Abstract
Transforming growth factor β (TGF-β) induces cell cycle arrest of most nontransformed epithelial cell lines. In contrast, many human carcinomas are refractory to the growth-inhibitory effect of TGF-β. TGF-β overexpression inhibits tumorigenesis, and abolition of TGF-β signaling accelerates tumorigenesis, suggesting that TGF-β acts as a tumor suppressor in mouse models of cancer. A screen to identify agents that potentiate TGF-β-induced growth arrest demonstrated that the potential anticancer agent rapamycin cooperated with TGF-β to induce growth arrest in multiple cell lines. Rapamycin also augmented the ability of TGF-β to inhibit the proliferation of E2F1-, c-Myc-, and V12H-Ras-transformed cells, even though these cells were insensitive to TGF-β-mediated growth arrest in the absence of rapamycin. Rapamycin potentiation of TGF-β-induced growth arrest could not be explained by increases in TGF-β receptor levels or rapamycin-induced dissociation of FKBP12 from the TGF-β type I receptor. Significantly, TGF-β and rapamycin cooperated to induce growth inhibition of human carcinoma cells that are resistant to TGF-β-induced growth arrest, and arrest correlated with a suppression of Cdk2 kinase activity. Inhibition of Cdk2 activity was associated with increased binding of p21 and p27 to Cdk2 and decreased phosphorylation of Cdk2 on Thr160. Increased p21 and p27 binding to Cdk2 was accompanied by decreased p130, p107, and E2F4 binding to Cdk2. Together, these results indicate that rapamycin and TGF-β cooperate to inhibit the proliferation of nontransformed cells and cancer cells by acting in concert to inhibit Cdk2 activity.
Previous studies of transgenic mouse model systems (11, 66, 78), as well as mutational analysis of human cancers (65, 91), indicate that the transforming growth factor β (TGF-β) signaling pathway is tumor suppressive. The observation that many human carcinomas are refractory to TGF-β-induced growth arrest (7, 14, 64) suggests that abolition of TGF-β-induced cell cycle arrest may be a requirement for tumorigenesis in many circumstances as suggested previously (26, 39, 54). TGF-β-induced growth arrest is initiated by TGF-β binding to the type II TGF-β receptor (TβRII), which leads to the recruitment, phosphorylation, and activation of TβRI. The activated serine/threonine kinase TβRI then phosphorylates the downstream effectors that mediate TGFβ signaling. The most extensively studied mediators of TGF-β signaling are the Smad proteins, which act as transcriptional regulators. Phosphorylation of Smad2 and -3 by TβRI triggers their oligomerization with Smad4 as well as their nuclear accumulation. A key role for the Smad proteins in TGF-β-induced cell cycle arrest is suggested by studies demonstrating Smad-dependent regulation of p15, p21, and c-Myc by TGF-β (14, 17, 24, 74, 76, 81, 86).
The loss of TGF-β growth-inhibitory responses in some cancers results from the functional inactivation of genes whose protein products mediate TGF-β signal transduction, including TβRI and TβRII and Smad2 and Smad4. These genetic alterations, however, explain only a small percentage of the TGF-β insensitivity observed in human carcinomas, suggesting that alternate mechanisms are responsible for the loss of TGF-β-induced cell cycle arrest in many tumors.
Oncogenes, including activated Ras, E2F1, and c-Myc, abolish TGF-β-induced growth arrest. Since the Ras and Myc oncogenes are commonly dysregulated in human cancers (2, 47, 59), and since activation and/or overexpression of receptor tyrosine kinases such as the epidermal growth factor receptor and Her2 may activate Ras and upregulate c-Myc, oncogenic transformation may explain the lack of TGF-β sensitivity of many cancer cell lines. A general observation in cells that are resistant to TGF-β-induced growth arrest is the lack of TGF-β-mediated c-Myc downregulation (68). The findings that c-Myc dysregulation alone is sufficient to abolish TGF-β-induced growth arrest (5) and that c-Myc dysregulation blocks TGF-β induction of the cyclin-dependent kinase inhibitors p15 and p21 (17, 24, 74, 76) suggest that agents capable of inducing c-Myc downregulation could restore TGF-β-induced growth arrest in transformed and cancer cells. It has been reported (43, 82) that the pathway of the mammalian homolog of the yeast target of rapamycin (TOR) (mTOR)/p70s6k regulates c-Myc protein levels. This suggests that inhibitors of the mTOR pathway may potentiate TGF-β-mediated growth arrest by inducing c-Myc downregulation.
We hypothesize that rapamycin may augment or restore TGF-β sensitivity in cancer cells that are refractory to TGF-β-mediated growth arrest due to oncogene activation by inducing c-Myc downregulation or by cooperating with TGF-β to inhibit E2F-dependent transcription. Such a hypothesis is particularly intriguing in light of the fact that the rapamycin derivatives RAD001 (Novartis) and CCI-779 (Wyeth-Ayerst) are in preclinical and clinical testing as anticancer agents with promising preliminary results (21, 25, 60, 88). We propose that the antitumor actions of rapamycin and its derivatives are in part due to their influence on TGF-β-induced growth arrest. Rapamycin was identified some time ago as a potent inhibitor of cell proliferation, yet little is known regarding the exact mechanisms by which rapamycin inhibits cell proliferation.
The observation that rapamycin arrests the proliferation of yeast cells by inhibiting TOR (30, 50) and blocks mammalian cell proliferation by inactivating mTOR (12, 31, 71) suggests that the TOR proteins play an evolutionarily conserved role in regulating the eukaryotic cell cycle. Subsequent studies demonstrated that rapamycin acts by binding to its intracellular receptor protein FKBP12 and that the rapamycin-FKBP12 complex binds and inhibits the function of the high-molecular-weight protein kinase mTOR. The two most extensively studied downstream effectors of mTOR are the protein kinase p70s6k and the inhibitor of cap-dependent translation, PHAS1, both of which play critical roles in regulating protein synthesis. D-type cyclins and c-Myc are also important targets of mTOR signaling that are downregulated in response to treatment with rapamycin (23, 27, 29, 43, 55, 82).
In this study we examined the interaction between TGF-β and rapamycin in arresting cell proliferation. The results demonstrate that rapamycin potentiates TGF-β-induced growth arrest in nontransformed cells and restores TGF-β-induced growth arrest in oncogene-transformed rodent cells and human carcinoma cells. Suppression of c-Myc expression is not required for G1 cell cycle arrest. Instead, TGF-β and rapamycin cooperated to increase binding of the Cdk inhibitors p21 and p27 to Cdk2 with inhibition of Cdk2 kinase activity.
MATERIALS AND METHODS
Cell culture and cell proliferation assays.
The Mv1Lu mink lung epithelial cells used (MLEC clone 32) were kindly provided by D. Rifkin (New York University, New York) and have been described previously (1). The HaCaT human keratinocyte, MDA-MB-231 human mammary carcinoma, DU145 human prostatic carcinoma, A549 human lung carcinoma, NMuMG mouse mammary epithelium, and NIH OVCAR-3 human ovarian carcinoma cell lines were obtained from the American Type Culture Collection and grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). MCF10A human mammary epithelial cells were also obtained from the American Type Culture Collection and were grown in a 1:1 mixture of DMEM and Ham's F12 medium supplemented with 20 ng of epidermal growth factor/ml, 100 ng of cholera toxin/ml, 0.01 mg of insulin/ml, 500 ng of hydrocortisone/ml, and 5% horse serum. Mouse NIH 3T3 cells (clone 7) were kindly provided by Doug Lowy (37) and were propagated in 10% FBS-90% DMEM. The Phoenix cells used for retroviral packaging were obtained from Gary Nolan (40) (Stanford University, Stanford, Calif.) and were grown in 10% FBS-90% DMEM. The origin and propagation of the BALB/MK mouse keratinocyte cell line and the NMuMG normal mouse mammary epithelial cell line were as described previously (9, 42). TGF-β was kindly provided by R&D Systems Inc., Minneapolis, Minn. Rapamycin was purchased from Sigma-Aldrich Co. (St. Louis, Mo.). The rapamycin derivative RAD001 was a kind gift from Heidi Lane (Novartis Pharma AG, Basel, Switzerland).
[3H]thymidine incorporation assays were performed as described previously (42). In all [3H]thymidine incorporation experiments, cells were plated at 20,000 cells per well in 24-well plates, incubated 24 h, treated as indicated in the figure legends, and pulsed for the last 2 h of the treatment time with [3H]thymidine. In sequential cell-counting experiments, MDA-MB-231 cells or DU145 cells were plated at 150,000 or 75,000 cells per well, respectively, in six-well plates and incubated for 24 h. Cells from three replicate wells were then counted (day 0) using a Coulter Counter (Beckman Coulter, Inc., Fullerton, Calif.). The remaining wells were treated in triplicate as indicated in the figure legend, and cells were counted in triplicate at 24-h intervals for the remainder of the experiment.
Flow cytometry was performed using a FACSCalibur instrument (Becton Dickinson, San Jose, Calif.), and the results were modeled using Winlist (Verity Software House, Topsham, Maine).
Retroviral gene transduction.
The retroviruses used were based on the pBabe vector with a puromycin selectable marker (53) and were made using the Phoenix packaging cell line. Viral vectors encoding c-Myc2 and c-Myc2(T58A) were supplied by S. Hann (Vanderbilt University, Nashville, Tenn.), and a viral vector encoding V12H-Ras was supplied by P. Liang (Vanderbilt University) and were described previously (17, 33, 36). The E2F1 retroviral vector was obtained by subcloning the E2F1 cDNA obtained from Srilata Bagchi (University of Illinois, Chicago) into pBabe.
Cell lines stably expressing the genes of interest were prepared by applying retrovirus-containing supernatants to rapidly growing MLEC-32 cells in the presence of 8 μg of Polybrene (Sigma Chemical Co.)/ml, followed by selection with 1.5 μg of puromycin/ml beginning 24 h postinfection. Individual clones were isolated and characterized. Clones expressing the highest levels of c-Myc2, c-Myc2(T58A), V12H-Ras, or E2F1 exhibited the least sensitivity to TGF-β-induced growth arrest and were used in subsequent experiments. Cell lines transformed by overexpression of c-Myc2, c-Myc2(T58A), V12H-Ras, or E2F1 formed colonies in soft agar, while cells transduced with the “empty” pBabe retroviral vector and selected with puromycin (Puro. Cont.) did not. The Puro. Cont. polyclonal cell line was used as the nontransformed control cell line in subsequent experiments. Derivation of the individual stable cell lines took approximately 6 weeks, and all experiments were performed using clones passaged less than 10 times. Similar results were obtained in at least three independent clonal cell lines expressing each oncogene.
Preparation of cell extracts and immunoblotting.
Whole-cell protein extracts were prepared as described before (42) with the exception that extracts were sonicated to insure the recovery of nuclear proteins. Immunoblotting was performed as described previously (42) using antibodies from Santa Cruz Biotechnology to detect p107 (sc-318), p130 (sc-317), BRCA1 (sc-646), cyclin A (sc-596), E2F1 (sc-193), E2F4 (sc-866), B-Myb (sc-725), Smad4 (sc-7966), p21 (sc-6246), cyclin E (sc-481), p70s6k (sc-230), Cdk2 (sc-163), Cdk4 (sc-601), and c-Myc (sc-764). β-Tubulin (T4026) and actin (A2066) antibodies were obtained from Sigma Chemical Co. The antibody recognizing total Rb levels (14001A) was obtained from BD-Pharmingen. Antibodies recognizing Rb phosphorylated on Thr821 (44-582Z) or Ser249/Ser252 (44-584Z) were obtained from Biosource International. Antibody specific for phosphorylated Smad2 (06-829) was obtained from Upstate Biotechnology, Inc. Antibodies specific for total levels of Smad2 (610842) p27 (K25020) were obtained from BD Transduction Laboratories, and antibody specific for Smad3 (51-1500) was obtained from Zymed Laboratories. Antibody to Cdk2 phosphorylated on Thr160 (no. 2561) was obtained from Cell Signaling Technologies, Inc. Cyclin D1 antibody (DCS-6) was obtained from Neomarkers. PHAS1 antibody was generously provided by John Lawrence, University of Virginia, Charlottesville (13). Protein assays were performed using the Bradford reagent (Bio-Rad Laboratories, Hercules, Calif.). β-Tubulin and actin were used as loading controls for immunoblotting. Since the level of neither protein was altered by the experimental treatments, β-tubulin and actin were used interchangeably as loading controls.
125I-TGF-β cross-linking.
125I-TGF-β1 was obtained from NEN Life Science Products, Inc. (Boston, Mass.). Nearly confluent MDA-MB-231 cells in 12-well plates were treated with 10 ng of TGF-β1/ml, 100 nM rapamycin, 10 ng of TGF-β1/ml plus 100 nM rapamycin, or dimethyl sulfoxide vehicle control. Twenty-four hours after treatment, the cells were washed three times over 30 min with 500 μl of ice-cold 0.1% bovine serum albumin-Dulbecco’s phosphate-buffered saline (D-PBS) containing Ca2+ and Mg2+. The cells were then affinity labeled with 100 pM 125I-TGF-β1 as previously described (22), with slight modifications. Briefly, after a 3-h incubation with 100 pM 125I-TGF-β1 at 4°C, the cells were washed with 500 μl of ice-cold D-PBS and the ligand-receptor complexes were cross-linked with 400 μl of 1 mM bis(sulfosuccinimidyl)suberate (BS3) (Pierce, Rockford, Ill.) for 10 min on ice. The cross-linking reaction was stopped with the addition of 100 μl of 500 mM glycine. Cells were washed twice with 500 μl of D-PBS and were solubilized with 125 μl of 20 mM Tris buffer, pH 7.4, containing 1% Triton X-100, 10% glycerol, 1 mM EDTA, and protease inhibitors. Solubilized material was recovered from each well and centrifuged for 10 min at 4°C to pellet cell debris, and the supernatants were transferred to 1/5 volume of fivefold-concentrated electrophoresis sample buffer, boiled, and vortexed. All samples were analyzed using sodium dodecyl sulfate-3 to 12% polyacrylamide gel electrophoresis and were visualized by autoradiography.
Luciferase assays.
The E2F-responsive luciferase reporter plasmid pE2F-TA-Luc is from the Mercury Cell Cycle Profiling System (Clontech Laboratories, Inc.). The (CAGA)12-luciferase reporter plasmid was provided by Jean-Michel Gauthier (20). NMuMG cells were transfected by the Ca3(PO4)2 precipitation method. DU145 cells were transfected using FuGENE 6 (Roche) according to the manufacturer's instructions. Twenty-four hours posttransfection the cells were treated as indicated in the figure legend and were incubated an additional 24 h. Cell extracts were prepared and subjected to luciferase assays and protein assays. Background readings were subtracted from the luciferase assay values, and the results were normalized to total protein concentration.
Kinase assays and coimmunoprecipitation.
Cdk2 and Cdk4 kinase assays were performed essentially as described previously (44). Briefly, 500 to 1,000 μg of total cell extract was precleared for 2 h with protein A-Sepharose; 2 or 3 μg of anti-Cdk2 or Cdk4 antibody was added to the precleared extracts and was mixed overnight at 4°C. The following day, 30 μl of protein A-Sepharose beads was added and mixed for an additional 2 h at 4°C. The beads were washed twice with extraction buffer and twice with kinase assay buffer (50 mM HEPES, pH 7.2, 10 mM MgCl2, 2.5 mM EGTA, 1 mM dithiothreitol, 0.1 mM NaF, and 0.1 mM Na3VO4). Kinase reactions were performed for 1 h at 30°C in kinase assay buffer and contained 20 μM [γ-32P]ATP at a specific activity of 10 Ci/mmol and 500 ng of glutathione transferase-Rb (Santa Cruz) for Cdk4 assays and 1 μg of histone H1 for Cdk2 assays. Reactions were stopped with fourfold-concentrated Laemmli sample buffer, and reaction products were separated into several portions and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 15% gels. The gels were either dried and exposed to phosphor screens to visualize and quantitate kinase assay results using a PhosphorImager (Molecular Dynamics) or were transferred to nitrocellulose membranes and analyzed by immunoblotting to verify uniform Cdk2 or Cdk4 immunoprecipitation and to detect p21 and p27 coimmunoprecipitation. The coimmunoprecipitation experiments depicted in Fig. 10 were performed similarly but were scaled up so that 3 mg of cell extract was immunoprecipitated with 4 μg of Cdk2 antibody. Normal rabbit immunoglobulin G (IgG) was used as a negative control in immunoprecipitation experiments.
Statistics.
P values were calculated on the GraphPad.com website using the unpaired Student t test.
RESULTS
TGF-β and rapamycin cooperate to induce a more complete G1 cell cycle arrest in nontransformed epithelial cells.
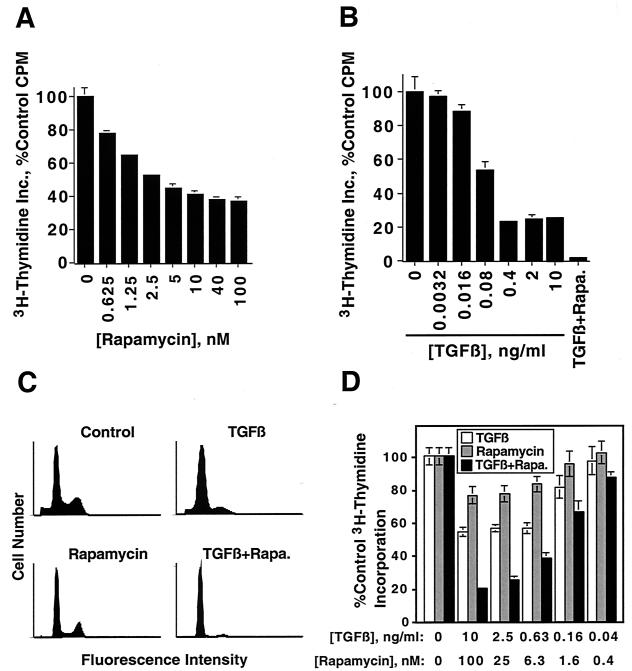
Mv1Lu mink lung epithelial cells were treated with increasing concentrations of TGF-β or rapamycin for 22 h to test the hypothesis that TGF-β and rapamycin cooperate to induce growth arrest. The cells were then pulsed with [3H]thymidine for 2 h, and [3H]thymidine incorporation was quantitated as a measure of the rate of DNA synthesis. Rapamycin induced a dose-dependent inhibition of DNA synthesis with a maximal inhibition to approximately 40% of control occurring at or above 100 nM rapamycin (Fig. 1A). TGF-β also inhibited DNA synthesis in a dose-dependent manner with a maximal inhibition to about 25% of control occurring at or above 0.4 ng/ml (Fig. 1B). Interestingly, combined treatment with 100 nM rapamycin and 10 ng of TGF-β/ml inhibited DNA synthesis to about 2% of control (Fig. 1B, last bar).
FIG. 1.
TGF-β and rapamycin cooperate to induce G1 cell cycle arrest of Mv1Lu cells and NMuMG cells. (A) Nontransformed Mv1Lu cells were treated for 22 h with the indicated concentrations of rapamycin. The cells were pulsed for an additional 2 h with [3H]thymidine, and thymidine incorporation (Inc.) was quantitated as described in Materials and Methods. Results are presented as the average percent control incorporation from six replicate wells ± standard deviation. (B) Nontransformed Mv1Lu cells were treated for 24 h with the indicated concentrations of TGF-β for 24 h and were processed, and the results were presented as in panel A. The last bar (TGF-β + Rapa.) represents treatment for 24 h with both 10 ng of TGF-β/ml and 100 nM rapamycin. (C) Nontransformed Mv1Lu cells were treated for 24 h with normal growth medium (Control), 10 ng of TGF-β/ml, 100 nM rapamycin, or 10 ng of TGF-β/ml plus 100 nM rapamycin. The cells were stained with propidium iodide and subjected to flow cytometry as described in Materials and Methods. (D) NMuMG cells were treated with the indicated concentrations of TGF-β1 and rapamycin either alone or in combination for 24 h, and [3H]thymidine incorporation assays were performed as for panel A.
Since TGF-β and rapamycin each induce a G1 cell cycle arrest, we examined whether combined treatment with TGF-β and rapamycin also inhibited cell proliferation by inducing a G1 cell cycle arrest. When flow cytometry analysis was used, both 10 ng of TGF-β/ml and 100 nM rapamycin were shown to induce a partial G1 cell cycle arrest when applied separately but to induce a more complete G1 arrest when applied in combination (Fig. 1C). The data demonstrate that TGF-β and rapamycin cooperate to induce a G1 cell cycle arrest in nontransformed Mv1Lu cells.
NMuMG normal mouse mammary epithelial cells were treated with increasing concentrations of TGF-β or rapamycin, either alone or in combination (Fig. 1D) to determine whether the cooperative growth-inhibitory effects of TGF-β and rapamycin apply to other cell types. The results indicate that TGF-β and rapamycin cooperate to inhibit the proliferation of NMuMG cells.
TGF-β and rapamycin cooperate to inhibit cell proliferation, induce Rb dephosphorylation, and downregulate E2F1 and B-Myb expression in NMuMG cells.
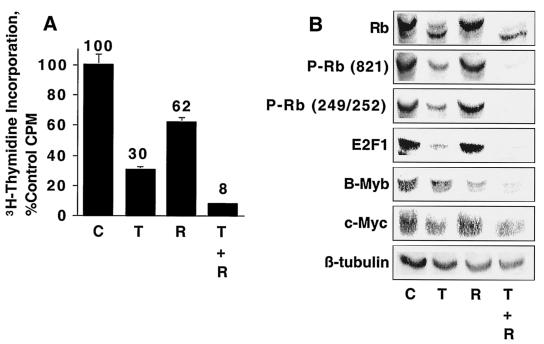
NMuMG cells were treated with TGF-β and rapamycin, either alone or in combination, and the effects were correlated with effects on Rb phosphorylation. Levels of E2F1, B-Myb, and c-Myc (Fig. 2) were examined to characterize the biochemical mechanisms of TGF-β-rapamycin-induced cooperative growth arrest. Rapamycin had a minimal effect on Rb phosphorylation when applied alone but cooperated with TGF-β to induce a more complete Rb dephosphorylation. Likewise, TGF-β and rapamycin cooperated to decrease levels of E2F1 and B-Myb. In contrast, while TGF-β induced a partial downregulation of c-Myc, rapamycin had no effect on c-Myc levels and did not cooperate with TGF-β to induce c-Myc downregulation. These results suggest that the ability of TGF-β and rapamycin to inhibit the proliferation of NMuMG cells correlates more closely with Rb dephosphorylation and E2F1 and B-Myb downregulation than with c-Myc downregulation.
FIG. 2.
TGF-β and rapamycin cooperate to induce Rb dephosphorylation in NMuMG cells. (A) NMuMG cells were treated for 24 h with either normal growth medium (C), 10 ng of TGF-β/ml (T), 100 nM rapamycin (R), or 10 ng of TGF-β/ml and 100 nM rapamycin (T + R) and were subjected to [3H]thymidine incorporation analyses as for Fig. 1. (B) NMuMG cells were treated as for panel A, and cell extracts were prepared and subjected to immunoblot analyses as described in Materials and Methods using antibody specific for total Rb, Rb phosphorylated on Ser249/Thr252 [P-Rb(S249/T252)], Rb phosphorylated on Thr821 [P-Rb(821)], E2F1, c-Myc, or β-tubulin as a loading control.
Rapamycin restores TGF-β-mediated arrest of cells rendered TGF-β resistant by oncogenic transformation.
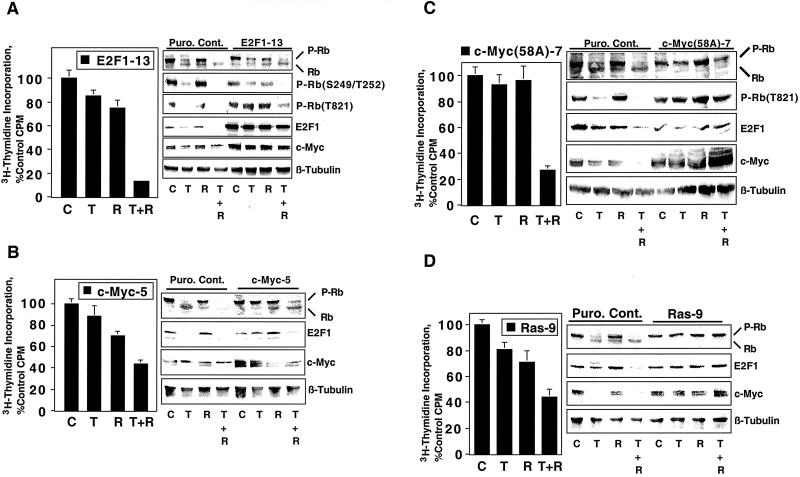
We examined whether rapamycin potentiates TGF-β-induced growth arrest of Mv1Lu cells transformed by the E2F1, c-Myc, and V12H-Ras oncogenes, since E2F1-, c-Myc-, and V12H-Ras-mediated transformation of epithelial cells abolishes the antiproliferative effects of TGF-β (5, 14, 38, 63, 73) and since oncogenic activation of c-Myc and Ras is a relatively common event during tumorigenesis (2, 47, 59). Neither TGF-β nor rapamycin strongly inhibited the proliferation of E2F1-transformed Mv1Lu cells, but TGF-β plus rapamycin cooperated to induce potent growth arrest of these cells (Fig. 3A, left panel). We examined several biochemical events associated with TGF-β-mediated growth arrest, including Rb dephosphorylation, E2F1 downregulation, and c-Myc downregulation to investigate the mechanisms by which TGF-β plus rapamycin induced growth arrest. The control cells used in these experiments (Puro. Cont.) were prepared by infecting Mv1Lu cells with the pBabe-Puro retrovirus (53), followed by selection with puromycin. When the control cells were treated with TGF-β, Rb dephosphorylation, E2F1 downregulation, and c-Myc downregulation were observed (Fig. 3A, right panel). Rapamycin did not significantly induce any of these changes. Interestingly, combined treatment with TGF-β plus rapamycin induced a more complete Rb dephosphorylation, E2F1 downregulation, and c-Myc downregulation than treatment with TGF-β alone. These results are consistent with rapamycin potentiating TGF-β-induced growth arrest. In the E2F1-transformed cell line, the inability of TGF-β alone to induce growth arrest correlated with the lack of Rb dephosphorylation, E2F1 downregulation, or c-Myc downregulation. As with the control cells, rapamycin alone had minimal effects on these biochemical endpoints. In contrast, TGF-β plus rapamycin induced potent Rb dephosphorylation in the E2F1-transformed cells. E2F1 was overexpressed under all four sets of treatment conditions relative to the control cells, as was c-Myc. E2F1-induced c-Myc overexpression is likely due to the presence of an E2F site within the c-Myc promoter (32, 52). The induction of growth arrest by TGF-β plus rapamycin in the absence of E2F1 and c-Myc downregulation suggests that these events are not essential to the antiproliferative effects of TGF-β plus rapamycin. We directly examined whether rapamycin could restore TGF-β-induced growth arrest in c-Myc transformed cells since c-Myc downregulation is essential for TGF-β-induced growth arrest (5).
FIG. 3.
Rapamycin cooperates with TGF-β to induce growth arrest of cells transformed by E2F1, c-Myc, and V12H-Ras. (A) Mv1Lu cells transformed with E2F1 (E2F1-13) were treated for 24 h with normal growth medium (C), 10 ng of TGF-β/ml (T), 100 nM rapamycin (R), or 10 ng of TGF-β/ml plus 100 nM rapamycin (T + R). The cells were then subjected to [3H]thymidine incorporation assays as for Fig. 1 (left panel), or cell extracts were subjected to immunoblot analyses using antibody to total Rb, Rb phosphorylated on Ser249/Thr252 [P-Rb(S249/T252)], Rb phosphorylated on Thr821[P-Rb(T821)], E2F1, or c-Myc or β-tubulin as a loading control (right panel). Results of [3H]thymidine incorporation assays are presented as the average percent control incorporation of six replicate determinations ± standard deviation. (B) Mv1Lu cells transformed with c-Myc (c-Myc-5) were subjected to [3H]thymidine incorporation analyses (left panel) or immunoblot analyses (right panel) as for panel A. (C) Mv1Lu cells transformed with c-Myc2(Ala58) [c-Myc(58A)-7] were subjected to [3H]thymidine incorporation analyses (left panel) or immunoblot analyses (right panel) as for panel A. (D) Mv1Lu cells transformed with V12H-Ras (Ras-9) were subjected to [3H]thymidine incorporation analyses (left panel) or immunoblot analyses (right panel) as for panel A.
c-Myc-transformed cells were largely resistant to TGF-β-induced growth arrest as observed previously (5) and were slightly inhibited by rapamycin treatment but were more strongly growth arrested by combined treatment with TGF-β plus rapamycin (Fig. 3B, left panel). Biochemical analyses indicated that, in the c-Myc-transformed cells, TGF-β did not induce Rb dephosphorylation, E2F1 downregulation, or c-Myc downregulation. Rapamycin did not induce a significant downregulation of c-Myc in nontransformed Mv1Lu cells, in contrast to previously published results in BALB/MK mouse keratinocytes (43). Interestingly, however, rapamycin consistently induced c-Myc downregulation in several different Mv1Lu clones overexpressing c-Myc, including clone 5 (Fig. 3B). These results suggest that rapamycin-induced c-Myc downregulation may depend on c-Myc expression level or the cellular context. We constructed Mv1Lu cells transformed by the Thr58-to-Ala (T58A) mutant of c-Myc to more conclusively rule out a role for c-Myc downregulation in TGF-β-rapamycin-induced growth arrest.
The c-Myc(T58A) mutation is observed in human lymphomas (8, 87). The Thr58-to-Ala mutation of c-Myc [c-Myc(T58A)] renders the mutant protein resistant to ubiquitin-mediated degradation by preventing phosphorylation of Thr58, producing a Myc protein with a significantly longer half-life (6, 69, 72). We reasoned that expression of the c-Myc(T58A) mutant might prevent rapamycin from cooperating with TGF-β to induce growth arrest since c-Myc overexpression may decrease sensitivity to rapamycin-induced growth arrest (34), as well as TGF-β-induced growth arrest (5), and since rapamycin regulates c-Myc at the level of protein abundance (82). As expected, transformation of Mv1Lu cells by c-Myc2(T58A) abolished TGF-β-induced arrest (Fig. 3C). Surprisingly, c-Myc2(T58A) expression blocked rapamycin-induced growth arrest but did not alter the ability of rapamycin to cooperate with TGF-β to induce growth arrest. Biochemical analyses indicated that, under conditions where TGF-β plus rapamycin cooperated to induce growth arrest, TGF-β plus rapamycin did not induce Rb dephosphorylation or E2F1 or c-Myc downregulation. These results indicate that Rb dephosphorylation and E2F1 and c-Myc downregulation are dispensable for growth arrest induced by TGF-β plus rapamycin in c-Myc2(T58A)-transformed cells and perhaps more generally.
Similar to c-Myc, the Ras oncogene is often activated during human tumorigenesis and abolishes TGF-β-induced growth arrest (35, 38, 63). It was important therefore to examine whether TGF-β and rapamycin cooperate to inhibit the proliferation of Ras-transformed cells. Transformation of Mv1Lu cells by V12H-Ras largely blocked TGF-β-mediated growth arrest, as well as partially overriding rapamycin-induced growth arrest (Fig. 3D). TGF-β plus rapamycin did not induce growth arrest as effectively in the V12H-Ras-transformed cells as in the E2F1- or c-Myc2(T58A)-transformed cells; however, TGF-β-rapamycin treatment inhibited proliferation to a greater extent than either treatment alone. No Rb dephosphorylation or downregulation of E2F1 or c-Myc was observed under conditions where TGF-β plus rapamycin inhibited proliferation by nearly 60%. Together, the results in Fig. 3C and D suggest that TGF-β plus rapamycin may induce growth arrest of transformed cells through a mechanism distinct from that employed by TGF-β in nontransformed cells, since TGF-β-rapamycin-mediated growth arrest can occur in the absence of classically observed TGF-β cell cycle effects. TGF-β did not increase the sensitivity of cells to rapamycin inhibition of the mTOR pathway. Rapamycin alone completely inhibited p70s6k phosphorylation and partially inhibited PHAS1 phosphorylation, and cotreatment with TGF-β plus rapamycin did not cause any further decrease in p70s6k or PHAS1 phosphorylation in all of the nontransformed and transformed cell lines examined (data not shown).
Rapamycin cooperation with TGF-β to induce growth arrest is mediated by rapamycin binding to FK506 binding protein(s).
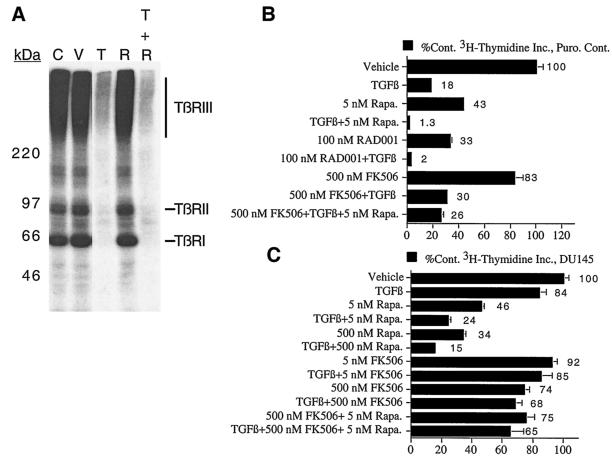
We next examined the effect of rapamycin on TGF-β-mediated signaling events to more fully understand the mechanisms by which rapamycin cooperates with TGF-β to induce growth arrest. We examined whether rapamycin alters 125I-TGF-β ligand binding to cell surface receptors in affinity cross-linking experiments (Fig. 4A), since many cancers exhibit decreased levels of TβRI or TβRII (64). Rapamycin did not significantly alter TGF-β binding to TβRI, TβRII, or TβRIII relative to either untreated controls or vehicle controls in MDA-MB-231 mammary carcinoma cells. Identical results were also obtained in similar experiments employing DU145 cells (data not shown), suggesting that rapamycin does not augment TGF-β signaling by potentiating TGF-β binding to its receptors.
FIG. 4.
Rapamycin potentiation of TGF-β-induced growth arrest is mediated by rapamycin binding to FK506 binding protein(s). (A) MDA-MB-231 cells were treated for 24 h with normal growth medium (C), dimethyl sulfoxide vehicle (V), 10 ng of TGF-β/ml (T), 100 nM rapamycin (R), or 10 ng of TGF-β/ml plus 100 nM rapamycin (T + R), and 125I-TGF-β affinity cross-linking experiments were performed as described in Materials and Methods. Puro. Cont. cells (B) or DU145 cells (C) were treated with 10 ng of TGF-β/ml and various combinations of rapamycin, the Novartis rapamycin derivative RAD001, or FK506 for 24 h as indicated, and [3H]thymidine incorporation assays were performed as for Fig. 2.
Previous studies indicating that FKBP12 binds and inhibits the function of TβRI and that FK506 and rapamycin induce the release of FKBP12 from the receptor (4, 15, 62, 77, 80) raise the possibility that rapamycin cooperates with TGF-β to induce growth arrest by reversing FKBP12-mediated inhibition of TβRI. If this hypothesis is correct, then FK506 should also cooperate with TGF-β to induce growth arrest to an extent similar to that observed with rapamycin. This hypothesis was directly tested by performing [3H]thymidine incorporation experiments in cells treated with various combinations of TGF-β, rapamycin, and FK506 (Fig. 4B and C). As expected, rapamycin cooperated with TGF-β to induce growth arrest in both the nontransformed Mv1Lu (Puro. Cont.) and DU145 human prostatic carcinoma cell lines. The Novartis rapamycin derivative RAD001, like rapamycin, cooperated with TGF-β to inhibit cell proliferation (Fig. 4B). FK506, however, did not cooperate with TGF-β to induce growth arrest in either the Mv1Lu cells or the DU145 cells. Rapamycin and FK506 are structurally similar and bind to the same binding pocket on FKBP12 and thus compete with each other for FKBP12 binding. Consequently, 500 nM FK506 not only did not cooperate with TGF-β to induce growth arrest in the Mv1Lu or DU145 cells but also blocked the ability of rapamycin to augment TGF-β-induced growth arrest. Together, the results in Fig. 4B and C suggest that rapamycin does not accentuate TGF-β-induced growth arrest by releasing FKBP12 from TβRI but that rapamycin cooperates with TGF-β signaling to induce growth arrest by binding to a FK506 binding protein(s).
TGF-β and rapamycin cooperate to induce growth arrest of human carcinoma cells.
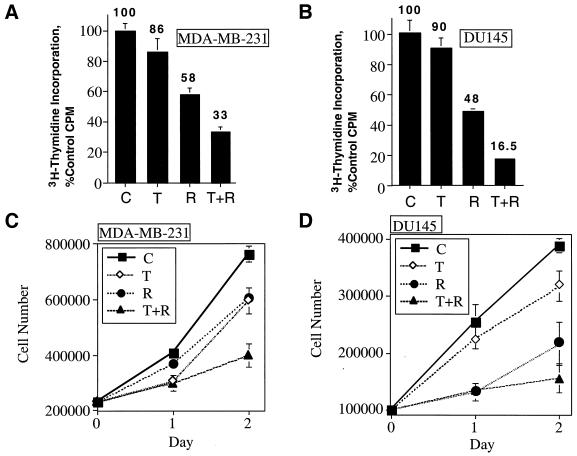
TGF-β and rapamycin cotreatment induced growth arrest of cells transformed by three different oncogenes, suggesting the possibility that TGF-β plus rapamycin may be effective in inhibiting the proliferation of tumor cells that harbor a variety of oncogenic lesions. MDA-MB-231 human mammary carcinoma cells and DU145 human prostatic carcinoma cells were therefore treated with TGF-β, rapamycin, or TGF-β plus rapamycin and were subjected to [3H]thymidine incorporation assays (Fig. 5A and B) or sequential cell-counting assays (Fig. 5C and D) to test the hypothesis that TGF-β and rapamycin cooperate to inhibit the proliferation of human carcinomas. The results of both assays indicate that TGF-β and rapamycin cooperate to inhibit the proliferation of these two human carcinoma cell lines.
FIG. 5.
TGF-β and rapamycin cooperate to inhibit the proliferation of human carcinoma cells. MDA-MB-231 (A) or DU145 human carcinoma (B) cells were subjected to [3H]thymidine incorporation assays as for Fig. 2. The values are shown over the bars. MDA-MB-231 (C) or DU145 (D) cells were plated as described in Materials and Methods and treated as for Fig. 4A and B, and cells were counted on sequential days using a Coulter counter. Results are presented as the average of triplicate determinations ± standard deviation.
We screened a number of different nontransformed cell lines and human carcinomas for their sensitivity to TGF-β, rapamycin, or TGF-β plus rapamycin (Table 1) to determine whether TGF-β and rapamycin cooperate to induce growth arrest in cells of epithelial origin in a more general way. TGF-β and rapamycin cooperated to induce a degree of growth arrest that was significantly greater than that induced by either agent alone in all epithelial cell lines tested. NIH 3T3 fibroblasts, however, which are not growth arrested by TGF-β, exhibited a similar level of growth inhibition in the presence of rapamycin or TGF-β plus rapamycin. Although the differences in thymidine incorporation observed were statistically significant (P = 0.017), the magnitude of the difference observed was small (51 versus 58% of control), suggesting that TGF-β and rapamycin do not cooperate to potently inhibit the proliferation of all cell types. Significantly, the combined effect of TGF-β and rapamycin was cytostatic, since no detectable cell death was apparent, either visually or as a sub-G0/G1 2N DNA content peak in flow cytometry experiments in any of the cell lines examined. In addition, the proliferation of Mv1Lu cells was not inhibited by BMP4, BMP6, or BMP7 alone. Further, BMPs did not cooperate with rapamycin to inhibit cell proliferation (data not shown). Together, these results indicate that rapamycin-mediated potentiation of growth arrest is not a general phenomenon observed with all TGF-β superfamily members.
TABLE 1.
TGF-β and rapamycin cooperate to induce growth arrest of multiple nontransformed epithelial cell lines and human carcinoma cell linesa
| Cell line (description) | % control [3H]thymidine incorporation
|
P | ||
|---|---|---|---|---|
| T | R | T + R | ||
| Mv1Lu (mink lung epithelial cells) | 20 | 41 | 1 | <0.0001 |
| HaCaT (human keratinocytes) | 53 | 54 | 19 | 0.03 |
| BALB/MK (mouse keratinocytes) | 34 | 28 | 1 | <0.0001 |
| NMuMG (mouse mammary epithelial cells) | 54 | 76 | 20 | <0.0001 |
| MCF10A (human mammary epithelial cells) | 58 | 49 | 11 | <0.0001 |
| MDA-MB-231 (human mammary carcinoma cells) | 86 | 58 | 33 | 0.0016 |
| DU145 (human prostatic carcinoma cells) | 90 | 48 | 17 | <0.0001 |
| A549 (human lung carcinoma cells) | 96 | 57 | 15 | 0.006 |
| NIH OVCAR-3 (human ovarian carcinoma cells) | 43 | 55 | 11 | <0.0001 |
| NIH 3T3 (mouse fibroblasts) | 106 | 58 | 51 | 0.017 |
The cell lines indicated were treated for 24 h with either 10 ng of TGF-β (T)/ml, 100 nM rapamycin (R), or 10 ng of TGF-β/ml plus 100 nM rapamycin (T + R). In each case, [3H]thymidine incorporation in normal growth medium was set at 100% (not shown) and the incorporation observed in the treated cells was presented as percentage of control [3H]thymidine incorporation. The results presented are the average of six replicate determinations. In all cases the differences between treatment with T + R and treatment with T or with R are statistically significant (P < 0.05) using Student's unpaired t test. The P values shown represent the difference between T + R treatment and the single-drug treatment (T or R) with the value closest to that of T + R treatment.
Effects of TGF-β and rapamycin on proliferation-related signaling pathways in DU145 cells.
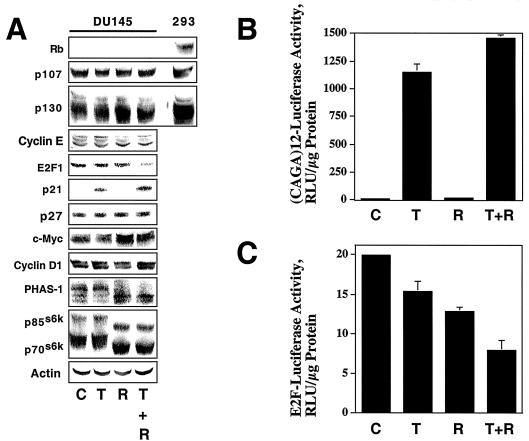
DU145 cells were treated with TGF-β and rapamycin, alone or in combination, and the resulting cell extracts were analyzed by immunoblotting (Fig. 6A) to determine the reasons for the lack of responsiveness of DU145 cells to TGF-β and the enhanced responsiveness to TGF-β in the presence of rapamycin. Surprisingly, TGF-β induced p21 expression even in the absence of rapamycin, but downregulation of E2F1, a well-characterized cell cycle response to TGF-β (46, 73), was observed only upon treatment with both TGF-β and rapamycin. Rapamycin treatment caused p70s6k and PHAS1 dephosphorylation as evidenced by shifts in electrophoretic mobility, and combined treatment with rapamycin and TGF-β had no additional effect on p70s6k or PHAS1 phosphorylation. These results suggest that, while rapamycin potentiates some biochemical responses to TGF-β, TGF-β does not potentiate rapamycin effects on p70s6k and PHAS1 phosphorylation. Interestingly, while rapamycin has been demonstrated (27, 29, 34, 43, 82) to induce cyclin D1 and c-Myc downregulation and while TGF-β induces c-Myc and cyclin D1 downregulation in some cell types (41, 56, 67, 68), neither rapamycin nor TGF-β altered c-Myc or cyclin D1 levels in DU145 cells, indicating that the growth-inhibitory effects of TGF-β plus rapamycin are independent of changes in c-Myc or cyclin D1 levels.
FIG. 6.
DU145 cells exhibit TGF-β-dependent activation of Smad signaling. (A) DU145 cells were treated for 24 h with normal growth medium (C), 10 ng of TGF-β1/ml (T), 100 nM rapamycin (R), or 10 ng of TGF-β1/ml plus 100 nM rapamycin (T + R), and immunoblotting experiments on whole-cell extracts were performed using antibodies specific for the indicated proteins. Total extracts from HEK 293 (293) cells were used as a positive control in Rb, p107, and p130 immunoblotting experiments. (B) DU145 cells were treated as for panel A, and (CAGA)12-luciferase assays were performed as described in Materials and Methods. Assays were performed in triplicate, and results are presented as relative luciferase units (RLU) normalized to micrograms of protein assayed. (C) Luciferase assays were performed using an E2F-luciferase reporter construct, and the results are presented as for panel B.
The observation that TGF-β treatment induced p21 suggested that TGF-β-stimulated signaling pathways are intact in the DU145 cells but are uncoupled from the cell cycle. Since TGF-β-dependent induction of p21 and p15 and repression of c-Myc are mediated in part by the Smad transcription factors (14, 86), we examined the effect of TGF-β and rapamycin on the Smad-dependent transcriptional reporter construct (CAGA)12-luc (20). As shown in Fig. 6, TGF-β potently stimulated Smad-dependent transcription, rapamycin was without effect, and the combination of TGF-β plus rapamycin stimulated transcription only slightly more than TGF-β alone. These results were similar to the effects observed with endogenous p21 levels.
We performed transcriptional reporter assays using an E2F-luciferase reporter construct (Fig. 6C) to verify that the decrease in E2F1 levels in Fig. 6A, lane T + R, correlated with a decrease in E2F-dependent transcription. The results suggest that, while TGF-β and rapamycin can each partially inhibit E2F-dependent transcription, the combination of both agents is more effective at inhibiting E2F-dependent transcription, consistent with their cooperative effect in downregulating endogenous E2F1. Additional experiments demonstrated that downregulation of the E2F1 protein correlated with a decrease in E2F1 mRNA (data not shown). Together, these results indicate that TGF-β activates the Smad signaling pathway in DU145 cells. Activation of this pathway in DU145 carcinoma cells is not sufficient to induce growth arrest, even though the Smad pathway mediates growth arrest in nontransformed cells (48).
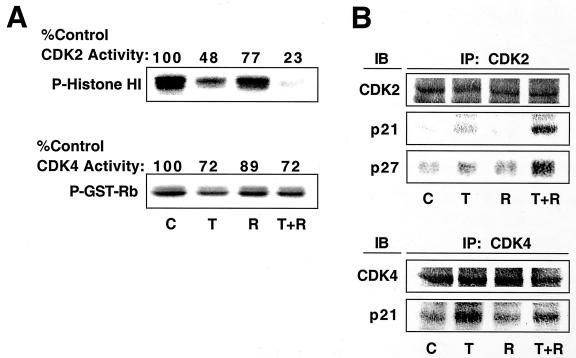
TGF-β-rapamycin-induced arrest of DU145 cells correlates with Cdk2 inhibition and increased p21 and p27 binding to Cdk2.
DU145 cells were previously shown to be refractory to growth arrest by TGF-β and to lack TGF-β inhibition of Cdk2 activity (16). We hypothesized that TGF-β and rapamycin would cooperate to inhibit Cdk2 activity since combined treatment of DU145 cells with TGF-β and rapamycin induced a growth arrest (Fig. 4). As demonstrated in Fig. 7A, TGF-β did not strongly inhibit Cdk4 activity in DU145 cells and only partially inhibited Cdk2 activity. In contrast, treatment with both TGF-β and rapamycin strongly inhibited Cdk2 but not Cdk4 activity. Analysis of the kinase assay immunoprecipitates by immunoblotting (Fig. 7B) showed that the increased Cdk2 inhibition observed upon treatment with both TGF-β and rapamycin correlated with an increased association with the cyclin-dependent kinase inhibitors p21 and p27. These data suggest that TGF-β and rapamycin cooperate to inhibit cell proliferation specifically by inhibiting Cdk2 kinase activity.
FIG. 7.
TGF-β and rapamycin cooperate to inhibit Cdk2 activity in DU145 cells. (A) DU145 cells were treated as for Fig. 6, and cell extracts were prepared and subjected to immune-complex kinase assays using Cdk2 or Cdk4 antibody as described in Materials and Methods. GST, glutathione transferase. (B) A portion of the kinase assays was subjected to immunoblot (IB) analysis using antibody specific for Cdk4, Cdk2, p21, or p27. IP, immunoprecipitation.
TGF-β-rapamycin inhibition of Cdk2 activity in NMuMG cells correlates with increased p27 binding to Cdk2 in the absence of changes in total p27 levels.
We performed similar experiments with the nontransformed NMuMG cell line to determine whether the cooperative inhibition of Cdk2 by TGF-β and rapamycin occurred in other cell types (Fig. 8). As with the DU145 cells, TGF-β partially inhibited Cdk2 activity and TGF-β plus rapamycin inhibited Cdk2 activity more effectively. Interestingly, immunoblots of whole-cell extracts indicated that, while none of the treatments significantly altered total p21 or p27 levels (Fig. 8B), TGF-β increased the amount of both p21 and p27 that coprecipitated with Cdk2 in the immune-complex kinase assays. Combined treatment with TGF-β and rapamycin further increased the amount of p27 but decreased the amount of p21 that coprecipitated with Cdk2, suggesting that, under these conditions, Cdk2 inhibition correlates better with p27 association than with p21 association.
FIG. 8.
TGF-β and rapamycin cooperate to inhibit Cdk2 activity in NMuMG cells. (A) NMuMG cells were treated as in Fig. 7, and cell extracts were prepared and subjected to immune-complex kinase assays using Cdk2 or Cdk4 antibody or normal rabbit IgG (Rab. IgG) as a control (top panel). A portion of the kinase assays was subjected to immunoblot (IB) analysis using antibody to Cdk2, p21, or p27 (lower panels). IP, immunoprecipitation. (B) Total protein extracts from cells treated as for panel A were analyzed by immunoblot with antibody specific for p21 or p27 or actin as a loading control.
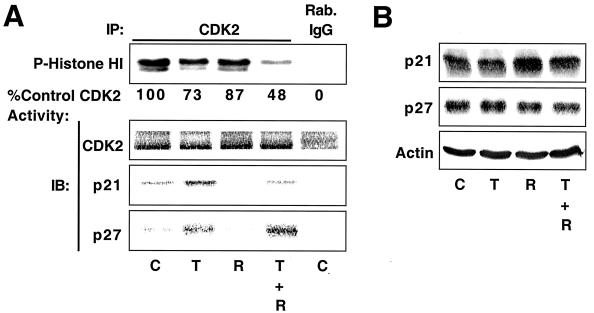
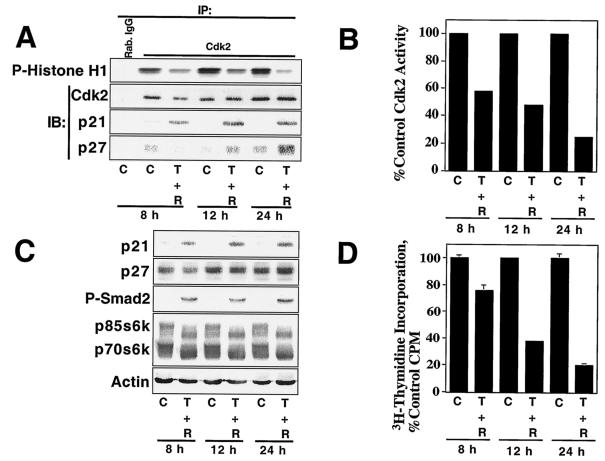
Time-dependent inhibition of DU145 cell proliferation by TGF-β plus rapamycin correlates with inhibition of Cdk2 activity and increased p27 binding to Cdk2.
We examined the time course of Cdk2 inhibition, since Smad2 phosphorylation induced by TGF-β and p70s6k and PHAS1 dephosphorylation induced by rapamycin occur within minutes, while cell cycle arrest generally takes several hours. As indicated in Fig. 9A and B, Cdk2 inhibition by TGF-β plus rapamycin was apparent at 8 h and increasingly more dramatic at 12 and 24 h. p21 association with Cdk2 was observed within 8 h, but the further inhibition of kinase activity observed at 12 and 24 h correlated with increased p27 binding, while the amount of p21 associated with Cdk2 increased only slightly.
FIG. 9.
TGF-β plus rapamycin inhibit Cdk2 activity in DU145 cells in a time-dependent manner. (A) DU145 cells were treated with normal growth medium (C) or 10 ng of TGF-β1/ml plus 100 nM rapamycin (T + R) for the indicated periods, and cell extracts were prepared and subjected to immune-complex kinase assays as for Fig. 7. IP, immunoprecipitation; IB, immunoblotting. The results of kinase assays were quantitated and plotted (B) with the control Cdk2 activity at each time point normalized to 100%. (C) Total protein extracts from cells treated as for panel A were analyzed by immunoblotting with antibodies specific for the indicated proteins. (D) Cells treated as for panel A were pulsed with [3H]thymidine for the last 2 h of each time point, and [3H]thymidine incorporation into DNA was quantitated and presented as the average of six replicate determinations ± standard deviation.
Immunoblot analysis of total cell extracts (Fig. 9C) showed that levels of p21- and TGF-β-receptor-phosphorylated Smad2 were near maximal at 8 h, demonstrating that TGF-β activates the Smad signaling pathway in DU145 cells in a rapid and sustained manner. In other experiments, TGF-β induced Smad2 phosphorylation within 15 min and p21 upregulation within 2 h and rapamycin-induced p70s6k and PHAS1 dephosphorylation was apparent within 15 min. Substantial inhibition of Cdk2 activity however, was not observed until later time points (≥8 h) (data not shown). Rapamycin-dependent dephosphorylation of p70s6k and p85s6k, which are alternatively translated protein products of the same mRNA (70), was near maximal at 8 h. Total p27 levels did not dramatically change over the time course of the experiment, but p27 association with Cdk2 increased at 12 and 24 h following treatment. These results suggest that the time-dependent inhibition of Cdk2 activity by TGF-β plus rapamycin correlates best with increased association of p27 with Cdk2 but does not correlate with changes in the total levels of p27 and does not occur with the same kinetics as p21 induction, Smad2 phosphorylation, and p70s6k dephosphorylation. Significantly, the inhibition of cell proliferation by TGF-β plus rapamycin, as measured by [3H]thymidine incorporation, correlated with inhibition of Cdk2 activity in a time-dependent manner (Fig. 9D).
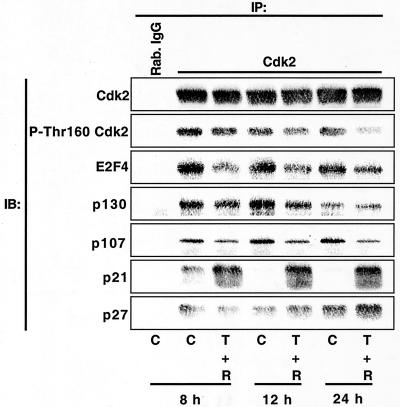
TGF-β plus rapamycin induces dissociation of E2F4:p107/p130 complexes from Cdk2 and dephosphorylation of Cdk2 at Thr160.
The results in Fig. 9 suggested that TGF-β-rapamycin-induced growth arrest results from a time-dependent change in the composition of Cdk2 complexes. Larger-scale immunoprecipitation experiments were performed followed by immunoblot analyses (Fig. 10) to more fully characterize changes in Cdk2 complexes. As in Fig. 9, TGF-β plus rapamycin induced a rapid association of p21 with Cdk2, followed by increased p27 association at later time points. Interestingly, TGF-β-rapamycin treatment induced dissociation of E2F4, p130, and p107 from Cdk2. These results are consistent with previous results showing that p107 binding to Cdk2 is mutually exclusive with p21 binding (3, 90) and that, while p130 and p107 are capable of inhibiting Cdk2 activity (18, 19, 83, 90), p130, p107, and E2F4 are present in Cdk2 complexes in rapidly growing DU145 cells. We examined the phosphorylation of Cdk2 on Thr160 using phosphorylation-state-specific antibodies, since p27 binding to cyclin A-Cdk2 complexes was previously shown to inhibit the activating phosphorylation of Cdk2 on Thr160 by cyclin-dependent kinase activating kinase. These experiments showed that the time-dependent inhibition of Cdk2 activity (Fig. 9) correlates with a time-dependent dephosphorylation of Cdk2 at Thr160. Thus, inhibition of Cdk2 activity by TGF-β plus rapamycin may result not only from direct inhibition by p21 and p27 but also indirectly from dephosphorylation at Thr160. Dephosphorylation at Thr160 could potentially result from (i) prevention of Cdk2 phosphorylation by p21 and/or p27 association, (ii) inhibition of cyclin-dependent kinase activating kinase toward Cdk2, as occurs in HepG2 cells in response to TGF-β treatment (57), or (iii) an increased rate of dephosphorylation at Thr160 by kinase-associated phosphatase. Together, these results suggest that combined treatment with TGF-β and rapamycin alters the composition of Cdk2 complexes in a complicated, time-dependent manner, ultimately culminating in the inhibition of Cdk2 kinase activity and inhibition of E2F-dependent transcription.
FIG. 10.
TGF-β plus rapamycin alter the composition of Cdk2 complexes in a time-dependent manner. DU145 cells were treated as for Fig. 9, and cell extracts were subjected to immunoprecipitation (IP) using Cdk2 antibodies or normal rabbit IgG (Rab. IgG). Immunoprecipitates were analyzed by immunoblotting (IB) with antibody specific for Cdk2, Cdk2 phosphorylated on Thr160, E2F4, p130, p107, p21, or p27.
DISCUSSION
Evidence has been accumulating for some time that TGF-β acts as an endogenous suppressor of tumorigenesis and that the tumor suppressor function of TGF-β is inactivated in carcinomas. The development of therapeutic approaches to reactivate or mimic the TGF-β tumor suppressor pathway in cancers, however, is in its infancy. In this report we demonstrate that rapamycin potentiates TGF-β-induced growth arrest in nontransformed cells, partially restores TGF-β-induced biochemical responses and growth arrest in oncogene-transformed cells, and cooperates with TGF-β to induce growth inhibition of several human carcinoma cell lines.
Rapamycin and TGF-β inhibit cell proliferation in an additive manner in many of the experiments presented. However, biochemical analyses indicate that rapamycin potentiates TGF-β responses, while TGF-β does not alter rapamycin responses. For example, TGF-β induces a partial dephosphorylation of Rb in NMuMG (Fig. 2) and combined treatment with TGF-β and rapamycin induces a more complete Rb dephosphorylation, while rapamycin alone has no effect on Rb phosphorylation status. In contrast, TGF-β does not augment the ability of rapamycin to induce p70s6k or PHAS1 phosphorylation (Fig. 6). Defining the interaction between TGF-β and rapamycin-sensitive signaling as either additive or cooperative is also complicated because rapamycin induces growth arrest through mechanisms independent of effects on TGF-β signaling. For example T47D human mammary carcinoma cells lack functional TGF-β receptors yet are still potently growth inhibited by rapamycin (data not shown). Thus, due to the biochemical complexity involved in the interactions between TGF-β and rapamycin and our incomplete knowledge of the mechanisms of TGF-β and rapamycin-induced cell cycle arrest, the interaction between TGF-β and rapamycin does not strictly fit the classical definition of an additive, synergistic, or cooperative interaction.
Our results indicate that the ability of rapamycin to augment TGF-β-induced antiproliferative responses is not due to alterations in TGF-β receptor levels or FKBP12 binding to TβRI. Rapamycin restores TGF-β-induced growth arrest to various degrees in cells transformed by the E2F1, c-Myc, c-Myc2(T58A), and V12H-Ras oncogenes. In E2F1-and c-Myc transformed cells, rapamycin restoration of TGF-β-induced growth arrest is associated with Rb dephosphorylation. Rapamycin cooperates with TGF-β to induce growth arrest in the absence of Rb dephosphorylation or E2F1 or c-Myc downregulation in c-Myc2(T58A)- and V12H-Ras-transformed cells. These observations, combined with the finding that TGF-β and rapamycin cooperate to inhibit the proliferation of DU145 cells, which lack functional Rb (10, 79, 89), suggest that Rb dephosphorylation is dispensable for TGF-β-rapamycin-induced growth arrest. The fact that TGF-β and rapamycin cooperate to inhibit the proliferation of c-Myc2(T58A)- and V12H-Ras- transformed Mv1Lu cells in the absence of the hallmarks of TGF-β-induced growth arrest, including Rb dephosphorylation and E2F1 and c-Myc downregulation, suggests that TGF-β plus rapamycin may inhibit cell proliferation through a novel mechanism. The data in Fig. 5 and Table 1 suggest that this putative novel mechanism of growth arrest may extend to human carcinoma cells.
Analysis of the mechanisms of TGF-β-rapamycin-induced growth arrest in DU145 carcinoma cells shows that growth inhibition is associated with inhibition of Cdk2 activity but not of Cdk4 activity. Inhibition of Cdk2 activity by TGF-βplus rapamycin is associated with increased p21 and p27 binding to Cdk2 and dephosphorylation of Cdk2 at Thr160. The ability of TGF-β and rapamycin to cooperate to inhibit Cdk2 activity is interesting in light of previous observations that the ability of Ras and Myc to cooperatively transform cells and induce tumorigenesis in mouse models correlates with their ability to stimulate Cdk2 activity and downregulate p27 (45).
Our results suggesting that rapamycin cooperation with TGF-β to inhibit Cdk2 activity involves the cyclin-dependent kinase inhibitor p27 in the TGF-β resistant DU145 carcinoma cells are consistent with previous reports demonstrating that rapamycin blocks interleukin-2-induced mitogenesis of T cells by preventing Cdk2 activation (61) and that the lack of TGF-β-induced growth arrest in Ras-transformed Mv1Lu cells is due to p27 mislocalization to the cytoplasm (49). In addition, p27 phosphorylation and localization were shown to differ between TGF-β- resistant and TGF-β-sensitive human mammary epithelial cell lines. Together these results suggest that p27 plays an important role in TGF-β-induced growth arrest; however, the exact part that p27 plays in TGF-β responses is likely to be complicated. T cells from p27-null mice respond normally to both TGF-β- and rapamycin-induced growth arrest (58). More recently, however it has been shown that, in the absence of both p27 and p21, p130 becomes upregulated and maintains normal cell cycle-dependent regulation of Cdk2 activity and in so doing compensates for the absence of p27 and p21 (19). Thus, the ability of TGF-β plus rapamycin to induce growth arrest in transformed cells and cancer cells suggests that the combined signaling effects of these agents may activate growth arrest mechanisms that are redundant in normal cells. Knowledge of the biochemical mechanisms by which TGF-β plus rapamycin induce growth arrest in carcinomas may be important in the development of novel therapeutic strategies against cancer, since one or more of the normal growth arrest pathways is lost in cancer cells (75).
The observation that TGF-β-rapamycin treatment of DU145 cells induces dissociation of p130, p107, and E2F4 from Cdk2 in parallel with increased p21 and p27 binding suggests that TGF-β plus rapamycin may regulate Cdk2 activity and E2F-dependent transcription simultaneously. It was proposed recently that p107 acts as a scaffolding protein facilitating the formation of a complex containing Cdk2, cyclin A, E2F4, and DP1 (90). This complex was proposed to sequester E2F4 in an inactive form until Cdk2 becomes activated, phosphorylates p107, and releases E2F4-DP1 complexes capable of activating transcription. If this model is correct, then the induction of increased p21 and p27 binding to Cdk2 may not only inhibit Cdk2 activity but may also keep E2F4 and DP1 in transcriptionally inactive or transcriptionally repressive complexes with p107 or p130. According to this model, TGF-β plus rapamycin would simultaneously inhibit Cdk2 activity and E2F-dependent transcription, both of which are required for cell cycle progression (28, 51, 84, 85).
Overall, the data presented here suggest that rapamycin treatment may be effective in restoring the TGF-β tumor suppressor pathway in a subset of tumors. Such a therapy might be particularly effective against tumors that secrete autocrine TGF-β but are resistant to TGF-β-induced growth arrest because of oncogene activation. Many breast cancers and prostate cancers secrete autocrine TGF-β, and autocrine TGF-β is thought to play an important role in tumor progression. Thus, it is possible that rapamycin in addition to directly inhibiting tumor cell proliferation may also inhibit tumor growth by potentiating autocrine or paracrine TGF-β-induced growth arrest.
Acknowledgments
This work was supported by Public Health Service Grants CA42572 and CA85492 (to H.L.M.) and Vanderbilt-Ingram Cancer Center Support Grant CA68485 from the National Cancer Institute.
We thank Heidi Lane and Novartis Pharma AG for supplying the rapamycin derivative RAD001. We thank S. Hann for supplying retroviral vectors encoding c-Myc and c-Myc2(T58A) and P. Liang for a retroviral vector encoding V12H-Ras. S. Bagchi kindly supplied E2F1 cDNA. D. Rifkin, D. Lowe, and G. Nolan generously provided various cell lines. Flow cytometry analysis and data analyses were performed by Jim Price (Veterans Administration Hospital, Nashville, Tenn.).
REFERENCES
- 1.Abe, M., J. G. Harpel, C. N. Metz, I. Nunes, D. J. Loskutoff, and D. B. Rifkin. 1994. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276-284. [DOI] [PubMed] [Google Scholar]
- 2.Adjei, A. A. 2001. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 93:1062-1074. [DOI] [PubMed] [Google Scholar]
- 3.Afshari, C. A., M. A. Nichols, Y. Xiong, and M. Mudryj. 1996. A role for a p21-E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. 7:979-988. [PubMed] [Google Scholar]
- 4.Aghdasi, B., K. Ye, A. Resnick, A. Huang, H. C. Ha, X. Guo, T. M. Dawson, V. L. Dawson, and S. H. Snyder. 2001. FKBP12, the 12-kDa FK506-binding protein, is a physiologic regulator of the cell cycle. Proc. Natl. Acad. Sci. USA 98:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrow, M. G., M. Kawabata, M. Aakre, and H. L. Moses. 1995. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor beta 1. Proc. Natl. Acad. Sci. USA 92:3239-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahram, F., N. von der Lehr, C. Cetinkaya, and L. G. Larsson. 2000. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95:2104-2110. [PubMed] [Google Scholar]
- 7.Barrack, E. R. 1997. TGF beta in prostate cancer: a growth inhibitor that can enhance tumorigenicity. Prostate 31:61-70. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia, K., K. Huppi, G. Spangler, D. Siwarski, R. Iyer, and I. Magrath. 1993. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat. Genet. 5:56-61. [DOI] [PubMed] [Google Scholar]
- 9.Bhowmick, N. A., R. Zent, M. Ghiassi, M. McDonnell, and H. L. Moses. 2001. Integrin beta 1 signaling is necessary for transforming growth factor-beta activation of p38MAPK and epithelial plasticity. J. Biol. Chem. 276:46707-46713. [DOI] [PubMed] [Google Scholar]
- 10.Bookstein, R., J. Y. Shew, P. L. Chen, P. Scully, and W. H. Lee. 1990. Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science 247:712-715. [DOI] [PubMed] [Google Scholar]
- 11.Bottinger, E. P., J. L. Jakubczak, D. C. Haines, K. Bagnall, and L. M. Wakefield. 1997. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor beta receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res. 57:5564-5570. [PubMed] [Google Scholar]
- 12.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 13.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. R., Y. Kang, and J. Massague. 2001. Defective repression of c-myc in breast cancer cells: a loss at the core of the transforming growth factor beta growth arrest program. Proc. Natl. Acad. Sci. USA 98:992-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Y. G., F. Liu, and J. Massague. 1997. Mechanism of TGFbeta receptor inhibition by FKBP12. EMBO J. 16:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cipriano, S. C., and Y. Q. Chen. 1998. Insensitivity to growth inhibition by TGF-beta1 correlates with a lack of inhibition of the CDK2 activity in prostate carcinoma cells. Oncogene 17:1549-1556. [DOI] [PubMed] [Google Scholar]
- 17.Claassen, G. F., and S. R. Hann. 2000. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta-induced cell-cycle arrest. Proc. Natl. Acad. Sci. USA 97:9498-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coats, S., P. Whyte, M. L. Fero, S. Lacy, G. Chung, E. Randel, E. Firpo, and J. M. Roberts. 1999. A new pathway for mitogen-dependent cdk2 regulation uncovered in p27(Kip1)-deficient cells. Curr. Biol. 9:163-173. [DOI] [PubMed] [Google Scholar]
- 19.Collado, M., R. H. Medema, I. Garcia-Cao, M. L. Dubuisson, M. Barradas, J. Glassford, C. Rivas, B. M. Burgering, M. Serrano, and E. W. Lam. 2000. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J. Biol. Chem. 275:21960-21968. [DOI] [PubMed] [Google Scholar]
- 20.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudkin, L., M. B. Dilling, P. J. Cheshire, F. C. Harwood, M. Hollingshead, S. G. Arbuck, R. Travis, E. A. Sausville, and P. J. Houghton. 2001. Biochemical correlates of mTOR inhibition by the rapamycin ester CCI-779 and tumor growth inhibition. Clin. Cancer Res. 7:1758-1764. [PubMed] [Google Scholar]
- 22.Dumont, N., M. D. O'Connor-McCourt, and A. Philip. 1995. Transforming growth factor-beta receptors on human endometrial cells: identification of the type I, II, and III receptors and glycosyl-phosphatidylinositol anchored TGF-beta binding proteins. Mol. Cell. Endocrinol. 111:57-66. [DOI] [PubMed] [Google Scholar]
- 23.Feng, L. X., N. Ravindranath, and M. Dym. 2000. Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. J. Biol. Chem. 275:25572-25576. [DOI] [PubMed] [Google Scholar]
- 24.Feng, X. H., Y. Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 25.Geoerger, B., K. Kerr, C. B. Tang, K. M. Fung, B. Powell, L. N. Sutton, P. C. Phillips, and A. J. Janss. 2001. Antitumor activity of the rapamycin analog CCI-779 in human primitive neuroectodermal tumor/medulloblastoma models as single agent and in combination chemotherapy. Cancer Res. 61:1527-1532. [PubMed] [Google Scholar]
- 26.Goustin, A. S., E. B. Leof, G. D. Shipley, and H. L. Moses. 1986. Growth factors and cancer. Cancer Res. 46:1015-1029. [PubMed] [Google Scholar]
- 27.Grewe, M., F. Gansauge, R. M. Schmid, G. Adler, and T. Seufferlein. 1999. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res. 59:3581-3587. [PubMed] [Google Scholar]
- 28.Guadagno, T. M., and J. W. Newport. 1996. Cdk2 kinase is required for entry into mitosis as a positive regulator of Cdc2-cyclin B kinase activity. Cell 84:73-82. [DOI] [PubMed] [Google Scholar]
- 29.Hashemolhosseini, S., Y. Nagamine, S. J. Morley, S. Desrivieres, L. Mercep, and S. Ferrari. 1998. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J. Biol. Chem. 273:14424-14429. [DOI] [PubMed] [Google Scholar]
- 30.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 31.Hentges, K. E., B. Sirry, A. C. Gingeras, D. Sarbassov, N. Sonenberg, D. Sabatini, and A. S. Peterson. 2001. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc. Natl. Acad. Sci. USA 98:13796-13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiebert, S. W., M. Lipp, and J. R. Nevins. 1989. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc. Natl. Acad. Sci. USA 86:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang, A. T., B. Lutterbach, B. C. Lewis, T. Yano, T.-Y. Chou, J. F. Barrett, M. Raffeld, S. R. Hann, and C. V. Dang. 1995. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol. Cell. Biol. 15:4031-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosoi, H., M. B. Dilling, L. N. Liu, M. K. Danks, T. Shikata, A. Sekulic, R. T. Abraham, J. C. Lawrence, Jr., and P. J. Houghton. 1998. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 54:815-824. [DOI] [PubMed] [Google Scholar]
- 35.Howe, P. H., S. F. Dobrowolski, K. B. Reddy, and D. W. Stacey. 1993. Release from G1 growth arrest by transforming growth factor beta 1 requires cellular ras activity. J. Biol. Chem. 268:21448-21452. [PubMed] [Google Scholar]
- 36.Jo, H., R. Zhang, H. Zhang, T. A. McKinsey, J. Shao, R. D. Beauchamp, D. W. Ballard, and P. Liang. 2000. NF-kappa B is required for H-ras oncogene induced abnormal cell proliferation and tumorigenesis. Oncogene 19:841-849. [DOI] [PubMed] [Google Scholar]
- 37.Ju, W. D., T. J. Velu, W. C. Vass, A. G. Papageorge, and D. R. Lowy. 1991. Tumorigenic transformation of NIH 3T3 cells by the autocrine synthesis of transforming growth factor alpha. New Biol. 3:380-388. [PubMed] [Google Scholar]
- 38.Kerr, D. I., J. A. Plumb, R. I. Freshney, M. Z. Khan, and D. A. Spandidos. 1991. The effect of H-ras oncogene transfection on response of mink lung epithelial cells to growth factors and cytotoxic drugs. Anticancer Res. 11:1349-1352. [PubMed] [Google Scholar]
- 39.Keski-Oja, J., E. B. Leof, R. M. Lyons, R. J. Coffey, Jr., and H. L. Moses. 1987. Transforming growth factors and control of neoplastic cell growth. J. Cell. Biochem. 33:95-107. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura, T., M. Onishi, S. Kinoshita, A. Shibuya, A. Miyajima, and G. P. Nolan. 1995. Efficient screening of retroviral cDNA expression libraries. Proc. Natl. Acad. Sci. USA 92:9146-9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko, T. C., W. Yu, T. Sakai, H. Sheng, J. Shao, R. D. Beauchamp, and E. A. Thompson. 1998. TGF-beta1 effects on proliferation of rat intestinal epithelial cells are due to inhibition of cyclin D1 expression. Oncogene 16:3445-3454. [DOI] [PubMed] [Google Scholar]
- 42.Law, B. K., P. Norgaard, L. Gnudi, B. B. Kahn, H. S. Poulson, and H. L. Moses. 1999. Inhibition of DNA synthesis by a farnesyltransferase inhibitor involves inhibition of the p70(s6k) pathway. J. Biol. Chem. 274:4743-4748. [DOI] [PubMed] [Google Scholar]
- 43.Law, B. K., M. E. Waltner-Law, A. J. Entingh, A. Chytil, M. E. Aakre, P. Norgaard, and H. L. Moses. 2000. Salicylate-induced growth arrest is associated with inhibition of p70s6k and down-regulation of c-Myc, cyclin D1, cyclin A, and proliferating cell nuclear antigen. J. Biol. Chem. 275:38261-38267. [DOI] [PubMed] [Google Scholar]
- 44.Lenferink, A. E., D. Busse, W. M. Flanagan, F. M. Yakes, and C. L. Arteaga. 2001. ErbB2/neu kinase modulates cellular p27(Kip1) and cyclin D1 through multiple signaling pathways. Cancer Res. 61:6583-6591. [PubMed] [Google Scholar]
- 45.Leone, G., J. DeGregori, R. Sears, L. Jakoi, and J. R. Nevins. 1997. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387:422-426. (Erratum, 387:932.) [DOI] [PubMed] [Google Scholar]
- 46.Li, J. M., P. P. Hu, X. Shen, Y. Yu, and X. F. Wang. 1997. E2F4-RB and E2F4-p107 complexes suppress gene expression by transforming growth factor beta through E2F binding sites. Proc. Natl. Acad. Sci. USA 94:4948-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao, D. J., and R. B. Dickson. 2000. c-Myc in breast cancer. Endocr. Relat. Cancer 7:143-164. [DOI] [PubMed] [Google Scholar]
- 48.Liu, X., Y. Sun, S. N. Constantinescu, E. Karam, R. A. Weinberg, and H. F. Lodish. 1997. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl. Acad. Sci. USA 94:10669-10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, X., Y. Sun, M. Ehrlich, T. Lu, Y. Kloog, R. A. Weinberg, H. F. Lodish, and Y. I. Henis. 2000. Disruption of TGF-beta growth inhibition by oncogenic ras is linked to p27Kip1 mislocalization. Oncogene 19:5926-5935. [DOI] [PubMed] [Google Scholar]
- 50.Lorenz, M. C., and J. Heitman. 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270:27531-27537. [DOI] [PubMed] [Google Scholar]
- 51.Meraldi, P., J. Lukas, A. M. Fry, J. Bartek, and E. A. Nigg. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88-93. [DOI] [PubMed] [Google Scholar]
- 52.Moberg, K. H., T. J. Logan, W. A. Tyndall, and D. J. Hall. 1992. Three distinct elements within the murine c-myc promoter are required for transcription. Oncogene 7:411-421. [PubMed] [Google Scholar]
- 53.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moses, H. L., R. J. Coffey, Jr., E. B. Leof, R. M. Lyons, and J. Keski-Oja. 1987. Transforming growth factor beta regulation of cell proliferation. J. Cell. Physiol. Suppl. 5(Suppl.):1-7. [DOI] [PubMed] [Google Scholar]
- 55.Muise-Helmericks, R. C., H. L. Grimes, A. Bellacosa, S. E. Malstrom, P. N. Tsichlis, and N. Rosen. 1998. Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J. Biol. Chem. 273:29864-29872. [DOI] [PubMed] [Google Scholar]
- 56.Munger, K., J. A. Pietenpol, M. R. Pittelkow, J. T. Holt, and H. L. Moses. 1992. Transforming growth factor beta 1 regulation of c-myc expression, pRB phosphorylation, and cell cycle progression in keratinocytes. Cell Growth Differ. 3:291-298. [PubMed] [Google Scholar]
- 57.Nagahara, H., S. A. Ezhevsky, A. M. Vocero-Akbani, P. Kaldis, M. J. Solomon, and S. F. Dowdy. 1999. Transforming growth factor beta targeted inactivation of cyclin E:cyclin-dependent kinase 2 (Cdk2) complexes by inhibition of Cdk2 activating kinase activity. Proc. Natl. Acad. Sci. USA 96:14961-14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, and D. Y. Loh. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 59.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18:3004-3016. [DOI] [PubMed] [Google Scholar]
- 60.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nourse, J., E. Firpo, W. M. Flanagan, S. Coats, K. Polyak, M. H. Lee, J. Massague, G. R. Crabtree, and J. M. Roberts. 1994. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 372:570-573. [DOI] [PubMed] [Google Scholar]
- 62.Okadome, T., E. Oeda, M. Saitoh, H. Ichijo, H. L. Moses, K. Miyazono, and M. Kawabata. 1996. Characterization of the interaction of FKBP12 with the transforming growth factor-beta type I receptor in vivo. J. Biol. Chem. 271:21687-21690. [DOI] [PubMed] [Google Scholar]
- 63.Park, B. J., J. I. Park, D. S. Byun, J. H. Park, and S. G. Chi. 2000. Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signaling pathway in human prostate cancer. Cancer Res. 60:3031-3038. [PubMed] [Google Scholar]
- 64.Pasche, B. 2001. Role of transforming growth factor beta in cancer. J. Cell. Physiol. 186:153-168. [DOI] [PubMed] [Google Scholar]
- 65.Pasche, B., P. Kolachana, K. Nafa, J. Satagopan, Y. G. Chen, R. S. Lo, D. Brener, D. Yang, L. Kirstein, C. Oddoux, H. Ostrer, P. Vineis, L. Varesco, S. Jhanwar, L. Luzzatto, J. Massague, and K. Offit. 1999. TbetaR-I(6A) is a candidate tumor susceptibility allele. Cancer Res. 59:5678-5682. [PubMed] [Google Scholar]
- 66.Pierce, D. F., Jr., A. E. Gorska, A. Chytil, K. S. Meise, D. L. Page, R. J. Coffey, Jr., and H. L. Moses. 1995. Mammary tumor suppression by transforming growth factor beta 1 transgene expression. Proc. Natl. Acad. Sci. USA 92:4254-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pietenpol, J. A., K. Munger, P. M. Howley, R. W. Stein, and H. L. Moses. 1991. Factor-binding element in the human c-myc promoter involved in transcriptional regulation by transforming growth factor beta 1 and by the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 88:10227-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pietenpol, J. A., R. W. Stein, E. Moran, P. Yaciuk, R. Schlegel, R. M. Lyons, M. R. Pittelkow, K. Munger, P. M. Howley, and H. L. Moses. 1990. TGF-beta 1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell 61:777-785. [DOI] [PubMed] [Google Scholar]
- 69.Pulverer, B. J., C. Fisher, K. Vousden, T. Littlewood, G. Evan, and J. R. Woodgett. 1994. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 9:59-70. [PubMed] [Google Scholar]
- 70.Reinhard, C., A. Fernandez, N. J. Lamb, and G. Thomas. 1994. Nuclear localization of p85s6k: functional requirement for entry into S phase. EMBO J. 13:1557-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 72.Salghetti, S. E., S. Y. Kim, and W. P. Tansey. 1999. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz, J. K., C. H. Bassing, I. Kovesdi, M. B. Datto, M. Blazing, S. George, X. F. Wang, and J. R. Nevins. 1995. Expression of the E2F1 transcription factor overcomes type beta transforming growth factor-mediated growth suppression. Proc. Natl. Acad. Sci. USA 92:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massague. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 75.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 76.Staller, P., K. Peukert, A. Kiermaier, J. Seoane, J. Lukas, H. Karsunky, T. Moroy, J. Bartek, J. Massague, F. Hanel, and M. Eilers. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392-399. [DOI] [PubMed] [Google Scholar]
- 77.Stockwell, B. R., and S. L. Schreiber. 1998. TGF-beta-signaling with small molecule FKBP12 antagonists that bind myristoylated FKBP12-TGF-beta type I receptor fusion proteins. Chem. Biol. 5:385-395. [DOI] [PubMed] [Google Scholar]
- 78.Tang, B., E. P. Bottinger, S. B. Jakowlew, K. M. Bagnall, J. Mariano, M. R. Anver, J. J. Letterio, and L. M. Wakefield. 1998. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat. Med. 4:802-807. [DOI] [PubMed] [Google Scholar]
- 79.Tricoli, J. V., P. H. Gumerlock, J. L. Yao, S. G. Chi, S. A. D'Souza, B. R. Nestok, and R. W. deVere White. 1996. Alterations of the retinoblastoma gene in human prostate adenocarcinoma. Genes Chromosomes Cancer 15:108-114. [DOI] [PubMed] [Google Scholar]
- 80.Wang, T., B. Y. Li, P. D. Danielson, P. C. Shah, S. Rockwell, R. J. Lechleider, J. Martin, T. Manganaro, and P. K. Donahoe. 1996. The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell 86:435-444. [DOI] [PubMed] [Google Scholar]
- 81.Warner, B. J., S. W. Blain, J. Seoane, and J. Massague. 1999. Myc downregulation by transforming growth factor β required for activation of the p15Ink4b G1 arrest pathway. Mol. Cell. Biol. 19:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.West, M. J., M. Stoneley, and A. E. Willis. 1998. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17:769-780. [DOI] [PubMed] [Google Scholar]
- 83.Woo, M. S.-A., I. Sánchez, and B. D. Dynlacht. 1997. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol. Cell. Biol. 17:3566-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu, C.-L., M. Classon, N. Dyson, and E. Harlow. 1996. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol. Cell. Biol. 16:3698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 86.Yagi, K., M. Furuhashi, H. Aoki, D. Goto, H. Kuwano, K. Sugamura, K. Miyazono, and M. Kato. 2002. c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277:854-861. [DOI] [PubMed] [Google Scholar]
- 87.Yano, T., C. A. Sander, H. M. Clark, M. V. Dolezal, E. S. Jaffe, and M. Raffeld. 1993. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene 8:2741-2748. [PubMed] [Google Scholar]
- 88.Yu, K., L. Toral-Barza, C. Discafani, W. G. Zhang, J. Skotnicki, P. Frost, and J. J. Gibbons. 2001. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr. Relat. Cancer 8:249-258. [DOI] [PubMed] [Google Scholar]
- 89.Zentella, A., F. M. B. Weis, D. A. Ralph, M. Laiho, and J. Massagué. 1991. Early gene responses to transforming growth factor-β in cells lacking growth-suppressive RB function. Mol. Cell. Biol. 11:4952-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu, L., E. Harlow, and B. D. Dynlacht. 1995. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 9:1740-1752. [DOI] [PubMed] [Google Scholar]
- 91.Ziv, E., J. Cauley, P. A. Morin, R. Saiz, and W. S. Browner. 2001. Association between the T29→C polymorphism in the transforming growth factor beta1 gene and breast cancer among elderly white women: the study of osteoporotic fractures. JAMA 285:2859-2863. [DOI] [PubMed] [Google Scholar]