Abstract
Previous studies have shown that changes in the affinity of the hamster Orc1 protein for chromatin during the M-to-G1 transition correlate with the activity of hamster origin recognition complexes (ORCs) and the appearance of prereplication complexes at specific sites. Here we show that Orc1 is selectively released from chromatin as cells enter S phase, converted into a mono- or diubiquitinated form, and then deubiquitinated and re-bound to chromatin during the M-to-G1 transition. Orc1 is degraded by the 26S proteasome only when released into the cytosol, and peptide additions to Orc1 make it hypersensitive to polyubiquitination. In contrast, Orc2 remains tightly bound to chromatin throughout the cell cycle and is not a substrate for ubiquitination. Since the concentration of Orc1 remains constant throughout the cell cycle, and its half-life in vivo is the same as that of Orc2, ubiquitination of non-chromatin-bound Orc1 presumably facilitates the inactivation of ORCs by sequestering Orc1 during S phase. Thus, in contrast to yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), mammalian ORC activity appears to be regulated during each cell cycle through selective dissociation and reassociation of Orc1 from chromatin-bound ORCs.
The mechanism for initiation of DNA replication is highly conserved among eukaryotes in that homologues of the proteins involved in assembly and activation of prereplication complexes (pre-RCs) in the budding yeast Saccharomyces cerevisiae are found in all other eukaryotes examined so far, and the sequence of events by which these proteins initiate DNA replication is remarkably similar throughout the eukaryotic kingdom (reviewed in references 4 and 20). Nevertheless, the first step in regulating this process appears to differ significantly between yeasts and mammals.
The first step in the initiation of eukaryotic DNA replication is the assembly of a six-subunit origin recognition complex (ORC) at specific sites distributed throughout the genome. For yeast, these sites coincide with genetically defined replication origins (replicons). Analogous sites have been identified for mammals, although they appear to be more complex (discussed in references 2, 10, 31, and 41). All six ORC subunits are required for the initiation of DNA replication of yeast, and Orc1 and Orc2 have been shown to be required for Drosophila and Xenopus (6, 24, 44, 45). In the yeasts S. cerevisiae (11, 17, 27) and Schizosaccharomyces pombe (22, 29), both DNA footprinting and immunoprecipitation analyses reveal that a complete ORC binds to the replication origins immediately after the initiation of replication occurs and it remains stably bound throughout the cell division cycle. Thus, the first step in regulating the assembly of pre-RCs in yeast is believed to occur by regulating the activity of Cdc6p/Cdc18p (20) and Cdt1p (51), two proteins that are required for loading Mcm proteins onto ORC-chromatin sites. However, the situation appears quite different in the Metazoa.
In multicellular eukaryotes such as frogs, flies, and mammals, the affinity of one or more ORC subunits for chromatin changes at specific times during the cell cycle. In Xenopus, Orc proteins in activated eggs bind to sperm chromatin, whereas Orc proteins in meiotic eggs do not (7, 16, 19, 46), and both Orc1 and Orc2 proteins are present on chromatin in cultured Xenopus cells during interphase but not during metaphase (44). In Drosophila, Orc2 remains bound to chromosomes throughout the cell cycle (36), whereas the amount of nuclear bound DmOrc1 is greatest during late G1 and S phases (3), suggesting a cell cycle-dependent, differential association of DmOrc proteins with chromatin. In mammals, the total amounts of Orc1 and Orc2 present throughout the cell cycle are constant (33, 34, 43, 47), but Orc1 is selectively released from chromatin during M phase (34) and S phase (23). This accounts for the fact that M-phase chromatin does not contain functional ORCs (34, 59) and for the disappearance of an ORC-like footprint at the human lamin B2 origin during mitosis (1). ORC activity is restored during the transition from M to G1 phase, concomitant with the appearance of Orc1 tightly bound to chromatin and the assembly of pre-RCs at specific genomic sites (25, 34). Therefore, it appears that multicellular eukaryotes regulate the initiation of DNA replication through cell cycle-dependent changes in ORC activity, the first step in the assembly of pre-RCs. In mammals, this appears to be accomplished through selective release of Orc1 from chromatin bound ORCs during the S-to-M phase transition, followed by reassembly of functional ORC-chromatin sites during the M-to-G1 phase transition. This would allow mammals to delay assembly of pre-RCs until both DNA replication and mitosis are completed and a nuclear membrane is assembled.
Here we provide further evidence in support of this hypothesis by showing that chromatin-bound Orc1 is selectively released as S phase begins in hamster cells and is targeted for ubiquitination. However, ubiquitination is limited to only one or two adducts, and these do not affect the stability of Orc1 in vivo. Orc1 can be degraded via the 26S proteasome but only when nuclei are permeabilized. These results strongly suggest that mammalian ORC activity is regulated during each cell cycle through selective dissociation and reassociation of Orc1 from chromatin-bound ORC, and that mono- or diubiquitination of non-chromatin-bound Orc1 is reversible and facilitates this dissociation by sequestering Orc1. This mechanism would not only prevent reinitiation at origins that already have been activated during S phase, but it would provide a mechanism by which the number and locations of initiation sites could be changed during animal development.
MATERIALS AND METHODS
Cells.
Exponentially proliferating CHOC 400 Chinese hamster ovary cells were cultured in Dulbecco’s modified Eagle’s medium plus 5% fetal bovine serum until 80% confluence was reached (34). Cells were synchronized in metaphase by culturing them in 0.05 μg of nocodazole/ml for 3 h and then releasing them into G1 phase as previously described (58). Cells were stained with Giemsa and scored for the absence of a nuclear envelope and the presence of condensed chromatin. Greater than 95% of the cells were in metaphase, and removal of nocodazole resulted in 85 to 95% of the cells undergoing DNA synthesis several hours later, as previously reported (18, 25, 34). In the presence of nocodazole, none of the cells entered S phase during the same time interval. Cells were also synchronized at their G1/S phase boundary by isoleucine and serum deprivation followed by arrest in aphidicolin, as previously described (method two in reference 21). This method was chosen because the replication origin (ori-β) identified in aphidicolin-synchronized CHO cells (21) was also identified in exponentially proliferating CHO cells (38).
GST-Ub assays.
For glutathione S-transferase-tagged ubiquitin (GST-Ub) labeling reactions, cells were lysed in modified CSK buffer (10 mM HEPES [pH 7.0], 150 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM ATP, and protease inhibitor cocktail [Boehringer Mannheim]) as described above or with M-PER mammalian protein extraction reagent (Pierce) plus a protease inhibitor cocktail and 0.1 mM Mg2+-ATP. About 50 μg of GST-Ub (Calbiochem) was added to 300 μl of cell extract and incubated at 25°C for 30 min in the presence of 10 μM clasto-lactacystin β-lactone (CLBL) or MG132. Stock 1 mM solutions of CLBL and MG132 (both from Calbiochem) were prepared in dimethyl sulfoxide. Reaction mixtures were then transferred to prewashed glutathione-Sepharose 4B (Pharmacia) and rotated at 4°C for 1 h. Beads were washed four times with 10 volumes of phosphate-buffered saline (PBS). Bound proteins were solubilized with sodium dodecyl sulfate (SDS) sample buffer and detected by immunoblotting with affinity-purified antisera.
Antibodies and immunoprecipitation.
Polyclonal, affinity-purified, anti-Ub antibody and monoclonal anti-FLAG antibody were purchased from Calbiochem and Sigma, respectively. Antiserum against Orc1 protein was produced by cloning the complete Chinese hamster Orc1 gene (34) into modified pFastBac (GibcoBRL) and producing N-terminal FLAG-tagged Orc1 protein that was isolated in its native form by affinity chromatography using 3X FLAG peptide (Sigma) as an eluent and then injecting it into rabbits (Covance Labs). Antiserum against Orc2 protein was produced as previously described (34).
Antibodies were purified first by affinity chromatography on protein A-Sepharose beads (Pharmacia) followed by dialysis against PBS. Either FLAG-Orc1 or FLAG-Orc2 was immobilized on a nitrocellulose membrane (Schleicher & Schuell) by transfer from an SDS-polyacrylamide gel electrophoresis (PAGE) gel in Tris-glycine buffer. Antibodies were then bound to their respective membrane-bound antigens and eluted with 0.2 M glycine (pH ∼2) and dialyzed against PBS (pH 7.4).
For immunoprecipitation of Orc proteins, antibodies were bound to protein A-Sepharose beads, and cells were extracted with M-PER mammalian protein extraction reagent (Pierce) containing complete protease inhibitor cocktail (Roche Molecular Biochemicals) and 0.1 mM Mg2+-ATP. Lysates were incubated with anti-Orc protein A-Sepharose beads for 1 h (at 4°C with rotation) and then washed five times with 10 volumes of extraction buffer. Pellets were resuspended either in Tris-glycine SDS sample buffer (Invitrogen) without a reducing agent, when proteins were fractionated by PAGE (34), or in the appropriate enzyme assay buffer.
Orc protein assays.
Total amounts of intracellular Orc proteins were determined from cells lysed with SDS sample buffer (34). To separate proteins into chromatin-bound and unbound fractions, cells were washed twice with PBS and then once with modified CSK buffer, resuspended in CSK buffer containing 0.1% Triton X-100 (about 40,000 cells/μl), incubated for 10 min on ice, and then centrifuged in an Eppendorf microcentrifuge for 4 min at 3,000 rpm (54). All reagents were on ice, and all steps were carried out at 4°C. Tightly bound Orc proteins were released by resuspending the pellet in modified CSK buffer containing 0.1% Triton X-100 and 0.3 M NaCl, incubating the mixture on ice for 10 min, and then centrifuging again. Proteins (∼20 μg/lane) were fractionated by SDS-PAGE and detected by immunoblotting as previously described (34). The amount of protein in each band was quantified by densitometry using National Institutes of Health Image software and normalized to the amount of protein present after Coomassie blue staining.
Pulse-chase assay for Orc1 stability.
Hamster cells were cultured until 40% confluent, washed twice with prewarmed PBS, and then cultured in the same medium without methionine (Gibco BRL) but supplemented with [35S]methionine (5 μCi/ml; Amersham) (>1,000 Ci/mmol). After 24 h, 10 to 15 million cells from a single flask were washed twice with prewarmed PBS and collected by trypsinization (zero hour chase). The remaining flasks were also washed with prewarmed PBS and then cultured in fresh medium containing 2× methionine for the indicated times before trypsinization. Trypsinized cells were lysed with CSK buffer plus 0.1% Triton X-100 and 0.3 M KCl in the presence of 10 μM CLBL and protease inhibitor cocktail. After centrifugation in an Eppendorf desktop centrifuge (4,000 rpm, 5 min) to remove debris, aliquots were taken from each lysate and fractionated by SDS-PAGE, and the proteins were stained with Coomassie blue. The remaining lysate was adjusted to 0.15 M KCl with CSK buffer and immunoprecipitated with either Orc1 or Orc2 affinity-purified antibodies.
RESULTS
Proliferating hamster cells contain a novel, high-molecular-mass form of Orc1.
Analysis of total Orc1 protein in SDS lysates of exponentially proliferating hamster cells revealed a single major band with a molecular mass of 99 kDa that reacted with affinity-purified Orc1 antibodies directed against the full-length protein as well as several minor bands of lower molecular masses (Fig. 1A, total lane). The amino acid sequence for hamster Orc1 predicts a molecular mass of 96 kDa (34). Therefore, based on its size, its antigenicity, and its mobility relative to cloned hamster Orc1 (data not shown), the 99-kDa protein was identified as Orc1.
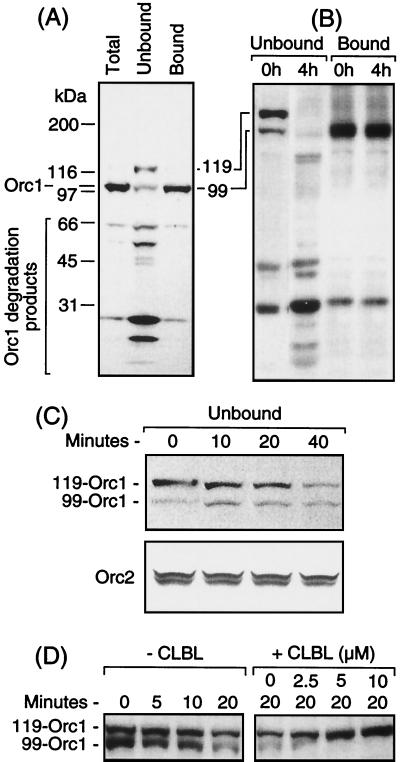
FIG. 1.
Proliferating hamster cells contained an ∼119-kDa form of Orc1 that was sensitive to degradation by the 26S proteasome. (A) CHOC 400 cells were lysed with SDS to identify all of the Orc1 cross-reacting peptides present (total) or proteins were first separated into chromatin-bound and unbound fractions before fractionating them by SDS-8% PAGE. Orc1 was detected by immunoblotting. SDS-PAGE protein standards (broad range; Bio-Rad) were fractionated in parallel on the same gel in order to calculate the molecular mass of Orc1. Each gel lane received ∼20 μg of protein (∼20,000 cell equivalents). (B) Chromatin-bound and unbound fractions of CHOC 400 cells were subjected to SDS-PAGE (4 to 12% gradient) either immediately after their preparation (0 h) or after fractions were kept on ice for 4 h, and Orc1 was assayed as described for panel A. (C) The unbound fraction was incubated at 35°C for the indicated times and then subjected to SDS-PAGE. Orc1 and Orc2 were detected by immunoblotting. (D) The unbound fraction was incubated at 35°C for up to 20 min either in the absence (−) or presence (+) of the indicated concentration of CLBL.
In an effort to identify the source of the other Orc1 cross-reacting proteins, cells were lysed in 0.1% Triton X-100 containing 0.15 M NaCl, to elute weakly bound proteins, and 1 mM Mg2+-ATP to stabilize the binding of ORC and Mcm proteins to chromatin (34). Nuclei (chromatin-bound proteins) were then separated from the cytosol (non-chromatin-bound proteins) by sedimentation and washed once in lysis buffer. Chromatin-bound and unbound proteins were then fractionated by SDS-PAGE.
Most of the Orc1 was in the chromatin-bound fraction, whereas most of the smaller Orc1 cross-reacting peptides were in the non-chromatin-bound fraction (Fig. 1A), suggesting that unbound Orc1 underwent degradation despite the presence of several protease inhibitors in the extraction buffer. In addition, the non-chromatin-bound fraction was enriched with a novel 119-kDa protein that cross-reacted with Orc1 antibodies (Fig. 1A, unbound lane) and was subsequently identified as a mono- or diubiquitinated form of Orc1.
Orc1 not bound to chromatin is degraded by the 26S proteasome in cell extracts.
The amount of Orc1 found in the unbound fraction depended on how rapidly these fractions were analyzed. Samples that were analyzed immediately contained a significant amount of both the 119- and 99-kDa forms of Orc1, while samples that had been stored for 4 h on ice had little or no full-length Orc1 but did have an increased amount of smaller Orc1 polypeptides (Fig. 1B). In contrast, Orc1 in the chromatin-bound fraction was stable over time (Fig. 1B). At 35°C, both the 119- and 99-kDa forms of Orc1 in the non-chromatin-bound fraction were rapidly degraded (Fig. 1C). However, the addition of either CLBL or MG132, two specific inhibitors of the 26S proteasome (15), resulted in the accumulation of the 119-kDa form of Orc1 with concomitant loss of the 99-kDa form of Orc1 (Fig. 1D). Since the 26S proteasome specifically degrades ubiquitinated proteins, these results suggested that the 119-kDa form of Orc1 in the non-chromatin-bound fraction was ubiquitinated, and its molecular mass suggested the presence of two 8.5-kDa Ub adducts. Moreover, since unbound Orc2 added to the non-chromatin-bound fraction was neither converted into higher-molecular-mass forms nor degraded (Fig. 1C), Orc1 appeared to be selectively modified. The possibility that this modification was phosphorylation was ruled out because incubating the 119-kDa form of Orc1 with bacteriophage lambda protein phosphatase did not alter its mobility during SDS-PAGE (data not shown).
Orc1 not bound to chromatin is selectively ubiquitinated.
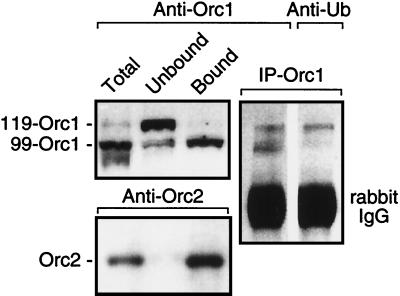
To confirm the presence of ubiquitinated Orc1 (Ub-Orc1), Orc1 was immunoprecipitated using affinity-purified Orc1 antibodies from a total-cell lysate prepared under conditions that solubilized all cellular proteins. The immunoprecipitate was then fractionated by SDS-PAGE and subjected to immunoblotting with either anti-Orc1 or anti-Ub serum. Only the 119-kDa form of Orc1 was recognized by anti-Ub serum (Fig. 2). In contrast, all of the Orc2 was bound to chromatin, and no evidence of a higher-molecular-mass form of Orc2 was detected (Fig. 2), suggesting that Orc1 was selectively ubiquitinated.
FIG. 2.
The 119-kDa form of Orc1 was linked to Ub. Cells were lysed either with SDS to display total intracellular Orc proteins or with Triton X-100-NaCl-ATP to separate proteins into chromatin-bound and unbound fractions. Samples were then fractionated by SDS-PAGE and stained with either anti-Orc1 or anti-Orc2 affinity-purified antiserum. A second sample containing both chromatin-bound and unbound proteins was immunoprecipitated with anti-Orc1 serum, and equal amounts of the aliquots of the immunoprecipitate (IP-Orc1) were fractionated by SDS-PAGE and then stained with either anti-Orc1 serum or anti-Ub serum.
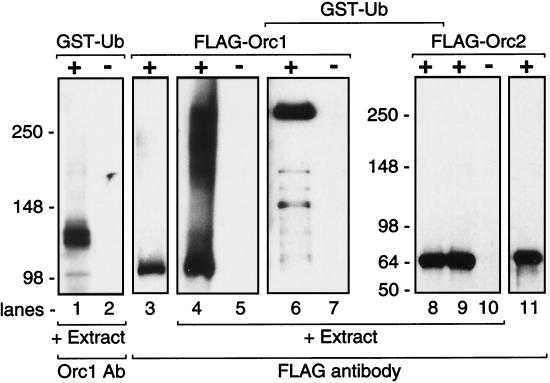
To confirm this conclusion, the non-chromatin-bound fraction was supplemented with GST-Ub (38.5 kDa) to label Orc1 and CLBL to prevent degradation of Ub-Orc1 by the 26S proteasome. GST-tagged proteins were isolated by affinity chromatography against glutathione-Sepharose beads and fractionated by SDS-PAGE, and Orc1 was identified by immunoblotting with Orc1 antiserum. The results revealed an accumulation of GST-Ub-Orc1 that was about 138 to 140 kDa in mass (Fig. 3, lane 1), consistent with a single Ub adduct per Orc1 molecule. The double band may reflect the existence of two GST moieties, one of which has a mass that is ∼1.5 kDa higher than that of the other (32), or the existence of two forms of hamster Orc1 that are sometimes detected by SDS-PAGE (Fig. 4). In the absence of CLBL, a series of lower-molecular-mass GST-tagged Orc1 fragments were observed, consistent with degradation by the 26S proteasome (data not shown).
FIG. 3.
Orc1, but not Orc2, protein was ubiquitinated in cell extracts. The non-chromatin-bound fraction (extract) was incubated either with (+) or without (−) GST-Ub (lanes 1 and 2). GST-tagged proteins were then isolated by affinity chromatography and fractionated by SDS-PAGE. Orc1 was detected with anti-Orc1 antiserum (Orc1 Ab). The non-chromatin-bound fraction (100 μl of extract) was also incubated with 1 μg of FLAG-tagged Orc1, fractionated by SDS-PAGE, and then immunoblotted with anti-FLAG antibody (lanes 4 to 7). In some assays (lanes 6 and 7), 50 μg of GST-Ub and 3 μg of FLAG-Orc1 were added to 300 μl of extract. In these assays, GST-tagged proteins were isolated first, then fractionated by SDS-PAGE, and then immunoblotted with anti-FLAG antibody. The same experiment was also carried out using FLAG-Orc2 (lanes 8 to 10). Purified FLAG-Orc1 (lane 3) and FLAG-Orc2 (lane 11) standards were fractionated in parallel.
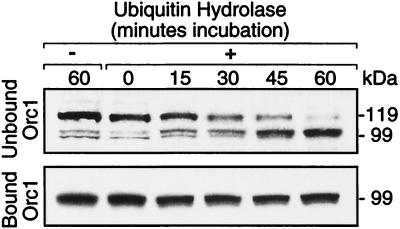
FIG. 4.
Ub hydrolase converted the 119-kDa form of Orc1 into Orc1 in a single step. Chromatin-bound and unbound fractions were incubated at 25°C for the times indicated, with Ub C-terminal hydrolase (2.5 μl of rabbit isopeptidase T at a rate of 0.4 μmol/min/mg and a concentration of 0.8 mg/ml) (catalog no. UW 8560; Biomol Research Laboratories) before adding SDS sample buffer. Assay mixtures (15 μl) contained 10 μl of cell extract, 25 mM HEPES (pH 7.5), 10 mM dithiothreitol, and 10 μM CLBL.
To determine whether or not exogenous, purified Orc1 behaved differently than the endogenous Orc1, N-terminal FLAG-tagged Orc1 purified from baculovirus-infected insect cells was added to a non-chromatin-bound fraction supplemented with CLBL to prevent protein degradation via the 26S proteasome. FLAG-tagged Orc1 was rapidly converted into a series of higher-molecular-mass forms consistent with polyubiquitination (Fig. 3, lane 4). When GST-Ub was also present, a series of distinct higher-molecular-mass forms was recovered by affinity chromatography that contained GST-Ub-FLAG-Orc1 (Fig. 3, lane 6). The major product had a molecular mass of ∼270 kDa, consistent with multiple Ub adducts. These results show that FLAG-tagged Orc1 was rapidly polyubiquitinated in hamster cell extract, whereas the endogenous Orc1 was only monoubiquitinated.
In contrast to FLAG-tagged Orc1, FLAG-tagged Orc2 was not converted into any higher-molecular-mass species, regardless of the presence or absence of GST-Ub (Fig. 3, lanes 8 and 9). This result, together with the absence of higher-molecular-mass forms of Orc1 in either the chromatin-bound (Fig. 2) or unbound fractions (Fig. 1), revealed that Orc2 was not a substrate for ubiquitination.
To further confirm that the 119-kDa form of Orc1 was ubiquitinated, and to determine the number of Ub adducts present, endogenous Orc1 was incubated with Ub C-terminal hydrolase, an enzyme that specifically removes Ub from the modified protein (48). Ub hydrolase was able to convert all of the 119-kDa form of Orc1 in the non-chromatin-bound fraction directly into the 99-kDa form of Orc1, but it did not alter the size of Orc1 in the chromatin-bound fraction (Fig. 4). The absence of any intermediate proteins between 119 and 99 kDa was consistent with the 119-kDa protein containing a single Ub adduct.
Orc1 is not selectively degraded in vivo.
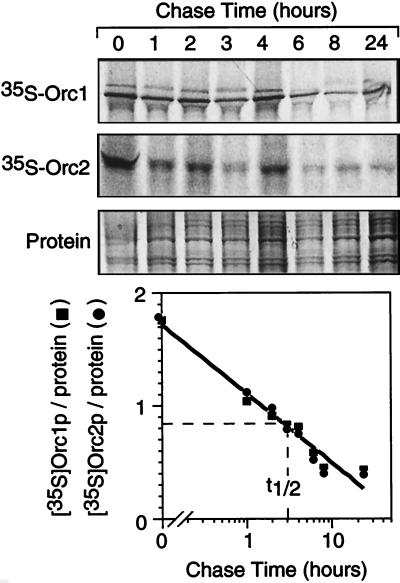
The ability of hamster cells to selectively ubiquitinate Orc1 and degrade it in vitro via the 26S proteasome pathway suggested that Orc1 may be selectively degraded in vivo. To test this hypothesis, hamster cells were cultured first in the presence of [35S]methionine to radiolabel newly synthesized proteins and then in the presence of excess unlabeled methionine to determine how long the 35S-labeled proteins remained. Both Orc1 (a substrate for ubiquitination and subsequent degradation) and Orc2 (which does not undergo ubiquitination) were degraded with the same half-life of ∼3 h (Fig. 5). Therefore, the ability of Orc1 to be selectively ubiquitinated did not result in its selective degradation in vivo.
FIG. 5.
Orc1 was not selectively degraded in vivo. Cellular proteins were radiolabeled in vivo with [35S]methionine, and then the label was chased for the times indicated before Orc1 and Orc2 were immunoprecipitated from total-cell lysates and processed as described in Materials and Methods. The amount of [35S]Orc1 and [35S]Orc2 in each band was quantified by densitometry, normalized to the amount of protein detected by Coomassie blue staining, and then plotted as a function of chase time.
One explanation for this result was that Ub-Orc1 might be retained within the nucleus and therefore protected from degradation by the 26S proteasome. To test this hypothesis, nuclei were prepared from exponentially proliferating hamster cells lysed with digitonin instead of Triton X-100. Digitonin leaves nuclei impermeable to large molecules but capable of initiating DNA replication in Xenopus egg extracts (25). Under these conditions, all of the Orc1 and Ub-Orc1 was retained in the nuclear fraction and remained stable when incubated either on ice or at 35°C, as described in the legend to Fig. 1 (data not shown). Therefore, degradation of Orc1 required the permeabilization of nuclei and the release of proteins into the cytosol.
Orc1 is released from chromatin during the S-to-M transition and ubiquitinated.
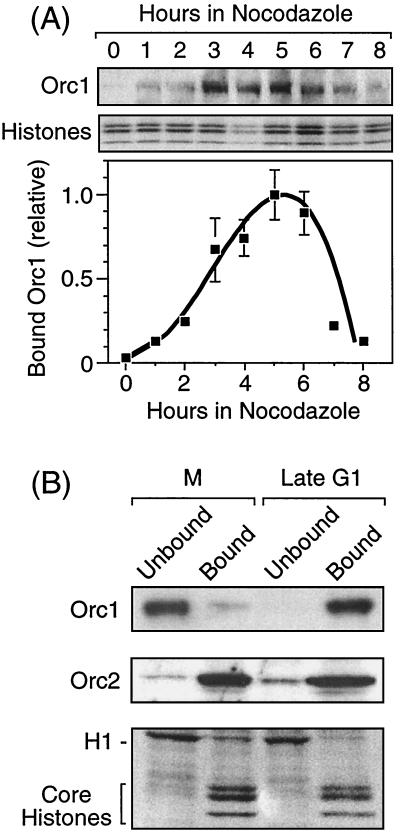
Previous studies in our laboratory reported that Orc1 underwent a dramatic change in its affinity for chromatin during the M-to-G1 transition of the cell division cycle while Orc2 remained tightly bound to chromatin (34). However, while the total amount of Orc1 in SDS-lysed cells in this study was constant, the amount detected in the non-chromatin-bound fraction varied considerably among experiments. The results described in the legend to Fig. 1 suggested that this variation may have resulted from the degradation of unbound Orc1. In fact, when cell fractions were analyzed immediately, most of the Orc1 in metaphase-arrested cells appeared in the non-chromatin-bound fraction that lacked core histones but was enriched for histone H1, whereas most of the Orc1 in late G1-phase cells appeared in the chromatin-bound fraction that contained the core histones (see Fig. 8B).
FIG. 8.
Changes in the affinity of Orc1 for chromatin in nocodazole-arrested metaphase cells. (A) The amount of Orc1 bound to metaphase chromatin depended on the length of time that cells were held in nocodazole. CHOC 400 cells were grown to ∼80% confluence and then nocodazole was added to the medium. Metaphase cells were collected at the times indicated. About 1% of the cells were recovered as M-phase cells in the absence of nocodazole and about 10% were recovered after 4 h in nocodazole. Chromatin-bound Orc1 was detected as described in the legend to Fig. 1, the amount bound was quantified by densitometry and normalized to the amount of protein detected by Coomassie blue staining, and the results were displayed relative to the maximum value observed. Each gel lane received ∼20 μg of protein (∼20,000 cell equivalents). The means ± standard errors of the means from three experiments are shown. (B) CHOC 400 cells were arrested in metaphase by culturing them with nocodazole for 3 h (M), and then they were released into G1 phase for 4 h (late G1) as described in Materials and Methods. Proteins were separated into chromatin-bound and unbound fractions and then fractionated by SDS-PAGE. The upper portions of the gel were stained with affinity-purified Orc1 and Orc2 antibodies, whereas the lower portion of the gel was stained with Coomassie blue.
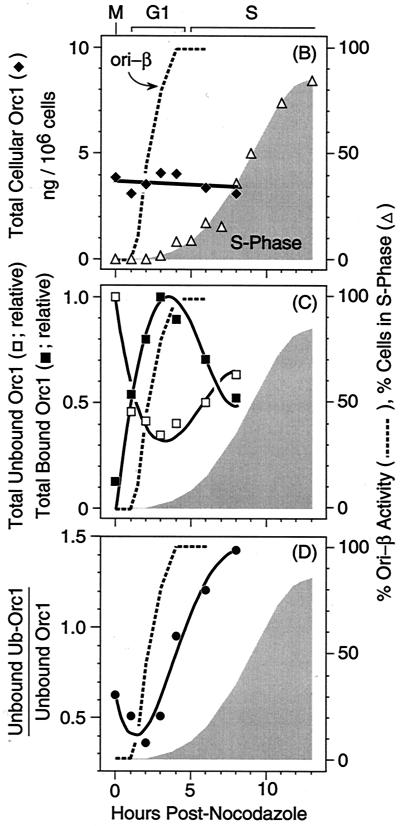
CHOC 400 cells were synchronized in metaphase with nocodazole and then isolated at various times after release of the nocodazole block. This method has been used extensively with these cells to characterize the time during G1 phase when pre-RCs appear at specific genomic loci (references 13, 25, 34, and 56 and references therein). Chromatin-bound and unbound fractions were isolated and then subjected to SDS-PAGE followed by immunoblotting to detect Orc1 (Fig. 6A), and these data were normalized against the amount of histones present in the same gel lane as previously described (34). Three conclusions were evident. First, the cellular concentration of Orc1, as determined by adding together the total amounts of Orc1 and Ub-Orc1 in each fraction, remained constant as cells progressed from M to S phase (Fig. 6B), as had been previously observed by lysing the same cell populations in SDS (34). Therefore, all of the intracellular Orc1 was recovered in these two fractions.
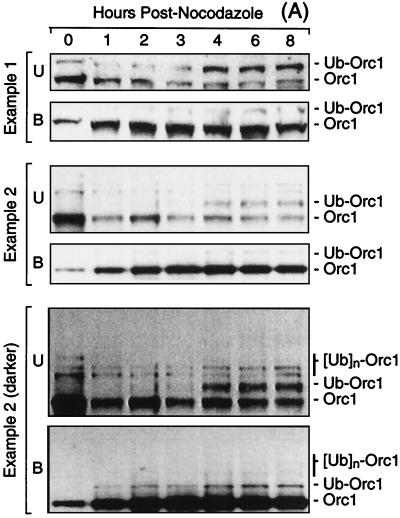
FIG. 6.
The fraction of Orc1 tightly bound to chromatin increased with time after release from metaphase, whereas Ub-Orc1 appeared during the G1-to-S transition. (A) Cells were arrested in nocodazole for 3 h and released into G1 phase, and then proteins were separated into chromatin-bound (B) and unbound (U) fractions at the times indicated, fractionated by SDS-PAGE, and immunoblotted with anti-Orc1. Two examples are shown, and there are two different exposures for example 2. (B) Data from four independent experiments were quantified by densitometry and normalized against the amount of histones present in the same gel lane as previously described (34). The sum of each form of Orc1 was determined (total cellular Orc1) and converted into nanograms of Orc1/106 cells as described in reference 34. The fraction of cells in S phase was determined by incorporating bromodeoxyuridine for 1 h and subsequent immunostaining with anti-bromodeoxyuridine antiserum (34). Data for the appearance of pre-RCs at ori-β in CHOC 400 cells (% ori-β activity) are from Fig. 3 of reference 34. These experiments were described in reference 25. The standard error of the mean for each point was between 5 and 15% of the mean. (C) Data from four independent experiments were quantified by densitometry, normalized against the amount of histones present in the same gel lane, and plotted relative to the maximum value observed. Unbound and bound Orc1 are the sums of Orc1 and Ub-Orc1 bands in these fractions. (D) The ratio of Ub-Orc1 to Orc1 in the non-chromatin-bound fraction was calculated from the data in panel C.
Second, the fraction of chromatin-bound Orc1 increased dramatically as cells went from metaphase to G1 phase. In metaphase-arrested cells, most of the Orc1 was recovered in the non-chromatin-bound fraction, and very little of it was ubiquitinated (Fig. 6A). In G1-phase cells (defined as those that contained a nucleus but had not yet begun to synthesize DNA), the amount of chromatin-bound Orc1 increased until 3 to 4 h postmetaphase, at which time it was 6- to 10-fold greater than in cells arrested in nocodazole (Fig. 6C). These changes paralleled the appearance of pre-RCs at a specific origin of bidirectional replication (ori-β) in the hamster genome (25, 34), consistent with the hypothesis that high-affinity binding of Orc1 to chromatin precedes site-specific assembly of pre-RCs in mammalian cells.
Third, the fraction of chromatin-bound Orc1 began to decrease as hamster cells passed from G1 to S phase, with a concomitant increase in the amount of Orc1 and Ub-Orc1 in the non-chromatin-bound fraction (Fig. 6A and C). This suggested that initiation of DNA replication resulted in the release of chromatin-bound Orc1 that was then targeted for ubiquitination. Consistent with this conclusion, the ratio of Ub-Orc1 to Orc1 in the non-chromatin-bound fraction increased in parallel with the onset of S phase (Fig. 6A and D). Ub-Orc1 was barely detectable in the chromatin-bound fractions from the same cells (Fig. 6A, compare unbound and bound Orc1). In some experiments, two or three higher-molecular-mass forms of Orc1 were also observed in addition to Ub-Orc1 (Fig. 6A, example 2).
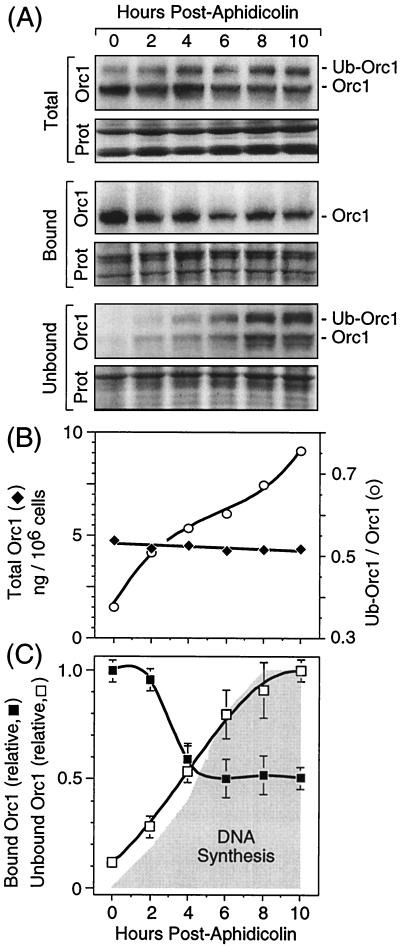
To confirm this hypothesis, cells were synchronized at their G1/S boundary with aphidicolin and then released into S phase. Cells were lysed with SDS at various times in order to measure the total intracellular levels of Orc1 and Ub-Orc1. The results confirmed that while the total intracellular Orc1 remained constant as cells progressed through S phase, the ratio of Ub-Orc1 to Orc1 increased dramatically (Fig. 7A and B). Moreover, most, if not all, of the Ub-Orc1 was in the non-chromatin-bound fraction (Fig. 7A). Conversely, the amount of chromatin-bound Orc1 decreased as cells progressed through S phase, although the level reached a plateau when about half of the Orc1 was released (Fig. 7C). This result suggests that Orc1 associated with ORCs assembled at newly replicated origins remained tightly bound to the newly replicated chromatin. In contrast to Orc1, the amount of Orc2 that remained bound to chromatin during S phase was constant (data not shown). These results suggested that initiation of DNA replication selectively reduced the affinity of Orc1 at activated ORC-chromatin sites, allowing Orc1 to be targeted for ubiquitination.
FIG. 7.
Ub-Orc1 appeared after initiation of DNA replication. (A) Cells were synchronized at the G1/S boundary using aphidicolin and then released into S phase (see Materials and Methods). At the times indicated, cells were either lysed with SDS to assay total cellular Orc1 or fractionated into chromatin-bound and non-chromatin-bound fractions and then assayed for Orc1. (B) Total intracellular Orc1 (SDS lysates) and the ratio of Ub-Orc1 to Orc1 in these lysates were determined as a function of time after release from aphidicolin. (C) The amounts of chromatin-bound and unbound Orc1 were determined as a function of time after release of cells into S phase (shaded area). Results from three experiments were averaged, and the standard errors of the means are indicated. With the exception of the method for cell synchronization, all other methods were as described in the legend to Fig. 6.
Changes in the affinity of Orc1 for chromatin are not coupled to microtubule assembly.
Previous studies using mitotic cells recovered in the absence of nocodazole (Fig. 7 in reference 34) suggested that M-phase cells isolated in the absence of nocodazole contained less Orc1p tightly bound to chromatin than was detected in M-phase cells that were arrested in nocodazole. This raised the issue of whether or not changes in Orc1 affinity for chromatin were coupled to the reassembly of microtubules when metaphase cells were released from their nocodazole block. To address this issue, hamster cells were cultured in nocodazole for increasing periods of time, and the amount of Orc1 tightly bound to metaphase chromatin was determined. As previously reported (34), when care was taken to avoid contamination with interphase cells, chromatin-bound Orc1 was virtually undetectable in metaphase cells isolated in the absence of nocodazole (Fig. 8A). However, the amount of Orc1 stably bound to metaphase chromatin increased during the first 5 h that cells were arrested in nocodazole and then decreased again (Fig. 8A). This time course was remarkably similar to that observed when nocodazole-arrested metaphase cells were released into G1 phase (Fig. 6C), revealing that Orc1 begins to bind stably to chromatin as soon as metaphase was completed, regardless of whether or not microtubule assembly occurs. Moreover, Orc1 was released from metaphase chromatin when S phase would have begun had nocodazole been absent. Therefore, the affinity of Orc1 for chromatin responded to cell cycle events that were not coupled to microtubule assembly. Furthermore, essentially all of the Orc1 in cycling cells passing through metaphase was weakly bound to chromatin and therefore could be selectively eluted with Triton X-100-0.15 M NaCl-1 mM Mg2+-ATP.
Three differences were detected between nocodazole-arrested metaphase chromatin and G1-phase chromatin that were relevant to the cell cycle-dependent affinity of Orc1 for chromatin. First, chromatin from M-phase cells that had been arrested in nocodazole for up to 4 h did not initiate DNA replication when incubated in a Xenopus egg extract depleted of Xenopus Orc proteins (25, 34, 59). Therefore, M-phase chromatin contains few, if any, active ORCs. Second, the amount of Orc1 bound to M-phase chromatin after 3 h in nocodazole was only ∼10% of the peak levels observed in G1-phase chromatin (Fig. 8B). Thus, the amount of Orc1 bound to metaphase chromatin never exceeded 10 to 20% of the level which bound to interphase chromatin. Higher levels were only observed when preparations of M-phase cells were visibly contaminated with interphase cells. Finally, hamster cells that were arrested in nocodazole for longer than 5 h failed to enter the cell cycle when released from the nocodazole block (18, 58), consistent with the inactivation of chromatin-bound ORCs by the release of Orc1.
DISCUSSION
Cell cycle-dependent selective release of chromatin-bound Orc1.
The results presented here and elsewhere support the existence of a novel regulatory pathway in the initiation of mammalian DNA replication (Fig. 9). In mammals, Orc1 and Orc2 are both tightly bound to chromatin during late G1 phase (33, 34, 43), and late G1-phase nuclei contain active ORC-chromatin sites by virtue of the fact that they can initiate DNA replication at specific genomic sites when incubated in an Orc-depleted Xenopus egg extract (25, 34, 59). In contrast to yeast in which all six ORC subunits are stably bound to chromatin throughout the cell cycle (11, 17, 22, 27, 29), the affinity of mammalian Orc1 for chromatin is selectively reduced during S phase, such that lysis of cells in 0.1% Triton X-100, 0.15 M NaCl, and 1 mM Mg2+-ATP releases Orc1, but not Orc2, into the non-chromatin-bound fraction (Fig. 6 and 7) (23). Moreover, the Orc1 that is released during S phase is rapidly ubiquitinated.
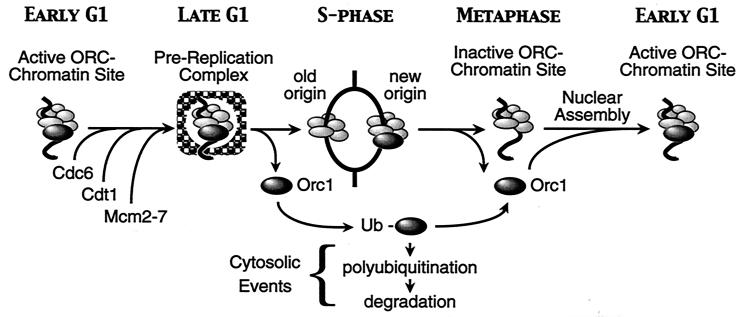
FIG. 9.
We propose that mammalian ORC activity is regulated through cell cycle-dependent changes in the affinity of Orc1 for ORC-chromatin sites. Orc1 is selectively released from chromatin as cells enter S phase, converted into a mono- or diubiquitinated form, and then deubiquitinated and rebound to chromatin during the M-to-G1 transition. In contrast, Orc2 (and presumably other Orc proteins) remains tightly bound to chromatin throughout the cell cycle and is not a substrate for ubiquitination. In vivo, monoubiquitination does not target Orc1 for degradation. Therefore, we propose that the role of monoubiquitination of non-chromatin-bound Orc1 is to sequester Orc1 during S phase and thus prevent its reassociation with the other ORC subunits. However, Orc1 is degraded by the 26S proteasome when the nuclear envelope is permeabilized, and modified forms of Orc1 (e.g., FLAG-tagged Orc1) are rapidly polyubiquitinated. Therefore, it is possible that DNA damage, cell quiescence, or cell differentiation could lead to the destruction of Orc1, which would trigger the loss of the remaining ORC subunits.
Although Orc2 was generally not detected in the non-chromatin-bound fraction (Fig. 2; also data not shown) (34), a small amount was sometimes observed (Fig. 8B). Whether this resulted from chromatin fragmentation or the release of a special Orc1/Orc2-containing complex, as suggested by others (23), is not known. However, the amount of Orc2 tightly bound to chromatin remains essentially constant throughout the cell cycle (33, 34, 43) (Fig. 8B and data not shown), and neither endogenous Orc2 (Fig. 2) nor FLAG-Orc2 (Fig. 3) was a substrate for ubiquitination, suggesting that only Orc1 is released from chromatin-bound ORCs. Orc4p also remains tightly bound to chromatin throughout the cell cycle (W.-S. Sun, unpublished data).
Since only half of the Orc1 was released during S phase (Fig. 7C), we postulate that it is released only from those ORC-chromatin sites where replication was initiated (old origins). However, since all of the Orc1 in metaphase cells (particularly those cells that have not been arrested in nocodazole) (Fig. 8A) (34) appears in the non-chromatin-bound fraction (Fig. 6) (34), the remaining Orc1 must be released from ORCs bound to newly replicated origins when S phase is completed. Since metaphase chromatin cannot initiate DNA replication in Xenopus egg extracts depleted of Xenopus Orc proteins (25, 34), the missing hamster Orc1 subunit does not allow the remaining hamster Orc proteins to form an active ORC-chromatin site. Therefore, release of hamster Orc1 during S phase would prevent reinitiation of DNA replication at those replication origins.
What triggers the release of Orc1 from ORC-chromatin sites? Since both Orc1 and Ub-Orc1 are rapidly released from chromatin when cells enter S phase and Ub-Orc1 is largely absent from mitotic cells, ubiquitination per se is not likely the cause of Orc1 release, but it is a mechanism to prevent its reassociation with ORC-chromatin sites during S phase. The affinity of Orc proteins for chromatin may depend on their phosphorylated state. For example, both XlOrc1 and XlOrc2 are hyperphosphorylated in metaphase-arrested eggs relative to activated eggs (5, 55), and Xenopus ORCs can be selectively released from chromatin by incubating chromatin either in a metaphase extract (46) or with Cdc2/cyclin A (16, 19). Thus, cyclin-dependent protein kinases may be responsible for altering the affinity of Orc1 for ORC-chromatin sites as cells transit from S to M phase.
Stable binding of Orc1 to chromatin occurs even when cells are arrested in nocodazole (Fig. 8A). Therefore, the affinity of Orc1 for chromatin is not linked to microtubule assembly. However, the amount of Orc1 bound to metaphase chromatin is much less than with interphase chromatin (Fig. 8B), and active ORC-chromatin sites are not detectable in metaphase chromatin (25, 34, 59), despite the presence of mammalian Cdc6p (33, 39) (data not shown). Mcm proteins may also begin binding to chromatin during this time period, but they do not produce active pre-RCs because active ORCs and pre-RCs do not appear until after cells have completed mitosis and formed an intact nuclear membrane (34, 59). In fact, chromatin-bound Cdc6p disappears during early G1 and reappears during late G1, consistent with assembly of functional pre-RCs during late G1 phase (33, 39).
Orc1 protein associates with chromatin in mammalian metaphase cells under several conditions (cells permeabilized with digitonin [34], cells overexpressing Orc1 [35], and cells arrested in TN16 for 24 h [52]). Differences in the amount of Orc1 eluted from chromatin in these experiments may reflect the presence or absence of ATP in the extraction buffer, the length of time cells are arrested in nocodazole, or the use of uncharacterized drugs, such as TN16. Nevertheless, chromatin-bound Orc1 in metaphase cells, relative to other Orc proteins, is salt sensitive (34, 35, 52), and ORC activity is absent (25, 34, 59). Therefore, whatever the nature of the association between Orc1 and metaphase chromatin, it does not produce functional ORCs. Restoration of ORC activity reappears only when Orc1 binds tightly to chromatin in early G1-phase cells (34).
ORC activity is restored during the 2-h transition from M to G1 phase (34, 59), concomitant with the appearance of tightly bound Orc1 (Fig. 6C) (34). Moreover, Mcm proteins, which are not bound to metaphase chromatin, begin to reassociate with chromatin during late telophase and reach their maximum level of association by 2 h after release from metaphase (14, 33, 34), consistent with the assembly of pre-RCs. Thus, the assembly of pre-RCs during the M-to-G1 transition is limited by the rate of assembly of functional ORCs on chromatin, which appears to be limited by the rate at which Orc1 becomes tightly bound to chromatin. These events are inversely correlated with the loss of cyclin B (33), which is required to drive cells into mitosis, and the formation of a nuclear envelope (12, 34), which is required for the initiation of DNA replication. Loss of cyclin B during the M-to-G1 transition would be expected to alter the phosphorylation pattern of proteins, one of which may be Orc1.
Cell cycle-dependent selective ubiquitination of Orc1.
A significant portion of the Orc1 that is released from chromatin is modified by ubiquitination (Fig. 1 to 4). However, the role of ubiquitination during the S-to-M transition is not to destroy Orc1 but to sequester it in order to prevent its reassociation with ORC-chromatin sites (Fig. 9). The rate of Orc1 degradation in vivo was the same as that of nonubiquitinated Orc2 (Fig. 5), and the intracellular concentration of Orc1 remained essentially constant throughout the cell cycle (Fig. 6B and 7B) (34). Since little if any of the Orc1 in mitotic cells is ubiquitinated, Ub-Orc1 must be converted back into unbound Orc1 sometime during the S-to-M transition, suggesting an equilibrium between ubiquitination and deubiqitination of unbound Orc1. Nonubiquitinated Orc1 then rebinds to chromatin during the subsequent G1 phase to produce active ORCs at specific chromosomal sites.
This conclusion is not surprising in view of the fact that ubiquitination is limited to one or possibly two adducts, whereas efficient degradation of ubiquitinated proteins requires chains of four or more Ub molecules. Despite the fact that Ub-Orc1 migrated during SDS-PAGE as a molecule 20 kDa larger than Orc1 (suggesting two Ub adducts), three other experiments detected only one Ub adduct. First, ubiquitination of endogenous Orc1 by GST-Ub produced a single major species whose molecular mass was consistent with only one GST-Ub adduct (Fig. 3). Second, Ub hydrolase (an enzyme that specifically removes Ub [48] and not Ub-related proteins such as SUMO-1 [26]) converted Ub-Orc1 into Orc1 (Fig. 4), whereas proteasome inhibitors such as CLBL and MG132 allowed conversion of Orc1 into the 119-kDa form of Orc1 (Fig. 1D). In both cases, no intermediate-sized proteins were detected, suggesting that the 119-kDa Orc1 contained a single Ub adduct.
While some unbound Orc1 is polyubiquitinated (Fig. 6A) and degraded by the 26S proteasome (Fig. 1), the bulk of Orc1 remains available for reassembly into an active ORC. Endogenous Ub-Orc1 was degraded only when nuclei were permeabilized using Triton X-100 and then either kept on ice for some time or incubated briefly at 30°C. The mono- or diubiquitinated form of Orc1 presumably is further ubiquitinated before undergoing degradation by the 26S proteasome. In some experiments (Fig. 6A, example 2), one can see additional bands consistent with 4 to 6 Ub adducts. Degradation was not observed when cell membranes were permeabilized with digitonin, and the nuclei remained impermeable to large proteins. Degradation was not observed when either CLBL or MG132 (specific inhibitors of the 26S proteasome) was included in the Triton X-100 lysate. In contrast, inhibitors of serine proteases, cysteine proteases, and metalloproteases that were already present in the cell lysis cocktail did not prevent degradation.
General protease inhibitors present in the lysis buffer did not prevent degradation. Only when Orc1 was modified by the addition of an eight-amino-acid N-terminal FLAG peptide was it rapidly polyubiquitinated (Fig. 3). Therefore, it is possible that Orc1 remains weakly associated with inactive ORC-chromatin complexes (Fig. 9), and the purpose of monoubiquitination is to prevent premature reassembly into an active ORC complex. In fact, both Orc1 and Orc2 were associated with metaphase chromatin in 50 mM KCl when cells were permeabilized with digitonin (34).
Other examples of mono- or diubiquitinated proteins have been reported, and all of them are involved in regulating cellular processes. For example, monoubiquitination on a single lysine is necessary and sufficient for rapid endocytosis of an activated signal-transducing receptor in yeast (53). Monoubiquitinated Gag protein is important for the release of viral particles from the host cell (37, 49). Monoubiquitination of Drosophila histone H1 is dependent on TAF250, and mutations in TAF250 that prevent synthesis of Ub-H1 are defective in transcription (40).
Polypeptide sequences enriched in proline, glutamic acid, serine, and threonine (PEST sequences) are strongly implicated in targeting proteins for degradation by the 26S proteasome (42). Orc1 contains only one potential PEST sequence (amino acids 396 to 427) (PESTFind program; www.at.embnet.org), and this PEST sequence had a score of +21. PEST sequences known to function as proteolytic signals have scores of +5 to +18. No additional signals were found in FLAG-Orc1, and no signals were found in either Orc2 or FLAG-Orc2. Analysis of PEST signals in human and mouse Orc proteins suggests that only Orc1 is targeted for ubiquitination, and therefore, the model proposed in Fig. 9 for hamster cells likely applies to all mammals.
The results presented here and in a previous study (34) are internally consistent and show an examination of the affinity of Orc1 and Orc2 proteins for chromatin under four different cell culture conditions. Exponentially proliferating cells were used to detect selective ubiquitination of Orc1 that was not bound to chromatin. The relationship between chromatin affinity, ubiquitination, and the cell division cycle was determined under three different conditions. Cells were synchronized in metaphase either with the help of nocodazole to prevent microtubule assembly or without nocodazole to examine cycling metaphase cells, and cells were synchronized at their G1/S boundary using isoleucine deprivation followed by aphidicolin to prevent DNA synthesis. Remarkably, the time courses for changes in the affinity of Orc1 protein for chromatin were essentially the same. Orc1 was weakly bound to chromatin in M phase. Cycling metaphase cells contained little, if any, detectable chromatin-bound Orc1 (Fig. 8A) (Fig. 7 in reference 34). Orc1 became tightly bound to chromatin during the M-to-G1 transition and then began to be released from chromatin during the G1-to-S transition. Release was completed somewhere during the S-to-M transition. Ub-Orc was detected primarily, if not exclusively, among the non-chromatin-bound proteins in S-phase cells.
Regulating initiation of DNA replication by regulating ORC activity.
One can envisage at least three advantages to regulating ORC activity in metazoan cells. The first is to prevent reassembly of pre-RCs during S phase by simply inactivating ORC-chromatin sites where initiation occurred through destabilization and subsequent ubiquitination of Orc1. Since mammalian Cdc6p binds Orc1 (47), selective inactivation of Orc1 may directly prevent recruitment of Cdc6p to the ORC-chromatin site. By applying this strategy to newly assembled ORC-chromatin sites as well as to those sites where initiation has occurred, the decision to reenter a proliferative cell cycle is delayed until mitosis is complete and a nuclear membrane has been reassembled. This leads directly to a second regulatory role for Orc1.
Reassembly of Orc1 into stable ORC-chromatin sites provides a mechanism for selecting which of the many potential initiation sites along the genome will be activated (the Jesuit model in references 8 and 9). Concurrent with the accumulation of stably bound Orc1 and Mcm proteins on mammalian chromatin is the appearance of pre-RCs in nuclei from G1-phase cells at the same specific initiation sites normally used in vivo (25, 34). This cell cycle-dependent event is referred to as the origin decision point (56). Prior to the origin decision point, Xenopus egg extract can initiate DNA replication randomly throughout the hamster genome (25, 34, 57, 59), a phenomenon that is dependent on Xenopus Orc proteins (25, 34). This phenomenon could account for the observation that both early and late replicating regions of the hamster genome replicate concurrently when early G1 nuclei (1 h postmetaphase) are incubated in a Xenopus egg extract (13). Thus, the absence of stably bound Orc1 during the S-to-M transition allows the number and locations of initiation sites to be reprogrammed during animal development.
Finally, cell cycle-dependent ORC instability offers a primary checkpoint control mechanism by which cells could disassemble ORC-chromatin sites and thereby prevent proliferation under special circumstances, for example, cells that have sustained DNA damage or that have entered a quiescent or terminally differentiated state. Cells entering a quiescent state have, in fact, been shown to lose their ability to establish pre-RCs at specific genomic sites (57) and to bind Cdc6 and Mcm proteins to their chromatin (30, 50). Moreover, terminally differentiated cells lack critical proteins required for the assembly of pre-RCs (28). While the mechanisms involved have not yet been elucidated, ubiquitination and destruction of Orc1 may well trigger a cascade of events that shuts down cell proliferation pathways.
Acknowledgments
We are indebted to Alex Vassilev and Xiaohong Zhang for preparing the FLAG-Orc1 and FLAG-Orc2 proteins and to the members of the DePamphilis laboratory for their helpful advice and suggestions.
REFERENCES
- 1.Abdurashidova, G., S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 1998. Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J. 17:2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, M., and R. P. Wharton. 1999. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 18:2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogan, J. A., D. A. Natale, and M. L. Depamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol. 184:139–150. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, P. B., and W. G. Dunphy. 1998. Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J. Biol. Chem. 273:24891–24897. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, P. B., P. R. Mueller, and W. G. Dunphy. 1996. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature 379:357–360. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87:53–63. [DOI] [PubMed] [Google Scholar]
- 8.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 62:29–63. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis, M. L. 1996. Origins of DNA replication, p. 45–86. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.DePamphilis, M. L. 1999. Replication origins in metazoan chromosomes: fact or fiction?. Bioessays 21:5–16. [DOI] [PubMed] [Google Scholar]
- 11.Diffley, J. F., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303–316. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., and D. M. Gilbert. 1998. Regulation of mammalian replication origin usage in Xenopus egg extract. J. Cell Sci. 111:2989–2998. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrova, D. S., and D. M. Gilbert. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4:983–993. [DOI] [PubMed] [Google Scholar]
- 14.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenteany, G., and S. L. Schreiber. 1998. Lactacystin, proteasome function, and cell fate. J. Biol. Chem. 273:8545–8548. [DOI] [PubMed] [Google Scholar]
- 16.Findeisen, M., M. El-Denary, T. Kapitza, R. Graf, and U. Strausfeld. 1999. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem. 264:415–426. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, M., Y. Hori, K. Shirahige, T. Tsurimoto, H. Yoshikawa, and C. Obuse. 1998. Cell cycle dependent topological changes of chromosomal replication origins in Saccharomyces cerevisiae. Genes Cells 3:737–749. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, D. M., H. Miyazawa, and M. L. DePamphilis. 1995. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol. 15:2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua, X. H., and J. Newport. 1998. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 140:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829–880. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong, D., and M. L. DePamphilis. 2001. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 21:8095–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreitz, S., M. Ritzi, M. Baack, and R. Knippers. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276:6337–6342. [DOI] [PubMed] [Google Scholar]
- 24.Landis, G., R. Kelley, A. C. Spradling, and J. Tower. 1997. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl. Acad. Sci. USA 94:3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, C. J., J. A. Bogan, D. A. Natale, and M. L. DePamphilis. 2000. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci. 113:887–898. [DOI] [PubMed] [Google Scholar]
- 26.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246–251. [DOI] [PubMed] [Google Scholar]
- 27.Liang, C., and B. Stillman. 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11:3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Z. H., H. Xu, and G. H. Leno. 1999. DNA replication in quiescent cell nuclei: regulation by the nuclear envelope and chromatin structure. Mol. Biol. Cell 10:4091–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lygerou, Z., and P. Nurse. 1999. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 112:3703–3712. [DOI] [PubMed] [Google Scholar]
- 30.Madine, M. A., M. Swietlik, C. Pelizon, P. Romanowski, A. D. Mills, and R. A. Laskey. 2000. The roles of the MCM, ORC, and Cdc6 proteins in determining the replication competence of chromatin in quiescent cells. J. Struct. Biol. 129:198–210. [DOI] [PubMed] [Google Scholar]
- 31.Malott, M., and M. Leffak. 1999. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19:5685–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McTigue, M. A., D. R. Williams, and J. A. Tainer. 1995. Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 246:21–27. [DOI] [PubMed] [Google Scholar]
- 33.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natale, D. A., C. J. Li, W. H. Sun, and M. L. DePamphilis. 2000. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 19:2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pak, D. T., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum, J. Marr, P. Romanowski, and M. R. Botchan. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311–323. [DOI] [PubMed] [Google Scholar]
- 37.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelizon, C., S. Diviacco, A. Falaschi, and M. Giacca. 1996. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16:5358–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357–2360. [DOI] [PubMed] [Google Scholar]
- 41.Phi-van, L., and W. H. Stratling. 1999. An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res. 27:3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267–271. [PubMed] [Google Scholar]
- 43.Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R. A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543–24549. [DOI] [PubMed] [Google Scholar]
- 44.Romanowski, P., M. A. Madine, A. Rowles, J. J. Blow, and R. A. Laskey. 1996. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6:1416–1425. [DOI] [PubMed] [Google Scholar]
- 45.Rowles, A., J. P. Chong, L. Brown, M. Howell, G. I. Evan, and J. J. Blow. 1996. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell 87:287–296. [DOI] [PubMed] [Google Scholar]
- 46.Rowles, A., S. Tada, and J. J. Blow. 1999. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112:2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha, P., J. Chen, K. C. Thome, S. J. Lawlis, Z.-H. Hou, M. Hendricks, J. D. Parvin, and A. Dutta. 1998. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18:2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein, R. L., Z. Chen, and F. Melandri. 1995. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry 34:12616–12623. [DOI] [PubMed] [Google Scholar]
- 49.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, W. H., M. Hola, N. Baldwin, K. Pedley, and R. F. Brooks. 2001. Heterogeneity in nuclear transport does not affect the timing of DNA synthesis in quiescent mammalian nuclei induced to replicate in Xenopus egg extracts. Cell Prolif. 34:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatsumi, Y., T. Tsurimoto, K. Shirahige, H. Yoshikawa, and C. Obuse. 2000. Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem. 275:5904–5910. [DOI] [PubMed] [Google Scholar]
- 53.Terrell, J., S. Shih, R. Dunn, and L. Hicke. 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell 1:193–202. [DOI] [PubMed] [Google Scholar]
- 54.Todorov, I. T., A. Attaran, and S. E. Kearsey. 1995. BM28, a human member of the MCM2–3–5 family, is displaced from chromatin during DNA replication. J. Cell Biol. 129:1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tugal, T., X. H. Zou-Yang, K. Gavin, D. Pappin, B. Canas, R. Kobayashi, T. Hunt, and B. Stillman. 1998. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem. 273:32421–32429. [DOI] [PubMed] [Google Scholar]
- 56.Wu, J. R., and D. M. Gilbert. 1996. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271:1270–1272. [DOI] [PubMed] [Google Scholar]
- 57.Wu, J.-R., and D. M. Gilbert. 1997. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol. Cell. Biol. 17:4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, J. R., G. Yu, and D. M. Gilbert. 1997. Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods 13:313–324. [DOI] [PubMed] [Google Scholar]
- 59.Yu, G., J. R. Wu, and D. M. Gilbert. 1998. Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts. Genes Cells 3:709–720. [DOI] [PubMed] [Google Scholar]