Abstract
Entamoeba histolytica is a protozoan parasite that causes colitis and liver abscesses. Several Entamoeba species and strains with differing levels of virulence have been identified. E. histolytica HM-1:IMSS is a virulent strain, E. histolytica Rahman is a nonvirulent strain, and Entamoeba dispar is a nonvirulent species. We used an E. histolytica DNA microarray consisting of 2,110 genes to assess the transcriptional differences between these species/strains with the goal of identifying genes whose expression correlated with a virulence phenotype. We found 415 genes expressed at lower levels in E. dispar and 32 genes with lower expression in E. histolytica Rahman than in E. histolytica HM-1:IMSS. Overall, 29 genes had decreased expression in both the nonvirulent species/strains than the virulent E. histolytica HM-1:IMSS. Interestingly, a number of genes with potential roles in stress response and virulence had decreased expression in either one or both nonvirulent Entamoeba species/strains. These included genes encoding Fe hydrogenase (9.m00419), peroxiredoxin (176.m00112), type A flavoprotein (6.m00467), lysozyme (6.m00454), sphingomyelinase C (29.m00231), and a hypothetical protein with homology to both a Plasmodium sporozoite threonine-asparagine-rich protein (STARP) and a streptococcal hemagglutinin (238.m00054). The function of these genes in Entamoeba and their specific roles in parasite virulence need to be determined. We also found that a number of the non-long-terminal-repeat retrotransposons (EhLINEs and EhSINEs), which have been shown to modulate gene expression and genomic evolution, had lower expression in the nonvirulent species/strains than in E. histolytica HM-1:IMSS. Our results, identifying expression profiles and patterns indicative of a virulence phenotype, may be useful in characterizing the transcriptional framework of virulence.
The protozoan parasite Entamoeba histolytica causes 50 million cases of invasive disease and approximately 100,000 deaths each year (55). The most common manifestations of amebic infection are dysentery and liver abscess, but infections of the lung, heart, and brain also occur (28). Only ∼10% of infections result in invasive disease, but the reasons behind this phenomenon remain largely unknown (27). The molecular mechanisms that regulate parasite invasion and pathogenesis are not well characterized.
Different species and strains of Entamoeba exhibit various levels of pathogenicity. Entamoeba histolytica and Entamoeba dispar are morphologically identical and highly similar species (their rRNAs are 98% identical), but they have vastly different virulence potentials in vivo (20). E. histolytica can cause invasive disease, whereas E. dispar, while able to colonize humans, appears to have no invasive potential in vivo. Key differences in a number of virulence determinants between the two species have been identified. Cysteine proteinases (CPs) are a family of cathepsin proteinases involved in the degradation of colonic mucin and extracellular matrix (47). Antisense inhibition of CPs in E. histolytica trophozoites results in reduced phagocytic activity (4), gut inflammation, and liver abscess formation (3). Two of the CP genes (CP1 and CP5) are either missing or highly degenerate (11, 58), and another (CP8), although conserved, has significantly lower expression in E. dispar (12). Amebapores, which are pore-forming peptides, lyse target cells and ingested bacteria (33, 34). Silencing of amebapore A in E. histolytica results in decreased cytotoxicity against nucleated cells and erythrocytes, as well as decreased liver abscess formation in vivo (10). Although highly conserved orthologs of the amebapore family are present in E. dispar (88 to 95% identical), the most abundant amebapore (AP-A) is ∼25-fold less active in E. dispar (42). KERP1, a lysine- and glutamic acid-rich protein postulated to be involved in attachment to host cells, was recently reported to be missing or divergent in E. dispar (49). The genetic element EhSINE1, a non-long-terminal-repeat (non-LTR) retrotransposon, and the Ariel1 gene are also missing or divergent in E. dispar (56, 57).
Entamoeba histolytica Rahman, isolated from an asymptomatic individual, has reduced cytotoxic capabilities for epithelial cells in vitro, does not form liver abscesses in animal models, and is classically referred to as a nonvirulent E. histolytica strain (2, 23). A few genetic differences between E. histolytica HM-1:IMSS and E. histolytica Rahman have been identified. One observation, which has been genetically proven to be related to virulence, is the decreased level of the light subunit of the Gal/Gal-NAc lectin in E. histolytica Rahman compared to that in E. histolytica HM-1:IMSS (2). Additionally, the proteophosphoglycans coating the surface of E. histolytica Rahmanhave truncated glucan side chains compared to those of the virulent E. histolytica HM-1:IMSS (40). The relationship of this observation to amebic virulence is not clear; however, the authors hypothesized that the glycans may protect parasite adhesion molecules from proteolysis or that the proteophosphoglycans may regulate the ability of parasite surface molecules to interact with host cell receptors.
The effort to sequence the genome of E. histolytica HM-1:IMSS has unveiled a number of unusual aspects of amebic biology (36). However, the essential differences between the virulent and nonvirulent species/strains and the factor(s) that determines the invasive potential in E. histolytica remain elusive. A recent comparative genomic hybridization approach identified a number of genomic differences between E. histolytica and E. dispar, with 67 genes out of 1,640 studied (4%) identified as highly or significantly divergent (50). Fewer genetic differences were identified between E. histolytica HM-1:IMSS and E. histolytica Rahman, with 5 out of 1,817 (0.3%) genes identified as highly or significantly divergent. Whether these genomic differences contribute to the various virulence phenotypes remains to be determined. Previous studies have shown that E. histolytica HM-1:IMSS is able to lyse colonic cell monolayers without major changes in its transcriptional profile, indicating that many of the genes important in host cell damage may be constitutively expressed under tissue culture conditions (37).
In order to identify the genes that are differentially expressed among the virulent and nonvirulent Entamoeba species/strains, we used a DNA microarray consisting of 2,110 unique genes to perform expression profiling of the virulent E.histolytica HM-1:IMSS and the nonvirulent E. histolytica Rahman and E. dispar SAW760. Using this technique, we have identified 415 genes that have lower expression in E. dispar SAW760 and 32 genes with lower expression in E. histolytica Rahman than in EH HM-1:IMSS. Interestingly, 29 genes showed decreased expression in both the nonvirulent species/strains E. dispar SAW760 and E. histolytica Rahman. These genes are of particular interest as their expression correlates with virulence, and it is interesting to speculate that they may play roles in amebic pathogenesis. Our work represents the first large-scale expression profiling of Entamoeba species/strains and opens the door to the investigation of genetic and expression differences which may relate to parasite virulence.
MATERIALS AND METHODS
Parasite culture.
E. histolytica HM-1:IMSS was originally isolated from a patient with colitis in 1967 (21). E. histolytica Rahman was isolated from an asymptomatic cyst passer in 1972 (21). Both were obtained from ATCC (http://www.atcc.org/) and grown under axenic conditions in Trypticase-yeast extract-iron-serum medium (TYI-S-33) with 15% adult bovine serum (Sigma), supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) (Gibco BRL), and 1× Diamond's vitamins (Biosource International) in 15-ml glass culture tubes at 36.5°C as previously described (21). E. dispar SAW760 was isolated from an adult human male in England in 1979 (http://www.atcc.org/). RNA from E. dispar SAW760 and E. histolytica HM-1:IMSS grown axenically in liver digest-yeast extract-iron-serum medium (LYI-S-2) were kindly provided by C. Graham Clark (15). The strains were proven to be E. histolytica HM-1:IMSS and E. histolytica Rahman by PCR analysis of the rRNA episome and the serine-rich Entamoeba histolytica protein gene by using previously described methods (16, 52).
RNA isolation and Northern blot analysis.
Amebae were harvested, and total RNA was extracted using TRIzol reagent (Invitrogen) and cleaned using the RNeasy cleanup kit (QIAGEN). Northern blot tests were performed using standard protocols (37). Briefly, 10 to 20 μg of total parasite RNA was separated by denaturing 1.2% agarose gel electrophoresis, transferred to membrane filters, and hybridized with radioactive probes using the ExpressHyb (Clontech, Palo Alto, CA) hybridization buffer. Probes were amplified by PCR from the appropriate clone or genomic DNA using M13F, M13R, or gene-specific primers, sequence verified, and labeled with [α-32P]dATP with the Random Primed DNA labeling kit (Roche, Germany). Primers used in the study are shown in Table 1. Blots were exposed to film, subjected to autoradiography, scanned, and prepared for publication using Adobe Photoshop (version 7; San Jose, CA). Blots were stripped using 0.5% sodium dodecyl sulfate and reused for subsequent hybridizations per the manufacturer's suggestions. For a loading control, EhActin (locus 8.m00351) was PCR amplified from E. histolytica HM-1:IMSS genomic DNA and labeled as described previously (50).
TABLE 1.
Primers used for generation of probesa
| Locus | Primer |
|---|---|
| 194.m00115 F | 5′-ATGGGCAATAGACCCAACAC-3′ |
| 194.m00115 R | 5′-TCCATAAATTTTTGCATGACCA-3′ |
| 2.m00567 F | 5′-ATGAACGCTATTAAACCTAAAACAAT-3′ |
| 2.m00567 R | 5′-CTATTATTTGTCTGAATTGTCTAAAGCTG-3′ |
| 2.m00567 putative promoter region F | 5′-AAAAGCCATTGAAAATGGATG-3′ |
| 2.m00567 putative promoter region R | 5′-TCCATCAACATCCAATACAATTG-3′ |
| 238.m00054 segment #1 F | 5′-ATGGCTGAACAAATCAGAGTCTGACA-3′ |
| 238.m00054 segment #1 R | 5′-CTAAAGTAATAATGCTGGAACATTTGGATGG-3′ |
| 238.m00054 segment #2 F | 5′-GAATCACCGAATGTTATTGCATCTGG-3′ |
| 238.m00054 segment #2 R | 5′-CTACTGTGAATCTTTAACACGGATGTTCGA-3′ |
| 238.m00054 segment #3 F | 5′-TCGAACATCCGTGTTAAAGATTCACAG-3′ |
| 238.m00054 segment #3 R | 5′-CTACATTGTCTTTCCTCCAATTTCATCTCC-3′ |
| 29.m00210 F | 5′-ATGCAACAAGAGACAGTTGTTGG-3′ |
| 29.m00210 R | 5′-CTATTATTTTTGTGCAAAGAATTTCTCT-3′ |
| 29.m00231 F | 5′-CCCCTTCATCGACAAACAAT-3′ |
| 29.m00231 R | 5′-TCGATGACGTCTTGATTTGG-3′ |
| 297.m00063 F | 5′-TCAGCATGGATTTGATTGGA-3′ |
| 297.m00063 R | 5′-CAGCAACACCTTTTTCAACG-3′ |
| 5.m00482 F | 5′-TCTGGTGCTTTTGATGTTGC-3′ |
| 5.m00482 R | 5′-CCACCGAAGGATCACACTCT-3′ |
| 51.m00161 F | 5′-GTCAAAGAGCTGTTGCATGG-3′ |
| 51.m00161 R | 5′-TTCTGCAACATTTCCTGGTG-3′ |
| 6.m00454 F | 5′-GATTCTTCATTTGCCGTGCT-3′ |
| 6.m00454 R | 5′-CCGAATGAAGCCCAATTATC-3′ |
| 6.m00467 F | 5′-AATGGGCACAACCTATTGCT-3′ |
| 6.m00467 R | 5′-TTTCCCCATTCAAAGCATGT-3′ |
| 9.m00419 F | 5′-AACCACCAAAAATTCCACCA-3′ |
| 9.m00419 R | 5′-AATTGGTGAACGGGCAGTAG-3′ |
| EhActin F | 5′-GCTGGTATGGGTCAAAAGGA-3′ |
| EhActin R | 5′-TTTCTGTGGACAATAGCTGGTC-3′ |
| EhLINE1 ORF1 F | 5′-GATCCTTTTCCAATGCAGGA-3′ |
| EhLINE1 ORF1 R | 5′-TGCTTTTCTCTTCGATTTCCA-3′ |
| EhLINE1 ORF2 F | 5′-TGAARATAGGGATTACTTCMGTGT-3′ |
| EhLINE1 ORF2 R | 5′-CCCATTAGACATGGTAAGTGGAA-3′ |
| M13 F | 5′-TGTAAAACGACGGCCAGT-3′ |
| M13 R | 5′-CAGGAAACAGCTATGACC-3′ |
Probes were used for Northern blot analyses and sequencing particular loci in E. histolytica Rahman. Tm values ranged between 56 and 58°C.
Microarray hybridizations and data analysis.
An 11,328-clone genomic DNA microarray was generated from E. histolytica HM-1:IMSS as described previously (50). Information from The Institute for Genomic Research (TIGR) gene annotation was used to identify clones that contained only one potential open reading frame (ORF) (defined as ≥98% identity over ≥200 bp). A total of 2,801 clones were in this category, representing 2,110 unique genes. Eight micrograms of total parasite RNA was prepared for array hybridizations as previously described (37). All experiments used at least two separate RNA samples, with each RNA sample isolated on different days. Microarrays were analyzed using the ScanAlyze program (Michael Eisen; http://rana.lbl.gov/EisenSoftware.htm) to determine the fluorescent intensities, and the data were then stored on the Stanford Microarray database (http://genome-www5.stanford.edu//). Data normalization and quality were assessed as previously described (37). Three arrays each were hybridized with RNA from E. histolytica HM-1:IMSS grown in TYI-S-33, E. histolytica HM-1:IMSS grown in LYI-S-2, E. histolytica Rahman grown in TYI-S-33, and axenic E. dispar SAW760 grown in LYI-S-2. In order to identify amebic genes that were differentially expressed between the different species/strains, the software Significance Analysis of Microarrays (SAM) version 1.21 (53) was used according to recommended procedures (http://www-stat.stanford.edu/∼tibs/SAM/) as described previously (37). Two-class unpaired sample analysis was used on log2-transformed data from E. histolytica HM-1:IMSS and either E. histolytica Rahman or E. dispar SAW760 using the K nearest-neighbor imputer. The user-defined delta value was assigned by maximizing the number of significant genes while maintaining a false-discovery rate of ≤5%. Additional filtering was performed to include only those genes that are likely to be expressed under trophozoite conditions.
PCR and sequence analysis.
Five genetic loci that displayed decreased expression in E. dispar SAW760 and E. histolytica Rahman were sequenced and analyzed. Primers were designed based on the E. histolytica HM-1:IMSS sequence and used to generate PCR products from E. histolytica Rahman genomic DNA. The PCR products (or several cloned PCR products) were sequenced using an ABI PRISM 310 genetic analyzer (PE Applied Biosystems). The primers used are listed in Table 1. The melting temperature for the primers ranged from 56 to 58°C.
BLAST analysis against the E. dispar sequence database.
Sequences of the genes of interest were downloaded from the GeneDB website (http://www.genedb.org/) and compared by BLASTN analysis to both the TIGR (http://www.tigr.org/tdb/e2k1/eha1/) and Sanger Institute (http://www.sanger.ac.uk/Projects/E_histolytica/) E. dispar databases (sequence data available as of March 2005), and the top hit for each locus was used in subsequent analyses. Since the current E. dispar sequence data are limited to twofold-genome coverage, we wanted to ascertain the likelihood of identifying orthologs in the E. dispar database. E. dispar genes encoding RabB (GenBank accession number AY882575), GEF2 (AY561277), Jacob (AF401985), peroxiredoxin (AB026184), cysteine synthase 2 (AB028632), cysteine synthase 1 (AB028631), pore forming protein (AF082529), GalNac lectin hgl (U73710.1), and GalNac lectin lgl1 (U85823.1) were analyzed by BLASTN analysis to see whether their full-length sequences could be identified in the current E. dispar databases. For these genes, a match in the E. dispar database could be identified with ≥90% identity over ≥50% of the locus. Thus, we used these criteria (≥90% identity over ≥50% of the locus size) as our cutoff to designate an E. histolytica gene as one that is highly conserved in E. dispar. For BLAST analyses involving EhLINEs and EhSINEs, we used the consensus sequence defined by Bakre et al. (5; A. Bakre and S. Bhattacharya, personal communication).
Statistical analysis.
Statistical analyses were performed using a two-tailed Student's t test. P values of <0.05 were deemed significant.
RESULTS
The E. histolytica DNA microarray accurately detected differences in message abundance.
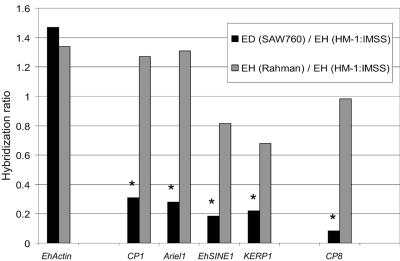
In order to find genes whose expression correlated with virulence, expression profiling of 2,110 amebic genes was performed for the virulent strain E. histolytica HM-1:IMSS, the nonvirulent strain E. histolytica Rahman, and the nonvirulent species E. dispar SAW760. We have previously shown that the microarray we utilized accurately detects message abundance and transcriptionally regulated genes (37). In order to ascertain whether we could accurately detect differences in message abundance from different species/strains, we examined genes previously published as being either absent or having decreased expression in E. dispar (Fig. 1). The ratio of hybridization intensities of E. dispar SAW760 and E. histolytica Rahman to E. histolytica HM-1:IMSS were calculated. As expected, genes previously shown to be missing or divergent (CP1, Ariel1, KERP1, and EhSINE1) or to have reduced expression (CP8) in E. dispar all had significantly less signal on the arrays in E. dispar SAW760 than in E. histolytica HM-1:IMSS (P value of <0.05) (12, 13, 49, 57). However, no such differences were observed for E. histolytica Rahman. Other genes with known differential expression between the species/strains (the amebapore and Lgl genes and CP5) were not analyzed, since the clones containing those also contained another potential ORF.
FIG. 1.
The hybridization ratios of E. dispar SAW760 and E. histolytica Rahman to E. histolytica HM-1:IMSS are shown for EhActin, CP1, Ariel1, EhSINE1, KERP1, and CP8. Actin has similar expression levels in all Entamoeba species/strains studied. CP1, Ariel1, EhSINE1, and KERP1, all previously shown to be missing at a genomic level in E. dispar SAW760, showed significantly less expression in E. dispar SAW760 than in E. histolytica HM-1:IMSS. CP8, present in E. dispar SAW760 but known to have less expression, shows significantly less hybridization for E. dispar SAW760 than E. histolytica HM-1:IMSS. E. histolytica Rahman had expression levels equivalent to those of E. histolytica HM-1:IMSS for all genes shown. Genes with significantly decreased (P value of <0.05) expression levels compared to those of E. histolytica HM-1:IMSS are denoted with an asterisk. ED, E. dispar; EH, E. histolytica.
Our arrays were able to detect differential expression levels between E. histolytica and E. dispar both for genes that are conserved at the genomic level and for those that are genomically divergent. For genes with sequence conservation between E. dispar and E. histolytica, the lack of array hybridization indicates that the gene is not expressed. In contrast, for genomic loci that are significantly divergent in E. dispar compared to E. histolytica, the lack of array hybridization is due to sequence differences, and expression data cannot be ascertained. Thus, even if that gene was functionally expressed in E. dispar, the significantly divergent sequence would mean that the signal would not cross-hybridize on the array. We and others have previously analyzed a number of genes that are divergent in E. dispar at the nucleotide level to assess whether they would make functionally similar proteins (50, 58). For the majority of the genes analyzed, a significantly divergent nucleotide sequence resulted in premature stop codons and/or a highly divergent protein. Overall, this indicates that genes with significant sequence divergence in E. dispar are likely to be nonfunctional in comparison to their orthologs in E. histolytica.
We used E. histolytica HM-1:IMSS parasites from two different laboratories grown in two different medium preparations. We did find some differences in gene expression between the two samples, which were confirmed by Northern blot analyses (data not shown). These differences are most likely due to differences between the media and/or growth conditions; however, the functional relevance of these differences has yet to be determined. Since we were less interested in genes that are specific to growth conditions or media, for our purposes the two E. histolytica HM-1:IMSS samples were analyzed together as a set.
Northern blot analysis confirmed the array data of differentially expressed genes.
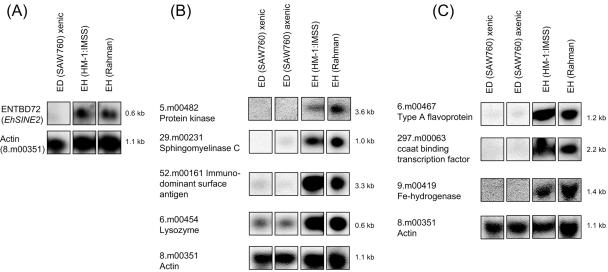
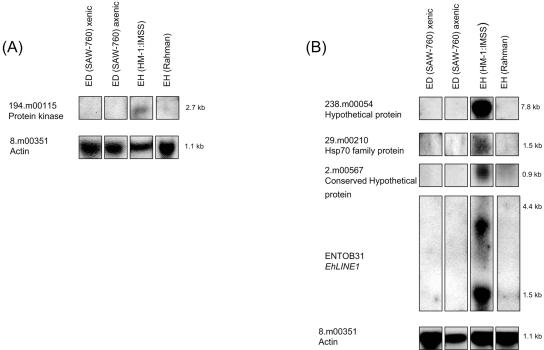
A subset of genes identified as differentially expressed between Entamoeba strains and species was confirmed by Northern blot analysis (Fig. 2 and 3). Eight genetic loci (ENTBD72 or EhSINE2, 5.m00482, 29.m00231, 52.m00161, 6.m00454, 6.m00467, 297.m00063, and 9.m00419) which exhibited significantly decreased hybridization in only the E. dispar SAW760 versus E. histolytica HM-1:IMSS comparison were confirmed by Northern blot analysis (Fig. 2). Five genetic loci (194.m00115, 238.m00054, 29.m00210, 2.m00567, and ENTOB31 or EhLINE1) which exhibited significantly decreased hybridization in both nonvirulent species/strains were confirmed by Northern blot analysis (Fig. 3). All of the probes gave results that matched the array data (absent or markedly reduced signal in the appropriate species/strains) and the expected transcript sizes based on the TIGR gene annotation.
FIG. 2.
Northern blot analysis for genes identified as differentially expressed between Entamoeba species. (A) Clone ENTBD72 was used to represent EhSINE2. (B) The genes 5.m00482, 29.m00231, 52.m00161, and 6.m00454 are shown. (C) The genes 6.m00467, 297.m00063, and 9.m00419 are shown. All of the genes in this figure exhibited significantly less expression in E. dispar (ED) SAW760 than in E. histolytica (EH) HM-1:IMSS by microarray analysis. Panels A, B, and C represent different blots. EhActin, which is equally expressed in all species/strains, is shown for each blot as a loading control.
FIG. 3.
Northern blot analysis for genes identified as differentially expressed in both nonvirulent species/strains. (A) The gene 194.m00115 is shown. (B) The genes 238.m00054, 29.m00210, 2.m00567, and EhLINE1, represented by the clone ENTOB31, are shown. All of the genes in this figure exhibited significantly less expression in both E. dispar (ED) SAW760 and E. histolytica (EH) Rahman than in E. histolytica HM-1:IMSS by microarray analysis. Panels A and B represent different blots. EhActin, which is equally expressed in all species/strains, is shown for each blot as a loading control.
Genes with lower expression in E. dispar SAW760 than in E. histolytica HM-1:IMSS.
In order to identify genes with lower expression in the nonvirulent E. dispar than in the virulent E. histolytica HM-1:IMSS, we used the Significance Analysis of Microarrays program (53). We found 415 unique genes expressed at significantly lower levels in E. dispar SAW760 than in E. histolytica HM-1:IMSS (Tables 2 and 3; see Table S1 in the supplemental material). Approximately two-thirds of the genes identified as having lower expression in E. dispar SAW760 did not have a highly conserved ortholog in the E. dispar database (Table 2; see Table S1 in the supplemental material). This subset contains genes already described as being divergent in E. dispar SAW760, including CP1 (242.m00078 and 79.m00156), Ariel1 (160.m00087), AIG1 (565.m00023) (50), and KERP1 (77.m00174). Other genes in this category included stress response genes such as those encoding a multidrug resistance protein (151.m00094), a DNA repair protein (151.m00093), and heat shock proteins (53.m00209, 92.m00150, and 136.m00105). Several members of the endoplasmic reticulum (ER)-associated translocon that were first identified in yeast secretory mutants (19), including Sec24 (1.m00597 and 178.m00100) and Sec6 (7.m00480), had decreased expression in E. dispar.
TABLE 2.
Subset of genes with lower expression in E. dispar SAW760 than in E. histolytica HM-1:IMSS and with loci not highly conserved in the E. dispar SAW760 databasea
| TIGR locusb | TIGR gene product | Microarray hybridization ratio |
Gene size (nt) | % Nucleotide identityc | % of locus found in databased | |
|---|---|---|---|---|---|---|
| E. dispar/ E. histolytica HM-1:IMSS | E. histolytica Rahman/ E. histolytica HM-1:IMSS | |||||
| 320.m00035 | Pseudogene, Gal/GalNAc lectin heavy subunit | 0.22 | 1.16 | 3,870 | 88 | 54 |
| 151.m00094 | Multidrug resistance protein, putative | 0.38 | 1.18 | 4,398 | 97 | 39 |
| 242.m00078 | Cysteine protease 1 | 0.31 | 1.27 | 948 | 85 | 60 |
| 79.m00156 | Cysteine protease 1 | 0.46 | 1.18 | 1,005 | 85 | 57 |
| 191.m00117 | Cysteine proteinase, putative | 0.09 | 1.07 | 957 | 82 | 65 |
| 13.m00317 | Sucrose transporter, putative | 0.15 | 1.10 | 1,386 | No hits | No hits |
| 116.m00130 | Ras GTPase activating protein, putative | 0.35 | 0.73 | 3,551 | 96 | 12 |
| 52.m00161 | Immunodominant variable surface antigen | 0.22 | 1.01 | 3,342 | 90 | 26 |
| 11.m00326 | tRNA intron endonuclease, putative | 0.54 | 0.77 | 492 | 86 | 28 |
| 5.m00482 | Protein kinase, putative | 0.31 | 4.52 | 3,594 | 88 | 34 |
| 114.m00128 | Rab family GTPase | 0.21 | 0.93 | 667 | 95 | 6 |
| 310.m00070 | BspA-like leucine-rich repeat protein, putative | 0.23 | 0.80 | 1,197 | 89 | 67 |
| 129.m00157 | DNA primase large subunit, putative | 0.14 | 0.73 | 1,374 | 90 | 47 |
| 501.m00019 | Cysteine protease 8 | 0.10 | 1.14 | 948 | 82 | 57 |
| 39.m00237 | Cortexillin, putative | 0.14 | 0.76 | 2,376 | 93 | 18 |
| 442.m00024 | Receptor protein kinase, putative | 0.36 | 1.70 | 6,617 | 96 | 28 |
| 151.m00093 | DNA repair protein, putative | 0.41 | 1.07 | 1,982 | 88 | 19 |
| 2.m00588 | Protein kinase, putative | 0.51 | 0.94 | 1,227 | 92 | 6 |
| 126.m00100 | Rho GTPase activating protein, putative | 0.10 | 0.75 | 1,242 | 93 | 46 |
| 565.m00023 | AIG1 family protein, putative | 0.07 | 0.57 | 1,095 | 86 | 47 |
| 7.m00480 | Sec6 protein, putative | 0.39 | 0.65 | 2,301 | 95 | 28 |
| 53.m00209 | Heat shock protein 70, putative | 0.44 | 0.82 | 1,898 | 88 | 62 |
| 16.m00343 | Ubiquitin carboxyl-terminal hydrolase, putative | 0.31 | 0.87 | 2,421 | 89 | 100 |
| 136.m00105 | 70-kDa heat shock protein, putative | 0.37 | 0.83 | 1,727 | 85 | 26 |
| 77.m00174 | Hypothetical protein (KERP1) | 0.22 | 0.68 | 552 | No hits | No hits |
| 338.m00048 | Leucyl-tRNA synthetase, putative | 0.48 | 0.71 | 3,219 | 96 | 32 |
| 130.m00115 | Importin beta subunit, putative | 0.25 | 1.27 | 3,115 | 90 | 25 |
| 92.m00150 | Heat shock protein 90, putative | 0.17 | 0.58 | 2,157 | 98 | 37 |
| 77.m00153 | Glycogen debranching enzyme, putative | 0.32 | 1.13 | 4,332 | 94 | 19 |
| 178.m00100 | Sec24 protein, putative | 0.35 | 0.59 | 2,283 | 93 | 47 |
| 14.m00281 | Pseudogene, beta-adaptin | 0.24 | 0.90 | 2,592 | 98 | 30 |
| 57.m00152 | CCR4/NOT complex transcription factor subunit 4 | 0.58 | 0.67 | 2,112 | 96 | 49 |
| 60.m00136 | Conserved hypothetical protein | 0.55 | 1.25 | 2,597 | 86 | 21 |
| 296.m00051 | Rho family GTPase | 0.35 | 0.86 | 746 | 97 | 41 |
| 1.m00597 | SEC-24 protein, putative | 0.51 | 0.95 | 1,944 | 94 | 19 |
| 309.m00046 | Phospholipid-transporting P-type ATPase, putative | 0.43 | 1.66 | 3,297 | 94 | 38 |
| 160.m00087 | Surface antigen ariel1 related | 0.28 | 1.31 | 648 | No hits | No hits |
Based on <90% nucleotide identity and/or <50% of the locus.
Genes confirmed by Northern blot analysis are shown in bold.
Compared to E. dispar SAW760 ortholog.
E. dispar SAW760 database.
TABLE 3.
Subset of genes with lower expression in E. dispar SAW760 than in E. histolytica HM-1:IMSS and with loci with significant orthologs in the E. dispar SAW760 databasea
| TIGR locusb | TIGR gene product | Microarray hybridization ratio |
Gene size (nt) | % Nucleotide identityc | % of locus found in databased | |
|---|---|---|---|---|---|---|
| E. dispar/ E. histolytica HM-1:IMSS | E. histolytica Rahman/ E. histolytica HM-1:IMSS | |||||
| 20.m00330 | CXXC-rich protein | 0.25 | 0.82 | 3,379 | 92 | 76 |
| 6.m00454 | Lysozyme, putative | 0.47 | 0.92 | 639 | 96 | 99 |
| 9.m00419 | Fe hydrogenase, putative | 0.39 | 1.42 | 1,407 | 91 | 65 |
| 18.m00300 | Protein kinase, putative | 0.45 | 2.42 | 1,188 | 94 | 99 |
| 171.m00098 | Ankyrin repeat protein, putative | 0.58 | 1.34 | 828 | 93 | 100 |
| 836.m00014 | ARP2/3 complex 21 kDa subunit, putative | 0.46 | 0.79 | 586 | 93 | 100 |
| 110.m00129 | Rho family GTPase | 0.56 | 0.67 | 639 | 96 | 96 |
| 32.m00230 | BspA-like leucine rich repeat protein, putative | 0.54 | 1.16 | 1,278 | 93 | 99 |
| 344.m00046 | Ser/Thr protein phosphatase, putative | 0.25 | 0.63 | 1,551 | 93 | 100 |
| 283.m00063 | DEAD/DEAH box helicase, putative | 0.37 | 1.20 | 1,260 | 94 | 100 |
| 94.m00134 | Glycogen synthase, putative | 0.34 | 0.96 | 4,128 | 96 | 76 |
| 50.m00199 | Sec61 protein, putative | 0.43 | 0.81 | 246 | 96 | 86 |
| 30.m00257 | Rab family GTPase | 0.31 | 0.57 | 648 | 92 | 101 |
| 1.m00628 | Protein disulfide isomerase, putative | 0.32 | 1.33 | 990 | 92 | 91 |
| 251.m00088 | Sir2 family transcriptional regulator, putative | 0.48 | 0.63 | 1,079 | 93 | 68 |
| 297.m00063 | CCAAT-box-binding transcription factor, putative | 0.40 | 1.07 | 2,157 | 94 | 100 |
| 87.m00163 | Potassium transporter, putative | 0.35 | 1.17 | 2,202 | 94 | 65 |
| 76.m00156 | Rab family GTPase | 0.50 | 0.98 | 760 | 95 | 68 |
| 176.m00112 | Pseudogene, peroxiredoxin | 0.42 | 0.68 | 696 | 94 | 91 |
| 8.m00352 | Phospholipid-transporting P-type ATPase, putative | 0.28 | 0.62 | 4,124 | 93 | 66 |
| 95.m00149 | Protein phosophatase 2C, putative | 0.32 | 1.35 | 2,925 | 91 | 66 |
| 143.m00082 | Protein kinase, putative | 0.53 | 0.93 | 1,686 | 96 | 100 |
| 16.m00300 | Gal/GalNAc lectin heavy subunit | 0.24 | 0.97 | 3,861 | 90 | 54 |
| 103.m00174 | Conserved hypothetical protein | 0.24 | 1.26 | 2,586 | 91 | 57 |
| 67.m00102 | Phosphatidylinositol 3-kinase, putative | 0.45 | 0.73 | 3,330 | 95 | 63 |
| 95.m00133 | Rab GTPase activating protein, putative | 0.48 | 0.92 | 1,954 | 94 | 60 |
| 29.m00231 | Sphingomyelinase C, putative | 0.17 | 0.51 | 975 | 95 | 100 |
| 6.m00467 | Type A flavoprotein | 0.23 | 0.87 | 1,221 | 92 | 99 |
| 15.m00331 | Peptidyl-prolyl cis-trans isomerase, putative | 0.41 | 0.78 | 1,185 | 93 | 69 |
Based on ≥90% nucleotide identity and/or ≥50% of the locus.
Genes confirmed by Northern blot analysis are shown in bold.
Compared to E. dispar SAW760 ortholog.
E. dispar SAW760 database.
We identified 146 genes that, although highly conserved in E. dispar SAW760, had significantly lower expression in E. dispar than in E. histolytica HM-1:IMSS (Table 3; see Table S1 in the supplemental material). Since the sequences of these genes in E. histolytica and E. dispar are highly similar, we can definitively ascribe the differences in the microarray signal to decreased transcript abundance. Importantly, these genes would not have been identified as being different between E. histolytica and E. dispar on the basis of comparative genomic hybridization or genome sequencing alone. This group includes genes potentially involved in virulence, including thoseencoding lysozyme (6.m00454), sphingomyelinase C (29.m00231), and several transcription regulators (251.m00088 and 297.m00063). Additionally, in this group were several putative stress response genes, including those encoding Fe hydrogenase (9.m00419), peroxiredoxin (176.m00112), and a type A flavoprotein (6.m00467). In other systems, these genes have been shown to be involved in a response to reactive oxygen species and may play similar roles in Entamoeba (9, 17, 24, 31). Additionally, a peptidyl-prolyl cis-trans isomerase gene (15.m00331) was identified. This gene (originally identified by accession number NP189160) has previously been shown to be induced in E. histolytica by exposure to high oxygen levels by differential-display PCR (1).
Genes with expression restricted to a virulent strain of E. histolytica.
We found 29 genes (∼1% of the total number examined) with significantly decreased expression in both the nonvirulent E. histolytica Rahman strain and E. dispar SAW760 (Table 4). Most genes in this category were genes for hypothetical proteins; however, genes encoding an endoplasmic reticulum-associated heat shock protein (29.m00210), a cell surface protein (80.m00165), and serine palmitoyltransferase (32.m00218) were also identified. The majority (∼80%) of genes in this category did not have highly conserved homologues in the E. dispar SAW760 database, and none of the genes had been identified in previous studies relating to amebic virulence. Interestingly, five of the identified genes (147.m00110, 543.m00021, 864.m00008, 296.m00047, and 460.m00024) showed homology to the open reading frame found in the SSE58 repeat region identified as encoding a stress-dependent polymorphic charged antigen (Ehssp1) (48). We confirmed a lack of expression in E. dispar SAW760 and E. histolytica Rahman for one gene family member (460.m00024) by Northern blot analysis (data not shown; P. Vanchinathan, personal communication). One of the hypothetical proteins identified as having decreased expression in both the avirulent strain and the species has homology to a Plasmodium STARP antigen as well as a hemagglutinin from Staphylococcus epidermidis. The gene for this protein (238.m00054) belongs to agene family consisting of three members (238.m00054, 21.m00228, and 312.m00036) which are each ∼7.8 kb in size. Two other loci (16.m00292 and 126.m00120), which may represent truncated versions (1.7 kb and 1.2 kb, respectively) of the gene, also exist and share homology with the 3′ region of the gene family.
TABLE 4.
Genes identified as having lower expression in E. dispar SAW760 and E. histolytica Rahman than in E. histolytica HM-1:IMSSa
| TIGR locusb | TIGR gene product | Microarray hybridization ratio |
Gene size (nt) | % Nucleotide identityc | % of locus found in databased | |
|---|---|---|---|---|---|---|
| E. dispar/ E. histolytica HM-1:IMSS | E. histolytica Rahman/ E. histolytica HM-1:IMSS | |||||
| 32.m00218 | Serine palmitoyltransferase | 0.37 | 0.39 | 2,649 | 94 | 62 |
| 2.m00567 | Conserved hypothetical protein | 0.62 | 0.40 | 912 | 91 | 85 |
| 343.m00074 | 26S proteasome regulatory subunit | 0.52 | 0.49 | 1,071 | 94 | 68 |
| 269.m00084 | Mitochondrial carrier protein | 0.25 | 0.44 | 831 | 95 | 77 |
| 143.m00082 | Protein kinase, putative | 0.23 | 0.35 | 1,686 | 96 | 100 |
| 312.m00036 | Hypothetical protein | 0.07 | 0.08 | 7,866 | 89 | 35 |
| 238.m00054 | Hypothetical protein | 0.10 | 0.09 | 7,815 | 89 | 35 |
| 196.m00089 | Conserved hypothetical protein | 0.31 | 0.47 | 863 | 89 | 56 |
| 296.m00047 | Hypothetical protein | 0.17 | 0.31 | 1,092 | 87 | 99 |
| 7.m00396 | Hypothetical protein | 0.04 | 0.13 | 621 | No hits | No hits |
| 209.m00109 | Hypothetical protein | 0.30 | 0.31 | 288 | 81 | 56 |
| 225.m00057 | Hypothetical protein | 0.18 | 0.16 | 828 | No hits | No hits |
| 394.m00058 | Hypothetical protein | 0.08 | 0.15 | 843 | 82 | 19 |
| 11.m00354 | Hypothetical protein | 0.24 | 0.28 | 1,242 | 91 | 7 |
| 120.m00092 | Hypothetical protein | 0.36 | 0.29 | 630 | 88 | 25 |
| 112.m00118 | Acyl-coenzyme A synthetase, putative | 0.27 | 0.44 | 1947 | 95 | 48 |
| 612.m00020 | Hypothetical protein | 0.10 | 0.23 | 678 | 89 | 26 |
| 29.m00210 | hsp70 family protein | 0.55 | 0.45 | 1,570 | 88 | 61 |
| 209.m00108 | Hypothetical protein | 0.50 | 0.32 | 243 | No hits | No hits |
| 864.m00008 | Conserved hypothetical protein | 0.55 | 0.30 | 1,172 | 87 | 39 |
| 80.m00165 | Hypothetical protein | 0.10 | 0.30 | 2,988 | 89 | 16 |
| 194.m00115 | Protein kinase, putative | 0.41 | 0.34 | 2,727 | 95 | 23 |
| 27.m00257 | Reverse transcriptase, putative | 0.21 | 0.16 | 1,011 | 89 | 85 |
| 393.m00037 | Hypothetical protein | 0.30 | 0.36 | 786 | No hits | No hits |
| 147.m00110 | Conserved hypothetical protein | 0.37 | 0.27 | 1,152 | 89 | 42 |
| 543.m00021 | Hypothetical protein | 0.44 | 0.44 | 321 | 90 | 85 |
| 244.m00078 | Hypothetical protein | 0.06 | 0.12 | 612 | No hits | No hits |
| 711.m00021 | Hypothetical protein | 0.24 | 0.33 | 342 | 84 | 56 |
| 460.m00024 | Conserved hypothetical protein | 0.10 | 0.32 | 612 | 85 | 78 |
The first five loci listed are highly conserved in E. dispar SAW760 (≥90% nucleotide identity over ≥50% of the locus).
Genes confirmed by Northern blot analysis are shown in bold.
Compared to E. dispar SAW760 ortholog.
E. dispar SAW760 database.
Genes with lower expression in the nonvirulent strain E. histolytica Rahman than in E. histolytica HM-1:IMSS.
In a comparison of E. histolytica Rahman to E. histolytica HM-1:IMSS, we found only three genes that had uniquely lower expression levels in E. histolytica Rahman (Table 5). These included the genes encoding the oxidoreductase aldo/keto reductase family of proteins (248.m00073), ubiquitin ligase (195.m00092), and a Rap/Ran GTPase-activating protein (putative 20.m00318). In addition, there were a number of clones that had homology to the non-LTR retrotransposable elements EhLINEs that also had lower expression in E. histolytica Rahman.
TABLE 5.
Genes with significantly lower expression in E. histolytica Rahman
| TIGR locus | TIGR gene product | Microarray hybridization ratio |
Gene size (nt) | |
|---|---|---|---|---|
| E. dispar/ E. histolytica HM-1:IMSS | E. histolytica Rahman/ E. histolytica HM-1:IMSS | |||
| 195.m00092 | Ubiquitin-protein ligase e3 component | 0.75 | 0.45 | 4,158 |
| 248.m00073 | Oxidoreductase, aldo/keto reductase family | 0.72 | 0.54 | 918 |
| 20.m00318 | Rap/Ran GTPase activating protein, putative | 1.28 | 0.40 | 2,012 |
A number of retrotransposons have decreased expression in the nonvirulent Entamoeba species/strains.
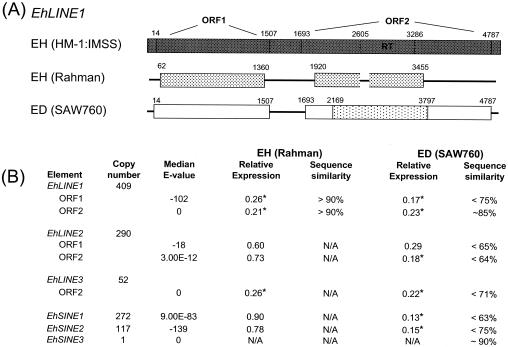
The E. histolytica genome has a number of repetitive elements (5, 6, 38). These include short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs) that represent non-LTR retrotransposons. Three SINE genes (EhSINE1, EhSINE2,and EhSINE3) and three LINE genes (EhLINE1, EhLINE2, and EhLINE3) have been identified, and each (with the exception of EhSINE3) is present in multiple copies in the genome. The consensus sequence for EhLINEs contains one or two ORFs: the protein encoded by ORF1 (1.5 kb) contains a putative coiled-coil domain, and the protein encoded by ORF2 (3 kb) contains endonuclease and reverse transcriptase domains (5, 6, 38). We used the consensus sequence for the conserved regions of each element to identify clones on our array that represent each of these elements (5). The 50 clones on the array with the greatest similarity to the consensus sequence for each LINE or SINE were used to determine the expression levels (Fig. 4). For each of the SINEs and LINEs, we had clones on the array with a high similarity to the consensus sequence (median E values, 9E-83 to E = 0), with the exception of EhLINE2, for which we could not find clones highly similar to the currently defined consensus sequences for ORF1 and ORF2 (median E values, E-18 and 3E-12). This is apparently due to the fact that the consensus sequence, as identified by Bakre, is significantly different from genomic copies, as we were unable to find many highly similar sequences even in the E. histolytica HM-1:IMSS genomic database (data not shown).
FIG. 4.
EhLINEs and EhSINEs have altered expression in the nonvirulent Entamoeba species/strains. (A) Diagrammatic representation of the sequence data for EhLINE1. E. histolytica Rahman data obtained by sequence analysis and E. dispar SAW760 obtained from BLASTN analysis versus genome sequence data. The numbers above the lines represent nucleotide positions, and the reverse transcriptase domain in ORF2 is noted. The shading is indicative of the nucleotide identity (▩, ≥95% identity; ░⃞, >90% nucleotide identity;  , >85% nucleotide identity; □, <80% nucleotide identity). No high-homology hit was found for ORF1 in E. dispar SAW760; however, a 1,600-bp region encompassing the reverse transcriptase domain showed 85% identity in E. dispar SAW760. (B) The average expression levels of EhLINE and EhSINE in E. histolytica Rahman and E. dispar SAW760 relative to those in E. histolytica HM-1:IMSS are shown. For each element, 50 clones with the highest similarity to the consensus sequence for each EhLINE or EhSINE were used. The copy number (adapted from Bakre et al. [5]), median BLASTN E-value for the 50 clones with highest homology, and genomic sequence similarity are also displayed. Expression levels that are significantly different from that of E. histolytica HM-1:IMSS are labeled by an asterisk and denote a P value of <0.05.
, >85% nucleotide identity; □, <80% nucleotide identity). No high-homology hit was found for ORF1 in E. dispar SAW760; however, a 1,600-bp region encompassing the reverse transcriptase domain showed 85% identity in E. dispar SAW760. (B) The average expression levels of EhLINE and EhSINE in E. histolytica Rahman and E. dispar SAW760 relative to those in E. histolytica HM-1:IMSS are shown. For each element, 50 clones with the highest similarity to the consensus sequence for each EhLINE or EhSINE were used. The copy number (adapted from Bakre et al. [5]), median BLASTN E-value for the 50 clones with highest homology, and genomic sequence similarity are also displayed. Expression levels that are significantly different from that of E. histolytica HM-1:IMSS are labeled by an asterisk and denote a P value of <0.05.
EhSINE1 has previously been shown to be divergent in E. dispar, and the array data confirmed those results (Fig. 1 and 4). Using a probe for E. histolytica EhSINE2, we determined that it had significantly decreased expression in E. dispar (Fig. 2Aand 4). Sequence analysis against the E. dispar databases revealed that EhSINE2 is missing or highly degenerate (75% identity over 54% of the locus) in E. dispar. Thus, the lower array hybridization and the absent signal on the Northern blot may be due to the inability of the E. histolytica-specific probe to cross-hybridize with the E. dispar gene. EhSINE3, originally identified as UEE1 in E. dispar, is present once in the E. histolytica genome (5, 51). Our arrays contained only one clone with similarity to EhSINE3, and it did not show significant hybridization for any of the species/strains tested.
Clones containing EhLINE1 consistently gave lower signals on the microarrays in both nonvirulent species/strains than in E. histolytica HM-1:IMSS (Fig. 3B and 4). Using clone ENTOB31 (with homology to EhLINE1) in a Northern blot analysis, we identified the expected 1.5-kb and 3-kb bands, and confirmatory to the array data, both bands showed dramatically less, if any, hybridization in both E. dispar SAW760 and E. histolytica Rahman (Fig. 3B). Additionally, EhLINE3 ORF2 (there is no ORF1 in EhLINE3) also displayed significantly lower expression in both E. histolytica Rahman and E. dispar SAW760 than in E. histolytica HM-1:IMSS. The expression levels of both ORFs of EhLINE2 were lower in E. dispar SAW760 (although not statistically significant for ORF1) than in E. histolytica HM-1:IMSS. EhLINE2 ORF1 and ORF2 were equally expressed in E. histolytica Rahman. Whether EhLINE2 is unique among the LINE genes and is expressed in E. histolytica Rahman or whether these data are misleading due to the lack of representative consensus sequences for EhLINE2 is not clear at present. Interestingly, both Northern blot and array data revealed that strains of E. histolytica HM-1:IMSS from different laboratories have various levels of expression of EhLINEs and EhSINEs (data not shown). Despite the variability, however, the expression levels for EhLINE1 and EhLINE3 for all E. histolytica HM-1:IMSS isolates were always higher than those of E. histolytica Rahman and E. dispar SAW760 (data not shown). Additionally, it has been postulated that some members of EhLINE1 contain a single ORF instead of two ORFs, and our Northern blots confirmed this hypothesis (data not shown) (5, 54). The roles of these LINEs and SINEs in E. histolytica are not clear at present, although diverse roles, including effects on genome structure and organization, gene expression (30, 43), and response to stress (32, 35), have been described for other systems.
Genomic characterization of genes with decreased expression.
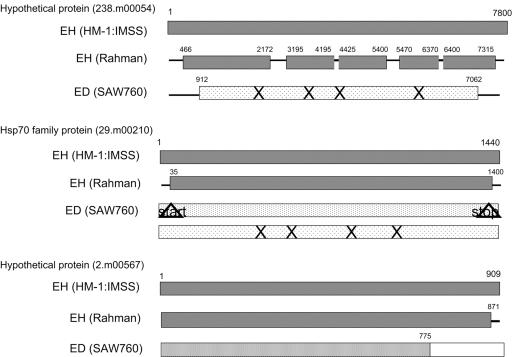
Five genetic loci identified as having lower expression in both nonvirulent species/strains were sequenced to determine if the differences in expression levels could be attributed to genetic loss, genetic drift, or other phenomena (Fig. 4 and 5). The hypothetical gene 238.m00054 was sequenced in three segments, covering the majority of the gene. In E. histolytica Rahman, the sequences were 95 to 99% identical for the regions sequenced, with a relatively conserved protein structure. For E. dispar SAW760, the sequence identity was significantly less (81% over 73% of locus), with a number of predicted stop codons. A gene for an Hsp70 family protein (29.m00210) was also sequenced; again, the sequence was nearly identical (96% identity) in E. histolytica Rahman to that in E. histolytica HM-1:IMSS. The primers used for this gene incorporated the start and stop codons, so the sequence identity in this region could not be ascertained. In E. dispar SAW760, two contigs (98673 and 98651) showing homology to this gene can be found. One contig (98673) showed 85% identity over 100% of the locus; however, the start and stop codons were both mutated. The other contig (98651) showed 82% identity over 100% of the locus and had both start and stop codons conserved; however, it contained many internal stop codons. The gene for the hypothetical protein (2.m00567) was also nearly identical to that in E. histolytica Rahman (99% identity); again, the primers used incorporated the stop codon, and thus, conclusive sequence data for the very end of the gene could not be determined. The promoter region of this gene in E. histolytica Rahman was sequenced and was identical to the E. histolytica HM-1:IMSS sequence, with the exception of a single nucleotide polymorphism (A→G change at position −442 relative to the start codon) (7). This single nucleotide polymorphism was also seen in another virulent strain of E. histolytica (200:NIH), and so its presence does not correlate with an avirulence phenotype. In E. dispar SAW760, the gene 2.m00567 had very high homology over the first 775 bp (contig 98778 with 91% identity); however, the remaining 134 bp had very poor homology (64%) to any E. dispar sequences. Sequence data from the two ORFs in EhLINE1 were also obtained (Fig. 4A). In E. histolytica Rahman, the sequence was highly conserved for both ORF1 and the reverse transcriptase domain of ORF2 (>90% identity). In E. dispar SAW760, ORF1 is missing or degenerate (74% identity over 24% of locus), whereas ORF2 is somewhat conserved (85% identity over 53% of locus). Our analysis revealed that the genes with decreased expression in E. histolytica Rahman were largely conserved at the nucleotide level; in contrast, a majority (∼65%) of the genes with decreased expression in E. dispar SAW760 had a divergent sequence.
FIG. 5.
Diagrammatic representation of the sequence data for the genes with differential expression levels in the nonvirulent species/strains. The shading is indicative of the nucleotide identity (▩, ≥95% identity; ░⃞, >90% nucleotide identity;  , >85% nucleotide identity;
, >85% nucleotide identity;  , >80% nucleotide identity; □, <80% nucleotide identity). ×, stop codons; ▵, mutated stop or start codons.
, >80% nucleotide identity; □, <80% nucleotide identity). ×, stop codons; ▵, mutated stop or start codons.
Comparison of genomic and expression differences identified by microarray analysis.
We have previously performed comparative genomic hybridizations of Entamoeba species and identified a number of loci that are absent, significantly divergent, or significantly decreased in copy number in E. dispar SAW760 compared to those in E. histolytica HM-1:IMSS. Overall, 22 genes were absent or highly divergent in E. dispar SAW760; of these, 8 genes are likely to be expressed in E. histolytica HM-1:IMSS under the trophozoite conditions tested. These eight genes had a median expression level in E. dispar SAW760 that was nearly 10-fold lower than that in E. histolytica HM-1:IMSS. Additionally, 45 genes were significantly divergent in E. dispar SAW760; of these, 18 are likely to be expressed in E. histolytica HM-1:IMSS. Again, the median expression level for these genes was significantly lower (eightfold) in E. dispar SAW760 than in E. histolytica HM-1:IMSS. Comparisons between E. histolytica Rahman and E. histolytica HM-1:IMSS using genomic DNA hybridizations yielded far fewer significantly divergent genes (only five), and none of these genes are likely to be expressed at appreciable levels in E. histolytica HM-1:IMSS. Therefore, none of the genes with decreased expression in E. histolytica Rahman should be attributed to genomic differences.
DISCUSSION
We have compared the transcriptional profiles of the nonvirulent strain Entamoeba histolytica Rahman and the nonvirulent species E. dispar SAW760 to that of a virulent E. histolytica strain, HM-1:IMSS, using an Entamoeba histolytica DNA microarray. We confirmed that the arrays are effective for identifying differential expression of genes that are missing or degenerate at a genomic level, as well as those genes that are conserved at a genomic level but expressed at differing levels. Additionally, we identified 415 genes with significantly decreased expression levels in E. dispar SAW760 and 32 genes with significantly decreased expression levels in E. histolytica Rahman compared to those in E. histolytica HM-1:IMSS. Importantly, we identified 29 genes with lower expression levels in both the nonvirulent strain E. histolytica Rahman and nonvirulent species E. dispar SAW760 than in the virulent E. histolytica HM-1:IMSS strain.
One interesting observation is that a number of genes that had decreased expression in one or both nonvirulent species/strains have roles in pathogenesis or stress response in other systems. Two hypothetical proteins with similarity to both a sporozoite threonine-asparagine-rich protein (STARP) antigen from Plasmodium and a hemagglutinin from Staphylococcus epidermidis (238.m00054 and 312.m00036) have decreased expression in both E. dispar SAW760 and E. histolytica Rahman. The STARP antigen from Plasmodium is located on the surface of sporozoites and is believed to be involved in pathogenesis, and antibodies against it inhibit invasion of hepatocytes (44). If the gene product functions as a streptococcal hemagglutinin, it may have a role in attachment or erythrophagocytosis. However, functional studies will have to be performed to confirm the roles of these genes in amebic pathogenesis. Serine palmitoyltransferase (locus 32.m00218), also with decreased expression in both nonvirulent Entamoeba species/strains, is a membrane-bound endoplasmic-reticulum-associated enzyme, which catalyzes the reaction of l-serine and palmitoyl coenzyme A to 3-ketodihydrosphingosine. This enzyme is essential in the production of ceramides and sphingolipids, which have diverse functions in endocytosis, stress response, adhesion, and trafficking of glycosylphosphatidylinositol-molecules (18, 26, 45). A decrease in the expression of this enzyme also may help explain the previously defined differences in lipophosphoglycans and proteophosphoglycans on the surface of virulent and nonvirulent Entamoeba organisms (40).
A number of genes involved in cytolysis were identified as having lower expression levels in E. dispar SAW760 than in E. histolytica HM-1:IMSS. These include the genes encoding sphingomyelinase C (29.m00231), a cytolytic factor involved in hemolysis in Listeria (25), Staphylococcus (39), and other pathogenic species, and a putative lysozyme (6.m00454) which has been shown to be involved in cell lysis in many systems, including Entamoeba (41).
Many genes classically involved in stress response, particularly the degradation of reactive oxygen species, had decreased expression in the nonvirulent Entamoeba species/strains. These included genes encoding a type A flavoprotein (6.m00467), which is potentially important in detoxifying nitric oxide and oxygen, and Fe hydrogenase (9.m00419), which has been shown in certain strains of bacteria to be involved in the response to high oxygen levels (17, 24). Additionally, a gene encoding peroxiredoxin (176.m00112), an important antioxidant involved in detoxifying peroxides, was also identified (9, 31). Choi et al. recently showed by enzyme-linked immunosorbent assay that E. histolytica contains as much as 50 times more peroxiredoxin than E. dispar (14). Peroxiredoxin is likely to be important in protection from the high oxygen content of the host after invasion and from reactive oxygen species from host immune cells. During colonization of the colon, the parasites are in an anaerobic environment; however, upon tissue invasion, they are exposed to aerobic conditions. Thus, decreased expression of genes involved in stress response or degradation of reactive oxygen species may potentially provide insights into the ability of certain parasite species/strains to colonize but not cause invasive disease. Five genes with similarity to a gene for stress-induced polymorphic charged antigen (296.m00047, 864.m00008, 147.m00110, 543.m00021, and 460.m00024), as well as a gene previously shown to be induced upon exposure to high oxygen levels (15.m00331), were also identified as having decreased expression levels in nonvirulent Entamoeba species/strains.
A number of the LINEs and SINEs, representing non-long-terminal-repeat retrotransposons, had altered expression in the nonvirulent Entamoeba species/strains. EhLINE1 and EhLINE3 had significantly lower expression in both E. histolytica Rahman and E. dispar SAW760. Additionally, two SINEs are nonfunctional in E. dispar SAW760. Notably, while E. histolytica HM-1:IMSS isolates from different labs displayed various levels of EhLINE expression, virulent E. histolytica species/strains always exhibited higher expression levels of EhLINE1 and EhLINE3 than nonvirulent species/strains. Interestingly, in a recent paper, Pritham et al. reported that the two human parasites Entamoeba histolytica and Entamoeba dispar possessed many copies of retrotransposons but very few DNA transposons, while the opposite was true for the reptilian parasite Entamoeba invadens and the free-living Entamoeba moshkovskii (46). The authors hypothesized that evolutionary pressures from their human host may be responsible for the phenomenon. A number of diverse roles for similar elements have been described in other systems, including transcriptional regulation, genome organization, and stress response. The roles of these elements in regulating amebic transcription are not well characterized. Interestingly, these LINEs and SINEs are frequently found in close proximity to coding regions in E. histolytica, which could allow these elements to influence gene expression (5). A recent observation of fortuitous silencing of the amebapore A gene, by expression driven by an adjacent SINE (10), strongly suggests that in E. histolytica, similar to other systems (30, 35), transcriptional regulation is controlled by the LINEs and SINEs. The roles of these elements in affecting transcriptional regulation and genome modulation and their potential roles in the transition of Entamoeba from a gut commensal to an invasive pathogen deserve further investigation. Genome-wide comparisons of virulent and nonvirulent species/strains have yielded important results in many pathogen systems (8, 22, 29, 50, 59, 60). In a recent study comparing invasive and noninvasive Staphylococcus epidermidis strains, certain genes (including streptococcal hemagglutinin and many transposases) were found to be lacking in noninvasive species/strains (60).
We have performed the first large-scale transcriptional profiling of E. histolytica and E. dispar and have found differences in the transcriptional profiles of virulent and nonvirulent Entamoeba species/strains. A number of genes with roles in pathogenesis and stress response had decreased expression in the nonvirulent Entamoeba species/strains. Some of these differentially transcribed genes may represent potential virulence determinants and are important targets for genetic studies.
Supplementary Material
Acknowledgments
This work was supported by a Cellular and Molecular Biology Training Program grant (NIH 5 T32 GM007276) to R.M., a Burroughs Wellcome Collaboration grant and a grant from the NIAID (AI-053724) to U.S., and a grant from the Stanford Digestive Disease Center (NIH P30 DK56339).
We gratefully acknowledge the help of Brendan Loftus, Neil Hall, and Iain Anderson (TIGR and Sanger Institute) for access to E. histolytica clones, sequence, and genome data. We thank Sudha Bhattacharya and Abhijeet Bakre for help in identifying EhSINE and EhLINE sequences, and Padmini Vanchinathan for unpublished data. We are indebted to C. Graham Clark for E. dispar and E. histolytica RNA and to Sandeep Jaggi for help with data analysis, Kevin Visconti for microarray printing, Janos Demeter and the members of the Stanford Microarray database staff for technical support, and all members of the laboratory for helpful suggestions and discussions. We thank Barbara Mann for critically reading the manuscript.
Editor: J. F. Urban, Jr.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akbar, M. A., N. S. Chatterjee, P. Sen, A. Debnath, A. Pal, T. Bera, and P. Das. 2004. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 133:187-196. [DOI] [PubMed] [Google Scholar]
- 2.Ankri, S., F. Padilla-Vaca, T. Stolarsky, L. Koole, U. Katz, and D. Mirelman. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol. 33:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Ankri, S., T. Stolarsky, R. Bracha, F. Padilla-Vaca, and D. Mirelman. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. [DOI] [PubMed] [Google Scholar]
- 5.Bakre, A. A., K. Rawal, R. Ramaswamy, A. Bhattacharya, and S. Bhattacharya. 2005. The LINEs and SINEs of Entamoeba histolytica: comparative analysis and genomic distribution. Exp. Parasitol. 110:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, A., S. Bhattacharya, and J. P. Ackers. 2003. Nontranslated polyadenylated RNAs from Entamoeba histolytica. Trends Parasitol. 19:286-289. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya, D., R. Haque, and U. Singh. 2005. Coding and noncoding genomic regions of Entamoeba histolytica have significantly different rates of sequence polymorphisms: implications for epidemiologial studies. J. Clin. Microbiol. 43:4815-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 9.Bozonet, S. M., V. J. Findlay, A. M. Day, J. Cameron, E. A. Veal, and B. A. Morgan. 2005. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J. Biol. Chem. 24:23319-23327. [DOI] [PubMed] [Google Scholar]
- 10.Bracha, R., Y. Nuchamowitz, and D. Mirelman. 2003. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot. Cell 2:295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruchhaus, I., T. Jacobs, M. Leippe, and E. Tannich. 1996. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol. Microbiol. 22:255-263. [DOI] [PubMed] [Google Scholar]
- 12.Bruchhaus, I., B. J. Loftus, N. Hall, and E. Tannich. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruchhaus, I., and E. Tannich. 1996. A gene highly homologous to ACP1 encoding cysteine proteinase 3 in Entamoeba histolytica is present and expressed in E. dispar. Parasitol. Res. 82:189-192. [DOI] [PubMed] [Google Scholar]
- 14.Choi, M. H., D. Sajed, L. Poole, K. Hirata, S. Herdman, B. E. Torian, and S. L. Reed. 2005. An unusual surface peroxiredoxin protects invasive Entamoeba histolytica from oxidant attack. Mol. Biochem. Parasitol. 143:80-89. [DOI] [PubMed] [Google Scholar]
- 15.Clark, C. G. 1995. Axenic cultivation of Entamoeba dispar Brumpt 1925, Entamoeba insolita Geiman and Wichterman 1937 and Entamoeba ranarum Grassi 1879. J. Eukaryot. Microbiol. 42:590-593. [DOI] [PubMed] [Google Scholar]
- 16.Clark, C. G., and L. S. Diamond. 1993. Entamoeba histolytica: a method for isolate identification. Exp. Parasitol. 77:450-455. [DOI] [PubMed] [Google Scholar]
- 17.Das, A., E. D. Coulter, D. M. Kurtz, Jr., and L. G. Ljungdahl. 2001. Five-gene cluster in Clostridium thermoaceticum consisting of two divergent operons encoding rubredoxin oxidoreductase-rubredoxin and rubrerythrin-type A flavoprotein-high-molecular-weight rubredoxin. J. Bacteriol. 183:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny, P. W., D. Goulding, M. A. Ferguson, and D. F. Smith. 2004. Sphingolipid-free Leishmania are defective in membrane trafficking, differentiation and infectivity. Mol. Microbiol. 52:313-327. [DOI] [PubMed] [Google Scholar]
- 19.Deshaies, R. J., and R. Schekman. 1987. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 105:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond, L. S., and C. G. Clark. 1993. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 21.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 22.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvorak, J. A., S. Kobayashi, T. Nozaki, T. Takeuchi, and C. Matsubara. 2003. Induction of permeability changes and death of vertebrate cells is modulated by the virulence of Entamoeba spp. isolates. Parasitol. Int. 52:169-173. [DOI] [PubMed] [Google Scholar]
- 24.Fournier, M., Z. Dermoun, M. C. Durand, and A. Dolla. 2004. A new function of the Desulfovibrio vulgaris Hildenborough [Fe] hydrogenase in the protection against oxidative stress. J. Biol. Chem. 279:1787-1793. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Zorn, B., G. Dominguez-Bernal, M. Suarez, M. T. Ripio, Y. Vega, S. Novella, A. Rodriguez, I. Chico, A. Tierrez, and J. A. Vazquez-Boland. 2000. SmcL, a novel membrane-damaging virulence factor in Listeria. Int. J. Med. Microbiol. 290:369-374. [DOI] [PubMed] [Google Scholar]
- 26.Gulbins, E., S. Dreschers, B. Wilker, and H. Grassme. 2004. Ceramide, membrane rafts and infections. J. Mol. Med. 82:357-363. [DOI] [PubMed] [Google Scholar]
- 27.Haque, R., P. Duggal, I. M. Ali, M. B. Hossain, D. Mondal, R. B. Sack, B. M. Farr, T. H. Beaty, and W. A. Petri, Jr. 2002. Innate and acquired resistance to amebiasis in Bangladeshi children. J. Infect. Dis. 186:547-552. [DOI] [PubMed] [Google Scholar]
- 28.Haque, R., C. D. Huston, M. Hughes, E. Houpt, and W. A. Petri, Jr. 2003. Amebiasis. N. Engl. J. Med. 348:1565-1573. [DOI] [PubMed] [Google Scholar]
- 29.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashkush, K., M. Feldman, and A. A. Levy. 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33:102-106. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. A., S. Park, K. Kim, S. G. Rhee, and S. W. Kang. 2005. Activity assay of mammalian 2-cys peroxiredoxins using yeast thioredoxin reductase system. Anal. Biochem. 338:216-223. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, R. H., P. V. Choudary, K. K. Stone, and C. W. Schmid. 2001. Stress induction of Bm1 RNA in silkworm larvae: SINEs, an unusual class of stress genes. Cell Stress Chaperones 6:263-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leippe, M., J. Andra, and H. J. Muller-Eberhard. 1994. Cytolytic and antibacterial activity of synthetic peptides derived from amoebapore, the pore-forming peptide of Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 91:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leippe, M., H. J. Sievertsen, E. Tannich, and R. D. Horstmann. 1995. Spontaneous release of cysteine proteinases but not of pore-forming peptides by viable Entamoeba histolytica. Parasitology 111:569-574. [DOI] [PubMed] [Google Scholar]
- 35.Li, T., J. Spearow, C. M. Rubin, and C. W. Schmid. 1999. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene 239:367-372. [DOI] [PubMed] [Google Scholar]
- 36.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane, R. C., P. H. Shah, and U. Singh. 2005. Transcriptional profiling of Entamoeba histolytica trophozoites. Int. J. Parasitol. 35:533-542. [DOI] [PubMed] [Google Scholar]
- 38.Mandal, P. K., A. Bagchi, A. Bhattacharya, and S. Bhattacharya. 2004. An Entamoeba histolytica LINE/SINE pair inserts at common target sites cleaved by the restriction enzyme-like LINE-encoded endonuclease. Eukaryot. Cell 3:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall, M. J., G. A. Bohach, and D. F. Boehm. 2000. Characterization of Staphylococcus aureus beta-toxin induced leukotoxicity. J. Nat. Toxins 9:125-138. [PubMed] [Google Scholar]
- 40.Moody, S., S. Becker, Y. Nuchamowitz, and D. Mirelman. 1997. Virulent and avirulent Entamoeba histolytica and E. dispar differ in their cell surface phosphorylated glycolipids. Parasitology 114:95-104. [DOI] [PubMed] [Google Scholar]
- 41.Nickel, R., T. Jacobs, and M. Leippe. 1998. Molecular characterization of an exceptionally acidic lysozyme-like protein from the protozoon Entamoeba histolytica. FEBS Lett. 437:153-157. [DOI] [PubMed] [Google Scholar]
- 42.Nickel, R., C. Ott, T. Dandekar, and M. Leippe. 1999. Pore-forming peptides of Entamoeba dispar. Similarity and divergence to amoebapores in structure, expression and activity. Eur. J. Biochem. 265:1002-1007. [DOI] [PubMed] [Google Scholar]
- 43.Ostertag, E. M., and H. H. Kazazian, Jr. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35:501-538. [DOI] [PubMed] [Google Scholar]
- 44.Pasquetto, V., D. A. Fidock, H. Gras, E. Badell, W. Eling, W. R. Ballou, J. Belghiti, A. Tartar, and P. Druilhe. 1997. Plasmodium falciparum sporozoite invasion is inhibited by naturally acquired or experimentally induced polyclonal antibodies to the STARP antigen. Eur. J. Immunol. 27:2502-2513. [DOI] [PubMed] [Google Scholar]
- 45.Perry, D. K. 2002. Serine palmitoyltransferase: role in apoptotic de novo ceramide synthesis and other stress responses. Biochim. Biophys. Acta 1585:146-152. [DOI] [PubMed] [Google Scholar]
- 46.Pritham, E. J., C. Feschotte, and S. R. Wessler. 2005. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol. Biol. Evol. 22:1751-1763. [DOI] [PubMed] [Google Scholar]
- 47.Que, X., and S. L. Reed. 2000. Cysteine proteinases and the pathogenesis of amebiasis. Clin. Microbiol. Rev. 13:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satish, S., A. A. Bakre, S. Bhattacharya, and A. Bhattacharya. 2003. Stress-dependent expression of a polymorphic, charged antigen in the protozoan parasite Entamoeba histolytica. Infect. Immun. 71:4472-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seigneur, M., J. Mounier, M. C. Prevost, and N. Guillen. 2005. A lysine- and glutamic acid-rich protein, KERP1, from Entamoeba histolytica binds to human enterocytes. Cell. Microbiol. 7:569-579. [DOI] [PubMed] [Google Scholar]
- 50.Shah, P. H., R. C. MacFarlane, D. Bhattacharya, J. C. Matese, J. Demeter, S. E. Stroup, and U. Singh. 2005. Comparative genomic hybridizations of Entamoeba strains reveal unique genetic fingerprints that correlate with virulence. Eukaryot. Cell 4:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma, R., A. Azam, S. Bhattacharya, and A. Bhattacharya. 1999. Identification of novel genes of non-pathogenic Entamoeba dispar by expressed sequence tag analysis. Mol. Biochem. Parasitol. 99:279-285. [DOI] [PubMed] [Google Scholar]
- 52.Tannich, E., and G. D. Burchard. 1991. Differentiation of pathogenic from nonpathogenic Entamoeba histolytica by restriction fragment analysis of a single gene amplified in vitro. J. Clin. Microbiol. 29:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Dellen, K., J. Field, Z. Wang, B. Loftus, and J. Samuelson. 2002. LINEs and SINE-like elements of the protist Entamoeba histolytica. Gene 297:229-239. [DOI] [PubMed] [Google Scholar]
- 55.WHO. 1997. Amoebiasis. Wkly. Epidemiol. Rec. 72:97-99. [PubMed] [Google Scholar]
- 56.Willhoeft, U., H. Buβ, and E. Tannich. 2002. The abundant polyadenylated transcript 2 DNA sequence of the pathogenic protozoan parasite Entamoeba histolytica represents a nonautonomous non-long-terminal-repeat retrotransposon-like element which is absent in the closely related nonpathogenic species Entamoeba dispar. Infect. Immun. 70:6798-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willhoeft, U., H. Buss, and E. Tannich. 1999. DNA sequences corresponding to the ariel gene family of Entamoeba histolytica are not present in E. dispar. Parasitol. Res. 85:787-789. [DOI] [PubMed] [Google Scholar]
- 58.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao, Y., D. E. Sturdevant, A. Villaruz, L. Xu, Q. Gao, and M. Otto. 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 73:1856-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.