Abstract
There are two alleles, m1 and m2, of the midregion of the vacuolating cytotoxin gene (vacA) of Helicobacter pylori which code for toxins with different cell specificities. Here we describe the construction of five chimeric strains in which regions of vacA were exchanged between the two genotypes. By analyzing the toxicity of these strains for HeLa and RK13 cells we have confirmed that a 148-amino-acid region determines the phenotypic differences between the two forms of the protein and that this entire region is important for cytotoxicity. Furthermore, we have used our chimeric strains to investigate whether variations in the midregion of VacA have an effect on phorbol 12-myristate 13-acetate (PMA)-induced VacA sensitivity in HL-60 cells. The PMA-induced VacA sensitivity of HL-60 cells has been previously associated with the appearance of the cell surface receptor protein tyrosine phosphatase beta (RPTPβ). Our data indicate that both the m1 and m2 forms of VacA are able to utilize RPTPβ, and the cell-specific phenotype of the midregion is independent of the presence of RPTPβ. It appears that another as-yet-unidentified receptor exists in HL-60 cells that accounts for the m2 phenotype in this cell line. Also, by studying the effect of PMA on levels of RPTPβ in other cell lines and toxicity of VacA in these cell lines we have shown that RPTPβ does not play a major role in the vacuolation of HeLa cells.
The gram-negative bacterium Helicobacter pylori colonizes the gastric mucosa of approximately 50% of the world's population and is the major cause of gastritis, peptic ulcer, and gastric cancer (3, 15). A major virulence factor secreted by the bacteria is the cytotoxin VacA, which induces intracellular vacuoles in vitro and gastric epithelial erosion in vivo and is associated with gastric ulcer risk in vivo (12, 14, 17). The secreted toxin is an 87-kDa polypeptide that can undergo proteolytic cleavage to yield two subunits of approximately 37 kDa and 58 kDa (p37 and p58, respectively) (13). The p37 subunit contains the functional vacuolating activity of the toxin, and the p58 subunit is responsible for binding to cell surface receptors (6, 21).
Two different VacA genotypes, m1 and m2, have been described which differ in a 300-amino-acid region, termed the midregion, of the p58 subunit of VacA (1). These two genotypes differ by about 50% in amino acid sequence in this region but share greater than 90% sequence identity in the rest of the protein (1). The m1 form of VacA is toxic to HeLa cells, but the m2 form is essentially nontoxic to these cells (1, 19). However, both forms are able intoxicate the rabbit kidney cell line RK13 and primary cultured human gastric cells (19). Also, the m2 phenotype is associated with duodenal ulcer and is prevalent in the Chinese population, in which there is high incidence of peptic ulcer and gastric cancer (10, 20). The lack of toxicity of the m2 form of VacA in HeLa cells is associated with the lack of cell surface binding, indicating differences in cell surface receptors for the two forms (19). Furthermore, analysis of the phenotypes of natural and artificial chimeras in this midregion have shown that the first N-terminal 35 amino acids of the midregion of the m1 form are essential for HeLa cell cytotoxicity and that the next 113 amino acids are required for full toxicity on HeLa cells (11).
The binding of VacA to specific high-affinity cell surface receptors has been shown by indirect immunofluorescence and flow cytometry and has been shown to be necessary for cell intoxication (9, 16). VacA can interact with target cells by binding to the 250-kDa receptor protein tyrosine phosphatase beta (RPTPβ) (18, 27). Also, the appearance of RPTPβ on HL-60 cells after treatment with phorbol 12-myristate 13-acetate (PMA) has been associated with the degree of vacuolation induced by VacA (17).
Here we describe the construction of five strains chimeric in the variable midregion of VacA and further define the amino acid sequences required for full toxicity in HeLa cells. These strains were used to investigate whether differences in toxicity defined by the midregion are associated with RPTPβ. We show that although both the m1 and m2 forms of VacA are able to utilize RPTPβ, the cell-specific cytotoxic phenotype of the midregion is independent of the appearance of RPTPβ and RPTPβ does not play a major role in the vacuolation of HeLa cells.
METHODS AND MATERIALS
Bacterial strains and growth conditions.
H. pylori m1 strain SPM326 (14) was used as the parent strain for the construction of all mutants in this study (Table 1). DNA sequences representative of the m2 allele were amplified from H. pylori 95-54 (19). H. pylori strains were grown on blood agar plates (Colombia agar with 5% horse blood) microaerobically at 37°C. Liquid cultures were grown in brain heart infusion broth containing 5% fetal calf serum and were cultured for 24 h microaerobically at 37°C. H. pylori culture supernatants were concentrated 10-fold with 50% ammonium sulfate, and the precipitated proteins were resuspended in phosphate-buffered saline (PBS) and dialyzed against the same buffer.
TABLE 1.
Primers used for the generation of chimeric PCR products
| Primer | Chimeric region(s) | Nucleotide sequence | Corresponding DNA sequence |
|---|---|---|---|
| DENIDAGS | All | 5′-ATC GCT CCT CCA GAA GGT GG-3′ | 970-988a |
| RAFL | All | 5′-CCTACGCTTAAGGTCGCTACACG-3′ | 2131-2154b |
| DSM2F | DSM233 | 5′-CGATATTAGTCTGGGAAAAGCGGTGAATTTAAGAGTGG-3′ | 1485-1522a |
| DSM2R | DSM233 | 5′-CCACTCTTAAATTCACCGCTTTTCCCAGACTAATATCG-3′ | 1470-1508b |
| DSGV | AB25 | 5′-AGTGGCGTTACAGACAAAGTC-3′ | 1588-1607a |
| RSGV | AB25 | 5′-GACTTTGTCTGTAACGCCACT-3′ | 1648-1669b |
| DAGSF1 | DAGS1 | 5′-GAATTGATTGTTACAACCCGTGTTCAGAG-3′ | 1669-1697a |
| DAGSR1 | DAGS1 | 5′-CTCTGAACACGGGTTGTAACAATCAATTCAATAATGTTG-3′ | 1719-1758b |
| DAGSF2 | DAGS2 | 5′-CAATCGCGCATCAATACCGTTAGTTTGCAAGCGGG-3′ | 1738-1773a |
| DAGSR2 | DAGS2 | 5′-CCCGCTTGCAAACTAACGGTATTGATGCGCGATTG-3′ | 1798-1834b |
| DAGSF3 | DAGS3 | 5′-GCGAAAAATTGGTTATAGATGAGATTTACCATG-3′ | 1814-1847a |
| DAGSR3 | DAGS3 | 5′-CATGGTAAATCTCATCTATAACCAATTTTTCGC-3′ | 1874-1907b |
Immunoblot analysis.
The presence of VacA in concentrated supernatants from H. pylori cultures was determined by immunoblot analysis using rabbit sera raised against a recombinant m1 form of the protein (24).
Construction of H. pylori chimeric strains.
To generate H. pylori chimeric strain SPM326/DSM233, a previously described counterselection approach was used (2). First, a recipient strain was created by the introduction of the sacB/kan cassette into the VacA gene of H. pylori. The sacB/kan cassette from pKSF was inserted into the EcoNI/XbaI sites of pBlueScriptKSp58, a plasmid containing a substantial portion of the vacA gene from the m1 strain SPM326. A kanamycin-resistant transformant (SPM326/KO), in which the sacB/kan cassette had integrated into the vacA chromosomal locus via allelic exchange, was selected.
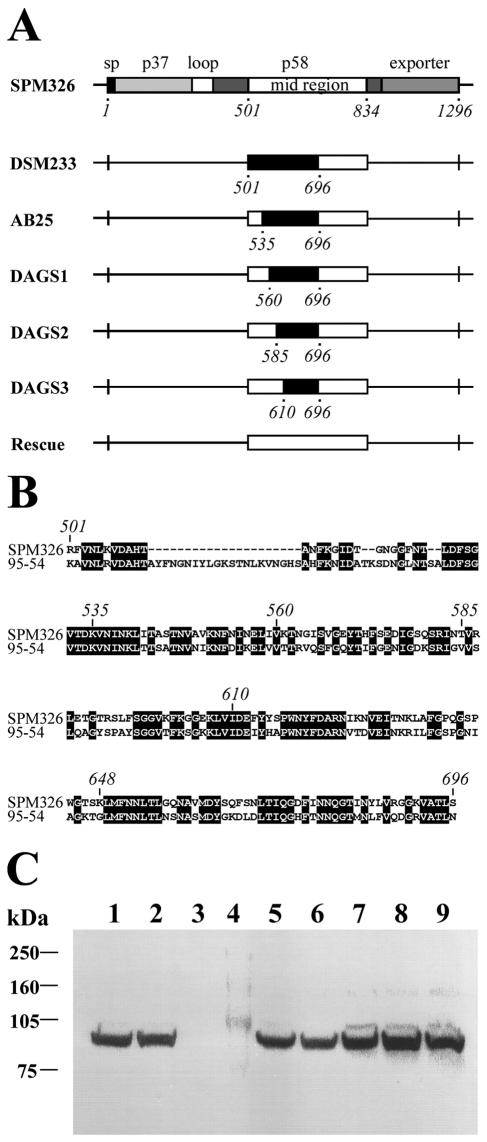
PCR products chimeric for the vacA midregion were created using the strategy described by Ji et al. (11). Briefly, the left and right parts of the m1 and m2 genes were amplified using primers based on their respective sequences but with the 3′ primer of the left part complementary to the 5′ primer of the right part. The resulting PCR products were purified and mixed in a second PCR containing only the 5′ primer of the left part and the 3′ primer of the right part such that the desired PCR product would be obtained through annealing of the complementary regions of each PCR. Primers used for the generation of chimeric PCR products are shown in Table 2. The chimeric PCR products were inserted into the unique EcoNI/AflII sites of pBlueScriptKSp58 such that they replaced the m1-type gene sequences in this plasmid. The resulting chimeric delivery constructs were used to transform the recipient strain SPM326/KO, and transformants were selected according to their sensitivity to sucrose. The presence of the desired chimeric sequences within the vacA midregion was confirmed by PCR. A total of five different H. pylori strains, each chimeric for the midregion of VacA, were isolated (Fig. 1A and B).
TABLE 2.
H. pylori strains and plasmids
| H. pylori strain | Relevant characteristicsa | Source or reference |
|---|---|---|
| 95-54 | Wild type; vacA s1/m2 | 19 |
| SPM326 | Wild type; vacA s1/m1 | 14 |
| SPM326/KO | As SPM326 except sacB/kan inserted in vacA between restriction sites | This study |
| EcoNI/Xbal; ntb 980-2273 | ||
| SPM326/DSM233 | As SPM326 except vacA nt 1501-2085 replaced with corresponding m2 type (nt 1487-2144) | This study |
| SPM326/AB25 | As SPM326 except vacA nt 1605-2085 replaced with corresponding m2 type (nt 1664-2144) | This study |
| SPM326/DAGS1 | As SPM326 except vacA nt 1680-2085 replaced with corresponding m2 type (nt 1730-2144) | This study |
| SPM326/DAGS2 | As SPM326 except vacA nt 1755-2085 replaced with corresponding m2 type (nt 1814-2144) | This study |
| SPM326/DAGS3 | As SPM326 except vacA nt 1830-2085 replaced with corresponding m2 type (nt 1889-2144) | This study |
| SPM326/Rescue | As SPM326 | This study |
Numbers designate nucleotides within the vacA coding region.
nt, nucleotide.
FIG. 1.
A, Schematic representation of the constructed chimeras. The dark boxes represent the regions of m1 vacA sequence replaced by m2 sequences. The VacA amino acid numbering system is based on the initiation codon of the vacA gene from strain SPM326. B, box shade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html) of the predicted amino acid sequences of the N-terminal 196 amino acids of the midregion. Identical amino acids are shaded black. C, immunoblot of concentrated supernatants from H. pylori control and chimeric VacA-expressing strains. Lane 1, wild-type SPM326; lane 2, 95-54; lane 3, SPM326/KO; lane 4, molecular mass marker; lane 5, SPM326/AB25; lane 6, SPM326/DAGS1; lane 7, SPM326/DAGS2; lane 8, SPM326/DAGS3; lane 9, SPM326/Rescue. The weights of the molecular mass standards are indicated on the left.
Cell vacuolation assay.
Concentrated supernatants from H. pylori cultures were low-pH activated, and their cytotoxic activity was tested on HeLa, RK13, and HL-60 cells as previously described (5, 7, 19, 11, 18). Briefly, the supernatants were incubated with HeLa, RK13, or HL-60 cells for 6 h. Where indicated in the text, the cells were previously incubated with 20 mM PMA for 24 h. Because PMA was dissolved in 0.1% (vol/vol) Me2SO, this solution served as a control for PMA. The extent of vacuolation was determined quantitatively by measuring the uptake of neutral red (5).
Cell sorter analysis.
HL-60 cells were incubated without or with 20 nM PMA for 24 h, washed with PBS, and fixed with 2% paraformaldehyde (15 min at room temperature). The cells were then permeabilized (20 min at room temperature) with permeabilizing solution (PS; PBS containing 1% bovine serum albumin and 0.5% saponin). After permeabilization the cells were washed with PS and incubated for 30 min at room temperature with anti-human RPTPβ mouse monoclonal antibody (1:10) (BD Transduction Laboratories, San Diego, CA) or irrelevant mouse immunoglobulin G (IgG) antibodies (control) in PS. The cells were then washed in PS, followed by incubation with second antibody (1:50) (R-phycoerythrin-conjugated goat anti-mouse IgG; Johnson ImmunoResearch Laboratories) for 30 min at room temperature. After a washing with PBS, the samples were analyzed using a FACSCalibur (Becton Dickinson, Mountain View, CA) machine equipped with an argon laser emitting at 488 nm.
Statistical analysis.
The paired t test was used to compare the differences in neutral red uptake between untreated and PMA-treated cells and between the different forms of VacA. P values are indicated. A P value of <0.05 was considered significant, and the Bonferroni adjustment was considered where appropriate.
RESULTS
The entire N-terminal 148 amino acids of the midregion is required for full toxicity in HeLa cells.
Ji et al. (11) established that the differences in target cell specificity between the m1 and m2 forms of the VacA protein were in fact determined by the first 148 amino acids of the midregion. Furthermore, within this region, the first 35 amino acids were found to be essential for toxicity of m1 VacA on HeLa cells (11). However, the subsequent 36-148 region remained undefined and may not be required in its entirety for full HeLa cell cytotoxicity. To further define this region, sequential 25-amino-acid sections at the N terminus of this 36-148 region were studied for their role in target cell specificity. Four chimeric strains have been created (as described in Materials and Methods) in which sequences coding for amino acids 535 to 695, 560 to 695, 585 to 695, and 610 to 695 of VacA from m1 strain SPM326 have been replaced with corresponding m2 sequences (Fig. 1A and B). In addition, the control strain SPM326/DSM233 was created to ensure that differences in cell specificity between these engineered chimeric strains and the m2 strain 95-54 are attributable solely to the midregion. In this control strain the first 195 amino acids (amino acids 501 to 695) from the midregion of m1 strain SPM326 have been replaced with the corresponding region from m2 strain 95-54. Another control strain, SPM326/Rescue, in which the vacA deletion strain used as host for the chimeric genes was rescued with a wild-type m1 gene, was created to ensure that unwanted changes in chromosomal background, which could affect cell specificity and vacuolating activity, were not created during chimeric strain construction.
Growth culture supernatants from these strains were prepared and concentrated 10-fold using 50% saturated ammonium sulfate. The presence of VacA in the concentrated supernatants was determined by immunoblot analysis using anti-rabbit sera raised against a recombinant m1 form of the protein. The results demonstrate that the chimeric strains expressed mature 87-kDa VacA protein (Fig. 1C). However, accurate quantification of the comparative levels of VacA is difficult, as antibodies to the m1 form of the protein recognize the m2 form less well (19). As expected, VacA was not expressed by strain SPM326/KO, which has the sacB/kan cassette integrated into the vacA chromosomal locus.
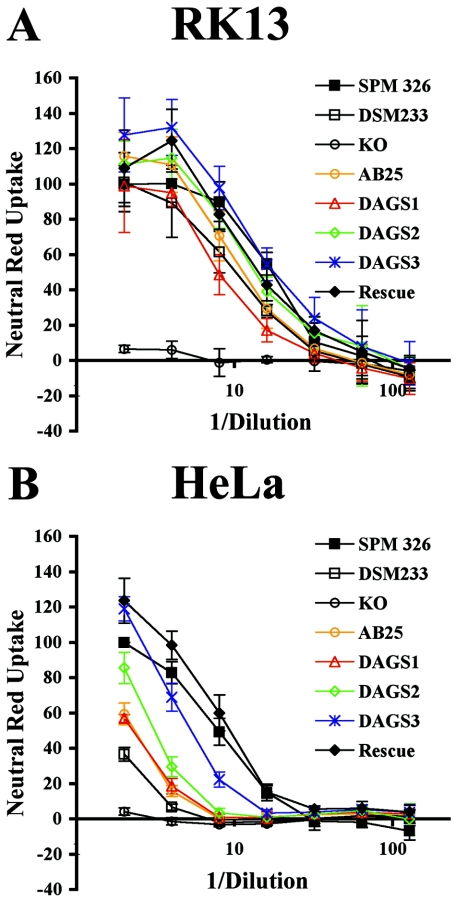
Concentrated growth culture supernatants from the chimeric and control strains were tested for vacuolating activity in an RK13 and HeLa cell assay. The titer of each supernatant was determined as the reciprocal of the dilution of each supernatant required to give 50% of the maximal neutral red activity of a twofold dilution of supernatant from wild-type SPM326. All strains displayed similar levels of cell vacuolation on RK13 cells, with titers from all strains falling within the range 0.125 to 0.056 (Fig. 2A). However, greater differences in vacuolating activity were observed on HeLa cells, with a range of titers from 0.667 to 0.105 (Fig. 2B). Statistical analysis of vacuolation by the least-diluted supernatants on RK13 cells indicated that only strains DAGS3 (P = 0.04) and KO (P = <0.01) were significantly different from SPM326 and strains AB25 (P = 0.02) and KO (P = <0.01) were significantly different from DSM233. This finding should be contrasted with HeLa cell results, where all strains of VacA were significantly different from SPM326 (Rescue P = 0.03) and DSM233 (data not shown). This reflects the target cell specificity of the m2 genotype. As expected, the highest titers were found for m1 strain SPM326 and strain SPM326/Rescue (0.125 and 0.105, respectively), which were both able to fully vacuolate HeLa cells. The lowest titer (0.667) was for m2 control strain SPM326/DSM233, with vacuolation only detected in the least-diluted preparations. Chimeric strain SPM326/DAGS3 with the smallest m2 replacement displayed a lower titer (0.196) and reduced vacuolation in HeLa cells compared to the parental SPM326 strain but only in concentrated supernatants diluted eightfold or more. Lesser toxicity again was observed for chimeric strain SPM326/DAGS2 (titer, 0.323), and toxicity was further reduced in the strains SPM326/DAGS1 (0.476) and SPM326/AB25 (0.476), which possess the largest m2 replacements of the chimeric strains. An inverse correlation appears to exist between the toxicity of VacA to HeLa cells and the size of the m2 coding sequence.
FIG. 2.
Neutral red uptake assay of RK13 (A) and HeLa (B) cells treated with growth culture supernatants from the VacA chimeric strains. Prior to dilution for the assay, growth supernatants were concentrated 10-fold using 50% saturated ammonium sulfate. Data are the averages of four individual assays and are normalized to the value of the lowest dilution of SPM326 in each experiment, which is taken as 100%. Error bars show standard deviation between assays.
The cell-specific phenotype of VacA is not associated with the appearance of RPTPβ.
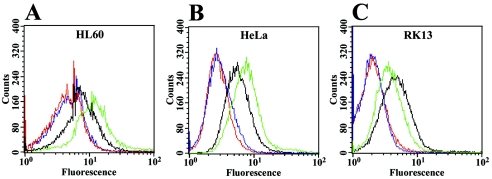
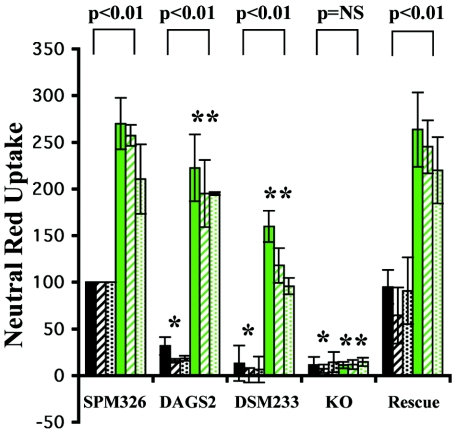
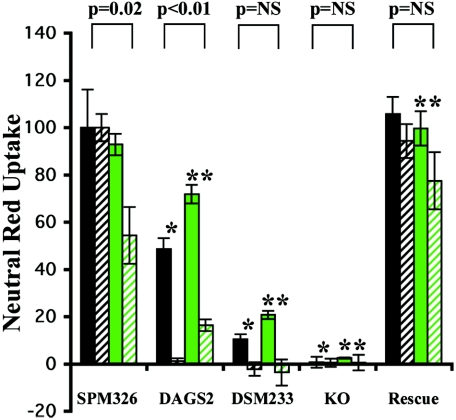
The differentiating agent, PMA, enhances the sensitivity of HL-60 cells to VacA (6). Padilla et al. (18) reported that this increase was associated with an increase in the expression of RPTPβ, a proposed receptor for the VacA cytotoxin. Our m1 and m2 control strains as well as chimeric strain SPM326/DAGS2 were tested for vacuolation on HL-60 cells both with and without treatment with PMA. To confirm the presence and increase of RPTPβ, fluorescence-activated cell sorter (FACS) analysis was performed using HL-60 cells and a monoclonal anti-RPTPβ antibody (Fig. 3A). HL-60 cells treated with PMA had greater fluorescence than untreated cells (Fig. 3A), confirming that the amount of RPTPβ was increased in PMA-stimulated cells. No difference in fluorescence was observed between PMA-treated and untreated cells when an irrelevant mouse IgG control antibody was used (Fig. 3A). As previously reported (6, 18) a significant increase in vacuolation of HL-60 cells after treatment with PMA was observed for m1 strain SPM326 (Fig. 4). This increase was also observed for the m2 control strain SPM326/DSM233 and chimeric strain SPM326/DAGS2, indicating that the increased sensitivity of PMA-treated HL-60 cells is not limited to the m1 form of VacA. However, levels of vacuolation detected on HL-60 cells treated with PMA were less for the chimeric strain SPM326/DAGS2 than for the parental SPM326 strain and lesser again for m2 control strain SPM326/DSM233. This decrease was mirrored in HL-60 cells not treated with PMA, indicating that it is independent of the PMA treatment and therefore independent of the increased expression of RPTPβ.
FIG. 3.
Effect of PMA on RPTPβ protein in HL-60 (A), HeLa (B), or RK13 (C) cells. Cells were incubated for 24 h with (green lines) or without (black lines) 20 nM PMA, and RPTPβ expression was quantified by indirect immunofluorescence and flow cytometry using anti-RPTPβ monoclonal antibodies. An irrelevant mouse IgG was used as a control on cells with (blue lines) or without (red lines) 20 nM PMA. Data are representative of three separate experiments.
FIG. 4.
Neutral red uptake assay of PMA-treated HL-60 cells for growth supernatants from selected VacA chimeric strains. Cells were incubated in the presence or absence of 20 nM PMA for 24 h. Prior to dilution for the assay, growth supernatants were concentrated 10-fold using 50% saturated ammonium sulfate. Data are the averages of at least three individual assays and are normalized in each experiment to the value of SPM326 on HL-60 cells not treated with PMA (taken as 100%). Error bars show standard deviation between assays. P values indicate the degree of significance between untreated and PMA-treated cell results. *, P <0.01 compared to SPM326 not treated with PMA. **, P <0.01 compared to SPM326 treated with 20 nM PMA. NS, not significant. Black bars, no PMA. Green bars, 20 nM PMA. Solid bars, supernatant diluted 1:4. Hatched bars, supernatant diluted 1:8. Dotted bars, supernatant diluted 1:16.
RPTPβ does not play a major role in the VacA-induced vacuolation of HeLa cells.
To investigate the role RPTPβ plays in the VacA-induced vacuolation of other cell lines, HeLa and RK13 cells were treated with PMA and their sensitivity to different forms of the toxin was evaluated. FACS analysis performed using HeLa cells and the monoclonal anti-RPTPβ antibody revealed an approximately 1.5-fold increase in RPTPβ fluorescence when cells were stimulated with PMA (Fig. 3B). PMA treatment of HeLa resulted in a small decrease in neutral red uptake in response to m1 VacA, but this was not reproduced in VacA from the rescue strain or the m2 or chimeric strains (Fig. 5). We conclude that PMA-induced RPTPβ does not play a major role in vacuolation of HeLa cells. FACS analysis performed on RK13 cells revealed no increase in the amount of RPTPβ when the cells were stimulated by PMA (Fig. 3C), and stimulation of these cells with PMA had no significant effect on vacuolation by the m1 and m2 control strains or by the chimeric strain SPM326/DAGS2 (not shown). However, from these data we cannot conclude that the RPTPβ receptor is not involved in vacuolation of RK13 cells.
FIG. 5.
Neutral red uptake assay of PMA-treated HeLa cells for growth supernatants from selected VacA chimeric strains. Cells were incubated in the presence or absence of 20 nM PMA for 24 h. Prior to dilution for the assay, growth supernatants were concentrated 10-fold using 50% saturated ammonium sulfate. Data are the averages of at least three individual assays and are normalized in each experiment to the value of SPM326 on HeLa cells untreated with PMA (taken as 100%). Error bars show standard deviation between assays. P values indicate the degree of significance between untreated and PMA-treated cells. NS, not significant. *, P <0.01 compared to SPM326 not treated with PMA. **, P <0.01 compared to SPM326 treated with 20 nM PMA. Black bars, no PMA. Green bars, 20 nM PMA. Solid bars, supernatant diluted 1:4. Hatched bars, supernatant diluted 1:8.
DISCUSSION
By analyzing the vacuolating activity of five strains chimeric in the N-terminal 195 amino acids of the midregion of VacA, we confirmed that the entire 148-amino-acid region defined by Ji et al. (11) is required for full toxicity. Within the 195-amino-acid region, the sizes of the m2 coding sequences in strains DSM233, AB25, DAGS1, DAGS2, and DAGS3 are 195, 161, 136, 111, and 86 amino acids, respectively. Furthermore, the amino acid differences between the m1 and m2 forms of VacA within these sequences are 86, 51, 43, 30, and 19 amino acids, respectively (Fig. 1B). These differences account for the observed differences in potency with respect to HeLa cells but do not affect the stability or functionality of the toxin, since all forms were fully toxic to RK13 cells.
RPTPβ has been identified as a cell surface receptor for VacA, and PMA-induced VacA sensitivity of HL-60 cells has been associated with the appearance of RPTPβ in these cells (18, 27). By investigating whether variations in the midregion have an effect on the PMA-induced sensitivity of HL-60 cells, we have shown that the appearance of RPTPβ increases the toxicity of both the m1 and m2 forms of VacA for this cell line. Furthermore, our data show that the cell-specific cytotoxic phenotype of the midregion seen with HeLa cells is also apparent for HL-60 cells but that it is independent of the appearance of RPTPβ. It is currently understood that the mechanism by which VacA induces cellular vacuolation involves the binding of VacA to the plasma membrane, internalization by the cell, oligomerization to form anion-selective channels in endosomal membranes, and the formation of vacuoles due to the swelling of endosomal compartments (4). While there is no evidence to suggest that m1 and m2 forms of the protein differ in any activity other than binding, we cannot exclude the possibility that minor differences in the cellular responses are due in part to functional differences between the toxins. However, it has been demonstrated that lack of toxicity of the m2 form of VacA in HeLa cells correlates with lack of cell surface binding and, furthermore, that the cell-specific phenotype is due to the different binding specificities of the m1 and m2 forms of the protein (19, 25). It would appear that another receptor (other than RPTPβ) exists in HL-60 cells which accounts for the greater sensitivity to the m1 form of the protein. A recent study found that VacA caused cellular vacuolation of primary gastric epithelial cells from RPTPβ−/− mice and suggested the presence of other receptors for VacA (8).
To elucidate the role RPTPβ plays in VacA cytotoxicity in HeLa and RK13 cells, the effect of PMA on RPTPβ levels in these cell lines was monitored and the subsequent toxicity of our chimeric strains measured. As HeLa cells are sensitive to m1 VacA but less sensitive to the m2 toxin and considering our findings that the differences in toxicity between the m1 and m2 forms are independent of RPTPβ, it would be unexpected if this protein were a principle VacA receptor on HeLa cells. Indeed, our results demonstrate that the appearance of RPTPβ on HeLa cells had little effect on vacuolation by m1, m2, and chimeric forms of VacA. RK13 cells are sensitive to both the m1 and m2 forms of the protein, and it is therefore possible that RPTPβ is the common receptor on these cells. This requires further investigation, as stimulation of RK13 cells with PMA had little effect on both RPTPβ levels detected by immunofluorescence and vacuolation by m1, m2, or chimeric strains. Although the interaction of VacA with RPTPβ has been characterized in the greatest detail, a number of other cell surface proteins have been proposed as receptors for VacA. Seto et al. (23) have proposed that the EGF receptor is the VacA receptor in HeLa cells. A more recent study has identified receptor protein tyrosine phosphatase alpha (RPTPα) as the VacA receptor in G401 cells, a cell line that does not express RPTPβ (26). As well as interaction of VacA with putative receptors, high-affinity interactions between VacA and glycosylphosphatidylinositol-anchored proteins within cholesterol-glycosphingolipid-enriched domains (“rafts”) have been identified (22). It appears that VacA has a number of cell surface receptors and may bind to multiple sites on the cell surface. Finding an association between these and the m1/m2 phenotype of VacA may provide useful insight into its mechanism of action and reasons why this polymorphism may have evolved.
Acknowledgments
This work was supported by Marie Curie Industry Host Fellowship QLK4 1999 50407 from the European commission.
We are grateful to S. Pasquini, L. Fini, and S. Magi for medium preparation. We thank A. Muzzi for synthesis of the oligonucleotides and S. Guidotti for automated sequencing. We also thank S. Nuti and S. Tavarini for their assistance with the cell sorter analysis.
Editor: J. D. Clements
REFERENCES
- 1.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 2.Copass, M., G. Grandi, and R. Rappuoli. 1997. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect. Immun. 65:1949-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 4.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 5.Cover, T. L., W. Puryear, G. I. Perez-Perez, and M. J. Blaser. 1991. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect. Immun. 59:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bernard, M., D. Burroni, E. Papini, R. Rappuoli, J. Telford, and C. Montecucco. 1998. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect. Immun. 66:6014-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bernard, M., E. Papini, V. de Filippis, E. Gottardi, J. Telford, R. Manetti, A. Fontana, R. Rappuoli, and C. Montecucco. 1995. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J. Biol. Chem. 270:23937-23940. [DOI] [PubMed] [Google Scholar]
- 8.Fujikawa, A., D. Shirasaka, S. Yamamoto, H. Ota, K. Yahiro, M. Fukada, T. Shintani, A. Wada, N. Aoyama, T. Hirayama, H. Fukamachi, and M. Noda. 2003. Mice deficient in protein tyrosine phosphatase receptor type Z are resistant to gastric ulcer induction by VacA of Helicobacter pylori. Nat. Genet. 33:375-381. [DOI] [PubMed] [Google Scholar]
- 9.Garner, J. A., and T. L. Cover. 1996. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect. Immun. 64:4197-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Go, M. F., L. Cissell, and D. Y. Graham. 1998. Failure to confirm association of vacA gene mosaicism with duodenal ulcer disease. Scand. J. Gastroenterol. 33:132-136. [DOI] [PubMed] [Google Scholar]
- 11.Ji, X., T. Fernandez, D. Burroni, C. Pagliaccia, J. C. Atherton, J. M. Reyrat, R. Rappuoli, and J. L. Telford. 2000. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect. Immun. 68:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 13.Lupetti, P., J. E. Heuser, R. Manetti, P. Massari, S. Lanzavecchia, P. L. Bellon, R. Dallai, R. Rappuoli, and J. L. Telford. 1996. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J. Cell Biol. 133:801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchetti, M., B. Arico, D. Burroni, N. Figura, R. Rappuoli, and P. Ghiara. 1995. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science 267:1655-1658. [DOI] [PubMed] [Google Scholar]
- 15.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 16.Massari, P., R. Manetti, D. Burroni, S. Nuti, N. Norais, R. Rappuoli, and J. L. Telford. 1998. Binding of the Helicobacter pylori vacuolating cytotoxin to target cells. Infect. Immun. 66:3981-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura, K., S. Maeda, M. Nakao, T. Watanabe, M. Tada, T. Kyutoku, H. Yoshida, Y. Shiratori, and M. Omata. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J. Exp. Med. 192:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padilla, P. I., A. Wada, K. Yahiro, M. Kimura, T. Niidome, H. Aoyagi, A. Kumatori, M. Anami, T. Hayashi, J. Fujisawa, H. Saito, J. Moss, and T. Hirayama. 2000. Morphologic differentiation of HL-60 cells is associated with appearance of RPTPβ and induction of Helicobacter pylori VacA sensitivity. J. Biol. Chem. 275:15200-15206. [DOI] [PubMed] [Google Scholar]
- 19.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. USA 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 21.Reyrat, J. M., S. Lanzavecchia, P. Lupetti, M. de Bernard, C. Pagliaccia, V. Pelicic, M. Charrel, C. Ulivieri, N. Norais, X. Ji, V. Cabiaux, E. Papini, R. Rappuoli, and J. L. Telford. 1999. 3D imaging of the 58 kDa cell binding subunit of the Helicobacter pylori cytotoxin. J. Mol. Biol. 290:459-470. [DOI] [PubMed] [Google Scholar]
- 22.Ricci, V., A. Galmiche, A. Doye, V. Necchi, E. Solcia, and P. Boquet. 2000. High cell sensitivity to Helicobacter pylori VacA toxin depends on a GPI-anchored protein and is not blocked by inhibition of the clathrin-mediated pathway of endocytosis. Mol. Biol. Cell 11:3897-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seto, K., Y. Hayashi-Kuwabara, T. Yoneta, H. Suda, H. Tamaki. 1998. Vacuolation induced by cytotoxin from Helicobacter pylori is mediated by the EGF receptor in HeLa cells. FEBS Lett. 431:347-350. [DOI] [PubMed] [Google Scholar]
- 24.Telford, J. L., P. Ghiara, M. Dell'Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, Z., E. Papini, C. Montecucco, L. Parente, and R. Rappuoli. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J. Exp. Med. 179:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, W. C., H. J. Wang, and C. H. Kuo. 2001. Two distinctive cell binding patterns by vacuolating toxin fused with glutathione S-transferase: one high-affinity m1-specific binding and the other lower-affinity binding for variant m forms. Biochemistry 40:11887-11896. [DOI] [PubMed] [Google Scholar]
- 26.Yahiro, K., A. Wada, M. Nakayama, T. Kimura, K. Ogushi, T. Niidome, H. Aoyagi, K. Yoshino, K. Yonezawa, J. Moss, and T. Hirayama. 2003. Protein-tyrosine phosphatase alpha, RPTPα, is a Helicobacter pylori VacA receptor. J. Biol. Chem. 278:19183-19189. [DOI] [PubMed] [Google Scholar]
- 27.Yahiro, K., T. Niidome, M. Kimura, T. Hatakeyama, H. Aoyagi, H. Kurazono, K. Imagawa, A. Wada, J. Moss, and T. Hirayama. 1999. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J. Biol. Chem. 274:36693-36699. [DOI] [PubMed] [Google Scholar]