Abstract
Ciliated protozoans present several features of chromosome segregation that are unique among eukaryotes, including their maintenance of two nuclei: a germline micronucleus, which undergoes conventional mitosis and meiosis, and a somatic macronucleus that divides by an amitotic process. To study ciliate chromosome segregation, we have identified the centromeric histone gene in the Tetrahymena thermophila genome (CNA1). CNA1p specifically localizes to peripheral centromeres in the micronucleus but is absent in the macronucleus during vegetative growth. During meiotic prophase of the micronucleus, when chromosomes are stretched to twice the length of the cell, CNA1p is found localized in punctate spots throughout the length of the chromosomes. As conjugation proceeds, CNA1p appears initially diffuse, but quickly reverts to discrete dots in those nuclei destined to become micronuclei, whereas it remains diffuse and is gradually lost in developing macronuclei. In progeny of germline CNA1 knockouts, we see no defects in macronuclear division or viability of the progeny cells immediately following the knockout. However, within a few divisions, progeny show abnormal mitotic segregation of their micronucleus, with most cells eventually losing their micronucleus entirely. This study reveals a strong dependence of the germline micronucleus on centromeric histones for proper chromosome segregation.
INTRODUCTION
All eukaryotic chromosomes rely on the fidelity of chromo-some segregation machineries to faithfully transmit genetic information from one generation to the next. Centromeres are the chromosomal sites of microtubule attachment that mediate this segregation. Despite the universal requirement of faithful chromosome segregation, different eukaryotic lineages have remarkable diversity in the individual components and steps that characterize mitosis and meiosis. First, centromere size can range from just ∼125 base pairs in budding yeast, to several hundred kilobases in mammals (Henikoff et al., 2001; Malik and Henikoff, 2002). In these “complex” centromeres, defining the centromeric boundaries is technically challenging and has only been accomplished in a few select organisms (Schueler et al., 2001; Sun et al., 2003; Nagaki et al., 2004). In some holokinetic organisms like nematodes, “mini-centromeres” organize through-out the length of interphase chromosomes, coalesce during prophase, and segregate as chromosome-long centromeres during anaphase (Buchwitz et al., 1999). Second, chromosome segregation is typically accompanied by a breakdown of the nuclear membrane in most eukaryotes. However, the nuclear envelope does not break down during cell division in yeasts and ciliated protozoans (“closed mitosis”), affecting the chronology of events from kinetochore assembly to microtubule attachment. For instance, yeast kinetochores maintain microtubule attachment for the bulk of the cell cycle, whereas in plants and animals, microtubule attachment only takes place after breakdown of the nuclear envelope. Third, eukaryotes undergo several different versions of meiosis. Animals and plants undergo both a traditional “male meiosis” (a symmetric process in which all meiotic products are retained) and “female meiosis” (an asymmetric process in which only one of four meiotic products is retained). Budding yeasts only have “symmetric meioses,” ciliated proto-zoans like Tetrahymena thermophila only have “asymmetric meioses” (Martindale et al., 1982), whereas asexual organisms like bdelloid rotifers appear to lack meiosis altogether (Welch and Meselson, 2000; Mark Welch et al., 2004).
In addition to these differences, a striking specialization to emerge in the eukaryotic kingdom is the invention and copropagation of two nuclei in ciliated protozoans (Katz, 2001). Germline function is restricted to the diploid, largely transcriptionally inert micronucleus, which undergoes “closed” meiosis and mitosis (Davidson and LaFountain, 1975). In contrast, the somatic macronucleus is highly polyploid and consists of amplified, highly rearranged segments of the micronuclear genome (Woodard et al., 1972; Yao et al., 1984). The macronucleus is transcriptionally active and is responsible for most gene expression in ciliates. Chromosome segregation in the macronucleus is believed to be amitotic and not subject to the same quality checks as the micronucleus. For instance, during the amitotic division of the macronucleus, DNA is randomly segregated (Orias and Flacks, 1975) and can lead to phenotypic assortment where an allele can be lost stochastically.
Ciliated protozoans like T. thermophila thus present several unique features of chromosome segregation in the eukaryotic lineage. Key to studying chromosome segregation in ciliates is the identification of a marker that unambiguously marks centromeres at all stages of the cell cycle. The chief determinants of centromere identity are the centromeric histones, which were first identified in mammals and yeast (Palmer et al., 1987, 1991; Stoler et al., 1995), and are now known to be present in all eukaryotic genomes (Henikoff and Malik, 2002; Malik and Henikoff, 2003). Centromeric histones (hereafter referred to as CenH3s) are expressed from a single-copy gene and bear strong homology to canonical histone H3 proteins, although CenH3s evolve much more rapidly (Henikoff et al., 2001; Malik and Henikoff, 2001; Talbert et al., 2002). Blocks of CenH3-containing nucleosomes (that lack canonical H3) physically identify the centromeric chromatin (Blower et al., 2002). These blocks recruit and organize kinetochore proteins, which will form micro-tubule attachment sites at mitosis and meiosis. The identification of centromeric histones in different eukaryotic lineages has greatly facilitated the study of centromere organization and function in diverse eukaryotes. Antibodies to centromeric histones have provided a direct means to look at chromosome segregation processes in various eukaryotes (Earnshaw et al., 1986; Schatten et al., 1988; Buchwitz et al., 1999; Henikoff et al., 2000; Talbert et al., 2002; Nagaki et al., 2004). Use of centromeric histones in chromatin immunoprecipitation experiments have led to the identification of satellite DNA repeats that are truly centromeric, separate from highly similar, adjacent heterochromatic counterparts in “complex” centromeres (Vafa et al., 1999; Schueler et al., 2001; Nagaki et al., 2004). Additionally, disruptions of centromeric histone genes or depletion of their products have delineated previously inscrutable processes of kinetochore assembly (Stoler et al., 1995; Buchwitz et al., 1999; Howman et al., 2000; Blower and Karpen, 2001; Moore and Roth, 2001; Oegema et al., 2001).
Here, we identify the T. thermophila centromeric histone from the recently released genome sequence. In keeping with the nomenclature guidelines for Tetrahymena researchers (Allen, 2000), we have named this gene CNA1 (for centromeric protein A like). Using specific antibodies raised to a CNA1p peptide, we describe in detail the cytology of mitosis and meiosis in the ciliate germline micronucleus. Most features of chromosome segregation are found to be similar to other eukaryotes, although striking differences in the localization of CNA1p to the meiotic chromosomes and spindle can be seen. In addition, we describe CNA1p localization to centromeres as one of the first steps that distinguish nascent micronuclei from macronuclei. We also show that germline knockouts of CNA1 have an exclusively micronuclear effect, with many cells losing their micronucleus entirely within 8-9 cell divisions. However, macronuclei in these cells remain unaffected despite the cell lacking most micronuclear chromosomes. Thus, the centromeric histones of ciliated protozoans provide unprecedented insight into the functional diversification of their two nuclei.
MATERIALS AND METHODS
Strains
The wild-type strains CU427PBC, CU428 PBN1 and B2086 II were obtained from Peter Bruns (Cornell University, Ithaca, NY). For conjugation, cells were washed in 10 mM Tris-HCl, pH 7.5, resuspended in 10 mM Tris-HCl, pH 7.5, at 2 × 105/ml, and incubated at 30°C for 20 h and then strains of different mating types were mixed.
Evolutionary Analyses
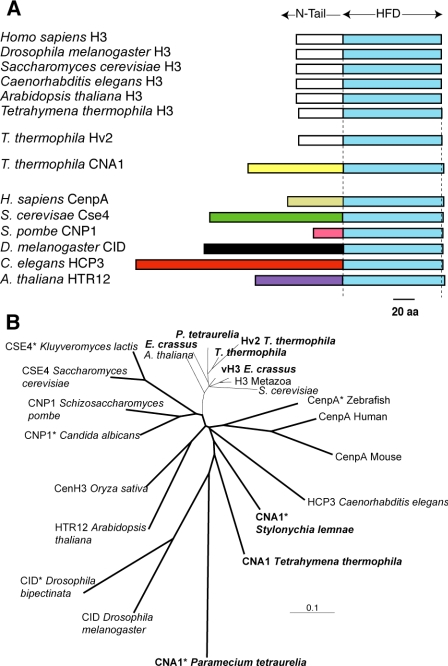
The CNA1 open reading frame was identified using TBLASTN searches against the Tetrahymena (macronucleus) genome database housed at The Institute for Genome Resources (TIGR). Preliminary sequence data for T. thermophila was obtained from The Institute for Genomic Research website at http://www.tigr.org and for Paramecium tetraurelia from http://www.geno-scope.cns.fr. The Stylonchia lemnae sequence (Bernhard, 1999) was obtained from GenBank. We used PCR amplification on genomic DNA to confirm the sequence of the gene from T. thermophila genomic DNA and RT-PCR using RNA derived from vegetatively growing cells to confirm the boundaries of the coding sequence. The predicted protein encoded by CNA1 is 187 amino acids in length and has a nonconserved N-terminal tail that cannot be aligned to canonical H3s (Figure 1A). However, the initial five amino acids (“MARKA... ”) are quite similar to canonical H3s (“MARTK... ”) as has been previously seen in the Arabidopsis CenH3 (Talbert et al., 2002), providing further support that we have correctly identified the coding boundaries of CNA1. The gene sequence has been deposited in GenBank under sequence accession no. DQ126145. The histone-fold domains of selected canonical and centromeric histones were aligned using ClustalX (Thompson et al., 1997) and a neighbor-joining phylogeny was constructed using the PAUP* suite of programs (Swofford, 2000).
Figure 1.
CNA1 is the T. thermophila centromeric histone gene. (A) Schematic comparison of selected canonical histone H3s from eukaryotes showing the conservation of the canonical histones in their histone-fold domains (HFD) and N-terminal tails. In contrast, the centromeric histones do not have a well-conserved N-terminal tail, and this evolves rapidly in both size and sequence. The three histone H3-like genes from T. thermophila are schematized, including the canonical H3, the H3.3 like replacement histone variant hv2 and the centromeric histone CNA1. (B) A neighbor-joining phylogeny of the histone-fold domains of selected canonical and centro-meric histone proteins (bold lines) is shown. Asterisks are used to indicate those proteins that, based on bioinformatic or evolutionary criteria, are thought to be the centromeric histone although this has not been directly tested. This includes putative centromeric histones from other ciliate species, P. tetraurelia and S. lemnae (ciliate histones are shown in bold lettering). In comparison to yeast, vertebrate and plant CenH3s, those from ciliates appear to be rapidly evolving.
Northern Analysis
RNA samples were collected using Qiagen RNeasy Mini Kit (Chatsworth, CA). DNA Probes were prepared from gel purified PCR products of the coding sequence of CNA1. A probe to the gene that codes the ribosomal rpL21 protein (Strausberg et al., 2002) was used as a loading control. PCR products were used for random prime labeling and unincorporated radionucleotides were removed with Amersham MicroSpin 325 columns (Piscataway, NJ). The membrane was hybridized at 65°C in Church's buffer, exposed to film, and developed.
Production of CNA1 Antibody
Previously, specific antibodies to CenH3s in Drosophila melanogaster, Caeno-rhabditis elegans, Arabidopsis thaliana, and Oryza sativa were successfully generated used polyclonal antibodies raised to a peptide at the amino-terminal end of the protein (Buchwitz et al., 1999; Henikoff et al., 2000; Talbert et al., 2002; Nagaki et al., 2004). We used a similar strategy to create an affinity-purified polyclonal antibody specific to CNA1p, using the amino-terminal peptide sequence ARKAYQPKRRSNSNQNQQC. Although this peptide sequence had lower than ideal sequence complexity, TBLASTN searches had indicated that it was likely to be very specific to CNA1p. Antibodies to CNA1p were made using the Premium Affinity protocol from Quality Controlled Biochemicals (QCB, Hopkinton, MA). The N-terminal peptide was produced and purified using HPLC, conjugated to the KLH carrier and used to immunize two rabbits. Bleeds from the rabbits were screened by ELISA assays and affinity-purified using the peptide.
To test the specificity of the antibody for CNA1p, we expressed the CNA1 coding sequence in an in vitro transcription-translation system. Because ciliates employ a slightly altered genetic code, in which codons TAA/TAG encode for glutamine (Horowitz and Gorovsky, 1985), it was necessary to change 11 TAA/TAG codons to CAA/CAG within the coding sequence before we could use the in vitro expression system. The CNA1 open reading frame was subcloned into the pCR T7/TOPO TA cloning vector. Untagged and unmodified CNA1 protein was produced from this plasmid (or no DNA mock control) using an in vitro-coupled transcription-translation kit (TnT Coupled Reticulocyte Lysate, Promega, Madison, WI).
Western Analysis
Tetrahymena cultures were collected during early log, 24 h in starvation, and at various time points during conjugation. Cell cultures were centrifuged and washed in 10 mM Tris-HCl, pH 7.5. Pellets were either frozen on dry ice or processed immediately by boiling for 5 min in ∼5 volumes of 2× SDS loading buffer (4% SDS, 160 mM Tris-HCl, pH 6.8, 20% glycerol, Bromophenol blue, 10% [vol/vol] 2-mercaptoethanol). Immediate processing of vegetatively growing Tetrahymena cells was essential to avoid protein degradation. Protein extracts were loaded onto either 12% or 4-12% gradient gels (Invitrogen, Carlsbad, CA) for SDS-PAGE analyses. Equivalent loading was monitored in all cases by Coomassie staining of duplicate gels (unpublished data). Gels were blotted onto nitrocellulose in blotting solution (20% methanol, 25 mM Tris-HCl, 192 mM glycine, pH 8.3). Filters were blocked in 10% milk, 0.3% Tween-20, 1× phosphate-buffered saline (PBS) and incubated with affinity-purified primary antibody (1: 1000 dilution in 1/10 blocking solution, 1× PBS). Blots were washed in 1× PBS, 0.3% Triton, 5 M NaCl. Secondary horseradish peroxidase-conjugated donkey anti-rabbit IgG antibody (Amer-sham) was used at 1:3000 dilution followed by washing as before and detection by ECL (Amersham). Blots were typically exposed for 2 s to reveal the modified form of CNA1 and for 1-2 min to visualize nonspecific bands as well as the unmodified CNA1 expressed in vitro, that is also present in Tetrahymena extracts.
Gene Disruption
The neomycin cassette under the control of the Metallothionein promoter, flanked by CNA1 flanking sequence was cloned into the pMNBL vector (gift from M. Gorovsky, University of Rochester, Rochester, NY). The vector was digested to release the neo fragment before biolistic transformation of Tetrahymena (Bruns and Cassidy-Hanley, 2000). The neo cassette was inserted into the CNA1 locus by homologous recombination replacing the entire CNA1 coding sequence. Transformed cultures were kept in 10 mM Tris overnight at room temperature then spun down and resuspended in SPP. After 6 h in SPP, 100 μg/ml paramomycin was added to the cultures and plated in 96-well plates. Heterozygous germline transformants were carried through genomic exclusion to create heterokaryons that are homozygous knockout in the germline nucleus and wild type in the macronucleus (Hai and Gorovsky, 1997). Insertion and deletion were verified by PCR and Southern analyses. After mating, nonmated cells were selected against using cycloheximide, as previously described (Bruns and Brussard, 1974; Orias et al., 2000).
For Southern analyses, genomic DNA was collected from wild-type and knockout progeny after >10 and 8 divisions, respectively (Blomberg et al., 1997). DNA was digested with XbaI overnight, run on 0.8% agarose gel, and transferred by pressure blotting to a nylon membrane. PCR products corresponding to the neo gene, the CNA1 coding sequence, and to the genomic region upstream of CNA1, were gel-purified using Qiagen QIAquick PCR purification, radioactively labeled as above, and hybridized in Denhardt's buffer. The membrane was exposed to film and developed.
Microscopy
Immunofluorescence using antibodies recognizing the CNA1p was adapted from previous studies (Stuart and Cole, 2000; Cole et al., 2002). Cells were grown to early log phase, ∼2 × 105/ml, 5 ml of cells was washed in 10 mM Tris, resuspended in 0.5 ml 10 mM Tris, and then 2 ml 1% paraformaldehyde in PHEM buffer was added to cells and left at room temperature for 10 min. After fixation cells were washed twice in PBS, resuspended in 2 ml PBS, and dropped onto coverslips. Coverslips were allowed to dry at room temperature and then stored at 4°C. Coverslips were allowed to warm to room temperature then set in PBS, 0.3% Tween-20 (PBS-T) for 45 min, blotted, and inverted onto 100 μl PBS-T with primary rabbit polyclonal CNA1p antibody (1:600) in a weighboat and incubated at 4°C overnight. After incubation at 4°C, coverslips were set in PBS-T for 45 min, blotted, and incubated with secondary goat anti-rabbit FITC conjugated antibody (Sigma, St. Louis, MO) at 4°C overnight in the dark. After incubation with secondary antibody, coverslips were set in PBS-T for 30 min and PBS-T with DAPI at room temperature in the dark for 10 min, PBS-T at room temperature in the dark for 10 min, and finally inverted onto 7 μl DABCO and sealed with nail polish. Slides were stored at 4°C.
Immunofluorescence of CNA1p along with α-tubulin was done as above except paclitaxel was added (final 10 μM) to fixation and PBS washing steps before dropping onto coverslips. The anti-α-tubulin antibody 12G10 developed by J. Frankel and E. M. Nelsen was obtained from the Developmental Studies Hybridoma Bank developed under the auspice of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences (Iowa City, IA). We used a 1:25 dilution of the 12G10 antibody and a 1:100 dilution of the secondary anti-mouse Texas Red-conjugated antibody (Molecular Probes, Eugene, OR; Invitrogen). The micronuclear specific linker histone δ and Pdd1p antibodies (gift from Emily Wiley and Dave Allis) and was used at a dilution of 1:100 along with the secondary goat anti-rabbit FITC-conjugated antibody at a dilution of 1:100.
Slides were viewed on an Olympus IX70 Deltavision microscope (Applied Precision, Issaquah, WA) with a 60× PlanApo 1.40 numerical aperture oil immersion lens. Images were collected with a Photometrics CH350 digital camera (Tucson, AZ), deconvolved using the Softworx program (Applied Precision), and processed using Photoshop (Adobe Systems, San Jose, CA).
RESULTS
Identification of CNA1
All eukaryotic genomes sequenced to date have a single variant histone H3 gene that encodes the centromeric histone (CenH3) protein (Malik and Henikoff, 2003). Bioinformatic criteria refined by using yeast, animal, and plant CenH3s (Malik and Henikoff, 2003) were used to screen the histone H3 homologues that were identified in the T. thermophila genome data-base. This allowed the unambiguous identification of a single gene, CNA1, which had all the hallmarks of being a CenH3 (Figure 1) in spite of the fact that ciliate histones appear to evolve more rapidly (Bernhard, 1999; Katz et al., 2004). The histone-fold domain of CNA1p is only ∼50% identical to other histone H3 variants in Tetrahymena, and like other CenH3 proteins, is evolving more rapidly than canonical H3 proteins (Figure 1B). Using TBLASTN searches, we have also obtained the sequence of the histone-fold domain from CenH3s of two other ciliates, P. tetraurelia and S. lemnae (Bernhard, 1999) to look at the evolution of CenH3s in ciliates. From this cursory analysis, the ciliate CNA1 proteins appear to be evolving more rapidly than most CenH3s, on par with what has been seen previously for the rapidly evolving Drosophila CID proteins (Malik and Henikoff, 2001; Malik et al., 2002).
CNA1 Expression
Northern analysis was used to determine the expression of CNA1 during vegetative growth and conjugation. Expression of CNA1 is not detectable in either growing cells, where only a small fraction of cells are dividing at any given moment, or in cells starved for 24 h. However, when starved cells of different mating types were mixed together to induce conjugation synchronously, we found that CNA1 was strongly expressed (Figure 2A). At 4 h in conjugation, at the time of meiosis, a strong band of ∼1 kb is apparent but expression steadily decreases by 6 h in conjugation, and is just detectable by 16 h in conjugation (Figure 2A).
Figure 2.
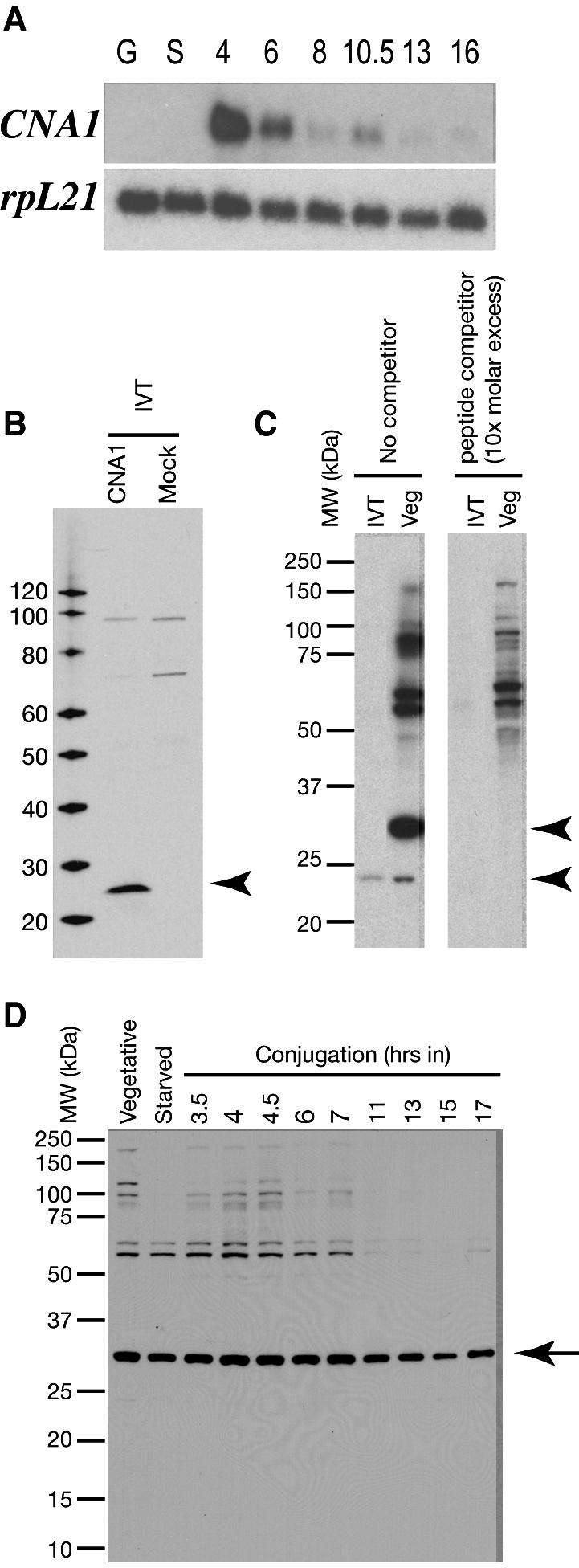
CNA1 expression. (A) On a Northern blot probed with the CNA1 coding sequence there is no detectable transcript during vegetative growth (G) or starvation (S) but high levels of expression are seen during meiosis (4 h) in conjugation. Expression continues to decrease as conjugation progresses. The rpL21 gene was used as a loading control. (B) Western blot shows that the polyclonal antibody raised to the CNA1 peptide specifically recognizes in vitro-translated (IVT) CNA1p, but no band is detected in the mock expression. (C) The CNA1 antibody recognizes two bands in vegetatively growing T. thermophila cells. The lower band corresponds to the unmodified protein as seen in the in vitro expression and can only be visualized by overexposure, whereas the predominant upper band is ∼5 kDa higher and corresponds to an unknown posttranslationally modified form of CNA1p. Neither band is detected when the antibody staining is competed with a 10-fold molar excess of peptide competitor. (D) CNA1p levels as detected by the Western blot appear stable in vegetative and starved cells, and during conjugation, in stark contrast to the Northern analysis in A.
A polyclonal rabbit ant-CNA1p antibody was generated and tested against in vitro-expressed CNA1p. Western blots show a single band at the expected ∼22 kDa from in vitro expressed CNA1p, but no bands in the mock in vitro expression (Figure 2B). We tested the anti-CNA1p antibody against protein extracts from Tetrahymena during vegetative growth (Figure 2C). In addition to the 22-kDa band that comigrates with the in vitro expressed protein, the antibody also recognizes a very prominent band ∼5 kDa larger than the expected size in all extracts with high specificity. The unmodified form of CNA1p is readily visible with an overexposure. However, the bulk of the CNA1p appears to be posttrans-lationally modified. The nature of this modification is un-known but posttranslational modifications have been previously seen in practically all CenH3s in Western analyses (Zeitlin et al., 2001; Talbert et al., 2002; Collins et al., 2004; Sullivan and Karpen, 2004). We could show that both these bands correspond to the CNA1p protein because their recognition was specifically outcompeted by the original peptide to which the antibody was generated, but nonspecific bands of higher molecular weight were not affected (Figure 2C). Thus, the polyclonal antibody has high specificity for the CNA1p protein. We then used this antibody to screen extracts of T. thermophila from vegetative growth, starvation, and conjugation in a Western blot (Figure 2D). Although we had expected to see a large increase in CNA1p during conjugation based on the Northern analysis (Figure 2A), we could detect no appreciable differences in standing levels of CNA1p. This implies that most of the increased transcription seen at the onset of meiosis does not lead to increased steady state protein levels, either due to translational or posttranslational control. Rigorous posttranslational control of CenH3 levels has been previously reported in the case of the budding yeast Cse4 (Collins et al., 2004; Pearson et al., 2004).
CNA1p Localization during Mitosis
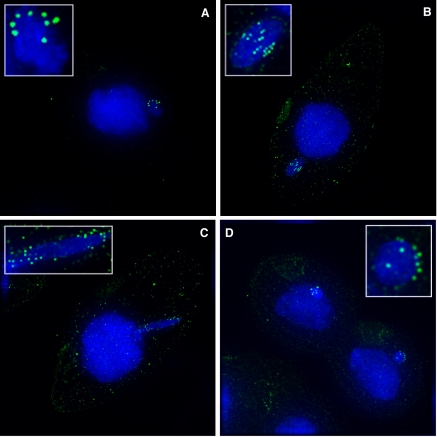
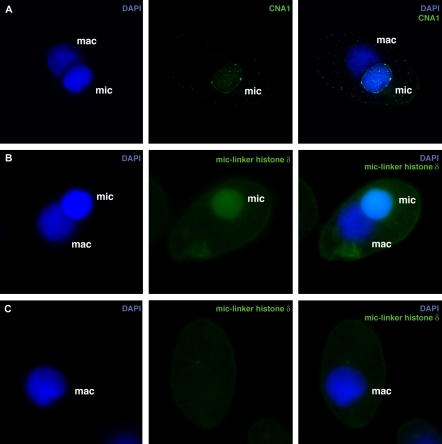
During vegetative growth, the germline micronucleus divides by mitosis, whereas the somatic macronucleus is believed to divide by an amitotic process that leads to random segregation of the macronuclear chromosomes. This random segregation of macronuclear chromosomes (Orias and Flacks, 1975) had led to the assumption that macronuclei do not contain centromeres. However, this was an indirect conclusion because of the lack of centromeric markers or centromeric satellite sequences. Localization of CNA1p gave us the first opportunity to verify this assumption. Using the antibody, we could show that CNA1p staining is completely absent from the somatic macronucleus (Figure 3A), confirming the absence of macronuclear centromeres.
Figure 3.
CNA1p localization during mitosis. (A) Immunofluorescence using CNA1p antibody during mitosis shows CNA1p is found only at the centromeres in the micronucleus. During interphase, the centromeres are clustered and peripheral. (B) Centromeres remain peripheral during metaphase when pairs of centromeres align along the middle of the chromatin. (C) The centromeres are on the leading edge of segregating chromosomes in anaphase. (D) By telophase the centromeres are peripheral again. CNA1p staining is shown in green, whereas DNA is stained with DAPI (blue). A weak background staining of basal bodies is seen with the CNA1 antibody, which occasionally outline the cellular boundaries.
CNA1p localizes to peripheral, clustered dots that mark the centromeres around the germline micronucleus during interphase. T. thermophila has five pairs of metacentric chromosomes in the micronucleus and we never see more than 10 “dots.” When micronuclei undergo closed mitosis (no nuclear envelope breakdown), CNA1p staining identifies pairs of centromeres that remain peripheral, aligning in a “ring” during metaphase (Figure 3B). Centromere pairs separate as anaphase progresses, and centromeres can be seen on the leading edge of the DNA (Figure 3C). By telophase and later, the centromeres have returned to a random, peripheral position with respect to the nuclear envelope (Figure 3D). Our results with the CNA1p antibody are in agreement with previous results based largely on electron microscopy (LaFountain and Davidson, 1979, 1980). The absence of a suitable centromere marker and lack of obvious chromosome morphology in mitosis had previously precluded the correct assignment of mitotic stages in Tetrahymena. The availability of the CNA1p antibody and assignment of centromere positions will now allow another means to reevaluate existing mutants to determine the exact stage of mitotic arrest (Kirk et al., 1997; Petcherskaia et al., 2003).
CNA1p Localization during Meiosis
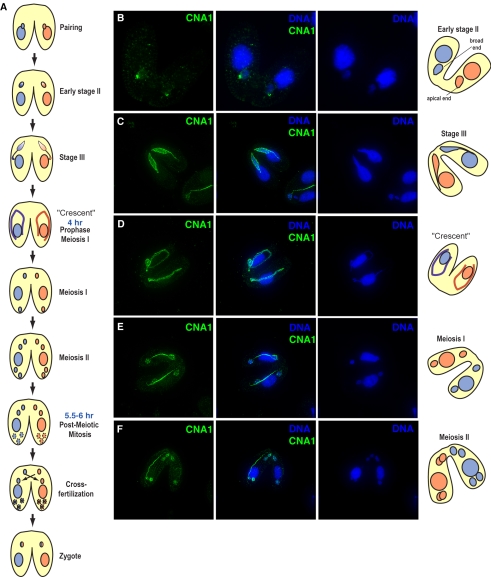
In Tetrahymena, conjugation is an extremely dynamic process involving (sequentially) nuclear elongation, meiosis, nuclear selection, and two rounds of postzygotic mitosis followed by genomic rearrangement (meiosis and fertilization are schematized in Figure 4A; Martindale et al., 1982). Unlike mitosis during vegetative growth, we found that localization of CNA1p during conjugation is highly dynamic. Soon after pairing of cells of opposite mating type, early meiotic prophase is distinguished by the micronucleus taking on a crescent shape that is almost twice the length of the cell. Early in crescent formation the micronucleus moves away from the macronucleus and is elongated to an egg shape, with the centromeres clustered at the apical end of the micronucleus (Figure 4B). Centromere location can be seen precisely coincident with microtubules, suggesting microtubule anchoring of centromeres determines their position. Previous work has shown that telomeric repeats cluster at the broad end of the micronucleus (Loidl and Scherthan, 2004), implying that centromeres and telomeres are on opposite ends at this stage of meiosis. During full crescent phase CNA1p is seen in a punctate pattern throughout the length of the crescent (Figure 4, C and D) and this pattern remains so throughout meiosis (Figure 4, C-G). During telophase of meiosis I and II, CNA1p localization is diffuse throughout the DNA as well as punctate along the spindle (Figure 4, E and F).
Figure 4.
Early events of conjugation. (A) Conjugation in Tetrahymena is schematized to highlight the major events starting from pairing of cells of different mating types (shown as blue or red) to the formation of the zygote. The diploid micronucleus undergoes meiosis, and one of the four haploid meiotic products is chosen, whereas the other three move to the bottom of the cell and are degraded. The chosen meiotic product divides mitotically and the two cells exchange one haploid nucleus each and fuse to give rise to the zygote (shown as a chimeric red/blue hereafter). (B) During prophase, the micronucleus goes through an egg shape (early stage 2). At this stage, centromeres are clearly visible at the apical end of the nucleus (telomeres have been shown to localize to the basal end at this stage). Micronuclei are then gradually stretched into stage III (C) and eventually the “crescent” shape (D). As the crescent elongates, CNA1p localization can be seen throughout the length of the crescent. (E and F) As meiosis progresses CNA1p remains punctate throughout the micronuclei and also localizes to the meiotic spindle. Distinct centromeres are visible in late anaphase of meiosis I and II. (CNA1p, green; DNA-DAPI, blue). Tubulin staining at these stages of meiosis are presented in Supplementary Information.
The localization of CNA1p during the closed meiosis of Tetrahymena is remarkable in two respects. The first novel feature of CNA1p deposition during meiosis is how strongly CNA1p appears to be “mislocalized” throughout the DNA. Such mislocalization has only been previously achieved by means of strong overexpression (Ahmad and Henikoff, 2002; Jager et al., 2005) or by eliminating turnover of the CenH3 (Collins et al., 2004). This deposition appears to be less stable than at centromeres, however, because the CNA1p is quickly displaced from noncentromeric chromatin as the “stretched” crescent chromosomes revert to normal in telophase. The second novel feature of CNA1p localization is that during meiosis, the CNA1 protein appears to move to the meiotic spindle during anaphase (Figure 4, E-G). Although other kinetochore proteins have been previously found as “passenger proteins” on the spindle (Wang, 2001; Vagnarelli and Earnshaw, 2004), this represents the first case where a centromeric histone has been found as a passenger protein on the meiotic spindle. This localization is not an artifact introduced by the antibody because we could see a similar pattern using a GFP-tagged CNA1 (gift from B. Cui and M. A. Gorovsky). The localization is also completely coincident with microtubules as stained by α-tubulin (tubulin staining for all stages is provided in Supplementary Material). We cannot formally rule out the possibility that there is a small amount of DNA remaining in the area between the divided nuclei that was not detectable by DAPI staining. However, the pattern persists up to nuclear separation when DNA segregation is complete, making this a highly unlikely possibility.
CNA1p Distinguishes Developing Micronuclei and Macronuclei in Late Conjugation
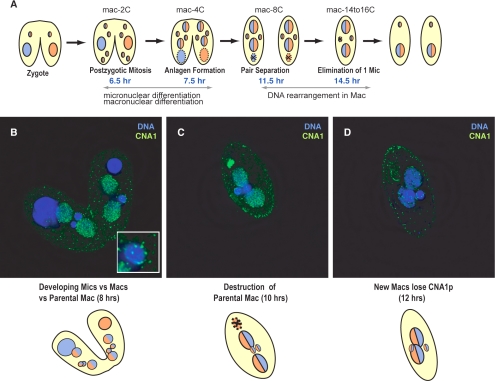
The completion of meiosis sets up a cascade of events that leads up to the formation and diversification of the germline micronucleus and somatic macronucleus (Figures 4A and 5A). One of four meiotic products is selected as a gamete and undergoes one round of mitosis. The paired cells exchange gametic nuclei and then the two gametic nuclei, one from each parent cell, fuse forming the diploid zygotic nucleus. After formation of the zygote, the new zygotic micro-nucleus undergoes two rounds of mitosis, producing four new diploid nuclei per cell. At this stage, at 8 h in conjugation there are five nuclei per cell: one parental macronucleus and four zygotic nuclei (Figure 5A). The parental macronucleus moves to the bottom of the cell and eventually goes through apoptotic degradation.
Figure 5.
CNA1 localization during late events in conjugation. (A) Conjugation events leading from zygote formation to progeny cells are schematized. The zygotic nucleus undergoes two rounds of mitosis producing four nuclei (6.5 h), two become micronuclei, whereas two become developing macronuclei (anlagen) and undergo genomic rearrangement and endoduplication. The active parental macronucleus remains in the middle of each cell until the new zygotic macronuclei become active, at ∼7.5 h, and then the parental macronucleus moves to the bottom of the cell and undergoes apoptotic degredation. (B) Eight hours into conjugation CNA1p localization reverts to the peripheral centromeres in the developing micronuclei (inset) but remains diffuse throughout the developing macronuclei. At this time cells remained paired, the parental or old macronucleus has migrated to the bottom of the cell and is beginning apoptotic degradation. (C) By 10 h, CNA1p is concentrated in the old macronucleus that has little DNA remaining. CNA1 localization is decreased in the developing macronuclei, whereas it remains constant at the centromeres in the micronuclei. (D) At the time of genomic rearrangement CNA1p is largely absent from the cell except at the centromeres in the micronuclei, similar to vegetative cells (Figure 3A) (CNA1p, green; DNA-DAPI, blue)
Of the four genetically identical zygotic nuclei, two become somatic macronuclei, whereas the other two remain germline micronuclei (Wenkert and Allis, 1984). Strikingly, at or very soon after this decision point, the CNA1p localization in the two nuclei destined to become micronuclei reverts to the localization pattern seen in interphase micro-nuclei. In these two nuclei, CNA1p is seen in fewer than 10 dots at the periphery of the nucleus (Figure 5B). However, in the nuclei destined to become macronuclei, CNA1p remains punctate throughout the nucleus, steadily decreasing as conjugation progresses until the time of genomic rearrangement when the staining disappears entirely within the developing macronuclei (Figure 5, C and D). Meanwhile, bright staining of CNA1p can be seen late in the apoptotic degradation of the parental macronucleus, when most of the DNA has been degraded (Figure 5C). It is possible that this intense CNA1p staining corresponds to CNA1p protein that moves from the developing macronucleus to the old, parental macronucleus to be degraded. Previous reports have shown that the Pdd1p protein that is involved in DNA elimination in the developing macronucleus, is similarly localized to the parental macronucleus before being degraded (Madireddi et al., 1996).
Thus, CNA1p joins a select list of early markers that can help distinguish between the two fates of zygotic nuclei in Tetrahymena. Previously, events that were known to contribute to this early differentiation included the removal of the micronuclear-specific linker histone, the appearance of a macronuclear histone H2A variant, and acetylation of histone H4 in those nuclei destined to become macronuclei (Chicoine et al., 1985; Lin et al., 1989; Pfeffer et al., 1989). All these events involved changes specific to those nuclei destined to become macronuclei, from the common stage. These results suggested that micronuclear fate was the default status of these zygotic nuclei, which was overwritten by epigenetic events only in macronuclei. Our results show that CNA1p localization is specifically altered only in those nuclei destined to become micronuclei, implying that micronuclei do not necessarily represent the default fate of zygotic nuclei. Thus, altered CNA1p localization is the first known identifying marker for micronuclear fate.
Knockouts of CNA1 Have Micronuclear Chromosome Segregation Defects
We next wanted to investigate the role of CNA1 in mediating chromosome segregation, by making gene knockouts. There are two means of introducing knockouts in the binucleate Tetrahymena, in either the somatic macronucleus or the germline micronucleus. Gene knockout in the somatic macronucleus affects the expression of the gene while leaving it unchanged in the germline. Because the somatic macronucleus contains ∼50 copies of each macronuclear chromosome, we replaced one copy of the CNA1 gene with a neomycin cassette. During division of the macronucleus the macronuclear chromosomes are segregated randomly which can lead to loss of one allele of a particular gene (Orias and Flacks, 1975; Merriam and Bruns, 1988). Growing cells in increasing amounts of the drug paramomycin will select for cells with more copies of the neomycin cassette and fewer wild-type copies of the CNA1 gene. For nonessential genes this “phenotypic assortment” can lead to complete loss of the wild-type gene. Despite several attempts, we could not obtain greater than 50% replacement of the CNA1 knockout. Even at this 50% replacement stage, there were defects in chromosome segregation during mitosis and cells would eventually cease division. When these “knockdown” cells were stained with DAPI, they showed lagging DNA during mitosis and many of the cells had smaller micronuclei than wild type (unpublished data). Thus the CNA1 gene, like CenH3s in other eukaryotes (Stoler et al., 1995; Buchwitz et al., 1999; Howman et al., 2000; Blower and Karpen, 2001), is essential for Tetrahymena viability. This further suggests that levels of CNA1 expression play a critical role in chromosome segregation, as we are seeing defects manifest in what is the macronuclear equivalent of a heterozygous state.
Because we could not accomplish a complete somatic knockout of the CNA1 gene, we decided to replace the CNA1 gene in the germline nucleus. The advantage of this approach is that because the germline nucleus is silent during vegetative growth, crosses can be engineered to create homozygous germline knockouts. Using biolistic transformation, we replaced the entire CNA1 coding region with a neomycin cassette. Two clones that segregated the neomycin cassette were taken through a mating of genomic exclusion where each clone is mated with a cell that does not contain a functional germline micronucleus leading to whole genome homozygotes (Kahn et al., 1993; Cassidy-Hanley et al., 1997; Hai and Gorovsky, 1997). These cells are phenotypically wild type, because gene expression is only from the wild-type macronucleus, whereas the homozygous knockout micronucleus is transcriptionally silent. This facilitates the study of the CNA1 knockout by mating two homozygous knockouts of different mating types. By mating two homozygous knockouts of different mating types we were able to observe the phenotype of the resulting CNA1 null progeny (hereafter simply referred to as ΔCNA1).
Conjugation was found to proceed normally in mating of two homozygous knockouts. No delay was detected in early events of meiosis and nuclear differentiation and markers of DNA elimination were identical to wild type and production of exconjugants was similar to a wild-type mating (unpublished data). In addition, the pattern of CNA1p localization throughout conjugation was identical in the knockout mating compared with wild type. Thus, any defects that arise from the CNA1 knockout only manifest in the cell divisions that follow conjugation, when all zygotic transcription relies solely on the new knockout macronucleus.
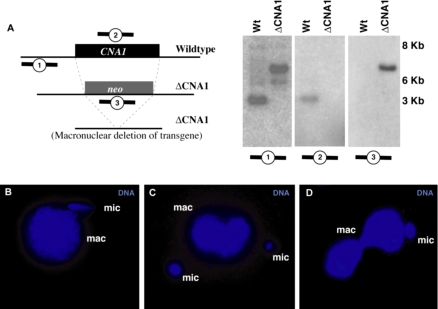
We ascertained whether the CNA1 gene had been completely replaced with the neo cassette in the progeny of the knockout matings (ΔCNA1) using Southern blots. After 2 d in growth media, genomic DNA was collected from progeny of both wild-type cells and those from knockout matings. Southern analysis was carried out to ascertain that the CNA1 gene had been completely replaced (Figure 6A). When we used a probe to the CNA1 gene, a band of the expected size was seen only in the wild-type sample. Similarly, a probe for the neo coding sequence was only seen in the knockout progeny. Finally, a probe to the genomic region upstream of the CNA1 gene confirmed that the CNA1 gene had been completely replaced with the neo cassette.
Figure 6.
Germline knockouts of CNA1 show micronuclear chromosome segregation defects. (A) Southern blots confirming the complete replacement of the CNA1 coding region by the neo cassette in ΔCNA1 cells. Genomic DNA from wild-type and ΔCNA1 cells was digested with XbaI and probed with the upstream genomic region (1), the CNA1 coding sequence (2), or the neo gene (3). The upstream probe detects the wild-type band of 4.1 kb only in wild type. In the ΔCNA1 cells the wild-type band is absent, the most prominent band is the expected 6.2-kb band for the knockout and a small amount of transgene deletion at 5.4 kb was apparent (Yao et al., 2003). The CNA1 gene was absent in the ΔCNA1 cells (middle panel), whereas the neo gene was only detectable in the ΔCNA1 cells (right panel). (B and D) A variety of micronuclear chromosome segregation defects are seen in ΔCNA1 cells, including asymmetric segregation of micronuclear DNA in early anaphase (B) and telophase (C). This asymmetry can lead to cells where the macronuclei divide even though the micronucleus has not (D), whereas typically macronuclear division occurs much later than micronuclear division in wild-type cells.
ΔCNA1 cells initially grew like wild-type cells, but they ceased to grow after 2 d. To further investigate the nature of defects in the ΔCNA1 cells, single conjugating pairs were cloned into drops and progeny scored for cytological defects using antibodies to CNA1p and a micronuclear-specific linker histone δ (Sweet et al., 1996). We found that the progeny retained parental CNA1p for at least eight divisions. The ΔCNA1 progeny cells completed ∼3-4 divisions normally but subsequent divisions showed unequal DNA segregation during mitosis, showing a range of defects that are represented in Figure 6, B-D. ΔCNA1 cells ceased dividing after ∼10 divisions. This limited number of divisions after a depletion of centromeric histones is in remarkable agreement with results from a number of diverse eukaryotes (Collins et al., 2004; Pearson et al., 2004; Regnier et al., 2005). By the time the ΔCNA1 cells stopped dividing, a few cells had micronuclei that were now the size of a macronucleus (Figure 7, A and B), whereas 70% of the cells did not have a detectable micronucleus (Figure 7C). The large micronuclei were found to stain with a micronuclear specific histone and still retain CNA1p although the localization patterns were clearly aberrant in being more peripheral and having more than ten dots (Figure 7, A and B). Cells without a detectable micronucleus (represented in Figure 7C) arise because micronuclear chromosome segregation is blocked but cytokinesis proceeds normally resulting in one daughter cell that does not inherit any micronuclear chromosomes. From these studies, we conclude that CNA1 is essential for normal micronuclear DNA segregation during mitosis. Such aberrant micronuclear chromosome segregation patterns have not been previously observed in T. thermophila (Shen et al., 1995).
Figure 7.
Germline knockouts of CNA1 lead to cells with either large micronuclei or no micronuclei. Micronuclear chromosome segregation defects in ΔCNA1 cells lead to cells that either have abnormally large micronuclei (A and B) or have completely lost their micronucleus (C). Large micronuclei have aberrant CNA1p localization (A). (B) In cells with two large nuclei, large micronuclei were unambiguously identified using antibodies to the micronuclear-specific linker histone δ (Sweet et al., 1996). (C) In cells lacking a cytologically visible micronucleus, no staining with the linker histone δ or CNA1p (unpublished data) was seen. (CNA1p or Linker histone δ, green; DNA-DAPI, blue)
Despite these dramatic defects in micronuclear division, we were not able to observe any obvious defects, such as lagging DNA or macronuclei being cut by the cleavage furrow of cytokinesis, in the somatic macronucleus in these cells. Previously, mutations that disturb macronuclear division have been shown to result in large extrusion bodies that contain macronuclear DNA (Wiley et al., 2005). We found no indication of extrusion bodies in the ΔCNA1 cells. Together, these results provide the strongest evidence to date, that macronuclear chromosome segregation in ciliates proceeds independent of any centromere function.
DISCUSSION
Centromeres in Closed Mitosis
Our identification of CNA1 as the T. thermophila centromeric histone has provided us with a unique tool to study centromeres in one of the earliest branches of eukaryotes. It affords us an unprecedented visualization of chromosome segregation in the ciliate micronucleus, a process that has been traditionally difficult to study because it is not accompanied by a nuclear envelope breakdown. Centromeres in the micronucleus are found to have a peripheral localization (as opposed to randomly dispersed in the interior of the nucleus) at practically all stages of the cell cycle (Figure 3). This localization pattern is similar to that of other organisms with closed mitoses, like budding and fission yeasts (Funabiki et al., 1993; Goshima and Yanagida, 2000; He et al., 2001; Pearson et al., 2004) but not similar to organisms known to have an “open” mitosis, like Drosophila and humans (Henikoff et al., 2000). In yeasts, it is believed that this peripheral localization is on account of the centromere attachment to the nuclear envelope by means of nuclear microtubules (Funabiki et al., 1993; Goshima and Yanagida, 2000; He et al., 2001; Pearson et al., 2004). Similar attachments between the kinetochores and nuclear envelope have been previously proposed for the Tetrahymena kinetochores based on electron microscopy (LaFountain and Davidson, 1979). Microtubule-based anchoring of kinetochores should not be a necessary prerequisite for closed chromosome segregation. Nonetheless, our results suggest that diverse eukaryotic lineages with closed mitosis appear to have adopted the same strategy of anchoring kinetochores to the nuclear membrane. Presumably, this obviates the requirement of microtubule “capture” and makes the process more efficient.
CenH3 Deposition Outside Centromeres
The intriguing localization of CNA1p during meiotic prophase also provides support to hypotheses concerning the determinants of CenH3 deposition. Previous work has highlighted the unique nucleosome and histone modifications present at centromeric regions. But the core issue of what targets CenH3s to centromeres has been a mystery. Even under normal expression levels, CenH3s can target euchromatin in addition to centromeres. In mammalian cells, CenH3 overexpression can lead to localization in euchromatin but this does not lead to the formation of a functional kinetochore at ectopic locations (Shelby et al., 1997; Van Hooser et al., 2001; Ahmad and Henikoff, 2002). Similarly, in instances where the turnover of centromeric histones is impaired in budding yeast, CenH3s can localize throughout the DNA. Under normal conditions, however, this localization appears to be transient and CenH3s are only stably maintained at centromeric regions. One determinant for this stability and increased deposition at centromeres has been proposed to be the tension resulting from microtubule attachments to the centromere (Mellone and Allshire, 2003; Henikoff and Dalal, 2005). The crescent phase of meiosis in Tetrahymena provides an unexpected albeit indirect insight in favor of this hypothesis.
The crescent phase of meiosis in Tetrahymena is a unique structure that stretches the nucleus up to 50-fold compared with interphase with the nucleus growing ∼0.8 μm/min (Wolfe et al., 1976). In contrast to the distinct centromeres seen during mitosis, CNA1p is distributed throughout the length of the micronuclear chromosomes as they are stretched in the crescent formation. Indeed, at full crescent we were not able to distinguish the position of the centromeres because of diffuse staining of CNA1p. Telomeres continue to be clustered at one end of the nucleus throughout prophase (Loidl and Scherthan, 2004). We can only assume that the centromeres remain clustered at the opposite end. It is reasonable to suggest that the high degree of “stretching” exerted on the meiotic chromosomes during the formation of the crescent generate torsional stresses analogous to the “tension” generated by the pulling forces on centromeres at anaphase, triggering the deposition of CNA1p along the length of the micronuclear chromosomes. At the later stages of conjugation, micronuclear fate is marked by the removal of CNA1p except for at the centromeres. Results obtained in budding yeasts (Collins et al., 2004; Pearson et al., 2004) and Tetrahymena suggests that although CenH3s may deposit in euchromatin, they are quickly replaced by canonical histones after telophase, except at centromeres where they are maintained by the anchoring of kinetochores to the nuclear envelope.
We do not believe that the “ectopic” CNA1p deposition implies that Tetrahymena meiotic micronuclear chromosomes are holocentric. Although we are not able to see individual chromosomes during mitosis or meiosis, the micronuclear chromosomes unambiguously behave as meta-centric chromosomes during mitosis and meiosis. CNA1p localization remains diffuse throughout meiosis but distinct centromeres are apparent at anaphase I. Other reports show chromosomes segregating in a “V” shape during meiosis as expected for metacentric chromosomes with centromeres leading (Cole, 1991). This is in contrast to the movement of holocentric chromosomes, which would segregate as an “|” shape. Furthermore, it is unlikely that the CNA1p along the chromosomes leads to the formation of a functional kinetochore. In mammalian cells when the centro-meric protein is overexpressed and localized to euchromatin, only a few kinetochore proteins are recruited to the ectopic loci but this does not produce a functional kinetochore (Van Hooser et al., 2001).
CNA1p: A Meiotic Passenger Protein?
Remarkably, in late anaphase I and II, CNA1p is also localized on the meiotic spindle akin to what has been previously described for passenger proteins. In mammals and yeast, the Aurora B kinase has been found to phosphorylate CenH3 (Buvelot et al., 2003; Parra et al., 2003; Vagnarelli and Earnshaw, 2004). Aurora is a chromosomal passenger protein that moves from positions on the chromosomes to the central spindle during late anaphase I. The functional significance of CNA1p's localization to the spindle is unclear. We suggest that CNA1p's association with another passenger protein contributes to its localization, rather than it directly being selected for some downstream role in nuclear division (at the spindle or mid-body), as is the case for “authentic” passenger proteins.
Depletion of CNA1p from Centromeres
Our finding that progeny of CNA1 germline knockouts are able to complete mitosis several times before we observe segregation defects is consistent with results from other organisms. When CenH3 expression is blocked either by mutation or RNAi, it takes several divisions until a defect in mitotic chromosome segregation is obvious (Collins et al., 2004; Regnier et al., 2005). In experiments using RNAi to deplete CenH3, it was found that as CenH3 is replaced by H3 at centromeric regions, centromere integrity is lost (Buchwitz et al., 1999; Oegema et al., 2001; Blower et al., 2002). However, the use of degron-associated Cse4 (the CenH3 is Saccharomyces cerevisiae) suggests that the effect of Cse4 depletion can be seen in the course of a single cell cycle in budding yeasts (Collins et al., 2005). The difference between diverse eukaryotes may be partly attributed to the number of CenH3 nucleosomes at centromeres. Budding yeasts have a simple 125-base pair centromere that is believed to package a single CenH3-nucleosome (Keith and Fitzgerald-Hayes, 2000), depletion of which would have instant repercussions for chromosome segregation. However, many higher eukaryotes have complex centromeres with blocks of CenH3 nucleosomes. Depletion of CenH3s in these organisms would lead to a dilution of CenH3 because of random assortment after DNA replication. Our analysis of the ΔCNA1 progeny supports the idea that there is a threshold of CenH3 required at the centromere for proper chromosome segregation, below which faithful chromosome segregation cannot be sustained. On the basis of this and the data on CNA1p localization in mitosis, we conclude that T. thermophila centromeres might be as complex as those in higher eukaryotes.
What leads to the arrest in ΔCNA1 cells? It has been previously shown in Tetrahymena that although cells with a diminished micronucleus can proceed through additional cell divisions, cells that lack a micronucleus will cease dividing. This paradigm is true for all but one Tetrahymena laboratory strain (Karrer et al., 1984) although the reasons behind this are still unclear. Nevertheless, as segregation defects become manifest in ΔCNA1 cells and as the majority of cells lose all micronuclear chromosomes, we suggest that this results in their arrest.
Absence of Centromeres in the Ciliate Macronucleus
The traditional model for chromosome segregation in the ciliate macronucleus has been that of amitotic division during vegetative growth, wherein chromosomes are subject to random segregation. The absence of requirements for centromeres in random segregation has supported the suggestion that centromeres are deleted from the developing macronucleus during genomic rearrangement. However, one unsatisfying aspect of this model had been the lack of direct evidence from known centromeric determinants. Our localization of CNA1p helps provide the most direct support to the hypothesis that the ciliate macronucleus does not employ centromeres at any stage of its development or propagation. CNA1p is not detected in the macronucleus during vegetative growth when centromeres are clearly visible in the micronucleus. The strongest piece of evidence comes from our somatic and germline CNA1 knockout experiments, where we can show that chromosome segregation in the micronucleus has clearly gone awry. Nonetheless, chromosome segregation in the macronucleus appears to proceed normally. Thus, the ciliate macronucleus, bereft of germline responsibilities, now lacks the means (and dependence on the means) adopted by all other eukaryotes to accurately transmit genetic information.
Supplementary Material
Acknowledgments
We are grateful to Josh Bayes, Sue Biggins, Steve Henikoff, Julie Kerns, Hisashi Tanaka, Toshi Tsukiyama, and anonymous reviewers for comments and helpful suggestions on the manuscript, and we thank members of the Yao laboratory for helpful discussions and Marty Gorovsky for generously sharing reagents and results before publication. This work was supported by National Institutes of Health Grants GM26210 (M.C.Y.) and GM074108 (H.S.M.), by startup funds from the Fred Hutchinson Cancer Research Center and a Scholar Award from the Sidney Kimmel Cancer Foundation (H.S.M.). H.S.M. is an Alfred P. Sloan Fellow in Computational and Evolutionary Molecular Biology.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0698) on October 26, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Ahmad, K., and Henikoff, S. (2002). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99 (Suppl 4), 16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, S. L. (2000). Genetic nomenclature rules for Tetrahymena thermophila. Methods Cell Biol. 62, 561-563. [PubMed] [Google Scholar]
- Bernhard, D. (1999). Several highly divergent histone H3 genes are present in the hypotrichous ciliate Stylonychia lemnae. FEMS Microbiol. Lett. 175, 45-50. [DOI] [PubMed] [Google Scholar]
- Blomberg, P., Randolph, C., Yao, C. H., and Yao, M. C. (1997). Regulatory sequences for the amplification and replication of the ribosomal DNA minichromosome in Tetrahymena thermophila. Mol. Cell. Biol. 17, 7237-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D., and Karpen, G. H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3, 730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D., Sullivan, B. A., and Karpen, G. H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, P. J., and Brussard, T. B. (1974). Positive selection for mating with functional heterokaryons in Tetrahymena pyriformis. Genetics 78, 831-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, P. J., and Cassidy-Hanley, D. (2000). Biolistic transformation of macro-and micronuclei. Methods Cell Biol. 62, 501-512. [DOI] [PubMed] [Google Scholar]
- Buchwitz, B. J., Ahmad, K., Moore, L. L., Roth, M. B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547-548. [DOI] [PubMed] [Google Scholar]
- Buvelot, S., Tatsutani, S. Y., Vermaak, D., and Biggins, S. (2003). The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160, 329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley, D., Bowen, J., Lee, J. H., Cole, E., VerPlank, L. A., Gaertig, J., Gorovsky, M. A., and Bruns, P. J. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146, 135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine, L. G., Wenkert, D., Richman, R., Wiggins, J. C., and Allis, C. D. (1985). Modulation of linker histones during development in Tetrahymena: selective elimination of linker histone during the differentiation of new macronuclei. Dev. Biol. 109, 1-8. [DOI] [PubMed] [Google Scholar]
- Cole, E. S. (1991). Conjugal blocks in Tetrahymena pattern mutants and their cytoplasmic rescue. I. Broadened cortical domains (bcd). Dev. Biol. 148, 403-419. [DOI] [PubMed] [Google Scholar]
- Cole, E. S., Stuart, K. R., Marsh, T. C., Aufderheide, K., and Ringlien, W. (2002). Confocal fluorescence microscopy for Tetrahymena thermophila. Methods Cell Biol. 70, 337-359. [DOI] [PubMed] [Google Scholar]
- Collins, K. A., Castillo, A. R., Tatsutani, S. Y., and Biggins, S. (2005). De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell 16, 5649-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K. A., Furuyama, S., and Biggins, S. (2004). Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14, 1968-1972. [DOI] [PubMed] [Google Scholar]
- Davidson, L., and LaFountain, J. R., Jr. (1975). Mitosis and early meiosis in Tetrahymena pyriformis and the evolution of mitosis in the phylum Ciliophora. Biosystems 7, 326-336. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W., Bordwell, B., Marino, C., and Rothfield, N. (1986). Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Invest. 77, 426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., Hagan, I., Uzawa, S., and Yanagida, M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619-633. [DOI] [PubMed] [Google Scholar]
- Hai, B., and Gorovsky, M. A. (1997). Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 94, 1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Rines, D. R., Espelin, C. W., and Sorger, P. K. (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106, 195-206. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H. S. (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098-1102. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J. S., and van Steensel, B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Dalal, Y. (2005). Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15, 177-184. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Malik, H. S. (2002). Centromeres: selfish drivers. Nature 417, 227. [DOI] [PubMed] [Google Scholar]
- Horowitz, S., and Gorovsky, M. A. (1985). An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. USA 82, 2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman, E. V., Fowler, K. J., Newson, A. J., Redward, S., MacDonald, A. C., Kalitsis, P., and Choo, K. H. (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97, 1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, H., Rauch, M., and Heidmann, S. (2005). The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma 113, 350-361. [DOI] [PubMed] [Google Scholar]
- Kahn, R. W., Andersen, B. H., and Brunk, C. F. (1993). Transformation of Tetrahymena thermophila by microinjection of a foreign gene. Proc. Natl. Acad. Sci. USA 90, 9295-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, K., Stein-Gavens, S., and Allitto, B. A. (1984). Micronucleus-specific DNA sequences in an amicronucleate mutant of Tetrahymena. Dev. Biol. 105, 121-129. [DOI] [PubMed] [Google Scholar]
- Katz, L. A. (2001). Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int. J. Syst. Evol. Microbiol. 51, 1587-1592. [DOI] [PubMed] [Google Scholar]
- Katz, L. A., Bornstein, J. G., Lasek-Nesselquist, E., and Muse, S. V. (2004). Dramatic diversity of ciliate histone H4 genes revealed by comparisons of patterns of substitutions and paralog divergences among eukaryotes. Mol. Biol. Evol. 21, 555-562. [DOI] [PubMed] [Google Scholar]
- Keith, K. C., and Fitzgerald-Hayes, M. (2000). CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics 156, 973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K. E., Harmon, B. P., Reichardt, I. K., Sedat, J. W., and Blackburn, E. H. (1997). Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275, 1478-1481. [DOI] [PubMed] [Google Scholar]
- LaFountain, J. R., Jr., and Davidson, L. A. (1979). An analysis of spindle ultrastructure during prometaphase and metaphase of micronuclear division in Tetrahymena. Chromosoma 75, 293-308. [DOI] [PubMed] [Google Scholar]
- LaFountain, J. R., Jr., and Davidson, L. A. (1980). An analysis of spindle ultrastructure during anaphase of micronuclear division in Tetrahymena. Cell Motil. 1, 41-61. [DOI] [PubMed] [Google Scholar]
- Lin, R., Leone, J. W., Cook, R. G., and Allis, C. D. (1989). Antibodies specific to acetylated histones document the existence of deposition-and transcription-related histone acetylation in Tetrahymena. J. Cell Biol. 108, 1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J., and Scherthan, H. (2004). Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 117, 5791-5801. [DOI] [PubMed] [Google Scholar]
- Madireddi, M. T., Coyne, R. S., Smothers, J. F., Mickey, K. M., Yao, M. C., and Allis, C. D. (1996). Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87, 75-84. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H. S., and Henikoff, S. (2002). Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 12, 711-718. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and Henikoff, S. (2003). Phylogenomics of the nucleosome. Nat. Struct. Biol. 10, 882-891. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., Vermaak, D., and Henikoff, S. (2002). Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99, 1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch, J. L., Mark Welch, D. B., and Meselson, M. (2004). Cytogenetic evidence for asexual evolution of bdelloid rotifers. Proc. Natl. Acad. Sci. USA 101, 1618-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale, D. W., Allis, C. D., and Bruns, P. J. (1982). Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp. Cell Res. 140, 227-236. [DOI] [PubMed] [Google Scholar]
- Mellone, B. G., and Allshire, R. C. (2003). Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet Dev. 13, 191-198. [DOI] [PubMed] [Google Scholar]
- Merriam, E. V., and Bruns, P. J. (1988). Phenotypic assortment in Tetrahymena thermophila: assortment kinetics of antibiotic-resistance markers, tsA, death, and the highly amplified rDNA locus. Genetics 120, 389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L. L., and Roth, M. B. (2001). HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153, 1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138-145. [DOI] [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A. A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias, E., and Flacks, M. (1975). Macronuclear genetics of Tetrahymena. I. Random distribution of macronuclear genecopies in T. pyriformis, syngen 1. Genetics 79, 187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias, E., Hamilton, E. P., and Orias, J. D. (2000). Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62, 189-211. [DOI] [PubMed] [Google Scholar]
- Palmer, D. K., O'Day, K., Trong, H. L., Charbonneau, H., and Margolis, R. L. (1991). Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88, 3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D. K., O'Day, K., Wener, M. H., Andrews, B. S., and Margolis, R. L. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, M. T., Viera, A., Gomez, R., Page, J., Carmena, M., Earnshaw, W. C., Rufas, J. S., and Suja, J. A. (2003). Dynamic relocalization of the chromosomal passenger complex proteins inner centromere protein (INCENP) and aurora-B kinase during male mouse meiosis. J. Cell Sci. 116, 961-974. [DOI] [PubMed] [Google Scholar]
- Pearson, C. G., Yeh, E., Gardner, M., Odde, D., Salmon, E. D., and Bloom, K. (2004). Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14, 1962-1967. [DOI] [PubMed] [Google Scholar]
- Petcherskaia, M., McGuire, J. M., Pherson, J. M., and Kirk, K. E. (2003). Loss of cap structure causes mitotic defect in Tetrahymena thermophila telomerase mutants. Chromosoma. 111, 429-437. [DOI] [PubMed] [Google Scholar]
- Pfeffer, U., Ferrari, N., Tosetti, F., and Vidali, G. (1989). Histone acetylation in conjugating Tetrahymena thermophila. J. Cell Biol. 109, 1007-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, V., Vagnarelli, P., Fukagawa, T., Zerjal, T., Burns, E., Trouche, D., Earnshaw, W., and Brown, W. (2005). CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25, 3967-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten, G., Simerly, C., Palmer, D. K., Margolis, R. L., Maul, G., Andrews, B. S., and Schatten, H. (1988). Kinetochore appearance during meiosis, fertilization and mitosis in mouse oocytes and zygotes. Chromosoma 96, 341-352. [DOI] [PubMed] [Google Scholar]
- Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K., and Willard, H. F. (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109-115. [DOI] [PubMed] [Google Scholar]
- Shelby, R. D., Vafa, O., and Sullivan, K. F. (1997). Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136, 501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., Yu, L., Weir, J. W., and Gorovsky, M. A. (1995). Linker histones are not essential and affect chromatin condensation in vivo. Cell 82, 47-56. [DOI] [PubMed] [Google Scholar]
- Stoler, S., Keith, K. C., Curnick, K. E., and Fitzgerald-Hayes, M. (1995). A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573-586. [DOI] [PubMed] [Google Scholar]
- Strausberg, R. L. et al. (2002). Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 99, 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K. R., and Cole, E. S. (2000). Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62, 291-311. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., and Karpen, G. H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Le, H. D., Wahlstrom, J. M., and Karpen, G. H. (2003). Sequence analysis of a functional Drosophila centromere. Genome Res. 13, 182-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, M. T., Jones, K., and Allis, C. D. (1996). Phosphorylation of linker histone is associated with transcriptional activation in a normally silent nucleus. J. Cell Biol. 135, 1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L. (2000). PAUP*:Phylogenetic Analysis Using Parsimony (*and Other Methods), Sunderland, MA: Sinauer.
- Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa, O., Shelby, R. D., and Sullivan, K. F. (1999). CENP-A associated complex satellite DNA in the kinetochore of the Indian muntjac. Chromosoma 108, 367-374. [DOI] [PubMed] [Google Scholar]
- Vagnarelli, P., and Earnshaw, W. C. (2004). Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113, 211-222. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A. A., Ouspenski, I. I., Gregson, H. C., Starr, D. A., Yen, T. J., Goldberg, M. L., Yokomori, K., Earnshaw, W. C., Sullivan, K. F., and Brinkley, B. R. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529-3542. [DOI] [PubMed] [Google Scholar]
- Wang, Y. L. (2001). The mechanism of cytokinesis: reconsideration and reconciliation. Cell Struct. Funct. 26, 633-638. [DOI] [PubMed] [Google Scholar]
- Welch, D. M., and Meselson, M. (2000). Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288, 1211-1215. [DOI] [PubMed] [Google Scholar]
- Wenkert, D., and Allis, C. D. (1984). Timing of the appearance of macro-nuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J. Cell Biol. 98, 2107-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, E. A., Myers, T., Parker, K., Braun, T., and Yao, M. C. (2005). Class I histone deacetylase Thd1p affects nuclear integrity in Tetrahymena thermophila. Eukaryot. Cell 4, 981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, J., Hunter, B., and Adair, W. S. (1976). A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosoma 55, 289-308. [DOI] [PubMed] [Google Scholar]
- Woodard, J., Kaneshiro, E., and Gorovsky, M. A. (1972). Cytochemical studies on the problem of macronuclear subnuclei in Tetrahymena. Genetics 70, 251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, M. C., Choi, J., Yokoyama, S., Austerberry, C. F., and Yao, C. H. (1984). DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36, 433-440. [DOI] [PubMed] [Google Scholar]
- Yao, M. C., Fuller, P., and Xi, X. (2003). Programmed DNA deletion as an RNA-guided system of genome defense. Science 300, 1581-1584. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S. G., Shelby, R. D., and Sullivan, K. F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.