Abstract
We describe the development of a novel protein expression system based on the industrial fermentation organism Ralstonia eutropha (formerly known as Alcaligenes eutrophus) NCIMB 40124. This new system overcomes some of the shortcomings of traditional Escherichia coli-based protein expression systems, particularly the propensity of such systems to form inclusion bodies during high-level expression. Using a proteomics approach, we identified promoters that can be induced by simple process parameters or medium compositions in high-density cell culture or shake flasks, respectively. By combining newly developed molecular biological tools with a high-cell-density fermentation process, we were able to produce high levels (>1 g/liter) of soluble, active organophosphohydrolase, a model enzyme prone to inclusion body formation in E. coli.
Advances in protein engineering, the completion of numerous bacterial and fungal genome sequencing projects, and the isolation of new genes from extremophiles have led to an increased number of useful proteins. However, to enable recombinant proteins to play a role in applications where larger quantities are required, such as tissue engineering or catalytic materials, production technologies that are more efficient and robust are desirable. Improving protein production is the primary goal of recombinant microbial process development and is a focus of our laboratory. Overall protein productivity can be improved by increasing the product of two variables: (i) the amount of recombinant protein per cell (specific productivity) and (ii) the amount of cell mass per unit of volume and time (cell productivity). In order to improve volumetric productivity in a cost-effective manner, recombinant proteins are often produced in high-cell-density fermentations. High-cell-density fermentations offer many advantages over traditional fermentations in that final product concentrations are higher and downtime and water usage are reduced, yet overall productivity is improved, resulting in lower setup and operating costs (3, 6).
In the absence of specific folding or posttranslational modification requirements, Escherichia coli is usually the expression host of choice. E. coli-based fermentation systems produce good yields at laboratory scale; however, scale up to industrial scale, where oxygen enrichment, dialysis, and sophisticated feeding algorithms become impractical and cost prohibitive, has been challenging (22). Unlike laboratory-scale fermentors, where the above-mentioned techniques are feasible, large-scale industrial fermentors are limited by mixing constraints and their ability to transfer oxygen and heat. It is well documented that oxygen-limiting conditions in E. coli result in the production of reduced carbon metabolites such as acetate, lactate, and formate (1, 20), which accumulate in high-cell-density cultures and ultimately inhibit further microbial growth. Titers of recombinant proteins in E. coli are limited by several factors including the final cell density (22), the tendency for inclusion bodies to be formed when strong promoters are used (15, 23), and proteolysis, which adversely affects the quality of the final product (14, 17).
Considerable efforts have been undertaken to overcome the limitations of E. coli as a recombinant-protein expression host, and improving the organism's fermentation performance has been the focus of many biochemical engineering research groups. The accumulation of organic acids during the fermentation process has been reduced by several approaches: (i) installation of inline dialysis membranes (12, 26), (ii) implementation of a controlled feeding regimen imposing a reduced growth rate on the microorganism (28), (iii) enrichment with pure oxygen (7), and (iv) engineering of a heme cofactor into the host organism (11). Inclusion body formation has been reduced by expression of a target protein in frame with a highly soluble protein, thereby creating a more soluble fusion protein, and/or by decreasing the growth rate of the organism by decreasing the temperature of the fermentation (39). Proteolytic activity has been reduced by the generation of protease-deficient mutants (18).
While considerable progress has been made in addressing some of these shortcomings, the performance of E. coli as a recombinant-protein expression host still leaves much to be desired. In this paper, we report the development of a novel prokaryotic protein expression system based on a nonpathogenic organism, Ralstonia eutropha (formerly Alcaligenes eutrophus). The described system permits high-cell-density culture in a defined minimal medium, does not require the addition of antibiotics to maintain plasmid stability, and does not require exogenous addition of inducers such as isopropyl-B-d-thiogalactopyranoside (IPTG) to initiate protein expression. A model protein that has traditionally been difficult to obtain in soluble form in E. coli (organophosphohydrolase [OPH] from Pseudomonas diminuta MG) was produced at high yield without any measurable formation of inclusion bodies.
R. eutropha has been used at a scale of several hundred thousand liters for the production of polyhydroxyalkanoate (PHA), a biodegradable polymer, by ICI/Zeneca and later Monsanto. The genome has been sequenced (http://jgi.doe.gov/JGI_microbial/html/ralstonia/ralston_homepage.html), andnumerous high-cell-density processes have been reported previously (13, 30). Unlike E. coli and other enterobacteria which preferentially metabolize hexose sugars through the Embden-Meyerhof-Parnas pathway, R. eutropha preferentially uses the Entner-Douderoff pathway to metabolize hexoses (21, 34). This results in a higher NADPH/NADH ratio, which causes a higher degree of biosynthetic as opposed to respiratory reducing equivalents. This, we believe, positively affects protein formation, since R. eutropha has the ability to balance the reducing equivalents generated from carbon metabolism by using the production of polyhydroxybutyrate (PHB) as a sink for reducing power (i.e., NADPH) in the absence of a final electron acceptor such as oxygen. Both E. coli and Bacillus spp. are forced to synthesize reduced organic acids to balance their redox state under oxygen-limiting conditions, and these acid metabolites have been shown to negatively affect overall fermentation performance. In contrast, PHB is a high-molecular-weight polymer that causes no osmotic stress on the cell and does not adversely affect overall bacterial growth. High-cell-density fermentations of R. eutropha (up to 230 g/liter) in large-scale reactors have been described previously by various groups (31, 37). Most of this work has been aimed at maximizing the PHB content; hence, the real cell mass (real cell mass = total biomass − PHB) was generally below 30 to 40 g/liter.
OPH, the model enzyme.
OPHs (EC 3.1.8.1) are enzymes that are able to hydrolyze and thereby substantially reduce the toxicity of cholinesterase inhibitors like sarin and VX. OPH is a 72-kDa homodimeric enzyme that requires a divalent cation such as cobalt or zinc (8). The enzyme catalyzes the hydrolysis of the organophosphorus compounds at a remarkable rate; hence, it has found applications in detoxification and decontamination of parathion-containing agricultural fields and chemical weapons stock (41). Other applications include its use as a biosensor for monitoring levels of these compounds and for protection against chemical warfare agents (29). Several attempts to express gram quantities of OPHs in E. coli and other organisms have been hampered by the formation of inclusion bodies and the inability to grow to high cell densities. Attempts to improve production by engineering a fusion protein with highly soluble proteins like green fluorescent protein have resulted in reduced specific productivity and require posttranslational cleavage of the fusion (40, 41). To date, there has been no report of a fermentation process that provides sufficient amounts of OPH for large-scale applications (9). OPH was chosen for this study because it has a well-documented history of forming inclusion bodies in E. coli and thus seemed to be a suitable model protein (32, 33).
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The various strains and plasmids that were used in this study are listed in Table 1. The oligonucleotides used in this study were PromUp (dCAGGAATTCCATCGCGCAGCATGC), PromDn (dGATGAGCTCTCTCCAGTGGTGAAC), ParUp (dCTCGAGCTCATGTCTATCGGTACC), and ParDn (dGGAAGGATCCAGATGGCGTCAT) (the restriction sites engineered in the oligonucleotides are underlined).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| R. eutropha strains | ||
| NCIMB 40124 | Wild type, gentamicin resistant | NCIMB |
| GS5 | NCIMB 40124 containing plasmid pKTPPCm | This study |
| SS14 | PHAP::OPH, phaPp-OPH translational fusion strain derived from NCIMB 40124 and pJQPPCm | This study |
| E. coli strains | ||
| TOP10 | Strain for ligation and cloning of genes | Invitrogen |
| S-17 | Host strain for pJQ plasmids | 10, 27 |
| Plasmids | ||
| pCR2.1-TOPO | High-copy-number plasmid for cloning; confers ampicillin and kanamycin resistance | Invitrogen |
| pJQ200mp18 | Homologous recombination plasmid conferring gentamicin resistance | 27 |
| pJQPPCm | phaPp-OPH translational fusion cloned into pJQ200mp18 | This study |
| pKNOCK-Cm | Plasmid for recombination, conferring chloramphenicol resistance | 2 |
| pKT230 | Broad-host-range plasmid | American Type Culture Collection |
| pUC19 | High-copy-number plasmid used for cloning; confers ampicillin resistance | New England Biolabs |
| pUCPPCm | phaPp-OPH translational fusion cloned into pUC19 | This study |
Growth media and antibiotics.
E. coli strains were grown in Luria-Bertani medium. R. eutropha strains were cultivated in one of the following media depending on the application: NR medium (36), PCT medium (Table 2), and Lee medium (25). NR is a complex medium, while both PCT and Lee media are minimal media. Antibiotics were added to the growth media in the following concentrations: for R. eutropha, 10 μg of gentamicin and 50 μg of chloramphenicol/ml; for E. coli, 100 μg of ampicillin, 10 μg of gentamicin, 25 μg of kanamycin, and 50 μg of chloramphenicol/ml (all antibiotics were obtained from Sigma, St. Louis, Mo.).
TABLE 2.
Composition of PCT mediuma
| Component | Concn (g/liter) |
|---|---|
| Glucose | 500 |
| Saltsb | |
| MgSO4 · 7H2O | 2.2 |
| K2SO4 | 3.0 |
| Na2SO4 | 0.18 |
| FeSO4 · 7H2O | 0.18 |
| H3PO4 (1.48 M) | 9.6c |
| Trace elements | |
| CuSO4 · 5H2O | 0.2 |
| ZnSO4 · 6H2O | 1.0 |
| MnSO4 · 4H2O | 1.0 |
| CaCl2 · 2H2O | 26.0 |
Glucose, salts, and trace elements were autoclaved separately, and 40 ml of glucose, 1 liter of salts, and 2.4 ml of trace elements were mixed at room temperature.
The pH was adjusted to 6.8 prior to autoclaving with 28% NH4OH.
Milliliters per liter.
Fermentation procedure.
Fed-batch fermentations of R. eutropha NCIMB 40124 were carried out in a 3-liter fermentor (Applikon, Foster City, Calif.) under the following conditions: 30°C, 1.5 liter of airflow/min, and a 630-rpm stirrer speed with an initial working volume of 1 liter (1× PCT medium with 5% [vol/vol] inoculum). An Applikon programmable logic controller (ADI1030) was used for maintaining temperature at 30°C and pH at 6.8. The pH of the medium was maintained with 85% phosphoric acid and aqueous ammonia (28%). The concentration of dissolved oxygen was maintained at 30% by controlling the stirrer speed up to a maximum speed of 1,250 rpm, at which point the dissolved-oxygen concentration dropped below 30% and became a function of the glucose feed rate. The fermentation was carried out in a batch mode until the initial glucose (20 g/liter) was consumed. Following the batch phase, a feed containing glucose, phosphoric acid, CoCl2 · 7H2O, and MgSO4 (600 g/liter, 220 mM, 0.15 mM, and 19 mM, respectively) was used to promote linear growth until a biomass concentration of approximately 100 g/liter was achieved. The glucose feed rate was initially maintained at 11.7 g/h and after 18 h of feeding was increased to 14 g/h. Induction was initiated by changing the feed solution to glucose (600 g/liter) containing 15 mM CoCl2. The second glucose feed solution contained no phosphate, leading to a gradual depletion in the medium, which in turn induced the expression of OPH. Offgas was analyzed in real time for O2 and CO2 concentrations with a model 3750 carbon dioxide and oxygen analyzer (Illinois Instruments, Ingleside, Ill.).
HPLC analysis.
Samples of supernatant from the fermentation were collected and analyzed by high-pressure liquid chromatography (HPLC) on an Aminex 87H column (Bio-Rad Laboratories, Hercules, Calif.) to determine the concentrations of phosphate, sulfate, glucose, acetate, lactate, and ethanol. Five millimolar sulfuric acid at a flow rate of 0.7 ml/min was used as the mobile phase. All six compounds were detected by measurement of the refractive index on a Waters 410 refractometer.
DNA preparation, manipulation, and Southern blot hybridization.
Preparation and manipulation of DNA, genomic DNA preparation, transformation, and hybridization were carried out using standard procedures or by following manufacturers' instructions. Sequencing was performed at the Molecular Biology Core Facility at Dartmouth College.
Construction of integration plasmid pJQ200mp18Cm.
pKNOCK-Cm (2) vector was digested with the restriction enzyme MluI and was separated by agarose gel electrophoresis. A 1.1-kb DNA fragment was purified from the gel, and the ends were blunted using T4 DNA polymerase (New England Biolabs, Beverly, Mass.). The fragment was inserted into the EcoRV site within the gentamicin resistance gene of plasmid pJQ200mp18. This plasmid was used for chromosomal integration into R. eutropha by homologous recombination.
Construction of the OPH expression cassette.
A 450-bp sequence of R. eutropha DNA containing the promoter along with the ribosomal binding site of the phaP gene (GA24) was amplified using PCR with oligonucleotides PromUp and PromDn and was inserted into pUC19 as an EcoRI/SacI insert. An 1,100-bp sequence corresponding to the coding region for OPH was amplified from pCMS75 with oligonucleotides ParUp and ParDn such that the ATG start site of the OPH gene was in the same position as that of phaP in the wild type. Plasmid pKNOCK-Cm was digested with the restriction enzyme MluI and separated by gel electrophoresis. A 1.1-kb DNA fragment was purified from the gel, and the ends were blunted using T4 DNA polymerase. The fragment was inserted into the HincII site of the pUC19 vector containing phaPp-OPH (phaP promoter carrying the coding region for OPH). The sequence of the construct was verified, and the plasmid was designated pUCPPCm.
Construction of OPH-producing R. eutropha.
The 2.4-kb EcoRI/PstI fragment containing phaPp-OPH-CAT (phaP promoter carrying the coding region for OPH together with the chloramphenicol acetyltransferase gene) of pUCPPCm was inserted into the broad-host-range plasmid pKT230, generating pKTPPCm. Similarly, the 2.4-kb FspI/PstI fragment was inserted into the EcoICRI/PstI-digested plasmid pJQ200mp18, generating pJQPPCm. Gene transformation from E. coli into R. eutropha was accomplished by adaptation of standard biparental-mating protocols (27). Colonies that grew on minimal medium and were resistant to gentamicin and chloramphenicol were selected and subjected to a qualitative test for OPH activity. A small amount of cells was scraped from the plates and was resuspended in a 10 mM Tris-HCl buffer containing 133 μM parathion. The appearance of a yellow color was used as an indicator of the presence of OPH.
Enzyme activity assays.
Cell pellets were resuspended in 150 mM CHES buffer [2-(cyclohexylamino)ethanesulfonic acid], pH 9.0, to an optical density at 600 nm (OD600) between 1 and 3. The samples were sonicated in a sonic dismembrator 550 (Fisher Scientific, Fair Lawn, N.J.) in two pulsed cycles (2 s on, 0.5 s off, 30-s duration, 5 min of cooling on ice between cycles). One milliliter of this cell extract was centrifuged at 16,000 × g for 5 min, and the supernatant was analyzed for enzyme activity against paraoxon (Sigma) as described previously (8, 32). An extinction coefficient of 17,000 M−1cm−1 for p-nitrophenol at pH 9.0 was used to calculate the activity. The total protein concentration was measured using a Bradford protein assay kit (Bio-Rad).
Western blots.
Whole-cell pellets were resuspended in water to a final concentration of 0.2 g (dry cell weight)/liter. The cell suspension (20 μl) was added to 20 μl of 2× sodium dodecyl sulfate (SDS) cracking buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 20% glycerol) and boiled for 10 min. Samples (15 μl) were then loaded on a Tris-HCl-SDS-12% polyacrylamide gel (Bio-Rad) and resolved at 100 V. The proteins were then transferred from the gel to a nitrocellulose membrane (0.2 μm; Schleicher & Schuell, Keene, N.H.) for 1 h at 100 V/350 mA. The membrane was then blocked in a TBST solution buffer (50 mM Tris base, 188 mM sodium chloride, 0.05% Tween 20, pH 7.5) containing 3% bovine serum albumin (Sigma) and incubated for 2 h at room temperature. The membrane was then exposed to a crude primary rabbit anti-OPH serum (generously supplied by Janet Grimsley, Texas A&M University) diluted in the TBST buffer. After being washed repeatedly with TBST buffer, the membrane was exposed to a secondary goat anti-rabbit antibody conjugated to horseradish peroxidase (Pierce, Rockford, Ill.) diluted in TBST buffer. After repeated washing with TBST buffer, color development was achieved in a Tris-buffered saline buffer containing 500 mg of DAB (di-amino benzinidine; Pierce)/liter and 100 μl of a 30% H2O2 solution (Fisher Scientific)/liter.
Quantification of PHB in R. eutropha cells.
PHB was quantified by the sulfuric acid-HPLC method of Karr et al. (19) with modifications (42).
2D SDS-PAGE and protein sequencing.
Samples from the fermentation were collected and centrifuged. Cell pellets were dissolved in osmotic lysis buffer (10 mM Tris [pH 7.4], 0.3% SDS) containing nuclease stock and protein inhibitors. The protein stock (10× stock) contained 50 mM MgCl2, 100 mM Tris (pH 7.0), 500 μg of RNase (RNase A from bovine pancreas type IIIA; Sigma)/ml, and 1,000 μg of DNase (DNase I, type II from bovine pancreas; Sigma)/ml. Protease inhibitors (100× stock) contained 20 mM AEBSF (4-[2-aminoethyl] benzenesulfonyl fluoride hydrochloride; Calbiochem, San Diego, Calif.), 1 mg of leupeptin (Sigma)/ml, 0.36 mg of E-64 (N-[N-{l-trans-carboxyoxiran-2-carbonyl}-l-leucyl]-agmatine; Sigma)/ml, EDTA (Calbiochem), and 5.6 mg of benzamidine (Sigma)/ml. One milliliter of each sample was lyophilized. Resulting residues were redissolved to a concentration of 5 mg/ml in a 1:1 dilution of SDS boiling buffer (5% SDS, 5% β-mercaptoethanol, 10% glycerol, 60 mM Tris [pH 6.8]) and heated in a boiling-water bath for 2 min. Two-dimensional (2D) SDS-polyacrylamide gel electrophoresis (PAGE) was performed by Kendrick Labs, Inc. (Madison, Wis.) according to a method modified from that of O'Farrell. Isoelectric focusing was carried out in glass tubes with an inner diameter of 2.0 mm by using 2.0% BDH ampholines (pH 4 to 8; Hoefer Scientific Instruments, San Francisco, Calif.) at 9,600 V · h. One microgram of an isoelectric focusing internal standard, tropomyosin protein, with a molecular weight (MW) of 33,000 and pI 5.2, was added to the samples. After equilibration for 10 min in buffer (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, 0.0625 M Tris [pH 6.8]), the tube gel was sealed to the top of a stacking gel on top of a 10% acrylamide slab gel (0.75 mm thick). SDS slab gel electrophoresis was then carried out for 4 h at 12.5 mA. The following proteins (Sigma) were added as MW standards to the agarose that was used to seal the tube gel to the slab gel: myosin (MW, 220,000), phosphorylase A (MW, 94,000), catalase (MW, 60,000), actin (MW, 43,000), carbonic anhydrase (MW, 29,000), and lysozyme (MW, 14,000). These standards appeared as horizontal lines on the silver-stained 10% acrylamide slab gel. After slab gel electrophoresis, the gel was transferred to the transfer buffer and transblotted onto polyvinylidene difluoride paper overnight at 200 mA and approximately 100 V per two gels. The blots were stained with Coomassie brilliant blue R250 in 50% methanol and destained in 50% methanol. Analysis of 2D gels for differentially expressed polypeptides was conducted by eye. The N terminus (10 amino acids) of the identified protein spots was sequenced by Edman degradation at the Protein Chemistry Core Facility, Howard Hughes Medical Institute, Columbia University.
RESULTS
High-cell-density fermentation.
We developed a simple, efficient, and reproducible high-cell-density fermentation protocol based on R. eutropha strain NCIMB 40124 that is aimed at maximizing the overall protein content in the reactor. We can routinely obtain cell densities of over 150 g (dry cell weight)/liter in a simple fed-batch mode (data not shown) in a minimal medium containing only inorganic salts and glucose. Previous studies have established that R. eutropha produces PHB under phosphate- or nitrogen-limited conditions when supplied with an appropriate carbon source such as glucose or fructose. We conducted a series of fermentation experiments to establish the amount of phosphate required to maximize cell mass formation while minimizing polymer accumulation. We found that by using various carbon/phosphate ratios in the feed, we were able to effectively control the PHB content of the cell in high-cell-density culture (data not shown). By using this feeding strategy, we were able to achieve a cell density of 182 g (dry cell weight)/liter, with low polymer content. R. eutropha has been shown to have a comparatively high protein content of 68% of total dry cell weight (16). Oxygen uptake rates were typically below 150 mmol · liter−1 · h−1, well within reach of commercial-scale fermentors (5). To determine the extent of overflow metabolism under these conditions, we performed HPLC analysis of the fermentation broths at various times during the fermentation process (data not shown). Throughout the process, acetate, lactate, and ethanol were below the detection limit of 0.1 g/liter (data not shown). It should be noted that glucose feed rates were held constant over extended periods of the process, which greatly simplified process control and improved the reproducibility of the process. With biomass increasing in a roughly linear fashion, the specific growth rates were therefore gradually reduced from μ = 0.25 h−1 to about μ = 0.025 h−1.
Identifying strong inducible promoters.
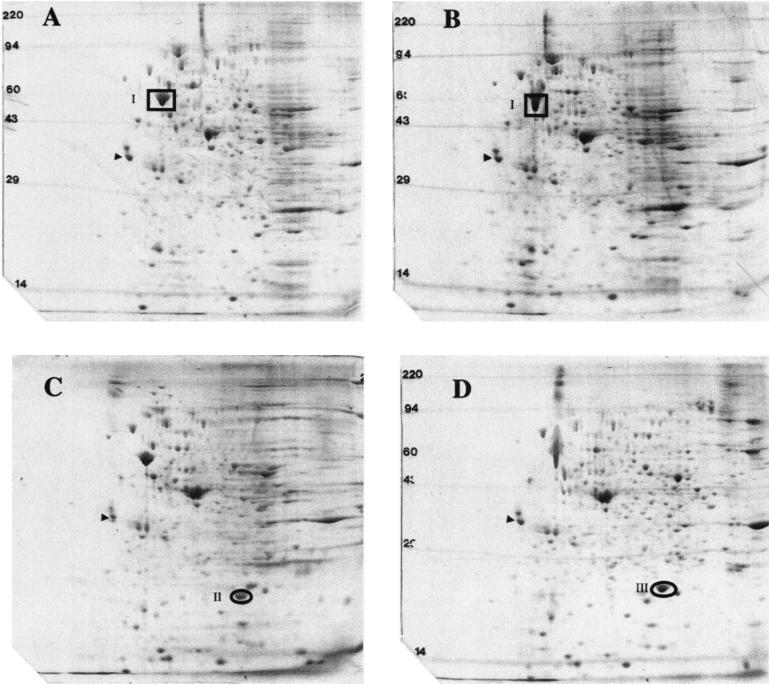
High-level expression of recombinant proteins requires the availability of a strong and preferably inducible promoter. On the large, commercial scale, the use of antibiotics (to ensure plasmid maintenance) and inducers such as IPTG (to induce expression) is not desirable for environmental and economic reasons. Thus, we sought to systematically identify genes that are up-regulated in R. eutropha during the fermentation process itself or are up-regulated in response to the variation of a simple process parameter. The induction events tested were as follows: (i) a change in temperature, (ii) the omission of phosphate in the medium feed, (iii) microaerobic conditions (oxygen limitation), (iv) change in pH, and (v) change in carbon source (glucose to fructose). In order to assess the change in the proteome in response to these induction events, we conducted several fermentations. A control fermentation without any induction event was used as the base case. In each of the fermentations, we grew the cells to an OD600 of approximately 300 and then altered one of the above-mentioned parameters and collected cell samples after imposing the specific changes. In three fermentation runs (pH change, temperature change, and oxygen limitation), high cell densities were not obtainable and thus were not considered further. In two of the five fermentation runs (phosphate limitation and change of glucose to fructose), we were able to achieve cell densities in excess of 150 g (dry cell weight)/liter. Cell samples from these fermentations were collected, and total cell protein was analyzed by 2D SDS-PAGE. By superimposing 2D SDS-PAGE gels containing protein samples obtained pre- and postinduction, we were able to identify two proteins (glucose-to-fructose change [Fig. 1C] and phosphate limitation [Fig. 1D]) that were up-regulated in response to the imposed conditions and one protein that appeared to be expressed at high constitutive levels under all conditions (Fig. 1). Table 3 describes the three proteins denoted in Fig. 1. One protein that was significantly induced by phosphate limitation was found to be GA24, a product of the phaP gene, a phasin previously described by Wieczorek and coworkers (38). GA24 is a protein found on the surface of PHA granules that regulates the size of the granule. Wieczorek et al. also found that GA24 was expressed at relatively high levels (5% of the total protein) in R. eutropha during PHB production under nitrogen-limiting conditions. Since phaPp was a promoter, inducible by a simple process parameter (phosphate limitation) and with low-level basal activity, it met our criteria for a suitable recombinant-protein expression promoter for high-cell-density culture. In order to determine whether quorum sensing or high cell density in itself affects a change in the proteome samples, we collected samples at OD600 values of 100 and 600 without changing any process parameters and then analyzed them by 2D SDS-PAGE (Fig. 1A and B).
FIG. 1.
2D gel electrophoresis of whole-cell extracts of R. eutropha from three separate fermentation experiments: fed-batch fermentation on glucose minimal medium. Samples were collected at OD600 values of 100 (A) and 600 (B), after changing the carbon source from glucose to fructose at an OD600 of 300 and sampling at an OD600 of 450 (C), and after changing the carbon source to a phosphate-free glucose feed at an OD600 of 300 and sampling at an OD600 of 450 (D). Samples shown in panels A and B were collected from the same fermentation experiment at different times; samples shown in panels C and D were collected from separate fermentation experiments. Cell pellets from the fermentation were collected and lysed. The samples were resuspended in SDS buffer, and 2D SDS-PAGE was run with each individual sample. The gels were visually analyzed, and three proteins of interest were identified: a constitutively expressed protein (I), a fructose-induced protein (II), and a protein induced by phosphate limitation (III). Molecular masses (in kilodaltons) are indicated. The small arrowheads denote an internal standard, the protein tropomyosin (MW, 33,000; pI 5.2).
TABLE 3.
N-terminal sequence and protein similarity
| Protein | N-terminal sequence | Protein similarity | Function |
|---|---|---|---|
| I | AAKDVVFGDA | GroEL (Burkholderia cepacia) | Chaperon |
| II | TQXTAEQCTK | No similarity identified | |
| III | VILTPEQVAA | GA24 (R. eutropha) (also known as PhaP protein) | Surface protein bound to PHB (an intracellular polymer)a |
OPH production in R. eutropha.
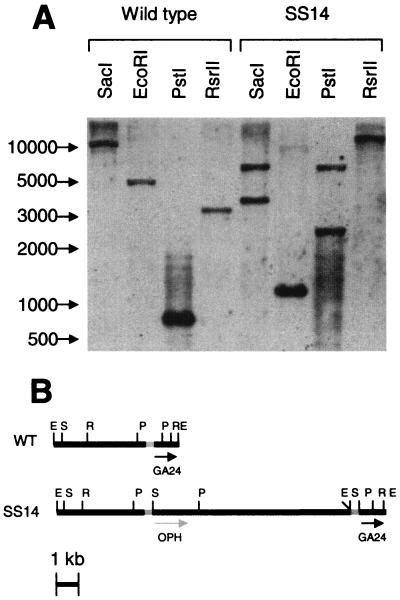
Plasmid pJQPPCm, containing the open reading frame of the OPH gene under the control of phaPp, was constructed and transformed into E. coli by electroporation. This plasmid was integrated into the phaP locus of R. eutropha by biparental mating, thereby providing a single stable chromosomal copy of the OPH gene. Three individual colonies were selected and analyzed for OPH production. PCR was performed with one internal and one external primer to verify the chromosomal location of integration. All three strains yielded PCR products consistent with a single chromosomal insertion (data not shown). In addition, Southern blotting was performed on one of the strains to confirm that only a single copy of the OPH open reading frame was received and that integration indeed occurred in the phaP locus (Fig. 2).
FIG. 2.
Southern blot analysis of wild-type R. eutropha and R. eutropha SS14. Genomic DNA from the wild type and integrant SS14 were extracted and digested with SacI, PstI, EcoRI, and RsrII. A 400-bp phaP promoter sequence (the FspI/SacI fragment of plasmid pUCPPCm, shown in grey) was used to probe the blot. (A) Results of Southern blot analysis of genomic DNA of the wild type and the integrant probed with a horseradish peroxidase-labeled phaP promoter. The sizes of the markers (base pairs) are indicated on the left. (B) Map of the analyzed genomic region in the wild type and the integrant, indicating the probe and the various restriction enzymes used. S, SacI; E, EcoRI; P, PstI; R, RsrII. The open reading frames of OPH and GA24 (phaP) are also indicated.
High-cell-density production of OPH.
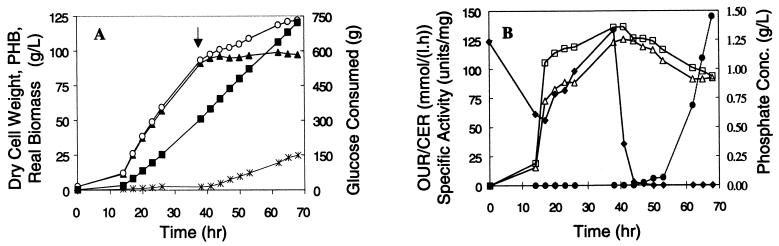
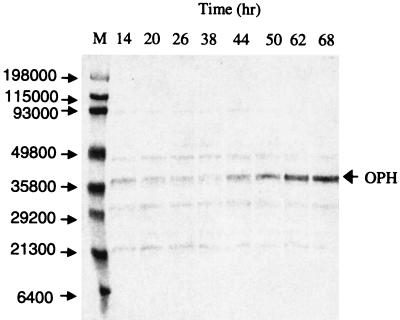
Although high OPH-specific activities have been obtained in E. coli in shake flask experiments, successful scale-up has failed due to inclusion body formation (33). In order to demonstrate that high levels of the enzyme can be produced in a simple and efficient fermentation process, we conducted a fermentation to produce approximately 100 g of biomass/liter and over 1 g of OPH/liter. The fermentation was conducted as a three-stage process: batch, fed batch, and induction. During the batch phase, the initial inoculum was allowed to grow such that the cells consumed the initial 20 g of glucose/liter. Glucose depletion (14 h after inoculation) was indicated by a sharp increase in the concentration of dissolved oxygen. After depletion of the initial glucose, a feed containing glucose, phosphate, and sulfate at a constant feed rate of 11.7 g of glucose/h was initiated. The cells continued to grow exponentially until a fixed growth rate corresponding to the glucose feed rate was achieved. The feed rate was then further increased to 14.0 g of glucose/h, thereby increasing the growth rate. During this phase, both phosphate and sulfate concentrations increased in the fermentation, whereas glucose as the rate-limiting nutrient remained undetectable due to its immediate consumption in the reactor. During this phase, the amount of PHB in the cell remained less than 5% of the total biomass. Despite some PHB production, the basal activity of the phaP promoter was low, as determined by enzyme assays (Fig. 3B) and Western blotting (Fig. 4). A biomass concentration of 93.4 g/liter was obtained 38 h after inoculation (24 h after initiating the glucose feed). At this point, the feed was changed to a glucose solution, containing 15 mM cobalt but lacking phosphate, to initiate the induction phase. OPH requires a divalent cation for enzymatic activity (25), so cobalt was added to the glucose feed. During the initial induction phase, the phosphate concentration rapidly decreased in the reactor (Fig. 3B), and upon depletion, OPH production was induced. After 30 h of induction (68 h after inoculation), the specific activity of the protein was found to be 145 U/mg (Fig. 3B). In addition to the results of the enzyme activity assays, the induction of OPH could also be seen in the appearance of a band corresponding to the MW of OPH (approximately 36,000) in the Western blot (Fig. 4, lanes corresponding to 44 to 68 h). In this fermentation, we achieved a concentration of 99.1 g of real biomass/liter. The specific activity of pure OPH (with cobalt as the divalent cation) has been reported to be 7,250 U/mg (29) and 8,020 U/mg (24) using the paraoxon enzyme activity assay; thus, the specific activity of 145 U/mg corresponds to 2.0 or 1.8% of total cellular protein, respectively. As previously noted, the protein content of R. eutropha has been found to be 68% of dry cell weight (15). The product of 99.1 g of biomass/liter, 0.68 g of protein/g of biomass, and 0.018 g of OPH/g of protein yields an OPH concentration of 1.2 g/liter.
FIG. 3.
OPH production in high-cell-density fermentation of R. eutropha. (A) The dry cell weight, real biomass, PHB, and glucose feeding profile for a typical high-cell-density fermentation. The arrow indicates the time of induction. Symbols: open circles, total dry cell weight; closed triangles, real biomass; crosses, PHB; closed squares, glucose consumed. (B) The oxygen uptake rate (OUR), the carbon dioxide evolution rate (CER), the phosphate concentration in the reactor, and the specific activity of OPH produced. Symbols: open squares, OUR; open triangles, CER; closed diamonds, phosphate concentration; closed circles; specific activity.
FIG. 4.
Western blot analysis of fermentation samples. Total cell pellets were diluted to approximately 0.2 g (dry cell weight)/liter and then boiled on SDS-PAGE cracking buffer and resolved on a 12% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane, and blotting was done with anti-OPH antibody. The figure shows the time course of OPH production. M, broad-range MW marker, with MWs indicated on the left.
DISCUSSION
Traditionally, the development of recombinant-protein production systems has focused on improving the fermentation processes of organisms for which molecular biological tools (e.g., transformation protocols, selection markers, inducible promoters, etc.) have already been established. In this study, we used an organism with robust fermentation characteristics and developed the molecular biological tools to produce recombinant proteins at high levels. We achieved cell densities of over 150 g/liter in a simple process that does not require complex medium components or the supplementation of air with pure oxygen. The process can be implemented manually or by a simple programmable logic controller. In the described fermentation process, growth-inhibiting organic acids were not synthesized although the levels of dissolved oxygen remained at 0% throughout most of the process.
Promoter traps have been used previously to identify strong promoters (4). However, the specific dynamic environment of a high-cell-density culture cannot be adequately reproduced on solid media. In particular, differentially expressed genes that respond to subtle environmental stimulants, or growth-phase-dependent genes, are not readily detected on solid media by use of promoter traps. Thus, we decided to isolate promoters that are up-regulated in the environment in which we expect to use these promoters, namely high-cell-density culture. By looking at the entire proteome before and after changing specific environmental conditions, we identified an inducible promoter in R. eutropha that could be regulated with a simple process parameter (i.e., phosphate limitation). Previous work using shake flasks has shown that phaPp is induced under PHB production conditions and that expression is controlled by a DNA binding protein, PhaR (43).
In this study, we were able to obtain 1.2 g of soluble active OPH/liter in a simple high-cell-density fermentation process. Although only a single chromosomal copy of the OPH gene was present, it represents the highest OPH titer reported to date (33). In a way very similar to the concept of man-hours, where the product of two variables determines overall productivity, R. eutropha fermentations allow more cells to produce protein at a lower specific rate while achieving higher overall productivities. This reduces the likelihood of inclusion body formation and thus allows for the higher recovery of soluble active protein. However, to further improve expression levels and to test the limits of soluble-protein production, we are currently investigating the influence of gene dosage and artificial promoters.
Acknowledgments
We thank Mikhail Alexeyev, Janet Grimsley, Gregory M. York, and Anthony J. Sinskey for providing us with the plasmids and the antibodies for the project. We thank Stefan Wildt and David Wood for productive discussions.
This project was supported by ARO grant DAAD19-00-1-0180, DOJ grant 2000-DT-CX-K001(S-1), and NIST grant 60NANB1D0064.
REFERENCES
- 1.Akesson, M., E. N. Karlsson, P. Hagander, J. P. Axelsson, and A. Tocaj. 1999. On-line detection of acetate formation in Escherichia coli cultures using dissolved oxygen responses to feed transients. Biotechnol. Bioeng. 64:590-598. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, L., L. Strandberg, L. Haggstrom, and S. O. Enfors. 1994. Modeling of high cell-density fed-batch cultivation. FEMS Microbiol. Rev. 14:39-44. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, S., T. Kondo, and M. Ishiura. 2002. A promoter-trap vector for clock-controlled genes in the Cyanobacterium synechocystis sp. PCC 6803. J. Microbiol. Methods 49:265-274. [DOI] [PubMed] [Google Scholar]
- 5.Charles, M., and J. Wilson. 1994. Fermentor design, p. 5-67. In B. K. Lydersen, N. A. D'Elia, and K. L. Nelson (ed.), Bioprocess engineering: systems, equipment and facilities. John Wiley & Sons, Inc., New York, N.Y.
- 6.Chen, H. C., C. F. Hwang, and D. G. Mou. 1992. High-density Escherichia coli cultivation process for hyperexpression of recombinant porcine growth hormone. Enzyme Microb. Technol. 14:321-326. [DOI] [PubMed] [Google Scholar]
- 7.Cutayar, J. M., and D. Poillon. 1989. High cell-density culture of Escherichia coli in a fed batch system with dissolved oxygen as substrate feed indicator. Biotechnol. Lett. 11:155-160. [Google Scholar]
- 8.Dave, K. I., C. E. Miller, and J. R. Wild. 1993. Characterization of organophosphorus hydrolases and the genetic manipulation of the phosphotriesterase from Pseudomonas diminuta. Chem. Biol. Interact. 87:55-68. [DOI] [PubMed] [Google Scholar]
- 9.DelaCruz, N., G. F. Payne, J. M. Smith, and S. J. Coppella. 1992. Bioprocess development to improve foreign protein production from recombinant Streptomyces. Biotechnol. Prog. 8:307-315. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey, A. D., J. E. Bailey, and P. T. Kallio. 2000. Expression of Alcaligenes eutrophus flavohemoprotein and engineered Vitreoscilla hemoglobin-reductase fusion protein for improved hypoxic growth of Escherichia coli. Appl. Environ. Microbiol. 66:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, C., D. Koster, S. Wiebusch, K. Mahr, G. Eisbrenner, and H. Markl. 2002. Scale-up of dialysis fermentation for high cell density cultivation of Escherichia coli. J. Biotechnol. 93:243-251. [DOI] [PubMed] [Google Scholar]
- 13.Gorenflo, V., G. Schmack, R. Vogel, and A. Steinbuchel. 2001. Development of a process for the biotechnological large-scale production of 4-hydroxyvalerate-containing polyesters and characterization of their physical and mechanical properties. Biomacromolecules 2:45-57. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, S. 1990. Minimizing proteolysis in Escherichia coli: genetic solutions. Methods Enzymol. 185:119-129. [DOI] [PubMed] [Google Scholar]
- 15.Han, K. G., S. S. Lee, and C. W. Kang. 1999. Soluble expression of cloned phage K11 RNA polymerase gene in Escherichia coli at a low temperature. Protein Expr. Purif. 16:103-108. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, R. A., and C. W. Jones. 1997. Physiology of poly-3-hydroxybutyrate (PHB) production by Alcaligenes eutrophus growing in continuous culture. Microbiology (Read.) 143:2361-2371. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, G. L., and S. W. Harcum. 2002. Characterization of up-regulated proteases in an industrial recombinant Escherichia coli fermentation. J. Ind. Microbiol. Biotechnol. 28:74-80. [DOI] [PubMed] [Google Scholar]
- 18.Jurgen, B., H. Y. Lin, S. Riemschneider, C. Scharf, P. Neubauer, R. Schmid, M. Hecker, and T. Schweder. 2000. Monitoring of genes that respond to overproduction of an insoluble recombinant protein in Escherichia coli glucose-limited fed-batch fermentations. Biotechnol. Bioeng. 70:217-224. [DOI] [PubMed] [Google Scholar]
- 19.Karr, D. B., J. K. Waters, and D. W. Emerich. 1983. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 46:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleman, G. L., and W. R. Strohl. 1994. Acetate metabolism by Escherichia coli in high-cell-density fermentation. Appl. Environ. Microbiol. 60:3952-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaf, T. A., and F. Srienc. 1998. Metabolic modeling of polyhydroxybutyrate biosynthesis. Biotechnol. Bioeng. 57:557-570. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 23.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omburo, G. A., J. M. Kuo, L. S. Mullins, and F. M. Raushel. 1992. Characterization of the zinc binding site of bacterial phosphotriesterase. J. Biol. Chem. 267:13278-13283. [PubMed] [Google Scholar]
- 25.Park, J. S., H. C. Park, T. L. Huh, and Y. H. Lee. 1995. Production of poly-β-hydroxybutyrate by Alcaligenes eutrophus transformants harbouring cloned phbCAB genes. Biotechnol. Lett. 17:735-740. [Google Scholar]
- 26.Portner, R., and H. Markl. 1998. Dialysis cultures. Appl. Microbiol. Biotechnol. 50:403-414. [DOI] [PubMed] [Google Scholar]
- 27.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 28.Riesenberg, D., and R. Guthke. 1999. High-cell-density cultivation of microorganisms. Appl. Microbiol. Biotechnol. 51:422-430. [DOI] [PubMed] [Google Scholar]
- 29.Rogers, K. R., Y. Wang, A. Mulchandani, P. Mulchandani, and W. Chen. 1999. Organophosphorus hydrolase-based assay for organophosphate pesticides. Biotechnol. Prog. 15:517-521. [DOI] [PubMed] [Google Scholar]
- 30.Ryu, H. W., K. S. Cho, B. S. Kim, Y. K. Chang, H. N. Chang, and H. J. Shim. 1999. Mass production of poly (3-hydroxybutyrate) by fed-batch cultures of Ralstonia eutropha with nitrogen and phosphate limitation. J. Microbiol. Biotechnol. 9:751-756. [Google Scholar]
- 31.Ryu, H. W., S. K. Hahn, Y. K. Chang, and H. N. Chang. 1997. Production of poly (3-hydroxybutyrate) by high cell density fed-batch culture of Alcaligenes eutrophus with phosphate limitation. Biotechnol. Bioeng. 55:28-32. [DOI] [PubMed] [Google Scholar]
- 32.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta. Bio/Technology 3:567-571. [Google Scholar]
- 33.Serdar, C. M., D. C. Murdock, and M. F. Rohde. 1989. Parathion hydrolase gene from Pseudomonas diminuta MG: subcloning, complete nucleotide sequence, and expression of the mature portion of the enzyme in Escherichia coli. Bio/Technology 7:1151-1155. [Google Scholar]
- 34.Shi, H. D., M. Shiraishi, and K. Shimizu. 1997. Metabolic flux analysis for biosynthesis of poly(beta-hydroxybutyric acid) in Alcaligenes eutrophus from various carbon sources. J. Ferment. Bioeng. 84:579-587. [Google Scholar]
- 35.Steinbuchel, A., K. Aerts, W. Babel, C. Follner, M. Liebergesell, M. H. Madkour, F. Mayer, U. Pieper-Furst, A. Pries, H. E. Valentin, et al. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can. J. Microbiol. 41(Suppl. 1):94-105. [DOI] [PubMed] [Google Scholar]
- 36.Taghavi, S., A. Provoost, M. Mergeay, and D. van der Lelie. 1996. Identification of a partition and replication region in the Alcaligenes eutrophus megaplasmid pMOL28. Mol. Gen. Genet. 250:169-179. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, K. 1997. Development of fermentation processes for autotrophic production of P(3HB) from CO2 under safe culture conditions. Seibutsu-Kogaku Kaishi-J. Soc. Ferment. Bioeng. 75:185-195. [Google Scholar]
- 38.Wieczorek, R., A. Steinbuchel, and B. Schmidt. 1996. Occurrence of polyhydroxyalkanoic acid granule-associated proteins related to the Alcaligenes eutrophus H16 GA24 protein in other bacteria. FEMS Microbiol. Lett. 135:23-30. [DOI] [PubMed] [Google Scholar]
- 39.Wu, C. F., H. J. Cha, G. Rao, J. J. Valdes, and W. E. Bentley. 2000. A green fluorescent protein fusion strategy for monitoring the expression, cellular location, and separation of biologically active organophosphorus hydrolase. Appl. Microbiol. Biotechnol. 54:78-83. [DOI] [PubMed] [Google Scholar]
- 40.Wu, C. F., H. J. Cha, J. J. Valdes, and W. E. Bentley. 2002. GFP visualized immobilized enzymes: degradation of paraoxon via organophosphorus hydrolase in a packed column. Biotechnol. Bioeng. 77:212-218. [DOI] [PubMed] [Google Scholar]
- 41.Wu, C. F., J. J. Valdes, G. Rao, and W. E. Bentley. 2001. Enhancement of organophosphorus hydrolase yield in Escherichia coli using multiple gene fusions. Biotechnol. Bioeng. 75:100-103. [DOI] [PubMed] [Google Scholar]
- 42.York, G. M., B. H. Junker, J. A. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]