Abstract
Class II histone deacetylases (HDACs) contain unique amino-terminal extensions that mediate interactions with members of the myocyte enhancer factor-2 (MEF2) family of transcription factors and responsiveness to kinases, including Ca2+/calmodulin-dependent kinase (CaMK). Despite intense investigation of class II HDACs, little is known of MEF2-independent mechanisms for transcriptional repression by these chromatin-modifying enzymes. Here, we demonstrate that class II HDACs 4 and 5 physically associate with ankyrin-repeat proteins ANKRA2 and RFXANK (RFX-B/Tvl-1/ANKRA1). ANKRA2 is a megalin- and BKCa potassium channel-interacting factor, whereas RFXANK is a positive regulator of major histocompatibility complex II (MHC II) gene expression. HDAC4 and HDAC5 interact with the ankyrin repeats of ANKRA2 and RFXANK and, through association with RFXANK, repress MHC II promoter activation. HDACs 4 and 5 also repress endogenous HLA-DRA gene expression induced by CIITA. Phosphorylation of class II HDACs by CaMK results in CRM1-dependent nuclear export of HDAC/RFXANK complexes. These results define a novel transcriptional pathway under the control of class II HDACs and suggest a role for these transcriptional repressors as signal-responsive regulators of antigen presentation.
INTRODUCTION
Posttranslational modification of lysine residues in nucleosomal histones provides a central mechanism for the control of gene expression (Jenuwein and Allis, 2001). Acetylation of the amino-terminal tails of core histones by histone acetyltransferases (HATs) results in chromatin relaxation and transcriptional activation, whereas deacetylation of histones by histone deacetylases (HDACs) is associated with transcriptional repression. Recruitment of HATs and HDACs to gene regulatory elements by sequence-specific DNA-binding transcription factors gives rise to multiprotein transcription regulatory complexes. There is also evidence that direct acetylation/deacetylation of certain transcription factors provides a mechanism for reversible regulation of transcription.
HDACs are divided into three classes, I, II, and III, on the basis of size, sequence homology, and formation of distinct complexes (Verdin et al., 2003). Class I includes HDACs 1, 2, and 3, which are expressed ubiquitously. Class II HDACs 4, 5, 7, and 9 exhibit a more restricted pattern of tissue distribution and contain a conserved domain of several hundred amino acids extending amino-terminal from the catalytic domain. These amino-terminal extensions contain two conserved serine residues that, upon phosphorylation, trigger nuclear export of class II HDACs. Class III HDACs are related to the yeast silent information regulator 2 (Sir2) and require NAD for deacetylase activity.
Class I HDACs interact with a plethora of transcription factors. In contrast, only a relatively small number of transcription factors have been shown to interact with class II HDACs. Most notably, class II HDACs associate with members of the myocyte enhancer factor-2 (MEF2) family of MADS-box transcription factors via their amino-terminal extensions, resulting in transcriptional repression (Miska et al., 1999; Sparrow et al., 1999; Wang et al., 1999; Lemercier et al., 2000; Lu et al., 2000; Yang and Gregoire, 2005). MEF2 factors play key roles in the control of muscle differentiation and growth through binding to A/T-rich elements in enhancer and promoter regions of a variety of muscle-specific genes (McKinsey et al., 2002). Thus, negative regulation of MEF2 activity by association with class II HDACs may inhibit growth and/or differentiation, depending on cell identity and the presence of other cofactors for MEF2 and HDACs. Indeed, genetic ablation of HDAC5 and 9 in mice revealed that they play important roles in cardiac gene regulation and development (Zhang et al., 2002; Chang et al., 2004). Recently, we also reported that another class II HDAC, HDAC4, governs chondrocyte hypertrophy and endochondral bone development by associating with Runx2, a master transcriptional regulator of these processes (Vega et al., 2004a).
In an effort to identify novel class II HDAC-interacting factors, we performed yeast two-hybrid screens using the amino-terminal extension of HDAC5 as bait. Here, we report that class II HDACs 5 and 4 associate with two related ankyrin-repeat proteins, termed ANKRA2 and RFXANK (ANKRA1). The function of ANKRA2 is largely unknown, although this protein has been shown to interact with the megalin receptor and BKCa, a calcium-regulated potassium channel, and thus may participate in receptor/channel signaling and/or metabolism (Rader et al., 2000; Lim and Park, 2005). In contrast, RFXANK has been shown to play a critical role in the assembly of a transcriptional complex containing the CIITA coactivator on the HLA-DRA major histocompatibility complex (MHC) II gene promoter, resulting in stimulation of MHC II expression and thus enhanced antigen presentation (Masternak et al., 1998; Nagarajan et al., 1999; Zhu et al., 2000; Zika and Ting, 2005). Mutations in RFXANK are associated with reduced levels of MHC II expression and immunodeficiency in humans referred to as bare lymphocyte syndrome (Reith and Mach, 2001; Nekrep et al., 2003). We demonstrate that class II HDACs 4 and 5 repress CIITA-mediated activation of the HLA-DRA promoter through association with RFXANK. HDACs 4 and 5 also repress endogenous HLA-DRA gene expression induced by CIITA. Phosphorylation of class II HDACs by Ca2+/calmodulin-dependent protein kinase (CaMK) results in CRM1-dependent nuclear export of RFXANK/HDAC complexes, providing means to control HLA-DRA expression in a signal-dependent manner. These results suggest novel roles for class II HDACs in the control of receptor-mediated signaling events through ANKRA2, and MHC class II-dependent antigen presentation via RFXANK.
MATERIALS AND METHODS
Yeast Two-hybrid Screen
A mouse 17-d embryo MATCHMAKER cDNA library (Clontech, Palo Alto, CA) was screened with a GAL4-HDAC5 bait in the yeast two-hybrid system. The bait was expressed from the pBD-GAL4-CAM vector and contained amino acids 1-664 of human HDAC5 fused to the GAL4 DNA-binding domain. Positive clones were subjected to specificity tests using the GAL4 DNA-binding domain alone as bait. Those clones that were specific for interaction with GAL4-HDAC5 bait were sequenced.
Cell Culture, Plasmids, and Transfections
10T1/2, COS, and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin. Transfections were performed using the lipid-based reagent Fugene 6 (Roche Molecular Biochemicals, Indianapolis, IN) and cells growing at a density of 5-10 × 105 cells/35-mm dish. Epitope-tagged derivatives of HDAC4, and HDAC5 containing amino-terminal FLAG or Myc tags were generated using the pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA). For GAL4-dependent reporter assays, 10T1/2 cells were cotransfected with a luciferase reporter plasmid under control of four GAL4 DNA-binding sites and the thymidine kinase (tk) promoter, pMH100-tk-luc (Nagy et al., 1997), pM1-based expression vectors encoding ANKRA2 fused to the GAL4 DNA-binding domain, and a CMV-lacZ (Invitrogen) plasmid to normalize for variable transfection efficiency. A luciferase reporter plasmid under control of the HLA-DRA promoter (DRA-luc), and an expression vector for human CIITA were kindly provided by J. Ting (University of North Carolina). For luciferase assays, cells were harvested 48 h after transfection, and luciferase and β-galactosidase activities were measured under conditions of linearity with respect to time and extract concentration.
Immunocytochemistry
COS cells were grown on six-well dishes, fixed in phosphate-buffered saline (PBS) containing 10% Formalin and stained with antibodies in PBS containing 3% bovine serum albumin and 0.1% Nonidet-P40. Primary antibodies against FLAG (M2; Sigma, St. Louis, MO) or Myc (polyclonal, A-14; Santa Cruz Biotechnology, Santa Cruz, CA) were used at a dilution of 1:500. Secondary antibodies conjugated to either fluorescein or Cy3 (Vector Laboratories, Burlingame, CA) were also used at a dilution of 1:1000.
Coimmunoprecipitation Assays
For coimmunoprecipitation experiments, transiently transfected COS or 293T cells were harvested 48 h after transfection in PBS containing 0.5% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors. After brief sonication and removal of cellular debris by centrifugation, FLAG-tagged proteins were immunoprecipitated from cell lysates using anti-FLAG affinity resin (Sigma) and washed five times with lysis buffer. Alternatively, Myc-tagged proteins were immunoprecipitated using polyclonal anti-Myc antibodies (Santa Cruz; A-14) and protein A Sepharose beads (Zymed Laboratories, South San Francisco, CA). Precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with either a monoclonal anti-Myc antibody (Santa Cruz; 9E10), a monoclonal anti-FLAG antibody (Sigma; M2), a goat polyclonal anti-CIITA antibody (Santa Cruz; N-20), a rat polyclonal anti-HDAC4 antibody (Santa Cruz), or a rat polyclonal anti-HDAC5 antibody (Upstate Biotechnology, Lake Placid, NY). Proteins were visualized with a chemiluminescence system (Santa Cruz).
RT-PCR Analysis of HLA-DRA mRNA Transcripts
HeLa cells were cotransfected with expression plasmids encoding CIITA (10 ng) and HDAC4 or 5 (100 ng). Forty-eight hours later, RNA was prepared from cells using Trizol reagent (Invitrogen) and 40 ng of total RNA was used for real-time RT-PCR of HLA-DRA mRNA transcripts and 18S rRNA. Real-time RT-PCR was performed using an ABI prism sequence detection system according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). A mixture of primers and probe for HLA-DRA and 18S RNA was purchased from Applied Biosystems. HLA-DRA expression was normalized to 18S values to control for differences in RNA loading. The relative value in control HeLa cells without ectopic CIITA was assigned as 1. *p < 0.05 versus CIITA alone.
RESULTS
Interaction between HDAC5 and ANKRA2 in Yeast
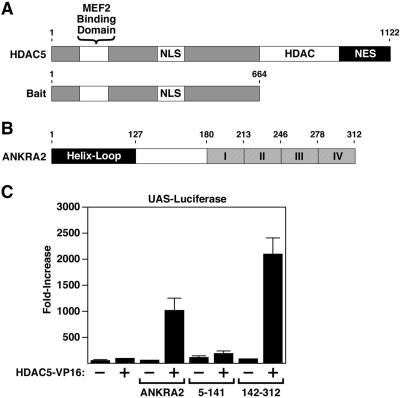
Class II HDACs 4, 5, 7, and 9 are bipartite, with an amino-terminal region that contains a MEF2-binding motif, and a carboxy-terminal catalytic domain (Figure 1A). The amino-terminal extensions of class II HDACs also serve as docking sites for the CtBP and HP1 corepressors and are subject to phosphorylation on two serine residues, which creates docking sites for the 14-3-3 chaperone protein. The subcellular distribution of class II HDACs is controlled by the opposing actions of nuclear localization and nuclear export sequences (McKinsey et al., 2002).
Figure 1.
Association of HDAC5 with ANKRA2 in yeast and mammalian cells. (A) Human HDAC5 is 1122 amino acids in length and contains a MEF2-binding domain, nuclear localization, and nuclear export sequences (NLS and NES, respectively) and a histone deacetylase catalytic domain (HDAC). The amino-terminal half of HDAC5 was used as bait in a yeast two-hybrid screen. (B) HDAC5 bait interacted with multiple clones of prey encoding ANKRA2. ANKRA2 is a 312-amino acid protein that contains an amino-terminal helix-loop domain and four carboxy-terminal ankyrin repeats (I-IV). (C) A mammalian two-hybrid system was used to confirm the interaction between HDAC5 and ANKRA2. COS cells were transfected with a luciferase reporter gene driven by five GAL4 DNA-binding sites (UAS-Luciferase), and the indicated combinations of vectors encoding amino acids 1-664 of HDAC5 fused to the VP16 transcriptional activation domain (HDAC5-VP16) and the GAL4 DNA-binding domain fused to either full-length ANKRA2 (ANKRA2), the amino-terminal half of ANRKA2 (5-141), or the carboxy-terminal half of ANKRA2 (142-312). Increased luciferase activity indicates that the GAL4-ANKRA2 construct recruited HDAC5-VP16 to the promoter of the luciferase reporter.
In an attempt to reveal novel functions of the amino-terminal extension of class II HDACs, we performed yeast two-hybrid screens using bait consisting of amino acids 1-664 of HDAC5 fused to the GAL4 DNA-binding domain (Figure 1A). Five positives obtained from a mouse 17-d embryo cDNA library encoded the GAL4 activation domain fused in-frame to ANKRA2. The ANKRA2:GAL4-activation domain fusion proteins interacted specifically with the HDAC5 bait and not with the GAL4-DNA-binding domain alone and did not stimulate reporter gene expression in the absence of the HDAC5 bait.
Association of HDACs 4 and 5 with ANKRA2 in Mammalian Cells
ANKRA2 contains an amino-terminal helix-loop domain and four carboxy-terminal ankyrin repeats (Figure 1B). To determine whether class II HDACs interact with ANKRA2 in mammalian cells, we used a modified two-hybrid system with constructs encoding full-length ANRKA2 (amino acids 1-312) fused to the GAL4 DNA-binding domain and the amino-terminal extension of HDAC5 (amino acids 1-664) fused to the VP16 transcriptional activation domain. The ability of ANKRA2 and HDAC5 to interact was assessed by monitoring expression of a luciferase reporter gene under the control of five GAL4 DNA-binding sites (5XUAS-luciferase). As shown in Figure 1C, activity of the reporter gene was unaffected by expression of either GAL4-ANKRA2 or HDAC5-VP16. In contrast, reporter gene expression was dramatically up-regulated upon coexpression of the two constructs, indicating that ANKRA2 efficiently recruited the HDAC5-tethered VP16 transcriptional activation domain to the gene promoter. Robust activation of the reporter gene was also observed upon coexpression of HDAC5-VP16 with an amino-terminal deletion mutant of ANKRA2 (amino acids 142-312), but not when coexpressed with a carboxy-terminal deletion mutant of the protein (amino acids 5-141) lacking the ankyrin repeats. These results suggest that HDAC5 associates with the ankyrin repeats of ANKRA2.
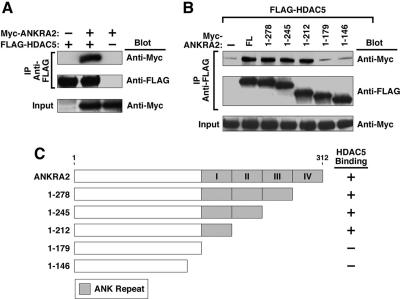
Coimmunoprecipitation studies were performed to further define the interaction between HDAC5 and ANKRA2. 293T cells were cotransfected with expression vectors encoding full-length HDAC5 and either full-length ANKRA2 or ANKRA2 deletion mutants. As shown in Figure 2, A and B, the interaction between ANKRA2 and HDAC5 was readily detected using this assay. HDAC5 interacted with ANKRA2 mutants lacking ankyrin repeats two through four, but not a mutant deficient in all ankyrin repeats (Figure 2, B and C). We conclude that a minimum of one ankyrin repeat in ANKRA2 is required for association with HDAC5.
Figure 2.
Physical interaction between HDAC5 and ANKRA2. (A) 293T cells were cotransfected with expression vectors encoding FLAG-tagged HDAC5 and Myc-ANKRA2. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibodies and proteins in the immune complexes were analyzed by Western blotting with the indicated antibodies (Blot). Crude lysates were analyzed by Western blotting to control for variability in protein expression (Input). (B) 293T cells were transfected with expression vectors encoding FLAG-HDAC5 and several deletion mutants of Myc-ANKRA2 (shown in C). Cell extracts were immunoprecipitated (IP) with anti-FLAG antibodies and proteins were analyzed by Western blotting as described above. (C) Shown are schematic depictions of the ANKRA2 deletion mutants used in B and a summary of the coimmunoprecipitation results.
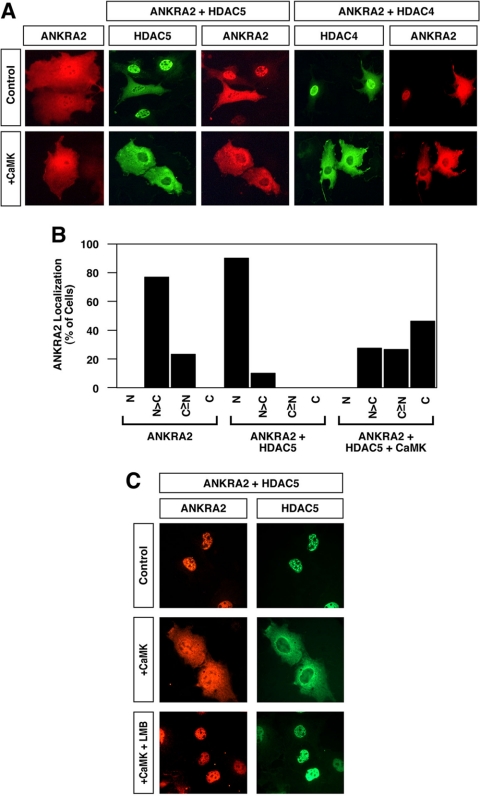
Indirect immunofluorescence studies were performed to determine whether ANKRA2 and HDAC5 were capable of associating in intact cells. ANKRA2 was distributed throughout the cell in the absence of ectopically expressed HDACs (Figure 3A, top panels). However, in cells overexpressing HDAC4 or HDAC5, ANKRA2 redistributed to the nucleus, where it colocalized with these transcriptional repressors. These results further suggest a biologically relevant interaction between ANKRA2 and class II HDACs.
Figure 3.
Colocalization of HDAC4 and 5 with ANKRA2. (A) COS cells were transfected with an expression vector encoding Myctagged ANKRA2 in the absence or presence of vectors for FLAG-tagged HDAC5 or HDAC4. As indicated, some cells also received a plasmid encoding constitutively active CaMKI. The subcellular localization of ANKRA2 and HDACs was determined by indirect immunofluorescence (red, ANKRA2; green, HDAC4 or 5). (B) Effects of HDAC5 and CaMKI overexpression on ANKRA2 subcellular distribution were quantified by microscopic examination of greater than 100 cells per condition. N, exclusive staining of ANKRA2 in the nucleus; N>C, nuclear ANKRA2 staining greater than cytoplasmic staining; C≥N, cytoplasmic ANKRA2 staining greater than or equal to nuclear staining; C, exclusive staining of ANRKA2 in the cytoplasm. (C) COS cells were cotransfected with expression vectors for ANKRA2 and HDAC5 in the absence or presence of a plasmid encoding activated CaMKI. Forty-eight hours after transfection, cells were exposed to leptomycin B (LMB; 10 nM) for 2 h. The subcellular localization of ANKRA2 and HDAC5 was determined by indirect immunofluorescence.
Signal Responsiveness of HDAC/ANKRA2 Complexes
One of the distinguishing characteristics of class II HDACs is their ability to undergo phosphorylation-dependent nucleocytoplasmic shuttling. To determine whether HDAC/ANKRA2 complexes might be signal-responsive, we examined the effect of CaMK signaling on the subcellular distribution of HDACs and ANKRA2. CaMKI has previously been shown to phosphorylate HDACs 4 and 5 and thereby promote their nuclear export via a 14-3-3 and CRM1-dependent mechanism (McKinsey et al., 2000a, 2000b; Harrison et al., 2004). As shown in Figure 3A (bottom panels), when HDACs and ANKRA2 were coexpressed, CaMKI activation resulted in their redistribution from the nucleus to the cytoplasm. In contrast, when the proteins were expressed separately, only HDACs displayed signal-dependent nuclear export. Quantification of the effects of coexpressing HDAC5 and CaMKI on ANKRA2 subcellular distribution is shown in Figure 3B.
To determine whether CaMK-dependent redistribution of HDAC/ANKRA2 complexes was due to inhibition of nuclear import or stimulation of nuclear export, we used leptomycin B (LMB), a fungal metabolite that suppresses the CRM1 exportin activity. As shown in Figure 3C, LMB completely inhibited CaMKI-mediated nuclear export of HDAC5/ANKRA2 complexes. These results suggest that class II HDACs and ANKRA2 form stable complexes that are subject to signal-dependent nuclear export via phosphorylation of HDAC residues.
Class II HDACs Associate with RFXANK
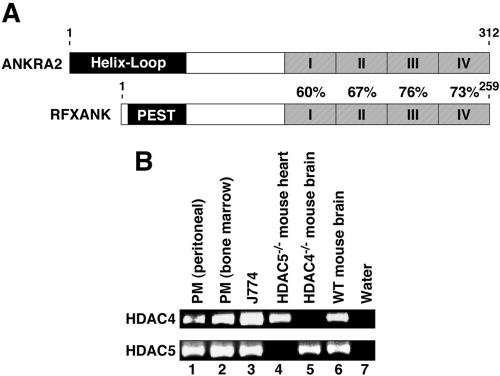
ANKRA2 has a paralogue gene product named RFXANK/RFX-B/Tvl-1/ANKRA1 (Long and Boss, 2005). The ankyrin repeat sequences of ANKRA2 are highly homologous to those of RFXANK (Figure 4A). RFXANK is a subunit of the RFX complex, a positive regulator of MHC II gene expression (Zika and Ting, 2005). Thus, we hypothesized that class II HDACs may associate with RFXANK and regulate MHC II gene expression. To begin to address this hypothesis, we analyzed the expression of HDACs 4 and 5 in macrophages, which are antigen-presenting cells that express MHC II. As shown in Figure 4B, mRNA transcripts for HDACs 4 and 5 were readily detected in primary macrophages purified from mouse bone marrow and peritoneum and in the macrophage cell line J774 (Singh et al., 2005). These results support the notion that these class II HDACs participate in the regulation of MHC II gene transcription.
Figure 4.
Expression of class II HDACs in macrophages. (A) The carboxy-terminal ankyrin repeats of ANKRA2 are highly homologous to those of RFXANK. Shown is the percentage of amino acid identity between the four ankyrin repeats of each protein. The amino-termini of ANRKA2 and RFXANK are distinct, with ANKRA2 containing a helix-loop domain and RFXANK, a prolinerich PEST domain. (B) RT-PCR for HDAC4 and HDAC5 mRNA transcripts was performed using total RNA extracted from mouse primary macrophages obtained from peritoneal or bone marrow or the J774 macrophage cell line. cDNAs synthesized from total RNA obtained from brain or heart of HDAC4- or HDAC5-null mice, respectively, was used to control for PCR primer specificity.
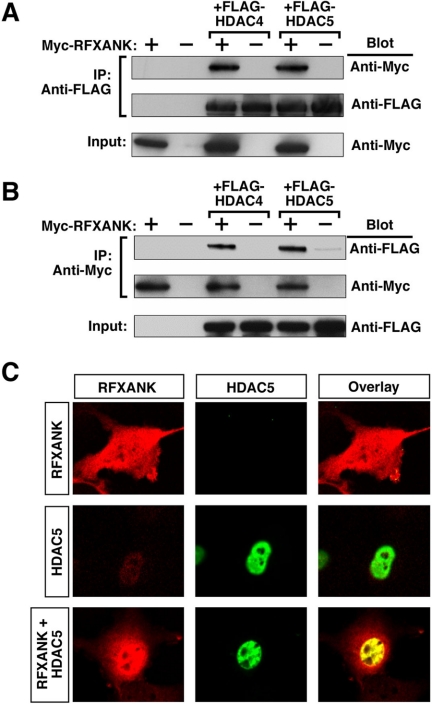
Coimmunoprecipitation studies were performed to determine whether RFXANK is capable of interacting with class II HDACs. As shown in Figures 5A, RFXANK was efficiently coimmunoprecipitated with HDAC4 or HDAC5 from protein lysates derived from transiently transfected 293T cells in which the relevant proteins were overexpressed. Identical results were obtained when employing an immunoprecipitating antibody directed against RFXANK (Figure 5B).
Figure 5.
Interaction of RFXANK with HDAC4 and HDAC5 in mammalian cells. (A) 293T cells were cotransfected with expression vectors encoding FLAG-tagged HDAC5 and Myc-RFXANK. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibodies and proteins in the immune complexes were analyzed by Western blotting with the indicated antibodies (Blot). Crude lysates were analyzed by Western blotting to control for variability in protein expression (Input). (B) 293T cells were transfected and protein lysates prepared as described in A. IP was performed with anti-Myc antibody and proteins in immune complexes were analyzed by Western blotting as described above. (C) Colocalization of HDAC5 with RFXANK. COS cells were transfected with expression vectors encoding either FLAG-tagged HDAC5 or Myc-tagged RFXANK. Some cells were cotransfected with these vectors (RFXANK + HDAC5). The subcellular localization of RFXANK and HDAC5 was determined by indirect immunofluorescence. Proteins were visualized with a confocal microscope and digital images captured (red, RFXANK; green, HDAC5; yellow, colocalized RFXANK and HDAC5).
Confocal microscopic studies were performed to further examine the interaction between HDAC5 and RFXANK. RFXANK was distributed throughout the cell in the absence of ectopic HDAC5 (Figure 5C). On coexpression of HDAC5, RFXANK redistributed to the nucleus where it colocalized with HDAC5.
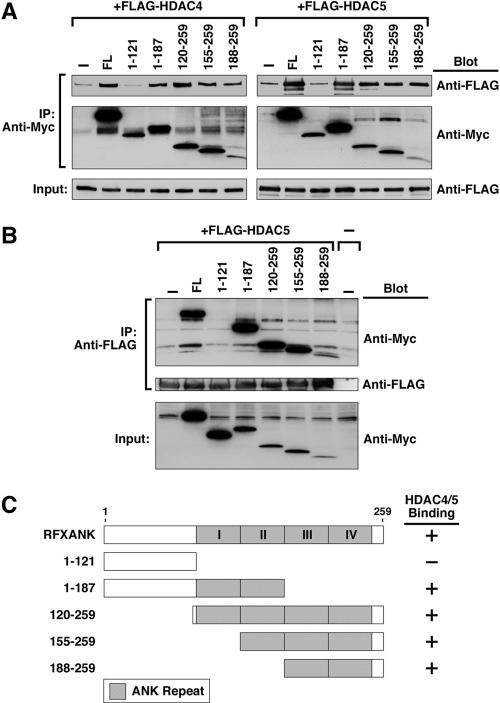
To further define the HDAC-binding region of RFXANK, coimmunoprecipitation studies were performed with fulllength HDAC4 or HDAC5 and a panel of RFXANK deletion mutants. As shown in Figure 6A, HDAC4 and HDAC5 were efficiently coimmunoprecipitated with RFXANK mutants harboring ankyrin repeats. In contrast, a deletion mutant of RFXANK lacking all ankyrin repeats failed to associate with either HDAC4 or HDAC5. Identical results were obtained when immunoprecipitating antibodies were directed against HDAC (Figure 6B). These results are summarized in Figure 6C.
Figure 6.
HDAC4 and HDAC5 bind to the ankyrin repeats of RFXANK. (A) 293T cells were cotransfected with expression vectors encoding FLAG-tagged HDAC4 or HDAC5 and Myc-tagged RFXANK. Cell extracts were immunoprecipitated (IP) with anti-Myc antibody and proteins in immune complexes were analyzed by Western blotting with the indicated antibodies (Blot). Crude lysates were analyzed by Western blotting to control for variability in protein expression (Input). (B) 293T cells were transfected and protein lysates prepared as described in A. IP was performed with anti-FLAG antibody and proteins in immune complexes were analyzed by Western blotting as described above. (C) Shown are schematic depictions of the RFXANK deletion mutants used in A and B and a summary of the coimmunoprecipitation results.
Functional Interaction between Class II HDACs and RFXANK
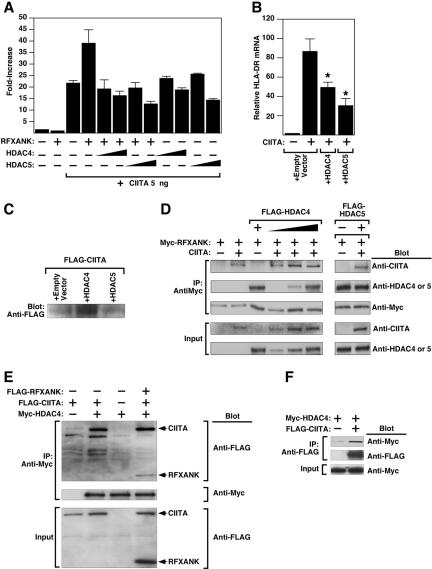
Promoter-reporter assays were performed to assess the functional significance of the interaction between class II HDACs and RFXANK. RFXANK stimulates MHC II gene transcription by coordinating the formation of a transcriptional complex containing CIITA (Zhu et al., 2000). CIITA possesses HAT activity and potently stimulates MHC II gene expression. We hypothesized that class II HDACs might repress MHC II promoter activity via interaction with RFXANK. To test this hypothesis, COS cells were cotransfected with a luciferase reporter under the control of the promoter from an MHC II gene, HLA-DRA (DRA-luc), in the absence or presence of vectors for CIITA, RFXANK, and HDAC4 or HDAC5. As shown in Figure 7A, expression of CIITA stimulated HLA-DRA promoter activity, and coexpression with RFXANK enhanced promoter activation. HDAC4 and HDAC5 repressed activation of the HLA-DRA promoter.
Figure 7.
HDAC4 and HDAC5 repress CIITA-inducible HLA-DRA promoter activity. (A) COS cells were transfected with a luciferase reporter controlled by the HLA-DRA promoter in the absence or presence of vectors for RFXANK (100 ng), CIITA (5 ng), or HDAC4 or HDAC5 (10 or 100 ng), as indicated. Cells were harvested for luciferase assay 48 h after transfection. (B) HeLa cells were transfected with expression plasmid encoding CIITA (10 ng) in the absence or presence of vectors for HDAC4 or HDAC5 (100 ng). Forty-eight hours later, RNA was prepared from the cells and HLA-DRA mRNA transcripts were detected by real-time RT-PCR. Values were normalized to those for 18S rRNA. (C) Hela cells were transfected as described in B. Protein lysates were analyzed by immunoblotting with anti-FLAG antibody to assess effects of HDAC overexpression on FLAG-CIITA levels. (D) 293T cells were transfected with the indicated combinations of expression vectors encoding CIITA, Myc-tagged RFXANK and FLAG-tagged HDAC4 or HDAC5. Cell extracts were immunoprecipitated (IP) with anti-Myc antibody and proteins in immune complexes were analyzed by Western blotting (Blot) with the indicated antibodies. Crude lysates were analyzed by Western blotting to control for variability in protein expression (Input). (E) 293T cells were transfected with expression vectors encoding FLAG-CIITA, FLAG-RFXANK, or myc-HDAC4 (1 μg each), as indicated. Cell extracts were immunoprecipitated with anti-Myc antibody and proteins were analyzed by immunoblotting with anti-FLAG antibody. (F) 293T cells were transfected with expression vectors encoding either FLAG-CIITA or Myc-HDAC4 (1 μg each), as indicated. Cell extracts were immunoprecipitated with anti-FLAG antibody and proteins were analyzed by immunoblotting with anti-Myc antibody.
We next determined whether class II HDACs are capable of suppressing endogenous HLA-DRA gene expression. Ectopic overexpression of CIITA stimulated HLA-DRA expression, as measured by quantitative RT-PCR analysis (Figure 7B). Coexpression of either HDAC4 or HDAC5 with CIITA significantly reduced the levels of endogenous HLA-DRA mRNA transcripts. Immunoblot analysis confirmed that HDACs did not reduce CIITA protein levels (Figure 7C), supporting the notion that HDACs 4 and 5 directly suppress MHC II gene expression.
Because CIITA is known to associate with the ankyrin repeat region of RFXANK, experiments were performed to determine whether HDACs repress CIITA-mediated transcriptional activation by interfering with the interaction of CIITA with RFXANK. As shown in Figure 7D, we found no evidence for disruption of RFXANK/CIITA interactions by HDAC4 or HDAC5, suggesting that HDACs form a ternary complex with RFXANK and CIITA. Indeed, CIITA and RFXANK were immunoprecipitated with HDAC4 (Figure 7E). In addition, we found that HDAC4 can interact with CIITA in the absence of RFXANK (Figure 7, E and F). These data demonstrate that class II HDACs interact with both RFXANK and CIITA to form complexes that regulate MHC II transcription.
Signal Responsiveness of HDAC/RFXANK Complexes
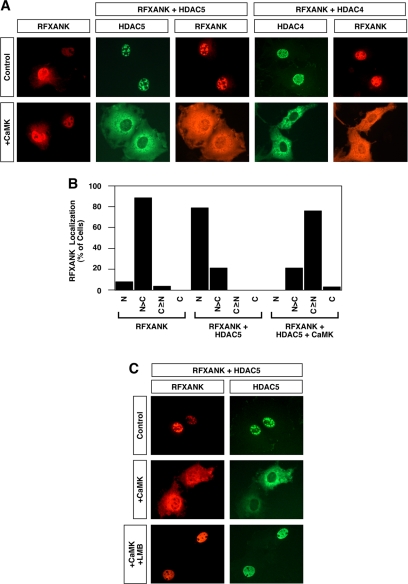
We next tested whether CaMK signaling stimulates nuclear export of HDAC/RFXANK complexes. As shown in Figure 8A, top panels, RFXANK colocalized with HDAC4 and HDAC5 in the nuclei of unstimulated cells. In response to activated CaMK, both HDAC and RFXANK were relocalized to the cytoplasm (Figure 8A, bottom panels). CaMK-mediated redistribution of RFXANK to the cytoplasm was dependent on coexpression of class II HDACs. The effects of coexpression of HDAC5 and CaMKI on RFXANK subcellular distribution were quantified and are summarized in Figure 8B. As seen with ANKRA2 (Figure 3), CaMKI-mediated redistribution of RFXANK from the nucleus to the cytoplasm was dependent on CRM1 exportin activity (Figure 8C). These results suggest that CRM1 triggers nuclear export of RFXANK via phosphorylation of associated class II HDACs.
Figure 8.
Nuclear export of class II HDAC/RFXANK complexes. (A) COS cells were transfected with an expression vector encoding Myc-tagged RFXANK in the absence or presence of vectors for FLAG-tagged HDAC5 or HDAC4. As indicated, some cells also received a plasmid encoding constitutively active CaMKI. The subcellular localization of RFXANK and HDACs was determined by indirect immunofluorescence (red, RFXANK; green, HDAC4 or HDAC5). (B) Effects of HDAC5 and CaMKI overexpression on RFXANK subcellular distribution were quantified by microscopic examination of greater than 100 cells per condition. N, exclusive staining of RFXANK in the nucleus; N>C, nuclear RFXANK staining greater than cytoplasmic staining; C≥N, cytoplasmic RFXANK staining greater than or equal to nuclear staining; C, exclusive staining of RFXANK in the cytoplasm. (C) COS cells were cotransfected with expression vectors for RFXANK and HDAC5 in the absence or presence of a plasmid encoding activated CaMKI. Forty-eight hours after transfection, cells were exposed to leptomycin B (LMB; 10 nM) for 2 h. The subcellular localization of RFXANK and HDAC5 was determined by indirect immunofluorescence.
DISCUSSION
ANKRA2 and RFXANK Are Novel Partners of Class II HDACs
In this study, we found that ANKRA2, a protein containing four ankyrin repeats, interacts with class II HDACs. ANKRA2 was originally identified as an interacting protein of megalin (Rader et al., 2000), a member of the low-density lipoprotein receptor superfamily, and recently has been shown to interact with the BKCa channel (Lim and Park, 2005). However, its role in the nucleus, where class II HDACs exert their effects on gene expression, is unknown.
Our results show that ANKRA2 displays a diffuse intracellular distribution in the absence of HDACs, but becomes localized to the nucleus in the presence of class II HDACs. Moreover, in response to CaMK signaling, ANKRA2 is redistributed from the nucleus to the cytoplasm in association with class II HDACs. The effects of CaMK signaling on ANKRA2 localization are mediated indirectly through phosphorylation of class II HDACs, which triggers their nuclear export via a CRM1- and 14-3-3-dependent mechanism (McKinsey et al., 2000b). Thus, ANKRA2 may function as a subunit of a signal-responsive complex in the nucleus of cells expressing class II HDACs, or class II HDACs may regulate the cytoplasmic activity of ANKRA2 in response to extracellular cues that stimulate CaMK activity.
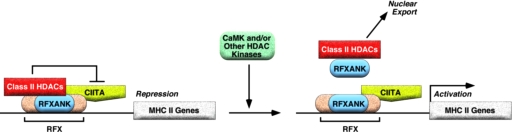
In contrast to ANKRA2, its paralogue, RFXANK, is well known as a subunit of the RFX complex, which binds to the promoters of MHC II genes and stimulates their expression through recruitment of the CIITA transcriptional coactivator (Zika and Ting, 2005). Because the ankyrin repeats of ANKRA2, which interact with class II HDACs, are highly conserved in RFXANK (Long and Boss, 2005), we then examined whether HDACs can interact with RFXANK. Coimmunoprecipitation and immunocytochemistry studies showed that HDAC4 and HDAC5 interact with RFXANK. Our demonstration that RFXANK acts as a partner of class II HDACs suggests that this ankyrin-repeat protein functions as an adaptor to link class II HDACs to downstream transcriptional effectors that control expression of antigen-presenting MHC II molecules (Figure 9).
Figure 9.
A model for regulation of MHC class II gene expression by class II HDACs. RFXANK stimulates MHC II gene expression through recruitment of the CIITA coactivator to the HLA-DRA promoter. Association of RFXANK with class II HDACs 4 or 5 results in repression of HLA-DRA promoter activity, presumably via deacetylation of local histones. CaMK, and possibly other kinases, phosphorylate class II HDACs and trigger CRM1-dependent nuclear export of HDAC/RFXANK complexes. Signal-dependent nuclear export of HDAC/RFXANK complexes may contribute to derepression of the HLA-DRA promoter in response to extracellular stimuli.
Ankyrin repeats are 33 amino acids in length and are found in a plethora of proteins with diverse biological functions. Of note, significant amino acid divergence among ankyrin repeats does exist (Mosavi et al., 2004), and homology searches indicate that the ankyrin repeats of ANKRA2 and RFXANK are only disparately related to those of other proteins (unpublished data). Thus, we propose that the interaction of class II HDACs with ANKRA2 and RFXANK is specific and does not represent a general affinity of these transcriptional regulators for ankyrin repeats.
Class II HDACs Repress the HLA-DRA Promoter through Interaction with RFXANK
RFXANK interacts with RFA-5, RFXAP to form the RFX complex (Burd et al., 2004). RFX, CREB, and NF-Y bind to X1, X2, and Y boxes in the promoter of MHC II genes, respectively, and thereby influence transcription of those genes. These three factors are ubiquitously expressed in all cell types. However, they are insufficient for MHC II gene expression. A transcriptional activator, CIITA, whose expression is regulated in a cell-type and developmental stage-specific manner, and also by cytokine stimulation, is required for MHC II gene activation (Zhu et al., 2000). In this study, we showed that HDAC4 and HDAC5 are expressed in macrophages, which are major MHC II-expressing cells, and that these HDACs repress CIITA-mediated HLA-DRA promoter activation and endogenous HLA-DRA gene expression by forming a complex with RFXANK and CIITA. These results suggest that class II HDACs regulate MHC II gene expression in antigen-presenting cells.
Of note, class I HDACs have also been shown to influence MHC II gene expression (Zika et al., 2003). Specifically, HDACs 1 and 2 were found to inhibit CIITA-mediated activation of the HLA-DRA promoter by disrupting CIITA association with the RFX complex. Thus, it is possible that full repression of MHC II gene expression relies on the coordinate action of both class I and class II HDACs. In this regard, prior studies have established a critical role for class I HDACs in the mechanism by which class II HDACs repress gene expression (Fischle et al., 2002).
A Potential Mechanism for Signal-dependent De-repression of Gene Expression
The results of this study demonstrate that CaMK signaling stimulates nuclear export of RFXANK/HDAC complexes. On the basis of the sensitivity of this process to LMB, we conclude that the CRM1 nuclear export receptor is a critical component of the export process. The coexport of RFXANK and HDAC5 in response to CaMK signaling indicates that that this protein-protein complex remains intact and that nuclear export is mediated by sequences in HDAC5, because RFXANK alone is insensitive to CaMK signaling.
The docking site for RFXANK on HDAC4 and HDAC5 is located within the amino-terminal extensions of these proteins. We and others have shown that signal-dependent nuclear export of HDAC4 and HDAC5 requires phosphorylation of two serine residues in these amino-terminal extensions, which creates binding sites for 14-3-3 chaperone proteins (Grozinger and Schreiber, 2000; McKinsey et al., 2000b; Wang et al., 2000). Binding of 14-3-3 to class II HDACs exposes a cryptic carboxy-terminal nuclear export sequence (NES), which is targeted by CRM1. Although the signal-responsive serines are contained in the RFXANK-binding domains of HDAC4 and HDAC5, they must be accessible to CaMK and to 14-3-3, because RFXANK complexed with a mutant form of HDAC5 lacking these sites is refractory to nuclear export (unpublished data). Because RFXANK is essential for CIITA recruitment to MHC II promoters, our model predicts that a pool of RFXANK is free from association with class II HDACs and thereby remains in the nucleus in the face of signals that stimulate nuclear export of HDAC5 (Figure 9).
Signal-dependent nuclear export of HDAC/RFXANK complexes could have implications for overriding the repressive influence of class II HDACs on MHC II gene promoters (Figure 9). This mechanism for derepression is predicted to be operative under conditions in which intracellular calcium levels have been elevated, which would result in CaMK activation. In addition, we have previously shown that class II HDACs 4, 5, and 7 are subject to nuclear export in response to signaling via protein kinase C and protein kinase D (Vega et al., 2004b; Chang et al., 2005; Parra et al., 2005). Thus, stimuli that lead to activation of these kinases may also influence MHC II expression via nuclear export of HDAC/RFXANK complexes.
While this work was in preparation, we became aware of a related manuscript by Wang et al. that is in press (Wang et al., 2005). These investigators also found that class II HDACs associate with RFXANK and ANKRA2. Our results further validate a role for these interactions and define a novel means of regulating HDAC/ankyrin repeat-containing protein complexes via a calcium-dependent signaling pathway.
CONCLUSIONS
Although we have shown that the association of class II HDACs with RFXANK modulates a well-characterized macrophage-specific gene promoter, the expression of class II HDACs and RFXANK in a wide range of cells suggests that this regulatory interaction may have important consequences in other cell types. Expression of most class II HDACs is enriched in heart, skeletal muscle, and brain. Thus, HDAC4 and HDAC5 may also contribute to the tissue-specific regulation of MHC II expression in those cells under some circumstances. It should be noted that changes in CIITA expression and activity are associated with reduced survival of cardiac allografts and increased susceptibility to myocardial infarction due to altered lymphocyte responses (Brickey et al., 2002; Swanberg et al., 2005). Thus, class II HDACs may play a role in the regulation of these processes via RFXANK/CIITA. In addition, it remains possible that RFXANK regulates genes other than MHC II and that its interaction with class II HDACs will influence these other targets. Further evaluation of the role of class II HDAC/RFXANK interactions should provide novel insights into the mechanisms of transcriptional regulation by RFXANK. In addition, the ability of class II HDACs to interact with ANKRA2 may have implications for as-yet-undefined functions of this ankyrin-repeat protein on receptor-mediated signaling events and/or gene regulation.
Acknowledgments
We are grateful to A. Tizenor for graphics and C. L. Zhang for advice. Work in Eric Olson's lab was supported by grants from the National Institutes of Health, The Donald W. Reynolds Clinical Cardiovascular Research Center, The Robert A. Welch Foundation, and the Texas Advanced Technology Program.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0612) on October 19, 2005.
References
- Brickey, J. W., Felix, N. J., Griffiths, R., Zhang, J., Wang, B., Piskurich, J. F., Itoh-Lindstrom, Y., Coffman, T. M., and Ting, J. P. (2002). Prolonged survival of class II transactivator-deficient cardiac allografts. Transplantation 74, 1341-1348. [DOI] [PubMed] [Google Scholar]
- Burd, A. L., Ingraham, R. H., Goldrick, S. E., Kroe, R. R., Crute, J. J., and Grygon, C. A. (2004). Assembly of major histocompatibility complex (MHC) class II transcription factors: association and promoter recognition of RFX proteins. Biochemistry 43, 12750-12760. [DOI] [PubMed] [Google Scholar]
- Chang, S., McKinsey, T. A., Zhang, C. L., Richardson, J. A., Hill, J., and Olson, E. N. (2004). Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart. Mol. Cell. Biol. 24, 8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., Bezprozvannaya, S., Li, S., and Olson, E. N. (2005). An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. USA 102, 8120-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Dequiedt, F., Hendzel, M. J., Guenther, M. G., Lazar, M. A., Voelter, W., and Verdin, E. (2002). Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45-57. [DOI] [PubMed] [Google Scholar]
- Grozinger, C. M., and Schreiber, S. L. (2000). Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97, 7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B. C., Roberts, C. R., Hood, D. B., Sweeney, M., Gould, J. M., Bush, E. W., and McKinsey, T. A. (2004). The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol. Cell. Biol. 24, 10636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- Lemercier, C., Verdel, A., Galloo, B., Curtet, S., Brocard, M. P., and Khochbin, S. (2000). mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275, 15594-15599. [DOI] [PubMed] [Google Scholar]
- Lim, H. H., and Park, C. S. (2005). Identification and functional characterization of ankyrin-repeat family protein ANKRA as a protein interacting with BKCa channel. Mol. Biol. Cell 16, 1013-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. B., and Boss, J. M. (2005). Evolutionary conservation and characterization of the bare lymphocyte syndrome transcription factor RFX-B and its paralogue ANKRA2. Immunogenetics 56, 788-797. [DOI] [PubMed] [Google Scholar]
- Lu, J., McKinsey, T. A., Nicol, R. L., and Olson, E. N. (2000). Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97, 4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak, K., Barras, E., Zufferey, M., Conrad, B., Corthals, G., Aebersold, R., Sanchez, J. C., Hochstrasser, D. F., Mach, B., and Reith, W. (1998). A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 20, 273-277. [DOI] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., Lu, J., and Olson, E. N. (2000a). Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2000b). Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97, 14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2, a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40-47. [DOI] [PubMed] [Google Scholar]
- Miska, E. A., Karlsson, C., Langley, E., Nielsen. S. J., Pines, J., and Kouzarides, T. (1999). HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18, 5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi, L. K., Cammett, T. J., Desrosiers, D. C., and Peng, Z. Y. (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan, U. M., Louis-Plence, P., DeSandro, A., Nilsen, R., Bushey, A., and Boss, J. M. (1999). RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity 10, 153-162. [DOI] [PubMed] [Google Scholar]
- Nagy, L., Kao, H. Y., Chakravarti, D., Lin, R. J., Hassig, C. A., Ayer, D. E., Schreiber, S. L., and Evans, R. M. (1997). Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89, 373-380. [DOI] [PubMed] [Google Scholar]
- Nekrep, N., Fontes, J. D., Geyer, M., and Peterlin, B. M. (2003). When the lymphocyte loses its clothes. Immunity 18, 453-457. [DOI] [PubMed] [Google Scholar]
- Parra, M., Kasler, H., McKinsey, T. A., Olson, E. N., and Verdin, E. (2005). Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280, 13762-13770. [DOI] [PubMed] [Google Scholar]
- Rader, K., Orlando, R. A., Lou, X., and Farquhar, M. G. (2000). Characterization of ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin J. Am. Soc. Nephrol. 11, 2167-2178. [DOI] [PubMed] [Google Scholar]
- Reith, W., and Mach, B. (2001). The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19, 331-373. [DOI] [PubMed] [Google Scholar]
- Singh, G., Singh, B., Trajkovic, V., and Sharma, P. (2005). Mycobacterium tuberculosis 6kDa early secreted antigenic target stimulates activation of J774 macrophages. Immunol. Lett. 98, 180-188. [DOI] [PubMed] [Google Scholar]
- Sparrow, D. B., Miska, E. A., Langley, E., Reynaud-Deonauth, S., Kotecha, S., Towers, N., Spohr, G., Kouzarides, T., and Mohun, T. J. (1999). MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 18, 5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanberg, M. et al. (2005). MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat. Genet. 37, 486-494. [DOI] [PubMed] [Google Scholar]
- Vega, R. B. et al. (2004a). Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555-566. [DOI] [PubMed] [Google Scholar]
- Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., and McKinsey, T. A. (2004b). Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin, E., Dequiedt, F., and Kasler, H. G. (2003). Class II histone deacetylases: versatile regulators. Trends Genet. 19, 286-293. [DOI] [PubMed] [Google Scholar]
- Wang, A. H., Bertos, N. R., Vezmar, M., Pelletier, N., Crosato, M., Heng, H. H., Thing, J. P., Han, J., and Yang, X. J. (1999). HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19, 7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. H., Kruhlak, M. J., Wu, J., Bertos, N. R., Vezmar, M., Posner, B. I., Bazett-Jones, D. P., and Yang, X. J. (2000). Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell Biol. 20, 6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. H., Gregoire, S., Zika, E., Xiao, L., Li, C. S., Li, H., Wright, K. L., Th'ng, J., and Yang, X. J. (2005). Identification of the ankyrin-repeat proteins ANKRA and RFXANK as novel partners of class IIA histone deacetylases. J. Biol. Chem. 280, 29117-29127. [DOI] [PubMed] [Google Scholar]
- Yang, X.-J., and Gregoire, S. (2005). Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25, 2873-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A., and Olson, E. N. (2002). Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. S., Linhoff, M. W., Li, G., Chin, K. C., Maity, S. N., and Ting, J. P. (2000). Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 20, 6051-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika, E., Greer, S. F., Zhu, X. S., and Ting, J. P. (2003). Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. Mol. Cell Biol. 23, 3091-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika, E., and Ting, J. P. (2005). Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 17, 58-64. [DOI] [PubMed] [Google Scholar]