Abstract
RTT107 (ESC4, YHR154W) encodes a BRCA1 C-terminal-domain protein that is important for recovery from DNA damage during S phase. Rtt107 is a substrate of the checkpoint protein kinase Mec1, although the mechanism by which Rtt107 is targeted by Mec1 after checkpoint activation is currently unclear. Slx4, a component of the Slx1-Slx4 structure-specific nuclease, formed a complex with Rtt107. Deletion of SLX4 conferred many of the same DNA-repair defects observed in rtt107Δ, including DNA damage sensitivity, prolonged DNA damage checkpoint activation, and increased spontaneous DNA damage. These phenotypes were not shared by the Slx4 binding partner Slx1, suggesting that the functions of the Slx4 and Slx1 proteins in the DNA damage response were not identical. Of particular interest, Slx4, but not Slx1, was required for phosphorylation of Rtt107 by Mec1 in vivo, indicating that Slx4 was a mediator of DNA damage-dependent phosphorylation of the checkpoint effector Rtt107. We propose that Slx4 has roles in the DNA damage response that are distinct from the function of Slx1-Slx4 in maintaining rDNA structure and that Slx4-dependent phosphorylation of Rtt107 by Mec1 is critical for replication restart after alkylation damage.
INTRODUCTION
Evolutionarily conserved signal transduction pathways termed checkpoints respond to DNA damage to facilitate cell cycle delay, promote DNA repair, and induce transcription when DNA lesions are present (Carr, 2002; Melo and Toczyski, 2002; McGowan and Russell, 2004). Absence of checkpoint pathways causes a failure to stably transmit the genome from one generation to the next, a hallmark of cancerous cells (Kastan and Bartek, 2004). Additionally, constitutive checkpoint activation may be an early event in cancer development (Bartkova et al., 2005; Gorgoulis et al., 2005).
In Saccharomyces cerevisiae, the DNA damage checkpoint is comprised of a signaling cascade that includes the essential protein kinase Mec1. Mec1 is thought to be a sensor of DNA damage (Carr, 2002; Melo and Toczyski, 2002). Mec1 localizes to sites of DNA damage with its binding partner Ddc2. This localization is independent of that of another check-point sensor protein complex, comprised of Mec3, Dcc1, and Rad17 (Edwards et al., 1999; Melo et al., 2001; Rouse and Jackson, 2002b; Zou et al., 2002). The full range of DNA damage that is recognized and bound by Mec1-Ddc2 is not clear, but at a minimum the Mec1-Ddc2 complex can bind to single-stranded DNA (ssDNA) coated with the ssDNA binding protein RPA (Zou and Elledge, 2003). Single-stranded DNA may be a common intermediate in the processing of diverse forms of DNA damage. Colocalization of the Mec1-Ddc2 and Mec3-Ddc1-Rad17 complexes at sites of damage, along with the activity of the checkpoint mediator protein Rad9, allows phosphorylation and activation of the Rad53 and Chk1 kinases downstream of Mec1 in the checkpoint signaling cascade (Carr, 2002; Melo and Toczyski, 2002; Rouse and Jackson, 2002a). After successful repair of the checkpoint-inducing lesions, the checkpoint presumably must be down-regulated to allow the resumption of cell cycle progression. Details of the mechanism of this recovery are scant, although several genes have been identified that are important for checkpoint inactivation during recovery from DNA damage (Vaze et al., 2002; Leroy et al., 2003). Despite the critical role of Mec1 in the checkpoint response to DNA damage, few effectors of Mec1 have been identified. Of the known Mec1 targets, five are downstream or parallel molecules in the checkpoint cascade: Ddc2 (Edwards et al., 1999; Paciotti et al., 2000), Rad9 (Emili, 1998; Vialard et al., 1998), Mrc1 (Osborn and Elledge, 2003), Rad53 (Sweeney et al., 2005), and Dun1 (Mallory et al., 2003). Others include the ssDNA binding protein RPA (Brush et al., 1996; Brush and Kelly, 2000; Bartrand et al., 2004) and Rtt107 (Rouse, 2004). RTT107 (also known as ESC4) was first identified in a genetic screen for increased Ty transposon mobility (Scholes et al., 2001). Deletion of RTT107 confers sensitivity to the DNA alkylating agent methyl methane sulfonate (MMS) and results in slower S-phase progression in the presence of MMS than is observed in wild-type cells, suggesting that Rtt107 is important for replication fork processivity in the presence of DNA damage (Chang et al., 2002). RTT107 exhibits synthetic genetic interactions with genes involved in DNA replication and repair, including SGS1 and RRM3 (Tong et al., 2001, 2004). Rtt107 is phosphorylated in response to DNA damage, and this phosphorylation requires the checkpoint kinase Mec1, and four Mec1 phosphorylation consensus sequences in the C terminus of Rtt107 (Rouse, 2004). Mutation of these four Mec1 phosphorylation consensus sequences in Rtt107 results in increased sensitivity to MMS and delays completion of DNA replication after DNA damage, indicating that Rtt107 is an important downstream effector of the Mec1 checkpoint protein kinase (Rouse, 2004).
Recent work in fission yeast indicates a genetic connection between the RTT107 homologue brc1+ and the slx1+ gene (Sheedy et al., 2005). Slx1 in both budding and fission yeasts forms a heterodimeric structure-specific DNA nuclease with the Slx4 protein (Mullen et al., 2001; Fricke and Brill, 2003; Coulon et al., 2004). Mutants in SLX1 or SLX4 are lethal when combined with mutations in SGS1 (Mullen et al., 2001; Tong et al., 2001; Coulon et al., 2004). This, along with the preference of the Slx1-Slx4 nuclease for the 5′ flap of fork structures, suggests that Slx1-Slx4 may cleave stalled replication forks that cannot be resolved by Sgs1-Top3 (Fricke and Brill, 2003; Coulon et al., 2004). The clearest in vivo function of the Slx1-Slx4 nuclease is in the maintenance of the rDNA repeats during DNA replication (Kaliraman and Brill, 2002; Coulon et al., 2004). Although Slx4 lacks an obvious nuclease domain, Slx4 enhances the nuclease activity of Slx1 in vitro (Fricke and Brill, 2003; Coulon et al., 2004). However, Slx4 is thought to have a role that is independent of Slx1 because slx4Δ mutants are more sensitive than slx1Δ mutants to DNA damage caused by MMS (Fricke and Brill, 2003) or by the topoisomerase I poison camptothecin (Deng et al., 2005).
Here, we report the identification of a physical interaction between Rtt107 and Slx4. Mutants in slx4 had DNA damage recovery phenotypes that were similar to those displayed by rtt107Δ mutants, including prolonged checkpoint activation, S phase delay, and cell cycle delay in anaphase. Of particular significance, Slx4, but not Slx1, was required for Mec1 phosphorylation of Rtt107 in vivo. Slx4 thus had a role in the DNA damage response that was independent of Slx1 and facilitated phosphorylation of a checkpoint effector protein, Rtt107, by Mec1.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast strains used in this study were derivatives of BY4741 (Brachmann et al., 1998) and are listed in Table 1. Nonessential haploid deletion strains marked with the kanamycin (G418) resistance gene were made by the Saccharomyces Gene Deletion Project (Winzeler et al., 1999). Deletion strains marked with nourseothricin (nat) resistance gene were constructed by switching the kanamycin resistance gene with the nourseothricin resistance gene as described previously (Tong et al., 2001). Standard yeast media and growth conditions were used (Moreno et al., 1991; Sherman, 1991).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| TRY30 | MATα RTT07-V5-VSV::KANMX6 leu2Δ0 his3Δ1 ura3Δ met15Δ0 | I. Stagljar |
| TRY38 | MATaRTT107-V5-VSV::KANMX6 SLX4-TAP::HISMX6 leu2Δ0 his3Δ1 ura3Δ | This study |
| TRY19 | MATaSLX4-TAP::HISMX6 leu2Δ0 his3Δ1 ura3Δ met15Δ0 | Ghaemmaghami et al. (2003) |
| TRY71 | MATaRTT107-V5-VSV::KANMX6 SLX4-TAP::HISMX6 slx1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ met15Δ0 | This study |
| TRY52 | MATaRTT107-V5-VSV::KANMX6 SLX4-TAP::HISMX6 rad53-11::URA3 leu2Δ0 his3Δ1 ura3Δ | This study |
| TRY64 | MATaRTT107-V5-VSV::KANMX6 SLX4-TAP::HISMX6 mec1Δ::LEU2 sml1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ met15Δ0 | This study |
| MKY51 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 RTT107-TAP::TRP1 | This study |
| MKY52 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 RTT107-TAP::TRP1 SLX4-3HA::KANMX6 | This study |
| MKY53 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 RTT107-TAP::TRP1 SLX1-3HA::KANMX6 | This study |
| MKY54 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 rtt107::HIS5MX6 SLX4-3FLAG-KANMX pVV221[4TAP TRP1 CEN/ARS] | This study |
| MKY55 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 rtt107::HIS5MX6 SLX4-3FLAG-KANMX pVV221[4TAP-RTT107aa1-1070 TRP1 CEN/ARS] | This study |
| MKY56 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 rtt107::HIS5MX6 SLX4-3FLAG-KANMX pVV221[4TAP-RTT107aa1-512 TRP1 CEN/ARS] | This study |
| MKY57 | W303 MATaade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 bar1 rtt107::HIS5MX6 SLX4-3FLAG-KANMX pVV221[4TAP-RTT107aa512-1070 TRP1 CEN/ARS] | This study |
| W3031A | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 | Thomas and Rothstein (1989) |
| JMY360 | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 slx1-10::TRP1 can1-100 | Mullen et al. (2001) |
| WFY1728 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 slx4-11::KAN::loxP can1-100 | This study |
| JMY1546 | MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 rtt107::his5+ can1-100 | This study |
| TRY49 | MATaRTT107-V5-VSV::KANMX6 slx4Δ::KANMX6 leu2Δ0 his3Δ1 ura3Δ lys2Δ0 | This study |
| TRY76 | MATα RTT07-V5-VSV::KANMX6 mec1Δ::LEU2 sml1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ | This study |
| TRY74 | MATaRTT07-V5-VSV::KANMX6 slx1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ | This study |
| BY4741 | MATaleu2Δ0 his3Δ1 ura3Δ0 met15Δ0 | Brachmann et al. (1998) |
| MCY252 | MATartt107Δ::KANMX6 leu2Δ0 his3Δ1 ura3Δ0 met15Δ0 | Giaever et al. (2002) |
| MCY282 | MATaslx4Δ::KANMX6 leu2Δ0 his3Δ1 ura3Δ0 met15Δ0 | Giaever et al. (2002) |
| W3792-4B | MATaDDC2-YFP ade2-1 can1-100 ura2-1 his3-11,15 leu2-3,112 trp1-1 | Lisby et al., (2004a) |
| MDY14 | MATartt107Δ::KANMX6 DDC2-YFP ade2-1 can1-100 ura2-1 his3-11,15 leu2-3,112 trp1-1 | This study |
| GBY637 | MATaslx4Δ::KANMX6 DDC2-YFP ade2-1 can1-100 ura2-1 his3-11,15 leu2-3,112 trp1-1 | This study |
| MCY283 | MATaslx1Δ::KANMX6 leu2Δ0 his3Δ1 ura3Δ0 met15Δ0 | Giaever et al. (2002) |
| Y3667 | MATα slx4Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ0 lys2Δ0 | C. Boone |
| Y3558 | MATα slx1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ0 lys2Δ0 | C. Boone |
| TRY9 | MATartt107Δ::KANMX6 slx4Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ lys2Δ0 met15Δ0 | This study |
| TRY55 | MATartt107Δ::KANMX6 slx1Δ::NATMX6 leu2Δ0 his3Δ1 ura3Δ met15Δ0 | This study |
Tandem Affinity Purification (TAP) and Mass Spectrometric Identification of Peptides
For large-scale purification of Rtt107-associated proteins, a yeast strain containing RTT107-TAP was grown to mid-log phase in 8 liters of YPD. After harvesting and washing, the cell pellet was flash frozen in liquid nitrogen and stored at -80°C. For the purification, the pellet was divided into four equal portions and purified in four parallel, identical reactions to minimize background binding. Pellets were broken in a Krupps coffee mill in the presence of dry ice, and this material was resuspended in 8 ml of TAP-B1 (50 mM Tris-Cl, pH 7.8, 200 mM NaCl, 1.5 mM MgAc, 10% glycerol, 1 mM dithiothreitol [DTT], 10 mM NaPPI, 5 mM EDTA, 5 mM EGTA, 0.1 mM Na3VO4, 5 mM NaF, and Complete protease inhibitor cocktail) after thawing. Extracts were centrifuged for 20 min at 14,000 rpm in an SS34 rotor. The supernatant was recovered and centrifuged for 60 min at 33,500 rpm in a Ti70 rotor. NP-40 was added to a final concentration of 0.15% (vol/vol), and the extract was incubated with 200 μl of IgG-Sepharose beads for 90 min at 4°C. Beads were washed four times with 2 ml of TAP-B2 (50 mM Tris-Cl, pH 7.8, 200 mM NaCl, 1.5 mM MgAc, 10% glycerol, 1 mM DTT, and 0.15% NP-40) at 4°C in a column by gravity flow. After washing, the TAP-tag was cleaved by 10 μl of 10 U/μl TEV protease (Invitrogen, Carlsbad, CA) in 400 μl of TAP-B2 overnight at 4°C, and cleaved material was eluted by gravity flow. CaCl2 was added to the eluate to a final concentration of 2 mM, and the eluate was incubated with 200 μl of calmodulin-agarose for 90 min at 4°C. Beads were washed three times with 1 ml of TAP-B4 (50 mM Tris-Cl pH 7.8, 200 mM NaCl, 1.5 mM MgAc, 10% glycerol, 1 mM DTT, 2 mM CaCl2, and 0.15% NP-40), followed by 1 ml of TAP-B5 (50 mM Tris-Cl, pH 7.8, 200 mM NaCl, 1.5 mM MgAc, 10% glycerol, 1 mM DTT, and 0.5 mM CaCl2). Finally, the protein complexes were eluted twice with 200 μl of TAP-EB (20 mM Tris-Cl, pH 7.8, and 5 mM EGTA) and processed for mass spectrometry.
Proteins were identified using shotgun tandem mass spectrometry essentially as described previously (Krogan et al., 2002). Briefly, the protein fractions were concentrated and denatured by trichloroacetic acid (TCA) precipitation. The pellets were resuspended in 100 mM NH4HCO3/1 mM CaCl2 buffer, pH 8.5, and digested by trypsin overnight at 37°C with 2 μl of immobilized trypsin beads (Poroszyme; PerSeptive/Applied Biosystems; Streetsville, ON, Canada). The digested peptides were loaded manually as described previously (Gatlin et al., 1998) and fractionated on a fused silica capillary microcolumn packed with ∼7 cm (150 μm i.d.) reverse phase C18 resin (Zorbax Eclipse XDB-C18; Agilent Technologies, Mississauga, ON, Canada). The peptides were eluted into an online LCQ Deca quadrupole ion trap tandem mass spectrometer (Thermo Electron, Waltham, MA) by a linear gradient of 5-60% solvent B (100% acetonitrile; solvent A consisted of 5% acetonitrile, 0.5% acetic acid, and 0.02% heptafluoro-butyric acid). The flow rate at the tip of the needle was set to ∼300 nl/min by programming the high-performance liquid chromatography pump and use of a split line. The mass spectrometer cycled through four scans (one full mass scan followed by three tandem mass spectrometry scans of the successive three most intense ions) as the gradient progressed. Peptide precursor ions were automatically selected, whereas a dynamic exclusion list was used to minimize collection of redundant spectra. All tandem mass spectra were searched using a distributed version of the SEQUEST algorithm (Eng et al., 1994) against a nonredundant yeast protein sequence database (6/2000). High-confidence matches (p value < 0.05) were detected using the STATQUEST probability filter algorithm (Kislinger et al., 2003).
Expression and Purification of Recombinant Proteins
All recombinant proteins were expressed in bacteria using the T7 expression system. His6-Slx4, His6-Slx4/Slx1, and His6-Slx4/Slx1/Rtt107 were expressed from plasmids pNJ6408, pNJ6125, and pKR6131, respectively, and purified essentially as described previously (Fricke and Brill, 2003). His6-Rtt107 and His6-Rtt107-FLAG/Slx4 were expressed from plasmids pNJ6653 and pJF6655, respectively, and were purified as follows: Escherichia coli BL21-RIL cells were transformed and grown at 37°C in 1 liter of LB containing 30 μg/ml kanamycin until the optical density (OD)600 was equal to 0.1. The culture was shifted to 15°C for 1 h, or until the OD600 was equal to 0.5, and protein production was induced by the addition of isopropyl β-d-thiogalactoside to 0.4 mM. Expression continued for 16 h after which the cells were pelleted, washed, and resuspended in 40 ml of buffer N (25 mM Tris-HCl, pH 8.1, 10% glycerol, 500 mM NaCl, 0.01% NP-40, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) containing the following protease inhibitors: 10 μg/ml pepstatin, 5 μg/ml leupeptin, 10 mM benzamidine, and 100 μg/ml bacitracin. The cells were then incubated with 0.1 mg/ml lysozyme on ice for 15 min, sonicated three times for 1 min each, and centrifuged at 30,600 × g for 30 min. The soluble portion was collected, passed through a 0.45-μm cellulose acetate filter, made 10 mM in imidazole, and applied to a 1-ml His-Trap column on an AKTA fast-performance liquid chromatography (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The affinity column was washed with 10 column volumes of buffer N containing 10 mM imidazole and eluted with a 6 ml of linear gradient of buffer N containing 10-500 mM imidazole. The peak fractions were identified by SDS-PAGE, pooled, made 1 mM in EDTA, and chromatographed on a 24-ml Superdex 200 sizing column in buffer C (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.01% NP-40, 1 mM DTT, and 0.1 mM PMSF) containing 250 mM NaCl. The peak fractions from this column were pooled and stored at -80°C.
Immunoprecipitation and Immunoblotting
Immunoprecipitation was performed essentially as described previously (Bellaoui et al., 2003). Purified rabbit IgG agarose (Sigma-Aldrich) was used to immunoprecipitate TAP-tagged proteins. Mouse anti-vesicular stomatitis virus (VSV)-G antibody (Roche Diagnostics, Indianapolis, IN) followed by protein G-agarose (GE Healthcare) was used to immunoprecipitate VSV-tagged proteins. Immunoprecipitates were resolved on 7.5% or 10% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and subjected to immunoblot analysis with rabbit anti-VSV-G (Bethyl Laboratories, Montgomery, TX), peroxidase anti-peroxidase-soluble complex (Sigma-Aldrich), or rabbit IgG (anti-hemagglutinin [HA]; Upstate Biotechnology, Lake Placid, NY) to detect the TAP-tag, mouse anti-HA antibodies (Covance, Berkeley, CA), mouse anti-FLAG antibodies (Sigma-Aldrich), or anti-phospho-[S/T]Q (Cell Signaling Technology, Beverly, MA) antibodies. Immunoblots were developed using SuperSignal ECL (Pierce Chemical, Rockford, IL).
Rad53 In Situ Kinase Assays, Contour-clamped Homogeneous Electric Field (CHEF) Gel Electrophoresis, and Microscopy
Cells were arrested in G1 by culturing in the presence of 2 μg/ml α mating factor for 2 h at 30°C in YPD, pH 3.9. Cells were released into the cell cycle by harvesting, washing, and resuspending in YPD containing 0.03% MMS for 1 h. Aliquots were removed at the indicated times and processed further for Rad53 in situ kinase assays, CHEF gel electrophoresis, or microscopy. Rad53 in situ kinase assays were carried out essentially as described previously (Pellicioli et al., 1999). The relative level of Rad53 activation was quantified by exposure to a storage phosphor screen, subsequent scanning on a Storm scanner (Molecular Dynamics, Sunnyvale, CA), and analysis with Image-Quant software (Molecular Dynamics). CHEF gel analysis was carried out as described previously (Kaliraman and Brill, 2002). To examine cellular and nuclear morphology, cells were harvested, washed with phosphate-buffered saline, resuspended in 70% ethanol, and stored at -20°C. Before examination, cells were resuspended in Vectashield mounting media with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). At least 200 cells were counted for each time point. Imaging of Ddc2-YFP using live cells was done essentially as described for Rad52-YFP (Chang et al., 2005).
MMS Sensitivity Measurements
Cells were grown in YPD, diluted serially, spotted onto plates, and incubated at 30°C. MMS (Aldrich Chemical, Milwaukee, WI) plates contained 0.03% (vol/vol) MMS in YPD and were used within 24 h of preparation. Viability after exposure to 0.04% MMS in liquid culture was determined as described previously (Bellaoui et al., 2003).
RESULTS
Rtt107 Physically Interacted with Slx4
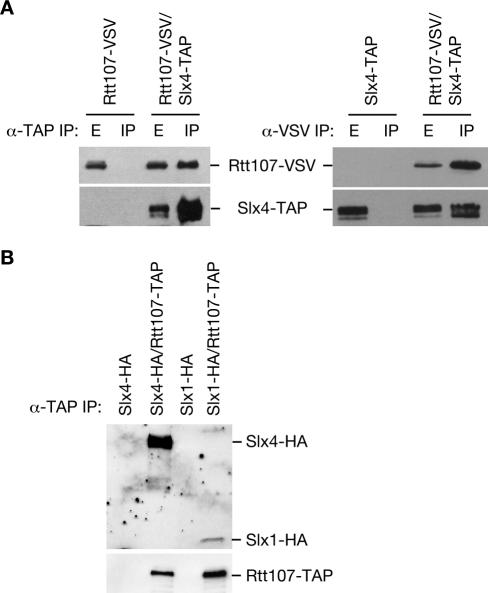
To identify proteins that interact with Rtt107, Rtt107-TAP was purified using a tandem affinity purification. The purified Rtt107-TAP and associated proteins were subjected to mass spectrometric analysis. Seven peptides were identified from Slx4, covering 3.3% of the protein, identifying Slx4 with 99.6% confidence (Table 2). To confirm the physical interaction between Rtt107 and Slx4, we used a strain expressing TAP-tagged Slx4 and VSV epitope-tagged Rtt107 from their respective genomic loci, and appropriate control strains. By immunoblot analysis of immunoprecipitations, Rtt107-VSV coprecipitated with Slx4-TAP and vice versa (Figure 1A). Slx4 is known to interact with the nuclease Slx1 (Mullen et al., 2001; Fricke and Brill, 2003). Like Slx4, Slx1 was specifically present in the Rtt107 precipitates (Figure 1B). Therefore, Rtt107 formed a complex with both Slx4 and Slx1. Additionally, Rtt107 was present in Slx4 immunoprecipitates from slx1Δ cells, indicating that the Rtt107-Slx4 interaction occurs independently of Slx1 (Figure 2A).
Table 2.
Mass spectrometric identification of Rtt107-interacting proteins
| Gene | Description | % coverage | % confidence | No. of peptides |
|---|---|---|---|---|
| GPM1 | Converts 3-phosphoglycerate to 2-phosphoglycerate | 4.9 | 99.6 | 1 |
| RPP0 | Homology to rat P0, human P0, and E. coli L10e | 3.5 | 99.6 | 1 |
| RPP1A | Homology to rat P1, human P1, and E. coli L12eIIA | 20.8 | 99.6 | 1 |
| RPP2A | Homology to rat P2, human P2, and E. coli L12eIB | 23.6 | 99.6 | 1 |
| RPS20 | Homology to rat S20, human S20, Xenopus S22, and E. coli S10 | 9.9 | 99.6 | 1 |
| RTT107 | Regulator of Ty1 Transposition | 8.8 | 99.6 | 4 |
| SLX4 | Nuclease subunit | 3.3 | 99.6 | 7 |
| TDH3 | Glyceraldehyde-3-phosphate dehydrogenase 3 | 2.1 | 99.6 | 1 |
| TEF2 | Translational elongation factor EF-1α | 3.9 | 99.6 | 4 |
| SSA1 | Stress-seventy subfamily A | 1.6 | 85.8 | 4 |
| IPT1 | Necessary for synthesis of mannose-(inositol-P)2-ceramide | 1.1 | 80.8 | 1 |
| RPS24A | Homology to rat S24 | 4.4 | 58.7 | 1 |
| SSB1 | Stress-seventy subfamily B | 1.5 | 58.7 | 2 |
Figure 1.
Rtt107 physically interacts with Slx4 and Slx1. (A) Extracts from yeast strains expressing the indicated epitope-tagged proteins were immunoprecipitated with IgG agarose or α-VSV followed by protein G agarose. Ten percent of the input extract (E) and the entire immunoprecipitate (IP) were fractionated by SDS-PAGE. Immunoblots were probed with α-VSV to detect Rtt107-VSV, peroxidase-anti-peroxidase to detect Slx4-TAP. (B) Extracts were treated as in A, and immunoblots were probed with α-HA to detect Slx4-HA and Slx1-HA or rabbit IgG to detect Rtt107-TAP.
Figure 2.
The Rtt107-Slx4 interaction is direct and is independent of Slx1, DNA damage, and DNA damage checkpoints. (A and B) Logarithmically growing cultures were treated with 0 or 0.03% MMS for 1 h. Extracts from yeast strains expressing the indicated epitope-tagged proteins were immunoprecipitated with IgG agarose. The immunoprecipitate was fractionated by SDS-PAGE, and immunoblots were probed with α-VSV to detect Rtt107-VSV and peroxidase-anti-peroxidase to detect Slx4-TAP. (C) Recombinant Rtt107, Slx1, and His6-Slx4 were coexpressed in E. coli, and the His6-Slx4 and associated proteins were purified and resolved on an SDS-polyacrylamide gel. The gel was stained with silver. Note that Rtt107 and His6-Slx4 have very similar mobility in this gel system. (D) Recombinant Slx4 and His6-RTT107-FLAG were coexpressed in E. coli and the His6-Rtt107-FLAG was purified. Purified proteins were fractionated on SDS-PAGE and stained with Coomassie blue. Note the slower mobility of the His6-Rtt107-FLAG compared with His6-Rtt107 and the faster mobility of Slx4 compared with His6-Slx4. (E) Truncated and full-length Rtt107-TAP were coexpressed with Slx4-FLAG. Rtt107-TAP and associated proteins were immunoprecipitated with rabbit IgG. Immunoprecipitates were fractionated by SDS-PAGE, and proteins were detected by probing the immunoblot with α-FLAG to detect Slx4-FLAG or rabbit IgG to detect Rtt107-TAP, Rtt107(1-511)-TAP, and Rtt107(512-1070)-TAP. The vector lane is a control strain in which no Rtt107-TAP is present.
The Rtt107-Slx4 Physical Interaction Was Independent of Checkpoint Activation
Because Rtt107 and Slx4 contribute to resistance to MMS treatment (Chang et al., 2002), we examined the Rtt107-Slx4 complex after treatment of cells with MMS (Figure 2A). The interaction between Rtt107 and Slx4 was unaffected by MMS induced DNA damage. In agreement with the lack of DNA damage dependence, the Rtt107-Slx4 interaction was not dependent on either the Rad53 or the Mec1 checkpoint kinases (Figure 2B). Thus, although Rtt107 is a target of Mec1 phosphorylation (Rouse, 2004) the interaction between Rtt107 and Slx4 was not regulated by DNA damage or by checkpoint response.
To determine whether the interaction between Rtt107 and Slx1-Slx4 was direct, we coexpressed recombinant proteins in Escherichia coli and purified Slx4 complexes, using a six-histidine (His)-tag fused to Slx4 (Figure 2C). Under these conditions, His6-Slx4 was associated with both Slx1 and Rtt107. To increase the resolution between Rtt107 and Slx4, which have similar mobilities on SDS-PAGE, we coexpressed untagged Slx4 with His6-Rtt107-FLAG alone and found that Slx4 associated with purified His6-Rtt107-FLAG (Figure 2D). Thus, Rtt107 bound directly to Slx4, independently of Slx1.
BRCA1 C-terminal (BRCT) homology domains mediate protein-protein interactions in a variety of proteins with roles in the response to DNA damage (Callebaut and Mornon, 1997). The BRCT domains reside in the amino-terminal half of Rtt107. Truncation mutants comprising the amino- or carboxy-terminal half of Rtt107, expressed from low-copy plasmids in an rtt107Δ SLX4::FLAG strain, were tested for coimmunoprecipitation with Slx4-FLAG (Figure 2E). The interaction between Rtt107 and Slx4 required the presence of the BRCT-containing N terminus of Rtt107 but not the carboxy-half, implicating the BRCT homology domains in this interaction.
Rtt107 and Slx4 Contributed to Recovery from MMS-induced DNA Damage
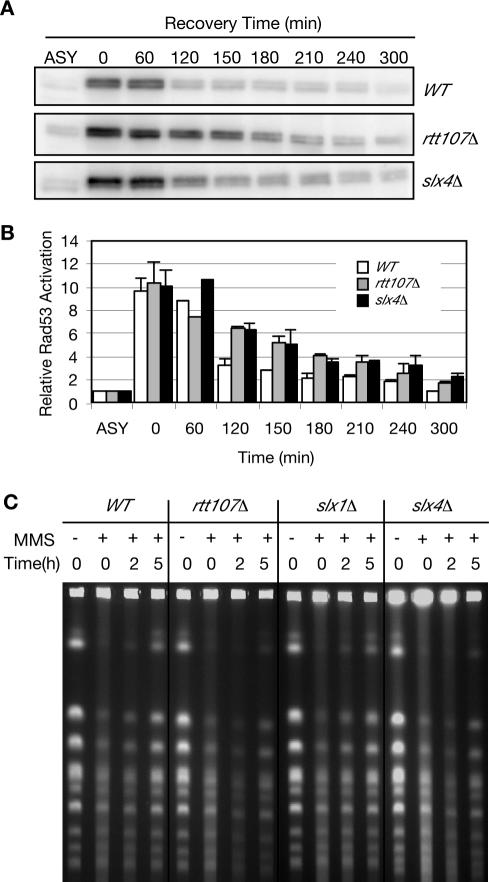
Rtt107 is involved in the recovery from MMS-induced DNA damage and rtt107Δ cells undergo prolonged Rad53 checkpoint activation after treatment with MMS (Chang et al., 2002; Rouse, 2004). Because Rtt107 and Slx4 formed a complex, we monitored Rad53 activity in wild-type, rtt107Δ, and slx4Δ strains during recovery from MMS damage using an in situ kinase assay (Pellicioli et al., 1999). Cells were synchronized in G1 and released into S phase in media containing MMS. Rad53 phosphorylation, which reflects Rad53 activity, was monitored after removal of MMS from the media (Figure 3, A and B). As reported previously (Rouse, 2004), rtt107Δ cells displayed prolonged Rad53 activation during recovery from MMS damage. slx4Δ cells also exhibited prolonged Rad53 activation, similar to that observed in rtt107Δ cells, indicating that like Rtt107, Slx4 is important for recovery from MMS-induced damage (Figure 3, A and B).
Figure 3.
Rtt107 and Slx4 are important for the recovery from MMS-induced DNA damage. (A) Cells were α-factor blocked in G1 and released into media containing 0.03% MMS for 1 h. Samples were taken at the indicated times, samples were fixed with TCA, and extracts were fractionated on SDS-PAGE for in situ kinase assay of Rad53. (B) Levels of Rad53 activity after treatment with MMS, relative to Rad53 activity in the asynchronous undamaged sample (ASY) are plotted. Average of two independent experiments is shown. (C) Chromosomes plugs were prepared from untreated cells, from cells treated with MMS, and from cells recovering from MMS damage. Chromosomes were separated by CHEF gel electrophoresis and detected by ethidium bromide staining.
Another measure of proficient recovery from MMS-induced DNA damage is the ability of cells to complete DNA synthesis during the recovery period. We assayed completion of DNA replication using CHEF gel electrophoresis (Figure 3C). Incompletely replicated chromosomes cannot be separated on CHEF gels because of the presence of replication intermediates (Hennessy et al., 1991). Chromosomes were prepared from wild-type, rtt107Δ, slx4Δ, and slx1Δ cells in the absence of MMS and during recovery from MMS treatment. Chromosomes from untreated cells are able to enter the gel; however, after treatment with MMS, the chromosomes remain trapped in the wells (Figure 3C). Wild-type cells recover quickly from MMS-induced damage because distinct chromosome bands begin to reappear by 2 h after removal of the drug, and chromosomes seem normal by 5 h. In contrast, chromosomes prepared from rtt107Δ and slx4Δ cells exhibited delayed completion of DNA replication after treatment with MMS, with chromosome bands remaining diffuse at 5 h after MMS removal (Figure 3C). Interestingly, chromosomes from slx1Δ cells behaved similarly to those from wild-type cells, indicating that Slx1 was less important than Rtt107 and Slx4 for recovery from MMS.
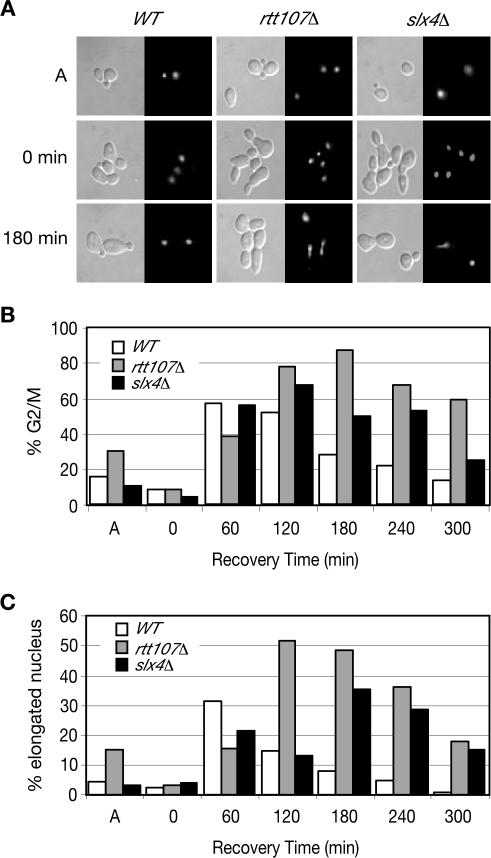
To examine the nuclear morphology of rtt107Δ and slx4Δ cells during recovery from MMS, cells were stained with the DNA binding dye DAPI (Figure 4, A-C). Within 60 min of removal of the MMS, wild-type, rtt107Δ, and slx4Δ strains accumulated large-budded cells with a single nucleus indicative of cells that are in G2. Wild-type cells proceeded through mitosis, as evidenced by the decrease in large-budded cells at 120 and 180 min. In contrast, rtt107Δ and slx4Δ strains continued to accumulate in G2/M, with an elongated nucleus spanning the bud neck (Figure 4A, 180 min; and C). This morphology was similar to that exhibited by cells with dicentric chromosomes, which delay at midanaphase (Yang et al., 1997). These results further implicated Rtt107 and Slx4 in the recovery from MMS-induced damage and suggested that rtt107Δ and slx4Δ mutants might accumulate in anaphase during recovery.
Figure 4.
rtt107Δ and slx4Δ mutants accumulate in anaphase during recovery from MMS-induced damage. (A) Logarithmically growing cultures were treated with 0.03% MMS for 1 h, samples were withdrawn at the indicated times, and stained with DAPI to examine nuclear morphology; differential interference contrast images (left), and corresponding DAPI images (right) are shown. (B and C) Cells treated as in A were sampled at the indicated times. The percentage of cells with a large bud (% G2/M) and percentage of cells with a large bud and an elongated nucleus spanning the bud neck (% elongated nucleus) are plotted.
Slx4 and Rtt107 Suppressed Spontaneous DNA Damage
The checkpoint protein Ddc2 relocalizes from a diffuse nuclear localization to punctate subnuclear foci when DNA damage is present (Melo et al., 2001; Lisby et al., 2004b). During an otherwise unperturbed cell cycle, both rtt107Δ and slx4Δ mutants displayed an increased fraction of cells with Ddc2 foci relative to wild-type cells (Figure 5, A and B), suggesting that rtt107Δ and slx4Δ mutants sustain higher levels of spontaneous DNA damage and/or replication fork stalling than wild-type cells during normal cell cycle progression. Furthermore, these results suggested that in addition to their roles in recovery from MMS induced DNA damage, Rtt107 and Slx4 functioned in the normal cell cycle to prevent spontaneous DNA lesions. rtt107Δ had a greater fraction of cells with Ddc2-YFP foci (28%) than did slx4Δ (11%), indicating a higher level of DNA damage in cells lacking Rtt107 than in cells lacking Slx4.
Figure 5.

Rtt107 and Slx4 suppress spontaneous DNA damage. (A) Logarithmically growing cells expressing Ddc2-YFP were visualized by fluorescence microscopy; representative fields are shown. (B) The percentage of cells with Ddc2-YFP foci in G1 (unbudded) and G2/S/M (budded) cells is plotted for the indicated strains.
Slx4 Was Necessary for Mec1 Phosphorylation of Rtt107 in Response to MMS-induced DNA Damage
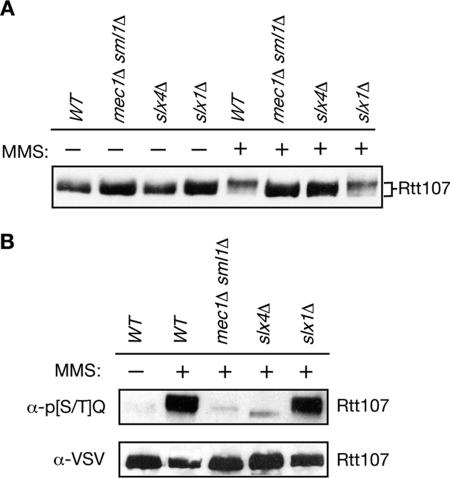
Rtt107 is phosphorylated by Mec1 in response to MMS-induced damage (Rouse, 2004). Rtt107 phosphorylation was evaluated in the absence and presence of DNA damage in wild-type, mec1Δsml1Δ, slx4Δ, and slx1Δ genetic backgrounds, detecting Rtt107 phosphorylation by the change in Rtt107 mobility on immunoblots of SDS-polyacrylamide gels (Figure 6A). Rtt107 underwent a Mec1-dependent mobility shift in response to MMS treatment, as reported previously (Rouse, 2004). In the absence of Slx4, the mobility of Rtt107 was not slowed to the same extent as in wild-type in the presence of DNA damage (Figure 6A, slx4Δ and WT, + MMS), suggesting that Rtt107 was not phosphorylated extensively in the absence of Slx4. In contrast, the mobility shift of Rtt107 was similar to wild-type in the absence of Slx1 (Figure 6A, slx1Δ+MMS). Thus, Slx4 played a role in Mec1 phosphorylation of Rtt107, but Slx1 did not. Mec1 phosphorylation occurs within [S/T]Q clusters and can be detected using an antibody specific for phospho-[S/T]Q. We examined the level of Mec1 phosphorylation of Rtt107 by immunoprecipitating Rtt107-VSV from mec1Δ, slx4Δ, or slx1Δ strains and by probing with anti-phospho-[S/T]Q antibodies on an immunoblot (Figure 6B). As expected, Mec1 phosphorylated Rtt107 in the presence of MMS-induced damage. However, in the absence of Slx4, the phosphorylation of [S/T]Q sites was eliminated. Slx1 was not required for this phosphorylation. Therefore, Slx4 was essential for Mec1 phosphorylation of Rtt107, and this role of Slx4 in the DNA damage response occurred independently of Slx1.
Figure 6.
Slx4 is required for Mec1 phosphorylation of Rtt107 in response to MMS induced damage. (A) Logarithmically growing cultures were treated with 0 or 0.03% MMS for 1 h. Samples were fixed with TCA, extracts were fractionated on SDS-PAGE, and immunoblots were probed for Rtt107-VSV. (B) Rtt107-VSV was immunoprecipitated from yeast extracts of the indicated strains, and the immunoprecipitates were fractionated by SDS-PAGE. The corresponding immunoblot was probed with α-phospho-[S/T]Q to detect phosphorylated Mec1 consensus sites, or with α-VSV to detect Rtt107-VSV.
RTT107 and SLX4 Made Independent Contributions to MMS Resistance
Although slx4Δ and rtt107Δ mutants display similar DNA damage response phenotypes, in all cases the phenotype of rtt107Δ was more severe than that of slx4Δ. Additionally, we identified functions of Slx4 in the DNA damage response that were independent of its binding partner Slx1. Thus, in addition to their shared roles in the DNA damage response, RTT107 and SLX4 could have independent functions. Indeed, the rtt107Δ slx4Δ double mutants were slightly more sensitive to MMS than either of the single deletion mutants (Figure 7, A and B), suggesting that in addition to their common roles in the DNA damage response, Rtt107 and Slx4 might also have distinct roles in MMS resistance.
Figure 7.
RTT107 and SLX4 make independent contributions to DNA damage resistance. (A) Ten-fold serial dilutions of cells were spotted onto YPD or YPD containing 0.03% MMS and incubated at 30°C for 3 d. (B) Logarithmically growing cultures were blocked in G1 with α-factor and released into media containing 0.04% MMS. Samples were withdrawn at the indicated times and plated on YPD to determine cell viability. The percentage of viable cells relative to the number of viable cells at t = 0 is shown. The average of three independent experiments is plotted.
DISCUSSION
We report a physical interaction between Rtt107 and Slx4 that was required for phosphorylation of Rtt107 after DNA alkylation damage by Mec1 kinase. Although Slx1 was present in the Rtt107-Slx4 complex, the physical interaction between Rtt107 and Slx4 occurred independently of Slx1. Furthermore, Slx1 was not required for phosphorylation of Rtt107 by Mec1, nor for the recovery from MMS-induced DNA damage. Consistent with a shared role in DNA damage repair, slx4Δ and rtt107Δ mutants shared a number of phenotypes, including MMS sensitivity, prolonged checkpoint activation, persistence of replication intermediates, and a mitotic delay. slx4Δ and rtt107Δ mutants also exhibited increased levels of spontaneous DNA damage, suggesting a common role in suppressing the occurrence of DNA lesions during normal cell cycle progression. These similar phenotypes were consistent with Rtt107 and Slx4 being members of the same complex.
Mec1 phosphorylation of Rtt107 is critical for the MMS recovery function of Rtt107 (Rouse, 2004), and we found that this phosphorylation did not occur in the absence of Slx4. Furthermore, the BRCT domains of Rtt107 were required for the interaction with Slx4 (Figure 2E), for MMS resistance, and for Mec1 phosphorylation (Rouse, 2004). The requirement for Slx4 binding to facilitate Mec1 phosphorylation of Rtt107 indicated that the Rtt107-Slx4 complex was the likely biologically relevant checkpoint target that is important for recovery from MMS damage.
In addition to their proposed common role in damage resistance, Rtt107 and Slx4 likely had roles independent of each other, because the rtt107Δ slx4Δ double mutant was somewhat more MMS sensitive than either of the single mutants. Mutations in rtt107 and slx4 might not be expected to have an additive effect in MMS sensitivity, because Slx4 was required for Mec1 phosphorylation of Rtt107 (Figure 6), and Mec1 phosphorylation of Rtt107 is important for MMS resistance (Rouse, 2004), which together suggest that Rtt107 and Slx4 function in the same MMS response pathway downstream of Mec1. On the other hand, there is phenotypic evidence that Rtt107 and Slx4 function differently in vivo. For example, slx4Δ is lethal when combined with sgs1Δ, whereas rtt107Δ sgs1Δ cells are viable (Mullen et al., 2001; Tong et al., 2001, 2004), and rtt107Δ exhibited stronger phenotypes than slx4Δ in most of our assays. One model that accounts for this apparent discrepancy is that Slx4 and Rtt107, in addition to the common role in DNA damage response that is suggested by the regulation of DNA damage checkpoint phosphorylation of Rtt107 by Slx4, also have independent roles in DNA damage resistance. Although we identified physical and functional interactions between Slx4 and Rtt107, we do not know the in vivo composition of this complex. For example, it is unclear whether these proteins function exclusively as an Slx4-Rtt107-Slx1 heterotrimer, or as part of a larger complex, or whether the composition of the complex changes throughout the cell cycle or in response to DNA damage. An intriguing possibility is that Rtt107, through its multiple BRCT domains, interacts with additional DNA damage response proteins to perform damage resistance functions independent of Slx4. Indeed, large-scale protein-protein interaction data sets indicate that Rtt107 may interact with Mms22 (Ho et al., 2002). The Rtt107-independent function of Slx4 likely involves Slx1. Slx4 and Slx1 form a complex that has nuclease activity (Fricke and Brill, 2003) and may be required to process DNA structures that remain in the rDNA in sgs1Δ cells (Kaliraman and Brill, 2002). Perhaps Rtt107 does not play a critical role in this activity. Thus, we propose that Rtt107 and Slx4 share a role in the DNA damage response, as evidenced by the regulation of Rtt107 phosphorylation by Slx4, but also carry out distinct roles, perhaps in combination with other binding partners.
Recent work in fission yeast indicates that Slx1 and Brc1 (the fission yeast Rtt107 homologue) function together in promoting repair of alkylation damage in smc6 mutants (Sheedy et al., 2005). Although we did not observe a requirement for Slx1 in phosphorylation of Rtt107 by Mec1 and in restart of replication after MMS damage, it was likely that Slx1 was present in the Rtt107-Slx4 complex and could therefore perform other DNA damage response functions in concert with Rtt107.
We propose that Slx4-mediated Mec1-phosphorylation of Rtt107 promotes the restart of replication forks stalled at sites of alkylation damage (and perhaps spontaneous damage) (Figure 8). In wild-type cells, alkylation damage causes activation of the checkpoint kinase Mec1 when the damage is encountered by replication forks. Activated Mec1 would then phosphorylate Rtt107-Slx4 complexes on the Rtt107 subunit (Figure 6) and perhaps the Slx4 subunit (Flott and Rouse, 2005). Phosphorylated Rtt107-Slx4 then promotes replication restart, resulting in timely completion of S phase. In the absence of Slx4, activated Mec1 was unable to phosphorylate Rtt107. Replication was not completed on schedule, as evidenced by prolonged Rad53 activation and the prolonged presence of replication intermediates suggested by CHEF gel analysis. Surprisingly, although replication seemed to be incomplete, the cells delayed with an elongated nucleus spanning the bud neck, a morphology reminiscent of cells with dicentric chromosomes that arrest in mid-anaphase in a Rad9-dependent manner (Yang et al., 1997). Alternatively, slx4Δ and rtt107Δ mutant cells could be accumulating preanaphase but with deregulated spindle elongation as seen in checkpoint mutants when DNA replication is blocked by hydroxyurea (Krishnan et al., 2004; Bachant et al., 2005). We favor the first possibility because, in contrast to the mec1-1 and rad53-21 checkpoint mutants that display anaphase-independent spindle elongation when replication is inhibited, slx4Δ and rtt107Δ have an intact DNA damage checkpoint, undergo extensive DNA replication, and display little loss of viability under conditions where the cell cycle delay is evident. There is precedent for incomplete replication and the resulting intrachromatid bridges causing DNA damage and cell cycle delay when sister chromatids separate at anaphase (Lengronne and Schwob, 2002). It will be of great interest to determine whether rtt107Δ and slx4Δ mutants are delayed in anaphase, whether the delay requires the DNA damage checkpoint, and why the prolonged Rad53 activation that we observed in the slx4Δ and rtt107Δ mutants was insufficient to mount and maintain a checkpoint arrest before spindle elongation.
Figure 8.
Model of the role of Rtt107 and Slx4 in the DNA damage response.
Acknowledgments
We thank Dan Durocher, Igor Stagljar, and Charlie Boone for reagents and strains. This work was supported by funds from the National Cancer Institute of Canada (to G.W.B.). G.W.B. is a research scientist of the National Cancer Institute of Canada. S.J.B. was supported by National Institutes of Health Grants GM-055583 and GM-067956. M.S.K. was partly supported by a longterm fellowship from the Human Frontier Science Program and also acknowledges start-up funding from the Child and Family Research Institute, University of British Columbia. J. R. was supported by National Institutes of Health Grant GM-31105, with core support from an National Institute of Environmental Health Sciences Mutagenesis Center Grant.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0785) on November 2, 2005.
Abbreviations used: BRCT, BRCA1 C-terminal; CHEF, contour-clamped homogeneous electric field; MMS, methyl methane sulfonate; TAP, tandem affinity purification.
References
- Bachant, J., Jessen, S. R., Kavanaugh, S. E., and Fielding, C. S. (2005). The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 168, 999-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova, J., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864-870. [DOI] [PubMed] [Google Scholar]
- Bartrand, A. J., Iyasu, D., and Brush, G. S. (2004). DNA stimulates Mec1-mediated phosphorylation of replication protein A. J. Biol. Chem. 279, 26762-26767. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Chang, M., Ou, J., Xu, H., Boone, C., and Brown, G. W. (2003). Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22, 4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Brush, G. S., and Kelly, T. J. (2000). Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res. 28, 3725-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush, G. S., Morrow, D. M., Hieter, P., and Kelly, T. J. (1996). The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl. Acad. Sci. USA 93, 15075-15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut, I., and Mornon, J. P. (1997). From BRCA1 to RAP 1, a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400, 25-30. [DOI] [PubMed] [Google Scholar]
- Carr, A. M. (2002). DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1, 983-994. [DOI] [PubMed] [Google Scholar]
- Chang, M., Bellaoui, M., Boone, C., and Brown, G. W. (2002). A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99, 16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M., Bellaoui, M., Zhang, C., Desai, R., Morozov, P., Delgado-Cruzata, L., Rothstein, R., Freyer, G. A., Boone, C., and Brown, G. W. (2005). RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 24, 2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon, S., Gaillard, P. H., Chahwan, C., McDonald, W. H., Yates, J. R., 3rd, and Russell, P. (2004). Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell 15, 71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, C., Brown, J. A., You, D., and Brown, J. M. (2005). Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170, 591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R. J., Bentley, N. J., and Carr, A. M. (1999). A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell. Biol. 1, 393-398. [DOI] [PubMed] [Google Scholar]
- Emili, A. (1998). MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 2, 183-189. [DOI] [PubMed] [Google Scholar]
- Eng, J. K., McCormack, A. L., and Yates, J. R., 3rd. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Am. Soc. Mass Spectrom. 5, 976-978. [DOI] [PubMed] [Google Scholar]
- Flott, S., and Rouse, J. (2005). Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem. J. 15, 325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke, W. M., and Brill, S. J. (2003). Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17, 1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin, C. L., Kleemann, G. R., Hays, L. G., Link, A. J., Yates, J. R., 3rd. (1998). Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography—microspray and nanospray mass spectrometry. Anal. Biochem. 263, 93-101. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003). Global analysis of protein expression in yeast. Nature 425, 737-741. [DOI] [PubMed] [Google Scholar]
- Giaever, G., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- Gorgoulis, V. G., et al. (2005). Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907-913. [DOI] [PubMed] [Google Scholar]
- Hennessy, K. M., Lee, A., Chen, E., and Botstein, D. (1991). A group of interacting yeast DNA replication genes. Genes Dev. 5, 958-969. [DOI] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- Kaliraman, V., and Brill, S. J. (2002). Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41, 389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan, M. B., and Bartek, J. (2004). Cell-cycle checkpoints and cancer. Nature 432, 316-323. [DOI] [PubMed] [Google Scholar]
- Kislinger, T., Rahman, K., Radulovic, D., Cox, B., Rossant, J., and Emili, A. (2003). PRISM, a generic large scale proteomic investigation strategy for mammals. Mol. Cell Proteomics 2, 96-106. [DOI] [PubMed] [Google Scholar]
- Krishnan, V., Nirantar, S., Crasta, K., Cheng, A. Y., and Surana, U. (2004). DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell 16, 687-700. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., Kim, M., Ahn, S. H., Zhong, G., Kobor, M. S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J. F. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22, 6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne, A., and Schwob, E. (2002). The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9, 1067-1078. [DOI] [PubMed] [Google Scholar]
- Leroy, C., Lee, S. E., Vaze, M. B., Ochsenbien, F., Guerois, R., Haber, J. E., and Marsolier-Kergoat, M. C. (2003). PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11, 827-835. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004a). Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699-713. [DOI] [PubMed] [Google Scholar]
- Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004b). Choreography of the DNA damage response; spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699-713. [DOI] [PubMed] [Google Scholar]
- Mallory, J. C., Bashkirov, V. I., Trujillo, K. M., Solinger, J. A., Dominska, M., Sung, P., Heyer, W. D., and Petes, T. D. (2003). Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair 2, 1041-1064. [DOI] [PubMed] [Google Scholar]
- McGowan, C. H., and Russell, P. (2004). The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16, 629-633. [DOI] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, T. (2002). A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14, 237-245. [DOI] [PubMed] [Google Scholar]
- Melo, J. A., Cohen, J., and Toczyski, D. P. (2001). Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., Kaliraman, V., Ibrahim, S. S., and Brill, S. J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, A. J., and Elledge, S. J. (2003). Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti, V., Clerici, M., Lucchini, G., and Longhese, M. P. (2000). The check-point protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14, 2046-2059. [PMC free article] [PubMed] [Google Scholar]
- Pellicioli, A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., Di Fiore, P. P., and Foiani, M. (1999). Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18, 6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J. (2004). Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 23, 1188-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., and Jackson, S. P. (2002a). Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547-551. [DOI] [PubMed] [Google Scholar]
- Rouse, J., and Jackson, S. P. (2002b). Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9, 857-869. [DOI] [PubMed] [Google Scholar]
- Scholes, D. T., Banerjee, M., Bowen, B., and Curcio, M. J. (2001). Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159, 1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy, D., Dimitrova, D., Rankin, J., Bass, K., Lee, K., Tapia-Alveal, C., Harvey, S., Murray, J., and O'Connell, M. (2005). Brc1-mediated DNA repair and damage tolerance. Genetics 171, 457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Sweeney, F., Yang, F., Chi, A., Shabanowitz, J., Hunt, D. F., and Durocher, D. (2005). Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15, 1364-1375. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., and Rothstein, R. (1989). Elevated recombination rates in transcriptionally active DNA. Cell 56, 619-630. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., et al. (2004). Global mapping of the yeast genetic interaction network. Science 303, 808-813. [DOI] [PubMed] [Google Scholar]
- Vaze, M. B., Pellicioli, A., Lee, S. E., Ira, G., Liberi, G., Arbel-Eden, A., Foiani, M., and Haber, J. E. (2002). Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373-385. [DOI] [PubMed] [Google Scholar]
- Vialard, J. E., Gilbert, C. S., Green, C. M., and Lowndes, N. F. (1998). The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17, 5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene delection and parallel analysis. Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- Yang, S. S., Yeh, E., Salmon, E. D., and Bloom, K. (1997). Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 136, 345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., Cortez, D., and Elledge, S. J. (2002). Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., and Elledge, S. J. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542-1548. [DOI] [PubMed] [Google Scholar]