Abstract
The pathway for biodegradation of benzothiazole (BT) and 2-hydroxybenzothiazole (OBT) by Rhodococcus pyridinovorans strain PA was studied in detail. The kinetics of biodegradation were monitored by in situ 1H nuclear magnetic resonance (NMR) in parallel with reversed-phase high-performance liquid chromatography (HPLC). Successive oxidations from BT to OBT and then from OBT to dihydroxybenzothiazole were observed. Further insight was obtained by using a mutant strain with impaired ability to grow on BT and OBT. The precise structure of another intermediate was determined by in situ two-dimensional 1H-13C NMR and HPLC-electrospray ionization mass spectrometry; this intermediate was found to be a ring-opening product (a diacid structure). Detection of this metabolite, together with the results obtained by 1H and 19F NMR when cells were incubated with 3-fluorocatechol, demonstrated that a catechol 1,2-dioxygenase is involved in a pathway for biodegradation of BTs in this Rhodococcus strain. Our results show that catechol 1,2-dioxygenase and catechol 2,3-dioxygenase activities may both be involved in the biodegradation of BTs depending on the culture conditions.
Benzothiazoles (BTs), a group of xenobiotic compounds containing a benzene ring fused with a thiazole ring, are manufactured worldwide for a variety of applications. Depending on the substituent of the thiazole ring, BTs are used as fungicides in lumber and leather production (30), as antialgal agents (6), as slimicides in the paper and pulp industry (27), and as chemotherapeutic agents (7). Methabenzthiazuron is used as a herbicide (25, 36). These applications indicate that BTs have a wide spectrum of biological activity. Aminobenzothiazole is an intermediate in dye production. Finally, mercaptobenzothiazole and its derivatives are used to promote vulcanizaton in rubber manufacture. This industrial process produces other BTs, such as benzothiazole-2-sulfonate and 2-hydroxybenzothiazole (OBT), as by-products. Released from rubber products or from BT production plants, these compounds have been detected in industrial wastewaters and in various environmental compartments (20, 30) and are of concern for the aquatic environment due to their limited biodegradability and potential toxicity (17, 23).
Only a few bacterial isolates have been shown to degrade BTs in pure culture (15, 16). Gaja and Knapp (21) described Rhodococcus strain PA, which grows on BT as a sole source of carbon, nitrogen, and energy, and strain TA, which grows on 2-aminobenzothiazole. Two strains, Rhodococcus erythropolis BTS1 and Rhodococcus rhodochrous OBT18, were shown to degrade BT and OBT (2, 17, 18). Benzothiazole-2-sulfonate is also degraded by R. erythropolis (18), and 2-aminobenzothiazole is degraded by R. rhodochrous (24).
There have been few reports on the metabolic pathways involved in biodegradation of BTs. Recently, we used 1H nuclear magnetic resonance (NMR) spectroscopy to elucidate the structures of intermediates in the biodegradation of BT and OBT by R. rhodochrous and R. erythropolis (2) and the structures of intermediates in the biodegradation of 2-aminobenzothiazole by R. rhodochrous (24). We showed that a common biodegradative pathway led to hydroxylation of the benzene ring at position 6 (2, 24). Various NMR techniques were used. (i) In situ 1H NMR was performed directly with culture media at natural abundance and allowed biodegradation kinetics to be monitored; this approach was used previously to study microbial biodegradation of xenobiotic compounds (3, 5, 8-10, 13, 14, 29). (ii) 1H-15N gradient heteronuclear multiple-bond correlation (HMBC) at natural abundance helped us to identify precisely the hydroxyl group position on the benzene ring by taking advantage of the information contained in 1H-15N scalar couplings (2, 24, 26).
In this paper we describe a detailed study of the pathway of biodegradation of BT and OBT by Rhodococcus pyridinovorans strain PA in which a mutant strain with impaired ability to grow on BT and OBT was used. Our data demonstrate that a catechol 1,2-dioxygenase is involved in the biodegradation of BTs by this Rhodococcus strain.
MATERIALS AND METHODS
Chemicals.
BT and OBT were purchased from Aldrich. Tetradeuterated sodium trimethylsilylpropionate (TSPd4) was purchased from Eurisotop (Saint Aubin, France), and 3-fluorocatechol (3FC) was purchased from Acros Organics (Geel, Belgium).
Dihydroxybenzothiazole (diOBT) was obtained as previously described (2).
Identification of R. pyridinovorans strain PA.
Rhodococcus strain PA, which was isolated and described by Gaja and Knapp (21), was identified here on the basis of 16S rRNA sequences kindly provided by Leonid Kulakov and Mike Larkin, Queens University, Belfast, Northern Ireland. For amplification of 16S rRNA genes, the following primers were used: forward primer Eubac27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer Eubac1492R (5′-AAGGAGGTGATCCAGCCGA-3′). The PCR regimen was as follows: denaturation at 95°C for 5 min; denaturation at 94°C for 45 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min for 32 amplification cycles; and a final step consisting of 8 min at 72°C. The products were allowed to stand at 8°C.
Mutagenesis.
Mutants of R. pyridinivorans strain PA were obtained by chemical mutagenesis performed with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) by the method of Delić et al. (12), except that the final concentration of MNNG was 40 μg/ml. Mutants with altered ability to utilize OBT were isolated on the basis of slow or no growth on OBT mineral salts agar.
Growth conditions.
Unless otherwise indicated, R. pyridinovorans strain PA and mutant strain N-120-8 were grown in 100-ml portions of Trypticase soy broth (TSB) (bioMérieux, Marcy l'Etoile, France) in 500-ml Erlenmeyer flasks incubated at 30°C and 200 rpm. The cells were harvested after 20 h of culture. Cells for enzyme assays and some NMR studies were grown for ca. 48 h on liquid mineral salts medium containing (per liter) 1 g of (NH4)2SO4, 1 g of K2HPO4, 1 g of KH2PO4, 4 mg of FeCl3 · 6H2O, and 40 mg of MgSO4 · 7H2O; the pH was adjusted to 6.7, and sodium acetate (10 mM), sodium benzoate (5 mM), or OBT (2 mM) was added as the sole source of carbon and energy.
Incubation with xenobiotic compounds.
Cells were centrifuged at 8,000 × g for 15 min at 5°C. The pellet was washed twice with buffer (1 g of K2HPO4 per liter, 1 g of KH2PO4 per liter, 4 mg of FeCl3 · 6H2O per liter, 40 mg of MgSO4 · 7H2O per liter; pH 6.7) and finally resuspended in this buffer or in water for mass analysis (5 g [wet weight] of cells in 100 ml of buffer). The resting cells were incubated with 3 mM OBT or 1 mM BT in 500-ml Erlenmeyer flasks at 30°C with agitation (200 rpm). Negative controls lacking substrate or cells were incubated under the same conditions. Samples (1 ml) were taken every 30 min, centrifuged at 12,000 × g for 5 min, and prepared for high-performance liquid chromatography (HPLC) or 1H NMR analysis.
HPLC analyses.
HPLC analyses were performed by using a Waters 600E chromatograph with a Waters 486 UV detector set at 295 nm and a reversed-phase column (Interchrom Nucléosil C18; 5 μm; 250 by 4.6 mm; Interchim) at room temperature. The mobile phase was acetonitrile-water (20:80, vol/vol), and the flow rate was 1 ml/min.
NMR spectroscopy.
All 1H and 13C NMR spectra were recorded with a Bruker Avance 300 spectrometer at 300.13 and 75.47 MHz, respectively, at 298 K by using a 5-mm triple-tuned 1H-13C-15N probe equipped with a z-gradient coil. The 1H and 13C 90° pulse lengths were 7.5 and 8.6 μs, respectively.
1H NMR spectroscopy.
Water resonance was suppressed by using the classical double pulsed-field gradient echo sequence WATERGATE. A total of 128 scans were collected (relaxation delay, 5 s; acquisition time, 3.64 s; spectral window, 3,420 Hz; 32,000 data points). A 1-Hz line-broadening procedure was applied before Fourier transformation, and baseline correction was performed on spectra before integration with Bruker software. TSPd4 was used as an internal reference for chemical shift (0 ppm) and for quantification of metabolites, as previously described (9).
1H-13C HSQC experiments.
Gradient-enhanced heteronuclear single quantum coherence (HSQC) was achieved by using 128 (t1) × 1,024 (t2) data points, and the number of free induction decays added for each t1 increment was 300. The acquisition time was 143 ms for spectral widths of 3,600 and 18,100 Hz in F2 and F1, respectively. Zero-filling to 512 points and π/2 shifted sine window function in t1 and π/3 shifted squared sine function in t2 were applied prior to two-dimensional Fourier transformation.
1H-13C gradient HMBC experiments.
A low-pass J filter (145 Hz) was used. The delay to allow nJCH correlations was set at 50 ms. Typically, 1,024 data points with 32 scans for each of 128 t1 increments were acquired with spectral widths of 3,600 Hz in F2 and 19,600 Hz in F1. The required acquisition time was 142 ms. A recycle delay of 2 s was used. Zero-filling to 512 points and an exponential filtering (2 Hz) window in t1 and an exponential filtering (1 Hz) window in t2 were applied prior to two-dimensional Fourier transformation. The 13C chemical shifts were referenced to TSPd4.
For both HSQC and HMBC experiments, water resonance was suppressed by using a 2-s presaturation period during the recycle delay.
19F NMR spectroscopy.
19F NMR measurements were obtained with a Bruker Avance 400 spectrometer at 376.49 MHz and 298 K by using a 5-mm QNP 13C/31P/19F-{1H} probe equipped with a z-gradient coil. The 19F 90° pulse length was 13.5 μs.
The spectral width used was 75,188 Hz. The number of data points used for acquisition was 131,072. About 19,500 scans were recorded. A 1.5-s recycle delay was used. No 1H decoupling was achieved. Chemical shifts were reported relative to CFCl3 used as an external reference.
Enzyme assays.
Catechol 1,2-dioxygenase and catechol 2,3-dioxygenase activities were assayed by the method of Gibson (22) by using cell extracts of R. pyridinivorans PA grown on TSB (bioMérieux), nutrient broth (Oxoid Ltd., Basingstoke, Hants, United Kingdom), acetate mineral salts, benzoate mineral salts, or OBT mineral salts. Cell suspensions were harvested by centrifugation, washed twice in distilled water, and resuspended in phosphate buffer (2 g of KH2PO4 per liter; pH 7) to which acetone (20%, vol/vol) was added. Cells were broken by ultrasonication (Sanyo Soniprobe150; amplitude, 10 to 15 μm) for 2.5 min; each 0.5-min burst was followed by a 2-min cooling period. Unbroken cells and cell fragments were removed by centrifugation (15 min at 12,000 × g and 4°C); the supernatant fluid was kept on ice until it was assayed. The protein concentrations of cell extracts were measured by the bicinchoninic acid method (Pierce, Rockford, Ill.).
MS.
HPLC-mass spectrometry (MS) analyses were performed with a system consisting of an HP 1100 LC and Quattro LC tandem mass spectrometer (Micromass, Manchester, United Kingdom) as described elsewhere (31). Analytes were separated by ion pair chromatography on a phenylhexyl column (length, 150 mm; inside diameter, 2 mm; 3 μm; Phenomenex) at 40°C with eluents A (methanol-H2O, 20:80) and B (methanol-H2O, 70:30), both containing 5 mM tributylamine (32). Gradient elution was as follows: 0 min with 20% eluent B, 13 min with 85% eluent B, 14 min with 85% eluent B, 15 min with 20% eluent B, and 24 min with 20% eluent B. The triple-stage quadrupole mass spectrometer was equipped with an electrospray probe and was operated in the negative ion mode with a cone voltage of 26 kV. For MS-MS experiments, a collision energy of 20 eV was used.
RESULTS AND DISCUSSION
Identification of R. pyridinovorans strain.
The Rhodococcus strain PA 16S rRNA sequence (1,384 bases) was obtained (GenBank database accession number AJ457068) and compared to 16S rRNA sequences in the GenBank databases (http://www.ncbi.nlm.nih.gov/GenBank/index.html) by using the program BLASTn 2.0 (1). The sequence of Rhodococcus strain PA was most similar to the sequence of R. pyridinovorans strain RO4 (accession number AF459741.1; 100% identical over the 1,384 bp sequenced for strain PA). On the basis of this similarity, we believe that strain PA belongs to the species R. pyridinovorans. Strain RO4 of this species (isolated on the basis of its ability to degrade pyridine) was described by Yoon et al. (37). Rhodococcus strain PA also showed high degrees of similarity to strains of Rhodococcus roseus (accession number X80624.1; 99.6% similar over the 1,384 bp sequenced, differing in only 5 bp) and Rhodococcus rhodochrous (accession number X79288.1; 97.2% similar over the 1,384 bp sequenced, differing in only 39 bp).
BT degradation by R. pyridinovorans strain PA.
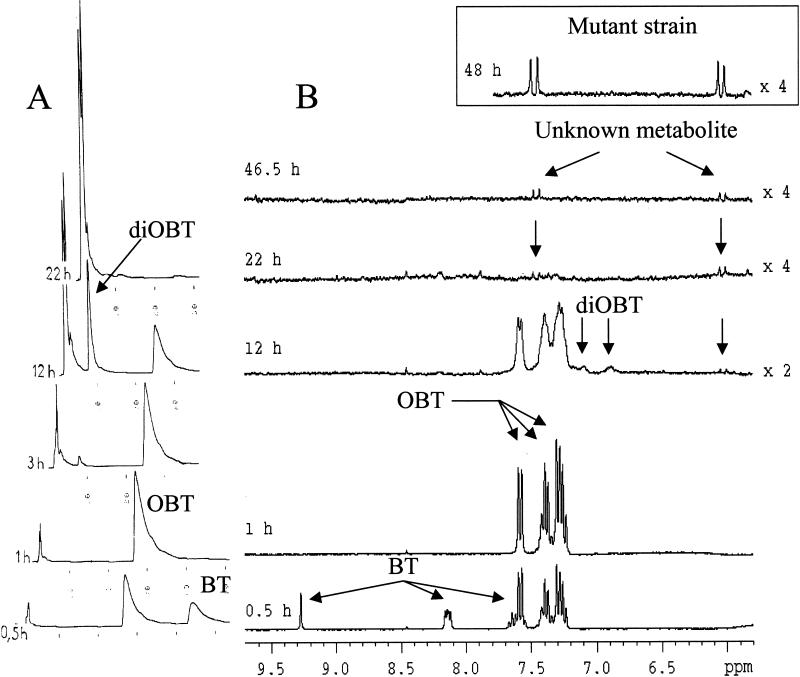
HPLC chromatograms and in situ 1H NMR spectra collected during the biodegradation of BT (3 mM) by TSB-grown resting cells of R. pyridinovorans are shown in Fig. 1. TSB was used as it allowed easy production of cells, resulted in very large amounts of biomass, and consequently permitted rapid transformation of BT. As Fig. 1A shows, the concentration of BT (retention time, 45 min) decreased with time, and BT completely disappeared after 1 h. A new metabolite appeared after 30 min of incubation; the retention time of this compound (25 min) was similar to that of OBT (2), and its identity was confirmed by coinjection with authentic OBT. OBT was not present after 22 h. After 3 h of incubation, a new product appeared. The retention time of this compound (8 min) was shorter than that of OBT, indicating greater polarity, and was identical to that of diOBT (2). This assignment was confirmed by coinjection of diOBT onto the HPLC column. The concentration of diOBT increased until 12 h and then decreased with time.
FIG. 1.
Degradation of 3 mM BT by resting cells of R. pyridinovorans strain PA. (A) HPLC chromatograms. (B) In situ 1H NMR spectra. The inset shows a 1H NMR spectrum of R. pyridinovorans mutant strain N-120-8 incubated with 3 mM OBT for 48 h on the same scale.
1H NMR signals corresponding to those of BT [δ ppm: 7.57 (t), 7.65 (t), 8.14 (2xd), 9.28 (s)] decreased with time (Fig. 1B), and new signals in the aromatic region appeared after 30 min; these new signals included two triplets at 7.24 and 7.38 ppm and two doublets at 7.29 and 7.58 ppm. They were similar to the signals observed previously during the degradation of BT by R. erythropolis and R. rhodochrous (2) and were assigned to OBT. The OBT intensity decreased, and OBT finally disappeared by 22 h. At 12 h, fleeting signals corresponding to those of diOBT (doublet at 7.08 ppm, doublet of doublet at 6.86 ppm, and doublet at 7.15 ppm) were detected in the NMR spectra; this 1H NMR fingerprint was fully analyzed by Besse et al. (2). The detection of OBT and diOBT by 1H NMR is consistent with the results obtained by HPLC.
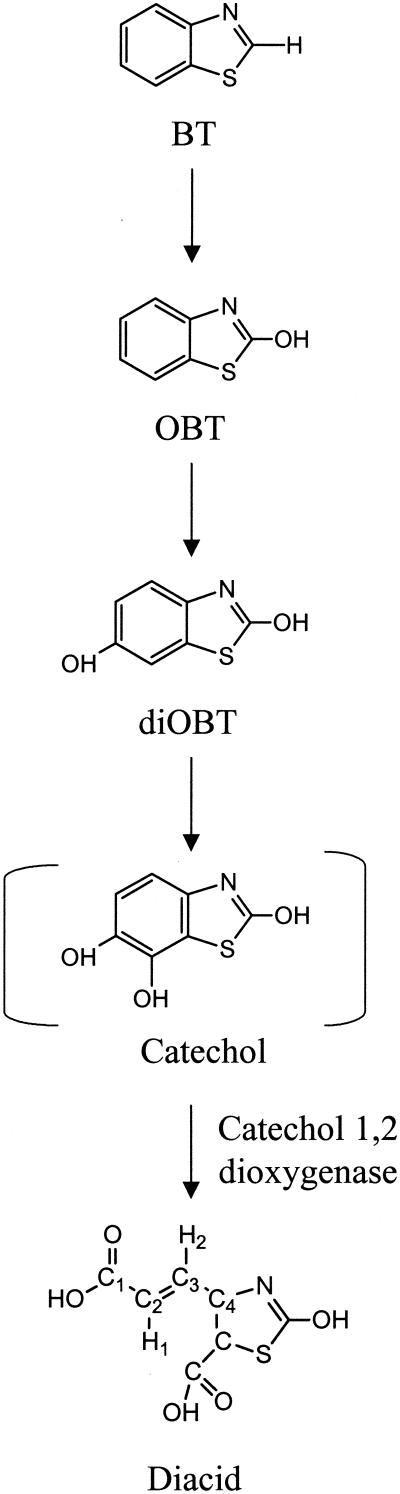
From 12 to 46.5 h, new, small 1H NMR signals that resonated at 6.03 and 7.45 ppm were observed, and they corresponded to the signals of an unknown metabolite. The coupling constant (J = 13.5 Hz) and the chemical shifts of these two protons are consistent with the presence of an ethylenic bond in the structure. Therefore, the compound could correspond to a ring-opening compound, such as a dicarboxylic acid compound resulting from an intradiol (ortho) cleavage of a catechol derived from BT (Fig. 2). However, the concentration of this metabolite was far too low to allow confirmation of this hypothetical structure. Attempts to isolate the compound were unsuccessful.
FIG. 2.
Pathway of biodegradation of BT by R. pyridinovorans strain PA.
Elucidation of the structure of the unknown metabolite.
In order to increase the concentration of the unknown metabolite, we constructed mutants of R. pyridinovorans strain PA by chemical mutagenesis with MNNG. Mutant isolate N-120-8 was chosen for further study; it was unable to degrade BT and grew more slowly than the wild-type strain when OBT was used as the sole carbon and nitrogen source. This indicates that the initial oxidation step (BT to OBT) is catalyzed by a different enzyme than the subsequent hydroxylation steps.
The kinetics of degradation of OBT (3 mM) by R. pyridinovorans strain PA and mutant strain N-120-8 were compared by performing 1H NMR and HPLC. For mutant strain N-120-8, HPLC showed that the OBT concentration decreased with time, while the concentration of diOBT increased (data not shown). The 1H NMR signals corresponding to those of OBT (two triplets at 7.24 and 7.38 ppm and two doublets at 7.29 and 7.58 ppm) decreased with time and were not present after 21 h of incubation; this is consistent with HPLC data. NMR could not detect diOBT due to its lower sensitivity than HPLC. Finally, the 1H NMR signals (δ = 6.03 and 7.45 ppm) of the unknown metabolite observed in the case of BT degradation were clearly detected (Fig. 1B, inset). It appears that the new metabolites observed during incubation of the wild-type and mutant strains with BT and OBT, respectively, were the same and that the unknown metabolite accumulated to a greater extent (2.5-fold) with the mutant strain, allowing complementary NMR and MS experiments to be performed.
Samples collected after 21 to 100 h of incubation of mutant strain N-120-8 with OBT were pooled and concentrated by freeze-drying (after 21 h OBT and diOBT were absent from the samples, simplifying further NMR and MS analyses).
First, in situ 1H-13C NMR experiments were performed with the concentrated sample without previous purification. On the 1H-13C HSQC spectrum two crosspeaks were detected, indicating that there were 1J1H-13C couplings between the protons resonating at δ = 6.03 and 7.45 ppm and the corresponding carbons resonating at 129.7 and 125.0 ppm, respectively. These data confirm that the protons are bound to ethylenic carbons. In the next experiment, 1H-13C HMBC was designed to select 1H-13C correlations between ethylenic protons and quaternary carbons (2J1H-13C, d6 = 50 ms). The long-range heteronuclear shift correlations seen on the two-dimensional NMR spectrum demonstrated that the proton resonating at 7.45 ppm is close to a carboxylic group (C-1, δ 13C = 177.2 ppm) and thus is assigned to H-1, while the proton resonating at δ = 6.03 ppm is two bonds distant from the quaternary carbon (C-4) resonating at 137.1 ppm and can be assigned to H-2 (Fig. 2).
The combined results of these two NMR experiments are consistent with the proposed diacid structure (Fig. 2). However, NMR data gave no indication about the structure of the thiazole ring; therefore, liquid chromatography-electrospray ionization tandem MS was performed with the same sample to elucidate this aspect of the molecule. The daughter ion spectrum of the metabolite (Rt = 7.1 min) showed five diagnostically important signals; besides a weak signal for the molecular anion, which is consistent with the molecular formula of the suggested product (m/z 214), we recorded fragment ions generated by single and twofold decarboxylation (m/z 170 and m/z 126, respectively), which clearly indicate the presence of two carboxylate groups in this metabolite. Loss of an ethylenic subunit likely results in an m/z of 98. Finally, the anion at m/z 42 is due to NCO− (31). These two final fragments confirm that the thiazole ring with the hydroxy group in position 2 was not altered during oxygenation.
OBT and diOBT were previously observed as intermediates in two other Rhodococcus strains able to degrade BT (and OBT), namely, R. erythropolis BTS1 and R. rhodochrous OBT18 (2, 18), suggesting that there is a common pathway in these various rhodococci. Successive hydroxylation steps are observed from BT to OBT and then from OBT to diOBT, both of which could result from action of monooxygenases, which are probably distinct, as the mutant strain cannot catalyze the first step.
Very importantly, the presence of a dicarboxylic acid intermediate derived from OBT was detected, demonstrating that there is opening of the benzene ring of the OBT (while it is still attached to the thiazole ring) by ortho (intradiol) catechol cleavage.
Incubation in the presence of 3FC.
Although the putative BT-catechol intermediate could not be detected by either HPLC or NMR because insufficient compound accumulated, the presence of the dicarboxylic intermediate is consistent with intradiol opening of the benzenic ring by a catechol 1,2-dioxygenase.
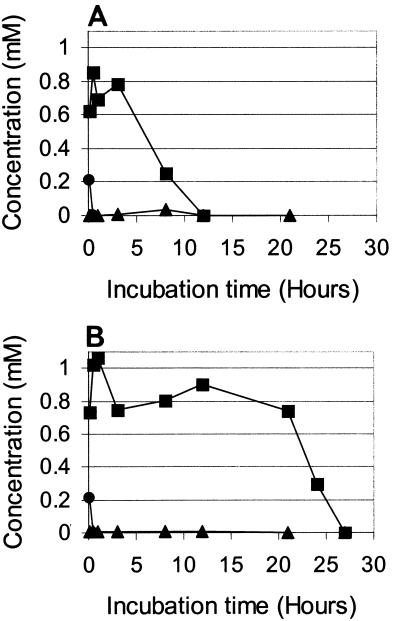
To confirm the activity of this enzyme, a specific inhibitor, 3FC (1 mM), was added to an incubation mixture containing R. pyridinovorans strain PA and BT (1 mM). The time courses of the BT, OBT, and diOBT concentrations (as determined by HPLC and in situ 1H NMR) in the absence and in the presence of 3FC are shown in Fig. 3A and B, respectively. The concentration of the diacid intermediate was too low to measure with the NMR spectra.
FIG. 3.
Degradation of BT by R. pyridinovorans strain PA in the absence of 3FC (A) and in the presence of 3FC (B). Time courses of the concentrations of BT (•), OBT (▪), and diOBT (▴) were determined from 1H NMR spectra.
3FC had a clear effect on the transformation of OBT into diOBT (Fig. 3B); OBT was exhausted within 27 h in the presence of the inhibitor and within only 12 h in its absence. Also, diOBT did not accumulate in the incubation medium in the presence of the inhibitor (Fig. 3B). The transformation of BT into OBT was so rapid (less than 30 min) that it was not possible to see the influence of 3FC (Fig. 3). In a complementary experiment performed with more BT (3 mM) and fewer cells (0.5 g/liter), a clear reduction in the BT transformation rate was observed in the presence of 3FC. In the absence of 3FC, BT was exhausted after 30 min, while BT disappeared only after 70 min in the presence of 3FC (data not shown).
When incubation of 3FC alone with R. pyridinovorans strain PA was monitored by in situ 1H NMR, the signals of 3FC (δ = 6.75 ppm, multiplet) decreased with time, while new signals (5.95 ppm, doublet; 6.95 to 7.30 ppm, unresolved signals) appeared; the chemical shifts of these signals and the coupling constant of the doublet (J1H-1H = 11.2 Hz) are consistent with the presence of 2-fluoromuconate. The structure of this compound was confirmed by 19F NMR. Two main signals were detected at −123.7 and −111.8 ppm in the 19F NMR spectrum recorded after 49 h of incubation; these signals were assigned to the fluoride anion (F−) (the presence of which proves that defluorination occurred) and 2-fluoromuconate, respectively. The signal corresponding to 3FC (−140.4 ppm) was no longer present. These assignments were based on the chemical shift data and on the coupling constant (3J19F-1H = 20 Hz; 2-fluoromuconate) reported by Boersma et al. (4). These data confirmed that there was an intradiol cleavage catechol dioxygenase activity in TSB-grown cells. 3FC inhibited BT degradation, but accumulation of the putative BT-catechol was not observed.
Enzyme assays.
As explained above, we worked with TSB-grown cells to get enough biomass and thus enough intermediates for analysis by NMR spectroscopy. Our data clearly revealed that there was catechol 1,2-dioxygenase activity; this finding appeared to conflict with previous reports (21) of a catechol 2,3-dioxygenase activity. Therefore, we tested the effects of various media (compared with TSB and OBT) on the catechol 1,2-dioxygenase and catechol 2,3-dioxygenase activities. As TSB contains aromatic compounds, like amino acids, that could induce dioxygenase activities, we also tested nutrient broth, a benzoate-containing medium, and an acetate-containing growth medium (without aromatic compounds).
Conventional enzyme assays were conducted with cell extracts of R. pyridinivorans strain PA grown on various media. The specific activities of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase acting on pyrocatechol as a substrate (Table 1) confirmed the presence of catechol 1,2-dioxygenase activity in cells grown on TSB, the medium used for growth of cells in all experiments on biotransformation of BTs reported so far. However, the activity in TSB-grown cells was low (similar to the activity in cells grown in nutrient broth or acetate mineral salts medium). This activity can be regarded as basal activity compared to the very high, induced levels of catechol 1,2-dioxygenase activity found in benzoate-grown cells. Interestingly, cells grown on OBT mineral salts medium did not contain detectable catechol 1,2-dioxygenase activity but did have catechol 2,3-dioxygenase activity, which was not observed in cells grown on any other substrate. The assays performed with OBT-grown cells confirmed previous findings (19, 21) obtained with OBT-grown rhodococci.
TABLE 1.
Specific activities of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase on pyrocatechol in cell extracts of wild-type R. pyridinivorans PAa
| Growth substrate | Protein concn in cell extract (mg/ml) | Sp act (nmol/min/mg of protein) ofa:
|
|
|---|---|---|---|
| Catechol 1,2-dioxygenase | Catechol 2,3-dioxygenase | ||
| Acetate | 0.9 | 31 | NDb |
| Benzoic acid | 4.7 | 870 | ND |
| Nutrient broth | 0.61 | 29 | ND |
| TSB | 2.7 | 7.9 | ND |
| OBT | 0.3 | ND | 0.48 |
The values are the means of at least three determinations, all of which gave very similar activities.
ND, no detectable activity.
Complementary NMR experiments were then performed with resting cells grown on TSB, acetate, and OBT mineral salts media and then incubated with OBT (3 mM). OBT was completely degraded after 24, 21, and 4 h, respectively. In resting cells grown on TSB, OBT, or acetate (data not shown), one-dimensional NMR spectra showed the presence of the signals assigned to the diacid compound resulting from catechol 1,2-dioxygenase activity. These experiments proved that this enzyme was active under these various conditions.
Enzyme assays could not detect catechol 1,2-dioxygenase activity in OBT-grown cells, but this could have been due to a problem of sensitivity or specificity, as pyrocatechol, not the putative BT-catechol, was the substrate for enzyme assays.
Both catechol 1,2-dioxygenase and catechol 2,3-dioxygenase have been reported in many Rhodococcus strains (33). Sometimes both activities are present in the same strain and can act simultaneously. For example, Warhurst et al. (34) showed that R. rhodochrous NCIMB 13259 could convert styrene to 3-vinylcatechol, which was metabolized by a catechol 1,2-dioxygenase to 2-vinyl-cis,cis-muconate, a dead-end product. At the same time a catechol 2,3-dioxygenase activity was also present, and it was proposed that this activity was part of a productive pathway by which 3-vinylcatechol was fully degraded. Similar observations were made by the same authors (35) with other aromatic compounds. More recently, Dean-Ross et al. (11) studied degradation of anthracene by an unidentified Rhodococcus species and proposed that degradation occurs via 1,2-dihydroxyanthracene, further metabolism of which proceeds via both ortho cleavage and meta cleavage. Moody et al. (28) obtained similar results for a related organism, Mycobacterium sp. strain PYR-1. Anthracene appears to be metabolized via three routes, but the product of the ortho cleavage [3-(2-carboxyvinyl)naphthalene-2-carboxylic acid] appears to accumulate, while the meta cleavage product (6,7-benzocoumarin) disappears from the culture medium and is assumed to be a transient intermediate and substrate for ring fission enzymes.
Conclusions.
The pathways for degradation of BTs are still largely unknown; we describe here a detailed study of the degradation of BT (and OBT) by R. pyridinovorans strain PA. Identification of various metabolites was made possible by the use of complementary analytical tools, such as HPLC, in situ 1H NMR, 1H-13C HSQC and HMBC, in situ 19F NMR, and HPLC-electrospray ionization MS. Finally, the use of a mutant strain allowed us to increase the concentration of one of the metabolites.
Considering the metabolites identified, a biodegradative pathway can be proposed for the degradation of BT (and OBT) by R. pyridinivorans strain PA (Fig. 2). A dicarboxylic acid produced by ortho cleavage of a putative BT-catechol has been identified, and the involvement of a catechol 1,2-dioxygenase in cleavage of the benzene ring of BT is reported here for the first time. However, in R. pyridinivorans PA it is not yet certain whether this dicarboxylic acid produced by ortho cleavage of OBT is part of a productive pathway for BT degradation or, in common with the examples described above, is a by-product of a basal activity of catechol 1,2-dioxygenase. The role in BT degradation of the catechol 2,3-dioxygenase activity, which is induced by growth on OBT, is similarly unclear. Both these questions are currently being investigated.
Acknowledgments
The sequence of the 16S rRNA of strain PA was kindly determined by L. Kulakov and M. J. Larkin of Queens University, Belfast, Northern Ireland, whose assistance is gratefully acknowledged. We also thank Lynsey Farrell for her participation in this work.
N. Haroune is a recipient of a grant from the Ministère de l'Education Nationale et de la Recherche. A. Diab is grateful to the Egyptian Ministry of Education for financial support.
REFERENCES
- 1.Altschul, S. F., W. M. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Besse, P., B. Combourieu, G. Boyse, M. Sancelme, H. De Wever, and A. M. Delort. 2001. Long-range 1H-15N heteronuclear shift correlation at natural abundance: a tool to study benzothiazole biodegradation by two Rhodococcus strains. Appl. Environ. Microbiol. 67:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besse, P., B. Combourieu, P. Poupin, M. Sancelme, N. Truffaut, H. Veschambre, and A. M. Delort. 1998. Degradation of morpholine and thiomorpholine by an environmental Mycobacterium involves a cytochrome P450. Direct evidence of the intermediates by in situ 1H-NMR. J. Mol. Biocatal. B Enzymes 5:403-409. [Google Scholar]
- 4.Boersma, M. G., T. Y. Dinarevia, W. J. Middelhoven, W. J. H. Van Berkel, J. Doran, J. Vervoort, and I. M. C. M. Rietjens. 1998. 19F nuclear magnetic resonance as a tool to investigate microbial degradation of fluorophenols to fluorocatechols and fluoromuconates. Appl. Environ. Microbiol. 64:1256-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brecker, L., and D. W. Ribbons. 2000. Biotransformations monitored in situ by proton nuclear magnetic resonance spectroscopy. Trends Biochem. 18:197-202. [DOI] [PubMed] [Google Scholar]
- 6.Bujdakova, H., K. Kralova, and E. Sidoova. 1994. Antifungal activity of 3-(2-alkylthio-6-benzothiazolylaminomethyl)-2-benzothiazolinethiones in vitro. Pharmazie 49:375-376. [PubMed] [Google Scholar]
- 7.Bujdakova, H., T. Kuchta, E. Sidoova, and A. Gvozdjakova. 1993. Anti-Candida activity of four antifungal benzothiazoles. FEMS Microbiol. Lett. 112:329-334. [DOI] [PubMed] [Google Scholar]
- 8.Combourieu, B., P. Besse, M. Sancelme, J. P. Godin, A. Monteil, H. Veschambre, and A. M. Delort. 2000. Common degradative pathways of morpholine, thiomorpholine, and piperidine by Mycobacterium aurum MO1: evidence from 1H-nuclear magnetic resonance and ionspray mass spectrometry performed directly on the incubation medium. Appl. Environ. Microbiol. 66:3187-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Combourieu, B., P. Besse, M. Sancelme, H. Veschambre, A. M. Delort, P. Poupin, and N. Truffaut. 1998. Morpholine degradation pathway of Mycobacterium aurum MO1: direct evidence of intermediates by in situ 1H nuclear magnetic resonance. Appl. Environ. Microbiol. 64:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Combourieu, B., P. Poupin, P. Besse, M. Sancelme, H. Veschambre, N. Truffaut, and A. M. Delort. 1998. Thiomorpholine and morpholine oxidation by a cytochrome P450 in Mycobacterium aurum MO1. Evidence of the intermediates by in situ 1H NMR. Biodegradation 9:433-442. [DOI] [PubMed] [Google Scholar]
- 11.Dean-Ross, D., J. D. Moody, J. P. Freeman, D. G. Doerge, and C. E. Cerniglia. 2001. Metabolism of anthracene by a Rhodococcus species. FEMS Microbiol. Lett. 204:205-211. [DOI] [PubMed] [Google Scholar]
- 12.Delić, V., D. A. Hopwood, and E. J. Friend. 1971. Mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine (NTG) in Streptomyces coelicolor. Mutat. Res. 9:167-182. [DOI] [PubMed] [Google Scholar]
- 13.Delort, A. M., and B. Combourieu. 2000. Microbial degradation of xenobiotics, p. 411-430. In J. N. Barbotin and J. C. Portais (ed.), NMR in microbiology: theory and applications. Horizon Scientific, Wymondham, Norfolk, United Kingdom.
- 14.Delort, A. M., and B. Combourieu. 2001. In situ 1H-NMR study of the biodegradation of xenobiotics: application to heterocyclic compounds. J. Ind. Microbiol. Biotechnol. 26:2-8. [PubMed] [Google Scholar]
- 15.De Wever, H., P. Besse, and H. Verachtert. 2001. Microbial transformations of 2-substituted benzothiazoles. Appl. Microbiol. Biotechnol. 57:620-625. [DOI] [PubMed] [Google Scholar]
- 16.De Wever, H., S. de Cort, I. Noots, and H. Verachtert. 1997. Isolation and characterization of Rhodococcus rhodochrous for the degradation of the wastewater component 2-hydroxybenzothiazole. Appl. Microbiol. Biotechnol. 47:458-461. [Google Scholar]
- 17.De Wever, H., and H. Verachtert. 1997. Biodegradation and toxicity of benzothiazoles. Water Res. 31:2673-2684. [Google Scholar]
- 18.De Wever, H., K. Vereecken, A. Stolz, and H. Verachtert. 1998. Initial transformations in the biodegradation of benzothiazoles by Rhodococcus isolates. Appl. Environ. Microbiol. 64:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Wever, H. 1995. Biodegradability of benzothiazoles. Ph.D. thesis. Katholieke Universiteit Leuven, Leuven, Belgium.
- 20.Fiehn, O., T. Reemtsma, and M. Jekel. 1994. Extraction and analysis of various benzothiazoles from industrial wastewater. Anal. Chim. Acta 295:297-305. [Google Scholar]
- 21.Gaja, M. A., and J. S. Knapp. 1997. The microbial degradation of benzothiazoles. J. Appl. Microbiol. 83:327-334. [Google Scholar]
- 22.Gibson, D. T. 1970. Assays of enzymes of aromatic metabolism. Methods Microbiol. 6A:463-478. [Google Scholar]
- 23.Gold, L. S., T. H. Slone, B. R. Stern, and L. Bernstein. 1993. Comparison of target organs of carcinogenicity for mutagenic and non-mutagenic chemicals. Mutat. Res. 286:75-100. [DOI] [PubMed] [Google Scholar]
- 24.Haroune, N., B. Combourieu, P. Besse, M. Sancelme, and A. M. Delort. 2001. 1H NMR: a tool to study the fate of pollutants in the environment. C. R. Acad. Sci. Paris Chim. 4:759-763. [Google Scholar]
- 25.Hartley, D., and H. Kidd. 1987. The agrochemical handbook. The Royal Society of Chemistry, Nottingham, United Kingdom.
- 26.Martin, G. E., and C. E. Hadden. 2000. Long-range 1H-15N heteronuclear shift correlation at natural abundance. J. Nat. Prod. 63:543-585. [DOI] [PubMed] [Google Scholar]
- 27.Meding, B., K. Toren, A. T. Karlberg, S. Hagberg, and K. Wass. 1993. Evaluation of skin symptoms among workers at a Swedish paper mill. Am. J. Ind. Med. 23:721-728. [DOI] [PubMed] [Google Scholar]
- 28.Moody, J. D., J. P. Freeman, D. G. Doerge, and C. E. Cerniglia. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poupin, P., N. Truffaut, B. Combourieu, P. Besse, M. Sancelme, H. Veschambre, and A. M. Delort. 1998. Degradation of morpholine by an environmental strain of Mycobacterium involves a cytochrome P450. Appl. Environ. Microbiol. 64:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reemtsma, T., O. Fiehn, G. Kalnowski, and M. Jekel. 1995. Microbial transformations and biological effects of fungicide-derived benzothiazoles determined in industrial wastewater. Environ. Sci. Technol. 29:478-485. [DOI] [PubMed] [Google Scholar]
- 31.Reemtsma, T. 2000. Determination of 2-substituted benzothiazoles of industrial use from water by liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 14:1612-1618. [DOI] [PubMed] [Google Scholar]
- 32.Storm, T., T. Reemtsma, and M. Jekel. 1999. Use of volatile amines as ion pairing agents for the high-performance liquid chromatographic-tandem mass spectrometric determination of aromatic sulfonates in industrial wastewater J. Chromatogr. A 854:175-185. [DOI] [PubMed] [Google Scholar]
- 33.Warhurst, A. M., and C. A. Fewson. 1994. Biotransformations catalysed by the genus Rhodococcus. Crit. Rev. Biotechnol. 14:29-73. [DOI] [PubMed] [Google Scholar]
- 34.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl. Environ. Microbiol. 60:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Production of catechols and muconic acids from various aromatics by the styrene-degrader Rhodococcus rhodochrous NCIMB 13259. Biotechnol. Lett. 16:513-516. [Google Scholar]
- 36.Wegler, R., and L. Eue. 1977. Chemie der Pflanzenschutz- und Schädlingsbekämpfungsmittel, vol. 5. Herbizide. Springer-Verlag, Berlin, Germany.
- 37.Yoon, J.-H., S.-S. Kang, Y.-G. Cho, S. T. Lee, Y. H. Kho, C.-J. Kim, and Y.-H. Park. 2000. Rhodococcus pyridinivorans sp. nov., a pyridine-degrading bacterium. Int. J. Syst. E vol. Microbiol. 50:2173-2180. [DOI] [PubMed] [Google Scholar]