Abstract
Protein kinase A (PKAi a cyclic AMP-dependent protein kinase) negatively regulates sexual development and gluconeogenesis in fission yeast by suppressing the transcription of ste11 required for the former and the transcription of fbp1 required for the latter. Here we show that Rst2p, a zinc finger protein that can bind to the upstream region of ste11 and fbp1 via the STREP motif, mediates the activity of PKA to transcription of these genes. A simple reporter system confirmed that PKA could cause its negative effect on transcription through the combination of Rst2p and STREP. Rst2p was phosphorylated by PKA in vitro at two consensus sequences on it. Substitution of the target threonine residues by alanine made the protein active even in the presence of high PKA activity. Rst2p underwent hyperphosphorylation in the medium lacking glucose, and PKA inhibited this hyperphosphorylation. Rst2p was mainly cytoplasmic under high PKA activity but was concentrated in the nucleus when this activity was lowered, suggesting that PKA might regulate ste11 and fbp1 negatively by excluding Rst2p from the nucleus. However, the shift of Rst2p localization was not perfect under physiological conditions, leaving the possibility that PKA inhibits Rst2p function in another way as well. Although the PKA-Rst2p-STREP pathway is apparently central to the regulation of ste11 and fbp1 transcription in accordance with nutritional conditions, some additional paths are likely to connect nitrogen to repression of ste11 and glucose to repression of fbp1. These paths may ensure the specificity between the type of nutrients in shortage and the type of genes to be expressed.
The level of intracellular cyclic AMP (cAMP) changes in accordance with environmental conditions in various organisms. In eukaryotic cells, the major target of cAMP is cAMP-dependent protein kinase A (PKA), and the activity of PKA plays a pivotal role in the regulation of adaptation to the external conditions and the induction of intrinsic cellular differentiation. In both fission yeast and budding yeast, the level of intracellular cAMP is kept high during mitotic growth. Conditions unfavorable for growth cause downregulation of PKA in these microbes, which triggers adaptation to the adverse conditions and the beginning of sexual reproduction. Gross changes in gene expression accompany these responses, indicating that many transcription factors are likely to be regulated by PKA either directly or indirectly.
The fission yeast ste11 gene encodes an HMG transcription factor, which is responsible for expression of many genes required for the initiation of sexual development (35; reviewed in reference 43). Expression of ste11 is under the regulation of PKA. A decrease in the PKA activity, which naturally results from starvation of environmental nutrients, triggers ste11 expression. Another fission yeast gene well characterized as a target of PKA is fbp1, which encodes fructose-1,6-bis-phosphatase (FBPase), a key enzyme in gluconeogenesis (39). Glucose starvation induces fbp1 expression via a reduction of the PKA activity (3, 14, 28). Glucose is known to repress expression of many genes besides the one encoding FBPase in various cell systems, which include genes involved in alternate sugar metabolism, the tricarboxylic acid cycle, respiration, gluconeogenesis, and mitochondrial functions. The entire mechanism of glucose repression remains to be elucidated in both budding yeast and fission yeast, as well as in other eukaryotes. However, it has been established in budding yeast that transcription of the genes for alcohol dehydrogenase II and invertase partly depends on Adr1p (5) and Msn2p (8), respectively. These transcription factors are both C2H2 Zn finger proteins and are negatively regulated by PKA (4, 12, 33, 37). In fission yeast, both the PKA pathway (3, 28) and the stress-responsive mitogen-activated protein kinase (MAPK) pathway (34, 36) have been shown to regulate transcription of fbp1 (27), which is also the case with ste11 (18, 19, 30, 36). PKA is an inhibitory factor for expression of both ste11 and fbp1, whereas Spc1/Sty1 MAPK is a stimulatory factor for it. It has been shown, however, that Ste11p is not involved in the control of transcription of fbp1 (24).
We have demonstrated previously that transcription of ste11 is regulated directly by the gene product of rst2, namely, Rst2p (20). We isolated the rst2 gene originally as a high-copy-number suppressor of the sterility of the cgs1 mutant, in which PKA is activated constitutively (6). Genetic analysis has indicated that Rst2p functions downstream of the PKA pathway (20). Rst2p has two C2H2 Zn finger motifs, similar to Saccharomyces cerevisiae Adr1p, Msn2p, and Mig1p, which are transcription factors involved in glucose repression. Rst2p binds to a STRE (stress response element)-like sequence in the promoter region of ste11 (CCCCTC), which is required for the full activation of ste11 (20). We hereafter call this 6-bp element STREP (stress-starvation response element of Schizosaccharomyces pombe). Cells missing Rst2p can grow mitotically but cannot initiate sexual development.
The above observations led us to a couple of intriguing questions. Is the activity of Rst2p controlled by phosphorylation by PKA? If so, how? Does Rst2p also regulate transcription of fbp1? We address these questions and present our findings here.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
S. pombe strains used in this study are listed in Table 1. Cells were routinely grown on the complete medium YES (26) or the synthetic medium SD at 30°C (29). Synthetic sporulation medium SSA (7) was used to induce mating and sporulation. Two kinds of liquid minimal medium (MM) (26), modified in the carbon source, were used: MMR, containing 8% glucose, as a medium to induce glucose repression and MMD, containing 3% glycerol and 0.1% glucose, as a medium for derepression (15). General genetic methods for S. pombe were described previously (13). Protoplast fusion was performed according to the method of Sipiczki and Ferenczy (32). Transformation and gene tagging replacement were performed as described previously (2). Specific mutations were introduced into the cloned rst2 gene according to the method developed by Kunkel (21). The rst2-M3 allele was integrated into the chromosome, replacing the rst2::ura4+ allele of the host strain, by selecting 5′-fluoroorotic acid-resistant transformants.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| JY333 | h−ade6-M216 leu1 |
| JY450 | h90ade6-M216 leu1 |
| JX222 | h−ade6-M216 leu1 ura4-D18 cgs1::ura4+ |
| JX231 | h90ade6-M216 leu1 ura4-D18 rst2::ura4+ |
| JX233 | h−ade6-M216 leu1 ura4-D18 rst2::ura4+ |
| JX261 | h−ade6-M216 leu1 ura4-D18 cgs1::ura4+rst2::ura4+ |
| JX282 | h−ade6-M216 leu1 ura4-D18 pka1::ura4+rst2::ura4+ |
| JX384 | h−ade6-M216 leu1 ura4-D18 pka1::ura4+ |
| JW352 | h−ade6-M216 leu1 ura4-D18 rst2-GFP(S65T)≪ura4+a |
| JW355 | h−ade6-M216 leu1 ura4-D18 rst2-GFP(S65T)≪ura4+cyr1::ura4+ |
| JW359 | h−ade6-M216 leu1 ura4-D18 rst2-GFP(S65T)≪ura4+cgs1::ura4+ |
| JW363 | h−ade6-M216 leu1 ura4-D18 rst2-GFP(S65T)≪ura4+pka1::ura4+ |
| JW367 | h−ade6-M216 leu1 ura4-D18 rst2-3HA≪ura4+ |
| JW373 | h−ade6-M216 leu1 rst2-M3 |
| JW374 | h−ade6-M216 leu1 ura4-D18 cgs1::ura4+rst2-M3 |
| JW377 | h−ade6-M216 leu1 ura4-D18 pka1::ura4+rst2-M3 |
| JW487 | h90ade6-M216 leu1 ura4-D18 cgs1::ura4+rst2::ura4+ |
The expression rst2≪ura4+ indicates that ura4+ is inserted in the vicinity of rst2 on the chromosome.
Reporter plasmids.
pLopT4lacZ, provided by N. Jones, was a derivative of pREP41X-lacZ (9) and carried the lexA operator instead of the thiamine-repressive cis element of the nmt1 promoter. pSTREP-lacZ was a modified version of pLopT4lacZ, in which the lexA operator was replaced by a 39-bp sequence (5′-GCT TCC CCT CAT ACA CCC CTC ATA CAC ACC CCT CAT GCA-3′) containing three tandem repeats of STREP (underlined), the Rst2p-binding core identified in the ste11 promoter (20). pSTREPM-lacZ was identical to pSTREP-lacZ except that the STREP motifs were mutated from CCCCTC to ACCCTC.
β-Galactosidase assay.
Several independent colonies of each S. pombe strain transformed with a reporter plasmid were cultured in MMR to 5 × 106 cells/ml. A portion of each culture was sampled, and the remainder was shifted to MMD and further cultured for 3 h before sampling. Cells were harvested by the filtration method described previously (31). The β-galactosidase activity was determined as described n Alfa et al. (1), although the scale of the reaction was reduced to 60% in volume. At least three independent measurements were done for each type of host strain because the copy number of the reporter plasmid might vary among transformants.
Northern blotting.
Cells were harvested by filtration (31). Total RNA was extracted and RNA blot analysis was performed as described previously (40). Poly(A) RNA was selected by using oligo(dT)-cellulose (Invitrogen). We used a 1.3-kb PvuII-PvuII DNA fragment as a probe to detect ste11+ mRNA (35) and a PCR-amplified DNA fragment covering the whole open reading frame to detect fbp1+ mRNA (39).
Detection of Rst2p by Western blotting.
We essentially used a previously described protocol (31), except for a few substantial modifications. Briefly, harvested cells were boiled and then disrupted with glass beads in modified buffer L, which contained 50 mM HEPES-KOH and 400 mM KCl instead of the 50 mM Tris-Cl and 400 mM NaCl used in the original protocol. Each suspension containing 20 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22). We used separating acrylamide gels (7.5 or 8.0%) with a mono/bis ratio of 29.8:0.2 to detect Rst2p and a gel (12%) with a mono/bis ratio of 29.2:0.8 to detect Cdc2p. Purified polyclonal anti-Rst2p antibodies (this study) and commercialized anti-Cdc2 antibodies (Santa Cruz Biotechnology) were used as the primary antibodies, and goat anti-rabbit immunoglobulin G (IgG) Fc fragments conjugated with horseradish peroxidase (Jackson Immunoresearch) were used as the secondary antibodies. Enhanced chemiluminescence (Amersham-Pharmacia) was used for immunodetection on the membrane.
Phosphatase treatment.
Rst2p was enriched in the precipitate after centrifugation of the cell suspension described above. A precipitated sample containing 30 μg of protein was washed four times with the alkaline phosphatase buffer recommended by the manufacturer (Takara Biomedicals) and suspended thoroughly in the same buffer. Each substrate suspension received 20 U of calf intestinal alkaline phosphatase (Takara Biomedicals) and was incubated at 37°C for 60 min, either with or without the inhibitor mixture (10 mM EGTA, 0.1 M Na3VO4, 50 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate). The suspension was then boiled with SDS-PAGE sample buffer and subjected to electrophoresis. Immunodetection of Rst2p was done as described above.
Phosphorylation assay in vitro.
Rst2p fused with maltose-binding protein (MBP), together with variants of this fusion protein carrying mutations in the PKA target sites, was produced in bacteria by using expression vector pMAL-cRI (New England Biolabs). The fusion proteins were extracted in buffer E, which contained 25 mM HEPES-KOH (pH 7.5), 600 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1 protease inhibitor cocktail tablet per 10 ml (Complete Mini; Roche Diagnostics). Affinity purification of the MBP fusion proteins was done according to the protocol provided by the manufacturer, except that buffer E was used as the solvent. S. pombe cells producing hemagglutinin (HA)-tagged PKA catalytic subunit (Pka1p) were disrupted with glass beads in buffer I (50 mM Tris-Cl [pH 7.5], 150 mM KCl, 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 25 mM β-glycerophosphate, 0.1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride, supplemented with the protease inhibitor cocktail). HA-Pka1p was immunoprecipitated with monoclonal anti-HA antibody 16B12 (Berkeley Antibody Co.) and protein G-Sepharose (Amersham-Pharmacia). The Sepharose beads were washed twice with buffer I and then washed twice with buffer K (20 mM HEPES-KOH [pH 7.6], 20 mM MgCl2, 1 mM dithiothreitol, 25 mM β-glycerophosphate, 0.1 mM Na3VO4). About 1 μg of each substrate (MBP-Rst2p) was added to the tube containing immunoprecipitated HA-Pka1p, and the phosphorylation reaction mix was adjusted to the scale of 20 μl with buffer K, which contained 20 μM cold ATP and 5 μCi of [γ-32P]ATP (ICN Biomedicals). The reaction mix was incubated for 20 min at room temperature and then boiled with SDS-PAGE sample buffer. The PKA inhibitor PKI was purchased from Sigma.
Fluorescence microscopy.
Methanol fixation and indirect immunostaining of S. pombe cells were done essentially according to the method of Gaits et al. (10). HA-tagged Rst2p was stained with anti-HA epitope monoclonal antibody 16B12 as the primary antibody and Alexa 568-conjugated anti-mouse IgG goat antibodies (Molecular Probes) as the secondary antibodies, whereas green fluorescent protein (GFP) was detected by its own fluorescence expressed in living cells. DNA was counterstained with Hoechst 33342. Fluorescence microscopy was performed by using an Axiophoto microscope (Carl Zeiss) with appropriate filters. To optimize expression of Rst2p-3HA from the nmt1 promoter on a multicopy plasmid, cells were first grown to the exponential phase in SD medium, which contained 1.2 μM thiamine, then washed twice with thiamine-free MMR, and finally resuspended in MMR at the concentration of 5 × 106 cells/ml. The suspension was diluted 32-fold with MMR, and cells were grown for five generations before fixation with methanol. A portion of the cells was further shifted to and incubated in the glycerol medium before fixation.
RESULTS
Rst2p is a common transcription factor for the cAMP-repressible genes ste11 and fbp1.
Rst2p is a transcription factor for ste11+, which functions downstream of the PKA pathway (20). We suspected that Rst2p might also regulate expression of fbp1+, another gene known to be controlled by PKA in fission yeast (3, 14, 28). We thus examined growth of the rst2Δ strain on nonfermentable carbon sources. Expression of fbp1+, encoding FBPase, is required to assimilate nonfermentable carbon sources and is induced sharply under such nutritional conditions (39). Without FBPase, cells cannot generate glucose from nonfermentable carbon sources via gluconeogenesis and hence fail to proliferate. The rst2Δ strain grew very poorly compared to the wild-type strain on medium containing 3% glycerol and 0.1% glucose (data not shown). A trace amount of glucose was added in this assay because S. pombe cells did not feed on glycerol alone for an unclear reason. A more obvious difference was seen on medium containing 3% gluconate, where the wild-type strain grew robustly but the rst2Δ strain did not grow at all (data not shown).
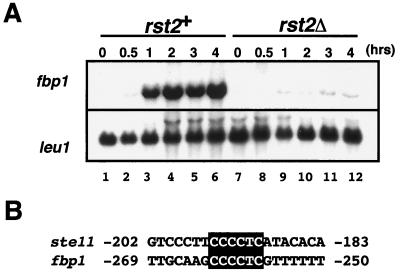
To demonstrate the necessity of Rst2p for transcription of fbp1 more directly, we measured the level of fbp1 mRNA in rst2+ and rst2Δ cells by Northern blotting. As shown in Fig. 1A, fbp1 mRNA accumulated significantly in wild-type cells 1 h after the shift from the repression medium (MMR; see Materials and Methods) to the derepression medium (MMD). In contrast, no fbp1 mRNA was detected in rst2Δ cells, indicating that Rst2p was essential to express fbp1. Scanning of the promoter region of fbp1 revealed one STREP (CCCCTC), the consensus motif for Rst2p binding, at nucleotides −262 to −257 (Fig. 1B). A recent study has shown that this sequence is positioned in a cis regulatory element of fbp1 designated UAS2 (27). We prepared a double-stranded oligonucleotide harboring this sequence (nucleotides −273 to −249) and showed that Rst2p could bind to it in a gel shift assay (data not shown), as was demonstrated with the ste11 upstream sequence (20). These observations strongly suggest that Rst2p is a transcriptional activator of both fbp1 and ste11 and functions via binding to STREP in both cases.
FIG. 1.
The rst2 gene function is required for transcription of the fbp1 gene. (A) Induction of fbp1 transcription by a medium shift. Cells, either rst2+ (JY333) or rst2Δ (JX233), were grown in MMR containing 8% glucose to a concentration of 5 × 106 cells/ml and then shifted to MMD containing 3% glycerol and 0.1% glucose. Cells were then harvested at the indicated times after the shift, and the expression of fbp1 and leu1 as an internal control was examined by Northern blotting. RNA (5 μg) was loaded in each lane after denaturation by formamide. (B) Alignment of STREP sequences, recognized by Rst2p, in the upstream activation regions of ste11 and fbp1. The numbers refer to positions relative to the transcriptional start.
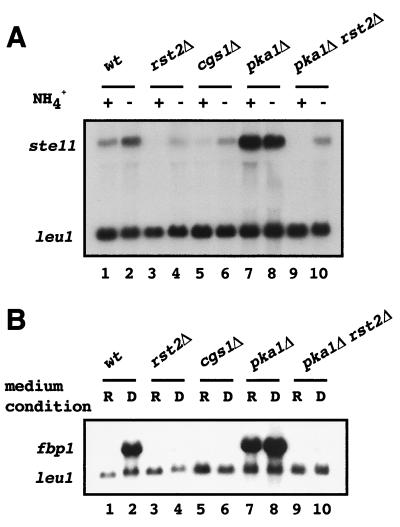
Figure 2 displays collectively the effects of the PKA-activated and the PKA-null mutations on expression of ste11 and fbp1, as assayed by Northern blotting. Some of these observations have been reported previously (3, 6, 20, 28, 35, 40). In Fig. 2, induction of ste11 expression was triggered by nitrogen starvation, whereas the induction of fbp1 expression was triggered by a shift of carbon sources. Both genes were expressed very poorly in the cgs1Δ strain, which lacked the regulatory subunit of PKA and hence retained high PKA activity (Fig. 2, lanes 5 and 6). In contrast, they were expressed constitutively in the pka1Δ strain, which lacked the catalytic subunit of PKA (Fig. 2, lanes 7 and 8). Disruption of rst2 in the pka1Δ strain abolished constitutive expression of both ste11 and fbp1. Thus, it is obvious that PKA affects expression of ste11 and fbp1 through Rst2p. However, it is also notable that the regulation of ste11 expression by the PKA-Rst2p system is not so thorough as is the case for fbp1. Expression of fbp1 was not detectable in cgs1Δ and rst2Δ cells, but a low level of residual ste11 expression was evident in them (Fig. 2A, lane 4). This may imply an involvement of a PKA-independent regulatory pathway in ste11 expression (see Discussion).
FIG. 2.
Effects of PKA-related mutations on expression of ste11 and fbp1. (A) To examine ste11 expression, cells of each genotype indicated (wild type [wt], JY333; rst2Δ, JX233; cgs1Δ, JX222; pka1Δ, JX384; pka1Δ rst2Δ, JX282) were grown in MM supplemented with ammonium chloride (+), and some of them were subjected to starvation of nitrogen for 2 h (−). Total RNA was extracted from each preparation and analyzed by Northern blotting. RNA (5 μg) was loaded in each lane after denaturation by formamide. Expression of ste11 and leu1 was detected by appropriate probes. (B) To examine fbp1 expression, cells of each strain (the same as in panel A) were grown in glucose-based MMR (R), and some of them were shifted to glycerol-based MMD (D) and incubated for 1 h. Expression of fbp1 was detected as described in panel A.
STREP can confer repression by cAMP and activation by Rst2p in a reporter gene system.
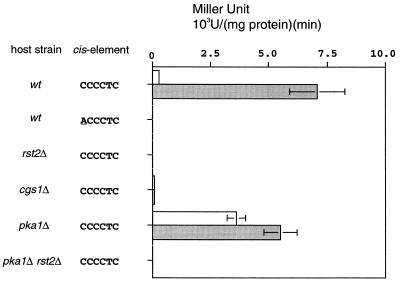
To assess the relation between Rst2p and STREP in a simpler system, we constructed a reporter plasmid, by connecting the open reading frame of Escherichia coli lacZ to a modified S. pombe nmt1 promoter. This nmt1 promoter lacked the element required for the repression by thiamine and instead carried a 39-bp sequence containing three tandem repeats of STREP (see Materials and Methods for details). Wild-type strain JY333 was transformed with this reporter construct, and transformed cells were grown in MMR. A portion of them was shifted to MMD. Cells were harvested at intervals and assayed for β-galactosidase activity. The enzymatic activity was low when cells were grown in MMR. It increased significantly after the shift to MMD, indicating that this reporter system could reproduce glucose repression (Fig. 3). Further analysis by using this system revealed that a single base change in STREP completely abolished expression of the reporter and that no expression was inducible if the host strain was rst2Δ (Fig. 3). The reporter was expressed poorly in the cgs1Δ strain cultured in MMD, whereas it was expressed strongly in the pka1Δ strain cultured in MMR (Fig. 3). Disruption of rst2 in the pka1Δ strain abolished expression of the reporter completely. Together, the observations obtained in this simple reporter system support the view that PKA regulates the activity of Rst2p negatively and that Rst2p controls expression of a target gene by interacting with a STREP motif located in its promoter.
FIG. 3.
An Rst2p- and STREP-dependent reporter system. The lacZ-reporter plasmids used here are constructed as described in Materials and Methods. Various strains as indicated (as defined in Fig. 2) were transformed with a reporter plasmid carrying either the perfect or a mutated STREP sequence. Lysates were prepared from cells grown in MMR (open bars) or derepressed in MMD for 3 h (shaded bars). An assay of β-galactosidase activity was done as described in Materials and Methods. An average of at least three independent measurements, performed with stable transformants, is shown for each case.
The activity of Rst2p is not regulated at the transcription level.
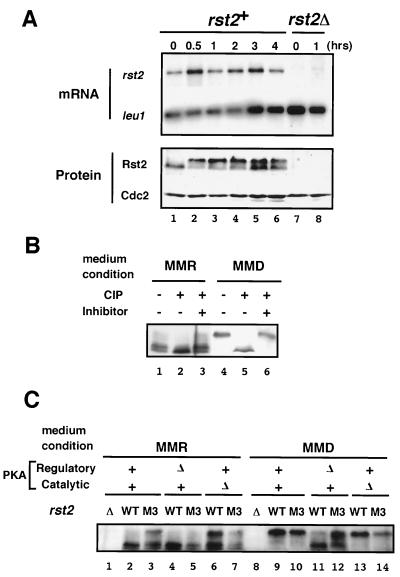
We investigated whether transcription of rst2 is affected by the medium shift from MMR to MMD. Because of its low transcription level, the amount of rst2 mRNA was not accurately quantified by blotting total cellular RNA. Therefore, we performed Northern analysis by using poly(A) RNA (Fig. 4A). The level of rst2 mRNA, normalized by the amount of leu1 mRNA as an internal control, did not appear to change significantly by the medium shift. Clearly, the extensive induction of fbp1 expression by the medium shift, as seen in Fig. 1A, was not attributable to the change of the level of rst2 mRNA. This suggested that the activity of Rst2p could be modified posttranscriptionally in response to the medium shift.
FIG. 4.
Rst2p activity is likely to be regulated posttranscriptionally. (A) Detection of rst2 mRNA and the gene product before and after a medium shift. To determine the level of rst2 mRNA, extracts were prepared from cells grown in MMD for the indicated duration, as detailed in Fig. 1. Poly(A) RNA was selected from each sample and subjected to gel electrophoresis after denaturation with formamide. We detected leu1 mRNA as an internal loading control (the time for exposure on X-ray film is much shorter than that for rst2 mRNA). For analysis of protein, boiled cell lysates were subjected to SDS-PAGE, followed by immunoblot analysis as described in Materials and Methods. (B) Phosphatase treatment of Rst2p. Protein precipitates prepared from JY333 cells either growing in MMR or incubated in MMD for 1 h were treated with calf intestinal alkaline phosphatase (CIP), with or without the addition of the inhibitor mix, and subjected to SDS-PAGE and immunoblot analysis. (C) Detection of wild-type and mutated Rst2p in PKA-related mutant cells under repressing or derepressing conditions. The M3 mutant form of Rst2p lacks phosphorylatable residues at the two putative PKA-target sites. Cells of JX233 (rst2Δ), JY333 (WT), JW373 (rst2-M3), JX222 (cgs1Δ), JW374 (cgs1Δ rst2-M3), JX384 (pka1Δ), and JW377 (pka1Δ rst2-M3) were grown in MMR. A portion of each culture was shifted to MMD and incubated for 1 h. Cell lysates were prepared from the cultures in MMR and MMD and then subjected to SDS-PAGE and immunoblot analysis.
Rst2p takes different phosphorylation states according to the medium conditions.
To see whether Rst2p undergoes modification at the protein level, we probed Rst2p by using affinity-purified anti-Rst2p antibodies in Western blotting experiments (Fig. 4A). The antibodies recognized a band (doublet; see below) of ca. 80 kDa in an extract prepared from wild-type cells grown in MMR (lane 1). This band was not detected in the extract of rst2Δ cells (lanes 7 and 8), confirming its identity as Rst2p. The shift of wild-type cells from MMR to MMD resulted in a reduction of the mobility of Rst2p in SDS-PAGE, suggesting that the protein underwent certain modification by the medium shift (lanes 2 through 6). The amount of Rst2p in a cell did not appear to change significantly by this shift. These observations led us to speculate that Rst2p might be phosphorylated under the derepressing conditions. To test this possibility, we treated crude preparations of Rst2p with nonspecific alkaline phosphatase, as described in Materials and Methods. Rst2p derived from derepressed cells regained a faster mobility after the phosphatase treatment, and this was blocked by the addition of phosphatase inhibitors (Fig. 4B, lanes 4 to 6), indicating that Rst2p was hyperphosphorylated under the derepressing conditions.
Rst2p derived from repressed cells was subjected to a similar phosphatase treatment (Fig. 4B, lanes 1 to 3). Analysis by SDS-PAGE by using a soft gel (7.5%), as shown here, revealed that Rst2p under the repressing conditions existed in two forms, giving two close but separable bands in electrophoresis (lanes 1 and 3). Rst2p treated by phosphatase appeared to give only the lower band (lane 2). Because the intracellular PKA activity was supposed to be high in the repression medium, we suspected the possibility that the upper band represented Rst2p phosphorylated by PKA. Two lines of evidence, however, denied this possibility. One was that we could detect these two bands in cgs1Δ (PKA-active) cells just as in wild-type cells (Fig. 4C, lane 2 versus lane 4). The other was that a mutant form of Rst2p that lacked the PKA target sites (M3; see below) still gave the two bands in both wild-type and cgs1Δ cells (lanes 3 and 5). Thus, we presume that the upper band represents a phosphorylated form of Rst2p but that PKA is not responsible for this phosphorylation.
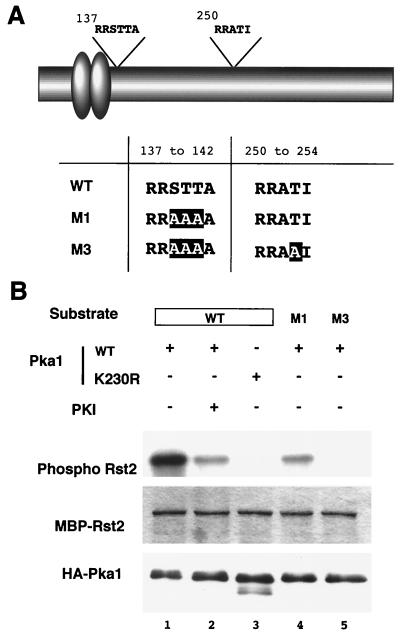
PKA phosphorylates Rst2p in vitro.
Rst2p carried putative PKA target sequences in the vicinity of the zinc finger motifs and in the central region (Fig. 5A). To determine whether PKA could phosphorylate Rst2p in vitro, we performed an assay with bacterially expressed recombinant Rst2p and the catalytic subunit of fission yeast PKA, namely, Pka1p. Pka1p used here was tagged with HA and purified from fission yeast cells by immunoprecipitation. We constructed MBP-Rst2p fusion proteins, bearing either the wild-type Rst2p sequence or the sequences mutated in the putative phosphorylation sites. One of the mutant forms (M1) carried two T-to-A substitutions and one S-to-A substitution in the target site close to the Zn fingers, and the other (M3) carried a T-to-A substitution in the central target site in addition to those in M1 (see Fig. 5A). As shown in Fig. 5B, the wild-type protein was efficiently phosphorylated by Pka1p in vitro (lane 1). This phosphorylation was inhibited largely by the addition of PKI peptide, a specific inhibitor of PKA (lane 2). Pka1p harboring a mutation in the kinase domain (K230R) was incapable of phosphorylation (lane 3). The M1 protein was phosphorylated to a lesser extent but still detectably (lanes 4), and the M3 protein was barely phosphorylated (lane 5). These results demonstrate that Pka1p can phosphorylate Rst2p and, furthermore, that the two presumed target sites on Rst2p assigned above are indeed the sites for phosphorylation by PKA.
FIG. 5.
Phosphorylation of Rst2p by PKA in vitro. (A) Schematic diagram of Rst2p showing consensus sequences for PKA phosphorylation and a table of mutations introduced into bacterially expressed Rst2p used for in vitro phosphorylation assay. Zinc finger motifs are shown as ovals. (B) Audoradiogram showing phosphorylation of bacterially expressed Rst2p, fused with MBP. The engineered proteins were incubated with [γ-32P]ATP and PKA immunoprecipitated from fission yeast cells. A kinase-negative PKA variant (K230R) was used in lane 3. The PKA inhibitor 5-24 (PKI) was added to lane 2.
PKA activity affects the phosphorylation state of Rst2p.
To examine a possible involvement of PKA in the hyperphosphorylation of Rst2p observed above, we analyzed Rst2p in cgs1Δ (PKA-active) and pka1Δ (PKA-null) cells grown under either the repressing or the derepressing conditions by SDS-PAGE (Fig. 4C). Rst2p from pka1Δ cells under the repressing conditions assumed a ladder of slow-moving bands, the top of which reached the position of the band observed in wild-type cells under the derepressing conditions (lanes 6 and 9). Because pka1 is the single gene encoding PKA in fission yeast (24), this suggested that the hyperphosphorylation of Rst2p in the derepression medium was brought about by a kinase(s) other than PKA. In contrast, Rst2p was not fully hyperphosphorylated in cgs1Δ cells under the derepressing conditions (lane 11), suggesting that high activity of PKA is inhibitory for the hyperphosphorylation.
We then investigated effects of the M3 mutation on hyperphosphorylation of Rst2p. Whereas the wild-type Rst2p was not hyperphosphorylated in the repression medium (Fig. 4A, lane 1, and Fig. 4C, lane 2), the M3 mutant was partially hyperphosphorylated in the same medium (Fig. 4C, lane 3). Notably, the pattern of slow-moving bands exhibited by the M3 mutant here was quite similar to those of the wild-type Rst2p and the M3 mutant in PKA-defective cells under the repressing conditions (lane 3 versus lanes 6 and 7). Furthermore, the M3 mutant was phosphorylated more extensively than the wild-type Rst2p in PKA-active cells under the derepressing conditions (lane 12 versus lane 11). These results suggest strongly that PKA suppresses hyperphosphorylation of Rst2p through phosphorylation of its target sites, which are mutated in M3. The data shown in Fig. 4C also suggest that, besides the absence of this direct phosphorylation by PKA, two conditions are essential for the kinase(s) responsible for hyperphosphorylation of Rst2p to become fully active (or the phosphatase to become fully inactive). One is depletion of glucose from the medium (as typically deduced from lane 7), and the other is reduction of the cellular PKA activity below a certain threshold (as typically deduced from lane 12).
Mutations in the phosphorylation sites activate Rst2p ectopically.
We investigated biological consequences of mutations introduced into the phosphorylation sites on Rst2p. Cells moderately expressing the rst2-M3 allele showed markedly slow growth and flocculation in a synthetic medium, which contained a sufficient amount of nitrogen to suppress sexual development of wild-type cells. The rst2-M3 strain grew even more slowly than the pka1Δ strain, which has been shown to grow poorly (24). Inhibition of cell growth by the rst2-M3 mutation was less severe in the cgs1Δ background, although the reason for this suppression is unclear.
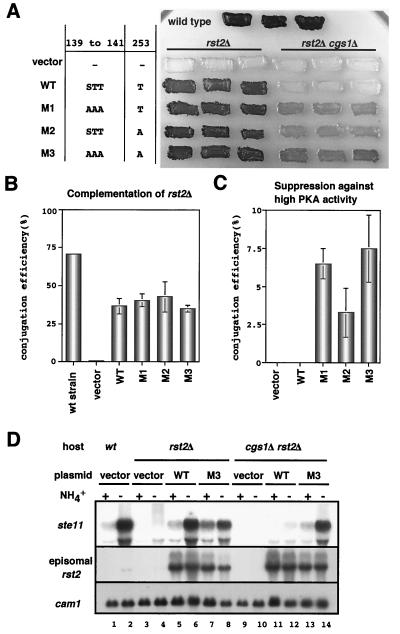
To see the effects of the M1 and M3 mutations on sexual development, we transformed homothallic rst2Δ cells, whose genetic background was either cgs1+ or cgs1Δ, with a multicopy plasmid carrying each rst2 allele connected to the weak nmt1 promoter. The M2 mutation, which harbored a single T-to-A substitution in the central target site, was also examined. Each transformant was grown on SD medium, which represses the nmt1 promoter, and a portion was restreaked on SSA medium, which induced sexual development due to the scarcity of the nitrogen source and also derepresses the nmt1 promoter due to the lack of thiamine. The results are summarized in Fig. 6. When the host cells were cgs1+, the wild-type and the mutant rst2 alleles were equally active in the overall induction of sporulation, as assayed by iodine staining (Fig. 6A). A microscopic measurement of the conjugation efficiency of the same samples confirmed that they were nearly equivalent (Fig. 6B). However, when the host cells were cgs1Δ, differences arose among them. The wild-type allele could recover neither overall sporulation (Fig. 6A) nor conjugation (Fig. 6C) in cgs1Δ rst2Δ cells. However, each mutant allele could do so, although not very strongly (Fig. 6A,C). The double-mutant allele, M3, was the most active, and the M1 allele appeared to be more active than M2 (Fig. 6A and C).
FIG. 6.
Unphosphorylatable mutations in the PKA target sites enhances the activity of Rst2p. (A) Iodine-staining assay of sporulation efficiency in various rst2 transformants. Cells of JX231 (h90 cgs1+ rst2Δ) and JW487 (h90 cgs1Δ rst2Δ) were transformed with pREP81-based plasmids carrying each allele as indicated on the left. They were incubated in patches on SSA plates for 4 days and stained with iodine vapor, which turns sporulated cells dark brown. JY450 cells transformed with the vector plasmid were used as a control. (B) JX231 transformants, shown in panel A, were examined microscopically for their conjugation efficiency. (C) JW487 transformants, shown in panel A, were examined microscopically for their conjugation efficiency. (D) Restoration of ste11 transcription in cgs1Δ cells by the rst2-M3 allele. Total RNA was prepared from heterothallic (h−) cgs1+ rst2Δ (JX233) or cgs1Δ rst2Δ (JX261) cells transformed with a pREP81-based plasmid carrying either the rst2+ or the rst2-M3 allele. Cells were either growing exponentially in MM (+) or were subjected to nitrogen starvation for 4 h (−). RNA (5 μg) was loaded in each lane after denaturation by formamide and analyzed by Northern blotting. Expression of cam1 and plasmid-borne rst2 was measured as an internal control and for confirmation, respectively.
These observations suggested that Rst2p not phosphorylatable by PKA was likely to stimulate unconditional expression of the target genes involved in sexual development. To examine this, we measured induction of ste11 expression in cgs1+ rst2Δ and cgs1Δ rst2Δ cells transformed with either the wild-type or the M3 mutant allele, driven by the weak nmt1 promoter. Because expression of the M3 allele in cgs1+ cells impaired their growth severely, all of the transformants were precultured in the presence of thiamine. They were transferred to thiamine-free medium, cultured for five rounds of cell division, and examined for expression of ste11. As shown in Fig. 6D, induction of ste11 expression by nitrogen depletion occurred in cgs1+ cells transformed with either the wild-type or the M3 allele. Expression of ste11 was not inducible in cgs1Δ cells carrying the wild-type rst2 allele under the experimental conditions. However, cgs1Δ cells transformed with the M3 allele expressed ste11 intensively in response to nitrogen starvation. From these observations we conclude that a mutant form of Rst2p unphosphorylatable by PKA overcomes hyperactivation of PKA with respect to the induction of ste11 expression. It is also conclusive, however, that this mutation does not represent the total effects brought by nitrogen starvation.
Rst2p undergoes nucleocytoplasmic shuttling.
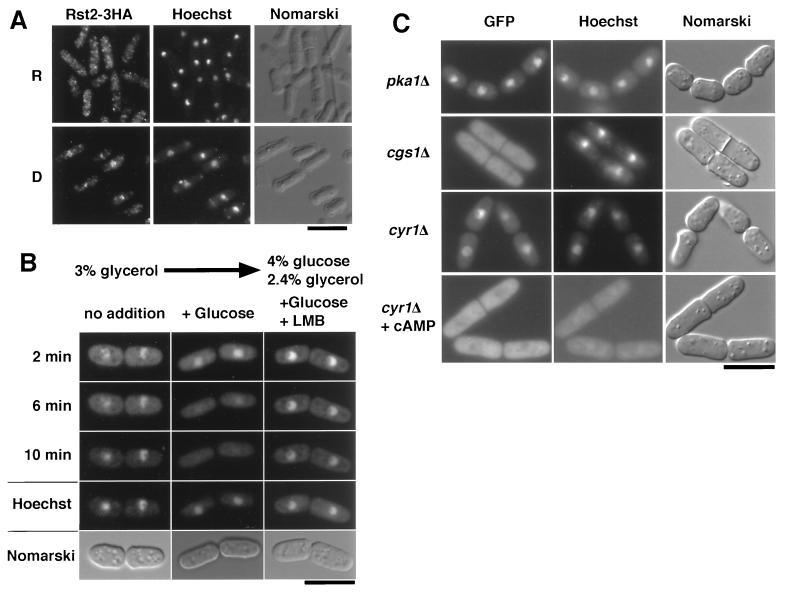
We investigated subcellular localization of Rst2p. To visualize Rst2p, we tagged this protein with three copies of the HA epitope, and cells producing HA-tagged Rst2p from the fusion gene on the chromosome were stained with anti-HA. The results indicated that Rst2p was cytoplasmic almost exclusively under the repressing conditions (Fig. 7A, R panels). However, it was mainly nuclear under the derepressing conditions (Fig. 7A, D panels). A more statistical discussion will be given below. Essentially the same results were obtained when Rst2p was visualized by the conjugation of the jellyfish GFP (data not shown). A dynamic nature of Rst2p was investigated by using a strain that carried the rst2-GFP fusion gene on the chromosome (JW352). JW352 cells were adapted in MMD for 1 h so that Rst2p-GFP was accumulated in the nucleus, and we applied glucose to these cells at the final concentration of 4%. This caused rapid disappearance of much of the fluorescence from the nucleus (Fig. 7B). This disappearance was blocked by the simultaneous application of leptomycin B, an inhibitor of nuclear export (reviewed in reference 38) (Fig. 7B). GFP alone was distributed throughout the cytoplasm and the nucleus and did not exhibit such a dynamic change in localization in response to glucose (data not shown). These results indicate that Rst2p can undergo nucleocytoplasmic shuttling and that glucose either promotes its export from the nucleus or inhibits its import to the nucleus.
FIG. 7.
Effects of glucose on subcellular localization of Rst2p. (A) Immunostaining of Rst2-3HAp. Cells (JW367) grown in MMR (R panels) were harvested by filtration, transferred to MMD, and incubated for 1 h (D panels). They were stained with anti-HA antibody and counterstained with Hoechst 33342 to visualize the DNA. (B) Addition of glucose promotes exclusion of Rst2p from the nucleus. JW352 cells expressing Rst2p-GFP were adapted in MMD for an hour (no addition). Glucose was added to a portion of the culture to the final concentration of 4% (+Glucose). To block exportin-dependent nuclear export, leptomycin B (0.1 μg/ml) was added simultaneously to a portion (+Glucose +LMB). DNA was stained with Hoechst 33342. Bar, 10 μm. (C) Subcellular localization of Rst2p-GFP in PKA-related mutants. Cells of JW363 lacking PKA (pka1Δ) and cells of JW359 with constitutively active PKA (cgs1Δ), both of which were grown in MM, were observed for GFP fluorescence. Cells of JW355 lacking adenylyl cyclase were first grown in MM containing 2 mM cAMP and 5 mM caffeine to block phosphodiesterase (cyr1Δ + cAMP). They were filtered, transferred to cAMP-free MM, and incubated for 10 min (cyr1Δ). DNA was stained with Hoechst 33342. The presence of caffeine somehow weakened staining of DNA by Hoechst 33342. Bars, 10 μm.
PKA activity affects subcellular localization of Rst2p.
To examine whether localization of Rst2p could be affected by the PKA activity, we expressed Rst2p-GFP in the pka1Δ, cgs1Δ, or cyr1Δ strains. The cyr1Δ strain lacks adenylyl cyclase and has no detectable level of cAMP (23, 44, 45). As summarized in Fig. 7C, Rst2p-GFP was almost exclusively nuclear in the pka1Δ and cyr1Δ strains and almost exclusively cytoplasmic in the cgs1Δ strain, when they were grown in the MM medium with 2% glucose. The nuclear localization of Rst2p-GFP in the cyr1Δ strains was canceled by the addition of 2 mM cAMP to the medium (Fig. 7C). Altogether, these results indicate that the PKA activity is a key determinant of the subcellular localization of Rst2p.
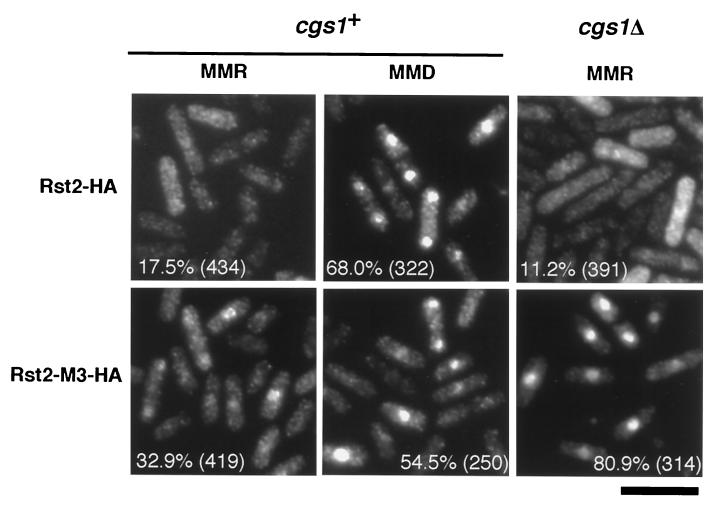
To see the effects of the M3 mutation on subcellular localization of Rst2p, we expressed HA-tagged Rst2-M3p from a multicopy plasmid in cgs1+ and cgs1Δ cells. HA-tagged Rst2p was expressed similarly as a control. When HA-tagged Rst2p was produced in cgs1+ cells, less than 20% of the cells exhibited nuclear accumulation of the protein under the repressing conditions (in MMR), whereas nearly 70% did so under the derepressing conditions (in MMD) (Fig. 8). In contrast, more than 30% of the cells exhibited nuclear accumulation of HA-tagged Rst2-M3p under the repressing conditions, and ca. 50% did so under the derepressing conditions (Fig. 8). These observations suggest that phosphorylation at the two PKA target sites on Rst2p, which are mutated in M3, may be pertinent with nuclear exclusion of the protein. However, the data also imply that PKA affects localization of Rst2p not only through phosphorylation of these sites but also in an additional way(s). Curiously, Rst2-M3p showed highly efficient nuclear accumulation in cgs1Δ cells placed under the repressing conditions (Fig. 8). The wild-type Rst2p showed extensive cytoplasmic localization in cgs1Δ cells under the same conditions (Fig. 8). These observations suggest that PKA regulates subcellular localization of Rst2p in a complex manner, probably involving as-yet-unidentified factors.
FIG. 8.
Quantitative measurements of subcellular localization of the wild-type Rst2p and the M3 mutant under the repressing and derepressing conditions. Rst2p and Rst2-M3p, both tagged with HA, were expressed from the expression vector pREP81 in cells that were either cgs1+ rst2Δ (JX 233) or cgs1Δ rst2Δ (JX 261). Cells cultured in MMR or shifted to MMD (30 min) were stained with anti-HA antibody, and the percentage of cells showing clear nuclear accumulation of Rst2p was culculated for each sample. Bar, 10 μm.
DISCUSSION
In this study we have demonstrated that fission yeast Rst2p regulates expression of both ste11 and fbp1 through interaction with the cis element STREP. Further, the obtained data suggest strongly that Rst2p is phosphorylated directly by PKA, thereby suppressing its transcription-activating activity. It is interesting that a single factor is used in order to regulate two mutually independent biological processes, namely, sexual development and gluconeogenesis. Induction of ste11 expression, a key step for the sexual development, can occur in response to starvation of a nitrogen source, whereas induction of fbp1 expression, which ensures growth on a nonfermentable carbon source, occurs in response to the consumption of glucose. Both types of nutritional starvation cause a decrease in the intracellular cAMP level (16, 25), and a role for PKA has been inferred in the induction of both ste11 and fbp1 expression (3, 14, 20, 27, 28, 35). The present study establishes Rst2p as a pivotal mediator between the activity of PKA and transcription of the ste11 and fbp1 genes.
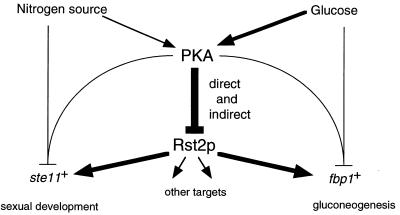
If fission yeast cells lose the PKA activity completely, which is an unnatural condition realized only in the pka1Δ or cyr1Δ strain, the expression of both ste11 and fbp1 becomes constitutive regardless of the nutritional conditions. On the contrary, their expression is always suppressed in the cgs1Δ strain sustaining high PKA activity. Our analysis has shown that Rst2p lacking the PKA target sites (M3) can cause effects similar to those of the loss of PKA activity with respect to hyperphosphorylation of the protein and can promote ste11 transcription in cgs1Δ cells starved for nitrogen. These observations strongly suggest that Rst2p is a substrate of PKA in vivo. However, the mutant form of Rst2p cannot induce ste11 transcription in the presence of ample nitrogen, although the complete loss of PKA activity can do so. In other words, the M3 mutant does not replace the loss of PKA completely in induction of ste11 expression. Furthermore, the results presented in Fig. 2, lanes 3 through 6, indicate that nitrogen can yet affect expression of ste11 to some extent independent of the PKA-Rst2 pathway, as has been inferred in previous studies (18, 20, 30, 36, 41). Taking these results together, we propose the scheme shown in Fig. 9 as the most rational based on current knowledge. In this scheme, phosphorylation of Rst2p by PKA serves as the major switch to turn off ste11 expression, but there are additional negative regulatory pathways that bypass Rst2p, one connecting the nitrogen source to ste11 expression and another connecting PKA to ste11 expression.
FIG. 9.
Diagram of the regulatory network involving the PKA-Rst2p pathway for transcriptional activation of ste11 and fbp1. The pathway established in this study is shown in thick lines. Other possible paths are shown in thin lines.
A similar scheme appears to apply to the relation of Rst2p to fbp1 expression. Our preliminary observations suggest that the M3 mutant form of Rst2p cannot activate fbp1 expression in the presence of glucose, nor can it do so under conditions of high PKA activity (unpublished results). This finding may parallel the observation that both depletion of glucose and reduction of cellular PKA activity are apparently required to hyperphosphorylate the M3 form of Rst2p completely (Fig. 4C). Furthermore, several factors other than Rst2p have been shown to be involved in the activation and repression of fbp1 transcription. Neely and Hoffman (27) have shown that the PKA pathway regulates transcription of fbp1 in a dual manner, through the STREP motif (UAS2) and the CRE motif (UAS1), the latter of which is recognized by a complex of Atf1/Gad7p and Pcr1p. These authors have also shown that at least four forms of protein complexes bind to STREP and that the stress-responsive MAPK cascade regulates transcription of fbp1 at both STREP and CRE sites. Janoo et al. (17) have reported that two homologs of the Saccharomyces transcriptional repressor Tup1p are involved in repression of fbp1 expression. Thus, combined with these observations, our results lead to a simplified scheme illustrating the regulation of fbp1 expression as shown in Fig. 9.
A portion of Rst2p is phosphorylated in wild-type cells grown in MMR, but PKA, which is supposed to be active in these cells, does not appear to be responsible for this phosphorylation, as discussed in Results. We have thus far been unsuccessful in detecting in vivo phosphorylation of Rst2p by PKA as a shift of the mobility of the protein in gel electrophoresis. However, the results of mutational analysis suggest strongly that PKA affects the activity of Rst2p through both direct phosphorylation and indirect modification. Thus, biochemical evidence is awaited to show critically whether Rst2p is phosphorylated by PKA in living cells.
An unexpected observation is that cells expressing rst2-M3 are more impaired in vegetative growth than pka1Δ cells in synthetic medium, whereas they grow better than pka1Δ cells in rich medium. Furthermore, cgs1Δ suppresses the slow growth of rst2-M3 cells in synthetic medium to some extent. These observations suggest that emergence of Rst2p unphosphorylated by PKA, not precisely harmonized with the state of the PKA activity, can cause certain adverse effects on cell growth. Because both ste11 and fbp1 are unlikely to be fully expressed in rst2-M3 cells under growing conditions, these observations indicate that Rst2p is likely to have a target(s) other than these two genes, which is more closely related to growth control.
This study has revealed a clear correlation between the subcellular localization of Rst2p and the activity of PKA in the cell. When the PKA activity is high, Rst2p tends to be located in the cytoplasm. When it is low, Rst2p tends to be nuclear. Nuclear exclusion of Rst2p in the presence of glucose apparently explains why high PKA activity is inhibitory to the transcription-activating activity of Rst2p. It is currently not conclusive whether phosphorylation by PKA accelerates nuclear export or inhibits nuclear import of Rst2p. Because leptomycin B blocks nuclear export of Rst2p, it appears that Rst2p is exported from the nucleus by the exportin-dependent system. However, our attempts to identify possible nuclear localization signal or nuclear export signal on Rst2p have not been successful so far.
Although phosphorylation by PKA apparently affects nucleocytoplasmic localization of Rst2p, the shift between the cytoplasm and nucleus is never all-to-none under our experimental conditions, suggesting that the phosphorylation may also impair other aspects of Rst2p required for its transcription-activating activity. Judging from the proposed ternary structure of Zn finger domain, as reviewed by Wolfe et al. (42), the N-terminal phosphorylation site, 10 residues apart from the DNA-binding domain (Fig. 6A and C and Fig. 8), does not seem to affect the ability of Rst2p to recognize either a base or a phosphate. Rather, phosphorylated Rst2p may be impaired in the ability to activate the transcription machinery, because there lies a glutamine stretch near the phosphorylation site, as is frequently found in the activating domain of transcription factors. Further studies are necessary to clarify these possibilities.
S. cerevisiae Msn2p and Msn4p, Zn finger proteins that recognize the STRE motif, have been shown to undergo hyperphosphorylation in response to a carbon source shift and heat shock, and high PKA activity blocks this hyperphosphorylation (11). It is suspected, though not proven, that some stress-responsive kinases may be involved in this hyperphosphorylation in S. cerevisiae. Garreau et al. hypothesized further that different stresses may cause different states of hyperphosphorylation (11). It is very intriguing that transcription activators that recognize similar motifs (STRE and STREP) undergo analogous modifications in S. cerevisiae and S. pombe, which are only distantly related in phylogeny. Although the identity of the kinase(s) (and possible antagonizing phosphatase) involved in the hyperphosphorylation of Rst2p remains to be clarified, it appears likely that, in S. pombe, Rst2p undergoes hyperphosphorylation differently under depletion of glucose and depletion of nitrogen. The full activation of fbp1 and that of ste11 may require different levels of hyperphosphorylation of Rst2p. These questions remain to be answered.
Acknowledgments
We thank N. Jones for helpful advice and plasmids.
This work was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Alfa, C., P. A. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, S. M., and C. S. Hoffman. 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherry, J. R., T. R. Johnson, C. Dollard, J. R. Shuster, and C. L. Denis. 1989. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 56:409–419. [DOI] [PubMed] [Google Scholar]
- 5.Denis, C. L., and E. T. Young. 1983. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol. Cell. Biol. 3:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVoti, J., G. Seydoux, D. Beach, and M. McLeod. 1991. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10:3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egel, R., and M. Egel-Mitani. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127–134. [DOI] [PubMed] [Google Scholar]
- 8.Estruch, F., and M. Carlson. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaits, F., G. Degols, K. Shiozaki, and P. Russell. 1998. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 12:1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garreau, H., R. N. Hasan, G. Renault, F. Estruch, E. Boy-Marcotte, and M. Jacquet. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146:2113–2120. [DOI] [PubMed] [Google Scholar]
- 12.Görner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395–446. In R. D. King (ed.), Handbook of genetics. Plenum Press, New York, N.Y.
- 14.Hoffman, C. S., and F. Winston. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5:561–571. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, C. S., and F. Winston. 1990. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics 124:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6:2455–2462. [DOI] [PubMed] [Google Scholar]
- 17.Janoo, R. T., L. A. Neely, B. R. Braun, S. K. Whitehall, and C. S. Hoffman. 2001. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157:1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanoh, J., Y. Watanabe, M. Ohsugi, Y. Iino, and M. Yamamoto. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1:391–408. [DOI] [PubMed] [Google Scholar]
- 19.Kato, T., Jr., K. Okazaki, H. Murakami, S. Stettler, P. A. Fantes, and H. Okayama. 1996. Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 378:207–212. [DOI] [PubMed] [Google Scholar]
- 20.Kunitomo, H., T. Higuchi, Y. Iino, and M. Yamamoto. 2000. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11+ gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 11:3205–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125–139. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 23.Maeda, T., N. Mochizuki, and M. Yamamoto. 1990. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87:7814–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, T., Y. Watanabe, H. Kunitomo, and M. Yamamoto. 1994. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 269:9632–9637. [PubMed] [Google Scholar]
- 25.Mochizuki, N., and M. Yamamoto. 1992. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol. Gen. Genet. 233:17–24. [DOI] [PubMed] [Google Scholar]
- 26.Moreno, S., A. Klar, and P. Nurse. 1990. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–826. [DOI] [PubMed] [Google Scholar]
- 27.Neely, L. A., and C. S. Hoffman. 2000. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol. Cell. Biol. 20:6426–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nocero, M., T. Isshiki, M. Yamamoto, and C. S. Hoffman. 1994. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein alpha subunit encoded by gpa2 (git8). Genetics 138:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman, F., G. Fink, and J. Hicks. 1986. Methods in yeast genetics: laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276–2288. [DOI] [PubMed] [Google Scholar]
- 31.Shiozaki, K., and P. Russell. 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 283:506–520. [DOI] [PubMed] [Google Scholar]
- 32.Sipiczki, M., and L. Ferenczy. 1977. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 151:77–81. [DOI] [PubMed] [Google Scholar]
- 33.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stettler, S., E. Warbrick, S. Prochnik, S. Mackie, and P. Fantes. 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109:1927–1935. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe, and M. Yamamoto. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5:1990–1999. [DOI] [PubMed] [Google Scholar]
- 36.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, W. E., and E. T. Young. 1990. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding Proc. Natl. Acad. Sci. USA 87:4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullman, K. S., M. A. Powers, and D. J. Forbes. 1997. Nuclear export receptors: from importin to exportin. Cell 90:967–970. [DOI] [PubMed] [Google Scholar]
- 39.Vassarotti, A., and J. D. Friesen. 1985. Isolation of the fructose-1,6-bisphosphatase gene of the yeast Schizosaccharomyces pombe: evidence for transcriptional regulation. J. Biol. Chem. 260:6348–6353. [PubMed] [Google Scholar]
- 40.Watanabe, Y., Y. Iino, K. Furuhata, C. Shimoda, and M. Yamamoto. 1988. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7:761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe, S. A., L. Nekludova, and C. O. Pabo. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29:183–212. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, M. 1996. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci. 21:18–22. [PubMed] [Google Scholar]
- 44.Yamawaki-Kataoka, Y., T. Tamaoki, H. R. Choe, H. Tanaka, and T. Kataoka. 1989. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:5693–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young, D., M. Riggs, J. Field, A. Vojtek, D. Broek, and M. Wigler. 1989. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86:7989–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]