Abstract
Grn1p from fission yeast and GNL3L from human cells, two putative GTPases from the novel HSR1_MMR1 GTP-binding protein subfamily with circularly permuted G-motifs play a critical role in maintaining normal cell growth. Deletion of Grn1 resulted in a severe growth defect, a marked reduction in mature rRNA species with a concomitant accumulation of the 35S pre-rRNA transcript, and failure to export the ribosomal protein Rpl25a from the nucleolus. Deleting any of the Grn1p G-domain motifs resulted in a null phenotype and nuclear/nucleolar localization consistent with the lack of nucleolar export of preribosomes accompanied by a distortion of nucleolar structure. Heterologous expression of GNL3L in a Δgrn1 mutant restored processing of 35S pre-rRNA, nuclear export of Rpl25a and cell growth to wild-type levels. Genetic complementation in yeast and siRNA knockdown in HeLa cells confirmed the homologous proteins Grn1p and GNL3L are required for growth. Failure of two similar HSR1_MMR1 putative nucleolar GTPases, Nucleostemin (NS), or the dose-dependent response of breast tumor autoantigen NGP-1, to rescue Δgrn1 implied the highly specific roles of Grn1p or GNL3L in nucleolar events. Our analysis uncovers an important role for Grn1p/GNL3L within this unique group of nucleolar GTPases.
INTRODUCTION
The nucleolus is the principal site for the generation of rRNA as also for ribosome assembly and maturation (see Venema and Tollervey, 1999; Tschochner and Hurt, 2003). The initial rRNA processing is concomitant with the formation of the 90S ribosomal precursor particle that separates into 40S and 60S presubunits. The pre-60S ribosomes undergo a series of RNA processing reactions that begin within the nucleolus followed by export of the ribosomes through the nuclear pore complex (NPC; Venema and Tollervey, 1999; Milkereit et al., 2001; Fatica and Tollervey, 2002; Nissan et al., 2002; Tschochner and Hurt, 2003). Nucleotide-binding proteins comprising several putative GTPases are known to be associated with pre-60S ribosomes on their journey from the nucleolus to the cytoplasm (Nissan et al., 2002; Tschochner and Hurt, 2003). However, their precise function at the molecular level is unclear. All studies to date point to their role(s) in rRNA processing/maturation and/or 60S ribosomal subunit export from the nucleolus/nucleus (Tschochner and Hurt, 2003). Although most of the nucleotide-binding GTPases possess both GDP/GTP-binding and GTP-hydrolyzing potential from the structural point of view, some members lack either one or both abilities in vivo (Takai et al., 2001). The YAWG/YlqF/HSR1_MMR1 GTP-binding protein subfamily of GTPases belongs to the TRAFAC (for Translation factor-related) subclass of GTPases characterized by a circular permutation of their GTPase signature motifs (G1-5) so that the G4, -5, and -6 subdomains are relocated from the C-terminal side of the protein to the N-terminus (Daigle et al., 2002; Leipe et al., 2002). Several members of this family from human (NGP-1 and Nucleostemin; Racevskis et al., 1996; Tsai and McKay, 2002, 2005), and yeast (Nug1p, Nug2p/Nog2p; Bassler et al., 2001; Saveanu et al., 2001) have been shown to localize to the nucleolus including some closely associated with ribosomal assembly and nucleolar/nuclear export. One member of this group, Nucleostemin (NS), was preferentially expressed in nucleoli of CNS stem cells, embryonic stem cells and several cancer cell lines and may control cell cycle progression in those cell lines (Tsai and McKay, 2002; Liu et al., 2004; Sijin et al., 2004).
There are at least four predicted HSR1_MMR1 nucleolar/nuclear GTP-binding proteins in the fission yeast. Fission yeast has served as an excellent genetically tractable model organism for the study of the molecular aspects of growth and regulation (Lee and Nurse, 1987; Lee and Nurse, 1988; Sheldrick and Carr, 1993; Hermeking et al., 1997; Nurse, 1997; Rustici et al., 2004). Given the growing awareness regarding the importance of nucleolar GTPases such as NS as modulators of cell growth and proliferation in cancer (Normile, 2002; Tsai and McKay, 2002; Bernardi and Pandolfi, 2003; Misteli, 2005; Tsai and McKay, 2005), we wanted to identify and characterize similar nucleolar/nuclear GTPases in the fission yeast S. pombe in order to understand their role in cell growth and regulation.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Schizosaccharomyces pombe strains used in this study are listed in Table 1. Yeast strains were maintained in YES (yeast extract plus supplements) or EMM (Edinburgh minimal media) medium supplemented with appropriate amino acids and ±15 μM thiamine routinely to repress (it must be noted that repression is not 100%) or induce, respectively, the nmt1 promoter (Moreno et al., 1991). Unless otherwise specified, yeast cultures were maintained or grown at 32°C and harvested at 0.4-1.0 OD600 for all experiments. YNB483 (leu1-32 ura4-D18 his3-Δ1) and YNB484 (leu1-32 ΔSPBC26H8.08c::ura4+ his3- Δ1) were the principal yeast strains used in this study and are referred to in the text as either wild type and null mutant or Δgrn1, respectively.
Table 1.
Yeast strains
| Strains (YNB) | Parent strain | Plasmid/Character/Genotype | Marker | Reference |
|---|---|---|---|---|
| YNB483 | leu1-32 ura4-D18 his3-Δ1 (Wild type) | This study | ||
| YNB484 | leu1-32 ΔSPBC26H8.08c::ura4+ his3-Δ1(Mutant) | This study | ||
| YNB544 | YNB484 | pBNB190 | Leu2 | This study |
| YNB545 | YNB484 | pBNB189 | Leu2 | This study |
| YNB546 | YNB484 | pCDL280 | Leu2 | This study |
| YNB566 | YNB484 | pBNB203 | Leu2 | This study |
| YNB567 | YNB484 | pBNB202 | Leu2 | This study |
| YNB568 | YNB484 | pBNB204 | Leu2 | This study |
| YNB611 | YNB484 | pBNB217 | Leu2 | This study |
| YNB631 | YNB484 | pBNB221 | Leu2 | This study |
| YNB795 | YNB484 | pBNB284 | Leu2 | This study |
| YNB805 | YNB484 | pBNB316 | Leu2 | This study |
| YNB858 | leu1-32 ura4-D18 his3-Δ1, ΔSPBC26H8.08c::GNL3L-FLAG KanMX6 h− | G418R | This study | |
| YNB859 | leu1-32 ura4-D18 his3-Δ1, SPBC26H8.08c::FLAG KanMX6 h− | G418R | This study | |
| YNB860 | YNB484 | pBNB395 | Leu2 | This study |
| YNB956 | YNB484 | pBNB412 | Leu2 | This study |
| YNB961 | YNB484 | pBNB494 | Leu2 | This study |
| YNB1003 | YNB484 | pBNB561 | Leu2 | This study |
| YNB1075 | YNB858 | pBNB221 | G418R, Leu2 | This study |
| YNB1076 | YNB859 | pBNB221 | G418R, Leu2 | This study |
G418R is the geneticin (G418) resistance gene.
Biochemical Methods
Standard laboratory techniques were used for extraction of DNA, total RNA, or protein.
Plasmid Constructions
Plasmids generated for this study are described in Table 2. DNA fragments used to create plasmids for this study were generated by PCR using high-fidelity enzyme Turbo Pfu (Stratagene, La Jolla, CA). All constructs were confirmed by DNA sequencing. Oligonucleotide primers are listed in Table 3.
Table 2.
Plasmids/Constructs
| Construct | Description | Markera | Reference |
|---|---|---|---|
| pCDL280 | pREP1 with GFP (S65T) | AmpR | Varadarajan et al. (2005) |
| pBNB168 | pBlueScript KS II-Ura4 | AmpR | Chen et al. (2004) |
| pBNB189 | ΔG1 (AA276-283): Nucleotides encoding AA1-275 and AA284-470 were generated by PCR using pBNB190 as template with NB380+NB379 and NB378+NB377 sets of respective primers. The two fragments were used in fusion PCR using primers NB380 and NB377. The PCR product was inserted into pCDL280 as in pBNB190. | Amp | This study |
| pBNB190 | The full-length Grn1 gene without its intron was amplified by PCR using S. pombe genomic DNA as template and primers NB380 and NB377. The resulting Grn1 was digested with SalI and NotI and inserted immediately upstream of the GFP gene in vector pCDL280. | AmpR | This study |
| pBNB202 | ΔCC (AA70-90): Nucleotides encoding AA1-69 and AA91-470 were generated by PCR using BNB190 as template with NB380+NB454 and NB453+NB377 sets of respective primers. The resulting fragments were used in fusion PCR with NB380 and NB377 as the primers. The PCR product was inserted into pCDL280 as in pBNB190. | AmpR | This study |
| pBNB203 | ΔG5 (AA164-175): Nucleotides corresponding to AA1-163 and AA176-470 were amplified by PCR using BNB190 as template with NB380+NB458 and NB457-NB377 sets of respective primers. The products were used for fusion PCR with primers NB380 and NB377. The resulting fragment was inserted in pCDL280 as in pBNB190. | AmpR | This study |
| pBNB204 | ΔRG (AA405-415): Nucleotides corresponding to AA1-404 and AA416-470 were amplified by PCR using BNB190 as template with NB380+NB456 and NB455+NB377 sets of respective primers. The PCR products were used for fusion PCR with primers NB380 and NB377. The resulting product was inserted in pCDL280 as in pBNB190. | AmpR | This study |
| pBNB217 | ΔG4 (AA195-208): Nucleotides corresponding to AA1-194 and AA209-470 were amplified by PCR using BNB190 as template with NB380+NB525 and NB524+NB377 sets of respective primers. The PCR products were used for fusion PCR with primers NB380 and NB377. The resulting product was inserted in pCDL280 as in pBNB190. | AmpR | This study |
| pBNB221 | The Rpl25a gene (SPBC106.18) was generated without its intron. The N- and C-terminal fragments were amplified by PCR using the genomic DNA as template with NB535+NB541 and NB540+NB536 sets of respective primers. The fusion product was obtained by PCR using N- and C-terminal products as template with NB535 and NB536 as primers. The resulting product was inserted into pCDL280 using SalI and NotI. | AmpR | This study |
| pBNB284 | The full-length human NGP-1 gene was amplified by PCR using a HeLa cDNA library as template, with primers NB712 and NB713. The PCR product was digested with SalI and NotI and inserted in pCDL280 similarly as in pBNB190. | AmpR | This study |
| pBNB316 | The full-length GNL3L gene was amplified by PCR using a HeLa cDNA library as template and primers NB762 and NB763. The PCR product was cloned into pCDL280 immediately upstream of GFP gene with SalI and NotI. | AmpR | This study |
| pCDNA3.1 | AmpR | Novagen | |
| pBNB335 | ΔG5-GFP fusion was amplified using pBNB203 as template and NB864+NB865 as primers. The PCR was cloned into pCDNA3.1 with KpnI and XhoI. | AmpR | This study |
| pBNB336 | ΔG4-GFP fusion gene was cloned into pCDNA3.1 following the same strategy as in pBNB335 except using pBNB217 as template for PCR. | AmpR | This study |
| pBNB337 | ΔG1-GFP fusion gene was cloned in pCDNA3.1 following the same strategy as in pBNB335 using pBNB189 as template. | AmpR | This study |
| pBNB338 | The Grn1-GFP fusion gene was cloned into pCDNA3.1 following the same strategy as in pBNB335 using pBNB190. | AmpR | This study |
| pBNB339 | ΔRG-GFP fusion gene was cloned into pCDNA3.1 following the same strategy as in pBNB335 using pBNB204. | AmpR | This study |
| pBNB340 | GFP was released from pBNB8 by BamHI/XhoI and inserted in pCDNA3.1 containing the same unique sites. | AmpR | This study |
| pBNB341 | GNL3L-GFP fusion gene was cloned into pCDNA3.1 following the same strategy as pBNB335 cloning using pBNB316 as template and NB883+NB865 as primers. | AmpR | This study |
| pBNB343 | pBlueScript KS II-KanMX6 | AmpR | Chen et al. (2004) |
| pBNB373 | Nucleotides encoding the FLAG epitope were annealed and cloned into pBNB341 with NotI and XhoI. FLAG replaces GFP in this vector. | AmpR | This study |
| pBNB376 | Primers NB907+NB908 were annealed to form a siRNA fragment specific for GNL3L at nt 1047-1065 and inserted in the pSIREN shuttle vector (BD Biosciences). | KanR | This study |
| pBNB377 | Primers NB909+NB910 were annealed to form a scrambled version of GNL3L siRNA (pBNB376) and inserted in pSIREN shuttle vector. | KanR | This study |
| pBNB378 | Oligonucleotides specific for the Luciferase gene were annealed to form a siRNA fragment and inserted in pSIREN shuttle vector. | KanR | This study |
| pBNB395 | Nucleotides encoding the FLAG epitope were annealed and cloned into pBNB316 with NotI and XhoI. FLAG replaces GFP in this vector. | AmpR | This study |
| pBNB396 | The KanMX6 cassette (BNB343) was inserted into pBNB373 using ApaI and XbaI. | AmpR | This study |
| pBNB412 | ΔG3 (AA326-329): Nucleotides encoding AA1-325 and AA330-470 were generated by PCR using BNB190 as template with NB380+NB1007 and NB1006+NB377 sets of respective primers. The two fragments were used in fusion PCR using primers NB380 and NB377. The resulting product was inserted in pCDL280 as in pBNB190. | AmpR | This study |
| pBNB417 | ΔG3-GFP fusion was cloned into pCDNA3.1 following the same strategy as in pBNB335 using pBNB412. | AmpR | This study |
| pBNB494 | The human nucleostemin (NS) gene was amplified by PCR using a HeLa cDNA library as template, with primers NB1160 and NB1161. The PCR product was cloned into pCDL280 following the same strategy as in pBNB190. | AmpR | This study |
| pBNB561 | The S.cerevisiae NUG1 gene was amplified by PCR using yeast genomic DNA as template, with primers NB1309 and NB1310. The PCR product was cloned into pCDL280 following the same strategy as in pBNB190. | AmpR | This study |
R, resistance.
Table 3.
Oligonucleotides
| Numbera | Sequence (5′-3′) |
|---|---|
| NB110F | GGTTAAAAAAGAATAATCGGTAATGTTTTTTCTCTAGACAACCAACTGTAAAATTTGTAACTACAGCATTTTTTACAATGCAACAGCTATGACCGGCTACCATTCACCCGCTCAACCCTCACTAAAGGGAAC |
| NB111R | GAAAAACCGCAACCGAAAACCAAATCCCAAAATATAAGCTCTAAGCAACAATAGCTTTTTTCGTAAGTTGAAAACTCATTGTAAAACGACGGCCCGTTCTGCCGAGCATGACGACACTATAGGGCGAATTGG |
| NB373F | GCATTGCTAAACTAAGGAAATCTTTCTAAATGTGAATATAAATTACTAATTAGCTTCAACTTTAAAAATAACGAGGGAATTCGAGCTCGTTTAAAC |
| NB375R | GTTCTTCTTTTACTCTTTTTTTCTTAAGAAATAAGTTAGAAATAGTTACGCGTGCATATACTTACTTAAGGAAACTTTGTATAGTTCATCC |
| NB377R | CATCTGCGGCCGCGGAAAATCATTAAGGTCAAA |
| NB378F | CTTACAGTCGGTGTAATTTCTGTTATTAACGCTCTT |
| NB379R | AAGAGCGTTAATAACAGAAATTACACCGACTGTAAG |
| NB380F | CATATGTCGACTATGGTTTCCTTAAAAAAAAAGAGTAAAAGAAG |
| NB453F | GATCGAAGAACAGAAGCGCGAAGACGCTGTTGATGAA |
| NB454R | TTCATCAACAGCGTCTTCGCGCTTCTGTTCTTCGATC |
| NB455F | GCTACTGATTTTTTAGTCAATATTATTCCAAATCTTAACGCTGC |
| NB456R | GCAGCGTTAAGATTTGGAATAATATTGACTAAAAAATCAGTAGC |
| NB457F | AAGTTGTTGAAGCGTCAGAAGGGACTCGTTCAAAAG |
| NB458R | CTTTTGAACGAGTCCCTTCTGACGCTTCAACAACTT |
| NB524F | GCATCTTCTGCTGAAGAATCAGAAGTACTCAACAAG |
| NB525R | CTTGTTGAGTACTTCTGATTCTTCAGCAGAAGATGC |
| NB629R | CCCAAAAAGTTAAAAGATGG |
| NB631R | TCGTTCAACACCTCATC |
| NB700R | TCGTTAGAGGTGAGACAA |
| NB702F | AGAAGTGGAAAAGGAGAC |
| NB712F | CATATGTCGACTATGGTGAAGCCCAAGTACAAAGG |
| NB713R | CATCTGCGGCCGCGGCTGCTTTTGTCTGAATTTT |
| NB762F | CATATGTCGACTATGATGAAACTTAGACACAA |
| NB763R | CATCTGCGGCCGCGGGTCACCAACACCATCATCAGC |
| NB864F | GACTCAGGTACCATGGTTTCCTTAAAAAAAAAG |
| NB865R | GACTCACTCGAGCTATTTGTATAGTTCAT |
| NB883F | GACTCAGGTACCATGATGAAACTTAGACACAA |
| NB907F | GATCCGCTATTATGGCGTCTCTGGGTTCAAGAGACCCAGAGACGCCATAATAGCTTTTTTGGTACCG |
| NB908R | AATTCGGTACCAAAAAAGCTATTATGGCGTCTCTGGGTCTCTTGAACCCAGAGACGCCATAATAGCG |
| NB909F | GATCCGCTATATTGCGGTCTGGTCGTTCAAGAGACGACCAGACCGCAATATAGCTTTTTTGGTACCG |
| NB910R | AATTCGGTACCAAAAAAGCTATATTGCGGTCTGGTCGTCTCTTGAACGACCAGACCGCAATATAGCG |
| NB963F | GGCCGCAGATGGACTACAAGGATGACGATGACAAATAAC |
| NB964R | TCGAGTTATTTGTCATCGTCATCCTTGTAGTCCATCTGC |
| NB1002F | GTTAAAAAAGAATAATCGGTAATGTTTTTTCTCTAGACAACCAACTGTAAAATTTGTAACTACAGCATTTTTTACAATGATGAAACTTAGACACAA |
| NB1004F | CGAAAAGAACTCATCTGAAGTTCAGGATACTCAAATCGTTACTGAGTGGGCCAAAGAATTTGACCTTAATGATTTTCCGCGGCCGCAGATGGACTAC |
| NB1006F | AACAAATTACGTTTGGTCATTGTTTTTCCTTCTAGT |
| NB1007R | ACTAGAAGGAAAAACAATGACCAAACGTAATTTGTT |
| NB1102F | GATATAATTAATTCAGAC |
| NB1125R | AACCGCAACCGAAAACCAAATCCCAAAATATAAGCTCTAAGCAACAATAGCTTTTTTCGTAAGTTGAAAACTCTCTAGAACTAGTGGATCTG |
| NB1149F | GACGCAGGTACCGTCGACGAGCTCCTTTATATTAAAAATTATTAATTGC |
| NB1150R | GCTATCTTTGAAATCATTAGGGATCCAT |
| NB1151F | ACTACAGCATTTTTTACAATGGTTTCCTTAAAAGCTGC |
| NB1152R | GCAGCTTTTAAGGAAACCATTGTAAAAAATGCTGTAGT |
| NB1153F | ACTACAGCATTTTTTACAATGGTTTCCTTAAAAAATCCGC |
| NB1154R | GCGGATTTTTTAAGGAAACCATTGTAAAAAATGCTGTAGT |
| NB1160F | CATATGTCGACTATGAAAAGGCCTAAGTTAAAG |
| NB1161R | CATCTGCGGCCGCGGCACATAATCTGTACTGAAGTC |
| NB1478F | TTTCGCTGCGTTCTTC |
| NB1160F | CATATGTCGACTATGAAAAGGCCTAAGTTAAAG |
| NB1161R | CATCTGCGGCCGCGGCACATAATCTGTACTGAAGTC |
| NB1309F | CATATGTCGACTATGAGAGTCAGAAAGCGCC |
| NB1310R | CATCTGCGGCCGCGGTTCCATCATTGTATCTTGATC |
F, forward; R, reverse.
Deletion of Grn1
PCR-based gene deletion of Grn1 was performed using previously described procedures (Bahler et al., 1998; Chen et al., 2004). The open reading frame (ORF) SPBC26H8.08c encodes the gene Grn1 (GTPase in Ribosomal export from the Nucleolus) expressing a putative GTPase. Briefly, the Ura4-marker cassette with SPBC26H8.08c flanking (5′ and 3′) homology regions was generated by PCR using the primers NB110 and NB111. This 2.2-kb fragment was directly used to transform the homozygous diploid YNB400 (ade6-M210/ade6-M216 leu1-32 ura4-D18 his3-Δ1, h+/h). Diploid transformants were selected on EMM-ura-ade plates. Sporulation of the heterozygous diploid and dissection of tetrads from at least 24 independent asci yielded four haploid spores/tetrad. On germination, two of the four spores from each tetrad grew extremely slowly on rich medium. Each one of these slow-growing colonies was confirmed as having the SPBC26H8.08c deletion by colony PCR using 5′ and 3′ primers flanking the gene.
Construction of Strains YNB858 and YNB859 by Gene Integration
PCR-based gene integration of GNL3L into the Grn1 locus using the KanMX6 marker was performed using previously described procedures (Bahler et al., 1998; Chen et al., 2004). Oligonucleotides representing the FLAG sequence (AGATGGACTACAAGGATGACGATGACAAATAA) were annealed and inserted into pBNB341 to replace the GFP ORF generating pBNB373. Next, the gene encoding KanMX6 (BNB343) was inserted into pBNB373 downstream of GNL3L-FLAG fusion in the reverse orientation (pBNB396). The GNL3L-FLAG KanMX6 cassette was amplified by PCR using pBNB396 as the template with the primers NB1004 (5′ end) + NB1125 (3′ end). FLAG KanMX6 cassette was amplified using the same template and 3′ end primer as for the GNL3L cassette, whereas the 5′ end primer (NB1002) corresponded to 76 nts of Grn1 ORF sequence immediately upstream of its stop codon. The integrations resulted in the replacement of wild-type Grn1 ORF with GNL3L-FLAG KanMX6 (YNB858) and the fusion of Grn1 with FLAG epitope (YNB859).
Fluorescence Microscopy
Fission yeast cells were prepared for DAPI, GFP fluorescence or indirect immunofluorescence as previously described (Balasundaram et al., 1999; Chen et al., 2004; Varadarajan et al., 2005). All epi-fluorescence microscopy were performed at 1000× magnification using a Leica DMLB microscope (Deer-field, IL) equipped with an Optronics DEI-750T coded CCD camera (Goleta, CA) with Leica Qwin proprietary software. For some experiments, samples were viewed with an upright Nikon E800 confocal microscope (Melville, NY), and images were acquired using a Nikon DXM1200 camera with Image Proplus 4.5 software (Media Cybernetics, Silver Spring, MD). Cos-7 cells in chamber culture slides (BD Biosciences, San Diego, CA) were infected with vaccinia virus vTF7-3 and transfected with GNL3L-GFP (BNB341) and Grn1-GFP (BNB338) expression plasmids using Lipofectin (Invitrogen, Carlsbad, CA). After 12 h, cells were fixed with 3% paraformaldehyde and mounted in mounting medium (Vector Laboratories, Burlingame, CA). Localization of GNL3L and Grn1p was determined by confocal microscopy. Nucleoli were revealed by immunostaining with monoclonal anti-nucleolin (Upstate Biotechnology, Charlottesville, VA). Adobe Photoshop software was used to process all image presentations (San Jose, CA).
Rpl25a Localization for Δgrn1, Grn1-FLAG, and Δgrn1::GNL3L-FLAG Strains
Strains expressing nmt1:Rpl25a:GFP were cultured in EMM medium supplemented with appropriate amino acids and 15 μM thiamine were grown to log-phase, after which the cells were washed in medium without thiamine and diluted to an OD595 0.1 in fresh medium with (nmt1 OFF) or without thiamine (nmt1 ON). Samples of cells were harvested at early log-phase (OD595 0.5-0.8), stained with DAPI, and examined for DAPI and GFP fluorescence.
Western Analysis
To analyze the expression of GFP- or FLAG-tagged Grn1p or GNL3L in yeast, 5-10 ml cultures were grown in appropriate medium and harvested at an OD595 of 0.5-1.0. Because wild-type and mutant strains grew at different rates, great care was taken to harvest strains at the same OD595. Unless otherwise specified, equal numbers of cells as judged by OD595 measurements were processed for Western blots. Proteins were extracted by resuspending cell pellets in 1% SDS phosphate-saline buffer (pH 8.0) and lysing them with an equal volume of glass beads in a bead-beater four times for 30 s at 4°C. In some instances, to concentrate protein, TCA was added to the above yeast lysate at 4°C to a final concentration of 25%. Precipitated proteins were resuspended in SDS/sample buffer containing 8 M urea and pH adjusted with 1.0 M Tris-base buffer. After separation on 12% SDS-PAGE gels, proteins were transferred to PVDF membranes (Millipore, Bedford, MA). Specific epitope-tagged proteins were visualized by their reaction either with polyclonal anti-GFP (Molecular Probes, Eugene, OR) or polyclonal anti-FLAG (Sigma, St. Louis, MO).
RNA Extraction and Northern Analysis
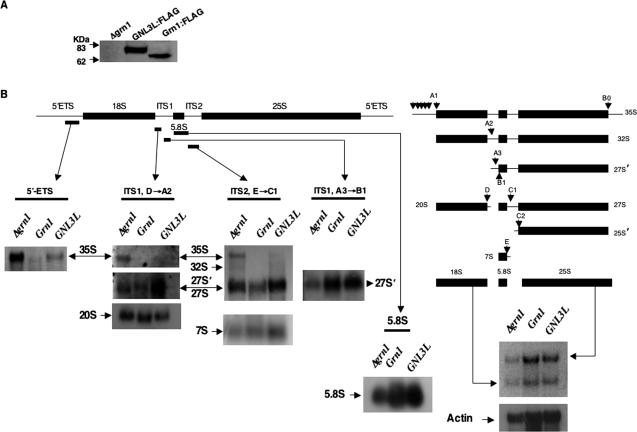
Total RNA was isolated by phenol:chloroform method following standard methods and was analyzed on a 1.2% agarose-acrylamide gel. After electrophoresis, ethidium bromide-stained-RNA bands were imaged to record 25S and 18S mature rRNA species and then transferred onto Hybond N+ membrane (Amersham Biociences, Bucks, United Kingdom). All oligonucleotide probes were more-or-less based on Good et al. (1997) and are shown in Figure 3. To identify 35S pre-rRNA species, a DIG-labeled PCR probe specific for 5′ externally transcribed spacers (ETS) was synthesized using S. pombe genomic DNA as template, DIG-DNA labeling Mix as substrate and primer sets of NB700 + NB702 (all commercial reagents for Northern analysis were from Roche, Mannheim, Germany). The resulting PCR product corresponded to the sequence -900 nucleotides upstream of the 18S rRNA ORF. The 5.8S probe corresponds to a sequence within the 5.8S ORF (NB1478), whereas the ITS1 oligonucleotide probes (NB629 and NB1102) corresponded to the D→A2 and A3→B1 cleavage sites, respectively, and the ITS2 oligonucleotide probe (NB631) corresponding to the sequence within E→C1 cleavage sites. All four were generated by 3′ end labeling using DIG-11-ddUTP according to manufacturer's instructions. Hybridization of RNA with the above probes was made with commercially available buffers. For 5′ ETS northern, hybridization was performed at 55°C and washes at 65°C, whereas for 5.8S, ITS1 and ITS2, hybridization and washes were performed at 30°C. The membrane containing RNA was hybridized with above probes and subsequently detected with anti-DIG AP fragment and developed using CSPD according to the manufacturer's instructions. A commercial actin RNA DIG-labeled probe was used to detect yeast actin mRNA levels as an internal control.
Figure 3.
Effect of Grn1p and GNL3L on processing of 35S pre-rRNA species. (A) Grn1:FLAG (YNB859) and GNL3L:FLAG (YNB858) were tested for genomically expressed FLAG-tagged Grn1p and GNL3L by Western analysis and probing with anti-FLAG. The null mutant (YNB484) was used as control. (B) Pre-rRNA and mature rRNA species were detected in the above strains by Northern hybridizational analysis. DNA probes specific for 5′ ETS, 5.8S, ITS1 or ITS2 are indicated by bars under the respective flanks. The rRNA processing pathway was adapted from Good et al. (1997). Downward pointed arrows indicate relative positions of processing sites.
Small Inhibitory RNA Knockdown
A unique sequence of GNL3L (nt1047-1065) was chosen as the target sequence for RNA interference. NB907/908 containing the target sequence, 5′-CTATTATGGCGTCTCTGGG-3′ and a scrambled version, 5′-CTATATTGCGGTCTGGTCG-3′ (NB909/910; used as a negative control) were cloned into the pSIREN vector (BD Biosciences) under the control of the human U6 promoter. A small inhibitory RNA (siRNA) targeted to the Luciferase gene was used as an additional negative control. All siRNA expressing constructs (9 μg of each) were cotransfected with pcDNA3 vector (0.9 μg) into HeLa cells. After 120 h selection in G418 (500 mg/ml), the cells were photographed and total RNA was isolated using spin minicolumns according to manufacturer's instructions. RT-PCR was performed by a standard protocol and the GNL3L-specific signal was amplified using the following primers: GNL3L forward: 5′ATGTGCGAATTCATGATGAAACTTAGACACAAAAATAAAAAGCC and GNL3L reverse: 5′CACCATGATATCCCGGATGAACTTGTCCAGGTAGAC. β-actin was amplified with the following primers: forward: 5′GGCGACGAGGCCCAGA and reverse: 5′CGATTTCCCGCTCGGC as an internal control to normalize the equal quantity of RT products used in PCR.
RESULTS
Grn1p Is a Member of a Novel G-Protein Family
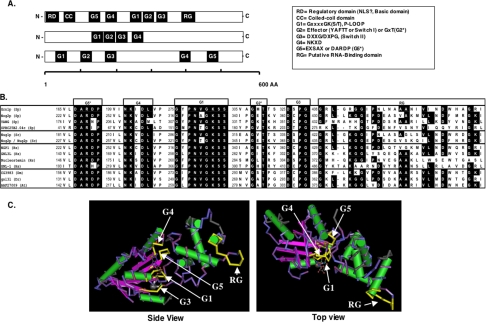
Grn1 encodes a predicted protein of 470 residues. PSORT analysis (Nakai and Horton, 1999) identifies a predicted coiled-coil domain and at least four GTPase-consensus motifs designated here as G1, G3, G4, and G5* that define a G protein (Takai et al., 2001; Leipe et al., 2002; Figure 1A). In addition, there is a putative RNA-binding domain at the C-terminus (RG-stretch). A BLASTp search (Altschul et al., 1997) of the predicted protein data banks showed that highly related sequences are found in yeast as well as in diverse eukaryotes with the “G”-domain displaying an extremely high degree of sequence homology (Figure 1B). A CD-search of conserved domain databases (CDD; Marchler-Bauer et al., 2005), Pfam (Bateman et al., 2004) and clusters of orthologous groups (COGs; Tatusov et al., 2000) revealed some very interesting aspects of this GTPase. All the diagnostic motifs of the putative GTPases described above were present albeit in altered juxtaposition to each other. Rather than the usual G1-G2-G3-G4-G5, found in the superfamily of regulatory GTP hydrolases (Takai et al., 2001; Leipe et al., 2002), the G1 motif or P-loop (GXXXXGK(S/T) is situated between the G4 (KXDL) and G3 (DXXG/DXPG) motifs as G5*-G4-G1-G2*-G3 (Figure 1A) in what has been described as a circularly permuted G-motif (Daigle et al., 2002; Leipe et al., 2002). Thus, the domain structure of the putative GTPase like other previously studied nucleolar GTPases, Nug1p (Bassler et al., 2001), Nog2p (Nug2p; Saveanu et al., 2001), NGP-1 (Racevskis et al., 1996), and NS (Tsai and McKay, 2002, 2005) conforms to that observed for the HSR1_MMR1 GTP-binding protein GNL-1, (Vernet et al., 1994) and members of the Ylqf/YawG family of GTPases (Leipe et al., 2002). The so-called G2* (YAFTT or Effector or Switch I is a less conserved motif and is not present in all GT-Pases) and G5* (EXSAX; Takai et al., 2001) appear to be ill defined in this group. However, in keeping with a previous report (Saveanu et al., 2001), the amino acid residues DARDP and GxT will be referred to as G5* and G2*, respectively. The predicted structure of the putative GT-Pase based on consensus motifs from the Ylqf/YawG family show the six-stranded β-sheet of the G-domain wherein the conserved sequence elements, G5*-G4-G1-G3 stack almost perfectly over each other (Figure 1C). Interestingly, except for a bacterial ancestor of this group of GTPases, YjeQ (Daigle et al., 2002), none of the eukaryotic members shown in Figure 1, A and B, have proven GT-Pase activity.
Figure 1.
Grn1p is member of a unique family of HSR1_MMR1-nucleolar GTPases with a highly conserved circularly permuted “G”-domain. (A) Schematic representation of three types of G-proteins illustrating the relative positions of the motifs that make up the G-domain. Top bar: GTPases with a circular permutation of the classic G-domain; middle bar: a small group of GTPases that belong to the Nog-subfamily; bottom bar represents the “classic” G-proteins as exemplified by the Ras, EF-2, and heterotrimeric G-protein families. (B) Alignment of the above circularly permuted G-domain showing individual motifs G5*, G4, G1, G2*, G3, and the putative RNA-binding domain (RG). Motifs G5* and G2* correspond to G5-like and G2-like, respectively (see text). Representatives were chosen from a broad selection of eukaryotes: S. pombe (Sp), S. cerevisiae (Sc), human (Hs), Drosophila melanogaster (Dm), Danio rerio (Dm), and Arabidopsis thaliana (At). Identical residues are shaded black (conservative substitutions are not indicated). Numbers to the left of each motif indicate the beginning of the amino acid motif. (C) 3D representation of the highly conserved circularly permuted GTPases based on the structure of Bacillus subtilis Ylqf GTPase as the consensus for the HSR1_MMR1 GTP-binding domain from the 3D-structure Entrez database, MMDB (http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml). Images were visualized and recorded using NCBI's Cn3D4.1 software. G-motifs and the RG-domain are indicated with arrows. Flat arrows represent β-sheets, whereas cylinders represent α-helices.
Grn1 Is Required for Optimal Growth of S. pombe and Localizes to the Nucleolus
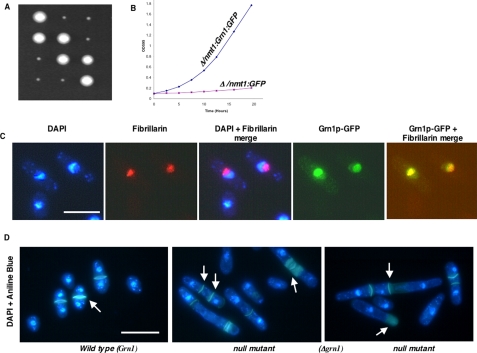
To study the function of this putative GTPase, the ORF SPBC26H8.08c was deleted and replaced with the Ura4 gene by homologous recombination using a previously described PCR-based deletion strategy (Bahler et al., 1998; Chen et al., 2004). Dissection of tetrads from at least 24 independent asci yielded two of the four spores from each tetrad that grew extremely slow on YES medium when compared with the wild-type strain at 32°C (Figure 2A) and similarly at 24 or 37°C (results unpublished data). Each one of these slow-growing colonies was confirmed as having the SPBC26H8.08c deletion. In contrast, genes encoding similar putative GTPases from yeast, Nug1 (YER006W) and Nug2/Nog2 (YNR053C; Bassler et al., 2001; Saveanu et al., 2001) were essential for viability. To investigate the subcellular localization of the Grn1p, its ORF, Grn1, was cloned as a C-terminal fusion to the green fluorescent protein (GFP) downstream from an inducible promoter, nmt1, and transformed into the null mutant. Figure 2B shows that the growth phenotype of the null mutant was rescued. To establish that Grn1p localizes to the nucleolus, we used as a nucleolar reference marker, Fibrillarin/Nop1p (Aris and Blobel, 1988; Henriquez et al., 1990). Monoclonal antibody staining detected Fibrillarin/Nop1p in a discrete region (Figure 2C, image showing red staining) that selectively excluded the DAPI-stained area of the nucleus establishing the nucleolar region (Figure 2C, DAPI + fibrillarin merge). The bulk of the Grn1p:GFP (green) signal was observed within the area colocalized by the Fibrillarin/Nop1p (red), resulting in the yellow merged signal shown in the last image of the panel in Figure 2C. However some signal was also evident in the nucleoplasm area. Our results therefore suggest that Grn1p accumulates primarily in the nucleolus in S. pombe. Because the episomally expressed gene is able to complement the growth defect in a null mutant, we confirm the involvement of this nucleolar protein in growth. In addition to Grn1, S. pombe has at least three other ORFs predicted to generate putative nuclear/nucleolar GTPases with an HSR1_MMR1-type domain (Figure 1B). We conclude the function of the protein encoded by Grn1 does not overlap directly with those of the other three putative GTPases. Our genetic data implies that the gene is not essential for viability. We did, however, observe that 20-40% of cells exhibited morphogenic aberrations represented by irregular, uneven, or overdeposition of septum material as well as septation and cell separation defects, often resulting in pseudofilamentous or multiseptated cells (Figure 2D), suggesting a failure of cytokinesis in those cells. Cultures of null mutant cells did not appear to exhibit a growth arrest. Because GTPases are known to be involved in nuclear-cytoplasmic transport (Saitoh et al., 1996; Moy and Silver, 1999; Seedorf et al., 1999; Bassler et al., 2001), we asked whether the Grn1 deletion would lead to defects in the nuclear pore complexes (NPC), nuclear import, and mRNA export. However, cultures of null mutant cells did not appear to have defects in either the NPC or nuclear-cytoplasmic transport (protein import or mRNA export; unpublished data).
Figure 2.
Grn1 is required for wild-type growth and encodes a nucleolar protein. (A) Grn1 (SPBC26H8.08c) was deleted as described in the text. Tetrad dissection yielded two fast-growing (Wild type) and two slow-growing (null mutant) colonies. (B) A null mutant expressing full-length Grn1p:GFP (YNB544) or an empty vector (YNB546) were in EMM-leu medium. Optical density (OD595) was determined at the indicated time points. (C) YNB544 (see above) was used to show the localization of Grn1p. Nuclear DNA was stained with DAPI. Nucleoli were revealed by indirect immunostaining with anti-Fibrillarin (AbCam, Cambridge, United Kingdom). Independent or merged images are indicated. (D) Wild-type (YNB483) and null mutant (YNB484) strains were stained with DAPI and Aniline blue for visualizing the nucleus and septum, respectively. Arrows indicate the septum. Bar, 10 μm.
Pre-rRNA Processing Is Impaired in Grn1-depleted Cells
Because the Saccharomyces cerevisiae GTPases Nug1p and Nug2p were linked closely with pre-rRNA processing (Bassler et al., 2001; Saveanu et al., 2001), we asked if the Δgrn1 mutant was defective in the processing of 35S pre-rRNA precursor to mature rRNA species by performing a Northern blot analysis. The yeast rDNA unit is made up of the 35S pre-rRNA operon and two nontranscribed spacers (NTS) interrupted by the 5S rRNA gene. The 35S pre-rRNA operon, flanked on either end by ETS, 5′ ETS and 3′ ETS, eventually gives rise to the mature 18S, 5.8S, and 25S rRNA species (Venema and Tollervey, 1999). The S. pombe rRNA processing pathway (Figure 3B) is similar to that of S. cerevisiae and several other eukaryotes although it may depart from the same in specific processing steps (Good et al., 1997). Probes corresponding to the 5′ ETS, 5.8S, ITS1 and ITS2 regions are indicated in Figure 3B. Our 5′ ETS probe beginning at -900 bp upstream of the 5′ end of 18S rRNA covers all the putative 5′ ETS processing sites and would thus identify all intermediates from 35S to 32S pre-rRNA (it will not detect 32S) species, whereas the ITS1 probes (D→A2) and (A3→B1) identify 35S, 32S, and 20S; and 35S, 32S, and 27S′, respectively. The ITS2 probe (E→C1) detects 35S, 32S, 27S′, 27S, and 7S species. The 27S′, 27S rRNA precursors are indistinguishable (Good et al., 1997). To detect 5.8S mature rRNA, the probe was based on a sequence within the 5.8S operon. To investigate rRNA processing and other experiments (see below) in wild-type and mutant strains, we constructed the strain YNB859 (Table 1), wherein the FLAG KanMX6 sequence was integrated at the 3′ end of Grn1 as described in Materials and Methods. YNB859 expressed a stable C-terminus tagged Grn1p:FLAG fusion protein when tested for protein expression along with the null mutant, YNB484 (Figure 3A). The results depicted in Figure 3B show the accumulation of the 35S pre-rRNA species in the null mutant (YNB484) when compared with the wild type (YNB859) with a concomitant decrease in the 25S, 18S, and 5.8S mature rRNA species. Under wild-type conditions it may not be possible to see the 35S pre-rRNA species because it is processed very rapidly (Good et al., 1997; Venema and Tollervey, 1999). The observations that mature rRNA species in the null mutant are significantly lower than that of the wild type, coupled with the increase in levels of the 35S pre-rRNA species, is suggestive of a significant inhibition or slowing down of the early pre-rRNA processing steps.
Grn1p Is Required for Nuclear Export of the Putative Ribosomal Protein Rpl25a
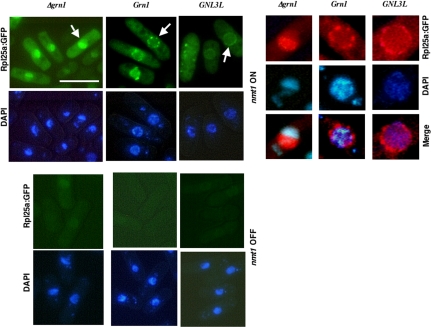
RNA export from the nucleus is linked to its proper processing and packaging into ribonucleoprotein complexes within the nucleus (Strasser and Hurt, 1999; Tschochner and Hurt, 2003). The use of functional GFP-tagged ribosomal protein reporters has greatly facilitated the elucidation of the large-subunit (Hurt et al., 1999; Stage-Zimmermann et al., 2000; Gadal et al., 2001) and small-subunit (Grandi et al., 2002; Milkereit et al., 2003) ribosome assembly and nucleolar/nuclear export pathway. L25 (Rpl25 in yeast and L23 in plant/mammal/human) is perhaps the most extensively studied and highly conserved eukaryotic ribosomal protein (Supplemental Figure S1). The S. cerevisiae RpL25:GFP, binds to pre-rRNA, assembles with 60S ribosomal subunits after its import into the nucleolus and is subsequently exported into the cytoplasm, thus allowing for monitoring of the localization of pre-60S and 60S particles by fluorescence microscopy (Hurt et al., 1999). We therefore used an in vivo assay (Hurt et al., 1999) exploiting the GFP-tagged version of the S. pombe putative ribosomal protein RpL25a encoded by the ORF SPBC106.18 that is homologous to ORF YOL127W of S. cerevisiae encoding Rpl25 (Supplemental Figure S1). We cloned the S. pombe putative Rpl25a into a vector and expressed nmt1:Rpl25a:GFP in wild-type (Grn1) and null mutant (Δgrn1) strains. When induced (nmt1 ON), Rpl25a:GFP was detected as punctate foci primarily outside the nucleolus at what appears to be the nuclear membrane of wild-type cells (Figure 4). However, in stark contrast, the null mutant consistently revealed a nucleolar accumulation of RpL25a: GFP (Figure 4, panels showing enlarged nucleus). The combination of our results regarding the impaired release of Rpl25a:GFP from the nucleolus in the Δgrn1 mutant coupled with its inability to efficiently process the 35S pre-rRNA transcript is suggestive of its involvement in early 5′-end pre-rRNA processing step(s) and in the assembly and export of Rpl25a/pre-ribosomal complexes from the nucleolus.
Figure 4.
Rpl25a localization in Δgrn1, Grn1-FLAG, and Δgrn1::GNL3L-FLAG strains. The null mutant (YNB484), Grn1:FLAG (YNB859), and GNL3L:FLAG (YNB858) were transformed with nmt1:Rpl25a:GFP (BNB221) to give YNB631, YNB1076, and YNB1075, respectively. GFP fluorescence was visually inspected in >100 cells for each of the indicated strains. Bar, 10 μm. In > 90% null mutant cells (YNB631) Rpl25a:GFP appeared inside nucleus with a significant accumulation within the nucleolus. For YNB1075 and YNB1076, >90% showed localization to the nuclear rim, with no accumulation within the nucleolus. A representative image of each strain is depicted. The top right panel depicts the GFP and DAPI images of a single nucleus (indicated by arrowhead) that were enlarged and digitally manipulated to convert the GFP-green fluorescence to red.
The Canonical G Domain and a Putative RNA-binding Domain (RG) Are Required for Grn1p Function
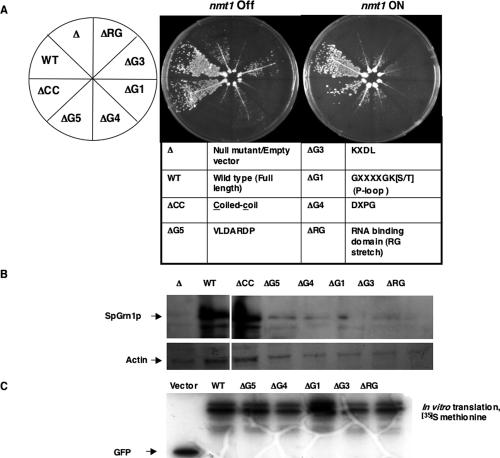
To dissect the molecular mechanism underlying Grn1p function and to assess the functional significance of the signature GTP-binding motifs and the RG domain, we tested the ability of Grn1p with deletions of those motifs/domains to complement the growth defect of the null mutant. Constructs were engineered so that C-terminal GFP fusion proteins were generated with deletions of the putative coiled-coil domain ΔCC (AA70-AA90), ΔG5* (AA164-AA175), ΔG4 (AA195-AA208), ΔG1 (AA276-AA283), ΔG3 (AA326-AA329), and the RNA binding domain ΔRG (AA405-AA415; Figure 5A). The Δgrn1 strain was transformed with plasmids bearing the above constructs (Tables 1 and 2). An nmt1 promoter drove transcription of wild-type and mutant versions of Grn1 in the absence of thiamine as described below. Independent transformants were struck for single colonies onto selective growth medium allowing for induction of gene expression. Figure 5A depicts growth of the various mutants in comparison to the wild-type and null mutant. The WT and the ΔCC mutant fully complement the null mutant. However, the ΔG5, ΔG4, ΔG1, ΔG3, and ΔRG mutants were unable to rescue the null growth defect, indicating that those domains or motifs were required for its function. We noted that even in the presence of thiamine (nmt1 turned off), WT and ΔCC grew very well. The nmt1 promoter is known to be “leaky,” and as a result, even a low expression is sufficient to rescue growth. Expression of WT and mutant GFP-tagged proteins was verified by Western analysis using anti-GFP (Figure 5B), after which the membrane was stripped and reprobed with actin antibody to visualize actin levels. Figure 5B shows that levels of ΔG5, ΔG4, ΔG1, ΔG3, and ΔRG deletion proteins are extremely low when compared with the WT or ΔCC levels of expression. Because equal amounts of cells were sampled for Western blots (Materials and Methods), the protein expression levels may in fact reflect the growth of the above strains. Actin levels are consistent with the above results. Our growth results show that deletion of any one of the ΔG- or ΔRG motifs results in a null phenotype and that the G5, G4, G1, G3, and RG motifs are equally critical for Grn1p function. Actin, here an example of a “house-keeping” protein, was used as an index of the protein levels of the cell. Actin levels in the above mutants appear to be similar to that of the null mutant. To make sure that the low levels of protein observed in these mutants was not because of defective constructs used, we detected the quantity and size of each protein by in vitro transcription-translation from a rabbit reticulocyte system. Figure 5C shows that an equivalent amount of each protein of the expected size shown was realized. We noted that in each case doublet bands were evident (including wild type) that were not present in the null mutant. Similar doublets were also observed in the Western blots in Figure 5B.
Figure 5.
The G-domain and RG-domain of Grn1p are required for growth. (A) The growth of all the indicated strains: WT (YNB544), Δgrn1 (YNB546), ΔRG (YNB568), ΔG3 (YNB956), ΔG1 (YNB545), ΔG4 (YNB611), ΔG5 (YNB566), and ΔCC (YNB567) was determined. Strains were struck for single colonies on EMM-leu plates with (nmt1 OFF) or without 15 μM thiamine (nmt1 ON). (B) The total proteins of all the strains were isolated and processed by Western using anti-GFP antibody. (C) GFP, Grn1:GFP, ΔG5-Grn1:GFP, ΔG4-Grn1:GFP, ΔG1-Grn1:GFP, ΔG3-Grn1:GFP, ΔRG-Grn1:GFP, from pBNB340, pBNB338, pBNB335, pBNB336, pBNB337, pBNB417, and pBNB339, respectively (Table 2), were transcribed and translated in the presence of l-[35S]methionine using a TNT-coupled Reticulocyte Lysate System (Promega, Madison, WI) according to the manufacturer's instructions. One microliter of each of product was analyzed on a 12% SDS-PAGE gel and exposed to x-ray film.
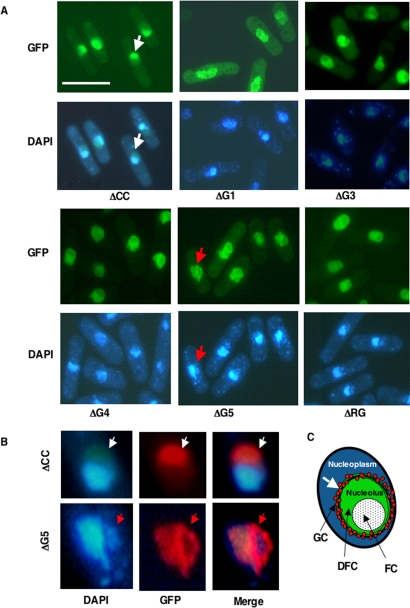
Mutations in the Functional Domains of Grn1p Alter Its Localization within the Nucleus
In Figure 2C, we established that full-length Grn1p:GFP localized to the nucleolus. We therefore wanted to know if the growth function of Grn1p and mutants observed in Figure 5A was correlated with their ability to localize to the nucleolus. GFP localization data are depicted in Figure 6A. The ΔCC mutant localized to the nucleolus like the wild type (Figure 2C), whereas ΔG1, ΔG3, ΔG4, ΔG5, and ΔRG exhibited an aggregation of GFP signal at the border of the nuclear and nucleolar regions (Figure 6, A and B). The evidence is clear from the enlarged images of the nucleus (Figure 6B) showing ΔCC (an example of wild-type nucleolar localization) and ΔG5 (a representative of the ΔG-domain and ΔRG-mutants). The ΔCC mutant exhibits GFP fluorescence evenly distributed throughout the nucleolus (area defined by DAPI exclusion), whereas in the ΔG5 mutant, the ΔG5:GFP fluorescence appears to be concentrated in a specific area rather than be evenly distributed (Figure 6B, bottom panel). This area may be consistent with the granular component (GC) region of the nucleolus (Figure 6C). We also noted significant distortion of nucleolar/nuclear morphology with the ΔG-motifs and ΔRG mutants when compared with the WT (Figure 2C) or ΔCC (Figure 6A).
Figure 6.
Effect of deletions in the G-domain and RG-domain on the localization of Grn1p. (A) Strains indicated in Figure 5A with plasmids containing GFP-tagged mutant versions of Grn1p expressed from an nmt1 inducible promoter were grown in EMM-leu medium with and without 15 μM thiamine. Only cells that were induced (nmt1 ON) are shown. Bar, 10 μm. (B). The GFP and DAPI images of a single nucleus from ΔG5 (YNB566) and ΔCC (YNB567), marked with colored arrowheads, were enlarged and digitally manipulated to convert the GFP-green fluorescence to red in order to render a sharper contrast against the blue DAPI fluorescence and thus render and delineate more vividly the nucleolar region from the extranucleolar region. (C) Cartoon shows a single nucleus with morphological subcompartments of the nucleolus. FC, fibrillar center; DFC, dense fibrillar component; GC, the granular component. White arrow indicates the accumulation of GFP-signal at the granular component on the nucleolus.
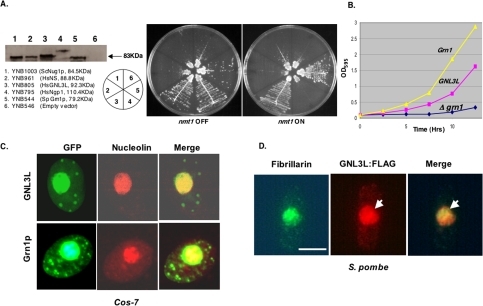
A Human Homolog FLJ10613 (GNL3L) Encoding a Hypothetical Protein Rescues the Δgrn1 Growth Defect in S. pombe
As mentioned earlier, BLASTp and CD-search searches of available databases identified several HSR1_MMR1 GTP-binding proteins similar to Grn1p (Figure 1, A-C). However, our attention was drawn to the association of these human proteins with cancer. NGP-1 (GNL2) was identified as a nucleolar breast tumor-associated autoantigen (Racevskis et al., 1996). NS, a nucleolar GTPase controlling stems cell proliferation, was found in several cancer cell lines (Tsai and McKay, 2002; Liu et al., 2004; Sijin et al., 2004; Tsai and McKay, 2005). GNL3L (also referred to as FLJ10613; Ota et al., 2004), on the other hand, is essentially an uncharacterized and hypothetical protein predicted to be a GTPase. To determine whether GNL3L, NGP-1, or NS could complement the growth defect in the S. pombe Δgrn1 mutant, we cloned their ORFs from a HeLa cell cDNA library into an inducible (nmt1) yeast expression vector and assayed growth of the yeast transformants (Figure 7A). GNL3L rescued the Δgrn1 deletion in a manner similar to that of Grn1. Similarly, the null growth phenotype is rescued when the GNL3L is expressed from the endogenous Grn1 promoter instead. For these experiments, the wild-type Grn1 ORF was replaced with GNL3L-FLAG KanMX6 (YNB858, Table 1) so the latter would be transcribed from the S. pombe Grn1 native promoter (see Materials and Methods). Therefore, strains with GNL3L-FLAG (YNB858) and Grn1::FLAG (YNB859) are isogenic. Figure 7B shows that though GNL3L-FLAG exhibited a longer lag, its growth rate became almost equal to that of Grn1. Thus, under identical endogenous promoter activities, GNL3L expression complements the Grn1 deletion. We noted that although all these proteins were expressed in fission yeast, NGP-1, NS, or the yeast ScNug1 were unable to complement SpGrn1 activity when induced. Surprisingly however, we observed that NGP-1 was able to fully complement SpGrn1 when induced very weakly (nmt1 OFF, Figure 7A) implying that nmt1-dependent overexpression of NGP-1 was toxic to cell growth. Our results suggest that these HSR1_MMR1 GTPases could have unique activities within the nucleolus/nucleus.
Figure 7.
Expression of a human gene GNL3L rescues the growth defect of the null mutant. (A) Yeast strains expressing the indicated proteins transcribed from an nmt1 promoter were tested for growth in EMM-leu medium in the presence (nmt1 OFF) and absence (nmt1 ON) of thiamine. (B) Growth of GNL3l:FLAG (YNB858) and Grn1:FLAG (YNB859) is compared with a null mutant (YNB484) in YES medium. (C) GNL3L and Grn1p colocalize with nucleolin in Cos-7 cells. Localization of GNL3L and Grn1p was determined by confocal microscopy. Nucleoli were revealed by immunostaining with anti-nucleolin. (D) S. pombe cells showing localization of GNL3L:GFP in S. pombe. Wild-type S. pombe was transformed with an expression vector containing GNL3L:FLAG (BNB395). Nucleoli were revealed by immunostaining with anti-fibrillarin and GNL3L with anti-FLAG. Independent or merged images are indicated. Bar, 4 μm.
To test whether GNL3L was also a nucleolar protein and that fission yeast Grn1p:GFP would similarly be targeted to the nucleolus in human cells, Cos-7 cells were transfected with GNL3L:GFP or Grn1p:GFP. The human nucleolar protein nucleolin was used as a marker for the nucleolus. Figure 7C shows that both putative GTPases are targeted to the mammalian nucleolus. Our genetic complementation thus identifies GNL3L as a homolog of Grn1p. Figure 7D show the localization of GNL3L:FLAG in S. pombe. Comparing the localization of fibrillarin and GNL3L (Figure 7D, see arrow), we noted the latter was not concentrated in the nucleolus to the same degree as Grn1p:GFP (compared with Figure 2C), suggesting that GNL3L and Grn1p may differ in their ability to either be targeted to, or retained by, the nucleolus.
GNL3L Functionally Complements Grn1p
Because GNL3L rescued the Δgrn1 growth defect and the GFP-tagged protein localized to the nucleolus, we wanted to know if its expression in fission yeast would 1) rescue the rRNA processing defect and prevent the accumulation of the 35S pre-rRNA and 2) rescue the Rpl25a:GFP nuclear export defect. Strains expressing GNL3L-FLAG or Grn1::FLAG from the endogenous Grn1 promoter and the null mutant were investigated for rRNA processing using 5′ ETS, 5.8S, ITS1 and ITS2 probes. Figure 3 shows that in the GNL3L-FLAG strain there is a marked reduction in accumulation of 35S pre-rRNA accompanied by a significant increase in the amounts of 5.8S, 18S, and 25S mature rRNA species when compared with the null mutant. 5′ ETS, ITS1 and ITS2 probing confirmed the reduction in accumulation of 35S pre-rRNA when GNL3L was expressed. Thus, expression of GNL3L rescues the pre-rRNA processing defect in the null mutant although it was not equivalent to the wild type. Because nuclear export of Rpl25a:GFP was blocked in the null mutant (Figure 4), we asked whether GNL3L could rescue that ribosome export defect. As shown in Figure 4, Rpl25a:GFP accumulated within the nucleoli of the grn1 null but was exported to the nuclear periphery and cytoplasm with equal efficiency in both GNL3L and Grn1p strains. Thus, GNL3L functionally complements Grn1p.
GNL3L Is Required for Proliferation of Mammalian Cells
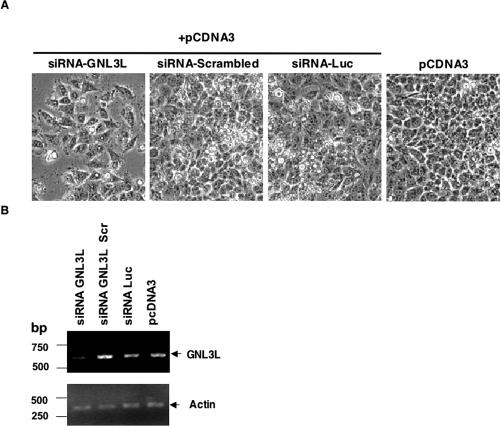
Our results imply an important and unique role for the putative GTPase Grn1p in fission yeast because it is required for wild-type growth despite the presence of three other HSR1_MMR1 putative nucleolar GTPases (see Figure 1B). In human cells, depletion or overexpression of NS causes a reduction in cell proliferation in CNS stem cells and transformed cells (Tsai and McKay, 2002). In another study, HeLa cells wherein NS expression was knocked down with siRNA could not complete DNA synthesis to pass through S phase and resulted in an increase of cells in the G0/G1 phase (Sijin et al., 2004). We therefore investigated if decreasing expression of GNL3L would result in adversely affecting cellular proliferation in HeLa cells. HeLa cells were transfected with GNL3L-siRNA, a scrambled version of the GNL3L-siRNA, Luciferase-specific siRNA, and an empty vector, pCDNA3. Cultures transfected with GNL3L-siRNA showed consistently a 30-40% decrease in the number of cells when compared with Luciferase siRNA or GNL3L nonspecific (scrambled sequence) siRNA-transfected cells used as negative controls (Figure 8A). RT-PCR analysis was performed using primers for a 600-bp GNL3L-specific product or 460-bp actin-specific product. Our analysis confirmed a reduction in GNL3L RNA in cultures transfected with GNL3L-specific siRNA when compared with cultures transfected with control siRNA (Figure 8B). Similar levels of a house-keeping gene, actin-specific PCR product from all above siRNA treatments ensured there was no bias for either RNA or the RT-PCR reaction. We demonstrate that the specific knockdown of GNL3L expression is consistent with a decrease in HeLa cell proliferation and is also consistent with the growth function of its homolog Grn1p in S. pombe.
Figure 8.
siRNA knockdown of GNL3L in HeLa cells. (A) Cultures of HeLa cells were transfected with the indicated siRNA sequences. After a 24-h transfection, the siRNA expression cells were selected in the presence of neomycin (500 μg/ml) for 120 h and photographed. (B) RT-PCR analysis of GNL3L transcript. Total RNA was isolated from the cells transfected with the indicated siRNA. RT-PCR analysis was performed as described in Materials and Methods. β-actin was used as internal control.
DISCUSSION
Role of Grn1p and GNL3L in Nucleolar/Nuclear Function
Because many aspects of ribosome biogenesis and nuclear export are conserved to a significant extent between yeast and humans (Venema and Tollervey, 1999; Tschochner and Hurt, 2003), we reasoned that should expression of GNL3L rescue the grn1 null mutant, it would complement rRNA processing and the Rpl25a export defect too. We have demonstrated in this article that was indeed the case. Our rRNA processing and Rpl25a export data suggest that Grn1p/GNL3L acts to reinstate efficient processing of the 35S pre-rRNA transcript to wild-type levels of mature RNA and the nucleolar export of the ribosomal protein Rpl25a in a grn1 null mutant. Our conclusions are in general, consistent with those of the S. cerevisiae paralogues, Nug1 and Nug2/Nog2p (Bassler et al., 2001; Saveanu et al., 2001) with one notable difference. Although Grn1p/GNL3L is required for processing of the 35S preribosomal rRNA, Nug1p's involvement in pre-rRNA cleavage is not apparent (Bassler et al., 2001), whereas Nog2p is required for processing of the 27S precursor to 25S and 5.8S rRNA (Saveanu et al., 2001). There is, however, a close and almost inseparable relationship within the nucleolus between the two processes: rRNA processing and preribosome/Rpl25 export (Venema and Tollervey, 1999; Tschochner and Hurt, 2003). Such a relationship is exemplified by other studies (also in S. cerevisiae) as well, where depletion of the ribosomal protein L25 (Rpl25) led to a severe reduction in the levels of the large subunit rRNAs with a concomitant accumulation of the 35S pre-rRNA (van Beekvelt et al., 2001). Interestingly, the same study also established a similar phenotype if binding of Rpl25 to the preribosome was abolished reiterating the notion that the assembly of Rpl25 with the 60S preribosome is required for rRNA processing (van Beekvelt et al., 2001). Furthermore, Rpl25 is incorporated into nascent pre-60S ribosomes and is known to bind both, 35S pre-rRNA as well as the 25S rRNA in yeast (el-Baradi et al., 1987; van Beekvelt et al., 2000; van Beekvelt et al., 2001). Therefore, because nuclear or nucleolar retention of Rpl25 has been observed for mutants defective in 60S ribosome biogenesis and/or nucleocytoplasmic transport, it is predicted that such a phenotype may stem from an inability to process rRNA properly resulting in ribosome maturation and transport defects (Tschochner and Hurt, 2003). As depicted in Figure 6C, mammalian nucleoli contain fibrillar centers (FC) known to house rDNA genes, surrounded by a layer called the dense fibrillar component (DFC) in which the maturation of pre-rRNA transcripts is said to take place, which in-turn is surrounded by a granular component (GC) wherein the assembly of preribosomes takes place (Carmo-Fonseca et al., 2000). Similar morphological nucleolar subcompartments are found in S. pombe (Leger-Silvestre et al., 1997) and S. cerevisiae (Trumtel et al., 2000). Though the presence of FC and DFC in yeast is currently a debated issue, the existence of a granular zone (GC?) is accepted as comprising preribosomes/ribosomes (Thiry and Lafontaine, 2005). Thus, accumulation of GFP signal at what appears to be the GC region and compared with nucleolar accumulation in a ΔCC mutant as shown in Figure 6B (here, ΔG5 is used as a representative because all of them have a similar phenotype) suggests that the absence of G- or RG-motifs may block release from the preribosome assembly from the nucleolus, thereby restricting cycling of the GTPase in and out of the nucleolar compartment or that the movement of preribosomes from the nucleolus-nucleus interface to the NPC is compromised. Because wild-type and the ΔCC mutant Grn1p proteins are nucleolar and those strains do not have any growth defect, we must conclude from Figures 5 and 6 that failure of Grn1p to either localize or relocate to the nucleolus is responsible for the null phenotype and that nucleolar localization was indeed essential for Grn1p function. We also noted a significant distortion of nucleolar structure in all our mutants except for the ΔCC and our observations are consistent with those for NS (Tsai and McKay, 2002), wherein mutations that affected its normal localization disrupted nucleolar stability and integrity. Such disruptions in nucleolar integrity may be regarded as signs of cellular stress (Olson, 2004). Studies with NS suggest that there exists a dynamic partitioning of the protein between the nucleolus and nucleoplasm (Tsai and McKay, 2002, 2005), possibly the driving force behind signal-mediated activities within nuclear subcompartments (Misteli, 2005). It would be difficult to reconcile our rRNA processing and Rpl25 data with the concept of “nucleolar retention rather than release,” as being the cornerstone of Grn1p/GNL3L activity as has been suggested for NS (Misteli, 2005) because it would be necessary for the putative GTPase to be physically present within the nucleolus to execute such functions. However, despite their similarities, the modus operandi of NS and Grn1p/GNL3L may be quite different (see below).
Grn1p and GNL3L as Members of a Novel Group of Putative Nucleolar GTPases with a Circularly Permuted G-Domain
The circularly permuted G-domain is conserved in evolution and is present in GTPases from prokaryotes (bacteria) to eukaryotes (mammals; Mans et al., 2004). Interestingly, bacterial permuted GTPases were probably involved in translation regulation and in ribosome assembly, whereas the presence of duplicated or multiple forms of permuted GTPases in eukaryotes correlates with the emergence of a nucleus/nucleolus (Mans et al., 2004). A recurrent theme emerging from recent proteomic analyses is that besides its traditional role(s) in ribosome biogenesis, the nucleolus serves as a repository for molecules involved in “nonribosomal” activities as well (Olson et al., 2000, 2002; Andersen et al., 2002; Dundr and Misteli, 2002; Scherl et al., 2002; Leung et al., 2003; Staub et al., 2004; Olson and Dundr, 2005). Could the existence of multiple nucleolar/nuclear GTPases as depicted in Figure 1B (including Grn1p or GNL3L) with a unique circularly permuted G-domain serve as a paradigm for understanding nucleolar ribosomal/nonribosomal-mediated regulation of cell growth? Of the 700-or-so human nucleolar proteins in the nucleolar protein database (http://lamondlab.com/nopdb/), four putative GTPases-NGP-1 (GNL2), GNL3L, NS, and GNL-1 possess a circularly permuted G-domain containing the so-called G5* motif [DARDP] (Figure 1B). According to AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly), these genes are expressed at 4.1 or very high (NGP-1), 3.2 or high (GNL3L), 0.8 or well expressed (NS) times the average gene (GNL-1 expression is moderate to low). We have examined three of them, NGP-1, NS, and GNL3L for their ability to complement Grn1. Despite the fact that all three were expressed in S. pombe, only GNL3L rescued the Δgrn1 mutant. The S. cerevisiae Nug1 did so only very weakly. NGP-1 complemented Grn1 at low levels of induction (nmt1 OFF) but inhibited growth at high levels of expression (nmt1 ON). Because NGP-1 localizes exclusively to the nucleolus and nucleolar organizer regions (NORs; Racevskis et al., 1996), it is possible that excess NGP-1 in fission yeast disrupts nucleolar function. However, applying the same analogy, it is intriguing that excess GNL3L is not toxic. It would be interesting to find the basis of such a dosage-dependent nucleolar response to GNL3L and NGP-1. Our work clearly demonstrates that, NS does not complement Grn1 implying that the former does not play a role in rRNA processing and/or ribosomal export. Besides GNL3L (this work) and NGP-1 (Racevskis et al., 1996), only NS appears to have been studied in some detail (Tsai and McKay, 2002; Baddoo et al., 2003; Schwartz et al., 2003; Cai et al., 2004; Liu et al., 2004; Sijin et al., 2004; Xu et al., 2004; Grasberger and Bell, 2005; Politz et al., 2005; Tsai and McKay, 2005; Yang et al., 2005). Although no well-defined molecular function has been attributed to NS, its ability to shuttle between the nuclear-nucleolar compartments in a GTP-dependent manner is a key factor in modulating growth of stem and cancer cells (Tsai and McKay, 2002; Misteli, 2005; Tsai and McKay, 2005). Additional support of our conclusion that GNL3L and NS may be acting via noninclusive or independent pathways comes from the high-resolution electron spectroscopic imaging analysis of NS localizing to the GC regions of the nucleolus having little or no rRNA, thus leading to the prediction that NS is not associated with ribosome biogenesis/rRNA processing (Politz et al., 2005). Furthermore, our finding that Grn1 is nonessential in fission yeast implies that its activity is redundant. Yet, because the null mutant has an acute growth defect despite the presence of at least three other circular permuted putative GTPases (in the fission yeast), it is appropriate to conclude that Grn1p/GNL3L has specialized functions or responds to molecular signals that are distinct from those of other similar G-domain GTPases.
Finally, although the circularly permuted and highly conserved G-domain of NS or Grn1p/GNL3L may be critical for function as Tsai and McKay (2002, 2005) and we (this report) have established, it is likely that the key to differences in their localization vis-à-vis activities lies in domain(s) with least homology (See Supplementary Figure S2). Because nuclear subcompartments are in a state of constant flux, it is anticipated that molecular components would likewise move from one subcompartment to another (Dundr and Misteli, 2001; Olson and Dundr, 2005). It is known that the nucleolar localization and nuclear shuttling of NS is dependent on its N-terminal basic domain (targeting sequence?) and regulation of the latter by its G1/GTP-binding state (Tsai and McKay, 2002, 2005). Though we have not investigated the phenomenon ourselves, GNL3L appears in the Nucleolar Proteome Database as a dynamic component of the nucleolus based on SILAC analysis (stable isotope labeling by amino acids in cell culture; http://lamondlab.com/nopdb/). Our preliminary analysis of the N-terminus of Grn1p identified a putative nuclear/nucleolar sequence similar but not identical to either GNL3L or NS, which when deleted, failed to concentrate Grn1p:GFP in the nucleolus (Supplementary Figure S3). Quite surprisingly, these mutants did not exhibit any growth defect compared with either the null mutant or G-motif/RG mutants tested. Absence of nucleolar sequestration may thus allow these cells to “override” the wild-type requirement for this particular pathway. Conversely, as envisaged for NS by Tsai and McKay (2005) and Misteli (2005), the function(s) of Grn1p are realized in shuttling between the nucleolus/nucleoplasm interface (GC) and the nucleoplasm. In the absence of any one of the G-motifs or RG, Grn1p activity was impeded at the GC/nucleolus/nuclear border as shown in Figure 6B, whereas deletion of the putative targeting sequence allowed some of the protein to be retained in the nucleoplasm where it could continue to function. Thus, should it be that differences in circularly permuted GTPase activities are related to subnucleolar/nuclear compartmentalization as exemplified by NS, their locales transient or static, must define specific metabolic areas (or activities) within the nucleus. In this context, such GTPases may help to shed light on key sites of nonribosomal or ribosomal activity in the nucleolus/nucleus and their respective roles in growth.
Supplementary Material
Acknowledgments
This study was funded by the Agency for Science, Technology and Research (A*STAR), Republic of Singapore. S.M. is supported by grants from the Department of Biotechnology, Government of India. M.R.K.S.R. is supported by a fellowship from The Council of Scientific and Industrial Research, Government of India. We gratefully acknowledge Edward Manser (Institute of Molecular and Cell Biology, Singapore) for his critical reading of the manuscript.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0848) on October 26, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung, A. K., Lam, Y. W., Steen, H., Mann, M., and Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1-11. [DOI] [PubMed] [Google Scholar]
- Aris, J. P., and Blobel, G. (1988). Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107, 17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddoo, M., Hill, K., Wilkinson, R., Gaupp, D., Hughes, C., Kopen, G. C., and Phinney, D. G. (2003). Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J. Cell. Biochem. 89, 1235-1249. [DOI] [PubMed] [Google Scholar]
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Balasundaram, D., Benedik, M. J., Morphew, M., Dang, V. D., and Levin, H. L. (1999). Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol. Cell. Biol. 19, 5768-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler, J., Grandi, P., Gadal, O., Lessmann, T., Petfalski, E., Tollervey, D., Lechner, J., and Hurt, E. (2001). Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8, 517-529. [DOI] [PubMed] [Google Scholar]
- Bateman, A. et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, R., and Pandolfi, P. P. (2003). The nucleolus: at the stem of immortality. Nat. Med. 9, 24-25. [DOI] [PubMed] [Google Scholar]
- Cai, J., Cheng, A., Luo, Y., Lu, C., Mattson, M. P., Rao, M. S., and Furukawa, K. (2004). Membrane properties of rat embryonic multipotent neural stem cells. J. Neurochem. 88, 212-226. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Mendes-Soares, L., and Campos, I. (2000). To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107-E112. [DOI] [PubMed] [Google Scholar]
- Chen, X. Q., Du, X., Liu, J., Balasubramanian, M. K., and Balasundaram, D. (2004). Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast 21, 495-509. [DOI] [PubMed] [Google Scholar]
- Daigle, D. M., Rossi, L., Berghuis, A. M., Aravind, L., Koonin, E. V., and Brown, E. D. (2002). YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41, 11109-11117. [DOI] [PubMed] [Google Scholar]
- Dundr, M., and Misteli, T. (2001). Functional architecture in the cell nucleus. Biochem. J. 356, 297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., and Misteli, T. (2002). Nucleolomics: an inventory of the nucleolus. Mol. Cell 9, 5-7. [DOI] [PubMed] [Google Scholar]
- el-Baradi, T. T., de Regt, V. C., Planta, R. J., Nierhaus, K. H., and Raue, H. A. (1987). Interaction of ribosomal proteins L25 from yeast and EL23 from E. coli with yeast 26S and mouse 28S rRNA. Biochimie 69, 939-948. [DOI] [PubMed] [Google Scholar]
- Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14, 313-318. [DOI] [PubMed] [Google Scholar]
- Gadal, O., Strauss, D., Kessl, J., Trumpower, B., Tollervey, D., and Hurt, E. (2001). Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21, 3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, L., Intine, R. V., and Nazar, R. N. (1997). The ribosomal-RNA-processing pathway in Schizosaccharomyces pombe. Eur. J. Biochem. 247, 314-321. [DOI] [PubMed] [Google Scholar]
- Grandi, P. et al. (2002). 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10, 105-115. [DOI] [PubMed] [Google Scholar]
- Grasberger, H., and Bell, G. I. (2005). Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int. J. Biochem. Cell Biol. 37, 1421-1437. [DOI] [PubMed] [Google Scholar]
- Henriquez, R., Blobel, G., and Aris, J. P. (1990). Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J. Biol. Chem. 265, 2209-2215. [PubMed] [Google Scholar]
- Hermeking, H., Lengauer, C., Polyak, K., He, T. C., Zhang, L., Thiagalingam, S., Kinzler, K. W., and Vogelstein, B. (1997). 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3-11. [DOI] [PubMed] [Google Scholar]
- Hurt, E., Hannus, S., Schmelzl, B., Lau, D., Tollervey, D., and Simos, G. (1999). A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144, 389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., and Nurse, P. (1988). Cell cycle control genes in fission yeast and mammalian cells. Trends Genet 4, 287-290. [DOI] [PubMed] [Google Scholar]
- Lee, M. G., and Nurse, P. (1987). Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327, 31-35. [DOI] [PubMed] [Google Scholar]
- Leger-Silvestre, I., Noaillac-Depeyre, J., Faubladier, M., and Gas, N. (1997). Structural and functional analysis of the nucleolus of the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 72, 13-23. [PubMed] [Google Scholar]
- Leipe, D. D., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41-72. [DOI] [PubMed] [Google Scholar]
- Leung, A. K., Andersen, J. S., Mann, M., and Lamond, A. I. (2003). Bioinformatic analysis of the nucleolus. Biochem. J. 376, 553-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. J., Cai, Z. W., Liu, Y. J., Dong, M. Y., Sun, L. Q., Hu, G. F., Wei, Y. Y., and Lao, W. D. (2004). Role of nucleostemin in growth regulation of gastric cancer, liver cancer and other malignancies. World J. Gastroenterol. 10, 1246-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans, B. J., Anantharaman, V., Aravind, L., and Koonin, E. V. (2004). Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3, 1612-1637. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A. et al. (2005). CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. (2001). Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105, 499-509. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Strauss, D., Bassler, J., Gadal, O., Kuhn, H., Schutz, S., Gas, N., Lechner, J., Hurt, E., and Tschochner, H. (2003). A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278, 4072-4081. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2005). Going in GTP cycles in the nucleolus. J. Cell Biol. 168, 177-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Moy, T. I., and Silver, P. A. (1999). Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13, 2118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34-36. [DOI] [PubMed] [Google Scholar]
- Nissan, T. A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. (2002). 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile, D. (2002). Cell proliferation. Common control for cancer, stem cells. Science 298, 1869. [DOI] [PubMed] [Google Scholar]
- Nurse, P. (1997). The Josef Steiner Lecture: CDKs and cell-cycle control in fission yeast: relevance to other eukaryotes and cancer. Int. J. Cancer 71, 707-708. [DOI] [PubMed] [Google Scholar]
- Olson, M. O. (2004). Sensing cellular stress: another new function for the nucleolus? Sci. STKE 2004, pe10. [DOI] [PubMed] [Google Scholar]
- Olson, M. O., and Dundr, M. (2005). The moving parts of the nucleolus. Histochem. Cell Biol. 123, 203-216. [DOI] [PubMed] [Google Scholar]
- Olson, M. O., Dundr, M., and Szebeni, A. (2000). The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10, 189-196. [DOI] [PubMed] [Google Scholar]
- Olson, M. O., Hingorani, K., and Szebeni, A. (2002). Conventional and non-conventional roles of the nucleolus. Int. Rev. Cytol. 219, 199-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, T. et al. (2004). Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40-45. [DOI] [PubMed] [Google Scholar]
- Politz, J. C., Polena, I., Trask, I., Bazett-Jones, D. P., and Pederson, T. (2005). A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol. Biol. Cell 16, 3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis, J., Dill, A., Stockert, R., and Fineberg, S. A. (1996). Cloning of a novel nucleolar guanosine 5′-triphosphate binding protein autoantigen from a breast tumor. Cell Growth Differ. 7, 271-280. [PubMed] [Google Scholar]
- Rustici, G., Mata, J., Kivinen, K., Lio, P., Penkett, C. J., Burns, G., Hayles, J., Brazma, A., Nurse, P., and Bahler, J. (2004). Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36, 809-817. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Cooke, C. A., Burgess, W. H., Earnshaw, W. C., and Dasso, M. (1996). Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol. Biol. Cell 7, 1319-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu, C., Bienvenu, D., Namane, A., Gleizes, P. E., Gas, N., Jacquier, A., and Fromont-Racine, M. (2001). Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20, 6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl, A., Coute, Y., Deon, C., Calle, A., Kindbeiter, K., Sanchez, J.C., Greco, A., Hochstrasser, D., and Diaz, J. J. (2002). Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, P. H., Bryant, P. J., Fuja, T. J., Su, H., O'Dowd, D. K., and Klassen, H. (2003). Isolation and characterization of neural progenitor cells from postmortem human cortex. J. Neurosci. Res. 74, 838-851. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Damelin, M., Kahana, J., Taura, T., and Silver, P. A. (1999). Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19, 1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick, K. S., and Carr, A. M. (1993). Feedback controls and G2 check-points: fission yeast as a model system. Bioessays 15, 775-782. [DOI] [PubMed] [Google Scholar]
- Sijin, L. et al. (2004). The effect of knocking-down nucleostemin gene expression on the in vitro proliferation and in vivo tumorigenesis of HeLa cells. J. Exp. Clin. Cancer Res. 23, 529-538. [PubMed] [Google Scholar]
- Stage-Zimmermann, T., Schmidt, U., and Silver, P. A. (2000). Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11, 3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, E., Fiziev, P., Rosenthal, A., and Hinzmann, B. (2004). Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays 26, 567-581. [DOI] [PubMed] [Google Scholar]
- Strasser, K., and Hurt, E. (1999). Nuclear RNA export in yeast. FEBS Lett. 452, 77-81. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153-208. [DOI] [PubMed] [Google Scholar]
- Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry, M., and Lafontaine, D. L. (2005). Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15, 194-199. [DOI] [PubMed] [Google Scholar]
- Trumtel, S., Leger-Silvestre, I., Gleizes, P. E., Teulieres, F., and Gas, N. (2000). Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell 11, 2175-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, R. Y., and McKay, R. D. (2002). A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16, 2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, R. Y., and McKay, R. D. (2005). A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168, 179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner, H., and Hurt, E. (2003). Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13, 255-263. [DOI] [PubMed] [Google Scholar]
- van Beekvelt, C. A., de Graaff-Vincent, M., Faber, A. W., van't Riet, J., Venema, J., and Raue, H. A. (2001). All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 29, 5001-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beekvelt, C. A., Kooi, E. A., de Graaff-Vincent, M., Riet, J., Venema, J., and Raue, H. A. (2000). Domain III of Saccharomyces cerevisiae 25 S ribosomal RNA: its role in binding of ribosomal protein L25 and 60 S subunit formation. J. Mol. Biol. 296, 7-17. [DOI] [PubMed] [Google Scholar]
- Varadarajan, P., Mahalingam, S., Liu, P., Ng, S. B., Gandotra, S., Dorairajoo, D. S., and Balasundaram, D. (2005). The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol. Biol. Cell 16, 1823-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., and Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261-311. [DOI] [PubMed] [Google Scholar]
- Vernet, C., Ribouchon, M. T., Chimini, G., and Pontarotti, P. (1994). Structure and evolution of a member of a new subfamily of GTP-binding proteins mapping to the human MHC class I region. Mamm. Genome 5, 100-105. [DOI] [PubMed] [Google Scholar]
- Xu, W., Qian, H., Zhu, W., Chen, Y., Shao, Q., Sun, X., Hu, J., Han, C., and Zhang, X. (2004). A novel tumor cell line cloned from mutated human embryonic bone marrow mesenchymal stem cells. Oncol. Rep. 12, 501-508. [PubMed] [Google Scholar]
- Yang, H., Zhang, J., Wu, J., and Shou, C. (2005). Preparation and characterization of monoclonal antibodies against nucleostemin, a protein that controls cell proliferation in stem cells and cancer cells. Hybridoma (Larchmt.) 24, 36-41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.