Abstract
Background
Minichromosome maintenance (MCM) proteins are essential for the initiation of DNA replication and have been found to be relevant markers for prognosis in a variety of tumours. The aim of this study was to assess the proliferative activity of diffuse large B-cell lymphoma (DLBCL) in tissue microarray (TMA) using one of the minichromosome maintenance proteins (Mcm2) and to explore its potential value to predict prognosis.
Methods
Immunohistochemistry for Mcm2 was performed on TMAs constructed from 302 cases of DLBCL. A monoclonal mouse antibody was used after heat induced antigen retrieval. Mcm2 expression was scored quantitatively. Positivity for Mcm2 was defined as presence of nuclear expression of Mcm2 in greater than or equal to 40 % of tumour cells. A statistical analysis was carried out of the association of Mcm2 and the clinico-pathological characteristics.
Results
Mcm2 expression was clearly evident in the nuclei of proliferating non-neoplastic cells and tumour cells. Positivity for Mcm2 was found in 46% (98/211) of analysable cases. A significant correlation existed between Mcm2 expression and presence of bulky disease (p = 0.003). Poor disease specific survival was observed in patients with DLBCL positive for Mcm2 expression in the univariate analysis (p = 0.0424).
Conclusion
Mcm2 expression can be used to assess tumour proliferation and may be useful as an additional prognostic marker to refine the prediction of outcome in DLBCL.
Background
In Western countries, diffuse large B-cell lymphoma (DLBCL) is the most common type of mature B-cell lymphomas with a frequency of approximately 30 to 40% [1]. DLBCL is an aggressive but potentially curable disease. However, only about 40 to 45% of patients are cured with combination chemotherapy [2]. Currently, the International Prognostic Index (IPI) is the generally accepted predictor of prognosis [3]. In order to avoid over-treatment of some patients and identify patients at high-risk for early relapse or poor response to standard treatment, individualised prediction of prognosis becomes more and more important.
A number of biological markers have been studied for their predictive potential, but none has become universally accepted [4-7]. Gene expression profiling has helped to identify at least two subgroups of DLBCL, one with a germinal centre signature and the other with an activated B-cell signature, which show distinct clinical outcomes [3,8].
The proliferative capacity of neoplastic cells is an important feature of growing tumours. Assessment of cell proliferation may provide both pathologists and clinicians with more objective prognostic information [9,10]. Expression of Ki-67 as assessed by immunohistochemistry has become the standard proliferation marker [11-13]. In lymphoid neoplasms, it is controversial whether Ki-67 is a reliable prognostic indicator [14-17]. In DLBCL, a high proliferation index has been associated with an unfavourable clinical outcome in some studies [18]. Despite the extensive use of Ki-67, its functional significance still remains unclear [19]. It has been suggested, that Ki-67 plays a role in the ribosome biosynthesis rather than being directly responsible for cell proliferation [20,21]. Therefore, detecting markers directly involved in DNA replication might be a more precise method to evaluate the proliferative behaviour of a tumour.
The minichromosome maintenance (MCM) protein family consists of six members of DNA-binding proteins [22]. MCM proteins stand at the end of many signalling pathways involved in cell proliferation [23]. They ensure that synthesis of DNA is initiated only once during each cell cycle. All six proteins are abundant throughout the cell cycle and are broken down rapidly on differentiation or more slowly in quiescence [24,25]. Expression is only observed in cycling cells, there is no expression in quiescent and differentiating cells [25-28].
Antibodies for detection of MCM proteins in routinely processed tissue specimen have been found superior to Ki-67 in defining the proliferative compartments in both normal and abnormal human tissues [27,29-32]. Immunohistochemical assessment of all six MCM proteins has been observed to produce similar results in a range of tissue sections [33]. MCM proteins have been promoted as markers for cancer screening, surveillance and prognosis [27,29-31,34-36]. We used a specific monoclonal antibody directed against Mcm2 and a previously validated tissue microarray (TMA) with tissue samples of a large number of DLBCL [37]. Tissue microarrays are highly efficient for the investigation of large series of neoplasms including lymphoma [37-39].
The aim of this study was to systematically investigate if the analysis of Mcm2 expression might provide a novel tool to assess the proliferation of DLBCL and predict clinical outcome in patients with this disease.
Methods
Construction of tissue microarrays and acquisition of clinico-pathologic data
Tissue microarrays were constructed as described previously [39,40]. The TMAs contained a total of 302 tissue samples from tumours, which had been classified prior to this study as diffuse large B-cell lymphomas according to the WHO classification [1]. All samples had been obtained at the time of diagnosis, before any treatment had been given. Four different TMAs were constructed, each containing tumour samples from different histopathologic institutions (Basel, Bologna, Innsbruck, Zurich). Clinical data of patients with DLBCL at time of primary diagnosis and follow-up had been obtained by reviewing the charts. Clinico-pathologic data of patients at time of diagnosis are detailed in table 1. Retrieval of tissue and clinical data was performed according to the regulations of the local institutional review board and data safety laws.
Table 1.
Details of clinico-pathologic data and expression of Mcm2 in diffuse large B-cell lymphomas
| Variable | Categorisation | n analysable | % |
| Clinico-pathologic data: | |||
| Age at diagnosis | |||
| median 67 years (interquartile range 54–75 years) | |||
| <65 years | 69 | 43.1 | |
| ≥65 years | 91 | 56.9 | |
| Gender | |||
| male | 97 | 53.3 | |
| female | 85 | 46.7 | |
| Ann Arbor stage | |||
| 1 | 27 | 17.3 | |
| 2 | 50 | 32.1 | |
| 3 | 34 | 21.8 | |
| 4 | 45 | 28.8 | |
| International prognostic index (IPI) | |||
| 0 | 2 | 1.3 | |
| 1 | 66 | 43.4 | |
| 2 | 29 | 19.1 | |
| 3 | 32 | 21.1 | |
| 4 | 23 | 15.1 | |
| Bone marrow involvement | |||
| yes | 44 | 28.6 | |
| no | 110 | 71.4 | |
| Extranodal involvement | |||
| yes | 54 | 35.8 | |
| no | 97 | 64.2 | |
| Bulky disease | |||
| yes | 94 | 64.4 | |
| no | 52 | 35.6 | |
| Chemotherapy | |||
| no therapy | 7 | 4.6 | |
| CHOP | 69 | 45.1 | |
| other | 77 | 50.3 | |
| Serum levels of LDH at diagnosis | |||
| median 306 U/l (interquartile range 185–443 U/l) | |||
| <300 U/l | 61 | 49.2 | |
| ≥300 U/l | 63 | 50.8 | |
| Immunohistochemistry: | |||
| Mcm2 immunoreactivity | |||
| median 38% (interquartile range 29%–48%) | |||
| <40% | 113 | 53.6 | |
| ≥40% | 98 | 46.4 | |
Immunohistochemistry
Four-micrometre sections of the TMA blocks were cut to adhesive-coated slides (Instrumedics Inc, Hackensack, NJ, USA) and stained using standard procedures. Briefly, immunohistochemical studies utilized an avidin-biotin peroxidase method with diaminobenzidine chromatogen. After heat induced antigen retrieval (microwave oven for 30 min at 250 W) immunohistochemistry was carried out in a NEXES immunostainer (Ventana, Tucson, AZ). The following primary antibody was used: BM28 (mouse monoclonal, clone 46, BD Biosciences, San Jose, US, final dilution 1:3000). The dilution had been established using adequate controls. Negative controls were obtained by omitting the primary antibody. The slides were evaluated without knowledge of clinical data. At least 10% of cases were re-evaluated by a second observer. Only cases containing clearly recognisable tumour tissue were analysed. One-hundred cells were assessed in each tumour core and the percentage of positive cells (i.e. cells with a distinct staining) was calculated. If a biopsy core contained less than 100 cells as many neoplastic cells as possible were evaluated. If more than one core from the same tumour was available the results were averaged. Cases were defined positive for Mcm2 if ≥40 % of tumours cells showed distinct nuclear staining. The determination of cut-off levels was based on the analysis of the area under the receiver operating characteristic curve (AUROC) as described below and as detailed in table 2.
Table 2.
Performance of metric variables in prediction of mortality using the AUROC (area under the receiver operating characteristic curve) method
| Mcm2 IHCc | LDHd | |
| Optimum cutoff value | 39.5% | 306 U/l |
| Sensitivityb | 0.512 | 0.620 |
| Specificityb | 0.696 | 0.476 |
| AUROCa | 0.582 | 0.553 |
| 95% confidence interval | 0.462–0.702 | 0.434–0.672 |
| p (asymtptotic significance) | 0.182 | 0.384 |
a AUROC, area under the receiver operating characteristic curve.
b sensitivity and specificity were calculated for the cutoff, which represented the best discrimination as derived from the ROC curves.
c IHC: immunohistochemistry
d LDH: lactate dehydrogenase
Statistical analysis
Statistical analyses were performed using SPSS version 10.0 (SPSS, Chicago, IL). Differences were considered statistically significant if P values were <0.05. Receiver operating characteristic (ROC) curves by plotting sensitivity versus (1-specificity) were used to evaluate the diagnostic performance of parameters at various cut-off points. An AUROC closer to 1 indicates greater discriminatory power, whereas an AUROC of 0.5 denotes no diagnostic potential. The optimum cut-off was calculated as the maximum value of sensitivity multiplied by specificity. Sensitivity and specificity were calculated according to standard formulas for the cut-off that represented the best discrimination derived from the ROC curves. The cut-off value representing the best discrimination derived from the ROC analysis was chosen for categorization of metric variables (LDH (lactate dehydrogenase) and expression of Mcm2). A statistical association between clinico-pathologic parameters and Mcm2 expression was tested using a two-sided Fisher's exact test. Disease specific survival (DSS) curves were calculated using the Kaplan-Meier method with significance evaluated by two-sided log-rank statistics. For DSS analysis, patients were censored at the time of their last clinical follow-up appointment or at their date of death not related to the tumour. A stepwise multivariable Cox regression model was adjusted, testing the independent prognostic relevance of Mcm2 positivity. The limit for reverse selection procedures was P = 0.1. The proportionality assumption for all variables was assessed with log-negative-log survival distribution functions.
Results
Clinico-pathologic data
Clinical follow-up data were available for 123 of 302 (40.7 %) patients with a median follow-up period of 23.5 months (range 1 to 177 months). The median follow-up for censored patients was 28.5 months. Clinico-pathologic parameters associated with poor disease specific survival were Ann Arbor stage (p = 0.0026), IPI (p < 0.0001), and serum LDH ≥300 U/l (p = 0.0133) according to the univariate analysis. The optimum cut-off level for LDH had been established at 306 U/l (AUROC method) and a cut-off of 300 U/l was used for further statistical analysis. Age at diagnosis and gender of patients did not make an impact on DSS. Neither the presence of bone marrow involvement, extranodal involvement nor bulky disease at presentation was statistically relevant for DSS.
The results of the univariate analysis are presented in table 3.
Table 3.
Univariate analysis of factors regarding disease specific survival
| Tumor-related death | ||||
| Variable | Categorisation | n | events | pa |
| Clinico-pathologic data: | ||||
| Age at diagnosis | ||||
| <65 years | 44 | 20 | 0.0805 | |
| ≥65 years | 77 | 43 | ||
| Gender | ||||
| male | 64 | 31 | 0.4220 | |
| female | 59 | 34 | ||
| Ann Arbor Stage | ||||
| 1 | 19 | 6 | 0.0026 | |
| 2 | 37 | 16 | ||
| 3 | 27 | 15 | ||
| 4 | 39 | 27 | ||
| International prognostic index | ||||
| 0 | 2 | 0 | <0.0001 | |
| 1 | 46 | 16 | ||
| 2 | 24 | 11 | ||
| 3 | 25 | 20 | ||
| 4 | 22 | 16 | ||
| Extranodal involvement | ||||
| no | 47 | 24 | 0.9997 | |
| yes | 70 | 36 | ||
| Bulky disease | ||||
| no | 73 | 36 | 0.3135 | |
| yes | 39 | 20 | ||
| Serum levels of LDH at diagnosis | ||||
| <300 U/l | 38 | 19 | 0.0133 | |
| ≥300 U/l | 54 | 31 | ||
| Immunohistochemistry: | ||||
| Mcm2 immunoreactivity | ||||
| <40% | 52 | 21 | 0.0424 | |
| ≥40% | 36 | 22 | ||
a Log rank test; bold face representing significant data.
Immunohistochemistry
Investigation of Mcm2 expression was informative in 69.9 % (211/302) of cases. Mcm2 expression in ≥40% of cells was detected in 46.4 % (98/211) of DLBCL. Mcm2 was clearly evident in the nuclei of tumour cells with low background staining. In non-neoplastic lymphoid tissue, which was used as a positive control and for establishing the appropriate staining protocols, expression of Mcm2 was mainly found in the germinal centres harbouring proliferating cells; expression in the mantle zone and parafollicular zone was negligible (Figure 1A). Identical results have previously been described [41].
Figure 1.
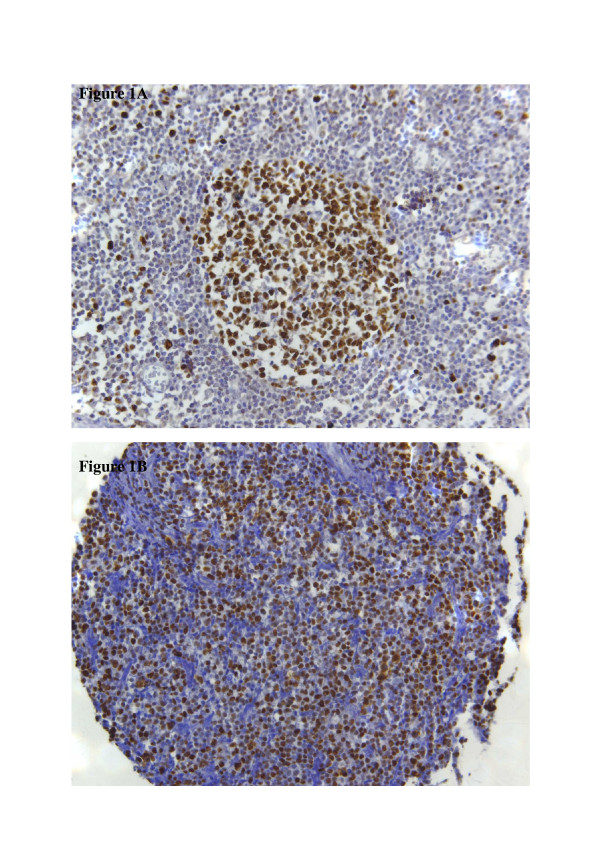
Immunohistochemistry. Figure 1A: Immunohistochemical staining of Mcm2 in a non-neoplastic lymph node. Mcm2-expression is found in the majority of cells in the germinal centre; only a few cells in the mantle zone are positive for Mcm2. Figure 1B: Immunohistochemical staining of Mcm2 in diffuse large B-cell lymphoma. This case is positive for Mcm2-expression with more than 40% of tumour cells expressing this marker.
The optimum cut-off for Mcm2 expression was calculated as 39.5 % of cells using the AUROC method. For practical reasons, a cut-off level of 40 % was chosen and patients were divided into two subgroups accordingly (positive: protein expression ≥40 % of cells; negative: protein expression <40 % of cells). For descriptive data analysis, all relevant variables were compared with Mcm2 expression (table 4). A representative image of immunohistochemical expression of Mcm2 in DLBCL is shown in figure 1B.
Table 4.
Clinico-pathologic and immunohistochemical parameters in relation to Mcm2 immunoreactivity
| Mcm2 immunoreactivity | |||||
| Variable | Categorisation | n analysable | negative (<40%) | positive (≥40%) | pa |
| Clinico-pathologic data: | |||||
| Age at diagnosis | |||||
| <65 years | 47 | 27 | 20 | 1.000 | |
| ≥65 years | 63 | 37 | 26 | ||
| Gender | |||||
| male | 72 | 39 | 33 | 0.593 | |
| female | 57 | 34 | 23 | ||
| Ann Arbor Stage | |||||
| 1 | 15 | 10 | 5 | 0.342 | |
| 2 | 34 | 15 | 19 | ||
| 3 | 23 | 14 | 9 | ||
| 4 | 35 | 22 | 13 | ||
| International prognostic index | |||||
| 0 | 2 | 2 | 0 | 0.122 | |
| 1 | 48 | 27 | 21 | ||
| 2 | 21 | 14 | 7 | ||
| 3 | 20 | 13 | 7 | ||
| 4 | 14 | 4 | 10 | ||
| Bone marrow involvement | |||||
| no | 72 | 39 | 33 | 0.414 | |
| yes | 35 | 22 | 13 | ||
| Extranodal involvement | |||||
| no | 40 | 27 | 13 | 0.103 | |
| yes | 62 | 31 | 31 | ||
| Bulky disease | |||||
| no | 62 | 44 | 20 | 0.003 | |
| yes | 35 | 13 | 22 | ||
| Serum levels of LDH at diagnosis | |||||
| <300 U/l | 42 | 23 | 19 | 0.505 | |
| ≥300 U/l | 41 | 26 | 15 | ||
a two-sided Fisher's exact test; bold type representing significant data.
Prognostic significance
Disease specific survival was compared between tumours positive and negative for Mcm2 by univariate log-rank statistics. Positivity for Mcm2 was associated with shorter DSS (p = 0.0424) (table 3; figure 2). However, when Mcm2 positivity was tested in the multivariate analysis in combination with LDH, IPI, and Ann Arbor stage it did not retain its prognostic value (p = 0.546). LDH was not found to be of prognostic relevance in the multivariate analysis as well (p = 0.063). Both IPI (p = 0.010) and Ann Arbor stage (p = 0.025) were significant prognostic markers in the multivariate analysis with respect to disease specific survival.
Figure 2.
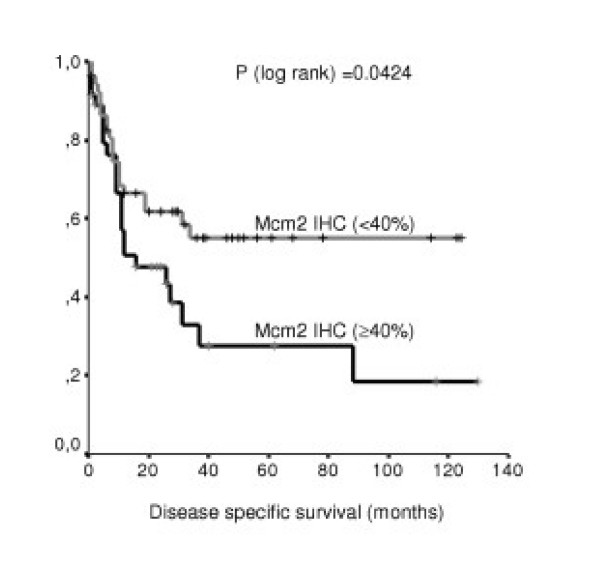
Disease specific survival. Distribution of time (months) to DLBCL-related deaths among patients with negative (<40 % of tumour cells) (n = 52) and positive (≥40% of tumour cells) (n = 36) expression of Mcm2 (Kaplan-Meier method). (IHC = immunohistochemistry)
Discussion
Cell proliferation is essential for the growth of any type of malignancy. Analysis of cell proliferation applying antibodies specifically detecting members of the MCM protein family has been proposed as a novel method for cell cycle assessment which may be of diagnostic and prognostic value in the histopathologic assessment of neoplasms. Other proliferation markers such as the widely used Ki-67 and proliferating cell nuclear antigen (PCNA) are probably not as effective as MCM proteins for this purpose, as Ki-67 is absent in early G1-phase, its function still remains unknown [42] and PCNA is less specific for determining proliferation since it is also present during DNA repair processes [43].
Analysis of MCM proteins by means of immunohistochemistry has been shown to be of prognostic value in a diverse range of human malignancies [30,34,44-51]; in lymphoma however, high expression of Mcm2 has been described in DLBCL, but so far data have never been evaluated with respect to clinical outcome [41].
Therefore we focused on the evaluation of Mcm2 expression in a large collective of clinically well documented patients with DLBCL using tissue microarray technology and immunohistochemistry. 46.4% of DLBCL showed positivity for Mcm2 defined as expression in ≥40% of tumour cells. Positivity was significantly associated with shorter disease specific survival in the univariate analysis (p = 0.0424). However, when tested in the multivariate analysis, it did not retain its prognostic value (p = 0.546). This finding may be attributable to the fact, that besides proliferation, a number of other factors not included into the clinico-pathological parameters of our analysis (such as concomitant morbidity) may influence disease specific survival. Therefore, assessment of Mcm2 expression may still be useful as an additional prognostic marker in conjunction with other established prognostic parameters.
Conclusion
In summary, expression of Mcm2 in ≥40% of tumour cells was found to be a negative prognostic marker for disease specific survival in this large series of DLBCL. Protein expression of MCM proteins can easily be evaluated in routinely processed tissue specimen using specific antibodies in daily routine practice. Assessment of MCM protein expression may be used as a marker of proliferation as well as a potentially prognostic indicator, and further work in this area is warranted.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
ECO conceived and designed the study, was responsible for immunohistochemical studies and drafted the manuscript. PW, AZ, AT and SAP were responsible for acquisition of material and clinical data as well as construction of tissue microarrays. PJW performed the statistical analysis, helped to prepare figures and tables and interpreted the data. RS revised the article critically. SD participated in the design and coordination of the study. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
The authors thank A Vielberth, H Weisskopf and R Epper for excellent technical assistance. Prof. F Hofstaedter, head of the Institute of Pathology, University of Regensburg, is acknowledged for his continuous support. Prof. G Jundt, head of the cancer registry Basel, is acknowledged for providing clinical data. Caroline Selai, PhD, University College London, is gratefully acknowledged; without her encouragement, this study would never have been undertaken. This study was supported by a grant of the University of Regensburg to ECO (Regensburger Forschungsförderung in der Medizin).
Contributor Information
Ellen C Obermann, Email: ellen.obermann@klinik.uni-regensburg.de.
Philip Went, Email: pwent@uhbs.ch.
Annette Zimpfer, Email: ANNZIM@gmx.de.
Alexandar Tzankov, Email: alexandar.tzankov@i-med.ac.at.
Peter J Wild, Email: p.wild@uke.uni-hamburg.de.
Robert Stoehr, Email: robert.stoehr@klinik.uni-regensburg.de.
Stefano A Pileri, Email: pileri@med.unibo.it.
Stephan Dirnhofer, Email: sdirnhofer@uhbs.ch.
References
- Jaffe ES, Harris NL, Stein H, Vardiman JW. Tumours of Haematopoietic and Lymphoid Tissue. Lyon: IARC Press; 2001. [Google Scholar]
- Fisher RI, Shah P. Current trends in large cell lymphoma. Leukemia. 2003;17:948–1960. doi: 10.1038/sj.leu.2403096. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Winter JN, Andersen J, Reed JC, Krajewski S, Variakojis D, Bauer KD, Fisher RI, Gordon LI, Oken MM, Jiang S, Jeffries D, Domer P. BCL-2 expression correlates with lower proliferative activity in the intermediate- and high-grade non-Hodgkin's lymphomas: an Eastern Cooperative Oncology Group and Southwest Oncology Group cooperative laboratory study. Blood. 1998;91:1391–1398. [PubMed] [Google Scholar]
- Moller MB, Nielsen O, Pedersen NT. Cyclin D3 expression in non-Hodgkin lymphoma. Correlation with other cell cycle regulators and clinical features. Am J Clin Pathol. 2001;115:404–412. doi: 10.1309/8KF0-0Y0C-2F4L-UHXL. [DOI] [PubMed] [Google Scholar]
- Moller MB, Kania PW, Ino Y, Gerdes AM, Nielsen O, Louis DN, Skjodt K, Pedersen NT. Frequent disruption of the RB1 pathway in diffuse large B cell lymphoma: prognostic significance of E2F-1 and p16INK4A. Leukemia. 2000;14:898–904. doi: 10.1038/sj.leu.2401761. [DOI] [PubMed] [Google Scholar]
- Gronbaek K, Nedergaard T, Andersen MK, thor SP, Guldberg P, Moller P, Zeuthen J, Ebbe HN, Hou-Jensen K, Ralfkiaer E. Concurrent disruption of cell cycle associated genes in mantle cell lymphoma: a genotypic and phenotypic study of cyclin D1, p16, p15, p53 and pRb. Leukemia. 1998;12:1266–1271. doi: 10.1038/sj.leu.2401090. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Hall PA, Levison DA. Review: assessment of cell proliferation in histological material. J Clin Pathol. 1990;43:184–192. doi: 10.1136/jcp.43.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CM, Wright NA. The clinical assessment of proliferation and growth in human tumours: evaluation of methods and applications as prognostic variables. J Pathol. 1990;160:93–102. doi: 10.1002/path.1711600202. [DOI] [PubMed] [Google Scholar]
- Burger PC, Shibata T, Kleihues P. The use of the monoclonal antibody Ki-67 in the identification of proliferating cells: application to surgical neuropathology. Am J Surg Pathol. 1986;10:611–617. doi: 10.1097/00000478-198609000-00003. [DOI] [PubMed] [Google Scholar]
- Brown DC, Gatter KC. Ki67 protein: the immaculate deception? Histopathology. 2002;40:2–11. doi: 10.1046/j.1365-2559.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- Cheung AN, Ngan HY, Collins RJ, Wong YL. Assessment of cell proliferation in hydatidiform mole using monoclonal antibody MIB1 to Ki-67 antigen. J Clin Pathol. 1994;47:601–604. doi: 10.1136/jcp.47.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czader M, Mazur J, Pettersson M, Liliemark J, Stromberg M, Christensson B, Tribukait B, Auer G, Ost A, Porwit A. Prognostic significance of proliferative and apoptotic fractions in low grade follicle center cell-derived non-Hodgkin's lymphomas. Cancer. 1996;77:1180–1188. doi: 10.1002/(SICI)1097-0142(19960315)77:6<1180::AID-CNCR26>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Bjorck E, Ek S, Landgren O, Jerkeman M, Ehinger M, Bjorkholm M, Borrebaeck CA, Porwit-MacDonald A, Nordenskjold M. High expression of cyclin B1 predicts a favorable outcome in patients with follicular lymphoma. Blood. 2005;105:2908–2915. doi: 10.1182/blood-2004-07-2721. [DOI] [PubMed] [Google Scholar]
- Martin AR, Weisenburger DD, Chan WC, Ruby EI, Anderson JR, Vose JM, Bierman PJ, Bast MA, Daley DT, Armitage JO. Prognostic value of cellular proliferation and histologic grade in follicular lymphoma. Blood. 1995;85:3671–3678. [PubMed] [Google Scholar]
- Llanos M, Alvarez-Arguelles H, Aleman R, Oramas J, Diaz-Flores L, Batista N. Prognostic significance of Ki-67 nuclear proliferative antigen, bcl-2 protein, and p53 expression in follicular and diffuse large B-cell lymphoma. Med Oncol. 2001;18:15–22. doi: 10.1385/MO:18:1:15. [DOI] [PubMed] [Google Scholar]
- Miller TP, Grogan TM, Dahlberg S, Spier CM, Braziel RM, Banks PM, Foucar K, Kjeldsberg CR, Levy N, Nathwani BN. Prognostic significance of the Ki-67-associated proliferative antigen in aggressive non-Hodgkin's lymphomas: a prospective Southwest Oncology Group trial. Blood. 1994;83:1460–1466. [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJ, Schlingemann RO, Boehmer AL, van Driel R, Brakenhoff GJ, Ramaekers FC. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J Cell Sci. 1989;92:123–130. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hall PA. The location of pKi67 in the outer dense fibrillary compartment of the nucleolus points to a role in ribosome biogenesis during the cell division cycle. J Pathol. 2000;190:537–544. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: MCMS, pre-replicative complexes and kinases. Trends Cell Biol. 1996;6:184–188. doi: 10.1016/0962-8924(96)10015-5. [DOI] [PubMed] [Google Scholar]
- Musahl C, Holthoff HP, Lesch R, Knippers R. Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp Cell Res. 1998;241:260–264. doi: 10.1006/excr.1998.4041. [DOI] [PubMed] [Google Scholar]
- Todorov IT, Werness BA, Wang HQ, Buddharaju LN, Todorova PD, Slocum HK, Brooks JS, Huberman JA. HsMCM2/BM28: a novel proliferation marker for human tumors and normal tissues. Lab Invest. 1998;78:73–78. [PubMed] [Google Scholar]
- Ritzi M, Baack M, Musahl C, Romanowski P, Laskey RA, Knippers R. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J Biol Chem. 1998;273:24543–24549. doi: 10.1074/jbc.273.38.24543. [DOI] [PubMed] [Google Scholar]
- Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci U S A. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Morris LS, Mills AD, Stoeber K, Laskey RA, Williams GH, Coleman N. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–2132. [PubMed] [Google Scholar]
- Rodins K, Cheale M, Coleman N, Fox SB. Minichromosome maintenance protein 2 expression in normal kidney and renal cell carcinomas: relationship to tumor dormancy and potential clinical utility. Clin Cancer Res. 2002;8:1075–1081. [PubMed] [Google Scholar]
- Chatrath P, Scott IS, Morris LS, Davies RJ, Rushbrook SM, Bird K, Vowler SL, Grant JW, Saeed IT, Howard D, Laskey RA, Coleman N. Aberrant expression of minichromosome maintenance protein-2 and Ki67 in laryngeal squamous epithelial lesions. Br J Cancer. 2003;89:1048–1054. doi: 10.1038/sj.bjc.6601234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WC, Khoo US, Ngan HY, Chan KY, Chiu PM, Tsao SW, Cheung AN. Minichromosome maintenance protein 7 expression in gestational trophoblastic disease: correlation with Ki67, PCNA and clinicopathological parameters. Histopathology. 2003;43:485–490. doi: 10.1046/j.1365-2559.2003.01728.x. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- Meng MV, Grossfeld GD, Williams GH, Dilworth S, Stoeber K, Mulley TW, Weinberg V, Carroll PR, Tlsty TD. Minichromosome maintenance protein 2 expression in prostate: characterization and association with outcome after therapy for cancer. Clin Cancer Res. 2001;7:2712–2718. [PubMed] [Google Scholar]
- Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH. Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett's mucosa. Gut. 2002;50:373–377. doi: 10.1136/gut.50.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RJ, Freeman A, Morris LS, Bingham S, Dilworth S, Scott I, Laskey RA, Miller R, Coleman N. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet. 2002;359:1917–1919. doi: 10.1016/S0140-6736(02)08739-1. [DOI] [PubMed] [Google Scholar]
- Went P, Dellas T, Bourgau C, Maurer R, Augustin F, Tzankov A, Dirnhofer S. [Expression profile and prognostic significance of CD24, p53 and p21 in lymphomas. A tissue microarray study of over 600 non-Hodgkin lymphomas] Dtsch Med Wochenschr. 2004;129:2094–2099. doi: 10.1055/s-2004-831850. [DOI] [PubMed] [Google Scholar]
- Schraml P, Bucher C, Bissig H, Nocito A, Haas P, Wilber K, Seelig S, Kononen J, Mihatsch MJ, Dirnhofer S, Sauter G. Cyclin E overexpression and amplification in human tumours. J Pathol. 2003;200:375–382. doi: 10.1002/path.1356. [DOI] [PubMed] [Google Scholar]
- Tzankov A, Zimpfer A, Pehrs AC, Lugli A, Went P, Maurer R, Pileri S, Dirnhofer S. Expression of B-cell markers in classical hodgkin lymphoma: a tissue microarray analysis of 330 cases. Mod Pathol. 2003;16:1141–1147. doi: 10.1097/01.MP.0000093627.51090.3F. [DOI] [PubMed] [Google Scholar]
- Tzankov A, Pehrs AC, Zimpfer A, Ascani S, Lugli A, Pileri S, Dirnhofer S. Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell-like types: a tissue microarray analysis of 90 cases. J Clin Pathol. 2003;56:747–752. doi: 10.1136/jcp.56.10.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann EC, Eward KL, Dogan A, Paul EA, Loddo M, Munson P, Williams GH, Stoeber K. DNA replication licensing in peripheral B-cell lymphoma. J Pathol. 2005;205:318–328. doi: 10.1002/path.1695. [DOI] [PubMed] [Google Scholar]
- Endl E, Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp Cell Res. 2000;257:231–237. doi: 10.1006/excr.2000.4888. [DOI] [PubMed] [Google Scholar]
- Toschi L, Bravo R. Changes in cyclin/proliferating cell nuclear antigen distribution during DNA repair synthesis. J Cell Biol. 1988;107:1623–1628. doi: 10.1083/jcb.107.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Xue WC, Khoo US, Ngan HY, Chan KY, Tam IY, Chiu PM, Ip PP, Tam KF, Cheung AN. Replicative MCM7 protein as a proliferation marker in endometrial carcinoma: a tissue microarray and clinicopathological analysis. Histopathology. 2005;46:307–313. doi: 10.1111/j.1365-2559.2005.02069.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez MA, Pinder SE, Callagy G, Vowler SL, Morris LS, Bird K, Bell JA, Laskey RA, Coleman N. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J Clin Oncol. 2003;21:4306–4313. doi: 10.1200/JCO.2003.04.121. [DOI] [PubMed] [Google Scholar]
- Hunt DP, Freeman A, Morris LS, Burnet NG, Bird K, Davies TW, Laskey RA, Coleman N. Early recurrence of benign meningioma correlates with expression of mini-chromosome maintenance-2 protein. Br J Neurosurg. 2002;16:10–15. doi: 10.1080/02688690120114174. [DOI] [PubMed] [Google Scholar]
- Wharton SB, Chan KK, Anderson JR, Stoeber K, Williams GH. Replicative Mcm2 protein as a novel proliferation marker in oligodendrogliomas and its relationship to Ki67 labelling index, histological grade and prognosis. Neuropathol Appl Neurobiol. 2001;27:305–313. doi: 10.1046/j.0305-1846.2001.00333.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Araki K, Osaki M, Nakamura H, Tomita K, Shimizu E, Ito H. MCM2 and Ki-67 expression in human lung adenocarcinoma: prognostic implications. Pathobiology. 2004;71:193–200. doi: 10.1159/000078673. [DOI] [PubMed] [Google Scholar]
- Kruger S, Thorns C, Stocker W, Muller-Kunert E, Bohle A, Feller AC. Prognostic value of MCM2 immunoreactivity in stage T1 transitional cell carcinoma of the bladder. Eur Urol. 2003;43:138–145. doi: 10.1016/S0302-2838(02)00580-8. [DOI] [PubMed] [Google Scholar]
- Kato H, Miyazaki T, Fukai Y, Nakajima M, Sohda M, Takita J, Masuda N, Fukuchi M, Manda R, Ojima H, Tsukada K, Asao T, Kuwano H. A new proliferation marker, minichromosome maintenance protein 2, is associated with tumor aggressiveness in esophageal squamous cell carcinoma. J Surg Oncol. 2003;84:24–30. doi: 10.1002/jso.10287. [DOI] [PubMed] [Google Scholar]
- Kodani I, Osaki M, Shomori K, Araki K, Goto E, Ryoke K, Ito H. Minichromosome maintenance 2 expression is correlated with mode of invasion and prognosis in oral squamous cell carcinomas. J Oral Pathol Med. 2003;32:468–474. doi: 10.1034/j.1600-0714.2003.00116.x. [DOI] [PubMed] [Google Scholar]


