Abstract
A standard for bioregulatory network diagrams is urgently needed in the same way that circuit diagrams are needed in electronics. Several graphical notations have been proposed, but none has become standard. We have prepared many detailed bioregulatory network diagrams using the molecular interaction map (MIM) notation, and we now feel confident that it is suitable as a standard. Here, we describe the MIM notation formally and discuss its merits relative to alternative proposals. We show by simple examples how to denote all of the molecular interactions commonly found in bioregulatory networks. There are two forms of MIM diagrams. “Heuristic” MIMs present the repertoire of interactions possible for molecules that are colocalized in time and place. “Explicit” MIMs define particular models (derived from heuristic MIMs) for computer simulation. We show also how pathways or processes can be highlighted on a canonical heuristic MIM. Drawing a MIM diagram, adhering to the rules of notation, imposes a logical discipline that sharpens one's understanding of the structure and function of a network.
INTRODUCTION
In recent years, we have been inundated with ever more detailed and comprehensive information on the molecular interactions that govern cell behavior, such as cell division, differentiation, and death. It will be a major task in coming years to organize this information in an accurate, complete, and comprehendible manner (Kohn, 1999; Pirson et al., 2000; Strogatz, 2001). To this end, there is an urgent need for a generally accepted graphical notation for diagrams of bioregulatory networks that could be used in a manner akin to electronic circuit diagrams. As noted by Ideker et al. (2001a), diagrams can be “a tremendous aid in thinking clearly about a model, in predicting possible experimental outcomes, and in conveying the model to others.” Kurata et al. (2003) point out that “without consistent and unambiguous rules for representation, not only is information lost but misinformation could be disseminated.” The diagrams commonly used to show bioregulatory schemes, however, are often incomplete and ambiguous (Pirson et al., 2000). Bioregulatory networks are much more difficult to diagram than classical metabolic pathways, because of the large role played by multimolecular complexes, protein modifications, and multidomain proteins (Kohn, 1999, 2001). To address this problem, a graphical notation for molecular interaction maps (MIMs) was devised (Kohn, 1999; Aladjem et al., 2004) and was used to create maps of several bioregulatory networks (Kohn, 1999, 2001; Kohn and Bohr, 2002; Kohn et al., 2003, 2004; Aladjem et al., 2004; Pommier et al., 2004; Kohn and Pommier, 2005; http://discover.nci.nih.gov/mim/). Here, we describe the notation in full detail with examples, and we review the published literature relating to this and other proposed notations.
A unique aspect of the MIM notation is that it can show all of the known interactions and allow the unknown contingencies (effects of one interaction on another) to be left unspecified until those details become available. In this sense, MIM diagrams are “heuristic.” A heuristic MIM therefore may not provide all the information required for computer simulation. Particular models for computer simulation can, however, be extracted from heuristic MIMs and formulated in “explicit” diagrams using a subset of the MIM symbols (Kohn, 2001). Heuristic MIMs are “canonical” in that they are not restricted to a particular cell type or cell state, and they do not indicate a particular sequence of events. Rather, they show the interactions that can occur if the relevant molecules are present in the same place at the same time. A diagram specific to a particular cell type or cell state can be derived from a canonical map by deleting the molecules that are not expressed as well as the interactions that do not occur because of lack of colocalization. A particular pathway or sequence of events can be depicted on a canonical map by numbering and/or highlighting the relevant interactions, as we describe and illustrate here.
Even when a network is depicted in a clear diagram, understanding how it functions may require computer simulation of plausible models (Ideker et al., 2001b). Paraphrasing E. O. Wilson (quoted by Strogatz, 2001), “the greatest challenge today in cell biology is the accurate and complete description of complex systems. The next task is to assemble mathematical models that capture the key system properties.” The MIM notation can be used both to describe what is known about a system and to define explicit models for computer simulation (Kohn, 1998, 2001; Kohn et al., 2004; http://discover.nci.nih.gov/mim/).
General Principles and Rules of the MIM Notation
A named molecular species generally occurs in only one place on a map. This makes the diagrams compact and shows all the interactions and modifications of a given molecule in one place on the map. (Exempt from this rule are molecules, such as GTP or ubiquitin, that act in a similar manner in many different reaction.)
Interactions between molecular species are shown by different types of connecting lines, distinguished by different arrowheads or other terminal symbols (Figure 1).
Interaction lines can change direction (but not by more than 90° at a corner; this restriction prevents ambiguities at branch points).
When lines cross, it is as if they do not touch.
Symbol definitions are not affected by color. Thus, the notation can be used as a convenient shorthand for sketching interaction schemes using ordinary pencil and paper. Color, however, can make different types of symbols more visually apparent in complicated maps. Red is used for interactions that have negative effects, so that the net effect of a sequence of interactions can be determined easily by counting the number of negative effects in the sequence: if the number is even, the net effect is stimulation; if odd, it is inhibition.
The consequence or product of an interaction is indicated by placing a small filled circle (“node”) on the interaction line (but not at the ends of the line). Thus, the consequence of binding between two molecules is production of a dimer, which is represented by a node on the binding interaction line (Figure 2d). The consequence of a modification (e.g., phosphorylation) event is production of the modified (e.g., phosphorylated) molecule; the phosphorylated product is represented by a node placed on the modification line (Figure 2e). Multiple nodes on an interaction line represent exactly the same molecular species (this can reduce crowding in a diagram). To avoid ambiguity, a node should not be placed at line crossings.
Nodes can be treated like named molecular species, thereby making the notation extensible. For example, if the node represents a dimer, a binding line connecting it to another molecular species shows the production of a trimer (node y in Figure 2d). The same principle applies to molecular modifications: a node on a modification line represents the modified molecule, which can participate in other interactions (Figure 2e).
Molecular interactions are of two types: reactions and contingencies, as listed in Figure 1. Reactions operate on molecular species; contingencies operate on reactions or on other contingencies.
Elementary molecular species are those that are named within or adjacent to a cartouche or box (Figure 2). Instead of species name, a cartouche may contain protein domain names (N- to C-terminal order from left to right) (Figure 2b); the molecular species name is then placed adjacent to the left end of the cartouche (Figure 2b).
Interactions of individual protein domains can be shown in the same way as interactions of molecular species (as shown for binding interactions in Figure 3, d–f). If its location is unknown, the interaction line points to the molecular species name adjacent to the cartouche, as in Figure 3d (node z).
Figure 1.
Molecular interaction symbols. Interactions are of two types: reactions (a– h) and contingencies (i–l). Reactions operate on (point to) molecular species, whereas contingencies operate on reactions or on other contingencies. (a) Noncovalent (reversible) binding is indicated by a line with barbed arrowheads at both ends. (b) Covalent modification is indicated by a single barbed arrowhead pointing to the modification site. (c) Stoichiometric reaction (reactants converted to a corresponding number of product molecules) is indicated by a filled triangle arrowhead. This symbol can be used also for translocation events, because molecules disappear from one location and reappear in another location, logically the equivalent of a stoichiometric reaction. (d) Production of a molecular species without loss of macromolecular reactants (as in transcription or translation) is indicated by an open triangle arrowhead pointing to a molecular species. (Small ubiquitous molecules, such as ATP, can usually be ignored.) (e) Production by transcription is indicated by an open triangle at the end of a hooked line. (f) Cleavage of a covalent bond is indicated by a zigzag symbol. (g) Degradation is indicated by a filled triangle pointing to a null symbol (stoichiometric conversion to debris). (h) Reaction between different molecules of the same type (reaction in trans) is indicated by a gap symbol in the interaction line. (i) Stimulation of a process is indicated by an open triangle arrowhead pointing to an interaction line. (j) If the stimulating agent is a requirement, a bar is added behind the open triangle arrowhead. (k) Inhibition is indicated by a bar at the end of the interaction line. (l) Enzymatic catalysis is indicated by an open circle at the end of the interaction line.
Figure 2.
Molecular species symbols. Elementary molecular species are those that are named (a– c). Complex molecular species are created as a consequence of interactions and are indicated by small circles (“nodes”) placed on an interaction line (d– g). (a) The name of a protein or RNA may occur within a cartouche. (b) Alternatively, the cartouche may contain domain names. Domain names are listed in N- to C-terminal order from left to right; the protein name then is placed adjacent to the left end of the cartouche. (c) Nucleotide sequence elements (e.g., promoter elements) are placed in a box. (d) Multimers are indicated by nodes placed on binding interaction lines: (e) Modified (e.g., phosphorylated) species are indicated by nodes placed on the modification line. Thus, a node on a phosphorylation line represents the phosphorylated molecule (i.e., the consequence of the interaction). (f) An isolated filled circle (node) at the end of an interaction line represents another copy of the molecular species represented at the other end of the line. Thus, x represents another copy of A, and y represents the A:A homodimer, which is the consequence of the interaction. (g) A node on a line without arrowheads indicates the consequence of the combined effect of the nodes at the ends of the line (see Figures 3d and 5i for examples).
Figure 3.

Binding interactions. (a) Multiple binding to the same molecule. B and C bind different sites on A. (b) Mutually exclusive binding (different sites, allosteric). (c) Competitive binding (same site). Nodes x and y represent the A:B and A:C dimers, respectively. Node z represents A bound to B or C. (The angle at the branch leading to B and C should be <90° to avoid suggesting that B can bind to C.) Node w represents D bound to A:B or A:C. (d) Domain-specific binding. B and C bind protein A at domains-1 and -2, respectively; D binds protein A, but it is not known to which domain. Node w represents A with B bound to domain 1 and C bound to domain 2. (e) Intramolecular binding. Binding between domains-1 and -2 within the same molecule (binding in cis). The node represents protein A containing this intramolecular bond. (f) Intermolecular binding between indicated domains in a homodimer: domains-1 and -2 in different molecules of the same type binding in trans to form a homodimer (represented by the node).
THE MIM NOTATION
Molecular Species
MIM diagrams have two kinds of molecular species: “elementary” and “complex” (Figure 2). Complex species are combinations or modifications of elementary species.
Elementary protein species are associated with a cartouche (a rectangle with rounded corners) and are named. The name may be inside the cartouche, as in Figure 2a. Alternatively, the cartouche may contain domain names, in which case the protein name is placed adjacent to the left end of the cartouche, as in Figure 2b. If several proteins are always considered together as a unit, they can be named within the same cartouche and treated as an elementary species.
DNA elements, such as promoters, are represented by a box. The name of the element or promoter can be inside the box, as in Figure 2c. Alternatively, the box may contain a consensus sequence, in which case the name of the element can be placed above or below the box.
Complex species are indicated by filled circles (“nodes”) placed on an interaction line. A node represents the molecular species that is produced as a consequence of the interaction. For example, binding interactions produce multimers (such as nodes x and y in Figure 2d or node y in Figure 2e) and modifications produce modified species (such node x in Figure 2e).
To indicate a homodimer, we use the isolated node convention (Figure 2f), which avoids having to represent the same elementary monomer twice. An isolated node is defined as another copy of the same species that is represented at the other end of the interaction line. Thus node x is another copy of A, and node y is the homodimer A:A.
We will explain the state-combination symbol (Figure 2g) in connection with Figure 3d.
Noncovalent Binding
Noncovalent (reversible) binding between molecular species is denoted by a line with barbed arrowheads at both ends (Figure 2d). The resulting dimer or multimer is denoted by a small filled circle or “node” placed on the line. Because nodes can be treated in the same way as elementary (named) molecular species, the notation is compact and extensible. In Figure 2d, for example, node x is the A:B dimer, and node y is C bound to A:B, i.e., the trimer (A:B):C. For an example of how this extensible notation can show the assembly of a multimolecular complex, see Aladjem et al. (2004) or http://discover.nci.nih.gov.
Figure 2d, however, does not tell us which of the two proteins in the dimer contains the binding site for the third protein. Figure 3a shows this detail: here A has two binding sites, one site for B and a different site for C (B does not bind directly to C). The default assumption is that these two bindings can coexist (B can bind indirectly to C through A). If the two bindings cannot coexist, an exclusivity symbol is applied (Figure 3b). The mutual exclusion here is due to allosteric interference between two different binding sites.
Mutual exclusion due to competition for the same site is shown using a branched binding line (Figure 3c). (The acute angle at the branch avoids the misinterpretaion that B could bind C; by convention, interaction lines do not change direction by more than 90° at a corner.) This notation provides a compact representation of alternative bindings that have the same function; for example, node w in Figure 3c represents two trimers: A:B:D and A:C:D; this convention can display multiple complexes in one symbol.
Regulatory proteins often are composed of domains that can function independently. The interaction details of individual domains can be shown as illustrated in Figure 3d. Node x represents B bound to domain 1 of A; y is C bound to domain 2 of A; z is D bound to A at an unknown location. Simultaneous binding is shown using the state-combination symbol (defined in Figure 2g): node w in Figure 3d represents the trimer in which domain 1 is bound to B and domain 2 is bound to C.
Binding between domains within the same molecule is represented as shown in Figure 3e. This intramolecular binding is called “binding in cis”, to distinguish it from intermolecular binding between different molecules of the same type (“binding in trans”). To indicate intermolecular binding between domain 1 of one molecule of A and domain 2 of another molecule of A (binding in trans), we insert a gap symbol in the state-combination line (Figure 3f). (The gap symbol is defined in Figure 1h.)
Contingencies of binding
Figure 1 defines symbols for four types of contingencies: stimulation, requirement, inhibition, and catalysis. Contingencies affect interactions or other contingencies; contingency lines, therefore, point to other interaction lines, not to molecular species. Note that the open arrowhead symbol has two different meanings (Figure 1, d and i): when it points to a line, it represents a stimulation contingency; when it points to a molecular species, it represents an increasing amount of that species (without consumption of specified reactants).
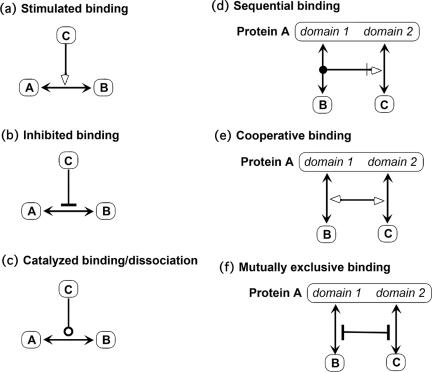
Figure 4 shows various types of contingencies that operate on binding interactions. Figure 4a shows stimulation of binding (or the equivalent effect produced by inhibition of dissociation); if the contingency is a requirement, a thin line is placed behind the arrowhead (Figure 1j). Figure 4b shows inhibition of binding. Figure 4c shows the case in which both binding and dissociation are stimulated (as in guanine nucleotide exchange factors, which stimulate the exchange between GTP and GDP at a binding site on G proteins; Figure 11). Because that interaction implies a reduced energy barrier of the reaction, we apply the catalysis (open circle) symbol.
Figure 4.
Contingencies of binding. (a) C stimulates binding between A and B (or inhibits dissociation of the A:B complex). (b) C inhibits A:B binding (mechanism unspecified). (c) C catalyzes the formation and dissociation of the A:B complex (reducing the energy barrier of the A:B interaction). (d) Sequential binding: B must bind to domain 1 before C can bind to domain 2. (e) Cooperative binding: binding of B to domain 1 increases the affinity of C for domain 2, and vice versa. (f) Mutually exclusive binding: protein A can bind B at domain 1 or C at domain 2, but not both at the same time (allosteric interference).
Figure 11.
Interactions at membranes: signaling via G-proteins. This classic signaling pathway is here shown in MIM notation. See text for detailed description and discussion.
Figure 4, d–f, shows contingencies involving specified domains. Figure 4d shows sequential binding: B must bind to domain 1 before C can bind to domain 2. Figure 4e shows cooperative binding: binding of either stimulates binding of the other. Figure 4f shows mutual interference: binding of either deters binding of the other.
Covalent Modifications and Their Contingencies
Covalent modification (phosphorylation, acetylation, myristoylation, ubiquitination, and so on) is represented by a line with a barbed arrowhead at one end pointing to the modification site (Figure 1b). Figure 5 shows how the symbols can be combined in various ways to represent a variety of circumstances. Figure 5a uses the catalysis symbol to show phosphorylation by a kinase. (An open circle symbol operating on a modification line implies catalysis that favors the modification.) Figure 5b uses the bond cleavage symbol (Figure 1f) to show dephosphorylation by a phosphatase. (The zig-zag symbol indicates a reaction that catalyzes bond cleavage.)
Figure 5.
Covalent modifications and their contingencies. (a) Phosphorylation by a protein kinase. Similarly for acetylation (Ac), methylation (Me), ubiquitination (Ub), myristoylation (Myr), and so on. The amino acid site of modification can be indicated as a superscript (as in this figure) or between the protein cartouche and the arrowhead (as in example Figure 5j). (b) Removal of a phosphate by a protein phosphatase. (c) Binding-contingent phosphorylation: B must bind to domain 1 before site-1 can be phosphorylated. (d) Inhibited phosphorylation: binding of B inhibits phosphorylation of site-1. (e) Phosphorylation of site-1 stimulates binding of B to domain 1. (f) Phosphorylation of site-1 inhibits binding of B to domain 1. (g) Sequential phosphorylation: site-1 must be phosphorylated before site-2 can be phosphorylated. (h) Phosphorylation of site-1 inhibits phosphorylation of site-2. (i) State-combination: x and y are protein A phosphorylated at sites-1 and -2, respectively; z is A phosphorylated at both sites. (j) Alternative modifications of the same site: lysine-100 (K100) can be either acetylated or ubiquitinated.
Figure 5, c–f, shows various contingencies between binding and modification within the same protein molecule. Figure 5g specifies that site-1 must be phosphorylated before site-2 can be phosphorylated. Figure 5h specifies that phosphorylation of site-1 prevents phosphorylation of site-2. In Figure 5i, node z represents the protein phosphorylated both at site-1 and at site-2.
Occasionally, the same site can be modified in different ways. For example, a given lysine in a protein might be either acetylated or ubiquitinated, as has been reported for a lysine in p53 (see Kohn and Pommier, 2005). This situation can be represented using the branched line convention (Figure 5j). (Note that the amino acid site of modification can be indicated in a superscript, as in Figure 5, a and b, or adjacent to the protein cartouche, as in Figure 5j.)
Covalent binding between proteins or between sites within the same protein sometimes require a symmetrical symbol, for which purpose we have recently adopted the double-line symbol shown in Figure 1b′ (see Figure 13 and associated text). (The new symbol may be used for all cases of covalent binding, and may eventually replace the current protein modification symbol.)
Figure 13.
Intramolecular covalent binding: reactions of SH groups in response to reactive oxygen (Temple et al., 2005). See text for description and discussion.
Kinase Phosphorylation Cascade: Contingency Notation and Compact Notations
Two ways to represent the effects of protein modification are illustrated with an example of a protein kinase phosphorylation cascade (Figure 6). A and B are protein kinases that are activated by phosphorylation. Phospho-A phosphorylates and thereby activates B; phospho-B phosphorylates C. Figure 6a shows this cascade using contingency notation; Figure 6b shows the same thing using compact notation.
Figure 6.
Kinase phosphorylation cascade. (a) Contingency notation. (b) Compact notation.
Compound Contingencies
When a contingency is controlled by multiple nodes, a complicated diagram can become excessively cluttered. As an alternative to the full representation of those situations (Figure 7, left), an abbreviated notation is often useful (Figure 7, right).
Figure 7.
Compound contingencies: full notation (left); abbreviated notation (right). A gap in a contingency line in the abbreviated notation is interpreted as if the line jumps over the subsequent nodes.
Transcription Control
Figure 8 illustrates the representation of transcription control. A DNA sequence element or promoter is indicated by a rectangle inserted in a heavy line that represents the DNA. Transcription to mRNA is indicated by a hooked line, similar to the way transcription is commonly represented. An open-triangle arrowhead points to the RNA, because the DNA is not consumed as RNA is produced. Similarly, an open-triangle arrowhead points to the translated protein, because the mRNA is not consumed. (As already mentioned, an open-triangle arrowhead pointing to a molecular species indicates an increase in the amount of that species without consumption of reactants.)
Figure 8.
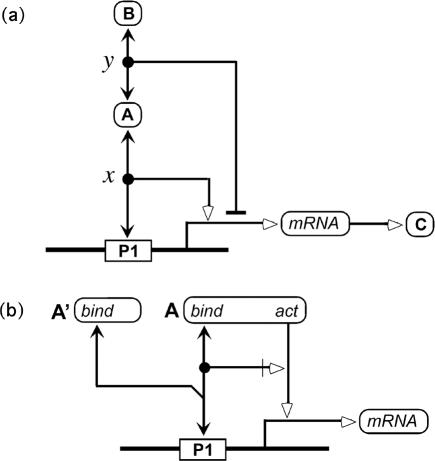
Control of transcription. (a) Stimulation of transcription by protein A and inhibition of transcription by protein B because its recruitment to the promoter via binding to protein A. (b) Actions of DNA binding and transcription activation domains of protein A. Protein A′ is a truncated variant of protein A that retains the DNA binding domain, but lacks the activation domain. It therefore competes with protein A for binding to the promoter but cannot activate transcription (thereby functioning as a transcription inhibitor).
Node x in Figure 8a represents protein A bound to promoter P1. The contingency line emerging from the node indicates stimulation of transcription. Node y represents protein A bound to protein B. The contingency line emerging from this node indicates inhibition of transcription. As it stands, Figure 8a does not tell us whether the inhibition of transcription requires binding of protein A to the promoter. The notation could be made explicit by adding a contingency line. To keep diagrams as simple as possible, however, we make a default assumption about contingencies that emerge from mutually bound entities and that operate on the same interaction: in the absence of contrary indicators, we assume that these interactions act in concert. The default interpretation of Figure 8a therefore is that protein B inhibits transcription by being recruited to the promoter via protein A.
Figure 8b illustrates the interactions of protein domains in regulation of transcription. The diagram shows a DNA-binding domain of protein A binding to promoter P1 and also an activation domain stimulating transcription of the gene controlled by this promoter. A contingency arrow shows that activation requires binding. A truncated variant, protein A′, is shown competing with protein A for promoter binding. Protein A′ retains the binding domain but lacks the activation domain; therefore, it can function as a transcription inhibitor. (Note that the acute angle at the competition branch point prevents the misreading that protein A binds protein A′.)
Translocation
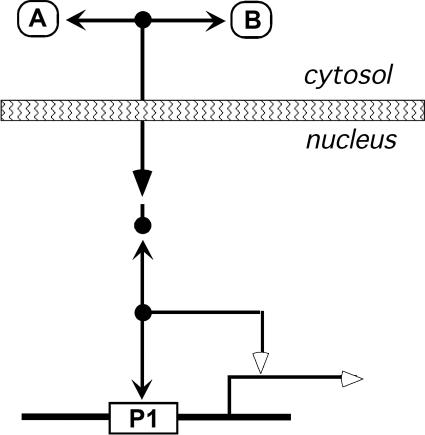
Translocation from one compartment of the cell to another is like a stoichiometric reaction in that molecules disappear from one place and an equal number of the same molecules appear at another place. We therefore represent translocation with the same symbol that is used for stoichiometric reactions: a filled triangle arrowhead. The example in Figure 9 shows the A:B dimer translocating from cytosol to nucleus. To avoid reproducing all the interactions in two different places, we invoke the isolated node convention: an isolated node represents the same species that is shown at the other end of the interaction line that points to it. Thus, the isolated node in Figure 9 represents the A:B dimer in the nucleus, which then can bind to promoter P1 and activate transcription. When two arrows point to the same isolated node, the diagram could be misread. In Figure 9, the isolated node might be interpreted to be another copy of the promoter. In most cases, including this one, alternative interpretations are untenable. To guard against accidental misreadings, one can add an optional short line to the node (as was done in Figure 9), directed toward the interaction line that defines the node.
Figure 9.
Translocation from cytosol to nucleus. A binds B in the cytosol. The A:B complex translocates to the nucleus (indicated by filled-triangle arrowhead). The translocated A:B, which is represented using the isolated node abbreviation, binds to the promoter and stimulates transcription. The optional short line attached to the isolated node can guard against misreading the diagram (see text).
Control by Protein Cleavage Induced by Specific Proteases
The function of one domain of a protein is sometimes regulated (stimulated or inhibited) by another domain in the same protein. Control is sometimes implemented by a specific protease that cuts the two domains apart, thereby abrogating the influence one domain upon the other. Classic examples are found in the control of apoptosis by caspases.
Figure 10 shows how we depict control by specific protein cleavage. Two not-quite-equivalent diagrams are shown. Figure 10a shows an inhibitory effect of domain 1 on domain 2. A specific protease can cut the protein between the two domains. The cleavage separates the two domains and prevents the inhibitory action of domain 1 on domain 2.
Figure 10.
Control by cleavage between interacting domains in the same protein. In this example, domain 1 inhibits the ability of domain 2 to bind protein B. The inhibition is abrogated by a specific protease that cleaves protein A at a site that separates the two domains. Two notations for this situation are shown. In a, the inhibitory effect of domain 1 is shown explicitly, and the effect of cleavage, namely, to reverse the inhibition, is implied by the fact that cleavage separates the two domains. In b, the cause of the inhibition of binding of protein B is not shown explicitly. Instead, the cleavage between the two domain is indicated to have the consequence of stimulating the binding of protein B to domain 2 (and may or may not have anything to do with domain 1).
The alternative in Figure 10b depicts the action of domain 2 being stimulated by the cleavage. This notation is consistent with the convention that a node on an interaction line represents the product or consequence of the interaction: thus the cleaved product stimulates the binding of B. This is not quite equivalent to Figure 10a, because it does not specify that domain 1 is what inhibits domain 2.
Interactions at the Plasma membrane: Signaling via G Proteins
Figure 11 illustrates interactions at membranes, using as an example G protein signaling, a process commonly shown in standard molecular cell biology textbooks in cartoon form (for example, Alberts et al., 1994, or Lodish et al., 1995, or later editions of these excellent textbooks). Each interaction is labeled with a number that can be used within descriptive text (as we do here), as a link to an annotation list (Kohn, 1999), or as an electronic link to hypertext (http://discover.nci.nih.gov/mim/). This example shows how the MIM notation organizes into a single diagram a process that previously required multiple panels in cartoon-like diagrams.
Figure 11 shows a G protein-coupled receptor (GPR) composed of an extracellular receptor domain, a transmembrane segment, and a cytoplasmic domain. The extracellular domain can bind a ligand, such as a hormone (interaction-1). The Gα subunit of the G protein binds to plasma membrane (interaction-2) and can bind either GDP (interaction-3)orGTP (interaction-4), which can exchange only very slowly unless the exchange is catalyzed. Gα(GDP) binds the Gβ:Gγ dimer (interaction-5). Gα(GDP):Gβγ binds the cytosolic domain of GPR (interaction-6), but only if the extracellular domain of GPR is bound to ligand (interaction-7: stimulatory contingency). Within the resulting complex, the exchange between GDP and GTP is facilitated (interaction-8). If there is more GTP than GDP in the cell, which is usually the case, GDP tends to be replaced by GTP. Gα(GTP) is released from binding to its partners (note the absence of binding interactions between the GTP-limb (interaction line-4) and either GPR or Gβγ). The freed Gα(GTP) binds adenylyl cyclase, an integral membrane protein (interaction-9). This binding stimulates (interaction-10) the enzymatic activity of the cyclase (interaction-11), which stoichiometrically converts ATP to cyclical-AMP (interaction-12). Gα(GTP) slowly converts to Gα(GDP) (interaction-13, due to an intrinsic GTPase activity), thus completing the cycle. As an additional control, a GTPase-activating protein (GAP) can stimulate the intrinsic GTPase activity of Gα(GTP) (interaction-14).
Whereas process diagrams (sometimes presented in cartoon-like panels) usually show a particular order of events, this is not the case for MIM diagrams. For example, process diagrams generally show Gα(GTP) binding to adenylyl cyclase before the GTPase step, whereas this need not be the case, for example if there is high GAP activity. Moreover, the exchange between GDP and GTP can go in either direction, the predominance of one direction over the other depending on the GTP/GDP concentration ratio. MIM diagrams do not specify order of events and therefore cover a greater range of circumstances in a canonical format.
Intramolecular Control: Calmodulin Kinase
A classic example of intramolecular control is calmodulin (CaM)-dependent protein kinase (CaMK) (Alberts et al., 1994, or a later edition of this textbook). This system is diagrammed in MIM notation in Figure 12. A molecular interaction map often is best examined starting from an end-effect and tracing the contingencies backward, as we will do here.
Figure 12.
Intramolecular control of protein kinase activity: CaMK, a classic pathway shown here in MIM notation. See text for description and discussion.
The end-effect is the phosphorylation of various substrates by the kinase domain of CaMK (interaction-1). This action is inhibited (interaction-2) by the intramolecular binding between the catalytic domain and the regulatory domain of CaMK (interaction-3). This intramolecular bond can be opened by the competitive binding of calmodulin (CaM) to the regulatory domain (interaction-4). Binding of CaM to the regulatory domain requires (interaction-5) that CaM be bound by calcium (interaction-6). The steps so far describe how calcium activates the kinase.
CaMK can autophosphorylate in trans (one molecule of CaMK phosphorylating another molecule of CaMK) (interaction-7). This phosphorylation prevents the binding between the kinase and the regulatory domain (interaction-8). Phosphorylated CaMK therefore retains its activity even when it dissociates from CaM. Eventually, CaMK is inactivated by dephosphorylation (interaction-9), which restores the ability of the regulatory domain to bind and block the kinase domain intramolecularly.
Another, more complex case of intramolecular control is that of the nonreceptor tyrosine kinase, Src. A molecular interaction map of that system has been published (Kohn, 2001), and an animated version of the process can be viewed at (http://discover.nci.nih.gov).
The activation of CaMK, Src, and G proteins behave similarly, in that they all exhibit amplified and prolonged action.
Intramolecular Covalent Binding: Reactions of SH Groups in Response to Reactive Oxygen
An interesting pathway involving intramolecular disulfide bond formation has recently been described for the response of budding yeast to oxidative stress (Temple et al., 2005). Figure 13 shows a molecular interaction map of this pathway. To represent an intramolecular covalent bond, we had to introduce a new symbol: an arrowless double line. (The single-arrowed line representing covalent modification was unsatisfactory, because it lacked symmetry. The new symbol can be used also for covalent modification and may in time replace the old symbol. We did not discard the old symbol at this time, because it has been used extensively in previous publications.)
We now describe the molecular interaction map of this system, as shown in Figure 13. We are indebted to Dr. Ian Dawes for a suggestion of how to represent the system properly (Temple et al., 2005). The transcription factor Yap1p in the budding yeast Saccharomyces cerevisiae is normally kept at low levels by rapid export from the nucleus (interaction-1). This export would be inhibited (interaction-2) by formation of an intramolecular disulfide bond in Yap1p (interaction-3, note new symbol for covalent bonds). This disulfide bond blocks the nuclear export signal in Yap1p. Intracellular reducing conditions however usually prevent the production of disulfide bonds. Oxidative stress can generate the Yap1p disulfide by the following mechanism. Reactive oxygen species add a hydroxyl to the Cys36 SH group of the peroxidase Gpx3p (interaction-4, covalent bond between OH and S), generating a sulfene. The activated Gpx3p reacts with Yap1p, producing the disulfide and concurrently converting the sulfene back to the sulfhydryl form of Gpx3p (interaction-5). (To show that these two conversions are stoichiometrically linked as parts of the same reaction, we have introduced a small circle at the branch point –for the moment, this is an ad hoc symbol, not yet formally adopted.) The disulfide form of Yap1p accumulates in the nucleus and retains its ability to stimulate transcription. One of its gene products is thioredoxin, which cleaves the Yap1p disulfide (interaction-6), thereby forming a negative feedback loop. This example illustrates how the MIM notation may evolve to accommodate new requirements.
Pathways within a Canonical Map: from Ataxia Telangiectasia Mutated (ATM) to p53
The MIM notation provides compact diagrams within which various reaction pathways and processes can be traced. As mentioned, heuristic MIM diagrams are canonical in the sense that they do not specify a particular process or sequence of events. A heuristic map may contain the ingredients for multiple processes or event sequences (pathways), which may function simultaneously or may be specific to particular conditions or cell types. Particular pathways however can be highlighted on a canonical map (http://discover.nci.nih.gov/mim/). Figure 14 shows a canonical map within which an effect is transmitted from one point (ATM) to another (p53) by four different pathways. The same canonical map is depicted in four panels, in each of which a different pathway is highlighted. Note that the actions by the four pathways are “coherent” in that they lead to the same effect; this may be a principle that makes bioregulatory networks robust.
Figure 14.
Pathways highlighted on a canonical map (MIM): from ATM to p53. Shown are four pathways by which ATM can increase the amount of transcriptionally active p53. (a) ATM phosphorylates p53; this phosphorylation blocks the binding of p53 to Mdm2, thereby preventing rapid MdM2-induced p53 degradation. (b) Another effect of ATM-induced p53 phosphorylation is to stimulate the binding of p53 to p300 (which probably competes with Mdm2 for p53 binding). p300 then acetylates p53, thereby stimulating p53 binding to the promoter and activating transcription. (c) ATM phosphorylates Mdm2, thereby blocking p53:Mdm2 binding and p53 degradation, as in a. (d) ATM phosphorylates Chk2 Thr68, thereby simulating Chk2 autophosphorylation of Ser516, which activates Chk2. Chk2 can then phosphorylate p53, with consequences similar to a and b.
p53 levels in cells are normally kept very low, by rapid degradation induced by Mdm2. In response to DNA damage, p53 increases in amount and activity, and functions to transcribe genes that arrest the cell cycle or that initiate programmed cell death (apoptosis). Certain types of DNA damage lead to increased levels of the ATM gene product. The four panels in Figure 14 highlight four pathways by which ATM can enhance the action of p53. In pathways a, c, and d, the effect is primarily inhibition of p53 degradation, due to abrogation of p53:Mdm2 binding. In pathway a, ATM phosphorylates p53; in pathway c, it phosphorylates Mdm2. Phosphorylation of either protein prevents their mutual binding. In pathway d, ATM phosphorylates Chk2, an amplification relay on the way to p53. Pathway b, on the other hand, rather than stabilizing p53, leads to increased promoter binding and increased transcriptional efficiency. Additional coherent pathways (not highlighted here) can go by way of c-Abl (Kohn and Pommier, 2005; http://discover.nci.nih.gov/mim/).
EXPLICIT DIAGRAMS FOR COMPUTER SIMULATION
To see why heuristic maps may not contain all the information needed for computer simulation, consider the following example, derived from Kohn et al. (2004). Suppose that molecules A and B bind to each other and that the resulting dimer binds to a promoter, thereby activating a gene. Suppose further that A can be phosphorylated, and that this phosphorylation causes A to be degraded. This heuristic description leaves open some questions for which a computer simulation model needs answers: Does phosphorylated A bind B? If it does, can the complex bind the promoter? If it can, does it activate the gene? Furthermore, if phosphorylated A binds B (either alone or promoter-bound), does this affect (stimulate or inhibit) A's degradation? For this simple heuristic map, there are 12 possible explicit models. Simulation studies require judgment about which explicit models are most plausible.
We have approached computer simulation studies from the point of view of “microworld models” (Kholodenko and Westerhoff, 1995), which are based solely on molecular interactions, avoiding arbitrary functions for stimulation or inhibition contingencies. Our explicit diagrams use the MIM notation, but without stimulation or inhibition symbols. These diagrams can be translated directly into an input file for computer simulation (Kohn, 1998, 2001; Kohn et al., 2004).
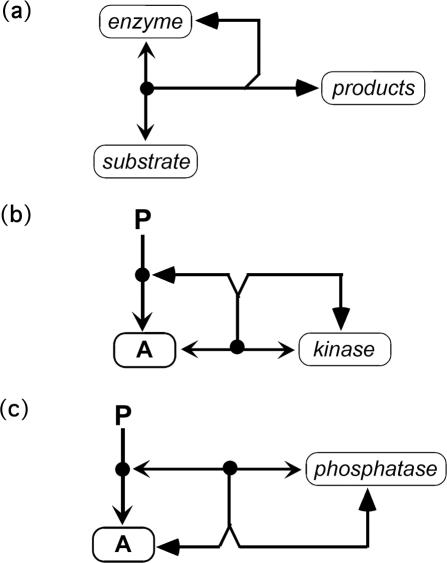
In this implementation, inhibition can be expressed simply by omitting the reactions that do not occur. Alternatively, inhibition may be represented by a mechanism, such as competitive binding by another molecular species or production of an inactive complex. Likewise, stimulation must be represented by a specific mechanism. Enzymatic reactions are represented in terms of the component reactions: enzyme–substrate association; enzyme–substrate dissociation; conversion of enzyme–substrate complex to products (Figure 15a). This avoids Michaelis–Menten approximations. Figure 15, b and c, shows how kinase and phosphatase reactions are represented in explicit notation.
Figure 15.
Explicit notation of enzymatic reactions. (a) General schema showing reversible production of enzyme-substrate complex and its conversion to products. The filled-triangle arrowheads signify a stoichiometric conversion. (b) Protein kinase mechanism. Protein A binds kinase reversibly, and the resulting complex is stoichiometrically converted to phospho-A and regenerated kinase. (c) Protein phosphatase mechanism. Phospho-A binds phosphatase reversibly, and the resulting complex is stoichiometrically converted to unphosphorylated-A and regenerated phosphatase.
ELECTRONIC MIMs
Large heuristic MIMs need an easy way to find any desired molecule on the map. This is accomplished in printed versions by a coordinate grid and an alphabetical list of molecules, analogous to the way towns are found on a roadmap (Kohn, 1999; Aladjem et al., 2004). Ancillary information is provided through numbering of the interactions; each number refers to an annotation that contains cogent information and references.
In electronic MIMs, the annotations are automatically brought up by clicking on an interaction number (http://discover.nci.nih.gov/mim/). Clicking on a molecular species name activates links to related databases. Electronic MIMs provide links to ancillary information and to other databases.
We are often asked what tools are available for generating MIMs. At this time it is not possible to generate satisfactory MIMs automatically. Moreover, we think there are significant advantages to preparing these maps manually (aided only by a symbols toolkit). The process of manual production encourages critical thinking about the structure and function of the network. New questions and possibilities emerge as one decides exactly how to arrange a map to make it easiest to comprehend and how best to group the interactions in a functionally integrated manner. In general, we think it unwise to assign too much responsibility to the computer, because today's software may insulate users from the objects they wish to understand.
BIOREGULATORY NETWORK DIAGRAMS: PROPOSALS AND CRITIQUE
Our MIM notation has been widely discussed (Pirson et al., 2000; Strogatz, 2001; Uetz et al., 2001; Kitano, 2003; Kurata et al., 2003). We now consider the critique and the alternative proposals. The two main limitations of MIMs are the absence of a fully automated way to produce them and the fact that some effort is required to learn the notation. These limitations are, for the most part, shared by all of the alternative notations that have been proposed.
Computer-generated diagram methods have been developed, such as BIOCARTA's connection diagrams (http://www.biocarta.com). However, the resulting diagrams lack important molecular details, such as protein phosphorylations. The graphical language described by Cook et al. (2001) may be more refined from an engineering standpoint, whereas the MIM language may be more intuitive from a biologist's perspective.
Kitano (2003) proposed a variant of the MIM notation in which interaction and modification sites of a protein are marked on the border of the protein's symbol instead of at the end of a line extending from the border. We retain the modification symbol at the far end of an external line, however, because a given site may be modified in different ways (for example, by acetylation or ubiquitination at the same lysine residue, as in Figure 5j). Kitano's notation marks intramolecular interactions within the border of the symbol representing the protein, instead of outside of it. As already mentioned, we reserve the interior of the protein's symbol for marking domain structure in N- to C-terminal order, thus allowing the interactions of individual protein domains to be depicted clearly.
Kitano and colleagues have also developed CellDesigner, a form of computer-aided design (CAD) for generating biomolecular network diagrams (Funahashi et al., 2003). It may be possible to develop an analogous facility for MIMs. The manual production of MIMs, however, is the best way to display networks in a functionally revealing manner, and it imposes a discipline of logic that often gives new insight and highlights gaps in our knowledge.
Kurata et al. (2003) used a slight modification of the MIM notation to develop a software suite called CADLIVE to design and simulate signal transduction models. They described notation for two types of models: their “semantic models” correspond to our heuristic maps; their “mechanistic models” correspond to our explicit diagrams and associated computer implementation. Following our approach, Kurata et al. (2003) start with the principle that “each molecular species should ideally occur only once in a diagram, and all interactions involving those species should emanate from a single symbolic object” and that an extensible representation of multimolecular assemblies is a fundamental requirement. They also note, as we did, that “the potential number of modification and/or multimerization combinations is tremendous, and the representation of all possible combinations of multimers and modifications in a single diagram is not practical.” Their symbol list is very similar to ours (http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html). Although they provide a computer implementation, its merits remain to be determined.
Protein interaction network diagramming methods based on large-scale data sets are receiving considerable attention (Kelley et al., 2003; Gagneur et al., 2004; Vazquez et al., 2004). However, such diagrams do not contain comprehensive information about protein modifications and their consequences. Koike et al. (2003) described a protein kinase database that includes protein interaction data, but does not include details at the level of modification sites.
Although different notations may in time find their optimal areas of application, we think that the MIM notation would be the most immediately useful for biologists.
To gather the information for a molecular interaction map, it is necessary to scan a large number of journal articles. Computer-assisted search programs have been developed (Tanabe et al., 1999; Corney et al., 2004), including MedMiner (http://discover.nci.nih.gov) from our own laboratory. However, the best up-to-date product requires direct culling of information from papers selected and scanned by knowledgeable persons, who can extract evidence for direct interaction between proteins and identify the domains and modification states that are involved.
MIMs have been faulted for not indicating dependence on cell type. The number of different cell types and cell states of interest, however, is very large. Heuristic MIMs are designed to show the molecular interactions that can occur if the interacting molecules are in the same place at the same time. We are developing tools to allow the user to delete molecules and pathways that may be absent in particular cases due to lack of expression of particular genes or protein species. In this way, maps specific to a particular cell type or state can be generated from a canonical map that includes all of the possible interactions.
Another criticism is that MIMs do not specify the order of events. Kurata et al. (2003), for example, state that “Kohn's diagram accurately describes the detailed relationships among components, but it does not provide the stepwise view of specific biological processes”. Similarly, Kitano (2003) states that “MIM is a good basis for a standard to represent interactions between molecular species, however, it does not explicitly show temporal sequences of biological events.”
However, MIMs intentionally avoid assumptions about order of events, because networks may operate in various ways involving different event sequences. Nevertheless, particular event sequences can be highlighted on a canonical map, as illustrated in Figure 14. Heuristic (canonical) MIMs provide a general framework from which specific process models can be extracted.
CONCLUSIONS AND PERSPECTIVES
There is an urgent, widely recognized need for standard notation capable of describing bioregulatory networks the way circuit diagrams describe electronic networks. Although several notations have been proposed, the molecular interaction map (MIM) notation is arguably the best suited to the purpose. It is the only extensively tested notation that can fully describe the known molecular details, such as the intricacies of protein modifications and complex formation, while allowing the unknown contingencies (of which there usually are many) to remain unspecified. Explicit, fully specified models for computer simulation can be extracted from the incompletely specified “heuristic” maps. Heuristic MIMs can encompass many explicit models and provide a foundation for testing these models. They are well suited to the biologist's perspective. We have found that, once it is mastered, the notation becomes invaluable as a diagrammatic shorthand that imposes a logical discipline and reveals the biologically relevant aspects of a network. MIMs often show the richness of interconnections that presumably confers extraordinary fluidity and robustness to bioregulatory networks.
In addition to their heuristic character, another attribute of MIMs is that they are canonical, in the sense that a single diagram can encompass schema for a variety of cell types and cell states. The maps describe the interactions that can occur when the relevant molecules exist at the same time in the same place. Diagrams for specific cell types and cell states are derived from canonical maps by deleting the molecules that are not expressed, as well as the interactions that do not occur due to lack of colocalization in time or place. We are developing on-line tools that will allow users to carry out these deletions. A toolbox is also being provided to assist in manual map production (http://discover.nci.nih.gov/mim/). The MIM notation may prove useful in other fields of study, such as ecologic systems, and could become a general rubric for systems biology.
Acknowledgments
We thank Drs. Silvio Parodi, Stephania Pasa, Sohyoung Kim, and Hiroaki Kitano for many valuable comments, suggestions, and discussion during the development of the MIM notation. We are grateful to David Kane, Margot Sunshine, and Hong Cao, on contract to J.W.'s group from SRA, International, for help in implementing electronic forms of MIMs (i.e., eMIMs). This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–09–0824) on November 2, 2005.
References
- Aladjem, M. I., Pasa, S., Parodi, S., Weinstein, J. N., Pommier, Y., and Kohn, K. W. (2004). Molecular interaction maps–a diagrammatic graphical language for bioregulatory networks. Sci STKE 2004, pe8. [DOI] [PubMed]
- Cook, D. L., Farley, J. F., and Tapscott, S. J. (2001). A basis for a visual language for describing, archiving and analyzing functional models of complex biological systems. Genome Biol. 2, RESEARCH0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney, D. P., Buxton, B. F., Langdon, W. B., and Jones, D. T. (2004). BioRAT: extracting biological information from full-length papers. Bioinformatics 20, 3206 –3213. [DOI] [PubMed] [Google Scholar]
- Funahashi, A., Morohashi, M., and Kitano, H. (2003). CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico 1, 159 –162. [Google Scholar]
- Gagneur, J., Krause, R., Bouwmeester, T., and Casari, G. (2004). Modular decomposition of protein-protein interaction networks. Genome Biol. 5, R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker, T., Galitski, T., and Hood, L. (2001a). A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372. [DOI] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R., and Hood, L. (2001b). Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929 –934. [DOI] [PubMed] [Google Scholar]
- Kelley, B. P., Sharan, R., Karp, R. M., Sittler, T., Root, D. E., Stockwell, B. R., and Ideker, T. (2003). Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl. Acad. Sci. USA 100, 11394 –11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko, B. N., and Westerhoff, H. V. (1995). The macroworld versus the microworld of biochemical regulation and control. Trends Biochem. Sci. 20, 52–54. [DOI] [PubMed] [Google Scholar]
- Kitano, H. (2003). A graphical notation for biochemical networks. Biosilico 1, 169 –176. [Google Scholar]
- Kohn, K. W. (1998). Functional capabilities of molecular network components controlling the mammalian G1/S cell cycle phase transition. Oncogene 16, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Kohn, K. W. (1999). Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol. Biol. Cell 10, 2703–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, K. W. (2001). Molecular interaction maps as information organizers and simulation guides. Chaos 11, 84 –97. [DOI] [PubMed] [Google Scholar]
- Kohn, K. W., Aladjem, M. I., Pasa, S., Parodi, S., and Pommier, Y. (2003). Molecular interaction map of mammalian cell cycle control. Encycl. Human Genome 1, 457– 474. [Google Scholar]
- Kohn, K. W., and Bohr, V. A. (2002). Genomic instability and DNA repair. In: Cancer Handbook, Vol. 1, London: Nature Publishing Group, Macmillan Publishing, 87–106. [Google Scholar]
- Kohn, K. W., and Pommier, Y. (2005). Molecular interaction map of the p53 and Mdm2 logic elements that switch on the response of p53 to DNA damage. Biochem. Biophys. Res. Commun. 331, 816–827. [DOI] [PubMed] [Google Scholar]
- Kohn, K. W., Riss, J., Aprelikova, O., Weinstein, J. N., Pommier, Y., and Barrett, J. C. (2004). Properties of switch-like bioregulatory networks studied by simulation of the hypoxia response control system. Mol. Biol. Cell 15, 3042–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, A., Kobayashi, Y., and Takagi, T. (2003). Kinase pathway database: and integrated protein-kinase and NLP-based protein interaction resource. Genome Res. 13, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata, H., Matoba, N., and Shimizu, N. (2003). CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle. Nucleic Acids Res. 31, 4071– 4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirson, I., Fortemaison, N., Jacobs, C., Dremier, S., Dumont, J. E., and Maenhaut, C. (2000). The visual display of regulatory information and networks. Trends Cell Biol. 10, 404–408. [DOI] [PubMed] [Google Scholar]
- Pommier, Y., Sordet, O., Antony, S., Hayward, R. L., and Kohn, K. W. (2004). Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene 23, 2934 –2949. [DOI] [PubMed] [Google Scholar]
- Strogatz, S. H. (2001). Exploring complex networks. Nature 410, 268 –276. [DOI] [PubMed] [Google Scholar]
- Tanabe, L., Scherf, U., Smith, L. H., Lee, J. K., Hunter, L., and Weinstein, J. N. (1999). MedMiner: an Internet text-mining tool for biomedical information, with application to gene expression profiling. Biotechniques 27, 1210 –1214, 1216 –1217. [DOI] [PubMed] [Google Scholar]
- Temple, M. D., Perrone, G. G., and Daws, I. W. (2005). Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15, 319 –326. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Ideker, T., and Schwikowski, B. (2001). Visualization and integration of protein-protein interactions. In: The Study of Protein–Protein Interactions - An Advanced Manual, ed. E. Golemis, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Vazquez, A., Dobrin, R., Sergi, D., Eckmann, J.P., Oltvai, Z. N., and Barabasi, A. L. (2004). The topological relationship between the large-scale attributes and local interaction patterns of complex networks. Proc. Natl. Acad. Sci. USA 101, 17940 –17945. [DOI] [PMC free article] [PubMed] [Google Scholar]