Abstract
Ribosome biogenesis requires the nuclear translocation of ribosomal proteins from their site of synthesis in the cytoplasm to the nucleus. Analyses of the import mechanisms have revealed that most ribosomal proteins can be delivered to the nucleus by multiple transport receptors (karyopherins or importins). We now provide evidence that ribosomal protein L12 (rpL12) is distinguished from the bulk of ribosomal proteins because it accesses the importin 11 pathway as a major route into the nucleus. rpL12 specifically and directly interacted with importin 11 in vitro and in vivo. Both rpL12 binding to and import by importin 11 were inhibited by another importin 11 substrate, UbcM2, indicating that these two cargoes may bind overlapping sites on the transport receptor. In contrast, the import of rpL23a, a ribosomal protein that uses the general ribosomal protein import system, was not competed by UbcM2, and in an in vitro binding assay, importin 11 did not bind to the nuclear localization signal of rpL23a. Furthermore, in a transient transfection assay, the nuclear accumulation of rpL12 was increased by coexpressed importin 11, but not by other importins. These data are consistent with importin 11 being a mediator of rpL12 nuclear import. Taken together, these results indicate that rpL12 uses a distinct nuclear import pathway that may contribute to a mechanism for regulating ribosome synthesis and/or maturation.
Assembly of the 40S and 60S subunits of the eukaryotic ribosome takes place within the nucleolar compartment of the nucleus. As a result, the ∼75 ribosomal proteins necessary for ribosomal assembly and maturation must be imported into the nucleus and targeted to the nucleolus following their synthesis in the cytoplasm. Although the sizes of these proteins are well below the diffusion limit of the nuclear pore, ribosomal protein import requires energy and soluble nuclear transport factors (15, 32) that include a class of import receptors, often called karyopherins or importins. It has been reasoned (32) that such rapid, active transport is necessary because of the short cytoplasmic half-lives (2 to 3 min) of many ribosomal proteins (38, 39).
Importins recognize specific nuclear localization signals (NLSs) encoded in the sequences of their cargo proteins. The importins also interact with nucleoporins, which are components of the nuclear pore complex. This interaction permits a facilitated diffusive translocation of the importin-cargo complex through the pores. Within the nucleus, the importins bind to Ran:GTP, which triggers cargo release. The importin-Ran:GTP complex then recycles back to the cytoplasm, where the GTP on Ran is hydrolyzed, and Ran:GDP dissociates from the importins (all reviewed in references 5, 10, 20, 21, 24, 26, 41, and 42). The classical nuclear import pathway mediates import of substrates containing basic NLSs via an importin-α-importin-β heterodimer (1, 2, 6, 9, 11). However, studies with the budding yeast Saccharomyces cerevisiae revealed that some ribosomal proteins are imported by distinct importins, Kap123p and Kap121p (32, 33). Importantly, these studies also demonstrated that a single import substrate could be translocated by multiple karyopherins independently. Interestingly, the same two karyopherins also mediate the import and nucleolar localization of the four core proteins of the signal recognition particle, and it has been proposed that they constitute a nucleolar import pathway (12).
Analysis of ribosomal protein import in mammalian cells corroborated and extended the findings from these yeast studies (15). Two importins, importin-β and transportin, that recognize distinct cargoes with unrelated NLSs and two less-well-characterized importins, importin 5 and importin 7, are each capable of importing certain ribosomal proteins. An isolated domain (termed the BIB domain) in one of the substrates, ribosomal protein L23a (rpL23a), can avidly bind all four receptors. Furthermore, transportin can bind in vitro to both its classical NLS (the M9 peptide) and the BIB domain simultaneously, suggesting that multiple cargoes can be coimported by a single karyopherin.
Taken together, these results support the existence of a mechanism we term the general ribosomal protein import pathway that makes use of multiple karyopherins. This redundancy may be required to support the high production rate of ribosomes in a proliferating cell. Nonetheless, it remains an open question as to whether all ribosomal proteins use this general pathway. It is conceivable that the provision of a specialized import pathway for key components could afford a useful level of control for ribosome synthesis and maturation.
In this report, we provide evidence that the nuclear import of mouse rpL12 differs from that of the bulk of ribosomal proteins and that importin 11 serves as a transport receptor for rpL12. In both in vitro and in vivo assays, rpL12 competes with UbcM2, an importin 11 substrate (28), for binding to the transport receptor, indicating that the two cargoes are not coimported by importin 11. In addition, our data suggest that importin 11 is not a principal contributor to the general ribosome protein import pathway.
MATERIALS AND METHODS
Cloning and recombinant protein expression.
pK-rpL12-GFP (green fluorescent protein)-GFP (L12-GG) was generated by PCR amplification of the coding sequence of mouse rpL12 and ligation of the PCR product into the XbaI site of pK-GFP-GFP. The bacterial vector pGEX-L12-ZZ was constructed by ligating the PCR-amplified coding sequence of mouse rpL12 into the BamHI site of pGEX-zz. pGEX-zz is a derivative of pGEX-2T (Pharmacia) that codes for two tandem Z domains fused in frame to the carboxy terminus of glutathione S-transferase (GST). The Z domain is the 60-amino-acid-residue domain of immunoglobulin G that binds to protein A (8). pK-L23-GFP was generated by ligating the PCR-amplified coding sequence of human rpL23a into the XbaI site of pK-GFP. Both pK-GFP-GFP and pK-GFP are derivatives of the mammalian expression plasmid pRK7 (19). The construction of all human importin 11 constructs used in these studies has been described previously (28). The importin-β open reading frame was cloned into the BamHI-EcoRI sites of the yeast expression vector pGBT10 (28). The open reading frame of importin 5 was subcloned from pQE-32 into pGBT10.
All recombinant proteins used in these studies, with the exception of GST-L12 and H6-S-importin 11, were expressed in Escherichia coli BL21 (DE3) by growing cultures to an optical density at 600 nm (OD600) of ∼0.8 at 37°C and overnight induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18°C in Terrific Broth (plus 2% ethanol). GST-L12 and H6-S-importin 11 were grown at room temperature to an OD600 of ∼1.0 and induced at room temperature with 2 mM IPTG for 4 h in Terrific Broth. GST-L12 and GST-L12-zz were purified with glutathione-Sepharose beads (Pharmacia). Following purification, L12-zz was generated by thrombin treatment of GST-L12-zz in thrombin-cleavage buffer (50 mM Tris-HCl [pH 8.0], 2.5 mM CaCl2, 150 mM NaCl, 0.1% β-mercaptoethanol). L12-GFP-GFP-H6 (L12-GGH6) and myc-UbcM2-H6 were purified with Ni2+-nitrilotriacetic acid (NTA) agarose beads (Qiagen). The techniques used for purification of H6-S-importin 11 and UbcM2-GFP-GFP-H6 (Ubc-GGH6) have been described previously (28).
Yeast two-hybrid and conjugation assays.
Wild-type importin 11 and importin 11(78-975) were used as baits in separate yeast two-hybrid screens of a random-primed, murine library created from a whole, 10-day embryo. From 1.2 × 106 transformants in the wild-type importin 11 screen, five independent positive clones were identified that included one full-length rpL12 open reading frame. From 5.4 × 106 transformants in the second screen, seven positive clones were identified, five of which corresponded to rpL12.
Yeast conjugation assays were done as described previously (25). Importin 11, importin 11(78-975), importin-β, and importin 5 were expressed in the S. cerevisiae HF7c (MATa) strain as carboxy-terminal fusions to the DNA-binding domain (DBD) of the GAL4 protein. rpL12 and Ran were expressed in the W303α (MATα) strain as carboxy-terminal fusions to the transactivation domain of the herpesvirus protein VP16.
Transient transfections and immunofluorescence.
Baby hamster kidney (BHK) cells were seeded onto polylysine-coated coverslips 20 to 24 h prior to transfection and transfected by a calcium phosphate precipitation procedure (27) or with the Effectene transfection reagent kit according to the manufacturer's directions (Qiagen). Cells were fixed for 20 min in 4% paraformaldehyde-phosphate-buffered saline (PBS), permeabilized for 2 min in −20°C methanol, and blocked for at least 1 h in 3% nonfat milk-PBS prior to antibody incubations. Hemagglutinin (HA)-tagged proteins were detected with monoclonal antibody 12CA5 (1:500) and Texas Red-conjugated, donkey anti-mouse secondary antibodies (1:1,500), and DNA was stained with 4",6-diamidino-2-phenylindole (DAPI; 0.01 μg/ml in PBS). Fluorescence microscopy was performed as described previously (28). Quantitative analyses were performed with Openlab software according to the following guidelines. (i) All fluorescence images were captured within the linear range of the camera. (ii) Nuclei were defined by DAPI staining. (iii) Exposure times were recorded for each image captured and factored into each data set to allow for a comparison of a range of expression levels. (iv) A separate background fluorescence measurement was captured and subtracted from each image. The statistical significance of all quantitative data was assessed with Student's t test (homoscedastic, two-tailed distribution).
Microinjections.
Microinjections were performed as described previously (27). L12-GGH6, GGNLS, and L23-GFP were injected at 3.75 μM (unless otherwise indicated in figure legends), wild-type Ran was injected at 100 μM, Q69L Ran was injected at 41 μM, and myc-UbcM2-H6 and zz-BIB-H6 were each injected at 50 μM. Cells were incubated for either 10 or 30 min (as indicated in the figure legends) at 37°C following injections, prior to being processed for fluorescence microscopy. For the time-lapse experiments, cells were maintained on a heated stage at either 25 or 30°C for L23-GFP or L12-GGH6 injections, respectively. Starting immediately prior to each injection, images were captured every 9 s for 3 min.
Binding assays.
All binding assays were carried out at 4°C. 35S-labeled importin 11 and importin-β were synthesized by in vitro transcription translation by using the pKH3-importin 11 and pKH3-importin-β vectors, respectively, as templates, according to the manufacturer's instructions (Promega). For the rpL12 competition experiment, 0.5 μg of Ubc-GGH6 was incubated in 60-μl reaction mixtures containing 35S-labeled importin 11, 20 μl of Ni2+-NTA agarose beads, increasing amounts of either recombinant rpL12-zz or zz (1, 5, or 10 μg), and binding buffer 1 (50 mM HEPES-KOH [pH 7.5], 100 mM LiCl, 0.25% Tween 20, 5 mM MgCl2, 5% bovine serum albumin [BSA]). Reaction mixtures were shaken at 1,400 rpm for 1 h, and bead-associated proteins were pelleted by centrifugation (Eppendorf microfuge, 4°C, 4 min, 16,000 × g) and rinsed with 2× 500 μl of binding buffer 1 lacking BSA and 1× 500 μl of PBS, prior to being solubilized with 20 μl of 2× concentrated Laemmli buffer. The bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by either Coomassie brilliant blue (CBB) staining or fluorography. For the BIB binding experiment, 3.6 μg of zz-BIB-H6 was incubated in 50-μl reaction mixtures containing either 35S-labeled importin 11 or 35S-labeled importin-β, 20 μl of Ni2+-NTA agarose beads, and binding buffer 2 (50 mM HEPES-KOH [pH 7.5], 100 mM NaCl, 0.5% Tween 20, 5 mM MgCl2, 5% BSA). Reaction mixtures were shaken at 1,400 rpm for 1 h. The bead-bound complexes were then rinsed twice with binding buffer 2 lacking BSA and combined with 7 μM GST-Ran preloaded with GTP, 7 μM GST-Ran(T24N), or binding buffer 2. After a 30-min incubation, the supernatant from each reaction was solubilized for SDS-PAGE, CBB staining, and fluorography. The remaining bead-bound complexes were then rinsed with 500 μl of PBS prior to being solubilized for SDS-PAGE, CBB staining, and fluorography. Binding assays to demonstrate a direct interaction between rpL12 and importin 11 were done as described previously (28).
RESULTS
As a strategy to identify candidate import cargoes for the transport receptor importin 11, we carried out separate yeast two-hybrid screens with wild-type importin 11 and an amino-terminally truncated importin 11, importin 11(78-975), as baits and a mouse cDNA library as the source of prey proteins. From these screens, we recovered multiple independent clones for two putative transport substrates, the ubiquitin-conjugating enzyme, UbcM2, and rpL12 (Table 1). No other mouse ribosomal proteins were recovered from the screens, suggesting that the interaction with rpL12 is specific. Previously, we demonstrated that importin 11 mediates the nuclear import of UbcM2 (28). In the present study, we examine the relationship between rpL12 and importin 11.
TABLE 1.
Results of two-hybrid screens with importin 11 proteins as baita
| Bait | Prey (no. of clones) |
|---|---|
| Importin 11 (wild type) | UbcM2 (2) |
| Npap60 (1) | |
| rpL12 (1) | |
| Ran (1) | |
| Importin 11(78-975) | UbcM2 (1) |
| rpL12 (5) | |
| KIAA312p (1) |
Two screens were carried out, one with human, wild-type importin 11 as the bait and a second with an amino-terminal deletion mutant lacking 77 amino acids, importin 11(78-975). A list of the mouse proteins (prey) recovered from each screen is shown along with the number of independent clones retrieved (in parentheses).
All other ribosomal proteins that have been tested to date can be imported by the general ribosomal protein import pathway, comprised of multiple karyopherins (15, 32, 33). To determine whether rpL12 belongs to this group of proteins and can access the general pathway, we tested whether rpL12 could interact with importin 5 or importin-β, in addition to importin 11, by using a yeast two-hybrid mating assay. The results of such an assay (Fig. 1A) show that rpL12 interacted with both wild-type importin 11 and importin 11(78-975), a mutant that lacks 77 amino acids from the N terminus and does not efficiently bind Ran:GTP (28). In contrast, it did not interact with either importin-β or importin 5. The expression and functional integrity of these two transporters were confirmed by their interaction with Ran. These data demonstrate that, in this assay, ribosomal protein rpL12 interacts with importin 11 and not with two other importins implicated in the general ribosomal protein import pathway.
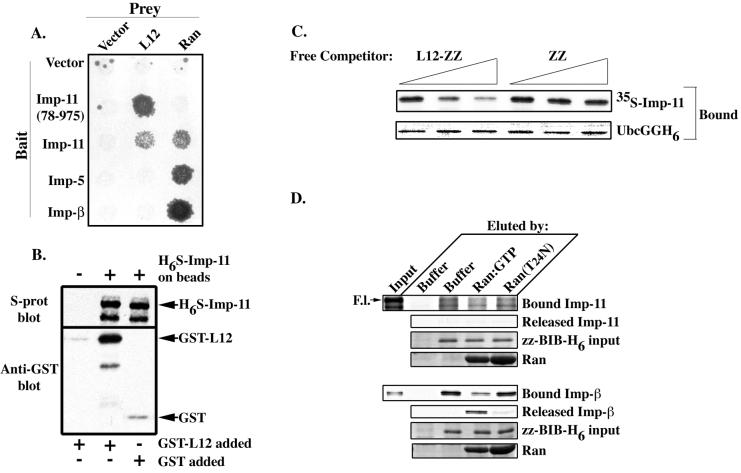
FIG. 1.
rpL12 binds specifically and directly to importin 11. (A) HF7c (MATa) yeast that express the GAL4 DBD alone (Vector) or the indicated transport receptors as GAL4 DBD fusions (Bait) were mated with the W303 (MATα) strain expressing either the VP16 TA domain alone (Vector) or as a fusion with Ran or rpL12 (L12) (Prey). Diploid yeasts were selected on Leu− Trp− plates and replica plated onto Leu− Trp− His− plates. Growth of yeast on the triple dropout plates indicates an interaction between the bait and prey proteins. (B) GST-L12 (83 nM) or GST (357 nM) was mixed with S-protein agarose beads plus H6-S-imp-11, and GST-L12 was also mixed with beads alone. Bound proteins were immunoblotted with an anti-GST antibody (Anti-GST blot) or with peroxidase-conjugated S-protein (S-prot blot). (C) 35S-labeled importin 11, expressed by in vitro transcription-translation, was incubated with recombinant UbcGGH6, Ni2+-agarose beads, and increasing amounts of either recombinant L12-zz or zz (free competitor). Proteins remaining associated with the beads were separated by SDS-PAGE and detected by CBB staining (UbcGGH6) or fluorography (35S-labeled Imp-11). (D) 35S-labeled importin 11 or importin-β was combined with recombinant zz-BIB-H6 immobilized on Ni2+-agarose beads or with beads alone. Bead-associated proteins were eluted by buffer (buffer), by GST-Ran loaded with GTP (Ran:GTP), or by GST-Ran(T24N), a mutant Ran that is defective in nucleotide binding [Ran(T24N)]. Proteins remaining associated with the beads (Bound Imp-11, zz-BIB-H6 input, and Bound Imp-β), and those eluted (Released Imp-11, Ran, and Released Imp-β) were resolved by SDS-PAGE and detected by either CBB staining (zz-BIB-H6 and Ran) or fluorography (Bound and Released Imp-11 and Imp-β). Ten percent of the starting material added to each reaction is shown (Input). Full-length (F.l.) 35S-importin 11 is indicated by an arrow. The bound fraction represents 50% of the bead-associated proteins, and the released fraction represents 37.5% of the eluted proteins.
To determine if rpL12 and importin 11 can interact directly, we carried out bead-binding assays with bacterially expressed forms of each protein. H6-S-tagged importin 11 (H6S-Imp-11) was immobilized on nickel-NTA beads and incubated with either GST-L12 or GST. Following several washes, the bead-associated proteins were solubilized, separated by SDS-PAGE, and detected by immunoblotting. The results of this experiment (Fig. 1B) show that rpL12 and importin 11 can associate directly in vitro.
We found previously that importin 11 mediates the nuclear import of a ubiquitin-conjugating enzyme, UbcM2 (28). Therefore, we next asked whether the binding of UbcM2 and the binding of rpL12 to importin 11 are mutually exclusive (i.e., substrates cannot coimport) or whether the two import cargoes bind to nonoverlapping sites (i.e., substrates may coimport). For this experiment, we performed a competition assay using recombinant UbcM2 bearing two, tandem GFP moieties and a six-His tag as a carboxy-terminal fusion (Ubc-GGH6) immobilized on nickel-NTA agarose beads. The beads were incubated with in vitro transcribed-translated, 35S-labeled importin 11 plus increasing concentrations of either recombinant rpL12 bearing a carboxy-terminal zz tag (L12-zz) or, as a control, zz alone. The amount of 35S-labeled importin 11 bound by Ubc-GGH6 was then assessed by SDS-PAGE and fluorography. We predicted that if rpL12 and UbcM2 bind an overlapping site on importin 11, then L12-zz should compete the binding of importin 11 to Ubc-GGH6, as indicated by a dose-dependent decrease in 35S-labeled importin 11 associated with the beads. Conversely, if rpL12 and UbcM2 bind to nonoverlapping sites, then L12-zz should not reduce the amount of 35S-labeled importin 11 bound by Ubc-GGH6. The results of this experiment (Fig. 1C) demonstrate that L12-zz effectively competes with Ubc-GGH6 for 35S-labeled importin 11 in a dose-dependent fashion. These data imply that both cargoes interact with the same or overlapping sites on importin 11 and cannot be imported into the nucleus as a trimeric complex with importin 11.
To examine if importin 11 plays a role in the general ribosomal protein import pathway, 35S-labeled importin 11 or importin-β was incubated with bacterially expressed zz-BIB-H6, immobilized on Ni2+-agarose beads, or with beads alone. zz-BIB-H6 is a fusion of two tandem Z domains to the BIB domain, the 43-amino-acid region of rpL23a that binds import receptors (15). Following multiple washes to remove unbound proteins, the bead-bound complexes were incubated with either buffer alone, GST-Ran preloaded with GTP, or GST-Ran(T24N), a mutant that is defective in nucleotide binding (17). Because the dissociation of importin-cargo complexes by Ran:GTP is a hallmark of nuclear transport pathways, disruption of the bead-bound complexes by Ran:GTP is indicative of a specific interaction in this assay (28). The eluted proteins, as well as those remaining associated with the beads, were solubilized for SDS-PAGE and detected by CBB staining or fluorography. The results from this experiment show that importin 11 only weakly and nonspecifically interacted with the BIB domain, and none of the receptor was specifically dissociated by Ran:GTP (Fig. 1D, top set of panels). However, and as shown previously (15), importin-β interacted with zz-BIB-H6 in a Ran:GTP-sensitive fashion (Fig. 1D, bottom set of panels). Because the BIB domain has been shown to avidly bind to four different importins (15), these data further bolster the notion that importin 11 may not be a major participant in the general ribosome protein import pathway.
To investigate the nuclear import of ribosomal protein rpL12, we expressed a carboxy-terminal fusion protein of rpL12 bearing two, tandem GFP moieties (L12-GG) by transient transfection in baby hamster kidney (BHK) cells and analyzed the intracellular distribution of this fusion protein by fluorescence microscopy. The double GFP tag was used because ribosomal protein rpL12 has a mass of only 20 kDa and is likely, therefore, to be able to diffuse freely between the cytoplasmic and nuclear compartments. The L12-GG fusion protein is ∼80 kDa in mass and thus exceeds the diffusion limit for the nuclear pore (estimated to be ∼50 to 60 kDa) (4). When expressed alone, L12-GG was distributed in both the nucleus and cytoplasm and concentrated within the nucleoli, the site of ribosome assembly (Fig. 2Aa). Because the reporterprotein was only minimally degraded, as assessed by anti-GFP immunoblotting (data not shown), its nuclear localization indicates that the import of rpL12 is carrier mediated.
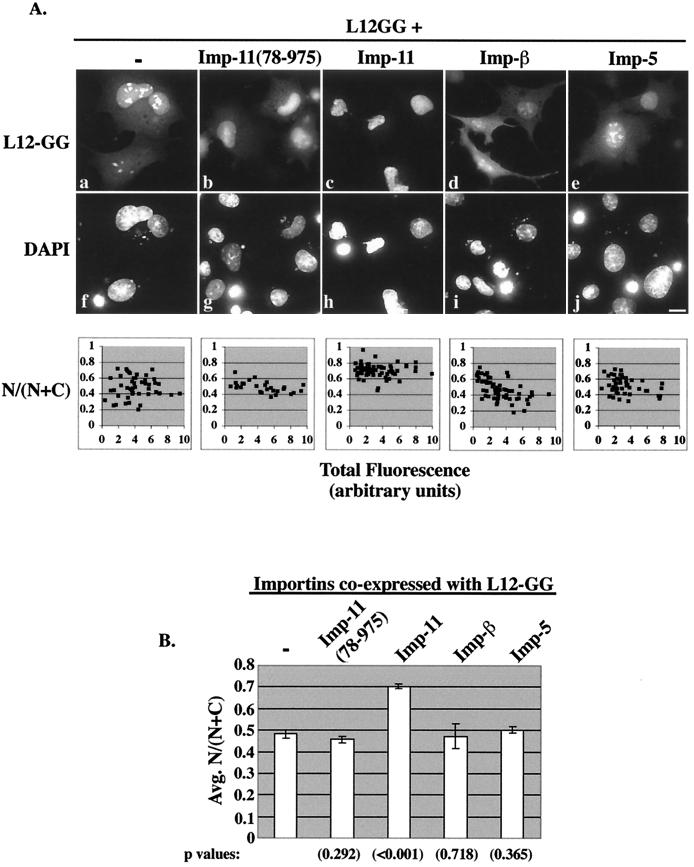
FIG. 2.
Importin 11 selectively increases the nuclear accumulation of rpL12. (A) Transiently transfected BHK cells ectopically expressing L12-GG alone or with the indicated HA-tagged importins were fixed, permeabilized, DAPI stained, and analyzed by fluorescence microscopy. Representative cells from each sample are shown. Panels a to e show the GFP fluorescence (L12-GG), and panels f to j show the DNA staining (DAPI). Bar, 10 μm. Corresponding graphs show fractional nuclear GFP fluorescence for a range of expression levels of L12-GG, in the presence or absence of the coexpressed importins. GFP images were captured such that no pixels were saturated and quantitated to obtain total fluorescence (N + C) and nuclear fluorescence (N) for each cell. The data were compiled from 25 to 50 cells for each sample. The L12-GG images shown were all captured with identical camera settings and exposure times and adjusted to the exact same settings using Photoshop software. As a result of these postquantitative adjustments, some pixels appear to be saturated, especially in the nucleoli. (B) Average N/(N + C) calculated from the data presented in the above scatter graphs. Standard error bars are indicated for each set of data, and statistical significance was determined by comparing the N/(N + C) values obtained for L12GG alone to those from each of the other conditions by using Student's t test. P values are indicated in parentheses below each bar of the graph. (C) BHK cells ectopically expressing a GFP fusion of ribosomal protein L23a (L23-GFP) alone or with the indicated HA-tagged importins. Cells were processed as described for panel A. The left panels show the GFP fluorescence (L23-GFP), and the right panels show the DNA staining (DAPI).
Coexpression of L12-GG with tagged importin 11 increased the nuclear accumulation of the fusion protein (Fig. 2Ac). Quantitation of the nuclear and cytoplasmic GFP fluorescence in these transfected cells revealed that, on average, cells transfected with L12-GG alone contained 48% of the fusion protein in the nucleus, whereas those coexpressing exogenous importin 11 contained 70% of the reporter within the nuclear compartment (Fig. 2A and B). This importin 11-dependent increase in nuclear L12-GG often made visualization of distinct nucleoli in these cells difficult (Fig. 2Ac). Importantly, this effect of importin 11 on L12-GG localization was specific, because coexpression of different transport receptors, importin-β or importin 5, did not influence the nucleocytoplasmic distribution of L12-GG [Fig. 2Ad and e and 2B; N/(N + C) = 0.47 and 0.50, respectively]. Quantitation of the tagged importins in these cells revealed that all three were expressed at approximately equivalent levels (data not shown). In addition, overexpression of wild-type importin 11 did not detectably influence the distribution of rpL23a, expressed as a GFP fusion (Fig. 2C). Similar to rpL12, rpL23a is a 60S subunit component that binds to the 28S rRNA (16, 37). The influence of importin 11 on the nuclear localization of rpL12 was further confirmed in cells overexpressing importin 11(78-975). As predicted, this mutant, which does not efficiently release cargo in response to high Ran:GTP concentrations (i.e., within the nucleus) (28), can interact with rpL12 (Fig. 1A), but did not increase the nuclear accumulation of L12-GG [Fig. 2Ab and 2B; N/(N + C) = 0.45]. Furthermore, quantitation of L12-GG nucleolar fluorescence revealed that importin 11(78-975), but not wild-type importin 11, decreased the average nucleolar accumulation of L12-GG by ∼35% (data not shown). This effect may result from the mutant importin having a reduced affinity for Ran and therefore being able to compete for L12-GG binding with rpL12 ribosomal binding partners in the nucleolus. Conversely, the ability of wild-type importin 11 to bind Ran:GTP within the nucleus should dissociate the importin 11-L12-GG complex and thus eliminate any such competition. In contrast to this reduction of nucleolar L12-GG, importin 11(78-975) had no detectable effect on the nucleolar accumulation of L23-GFP (Fig. 2C).
Collectively, these transfection data show that importin 11 specifically increases the nuclear accumulation of L12-GG and are consistent with the hypothesis of importin 11 as a candidate import receptor for rpL12. A similar effect on nuclear accumulation, obtained with this assay, had been observed for UbcM2 (28). An alternative explanation, which cannot be ruled out, is that exogenous importin 11 increases L12-GG nuclear accumulation by decreasing the incorporation of L12-GG into ribosomes and/or reducing 60S subunit nuclear export.
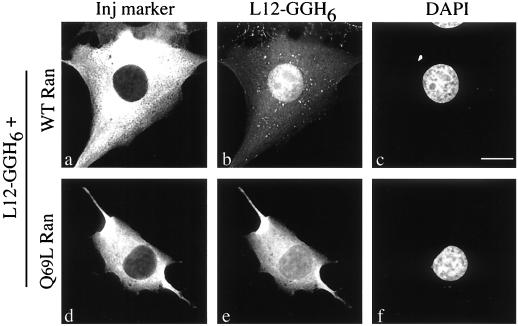
The role of the small GTPase, Ran, in nuclear transport is well established (reviewed in references 5, 10, 20, 21, 24, 26, 41, and 42). We examined the effect of Ran on rpL12 import by microinjection of bacterially expressed rpL12 bearing two, tandem GFP moieties and a six-His tag fused at its carboxy terminus (L12-GGH6). The L12-GGH6 was mixed with injection marker and either wild-type Ran or Q69L Ran:GTP (a GTPase-defective mutant that is constitutively GTP bound) (3, 17) and injected into the cytoplasm of BHK cells. After a 30-min incubation, the cells were fixed and permeabilized, and the distribution of L12-GGH6 was assessed by fluorescence microscopy. Because Ran:GTP promotes dissociation of cargo proteins from their import receptors (22, 31), this experiment tests whether rpL12 import is karyopherin and Ran mediated. The results (Fig. 3) demonstrate that the import of L12-GGH6 is Ran dependent, because Q69L Ran:GTP (Fig. 3d to f), but not wild-type Ran (Fig. 3a to c), prevented the fusion protein from accumulating in the nucleus. L12-GGH6 did not efficiently localize to nucleoli in these microinjection assays, suggesting that nucleolar accumulation is a slow process for this protein. Taken together, these data demonstrate that L12-GGH6 is functional for nuclear import and that rpL12 import is a Ran-mediated process.
FIG. 3.
Nuclear import of rpL12 is Ran dependent. BHK cells were microinjected into the cytoplasm with mixtures of L12-GGH6 (9.3 μM) and TRITC-labeled dextran (1 mg/ml) plus either wild-type Ran (a to c) or mutant Ran(Q69L) (d to f). Cells were incubated for 30 min at 37°C before fixation. Panels a and d show TRITC fluorescence (Inj marker), panels b and e show GFP fluorescence (L12-GGH6), and panels c and f show DNA staining (DAPI). Forty to 50 cells/sample were injected with similar results. Bar, 10 μm.
We next set out to obtain further evidence that the increased nuclear accumulation of L12-GG observed in cells overexpressing wild-type importin 11 was due to importin 11-mediated import. Initially, we carried out in vitro import assays in permeabilized cells, but these experiments were complicated by the nonspecific binding of bacterially expressed L12-GGH6 to cytoskeletal components. We therefore proceeded with a series of microinjection experiments in BHK cells to determine if L12-GGH6 import could be specifically competed in intact cells by another importin 11 transport substrate, UbcM2. For these studies, L12-GGH6 was introduced into the cytoplasm of cells in the presence or absence of a 13-fold molar excess of bacterially expressed, myc-tagged UbcM2 (myc-UbcM2-H6). After a short incubation, the intracellular distribution of L12-GGH6 was determined by fluorescence microscopy. In the absence of myc-UbcM2-H6, L12-GGH6 injected into the cytoplasm of cells localized efficiently into the nucleus (Fig. 4Aa to c). However, coinjection of myc-UbcM2-H6 drastically reduced the nuclear import and accumulation of L12-GGH6 (Fig. 4Ad to f), indicating that both rpL12 and UbcM2 are imported by the same transport pathway(s) and that saturation of the import receptor(s) with one cargo (i.e., myc-UbcM2-H6) prevents import of the other (i.e., L12-GGH6). These results also corroborate the in vitro competition data (Fig. 1C), indicating that rpL12 and UbcM2 bind to overlapping sites on importin 11.
FIG. 4.
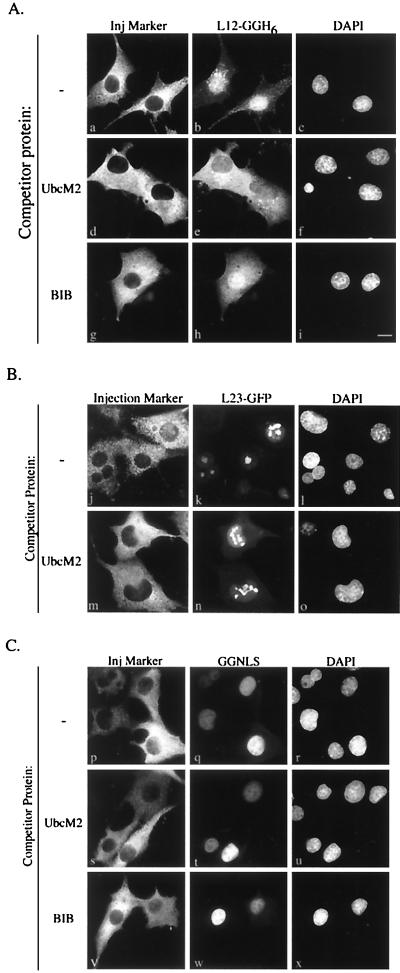
Nuclear accumulation of rpL12 is competed specifically by an importin 11 transport substrate. BHK cells were microinjected into the cytoplasm with mixtures containing a GFP-tagged import cargo (3.75 μM), TRITC-labeled dextran (1 mg/ml), and, where indicated, a competitor protein (50 μM). Cells were incubated for 10 min at 37°C before fixation. Thirty to 40 cells/sample were injected with similar results. (A) Panels a, d, and g show TRITC fluorescence (Inj marker); panels b, e, and h show GFP fluorescence from L12-GGH6; and panels c, f, and i show DNA staining (DAPI). Competitor proteins used were myc-UbcM2-H6 (UbcM2) and zz-BIB-H6 (BIB). Bar, 10 μm. (B) Panels j and m show TRITC fluorescence (Inj marker); panels k and n show GFP fluorescence from L23-GFP; and panels l and o show DNA staining (DAPI). The competitor protein used was myc-UbcM2-H6 (UbcM2). (C) Panels p, s, and v show TRITC fluorescence (Inj marker); panels q, t, and w show GFP fluorescence from GGNLS; and panels r, u, and x show DNA staining (DAPI). The competitor proteins used were myc-UbcM2-H6 (UbcM2) and zz-BIB-H6 (BIB).
To test the specificity of the competition by myc-UbcM2-H6, several sets of control injections were done. First, cells were coinjected with L12-GGH6 and a 13-fold molar excess of bacterially expressed zz-BIB-H6. In this experiment, zz-BIB-H6 represents an artificial import substrate for the general ribosomal protein import pathway (15). zz-BIB-H6 had only a marginal effect on the nuclear import and accumulation of L12-GGH6 (Fig. 4Ag to i). A second series of control injections were carried out with a GFP fusion of rpL23a (L23-GFP) ± the myc-UbcM2-H6 competitor protein. In contrast to L12-GGH6, L23-GFP localized to the nuclei (and nucleoli) equally efficiently in the absence or presence of myc-UbcM2-H6 (Fig. 4B). Attempts to inhibit L23-GFP import with zz-BIB-H6 were hindered by immediate precipitation of the L23-GFP when the two proteins were mixed together, under all buffer conditions tested. A third set of controls, obtained with a nonribosomal import substrate, confirmed that the competitor proteins were not affecting global nuclear import in a nonspecific fashion. Cells were injected with an artificial import substrate (GGNLS) comprised of GST, GFP, and the NLS from the simian virus 40 large-T antigen; this NLS has been shown to be imported by the importin-α-importin-β heterodimer (1, 6, 9, 14, 23, 30, 40). The results of these injections (Fig. 4C) demonstrate that GGNLS import was not diminished by coinjection of either myc-UbcM2-H6 or zz-BIB-H6.
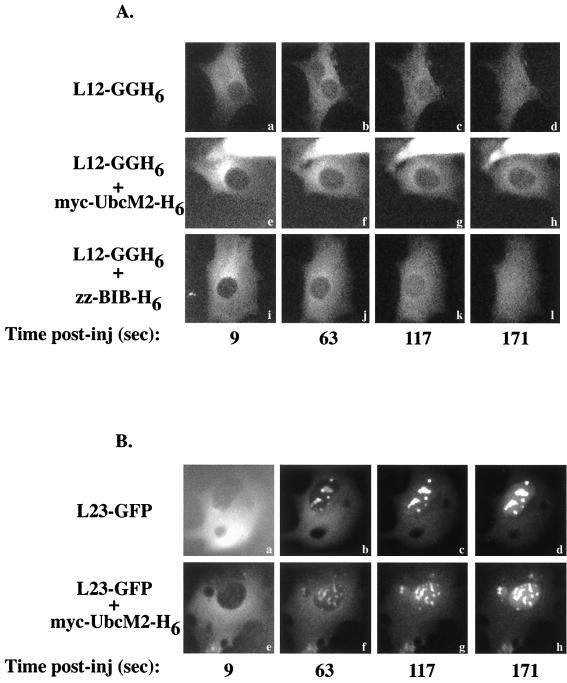
To further confirm that UbcM2 could specifically compete the import of rpL12, time-lapse experiments were carried out under conditions that permitted the initial stages of L12-GGH6 import to be analyzed. BHK cells were maintained on a heated stage at 30°C for these experiments to reduce the rate of L12-GGH6 import such that translocation of the reporter protein was less than 50% completed within 3 min. Furthermore, because L12-GGH6 is too large to diffuse through the nuclear pores and virtually none is exported from control cells injected within nuclei over the same time course (data not shown), these studies monitored unidirectional translocation of the GFP reporter protein from the cytoplasm to the nucleus. Images were captured every 9 s for 3 min, beginning immediately prior to microinjection. As can be seen in Fig. 5A, L12-GGH6 was imported equally efficiently into nuclei when injected alone (Fig. 5Aa to d) or with a 13-fold molar excess of zz-BIB-H6 competitor (Fig. 5Ai to l). However, in the presence of the myc-UbcM2-H6 competitor protein, movement of L12-GGH6 into the nucleus was greatly diminished (Fig. 5Ae to h). The lack of inhibition of L23-GFP import by myc-UbcM2-H6 competitor protein in this assay (Fig. 5B, compare results without competitor [panels a to d] to those with competitor [panels e to h]) further confirmed that rpL12 and rpL23a access distinct pathways. It should be noted that for the L23-GFP time-lapse experiments, cells were maintained at 25°C rather than 30°C, because the rate of import at 30°C was too high to allow for imaging over several minutes. We interpret these time-lapse data to indicate that rpL12 and UbcM2 share a common transport receptor pathway.
FIG. 5.
myc-UbcM2-H6 reduces L12-GGH6 nuclear accumulation by competing L12-GGH6 nuclear import. (A) BHK cells were maintained at 30°C on a heated stage and injected in the cytoplasm with L12-GGH6 (3.75 μM) alone (a to d) or with myc-UbcM2-H6 (e to h) or zz-BIB-H6 (i to l). GFP images were captured with 2-s exposures every 9 s for 3 min. Shown here are several time points for each condition tested from a representative time-lapse experiment. Although the absolute amounts of nuclear accumulation varied between groups of injected cells, the relative differences observed ± myc-UbcM2-H6 were consistent. Note that the starting mean fluorescence intensities for the three cells shown are similar, all images were manipulated identically, and the bright fluorescence at the top of panels e to h is due to a neighboring cell injected with a large amount of L12-GGH6. Also, panels a, e, and i have higher background fluorescence than the other panels due to residual L12-GGH6 in the solution bathing the cells. (B) BHK cells were maintained at 25°C on a heated stage and injected in the cytoplasm with L23-GFP (3.75 μM) alone (a to d) or with myc-UbcM2-H6 (e to h). Time-lapse conditions were as in panel A.
Because UbcM2 has been shown to be transported by importin 11 (28), these microinjection data, together with our other experiments, support the hypothesis that the importin 11 pathway is a main route for rpL12 import. Furthermore, the inability of myc-UbcM2-H6 to detectably reduce the nuclear import of L23-GFP (Fig. 4B and 5B) or of zz-BIB-H6 to efficiently prevent L12-GGH6 import (Fig. 4A and 5A) implies that the importin 11 pathway does not contribute significantly to rpL23a translocation (i.e., the general nuclear import pathway for ribosomal proteins). However, our data do not rule out that importin 11 may play a secondary role in the nuclear import of other ribosomal proteins. The results of the L23-GFP control injections would also be predicted if rpL23a interacted with a domain of importin 11 that does not overlap with the rpL12-UbcM2 binding domain on the receptor. This alternate interpretation is not likely, since importin 11 did not bind to the BIB domain in a Ran-sensitive fashion (Fig. 1E). Nonetheless, these experiments do not rule out that importin 11 may bind rpL23 through a domain other than the BIB domain.
DISCUSSION
The assembly and maturation of the 40S and 60S ribosomal subunits are a complex, multistep, nuclear process. The protein components of these structures are synthesized in the cytoplasm and efficiently transported into the nucleus for incorporation into nascently forming, preribosomal particles. Multiple importins have been shown to independently promote the nuclear translocation of a handful of ribosomal proteins (15, 32, 33), and in vitro, import of bulk, purified ribosomal proteins (15) and overlay assays (32) collectively indicate that the majority of ribosomal proteins likely access this general ribosomal protein import pathway. In this study, we present data that the 60S ribosomal subunit protein rpL12 is imported into the nucleus by a separate pathway mediated by the karyopherin importin 11. Another ribosomal protein, rpL23a, does not appear to access this transport carrier. Why then does rpL12 utilize this distinct import pathway? The answer to this question may be based in kinetic analyses of 60S subunit assembly (18). In these studies, the rpL12 of S. cerevisiae was found to associate with the 60S subunit late in the assembly process, which supports the hypothesis that rpL12 could play a role in signaling the final maturation and/or nuclear export of the 60S subunit. Such a role might necessitate a distinct import pathway to prevent rpL12 from having to compete with the bulk of ribosomal (and nonribosomal) import traffic or to allow control of the maturation rate.
Correlative evidence for this hypothesis might be taken from recent studies (36) with S. cerevisiae that describe a null mutant of KAP120, the gene coding the apparent yeast homolog of importin 11 (28). A strain with deletion of KAP120 was found to accumulate GFP-tagged rpl11b (a late-assembling 60S subunit ribosomal protein) in the nucleus. The KAP120 disruption also caused a significant deficit in the production of mature 60S subunits, but did not lead to trapping of the 40S subunit in the nucleus or to global defects in nuclear import. The authors of this study speculated that Kap120p may be an importer of ribosomal proteins or of factors necessary for ribosome biogenesis or maturation, or, alternatively, may be involved in export of the 60S subunit. However, the exporter Xpo1p and an adapter protein, Nmd3p, were recently identified as the mediators of 60S subunit export (7, 13). Taken together, these studies are consistent with the notion that Kap120p, like its human counterpart, importin 11, is involved in transporting a late-assembling ribosomal component necessary for efficient maturation and subsequent export of the 60S subunit.
Our data demonstrate that the import of rpL12 is distinguished from the import of other characterized ribosomal proteins. The primary distinction is that rpL12 accesses the importin 11 pathway as a main route for entering the nucleus, whereas the bulk of yeast (32, 33) and mammalian (15) ribosomal proteins can be imported efficiently by multiple other karyopherins. Two independent lines of in vivo data have led to this conclusion. In transfection assays (Fig. 2), we found that wild-type importin 11 specifically increased the nuclear accumulation of an rpL12-GFP reporter protein (L12GG), whereas neither a mutant importin 11, nor other importins, had such an effect. Moreover, the most convincing evidence derives from our microinjection assays (Fig. 4 and 5), in which import is mediated by endogenous levels of importins. In these experiments, we found that a different importin 11 cargo, UbcM2, specifically competed L12-GGH6 nuclear translocation or accumulation, but did not reduce the localization of either a control ribosomal protein-GFP fusion, L23-GFP, or a substrate that utilizes the classical importin-α-importin-β pathway. Time-lapse analysis of the initial stages of L12-GGH6 translocation from the cytoplasm to the nucleus revealed that the ability of myc-UbcM2-H6 to compete L12-GGH6 nuclear accumulation (Fig. 5A) results from a reduction in L12-GGH6 import. Of course, based on the data presented, we cannot conclude that rpL12 only accesses the importin 11 pathway, but the failure of zz-BIB-H6 to efficiently block L12-GGH6 import (Fig. 4A and 5) suggests that the general ribosomal protein import pathway may only play a minor role in rpL12 import. rpL12 was also shown to compete in vitro (Fig. 1C) and in vivo (Fig. 4A and 5A) with UbcM2 for binding to importin 11, indicating that the two cargoes are not coimported by importin 11. In contrast, the karyopherin-binding site (i.e., BIB domain) of a prototypical ribosomal protein, rpL23a, can form a ternary complex with a karyopherin (transportin) and the M9 domain of hnRNP A1, a transportin-specific cargo (29, 34, 35), implying that transportin can coimport rpL23a and hnRNP A1 (albeit, such coimport in vitro was very inefficient) (15). Furthermore, the BIB domain was found to interact with importin-β at a domain that is distinct from the importin-α binding region of importin-β.
In summary, we have provided multiple lines of evidence that importin 11 is a major transport receptor for rpL12 import. These findings are important because they distinguish rpL12 import from that of other characterized ribosomal proteins that utilize the general ribosome protein import pathway. Thus, our data suggest a novel, importin-mediated mechanism for regulating ribosome biogenesis and highlight the functional significance of having a cellular transport system comprised of multiple transport receptors.
Acknowledgments
We thank members of the Macara laboratory for helpful advice and Lucy Pemberton for critical reading of the manuscript. We are also grateful to Dirk Görlich for the zz-BIB-H6 expression vector and the pQE32-importin 5 expression vector. Special thanks are extended to Kendra Plafker for providing bacterially expressed L23-GFP and the pK-L23-GFP mammalian expression plasmid.
This work was supported by a grant from the National Institutes of Health to I.G.M. (GM50526). S.M.P. is the recipient of a postdoctoral training fellowship from the National Institutes of Health (F32 GM20478).
REFERENCES
- 1.Adam, S. A., and L. Gerace. 1991. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66:837-847. [DOI] [PubMed] [Google Scholar]
- 2.Adam, S. A., T. J. Lobl, M. A. Mitchell, and L. Gerace. 1989. Identification of specific binding proteins for a nuclear location sequence. Nature 337:276-279. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, F. R., C. Klebe, J. Kretschmer, A. Wittinghofer, and H. Ponstingl. 1994. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonner, W. M. 1978. Protein migration and accumulation in nuclei, p. 97-148. In H. Busch (ed.), The cell nucleus, vol. 6, part C. Academic Press, New York, N.Y. [Google Scholar]
- 5.Corbett, A. H., and P. A. Silver. 1997. Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev. 61:193-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enenkel, C., G. Blobel, and M. Rexach. 1995. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 270:16499-16502. [DOI] [PubMed] [Google Scholar]
- 7.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlich, D., S. Kostka, R. Kraft, C. Dingwall, R. A. Laskey, E. Hartmann, and S. Prehn. 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5:383-392. [DOI] [PubMed] [Google Scholar]
- 10.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 11.Gorlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246-248. [DOI] [PubMed] [Google Scholar]
- 12.Grosshans, H., K. Deinert, E. Hurt, and G. Simos. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153:745-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamoto, N., T. Tachibana, M. Matsubae, and Y. Yoneda. 1995. A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J. Biol. Chem. 270:8559-8565. [DOI] [PubMed] [Google Scholar]
- 15.Jakel, S., and D. Gorlich. 1998. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeeninga, R. E., J. Venema, and H. A. Raue. 1996. Rat RL23a ribosomal protein efficiently competes with its Saccharomyces cerevisiae L25 homologue for assembly into 60 S subunits. J. Mol. Biol. 263:648-656. [DOI] [PubMed] [Google Scholar]
- 17.Klebe, C., F. R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34:639-647. [DOI] [PubMed] [Google Scholar]
- 18.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 19.Lounsbury, K. M., and I. G. Macara. 1997. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran Gtpase-activating protein (RanGAP) inhibition by karyopherin beta. J. Biol. Chem. 272:551-555. [DOI] [PubMed] [Google Scholar]
- 20.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 21.Melchior, F., and L. Gerace. 1998. Two-way trafficking with Ran. Trends Cell Biol. 8:175-179. [DOI] [PubMed] [Google Scholar]
- 22.Moroianu, J., G. Blobel, and A. Radu. 1996. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing alpha from an overlapping binding site on beta. Proc. Natl. Acad. Sci. USA 93:7059-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moroianu, J., G. Blobel, and A. Radu. 1995. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92:2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 25.Neudauer, C. L., G. Joberty, N. Tatsis, and I. G. Macara. 1998. Distinct cellular effects and interactions of the Rho-family GTPase TC10. Curr. Biol. 8:1151-1160. [DOI] [PubMed] [Google Scholar]
- 26.Pemberton, L. F., G. Blobel, and J. S. Rosenblum. 1998. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10:392-399. [DOI] [PubMed] [Google Scholar]
- 27.Plafker, K., and I. G. Macara. 2000. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol. Cell. Biol. 20:3510-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plafker, S. M., and I. G. Macara. 2000. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 30.Radu, A., G. Blobel, and M. S. Moore. 1995. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA 92:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83:683-692. [DOI] [PubMed] [Google Scholar]
- 32.Rout, M. P., G. Blobel, and J. D. Aitchison. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell 89:715-725. [DOI] [PubMed] [Google Scholar]
- 33.Schlenstedt, G., E. Smirnova, R. Deane, J. Solsbacher, U. Kutay, D. Gorlich, H. Ponstingl, and F. R. Bischoff. 1997. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 16:6237-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siomi, M. C., P. S. Eder, N. Kataoka, L. L. Wan, Q. Liu, and G. Dreyfuss. 1997. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138:1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, K., and I. G. Wool. 1993. The primary structure of rat ribosomal protein L23a. J. Biol. Chem. 268:2755-2761. [PubMed] [Google Scholar]
- 38.Warner, J. R. 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53:256-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner, J. R., G. Mitra, W. F. Schwindinger, M. Studeny, and H. M. Fried. 1985. Saccharomyces cerevisiae coordinates the accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 5:1512-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 41.Wente, S. R. 2000. Gatekeepers of the nucleus. Science 288:1374-1377. [DOI] [PubMed] [Google Scholar]
- 42.Wozniak, R. W., M. P. Rout, and J. D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8:184-188. [DOI] [PubMed] [Google Scholar]