Abstract
Helicobacter pylori, a gram-negative spiral-shaped bacterium, specifically colonizes the stomachs of humans. Once established in this harsh ecological niche, it remains there virtually for the entire life of the host. To date, numerous virulence factors responsible for gastric colonization, survival, and tissue damage have been described for this bacterium. Nevertheless, a critical feature of H. pylori is its ability to establish a long-lasting infection. In fact, although good humoral (against many bacterial proteins) and cellular responses are observed, most infected persons are unable to eradicate the infection. A large body of evidence has shown that the interaction between H. pylori and the host is very complex. In addition to the effect of virulence factors on colonization and persistence, binding of specialized bacterial proteins, known as receptins, to certain host molecules (ligands) could explain the success of H. pylori as a chronically persisting pathogen. Some of the reported interactions are of high affinity, as revealed by their calculated dissociation constant. This review examines the binding of host proteins (serum and extracellular matrix proteins) to H. pylori and considers the significance of these interactions in the infectious process. A more thorough understanding of the kinetics of these receptin interactions could provide a new approach to preventing deeper tissue invasion in H. pylori infections and could represent an alternative to antibiotic treatment.
INTRODUCTION
Helicobacter pylori is a gram-negative spiral-shaped bacillus that colonizes the gastric mucosa of humans (57). This bacterium is microaerophilic and neutralophilic and is capable of neutralizing the acidic environment owing to the production of a urease (about 6% of the cellular protein content) that is released by cell lysis (53). The production of catalase is also important because catalase-negative mutants survive poorly (71). This bacteria is extremely motile, even in viscous environments like the gastric mucus, by virtue of one to six polar sheathed flagella (73). When exposed to a hostile environment or other high-stress conditions, the bacterium transforms from its spiral highly motile form to a coccoid form (39). H. pylori was first cultured in vitro and shown to be associated with gastritis and peptic ulcers by Marshall (54) and Warren (109). Once established in the host stomach, it remains in the mucosal environment virtually for the host's entire life (25). Infection with H. pylori remains one of the most common bacterial infections worldwide (29, 70), affecting over 50% of humans (91). The prevalence of H. pylori infection reaches 90% or more in developing countries (96). The vast majority of infections persist for life with the same colonizing strain unless an eradication attempt is successful (62). Oral-to-oral, fecal-to-oral, and gastric-to-oral transmission account for most infections (29, 70).
The majority of infections are acquired early in life, probably in childhood from close contact with parents (most probably from the mother) or other children (23, 52, 85, 97). In fact, the major risk factors for acquisition of this long-lasting infection in all countries are childhood and low socioeconomic status. The observed differences in the prevalence of the infection are probably related to hygiene conditions and sanitation (70).
H. pylori infection is associated with chronic gastritis, peptic ulcers, atrophic gastritis, intestinal metaplasia, gastric adenomas, gastric hyperplastic polyps, adenocarcinomas of the distal part of the stomach, and lymphomas of mucosa-associated lymphoid tissue (29, 33, 46, 61, 70, 74, 91). These diseases, in most instances, develop many years after the host colonization.
The sequences of the entire genomes of two unrelated pathogenic strains of H. pylori (J99 and 26695) have been reported (1, 98). This work demonstrated that even though the chromosomes are organized differently in a limited number of discrete regions, the genome size, genetic content, and gene order of the two strains examined are surprisingly conserved (35).
To date, numerous virulence factors responsible for gastric colonization, survival, and tissue damage have been described and characterized for H. pylori (for a review, see reference 65) (Table 1). The critical feature of H. pylori may, however, reside not in its ability to damage host tissues but rather in its special ability to persist within the host for such a long period. Many of the pathogenic events seen during H. pylori infection may be due to the alteration of the host response by the bacterium. This explains why H. pylori flourishes in the human stomach despite an important local immune response and can restore colonization with only few bacteria surviving after failure of therapeutic eradication. Underscoring the virulence of microbial pathogens is their capacity for adaptation and survival in variable and often hostile environments encountered in the host.
TABLE 1.
Virulence factors of H. pyloria
| Factor | Function | Distribution | Reference |
|---|---|---|---|
| Urease | Buffering of stomach acidity | All strains | 38 |
| NixA | Nickel uptake for urease | All strains | 63 |
| Flagella | Motility | All strains | 40 |
| Superoxide dismutase | Resistance to killing by phagocytes | All strains | 93 |
| Catalase | Resistance to killing by phagocytes | All strains | 71 |
| NAP | Neutrophil activation | All strains | 27 |
| BabA | Adhesin for Lewis b antigen | Type I strains | 10 |
| VacA | Cytotoxicity | Most strains | 20 |
| CagA | Immunodominant antigen | Type I strains | 19 |
| PAI | Genes (31) encoding type IV secretion system | Type I strains | 13 |
| Lewis X and Y | Molecular mimicry | Many strains | 4 |
H. pylori infection is associated with a marked infiltration of the gastric epithelium by neutrophils, macrophages, lymphocytes, and plasma cells. Despite the presence of phagocytes in close vicinity to the bacteria, a significant number of infected people are unable to eradicate the microbe, since the fate of H. pylori that comes into contact with phagocytes also depends on which molecules are bound to its cell surface. Interaction with fetuin, heparan sulfate, vitronectin, and hyaluronic acid, for example, in the presence of complement was shown to inhibit phagocytosis by macrophages (14).
In this review, we focus on the interaction (specific and less specific) of cell surface molecules of H. pylori with soluble serum proteins and extracellular matrix (ECM) proteins, of which many are also present in host serum, of the human host with which the bacterium comes into contact once the tissues have been damaged through the action of toxic factors.
We know that H. pylori binds host proteins (immunoglobulin, albumin, heparin, etc.) and that some of these interactions are of high affinity: they could thus play an important role in the infectious process. Microbial proteins with binding properties for mammalian proteins are now called receptins (41), and this term is used throughout this review. This term excludes the first step of adherence to the mammalian cell surface required for a microorganism to establish itself in a host; rather, it refers to the later stages that permit the persistence of a long-lasting infectious process through binding to soluble host cell proteins. Thus, this interplay of binding of H. pylori with host proteins represents a way to adapt to an evolving infection and to evade the host immunological response, which would normally tend to establish protective immunity. This review summarizes current knowledge of H. pylori binding to serum and ECM proteins and on the biological significance of these interactions during the infectious process.
RATIONALE FOR BACTERIAL ADHESINS AND RECEPTINS
To be infectious, pathogenic bacteria have to adhere to tissues they encounter in order to overcome removal by the natural (physical) mechanisms in place to get rid of invaders. Adherence is the first step of the pathogenic process that permits colonization by the microorganism. The persistence of the bacterium and growth in a particular niche within the host can occur without the production of any detrimental effect on the host. Adhesins are bacterial proteins, glycoconjugates, or lipids involved in the initial stages of colonization mediating the interaction between the bacterium and the host cell surface. Adherence of bacteria to host cell receptors (composed of lipids, proteins, glycolipids or glycoproteins) triggers cellular changes that include signal transduction cascades, leading to infiltration of inflammatory cells (neutrophils and monocytes) and eventually to persistence of the microorganism (95). A receptin, however, does not necessarily induce a secondary effect (41). It has become evident over the past decade that many microorganisms express cell surface receptins mediating microbial adherence to host tissues. These ligand interactions are important steps in the molecular pathogenesis of many infections (76, 110). Bacteria use, as well as nonspecific hydrophobic and electrostatic forces, surface proteins with specific affinity for plasma proteins and for components of the ECM. In addition to being surface exposed, some of these proteins are also released by the bacterium. In a healthy, normal human the ECM is not exposed and thus is not accessible for interaction with bacteria. However, these molecules could become exposed after a tissue trauma following a mechanical or chemical injury or after an infection.
H. pylori has evolved adherence and colonization mechanisms to maintain itself specifically in the gastric mucosa. It is highly adaptable, as evidenced by the fact that it can infect a host for decades. It can physically interact with host cells and mucins via a number of different adhesins displaying a variety of unique receptor specificities (for a review, see reference 95). The adherence to gastric epithelial cells is readily demonstrated in tissue culture and in gastric biopsy specimens. It is noteworthy that H. pylori possesses a fibrillar hemagglutinin that binds to N-acetlyneuraminyl-(alpha-2-3)-lactose (95). This property permits us to divide H. pylori strains into hemagglutinating and non-hemagglutinating strains (28).
The bacteria following an oral infection probably reach the stomach covered with salivary mucins containing both sialylated and sulfated components that are known to bind to H. pylori (60). After ingestion, H. pylori colonizes the human gastric mucus and epithelium by several specific adhesins (95). No cell invasin has been identified for H. pylori, and most researchers agree that cell invasion is a rare event (9, 12), although a very recent in vitro study suggested that H. pylori is able to invade cultured gastric epithelial cells via a zipper-like mechanism (43). This microorganism seems, rather, to gain access to the submucosal layer by passing through the intercellular tight junctions (107). The process is probably facilitated by expression of binding molecules for host serum and ECM proteins after gastric cell lesions are induced by ammonia as a result of urease activity and H. pylori toxins (107). It is likely that H. pylori uses binding to one or more of these ECM and/or serum proteins to penetrate the intercellular junctions of gastric epithelial cells. These adherence-penetration phenomena also involve coating with host proteins to escape the immune system and initiate a chronic lifelong infection (107). In addition, H. pylori cells coat themselves with host molecules to prevent attacks by professional phagocytes and strong immune reactions and to promote colonization and survival in the host. Thus, microbial receptins as such represent an attractive target for the development of new antimicrobial therapeutics.
INTERPLAY OF H. PYLORI VIRULENCE FACTORS RESULTING IN CELL DAMAGE
Urease, cytotoxin-associated gene A (CagA), vacuolating cytotoxin A (VacA), and neutrophil-activating protein (NAP) are important determinants of H. pylori stomach colonization and pathogenicity (65). They also represent major antigens implicated in the human response to H. pylori infection; some are being used as components of vaccine preparations (22).
Briefly, from what we know now of the pathogenesis of this microorganism, H. pylori enters the gastric lumen and, through its urease activity, survives in this highly acidic, hostile environment. Ammonia molecules are produced that buffer the acidic pH (38). Because of the helical shape, the bacterium is efficiently propelled through the mucus layer, covering the gastric epithelium, via polar sheated flagella (72, 73). Reaching the apex of the gastric epithelial cells, it sticks to them through specialized adhesins (95). CagA protein is then injected into the host cells by using a type IV secretion system (18). CagA is part of a pathogenicity island (PAI) and is associated with type I strains, which are responsible for severe forms of gastroduodenal disease (13). Other toxic factors are released as well, including NAP and VacA. VacA induces alterations of the tight junctions and formation of vacuoles (20, 21); it was recently shown to promote urea diffusion across the epithelia, possibly favoring H. pylori infectivity in vivo by optimizing urease activity (99). NAP crosses the epithelial lining and recruits neutrophils, monocytes, and mast cells, causing tissue damage through the release of oxygen radicals and other factors (67, 88). Injection of CagA causes cell cytoskeleton alteration and pedestal formation and signals the nucleus to release proinflammatory lymphokines. Taken together, the direct toxic activities of VacA and of the oxygen radicals and the proinflammatory activity and indirect activities mediated by other bacterial molecules lead to tissue damage that is enhanced by loosening of the protective mucus layer and acid permeation.
The interplay of H. pylori virulence factors leads to host tissue damage that exposes the ECM proteins as well as serum proteins with which the bacterium can now come into contact. H. pylori causes chronic infection during which several mechanisms lead to damage of the epithelial cell integrity. At different stages of the infection process, it is expected that the bacterium interacts with serum and ECM proteins which would otherwise be “hidden” from the pathogen. These additional interactions would be responsible for an even more complex dynamic between the bacterium and the host, causing a lifelong persistence of the infection.
SERUM PROTEINS BINDING TO H. PYLORI
Immunoglobulin
Several bacterial species express surface proteins that interact with immunoglobulins (Igs) in a nonimmune fashion [i.e., not through the specific binding site of F(ab′)2]. These molecules provide pathogens with the potential to evade or elude the host defenses by interfering with opsonization, phagocytosis, or complement activation and consumption (111).
Tosi and Czinn (100) reported that binding of IgG to H. pylori promoted phagocytosis and killing in vitro by polymorphonuclear leukocytes. H. pylori strains are susceptible to complement and activate either the classical pathway, even in the absence of specific antibodies, or the alternative pathway (7, 32). Nevertheless, most patients with chronic active type B gastritis show a high titer of specific antibodies to H. pylori yet fail to resolve the infection. Mattsson et al. (56) demonstrated that in H. pylori infection, a strong antibody response is generated in the gastric mucosa both in asymptomatic carriers and in patients with duodenal ulcer. No difference in either antigen specificity or magnitude of the B-cell response in the stomach could be detected in these two groups of subjects. Thus, H. pylori seems capable of evading host opsonic phagocytosis mechanisms in vivo. Even though those individuals develop a seemingly vigorous humoral response to the infection, they do not eliminate the bacteria. This is why we now think that a vaccination strategy against this pathogen needs to be capable of stimulating cellular immunity (26). Reasons for this observation may be that H. pylori either could react in a nonimmune way with Igs, thus affecting the normal opsonization process, or could enzymatically degrade the specific Igs produced in response to the infection.
In a descriptive study, Amini et al. (3), using a particle agglutination assay in which IgG was immobilized on latex beads, indicated that H. pylori strains commonly expressed IgG-binding proteins, since 24 of the 32 strains studied had bound human IgG. In a subsequent investigation, the protein that was binding human IgG in a nonimmune way was identified as a 60-kDa cell surface-associated heat shock protein (Hsp) (2). This protein could bind human IgG1, IgG3 and IgM. Binding was inhibited by the kappa chain of human IgG but not by the Fc fragment, suggesting that the interaction occurs through the Fab region of the IgG molecule. No reaction was noted with human IgA, rabbit IgG, or mouse IgG. Thus, the characteristics of the H. pylori IgG-binding activity seem to differ from those of Staphylococcus and Streptococcus species (11).
The uptake of host Igs by bacterial strains expressing IgG-binding proteins may, in theory, interfere with the defense mechanisms. This reaction at the cell surface of the pathogen could influence the coexistence of the host and the parasite and may help the microorganism to evade the host defense by a mechanism of molecular mimicry (37). In addition, Hsp60, identified as the Ig-binding protein in H. pylori, represents a family of heat shock proteins that are immunodominant antigens of many microbial pathogens that could induce immune tolerance in the host (80).
Immunization with H. pylori heat shock proteins induces protective immunity in a murine model of Helicobacter felis infection (30). Since H. pylori Hsp does not bind mouse IgG, in contrast to human IgG, as demonstrated by Amini et al. (2), one could hypothesize that in this animal model, protection is achieved through the specific immune responses and interactions between the Hsp and the specific binding sites of the antibodies induced. In contrast, natural infection of humans with Helicobacter is able to establish itself and be maintained as a persistent infection despite the production of antibodies to a number of H. pylori proteins, including Hsp (45, 51). This is probably because of the higher propensity of Hsp to bind to Igs in a nonimmune manner.
One other conceivable mechanism to elude the antibody-mediated protection could be due to the direct alteration of the Igs produced by the host in response to the invading pathogen. A study by Berstad et al. (8) of the mucosal production of IgA in response to H. pylori gastritis reported that a substantial increase in the number of Ig-producing cells occurred, particularly of the IgA1 isotype. H. pylori did not produce IgA1-specific or nonspecific IgA-degrading protease activity, ruling out the hypothesis that H. pylori is capable of enzymatically degrading IgA. Nevertheless, a recent study showed that an unidentified cell wall protein of H. pylori was reported to bind enzymatically deglycosylated human IgG (55). The IgG of patients infected with H. pylori also bound to this protein and consequently lost the reactivity of the sugar chain structure. These observations suggested to the authors that H. pylori infection was related to human IgG deglycosylation.
Albumin
Albumin is the most abundant protein in human serum. It interacts reversibly with a wide variety of endogeneous (long- and medium-chain fatty acids, bilirubin, and hemin) and exogeneous compounds and serves as an important depot and transport protein (78).
An exploratory investigation using human and bovine serum albumin immobilized on latex beads was conducted. Of the 32 strains of H. pylori tested, 16 could interact with bovine serum albumin and 12 could react with human serum albumin (3). This binding was probably due to nonspecific and hydrophobic interactions, since the binding was inhibited by chaotropic agents. In addition, the albumin-binding activity could not be extracted from the bacterial surface by using either glycine buffer or phosphate buffer or by sonication. Nevertheless, the hydrophobic interaction of albumin and H. pylori could have implications in the pathogenesis process. In fact, covering H. pylori with albumin found in serum could help the microbe to evade the defense mechanisms of the host by a mimicry stratagem. In addition, we cannot exclude the possibility that albumin, as a major transport protein, could, after binding to bacterial surface, provide substances to be used by the bacterium as nutrients in a similar manner to that suggested to occur in group G streptococci (89). It is not yet clear, however, why some H. pylori strains bind albumin better than others and whether this is associated with an increased virulence. In vivo experiments should be conducted to answer these intriguing questions.
EXTRACELLULAR MATRIX COMPONENTS
ECM is a biologically active tissue comprising glycoproteins and glycosaminoglycans, which form a network through a number of specific interactions between different ECM components. The composition of ECM differs in various organs, but fibronectin, collagen types I to XV, laminin, and glycosaminoglycans such heparan sulfate, chondroitin sulfate, and others are prevalent molecules (49). The ECM serves a structural function but also affects eukaryotic cell adhesion, differentiation, migration, and proliferation. Laminin, type IV collagen, and heparan sulfate are major constituents of basement membranes. Several ECM components are also found in plasma and other body fluids.
Interaction of microorganisms with ECM could play an important role in the establishment and maintenance of long-lasting deep tissue infection. Whether the infection remains superficial or proceeds into subepithelial tissues depends on the strain characteristics as well as the host immune response. Bacteria come in contact with the ECM after either physical or chemical alterations have occurred and proteolytic enzymes and/or toxins have disrupted the skin or epithelial integrity. The normal shedding of epithelia (18 to 24 h for gastric epithelium) could also expose ECM components. Coating of bacterial cells with some of the molecules exposed could represent a mechanism to evade host immune defense mechanisms, for example.
Expression of receptins for various ECM components is influenced by bacterial culture conditions and may appear at different stages of growth. For example, H. pylori expresses ECM-binding molecules during the stationary phase of growth. For specific ECM molecules, it was observed that both coccoidal and spiral forms of the organism express binding to a similar extent (39). When studying the interactions of bacteria with the ECM, it is important to grow the bacteria under in vivo-like conditions since pH, redox potential, divalent cations, and iron availability could represent important factors for their expression (49). A large number of intracellular and extracellular human pathogens adhere to the mammalian ECM and have been shown to contribute to the virulence of the microorganisms (77, 110).
Heparin and Heparan Sulfate
Heparin and heparan sulfate constitute a class of glycosaminoglycans with common structural features. Heparin is a sulfated glycosaminoglycan best known for its anticoagulant activity. It is produced by mast cells and is stored in intracellular granules; it does not represent a normal constituent of the ECM. It is found in various animal tissues, particularly the liver, lungs, and gut. A close structural relative, heparan sulfate, is found in the ECM on the surface of most animal cells. This acidic glycosaminoglycan, a component of cell surface- or basement membrane-associated proteoglycans, is produced by both epithelial and mesenchymal cells. Heparin is more extensively sulfated than is heparan sulfate. Many microorganisms express heparin- or heparan sulfate-binding surface proteins (76). Recent studies indicate that H. pylori may actively favor this interaction by enhancing the availability of heparin in the epithelial and subepithelial environment. In fact, NAP is able to cross the epithelial layer to activate mast cells with consequent degranulation and release of the granule content (67). Recently, VacA has also been shown to recruit and activate mast cells (94).
H. pylori was screened for binding to the ECM components heparan sulfate and heparin (5). Heparan sulfate could bind H. pylori strains isolated from patients with gastroduodenal ulcer-related diseases. Of the 20 strains studied, 90% bound 125I-heparan sulfate. Another study using H. pylori strain CCUG 17874 and strain 25 indicated high-level binding of 125I-labeled heparin (108). This common property involves a protein receptor-ligand binding interaction. The low binding at high pH (pH 8) and high binding at low pH (pH 4), as well as the binding inhibition at high ionic strength (2 M NaCl), suggested that charge interactions were important for optimal binding. The observations that pepsin treatment of H. pylori did not affect the capacity for binding to heparan sulfate and that binding had a low pH optimum (pH 4 to 5) could be significant in the pathogenesis of the gastric infection caused by H. pylori. This is because in vivo, the pepsin and acidic pH found in the gastric environment of H. pylori may not interfere with binding to heparan sulfate. This interaction was inhibited by various sulfated polysaccharides at high ionic strength and high pH but was not inhibited by carboxylated or nonsulfated compounds (36). The inhibition was related to the sulfate content of the molecule but was not related to the carbohydrate polymer backbone.
Utt and Wadström (103) confirmed that binding of heparan sulfate by H. pylori at pH 4 to 6 is very common among H. pylori strains. As the previous study had indicated, inhibitory assays with various compounds such as dextran sulfate and carrageenans revealed that inhibition was related to the sulfate content of the molecule, not to the carbohydrate polymer backbone. The calculated 50% inhibitory concentration (IC50) was 3.55 × 10−7 M heparin and 5.01 × 10−6 M dextran sulfate, respectively, for H. pylori strain 25. The heparin-binding protein is located at the cell surface, as revealed by biotinylation of cell surface proteins and an indirect-immunofluorescence assay; it could correspond to a polypeptide of 55 to 60 kDa.
Proteins with heparan sulfate affinity were recently isolated from H. pylori culture supernatants by affinity chromatography on heparin-Sepharose (86). Two major extracellular proteins with heparan sulfate affinity were identified by preparative isoelectrofocusing, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and blotting experiments using peroxidase-labeled heparan sulfate. One protein had a molecular mass of 66.2 kDa and a pI of 5.4, while the other was a 71.5-kDa protein with a pI of 5.0. NH2-terminal amino acid sequencing was performed on those proteins. The 66.2-kDa heparan sulfate-binding molecule showed a high homology to Escherichia coli chaperon proteine and equine hemoglobin. A third protein binding heparan sulfate was isolated from an outer membrane protein fraction of H. pylori and shown to have a molecular mass of 47.2 kDa.
Utt et al. (102) reported that the C-terminal domain of the 58-kDa subunit of H. pylori VacA was also binding to heparin and heparan sufate. Using surface plasmon resonance and synthetic VacA toxin peptides corresponding to predicted binding peptides as inferred by using bioinformatic tools, they showed that heparan sulfate might represent a receptor/coreceptor for VacA cytotoxin. These findings supported an earlier study indicating that the binding site for the toxin receptor is located in the 58-kDa C-terminal domain (81). Although the observed binding has moderate affinity (Ka = 4.89 × 105 M−1), the multimeric form of VacA cytotoxin may bind heparan sulfate through several domains and hence may contribute to a much stronger interaction. Experiments using cells in culture showed that inhibition of cell vacuolation was observed following treatment of H. pylori cells with heparin (92), confirming the surface plasmon resonance study.
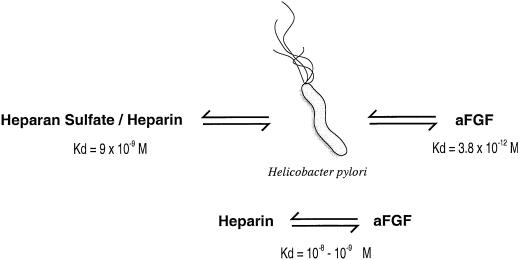
As a pathogenic mechanism, H. pylori could also interfere with growth factors and growth factor receptors important for the gastroduodenal mucosa integrity. In fact, Ascencio et al. (6) demonstrated that H. pylori not only binds to heparin-binding fibroblast growth factors (FGFs) with an extremely strong affinity (Kd = 3.8 × 10−12 M), but also could bind to heparan sulfate and heparin with higher affinity (9 × 10−9 M) (Table 2) than FGFs bind to heparin (10−8 to 10−9 M) (Fig. 1). H. pylori binds two heparin-binding growth factors (i.e., acidic and basic FGFs). Heparan sulfate-binding proteins are exposed on the surface of the microbe as well as it is shed from the surface; they are often localized close to epithelial stem cells in the gastroduodenal glands (5). Since FGFs are dependent on heparin for activation, H. pylori could efficiently interfere with growth factors and growth factor receptors, resulting in disturbances of the balance controlling the renewal, maintenance, and repair of the gastroduodenal mucosa, which are commonly observed during H. pylori infections. The high binding affinity to heparin and heparan sulfate of H. pylori could reduce the availability of FGF and result in its injurious effect on the human mucosa by inhibitory or repressive effects on the renewal and differentiation of gastroduodenal epithelial cells as well as the restoration of the tissue after damage. Heparin-binding proteins could play a role in the development of peptic ulcers by interfering with the availability and bioactivity of heparin-binding growth factors as well as of heparan sulfate.
TABLE 2.
Host serum and ECM proteins shown to bind H. pylori cells in vitro, the receptins involved, and the affinity of the interaction
| Serum and ECM proteins | Receptin(s) | Affinity (Kd)a | Referencesc |
|---|---|---|---|
| Albumin | Not identified | NDb | Amini et al. (3) |
| Human IgG1, IgG3, and IgM | 60-kDa Hsp | ND | Amini et al. (3) |
| Amini et al. (2) | |||
| Collagen (Type IV) | Not identified | 1.6 × 10−10 M | Trust et al. (101) |
| Khin et al. (39) | |||
| Ringnér et al. (82) | |||
| Fibronectin and fibrinogen | Not identified | ND | Trust et al. (101) |
| Heparin and heparan sulfate | 47.02-, 66.2- and 71.5-kDa proteins and the 58-kDa subunit of VacA cytotoxin | 9 × 10−9 M | Ascencio et al. (5) |
| Ruiz-Bustos et al. (86) | |||
| Utt et al. (102) | |||
| Wadström et al. (108) | |||
| Ljungh et al. (48) | |||
| Slomiany et al. (90) | |||
| Laminin | LPS and/or 25- and 67-kDa proteins | 7.9 × 10−9 M | Trust et al. (101) |
| Valkonen et al. (104) | |||
| 8.5 × 10−12 M | Valkonen et al. (105) | ||
| 2.5 × 10−9 M | Valkonen et al. (106) | ||
| Khin et al. (39) | |||
| Plasminogen and plasmin | 42-, 57-, and 58.9-kDa proteins | 7 × 10−7 M | Ljungh (47) |
| Pantzar et al. (75) | |||
| Ringnér et al. (83) | |||
| Ringnér et al. (82) | |||
| Vitronectin | Not identified | ND | Trust et al. (101) |
The affinity of the serum and ECM proteins toward H. pylori cells.
ND, not determined.
References in bold are those from which the affinity dissociation constants were taken.
FIG. 1.
Comparison of the dissociation constant between H. pylori, acidic FGF (aFGF), and heparan sulfate or heparin. Adapted from reference 6.
Recently, heparan sulfate-binding proteins were shown to be involved in the adherence of H. pylori cells to HeLa S3 and Kato III cells (34). A 71.5-kDa extracellular heparan sulfate-binding protein was involved in promoting attachment to those cells, and rabbit polyclonal antibodies against this molecule could inhibit the adherence of H. pylori.
Despite recruitment of numerous phagocytes to the inflammatory foci in the gastric mucosa, infected people are unable to eliminate the bacteria. It has been established that strains expressing heparan sulfate-binding proteins interact with phagocytes via those molecules (15, 17). Human and murine neutrophils and macrophages seem to recognize mainly the hemagglutinins on the surface of H. pylori strains and also heparan sulfate-binding proteins on other strains (16). Treatment of H. pylori with heparin caused a dramatic decrease in phagocytosis, probably due to a decrease of the surface hydrophobicity (14). Preincubation of the bacteria with heparin inhibited the ingestion process by 55 to 70%. Deposition of sialic acid-containing compounds, heparin, and vitronectin in the presence of complement on the bacterial surface rendered the bacteria resistant to phagocytosis (14). It is probable that in an inflamed tissue, binding of these compounds by H. pylori allows the bacteria to avoid phagocytosis.
Duensing et al. (24) describe a novel, extragenomic mechanism of bacterial surface modulation that could amplify the adaptive and pathogenic potential of numerous bacterial species, including H. pylori. The mechanism described involved specific bacterial recruitment of molecules found in the immediate environment during the infection process, such as heparin, glycosaminoglycans, and related sulfated polysaccharides. These molecules could then serve as universal binding sites for a diverse array of mammalian heparin-binding proteins. The recruited molecules could bind to adhesive glycoproteins (vitronectin and fibronectin), inflammatory (monocyte chemotactic protein 3, platelet factor 4, and macrophage inflammatory protein 1α), and immunomodulatory (gamma interferon) factors, and FGF. A study by Ascencio et al. (6) had clearly shown that preincubation of H. pylori cells with heparan sulfate and other sulfated polysaccharides resulted in enhanced FGF binding, supporting the novel microbial pathogenesis strategy proposed by Duensing et al. (24). This strategy may have a major impact on key aspects of microbial pathogenicity, as revealed by the observed increase in bacterial invasion of epithelial cells and inhibition of chemokine-induced chemotaxis (24).
Fibronectin and Fibrinogen
Fibronectin is a 440-kDa glycoprotein found in the ECM and body fluids of animals. It was the first ECM protein shown to act as a substrate for adherence of microorganisms to eukaryotic cells (42). Fibrinogen is a 340-kDa glycoprotein that is found in high concentrations in plasma, where it plays important roles in blood coagulation and wound-healing processes. It forms the structure of the blood clot and becomes deposited in the wound matrix primarily as a consequence of its proteolytic conversion to fibrin. Fibrin or fibrinogen could serve as a substrate for microbial receptins. Although fibrinogen has been shown for S. aureus to be an important molecule to which the bacteria bind (31), for H. pylori only a low level of binding was observed (between 5 and 7%) (83). Fibronectin also binds weakly (3 to 23%) to H. pylori (39, 101).
Collagen
A key feature of certain successful gram-positive and gram-negative bacteria is the ability after binding to cutaneous or mucosal surfaces to invade deep tissues. Collagen proteins are the major constituents of the ECM and as such may represent a major target site for microorganisms. Following attachment of H. pylori to the gastric epithelial cells, as discussed above, toxicity is exhibited. In certain cases, this leads to the destruction of the cells, loss of tissue integrity, and ultimately complete erosion of the epithelium, leaving the underlying basement membrane completely bare. Thus, in ulcerated tissue, the various connective tissue proteins, including those of the basement membrane, become exposed and available to colonizing bacteria. Therefore, the capacity to bind to such matrix proteins may be extremely important for H. pylori to colonize and damage the basement membrane before invading the lamina propria in the process of eliciting ulcerative lesions.
Collagen is a major protein of the ECM. Type IV collagen forms a two-dimensional reticulum and is a major component of the basal lamina. Type IV collagen binds at high levels to H. pylori (average binding, 27%; highest binding, 60%), while the bacterium is also capable of binding low levels of type I and II collagens (101). The ability of 16 isolates of H. pylori to bind125I-collagen was quantitated using a liquid- phase assay. Results showed that binding is rapid, not affected by pH, and saturable, indicating a limited number of receptors. The number of available binding sites on the bacterial surface was calculated to be approximately 3,000. Once bound, radiolabeled type IV collagen was not displaced by an excess of unlabeled molecule, indicating that the interaction was of high affinity (Kd = 16 nM). The ability to specifically bind to this component of the basement membrane is likely to represent an important virulence property for this gastric pathogen. The molecule(s) responsible for the interaction with collagen has not been identified yet.
Laminin
Laminin is one of the major glycoproteins of basement membranes (with type IV collagen) that plays a role in cellular adhesion processes. It anchors the cell surface to the basal lamina. It is a complex noncollagenous glycoprotein, which is important for the structure of the basement membrane by formation of networks with type IV collagen, entactin/nidogen, and heparan sulfate proteoglycans. It is involved in cell adhesion to basement membranes by normal cells, metastiting tumor cells, and some pathogenic bacteria (104). A characteristic feature of laminin is its high carbohydrate content (12 to 27%), most of which is present in complex-type oligosaccharides. Hence, it is not surprising that H. pylori as well as other microbes bind to this molecule by lectin interactions (48). The capacity of H. pylori strains to bind to laminin is unlikely to be involved in the initial colonization of gastric mucosal cells, since specific primary adherence is done through receptors in the mucus layer and on the epithelial cell surface (95). Nevertheless, this property would assume major importance once the basement membrane becomes exposed. The bacteria appear to undermine the mucosal integrity by penetrating the tight junctions of epithelial cells (107). Laminin binds H. pylori via surface proteins (104) and lipopolysaccharide (LPS) (105).
In a study by Valkonen et al. (104), N-acetylneuraminyllactose decreased laminin binding by 70% and neuraminidase treatment of laminin decrease binding by 50%, while a recombinant B1 chain of laminin, containing high-mannose-type oligosaccharides, inhibited binding by only 25%, suggesting that terminal sialic acids on laminin were competing for a specific sugar-binding protein on H. pylori cells. The receptor involved in this binding appeared to be proteinaceous since it was heat and protease sensitive. Scatchard analysis of specific binding indicated that approximately 2,000 sites per cell were present (104). The interaction of H. pylori with laminin involves a bacterial receptin recognizing certain sialylated oligosaccharides of the glycoprotein (105). Since the carbohydrate moiety of laminin is thought to play a role in eukaryotic cellular adhesion processes such an interaction of H. pylori with laminin could further disorganize the gastric epithelium. Trust et al. (101) showed that the interaction of H. pylori with laminin was rapid (15 to 30 min), of high affinity (Kd = 7.9 nM), and partially reversible. Since the binding was affected by salt, it suggested the presence of a hydrophobic component in this interaction (101).
Valkonen et al. (105) indicated that although all H. pylori strains tested bound laminin, hemagglutinating strains were good binders of laminin (maximum, 31%) compared to nonhemagglutinating strains (minimum, 6%). In their study, H. pylori purified LPS could inhibit the binding of laminin to H. pylori. Laminin binding was inhibited by homologous and also heterologous smooth-form LPS. Interestingly, nonhemagglutinating strains bind laminin with a higher affinity (Kd = 4.1 pM) than do hemagglutinating strains (Kd = 8.5 pM) but more binding sites are available on the surface of hemagglutinating strains, thereby explaining the higher extent of laminin binding by these strains (104). Thus, the hydrophobic surface- exposed component responsible for laminin binding previously described by Trust et al. (101) was shown to be attributable in part to LPS. The ability of heterologous rough LPS to produce inhibition comparable to that caused by smooth LPS indicated that the O side chain was not involved in the interaction. In fact, using dephosphorylated LPS, isolated core oligosaccharides, and free lipid A in inhibitory experiments for the studied hemagglutinating and poorly hemagglutinating strains, a phosphorylated structure and a conserved nonphosphorylated structure in the core oligosaccharide mediated the interaction, respectively. Scatchard plot analysis of data for the laminin binding by LPS indicated the presence of a single class of binding molecules with a dissociation constant of 2.5 nM. The authors postulated that the initial recognition and binding of laminin by H. pylori could occur through LPS and that subsequently a more specific interaction involving a lectin-like protein present on the bacterial cell surface could take place.
Slomiany et al. (90) demonstrated that H. pylori cells, in particular H. pylori LPS, inhibited the interaction between a laminin receptor in gastric epithelial cells and laminin. They isolated from rat gastric epithelial cell membranes a protein of 67 kDa that was shown to bind laminin. This protein receptor incorporated in liposomes displayed specific binding toward laminin, and the reaction was inhibited by LPS from H. pylori. A similar process may account for the loss of mucosal integrity in the pathogenesis of H. pylori-associated gastric disease. The subsequent disruption of epithelial cell- basement membrane interaction by LPS would explain the development and gradual loss of mucosal integrity during H. pylori infection (68).
A laminin-binding protein was identified on H. pylori (106). Inhibition of laminin binding by fetuin, but not asialofetuin, and reduced bacterial binding to periodate- or sialidase-treated laminin confirmed earlier results indicating that glycosylation and particularly sialylation were important for laminin binding by H. pylori. In fact, by using mono, di, and trisaccharides in inhibition experiments, it was shown that H. pylori bound to a region spanning a trisaccharide, in particular the trisaccharide N-acetylneuraminyl-(alpha-2-3)-lactose. Although LPS is involved in binding to laminin, inhibition experiments and heat and protease treatment of H. pylori cells indicated that a protein receptor, rather than LPS, on H. pylori bound this trisaccharide. Using Western blotting, a 25-kDa outer membrane protein was identified as responsible for the laminin-binding activity by both hemagglutinating and poorly hemagglutinating strains. Thus, it appears that this 25-kDa outer membrane protein acts as a lectin-like molecule together with H. pylori LPS to mediate attachment to laminin.
As stated above, the lectin-binding activity to laminin may disrupt epithelial cell-basement membrane interactions and may be responsible for the epithelial leakiness typically observed in H. pylori infections. The capacity of H. pylori to rapidly, avidly, and largely irreversibly bind both type IV collagen and laminin should facilitate both initial and continued colonization of any basement membrane that becomes exposed as a result of either H. pylori-induced disruption of mucosal integrity or the loss of epithelial cells resulting from eating and drinking, for example. Furthermore, the capacity to bind those proteins at both acidic and alkaline pH should promote the continued colonization of any ulcerative lesions formed in either the acidic stomach or the alkaline duodenum. Laminin binding may enable the pathogen to survive in chronically inflamed tissue.
Plasminogen and Plasmin
Plasminogen is a plasma and ECM glycoprotein composed of a 90-kDa single chain proenzyme in its native form. It is the main component of the fibrinolytic system (112) and circulates in plasma as an inactive zymogen, which under certain conditions is activated, by proteolytic cleavage, to the active form, plasmin (a serine protease). Activators such as urokinase and tissue type plasminogen activator convert plasminogen to plasmin. Plasminogen is a component of the fibrinolytic system, which is present in the plasma, subepithelial, and subendothelial basement membranes. It can be converted to a serine proteinase by a number of activators of eukaryotic and bacterial origin (44). The main function of plasminogen is to mediate fibrinolysis in normal hemostasis, a process in which fibrin is degraded to fibrin fragments. Plasmin is a potent enzyme that can dissolve blood clots. In addition, it can directly hydrolyze matrix proteins such as laminin, fibronectin, and collagens.
Plasminogen receptors are present on leukocytes, platelets, and the cell surface of several bacterial pathogens (50). Cell surface-bound plasminogen is easily activated to plasmin. Binding and activation of human plasminogen on the surface of bacterial cells is a common mechanism used by certain invasive bacteria to facilitate movement through normal tissue barriers (50). This provides the bacterium with a potent serine protease that has a broad enzyme specificity and may contribute to tissue breakdown, facilitating the invasion of H. pylori through the gastric epithelium as well as providing nutrients. It could enable bacterial pathogens to utilize the ECM proteins to penetrate infected tissue and be disseminated. In fact, degradation of matrix proteins is required to facilitate the movement of cells through tissue planes in various biological processes such as wound repair, tissue remodeling, and tumor cell metastasis (84). Bacteria able to acquire the host's enzymes with broad substrate specificity could dramatically alter the dynamics of the host-microbe relationship to favor the establishment of a long-lasting infection.
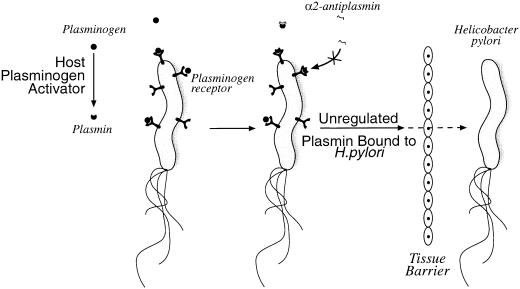
H. pylori binds to plasminogen, and the interaction is not sialic acid dependent (83). Radiolabeled plasminogen binds to hemagglutinating H. pylori strain 17874 at a high level (32%). The interaction is specific since it could not be inhibited by nonlabeled plasminogen. This interaction seems to be a common phenomenon among H. pylori strains (47). Plasminogen binding to H. pylori seems to be independent of the presence of CagA (75). The inhibition of binding by lysine and lysine analogues indicates that lysine residues are important for binding to plasminogen. It was suggested that H. pylori binds the fifth kringle structure of the plasminogen molecule. Two proteins (of 42 and 57 kDa) binding to plasminogen were identified by Pantzar et al. (75). Scatchard plot analysis revealed a unique molecule binding plasminogen with a dissociation constant of 7 × 10−7 M. No significant difference was found between binding to the spiral and the coccoid form of H. pylori (39). Conversion of H. pylori cell-bound plasminogen to plasmin in the presence of a tissue type plasminogen activator was demonstrated in vitro (Fig. 2). At the same time, receptor-bound plasmin is protected from inactivation by α2-antiplasmin. In essence, plasminogen receptors and activators turn bacteria into proteolytic organisms using a host-derived system. For H. pylori, binding is pH dependent with an optimum at pH 7. Although H. pylori survives the obligatory passage through the low pH in gastric juice, it prefers an environment close to the epithelial surface, embedded in mucus, where the pH is higher, this may account for the optimum pH observed. No activation was noted when plasminogen or tissue- type plasminogen activator was incubated with H. pylori cells alone. This indicates that H. pylori requires a host activator since it does not produce its own bacterial plasminogen activator. By using a different approach, Yarzábal et al. (112) identified a 58.9-kDa plasminogen-binding protein in a water extract of surface protein. The identified protein had a molecular mass similar to those of the proteins found in another strain of H. pylori (75).
FIG. 2.
H. pylori invasion model stressing the potential role of plasminogen-binding protein in facilitating movement through the gastric epithelium. Plasmin has the potential to degrade ECM protein and could permit migration of the bacteria into tissue. Cell-bound plasminogen is activated to plasmin and can no longer be inhibited by α2-antiplasmin.
However, a recent study demonstrated that H. pylori NAP could increase the production of tissue factor and of plasminogen activator inhibitor type 2 but not of urokinase type plasminogen activator by human blood mononuclear cells (66). Clotting assays established that NAP-induced tissue factor is functionally active, thus triggering blood-clotting activation and promoting fibrin formation. The up-regulation of plasminogen activator inhibitor type 2 results in impaired proteolysis in the cell environment. Thus, NAP, by inducing the coordinate expression of cell procoagulant and antifibrinolytic activities, could favor fibrin deposition and contribute to the inflammatory reaction of the gastric mucosa elicited by H. pylori.
The plasminogen binding and conversion to plasmin is the only known proteolytic activity of H. pylori (69). This proteolytic activity may enhance tissue penetration and be involved in carcinogenesis. For now, in vivo evidence for a role of plasminogen activation in pathogenesis is limited to Yersinia pestis, Borrelia, and group A streptococci (44). Experiments with H. pylori should now be conducted in animal models to evaluate the role of plasminogen binding in the pathogenesis.
Vitronectin
Vitronectin (also called serum-spreading factor or S-protein) is a serum and ECM glycoprotein known to promote the attachment and spreading of cells in vitro. It also interacts with the C5b-9 complex inhibiting cell lysis by the complement system and protects thrombin from inactivation by antithrombin III. Vitronectin binds to H. pylori (53%) (82, 83), and this reaction is fast, saturable, and reversible. Binding is heat and protease sensitive, suggesting that it is mediated by bacterial cell surface proteins. Cell surface protein extracts partly inhibited the binding, indicating that the vitronectin- binding structures are easily extracted surface proteins. Sialic acid-specific hemagglutinins were suspected to mediate this reaction since the reaction was inhibited by fetuin and orosomucoid but not by asialofetuin. Low (5 to 8%) and high (>50%) vitronectin binders were observed among hemagglutinating strains. Purified vitronectin, which includes an urea-activation step of vitronectin, binds to H. pylori in a fast and saturable way (82). On the other hand, Trust et al. (101) reported that only small amounts of vitronectin, purified by another method, bound to H. pylori. This discrepancy could be explained by the fact that vitronectin exists in at least two structurally and functionally distinct forms in human plasma.
Binding of vitronectin, type IV collagen, and laminin, all ECM molecules, may be important for H. pylori to protect itself from cell lysis by complement or from other immunological effects. Ringnér et al. (83) also showed a specific, saturable, and dose- dependent binding of 125I-vitronectin to hemagglutinating H. pylori strain 17874. The reaction was pH dependent, and maximal binding was attained at pH 4. Growth conditions influenced the expression of vitronectin-binding properties, since H. pylori grown on agar bound 50% more vitronectin than did broth-cultured bacteria. Competition experiments using 125I-labeled vitronectin and plasminogen indicated that the binding sites for those molecules were different. However, they are probably close together, since nonlabeled vitronectin could inhibit the binding of 125I-plasminogen but nonlabeled plasminogen had no effect on 125I-vitronectin binding.
Although vitronectin treatment of H. pylori slightly reduced the uptake of bacteria by macrophages, the simultaneous treatment with vitronectin and complement rendered them resistant to phagocytosis (14). Whether the cell surface-bound plasmin is of any importance in the development of gastric cancer by H. pylori remains to be investigated.
CONCLUDING REMARKS
The study of bacterial interactions with serum and ECM proteins aims first at understanding the infection process and the settling of a long-lasting infection and second at establishing therapy methods based on preventing receptin binding and on attempts to prevent deeper tissue invasive infections. With the increasing prevalence of antibiotic-resistant strains, this new approach could provide alternatives to antibiotic treatment. This path could also be considered a complementary or alternative treatment of peptic ulcer induced by H. pylori. Before any antibacterial surface-coating strategy can be optimized for application in body compartments, the receptin-binding mechanisms will have to be elucidated in detail. As stated above, the receptins would act in conjunction with the virulence (toxic) factors described for H. pylori and would help establish this frequently observed lifelong infection.
For certain microorganisms, receptins binding to host proteins have recently been shown to contribute to virulence in animal models. For example, a fibrinogen-binding protein of Streptococcus equi subsp. equi was protective against a lethal challenge in mice (59). This protein was shown to bind equine fibrinogen as well as IgG and to contribute to virulence in a mouse model, since a knockout mutant, which did not express fibronectin-binding protein on the cell surface, showed a greatly decreased virulence (58). In addition, the molecule had antiphagocytic properties that could account for the virulence of S. equi subsp. equi. For Mycobacterium tuberculosis, a heparin-binding haemagglutinin adhesin markedly affected interactions of the bacteria with epithelial cells but not with macrophage-like cells (79). When administered intranasally to mice, a strain mutated for the heparin-binding protein was severely impaired in spleen colonization but not in lung colonization. Coating of wild-type mycobacteria with antibodies directed against the heparin-binding protein also prevented dissemination after intranasal infection. Thus, it appears that such a heparin- binding hemagglutinin is required for extrapulmonary dissemination of the microorganism, as revealed in the mouse model. Concerning H. pylori, it was observed that intragastric vaccination with heparan sulfate-binding protein covalently coupled to the beta-subunit of cholera toxin protected mice against a challenge with H. pylori, as revealed by microbiological, histopathological, and molecular methods (87).
The numerous receptins described for H. pylori should not be surprising since this bacterium, in order to be a long-term resident in the human host, must have the potential to express many binding molecules at different times during the infectious process and in different combinations (Fig. 3). As we have observed for this pathogen, the interactions with certain host proteins are of high affinity and therefore are most probably not negligible as a pathogenicity attribute. The multiplicity of both receptins and their respective binding molecules (ligands) allows the bacteria to survive in a very dynamic microenvironment as the infection evolves, in some instances, to an invasive stage. These adherence-penetration phenomena involve coating of the microbe with host proteins to escape the host immune system and initiate a chronic lifelong infection process.
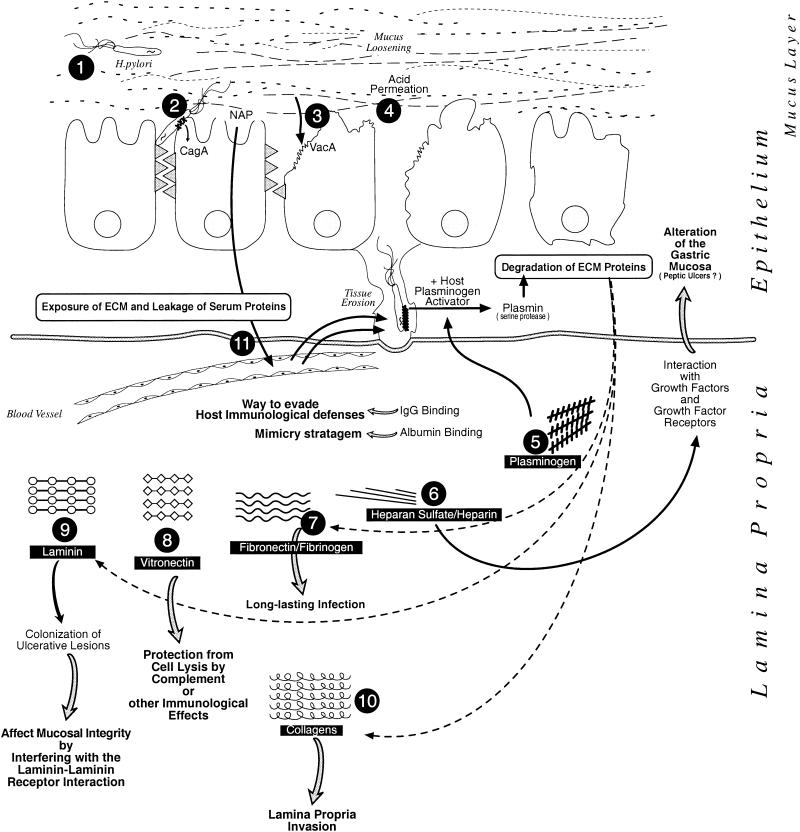
FIG. 3.
Schematic representation of the binding events that could take place between H. pylori and certain host serum and ECM proteins and the inferred resulting effects on the infectious process. Once the bacterium has entered the gastric lumen, its flagella propel it into the mucus layer (step 1) and allow it to reach the apex of the gastric epithelial cells, to which it sticks via specialized adhesins (step 2). H. pylori can, using a type IV secretion system, inject CagA protein into the host cells and also release other toxic factors such as NAP and VacA. Alterations of the tight junctions and formation of large vacuoles result from VacA action. Toxic activity of VacA and of reactive oxygen intermediates produced as a result of NAP leads to a loss of integrity of the epithelial mucosal surface, by exposing molecules constituting the ECM as well as leaking of serum proteins (step 3). Tissue damage is enhanced due to loosening of the protective mucus layer and acid permeation (step 4). As in vitro studies have shown, plasminogen, after binding to the H. pylori cell surface, could, in the presence of host plasminogen activator, be transformed into plasmin (a serine protease) that could be directly involved in the degradation of various ECM proteins (including fibronectin, fibrinogen, laminin, and collagens). This host-derived proteolytic activity could be involved directly in tissue erosion (step 5). H. pylori could bind heparan sulfate (and probably heparin after release from intracellular granules) and interfere with growth factors and growth factor receptors affecting the gastric mucosa. This interaction could be involved in peptic ulcer production (step 6). The bacterium could also bind weakly to fibronectin and fibrinogen molecules and permit the establishment of a long-lasting infection (step 7). Binding to vitronectin could result in protection from complement cell lysis or other immunological effects (step 8). Laminin- binding could promote continuous colonization of basement membrane and of ulcerative lesions. It could also account for the loss of mucosal integrity observed in H. pylori infection by interfering with the reaction between the laminin receptor and laminin that accounts for the basement membrane structure (step 9). In ulcerated tissue, collagen molecules become exposed and are available to H. pylori, probably helping invasion of the lamina propria (step 10). Tissue alteration and ulceration could also lead to leakage of serum proteins following blood vessel disruption. Binding to immunoglobulins could provide the pathogen with a way to evade the host immunological defenses by interfering with opsonization, phagocytosis, and/or complement activation and comsumption. Albumin coating could also provide a mimicry stratagem so the invader is recognized as self (step 11). Taken together, those numerous reactions with host proteins would permit the establishment of a long-lasting infection, as observed for H. pylori.
Vaccines based on recombinant ECM-binding proteins were shown for S. aureus, for example, to be much more protective than those based on whole bacteria (31). Binding of large amounts of a soluble host protein to the bacterial surface may result in a protective coat, allowing the bacteria to evade recognition by host defense mechanisms. An advantage for the microorganism would be the ability to up-regulate the expression of receptins when it senses attachment to serum or ECM molecules, allowing more effective colonization, invasion, and destruction of host tissues. Thus, a study of the environmental control of bacterial receptins should be pursued in order to better understand the signal(s) triggering the regulation of receptins elaborated by H. pylori. Potential targets for the development of novel strategies against H. pylori could include subunit vaccines as well as receptin blockers. Although the pieces of the puzzle are not all in place yet, we can recognize the potential importance of binding to host proteins by pathogenic microorganisms like H. pylori during the infectious process.
Acknowledgments
This review was written at IRIS Research Center during a sabbatical leave from l'Université de Montréal.
We thank Giorgio Corsi for the artwork.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Iyes, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Amini, H.-R., F. Ascencio, A. Cruz-Villacorta, E. Ruiz-Bustos, and T. Wadström. 1996. Immunochemical properties of a 60 kDa cell surface- associated heat shock-protein (Hsp60) from Helicobacter pylori. FEMS Immunol. Med. Microbiol. 16:163-172. [DOI] [PubMed] [Google Scholar]
- 3.Amini, H.-R., F. Ascensio, A. Ljungh, and T. Wadström. 1995. Particle agglutination assay for detection of albumin and IgG binding cell surface components of Helicobacter pylori. Zentbl. Bacteriol. Mikrobiol. Hyg. 282:255-264. [PubMed] [Google Scholar]
- 4.Appelmelk, B. J., I. Simoons-Smit, R. Negrini, A. P. Moran, G. O. Aspinall, J. G. Forte, T. De Vries, H. Quan, T. Verboom, J. J. Maaskant, P. Ghiara, E. J. Kuipers, E. Bloemena, T. M. Tadema, R. R. Townsend, K. Tyagarajan, J. M. Jr. Crothers, M. A. Monteiro, A. Savio, and J. De Graaff. 1996. Potential role of molecular mimicry between Helicobacter pylori lipopolysaccharide and host Lewis blood group antigen in autoimmunity. Infect. Immun. 64:2031-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ascencio, F., L. A. Fransson, and T. Wadström. 1993. Affinity of the gastric pathogen Helicobacter pylori for the N-sulphated glycosaminoglycan heparan sulphate. J. Med. Microbiol. 38:240-244. [DOI] [PubMed] [Google Scholar]
- 6.Ascensio, F., L. A. Hansson, O. Larm, and T. Wadström. 1995. Helicobacter pylori interacts with heparin and heparin-dependent growth factors. FEMS Immunol. Med. Microbiol. 12:265-272. [DOI] [PubMed] [Google Scholar]
- 7.Berstad, A. E., K. Holbjorn, G. Bukholm, A. P. Moran, and P. Brandtzaeg. 2001. Complement activation directly induced by Helicobacter pylori. Gastroenterology 120:1108-1116. [DOI] [PubMed] [Google Scholar]
- 8.Berstad, A. E., M. Kilian, K. N. Valnes, and P. Brandtzaeg. 1999. Increased mucosal production of monomeric IgA1 but not IgA1 protease activity in Helicobacter pylori gastritis. Am. J. Pathol. 155:1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bode, G., P. Malfertheiner, and H. Ditschuneit. 1988. Pathogenic implications of ultrastructural findings in Campylobacter pylori related gastroduodenal disease. Scand. J. Gastroenterol. 142:25-39. [PubMed] [Google Scholar]
- 10.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 11.Boyle, M. D. P. (ed.). 1990. Bacterial immunoglobulin binding proteins, vol. 1. Academic Press, Inc., San Diego, Calif.
- 12.Buck, G. E., W. K. Gourley, W. K. Lee, K. Subramanyam, J. M. Latimer, and A. R. DiNuzzo. 1986. Relationship of Campylobacter pyloridis to gastritis and peptic ulcer. J. Infect. Dis. 153:664-669. [DOI] [PubMed] [Google Scholar]
- 13.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenecity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chmiela, M., E. Czkwianianc, T. Wadström, and W. Rudnicka. 1997. Role of Helicobacter pylori surface structures in bacterial interaction with macrophages. Gut 40:20-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chmiela, M., J. Lelwala-Guruge, and T. Wadström. 1994. Interaction of cells of Helicobacter pylori with human polymorphonuclear leukocytes: possible role of haemagglutinins. FEMS Immunol. Med. Microbiol. 9:41-48. [DOI] [PubMed] [Google Scholar]
- 16.Chmiela, M., B. Paziak-Domanska, W. Rudnicka, and T. Wadström. 1995. The role of heparan sulphate-binding activity of Helicobacter pylori bacteria in their adhesion to murine macrophages. APMIS 103:469-474. [DOI] [PubMed] [Google Scholar]
- 17.Chmiela, M., B. Paziak-Domanska, and T. Wadström. 1995. Attachment, ingestion and intracellular killing of Helicobacter pylori by human peripheral blood mononuclear leukocytes and mouse peritoneal inflammatory macrophages. FEMS Immunol. Med. Microbiol. 10:307-316. [DOI] [PubMed] [Google Scholar]
- 18.Covacci, A., and R. Rappuoli. 2000. Tyrosine-phosphorylated bacterial proteins: trojan horses for the host cell. J. Exp. Med. 191:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 21.De Bernard, M., B. Arico, E. Papini, R. Rizzuto, G. Grandi, R. Rappuoli, and C. Montecucco. 1997. Helicobacter pylori toxin VacA induces vacuole formation by acting in the cell cytosol. Mol. Microbiol. 26:665-674. [DOI] [PubMed] [Google Scholar]
- 22.Del Giudice, G., A. Covacci, J. L. Telford, C. Montecucco and R. Rappuoli. 2001. The design of vaccines against Helicobacter pylori and their development. Annu. Rev. Immunol. 19:523-563. [DOI] [PubMed] [Google Scholar]
- 23.Deltenre, M., and E. de Koster. 2000. How come I've got it? (A review of Helicobacter pylori transmission). Eur. J. Gastroenterol. Hepatol. 12:479-482. [DOI] [PubMed] [Google Scholar]
- 24.Duensing, T. D., J. S. Wing, and J. P. M van Putten. 1999. Sulfated polysaccharide-directed recruitement of mammalian host proteins: a novel strategy in microbial pathogenesis. Infect. Immun. 67:4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton, K. A., and M. E. Mefford. 2001. Cure of Helicobacter pylori infection and resolution of gastritis by adoptive transfer of splenocytes in mice. Infect. Immun. 69:1025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans, D. J., Jr., D. G. Evans, T. Takemura, H. Nakano, H. C. Lampert, D. Y. Graham, D. N. Granger, and P. R. Kvietys. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans, D. G., D. J. Jr. Evans, Jr., J. J. Moulds, and D. Y. Graham. 1988. N- Acetyl-neuraminyllactose-binding fibrillar hemagglutinin of Campylobacter pylori: a putative colonization factor antigen. Infect. Immun. 56:2896-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everhart, J. E. 2000. Recent developments in the epidemiology of Helicobacter pylori. Gastroenterol. Clin. North Am. 29:559-578. [DOI] [PubMed] [Google Scholar]
- 30.Ferrero, R. L., J. M. Thiberge, I. Kansau, N. Wuscher, M. Huerre, and A. Labigne. 1995. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc. Natl. Acad. Sci. USA 92:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flock, J.-I. 1999. Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol. Med. Today 5:532-537. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Valencia, G., G. I. Perez-Perez, R. G. Washburn, and M. J. Blaser. 1996. Susceptibility of Helicobacter pylori to the bactericidal activity of human serum. Helicobacter 1:28-33. [DOI] [PubMed] [Google Scholar]
- 33.Gotoda, T., D. Saito, H. Kondo, H. Ono, I. Oda, M. Fujishiro, and H. Yamaguchi. 1999. Endoscopic and histological reversibility of gastric adenoma after eradication of Helicobacter pylori. J. Gastroenterol. 34:91-96. [PubMed] [Google Scholar]
- 34.Guzman-Murillo, M. A., E. Ruiz-Bustos, B. Ho, and F. Ascencio. 2001. Involvement of the heparan sulphate-binding proteins of Helicobacter pylori in its adherence to HeLa S3 and Kato III cell lines. J. Med. Microbiol. 50:320-329. [DOI] [PubMed] [Google Scholar]
- 35.Hancock, R. E. W., R. Alm, J. Bina, and T. J. Trust. 1998. The complete genome sequence of the gastric pathogen Helicobacter pylori: a surprisingly conserved bacterium. Nat. Biotechnol. 16:216-217. [DOI] [PubMed] [Google Scholar]
- 36.Hirmo, S., M. Utt, M. Ringnér, and T. Wadström. 1995. Inhibition of heparan sulphate and other glycosaminoglycans binding to Helicobacter pylori by various polysulphated carbohydrayes. FEMS Immunol. Med. Microbiol. 10:301-306. [DOI] [PubMed] [Google Scholar]
- 37.Hoepelman, A. I., and E. I. Tuomanen. 1992. Consequences of microbial attachment: direct host cell function with adhesins. Infect. Immun. 60:1729-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu, L.-T., and H. L. T. Mobley. 1990. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 58:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khin, M. M., M. Ringnér, P. Aleljung, T. Wadström, and B. Ho. 1996. Binding of human plasminogen and lactoferrin by Helicobacter pylori coccoid forms. J. Med. Microbiol. 45:433-439. [DOI] [PubMed] [Google Scholar]
- 40.Kostrzynska, M., J. D. Betts, J. W. Austin, and T. J. Trust. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kronvall, G., and K. Jönsson. 1999. Receptins: a novel term for an expanding spectrum of natural and engineered microbial proteins with binding properties for mammalian proteins. J. Mol. Recognit. 12:38-44. [DOI] [PubMed] [Google Scholar]
- 42.Kuusela, P. 1978. Fibronectin binds to Staphylococcus aureus. Nature 276:718-720. [DOI] [PubMed] [Google Scholar]
- 43.Kwok, T., S. Backert, H. Schwarz, J. Berger and T. F. Meyer. 2002. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70:2108-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lähteenmaki, K., P. Kuusela, and T. M. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531-552. [DOI] [PubMed] [Google Scholar]
- 45.Lelwala-Guruge, J., C. Schalén, I. Nilsson, A. Ljungh, T. Tyszkiewicz, M. Wikander, and T. Wadström. 1990. Detection of antibodies to Helicobacter pylori cell surface antigens. Scand. J. Infect. Dis. 22:457-465. [DOI] [PubMed] [Google Scholar]
- 46.Ljubicic, N., M. Banic, M. Kujundzic, Z. Antic, M. Vrkljan, I. Kovacevic, D. Hrabar, M. Doko, M. Zovak, and S. Mihatov. 1999. The effect of eradicating Helicobacter pylori infection on the course of adenomatous and hyperplastic gastric polyps. Eur. J. Gastrenterol. Hepatol. 11:727-730. [DOI] [PubMed] [Google Scholar]
- 47.Ljungh, A. 2000. Helicobacter pylori interactions with plasminogen. Methods 21:151-157. [DOI] [PubMed] [Google Scholar]
- 48.Ljungh, A., A. P. Moran, and T. Wadström. 1996. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol. Med. Microbiol. 16:117-126. [DOI] [PubMed] [Google Scholar]
- 49.Ljungh, A., and T. Wadström. 1996. Interaction of bacterial adhesins with the extracellular matrix. Adv. Exp. Med. Biol. 408:129-140. [DOI] [PubMed] [Google Scholar]
- 50.Lottenberg, R., D. Minning-Wenz, and M. D. P. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20-24. [DOI] [PubMed] [Google Scholar]
- 51.Macchia, G., A. Massone, D. Burroni, A. Covacci, S. Censini, and R. Rappuoli. 1993. The Hsp60 protein of Helicobacter pylori: structure and immune response in patients with gastroduodenal diseases. Mol. Microbiol. 9:645-652. [DOI] [PubMed] [Google Scholar]
- 52.Malaty, H. M., T. Kumagai, E. Tanaka, H. Ota, K. Kiyosawa, D. Y. Graham, and T. Katsuyama. 2000. Evidence from a nine-year birth cohort study in Japan of transmission pathways of Helicobacter pylori infection. J. Clin. Microbiol. 38:1971-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcus, E. A., and D. R. Scott. 2001. Cell lysis is responsible for the appearance of extracellular urease in Helicobacter pylori. Helicobacter 6:93-99. [DOI] [PubMed] [Google Scholar]
- 54.Marshall, B. J. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1:1273-1275. [PubMed] [Google Scholar]
- 55.Masuda, Y., and T. Sugiyama. 2001. A protein in the cell wall of Helicobacter pylori binds human IgG deprived of sugar chains. Tohoku J. Exp. Med. 194:65-69. [DOI] [PubMed] [Google Scholar]
- 56.Mattsson, A., M. Quiding-Jarbrink, H. Lonroth, A. Hamlet, I. Ahlstedt, and A. Svennerholm. 1998. Antibody-secreting cells in the stomachs of symptomatic and asymptomatic Helicobacter pylori-infected subjects. Infect. Immun. 66:2705-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNulty, C. A. 1999. The discovery of Campylobacter-like organisms. Curr. Top. Microbiol. Immunol. 241:1-9. [DOI] [PubMed] [Google Scholar]
- 58.Meehan, M., Y. Lynagh, C. Woods, and P. Owen. 2001. The fibrinogen- binding protein (FgBP) of Streptococcus equi subsp. equi additionally binds IgG and contributes to virulence in a mouse model. Microbiology 147:3311-3322. [DOI] [PubMed] [Google Scholar]
- 59.Meehan, M., P. Nowlan, and P. Owen. 1998. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology 144:993-1003. [DOI] [PubMed] [Google Scholar]
- 60.Mentis, A., L. Tzouvelekis, C. Spiliadis, C. C. Blackwell, and D. M. Weir. 1990. Inhibition of Helicobacter pylori haemagglutination activity by human salivary mucins. FEMS Microbiol. Immunol. 2:125-127. [DOI] [PubMed] [Google Scholar]
- 61.Meucci, G., M. Tatarella, M. Vecchi, M. L. Ranzi, E. Biguzzi, G. Beccari, E. Clerici, and R. de Franchis. 1997. High prevalence of Helicobacter pylori infection in patients with colonic adenomas and carcinomas. J. Clin. Gastroenterol. 25:605-607. [DOI] [PubMed] [Google Scholar]
- 62.Miehlke, S., R. Thomas, O. Gutierrez, D. Y. Graham, and M. F. Go. 1999. DNA fingerprinting of single colonies of Helicobacter pylori from gastric cancer patients suggests infection with a single predominant strain. J. Clin. Microbiol. 37:245-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mobley, H. L., R. M. Garner, and P. Bauerfeind. 1995. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 1:97-109. [DOI] [PubMed] [Google Scholar]
- 64.Montecucco, C., E. Papini, M. de Bernard, and M. Zoratti. 1999. Molecular and cellular activities of Helicobacter pylori pathogenic factors. FEBS Lett. 452:16-21. [DOI] [PubMed] [Google Scholar]
- 65.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: How Helicobacter pylori survives in the human stomach. Nat. Rev. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 66.Montemurro, P., G. Barbuti, W. G. Dundon., G. Del Guidice, R. Rappuoli, M. Colucci, P. De Rinaldis, C. Montecucco, N. Semeraro, and E. Papini. 2001. Helicobacter pylori neutrophil-activating protein stimulates tissue factor and plasminogen activator inhibitor-2 production by human blood mononuclear cells. J. Infect. Dis. 183:1055-1062. [DOI] [PubMed] [Google Scholar]
- 67.Montemurro, P., H. Nishioka, W. G. Dundon, M. de Bernard, G. Del Giudice, R. Rappuoli, and C. Montecucco. 2002. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a potent stimulant of mast cells. Eur. J. Immunol. 32:671-676. [DOI] [PubMed] [Google Scholar]
- 68.Moran, A. P., P. Kuusela, and T. U. Kosunen. 1993. Interaction of Helicobacter pylori with extracellular matrix proteins. J. Appl. Bacteriol. 75:184-189. [Google Scholar]
- 69.Nilius, M., M. Pugliese and P. Malfertheiner. 1996. Helicobacter pylori and proteolytic activity. Eur. J. Clin. Investig. 26:1103-1106. [DOI] [PubMed] [Google Scholar]
- 70.Oderda, G. 1999. Transmission of Helicobacter pylori infection. Can. J. Gastroenterol. 13:595-597. [DOI] [PubMed] [Google Scholar]
- 71.Odenbreit, S., B. Wieland, and R. Haas. 1996. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase- deficient mutant strain. J. Bacteriol. 178:6960-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Toole, P. W., M. C. Lane, and S. Porwollik. 2000. Helicobacter pylori motility. Microbes Infect. 2:1207-1214. [DOI] [PubMed] [Google Scholar]
- 74.Pakodi, F., O. M. Abdel-Salam, A. Debreceni, and G. Mozsik. 2000. Helicobacter pylori: one bacterium and a broad spectrum of human disease An overview. J. Physiol. (Paris) 94:139-152. [DOI] [PubMed] [Google Scholar]
- 75.Pantzar, M., A. Ljungh, and T. Wadström. 1998. Pasminogen binding and activation at the surface of Helicobacter pylori CCUG 17874. Infect. Immun. 66:4976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM- Mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 77.Patti, J. M., and M. Höök. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6:752-758. [DOI] [PubMed] [Google Scholar]
- 78.Peters, T., Jr. 1996. All about albumin: biochemistry, genetics, and medical applications, Academic Press, Inc., New York, NY.
- 79.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190-194. [DOI] [PubMed] [Google Scholar]
- 80.Polla, B. S., S. Baladi, K. Fuller, and G. Rook. 1995. Presence of hsp65 in bacterial extracts (OM-89): a possible mediator of orally induced tolerance? Experientia 51:775-779. [DOI] [PubMed] [Google Scholar]
- 81.Reyrat, J. M., S. Lanzavecchia, P. Lupetti, M. de Bernard, C. Pagliaccia, V. Pelicic, M. Charrel, C. Ulivieri, N. Norais, X. Ji, V. Cabiaux, E. Papini, R. Rappuoli, and J. L. Telford. 1999. 3D imaging of the 58 kDa cell binding subunit of the Helicobacter pylori cytotoxin. J. Mol. Biol. 290:459-470. [DOI] [PubMed] [Google Scholar]
- 82.Ringnér, M., M. Paulsson, and T. Wadström. 1992. Vitronectin binding by Helicobacter pylori. FEMS Microbiol. Immunol. 5:219-224. [DOI] [PubMed] [Google Scholar]
- 83.Ringnér, M., K. H. Valkonen, and T. Wadström. 1994. Binding of vitronectin and plasminogen to Helicobacter pylori. FEMS Immunol. Med. Microbiol. 9:29-34. [DOI] [PubMed] [Google Scholar]
- 84.Romer, J., T. H. Bugge, C. Pyke, L. R. Lund, M. J. Flick, J. L. Degen and K. Dano. 1996. Impaired wound healing in mice with a disrupted plasminogen gene. Nat. Med. 2:287-292. [DOI] [PubMed] [Google Scholar]
- 85.Rothenbacher, D., G. Bode, G. Berg, U. Knayer, T. Gonser, G. Adler, and H. Brenner. 1999. Helicobacter pylori among preschool children and their parents: evidence of parent-child transmission. J. Infect. Dis. 179:398-402. [DOI] [PubMed] [Google Scholar]
- 86.Ruiz-Bustos, E., J. L. Ochoa, T. Wadström, and F. Ascencio. 2001. Isolation and characterization of putative adhesions from Helicobacter pylori with affinity for heparan sulphate proteoglycan. J. Med. Microbiol. 50:215-222. [DOI] [PubMed] [Google Scholar]
- 87.Ruiz-Bustos, E., A. Sierra-Beltran, M. J. Romero, C. Rodriguez-Jaramillo, and F. Ascencio. 2000. Protection of BALB/c mice against experimental Helicobacter pylori infection by oral immunisation with H. pylori heparan sulphate-binding proteins coupled to cholera toxin beta-subunit. J. Med. Microbiol. 49:535-541. [DOI] [PubMed] [Google Scholar]
- 88.Satin, B., G. Del Giudice, V. Della Bianca, S. Dusi, C. Laudanna, F. Tonello, D. Kelleher, R. Rappuoli, C. Montecucco, and F. Rossi. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sjobring, U. 1992. Isolation and molecular characterization of a novel albumin binding protein from group G streptococci. Infect. Immun. 60:3601-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Slomiany, B. L., J. Piotrowski, S. Sengupta, and A. Slomiany. 1991. Inhibition of gastric mucosal laminin receptor by Helicobacter pylori lipopolysaccharides. Biochem. Biophys. Res. Commun. 175:963-970. [DOI] [PubMed] [Google Scholar]
- 91.Smith, V. C., and R. M. Genta. 2000. Role of Helicobacter pylori gastritis in gastric atrophy, intestinal metaplasia, and gastric neoplasia. Microsc. Res. Tech. 48:313-320. [DOI] [PubMed] [Google Scholar]
- 92.Sommi, P., V. Ricci, M. Romano, K. J. Ivey, E. Solcia, and U. Ventura. 1997. H. pylori-induced cell vacuolation in vitro: inhibitory action of heparin. Gastroenterology 112 (Suppl. S):A294. [Google Scholar]
- 93.Spiegelhalder, C., B. Gerstenecker, A. Kersten, E. Schiltz, and M. Kist. 1993. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect. Immun. 61:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Supajatura, V., H. Ushio, A. Wada, K. Yahiro, K. Okumura, H. Ogawa, T. Hirayama, and C. Ra. 2002. VacA, a vacuolating cytotoxin of Helicobacter pylori, directly activates mast cells for migration and production of proinflammatory cytokines. J. Immunol. 168:2603-2607. [DOI] [PubMed] [Google Scholar]
- 95.Testerman, T. L., D. J. McGee, and H. L. T. Mobley. 2001. Adherence and colonization. p. 381-417. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell, (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 96.Thomas, J. E., A. Dale, M. Harding, W. A. Coward, T. J. Cole, and L. T. Weaver. 1999. Helicobacter pylori colonization in early life. Pediatr. Res. 45:218-223. [DOI] [PubMed] [Google Scholar]
- 97.Tindberg, Y., C. Bengtsson, F. Granath, M. Blennow, O. Nyrén, and M. Granström. 2001. Helicobacter pylori infection in Swedish school children: Lack of evidence of child-to-child transmission outside the family. Gastroenterology 121:310-316. [DOI] [PubMed] [Google Scholar]
- 98.Tomb, J.-F., O. White, A. Kerlavage, R. Clayton, G. Sutton, R. Fleischmann, K. Ketchum, H. Klenk, S. Gill, B. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. Khalak, A. Glodek, K. McKenney, L. Fitzegerald, N. Lee, M. Adams, E. Hickey, D. Berg, J. Gocayne, T. Utterback, J. Peterson, J. Kelley, M. Cotton, J. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. Hayes, M. Borodovsky, P. Karp, and J. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 99.Tombola, F., L. Morbiato, G. Del Giudice, R. Rappuoli, M. Zoratti, and E. Papini. 2001. The Helicobacter pylori VacA toxin is a urea permease that promotes urea diffusion across epithelia. J. Clin. Investig. 108:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tosi, M. F., and S. J. Czinn. 1990. Opsonic activity of specific human IgG against Helicobacter pylori. J. Infect. Dis. 162:156-162. [DOI] [PubMed] [Google Scholar]
- 101.Trust, T. J., P. Doig, L. Emödy, Z. Kienle, T. Wadström, and P. O'Toole. 1991. High-affinity binding of the basement membrane proteins collagen type IV and laminin to the gastric pathogen Helicobacter pylori. Infect. Immun. 59:4398-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Utt, M., B. Danielsson, and T. Wadström. 2001. Helicobacter pylori vacuolating cytotoxin binding to a putative cell surface receptor, heparan sulfate, studied by surface plasmon resonance. FEMS Immunol. Med. Microbiol. 30:109-113. [DOI] [PubMed] [Google Scholar]
- 103.Utt, M., and T. Wadström. 1997. Identification of heparan sulphate binding surface proteins of Helicobacter pylori: inhibition of heparan sulphate binding with sulphated carbohydrate polymers. J. Med. Microbiol. 46:541-546. [DOI] [PubMed] [Google Scholar]
- 104.Valkonen, K. H., M. Ringnér, A. Ljungh, and T. Wadström. 1993. High-affinity binding of laminin by Helicobacter pylori. Evidence for a lectin- like interaction. FEMS Immunol. Med. Microbiol. 7:29-38. [DOI] [PubMed] [Google Scholar]
- 105.Valkonen, K. H., T. Wadström, and A. P. Moran. 1994. Interaction of lipopolysaccharides of Helicobacter pylori with basement membrane protein laminin. Infect. Immun. 62:3640-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Valkonen, K. H., T. Wadström, and A. P. Moran. 1997. Identification of the N-acetylneuraminyllactose-specific laminin-binding protein of Helicobacter pylori. Infect. Immun. 65:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wadström, T., S. Hirmo, and T. Boren. 1996. Biochemical aspects of Helicobacter pylori colonization of the human gastric mucosa. Aliment. Pharmacol. Ther. Suppl. 1:17-27. [DOI] [PubMed] [Google Scholar]
- 108.Wadström, T., S. Hirmo, H. Novak, A. Guzman, M. Ringnér- Pantzar, M. Utt, and P. Aleljung. 1997. Sulfatides inhibit binding of Helicobacter pylori to the gastric cancer Kato III cell line. Curr. Microbiol. 34:267-272. [DOI] [PubMed] [Google Scholar]
- 109.Warren, J. R. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273. [PubMed]
- 110.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]
- 111.Widders, P. R. 1990. Fc receptors and the pathogenesis of bacterial infections in animals, p. 375-396. In M. D. P. Boyle (ed.) Bacterial immunoglobulin binding proteins, vol. 1. Academic Press, Inc., San Diego, Calif.
- 112.Yarzábal, A., L. Avilán, K. Hoelzl, M. de Munoz, J. Puig, and I. Kansau. 2000. A study of the interaction between Helicobacter pylori and components of the human fibrinolytic system. Braz. J. Med. Biol. Res. 33:1015-1021. [DOI] [PubMed] [Google Scholar]