2/Δrrp6 cultures temperature shifted to 37°C for 15 or 60 min. DNAs were stained with DAPI. (B) Northern analysis of HSP104 mRNA (or U4 snRNA as a control) isolated from rna14-3 and rna14-3/Δrrp6 cultures harvested at the indicated time points after a 37°C temperature shift. Prior to gel electrophoresis, samples were treated with the DL163 oligonucleotide complementary to nt 2583 to 2606 of the HSP104 gene and RNase H. Approximate size markers are indicated. (C) Northern analysis of HSP104 RNA isolated from W303 and rna14-3/Δrrp6 cultures 15 min after a 37°C temperature shift. Prior to gel electrophoresis, samples were treated with RNase H and the indicated oligonucleotide [none (−), oligo(dT), D1, or D2] complementary to the region of the HSP104 gene (as schematized above the gel) for read-through as well as polyadenylated transcripts. Migration of bands corresponding to oligo(dT), D1, and D2 cleavage is shown. (D) Northern analysis of read-through as well as polyadenylated HSP104 RNA content in rna14-3 and rna14-3/Δrrp6 strains at 15- and 30-min time points after a 37°C temperature shift. To facilitate concomitant detection of read-through and polyadenylated HSP104 RNA, samples were treated with RNase H and a mix of oligonucleotide D1 and oligo(dT) prior to gel electrophoresis. (E) Quantitation of the result described for panel D. Signals were normalized to the amount of U4 RNA and expressed in comparison to the percentage of RNA produced in the rna14-3/Δrrp6 strain (set to a value of 100) at the same time point.
Interestingly, the rna14-3 growth phenotype was slightly suppressed by deletion of the RRP6 gene (Table 2). This mirrors data in the literature on the partial suppression of the pap1-1 phenotype by an RRP6 deletion (5).
Formation of free 3′ ends in a wild-type strain results in transcription site retention.
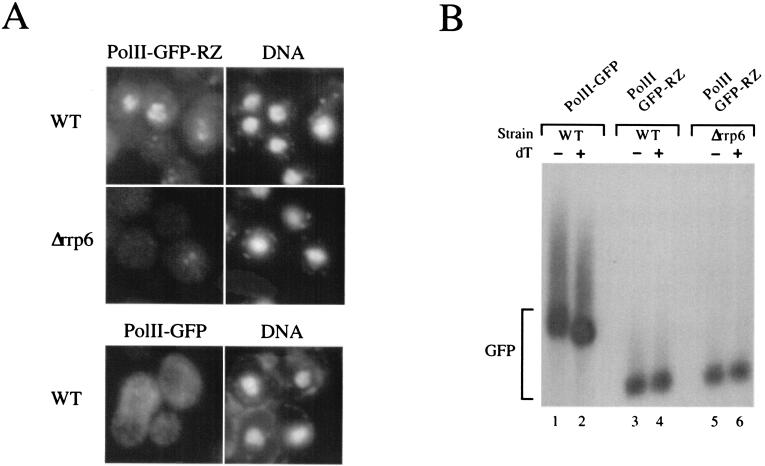
To generate transcripts with free 3′ ends without ts mutants and the consequent temperature change, we constructed a plasmid expressing a GFP RNA with a 3′ end formed by a self-cleaving hammerhead ribozyme (33). The DNA was transformed into wild-type or Δrrp6 cells, and the GFP RNA localization at 30°C was analyzed by using RNA FISH with probes specific for the GFP sequence. In wild-type cells, GFP RNA accumulated in nuclear granules, reminiscent of the signal obtained when transcription site-restricted transcripts are expressed from high-copy-number 2μm reporter plasmids (11, 19). The sequestration was dependent on the presence of Rrp6p, as the nuclear signal was significantly reduced in the Δrrp6 strain (Fig. 5A, middle row). Furthermore, mRNA accumulation in nuclear granules was dependent on the ribozyme-produced 3′ ends, as substitution of the hammerhead ribozyme sequence with a conventional yeast polyadenylation signal resulted in a GFP RNA signal dispersed throughout the cell (Fig. 5A, lower row). Northern analysis confirmed that the polyadenylation signal-containing construct produced RNAs with poly(A) tails of the expected length, whereas the ribozyme construct led to the synthesis of unadenylated transcripts (Fig. 5B). We conclude that Rrp6p is linked to the nuclear retention of transcripts with unadenylated 3′ ends, regardless of their origin.
FIG. 5.
Transcripts whose 3′ end is generated by a self-cleaving hammerhead ribozyme accumulate in Rrp6p-dependent transcription site foci. (A) GFP RNA FISH analysis of wild-type (top row) and Δrrp6 (middle row) strains transformed with a 2μm high-copy-number plasmid construct expressing GFP RNA terminated by a self-cleaving hammerhead ribozyme (polII-GFP-RZ) and wild-type cells transformed with a 2μm plasmid expressing GFP RNA terminated by a conventional 3′-end processing signal (polII-GFP) (bottom row). Δrrp6 cells were cotransformed with either a plasmid expressing the Rrp6p protein (top and bottom rows) or empty vector (middle row). Log-phase cultures growing at 30°C were fixed and subjected to RNA FISH utilizing probes specific for the GFP ORF. DNA was stained with DAPI. dT, oligo(dT). (B) Northern analysis of GFP RNA from the cultures described above. WT, wild type.
DISCUSSION
The DECD-box helicase Sub2p has been implied in splicing, polyadenylation, and nuclear export (13, 14, 18, 23, 27, 28, 38, 42). A connection between Sub2p function and transcription was suggested by the genetic and functional interactions of SUB2 with RAD3 and HPR1, whose protein products associate with the transcription machinery (8, 12, 18). Hpr1p belongs to the heterotetrameric THO complex, which was first described on the basis of genetic and physical interactions and was assigned a role in transcription elongation and mitotic recombination (9). More recently, Sub2p and Yra1p have been found to be associated with the THO complex in a large-scale yeast protein complex analysis (17), which suggested that this complex functions in nuclear mRNA export. Support for this notion was recently presented by Strasser et al. (39) while the manuscript for the present report was in preparation.
Data presented in this paper strengthen the functional interactions between export factors and members of the THO complex. Not only is the sub2-201 mutation synthetically lethal with deletion of the HPR1 gene, but a similar relationship also extends to the two other THO complex genes tested, THO2 and MFT1. Moreover, Sub2p interacts directly with Hpr1p and Yra1p, which indicates that the observed genetic interaction between SUB2, YRA1, and the THO complex is due to direct protein-protein interactions (39, 41).
Deletion of all tested members of the THO complex leads to sequestration of newly synthesized HSP104 transcripts in transcription site foci. This mRNA retention is Rrp6p dependent, which is a finding identical to the previously reported finding for inactivation of SUB2 (18). The exact nature of sequestered HSP104 transcripts in sub2 and THO complex mutants remains undetermined, as we cannot ascertain whether 3′-end-truncated transcripts are retained in transcription site foci or not. However, we note that HSP104 RNA is detectable with an in situ probe complementary to a sequence immediately upstream of the stop codon. Thus, complete, or nearly complete, transcripts are retained. Moreover, polyadenylated RNAs accumulate in the nucleus in THO complex mutants (39; this study). It is therefore likely that the HSP104 RNAs, which can be detected in a nuclear dot, belong to the class of transcripts that either have not yet been degraded (see below) or escaped degradation and await further processing steps (e.g., formation of export-competent mRNP).
Read-through HSP104 RNAs produced in rna14-3 and rna15-2 mutants are also retained in Rrp6p-dependent transcription site foci. These transcripts harbor extended 3′ untranslated regions, which are presumably unadenylated. Furthermore, transcripts generated by a self-cleaving hammerhead ribozyme are also subject to transcription site retention by Rrp6p. All of our results show that detection by Rrp6p, and possibly by the nuclear exosome, controls the release of several different incompletely and/or improperly processed mRNAs. Although it is presently unclear how recognition by Rrp6p is achieved, our results with the self-cleaving ribozyme imply that recognition of aberrant mRNA can occur independently of a polyadenylation signal and thus without binding of all factors normally involved in 3′-end formation.
THO complex mutants were originally described as defective in transcription of the bacterial LacZ gene, as well as of long and/or GC-rich yeast ORFs (9, 10). In the present study, we observed 3′-end-truncated HSP104 transcripts in THO complex and sub2-201 mutants. However, deletion of RRP6 leads to almost complete restoration of full-length HSP104 transcripts. Consistent with our results, LacZ transcript levels are also restored in a yra1 mutant upon deletion of RRP6 (41). Although an inhibitory role of Rrp6p on transcriptional elongation cannot be excluded, the specific restoration of HSP104 3′ ends (as opposed to an overall stabilization of HSP104 RNA) supports the hypothesis that incomplete HSP104 transcripts derive from partial degradation by Rrp6p. It is unclear why the exosome does not degrade HSP104 RNA to completion, but this might be due to inhibitory features of the substrate mRNP.
What is the substrate for Rrp6p? Oligo(dT)/RNase H digestion showed that the majority of restored HSP104 RNAs in double mutants are polyadenylated. Thus, either RNAs degraded in the presence of Rrp6p were originally polyadenylated, or RNAs produced in a THO-mutant environment can undergo normal polyadenylation only in the absence of competitive Rrp6p degradation. Consistent with these possibilities, we found that polyadenylated as well as read-through transcripts are stabilized when RRP6 is deleted in an rna14-3 mutant background (Fig. 4). This suggests that a nuclear degradation process might outcompete a defective mRNP formation step, in the THO and sub2 as well as the rna14-3 mutant backgrounds. Alternatively, polyadenylation took place in these mutants before degradation. Inefficiently spliced RNAs are also substrates of the nuclear exosome, and nuclear degradation has been shown to compete with inefficient splicing (3). Although these observations suggest that nuclear RNA degradation is a general and important way of controlling aberrant (and maybe even normal) transcript abundance, it is not known how this is controlled.
A distinctive feature of SUB2, YRA1, and THO complex genes is their strong genetic interaction with the nuclear exosome component RRP6. This appears to be specific, as deletion of cytoplasmic mRNA degradation factors XRN1 and SKI2 does not affect growth of Δhpr1 or sub2-201 strains. Furthermore, growth of a RRP6 deletion strain is unaffected when combined with lesions in three other classes of genes involved in mRNA metabolism: (i) conventional mRNA export factors MTR2 and RAT7, (ii) 3′-end formation components RNA14, RNA15, and PAP1, and (iii) bona fide transcriptional elongation factors GCN5, PPR2, and RPB9. The latter result suggests that a transcriptional elongation failure per se is insufficient to explain the genetic interaction of the THO complex with the nuclear exosome. The synthetic growth defects between mutants of yra1, sub2, or the THO complex with mutations of the nuclear exosome might be due to transcription site release or to reduced degradation. However, mRNA release is not toxic in the context of tested polyadenylation, transcription termination, or mRNA export mutants. This suggests that the released mRNP in THO complex, sub2, and yra1 double mutants has a different composition and/or might improperly sequester an important subset of factors.
The hypothesis of involvement of the THO complex in transcription is based on the assessment of transcript abundance in mutants and on run-on experiments. In light of the results presented in this report, we suggest that transcript abundance in THO complex and sub2 mutants is at least partially dependent on the extent of nuclear degradation. This, in turn, is probably triggered by defects in late events in transcription, 3′-end processing, and/or mRNP formation. Chromatin immunoprecipitation experiments indeed suggest a role of the THO complex in mRNP formation and specifically in the recruitment of Sub2p and Yra1p to nascent RNA (39, 41). Interestingly, LacZ sequences affect transcript abundance in mutants more markedly when located at the 3′ end of chimeric ORFs (10), which is consistent with the progressive cotranscriptional constitution of a functional mRNP. This hypothesis does not exclude functional feedback of mRNP formation on transcription elongation efficiency or pausing, possibly implicating factors recruited by the THO complex in one or more aspects of these processes. In this view, the primary effect of the THO complex is on nascent mRNP formation.
Acknowledgments
We thank T. McCarthy for excellent technical assistance. Furthermore, we thank H. Klein, A. Aguilera, P. Thuriaux, S. Denome, E. Hurt, C. Cole, F. Lacroute, M. Minet, and R. Parker for providing yeast strains. J. Kjems is thanked for critical reading of the manuscript and F. Stutz is thanked for communicating unpublished data.
This work was supported by the Centre National de la Recherche Scientifique (D.L.), the National Institutes of Health (M.R.), and the Danish Research Council and Novo Nordisk Foundation (T.H.J.).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bachi, A., I. C. Braun, J. P. Rodrigues, N. Pante, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Gorlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky, A. S., and P. A. Silver. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkard, K. T., and J. S. Butler. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20:604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12:90-96. [DOI] [PubMed] [Google Scholar]
- 7.Chang, M., D. French-Cornay, H.-Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavez, S., M. Garcia-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dower, K., and M. Rosbash. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA export. RNA 8:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, H.-Y., R. J. Merker, and H. L. Klein. 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 21:5459-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 14.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716-1721. [DOI] [PubMed] [Google Scholar]
- 15.Gorsh, L. C., T. C. Dockendorff, and C. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT/NUP159 causes both rapid cessation of mRNA export and reversible clustering of the nuclear pore complexes. J. Cell Biol. 129:939-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 17.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, T. H., J. Boulay, M. Rosbash, and D. Libri. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11:1711-1715. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, T. H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 20.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katahira, J., K. Straesser, T. Saiwaki, Y. Yoneda, and E. Hurt. 2002. Complex formation between Tap and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J. Biol. Chem. 277:9242-9246. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 23.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libri, D., N. Graziani, C. Saguez, and J. Boulay. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 15:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 31.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samarsky, D. A., G. Ferbeyre, E. Bertrand, R. H. Singer, R. Cedergren, and M. J. Fournier. 1999. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. USA 96:6609-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos-Rosa, H., H. Moreno, G. Simos, A. Segref, B. Fahrenkrog, N. Panté, and E. Hurt. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18:6826-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strasser, K., J. Bassler, and E. Hurt. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 39.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 40.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenklusen, D., P. Vinciguerra, J.-C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export requires cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed]
- 42.Zhang, M., and M. R. Green. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]