Abstract
Murine CDP/Cux, a homologue of the Drosophila Cut homeoprotein, modulates the promoter activity of cell cycle-related and cell-type-specific genes. CDP/Cux interacts with histone gene promoters as the DNA binding subunit of a large nuclear complex (HiNF-D). CDP/Cux is a ubiquitous protein containing four conserved DNA binding domains: three Cut repeats and a homeodomain. In this study, we analyzed genetically targeted mice (Cutl1tm2Ejn, referred to as ΔC) that express a mutant CDP/Cux protein with a deletion of the C terminus, including the homeodomain. In comparison to the wild-type protein, indirect immunofluorescence showed that the mutant protein exhibited significantly reduced nuclear localization. Consistent with these data, DNA binding activity of HiNF-D was lost in nuclear extracts derived from mouse embryonic fibroblasts (MEFs) or adult tissues of homozygous mutant (ΔC−/−) mice, indicating the functional loss of CDP/Cux protein in the nucleus. No significant difference in growth characteristics or total histone H4 mRNA levels was observed between wild-type and ΔC−/− MEFs in culture. However, specific histone genes (H4.1 and H1) containing CDP/Cux binding sites have reduced expression levels in homozygous mutant MEFs. Stringent control of growth and differentiation appears to be compromised in vivo. Homozygous mutant mice have stunted growth (20 to 50% weight reduction), a high postnatal death rate of 60 to 70%, sparse abnormal coat hair, and severely reduced fertility. The deregulated hair cycle and severely diminished fertility in Cutl1tm2Ejn/tm2Ejn mice suggest that CDP/Cux is required for the developmental control of dermal and reproductive functions.
CDP/Cut (CCAAT displacement protein) is a transcription factor involved in the regulation of cell growth and differentiation-related genes (25). The role of CDP/Cut in mammalian cell growth control is reflected by its functional interaction with the promoters of the five major classes of histone genes (H1, H2A, H2B, H3, and H4), as well as genes encoding modulators of proliferation including c-myc and p21 (7, 11, 42, 43). CDP/Cut DNA binding activity is highest during S phase (14, 20, 40, 46), when p21 expression is downregulated (46) and histone H4 expression is maximal (7, 43). The interaction of CDP/Cut with the promoters of all major histone classes during Xenopus development implicates CDP/Cut in the coordinate control of histone gene expression during the cell cycle and early development (13). Moreover, eliminating CDP/Cut binding to the human histone H4 promoter alters the timing of maximal transcription during S phase (2). Although CDP/Cut may have a bifunctional role, it is generally considered a repressor, possibly exerting its regulatory activity in conjunction with other nuclear proteins (7, 21, 23, 39). For example, transcriptional regulation of human histone H4 is mediated in part by the promoter complex HiNF-D, which is composed of CDP/Cut, the retinoblastoma protein family (pRb), cyclin A, and CDC2 (38).
Human CDP/Cut, its canine (Clox), murine (Cux-1), and rat (CDP2) homologues, and the Drosophila Cut protein have four DNA binding domains in common (1, 25, 26, 36, 50): a unique homeodomain and three Cut repeats, which are similar regions of 70 amino acids. CDP/Cut is expressed in most tissues (37), and its DNA binding activity is ubiquitous among mammalian cell lines (22). Phosphorylation by protein kinase C and casein kinase II or PCAF-mediated acetylation of the CDP/Cux homeodomain inhibits DNA binding activity (6, 8, 21). Stable CDP/Cux-DNA complexes are detected in proliferating cells but disappear upon cellular differentiation of HL60 promyelocytic leukemia cells and fetal rat calvarial cells (22, 27, 28). CDP/Cut has been shown to bind to a variety of promoters or enhancer sequences of genes involved in differentiation, including myeloid cytochrome gp91-phox (28), dog heart myosin heavy chain (1), rat tyrosine hydroxylase (50), human γ-globin (33), Xenopus β-globin, and mouse N-CAM (36). Transfection experiments suggest that CDP/Cut functions as a repressor of these target genes in proliferating precursor cells. Upon terminal differentiation, these target genes are induced when CDP/Cut DNA binding activity is downregulated. Two mechanisms of repression by CDP/Cut have been proposed: passive repression by competition with activators for occupancy of binding sites (4, 26, 28) and active repression possibly through the interaction of CDP/Cut with HDAC1 (23).
The role of Drosophila Cut in cell fate determination has been well defined by the phenotypes of various mutant flies. Mutations within the Cut locus which disrupt the coding region cause embryonic lethality, whereas some mutations in the enhancer regions result in viable mutant flies with malformations in the leg and wing where Cut fails to express (16-18). There is limited insight into the functions of mammalian CDP/Cut in vivo. Cutl1tm1Ejn mice with a deletion of Cut repeat 1 exhibit a mild phenotype, including curly vibrissae, wavy hair, and high postnatal lethality in litters born to homozygous mutant mothers due to the mothers' impaired lactation (34). In this study, we used insertional mutagenesis to inactivate CDP/Cut by targeting its homeodomain DNA binding region. Our results show that this mutation (Cutl1tm2Ejn) prevents CDP/Cut from accumulating in the nucleus as a functional DNA binding complex (i.e., HiNF-D). Homozygous mutant mice (Cutl1tm2Ejn/tm2Ejn), hereafter referred to as CDP/Cux ΔC−/− mice, display high postnatal lethality, growth retardation, nearly complete hair loss, and severely reduced male fertility. Despite the ubiquitous expression of CDP/Cux, these data indicate that its absence from the nucleus results in limited disturbance of normal tissue development.
MATERIALS AND METHODS
Targeting construct.
A targeting vector was constructed which contains an simian virus 40 early promoter-neomycin (neo) resistance cassette (Invitrogen, Carlsbad, Calif.) for positive selection and a phosphoglycerate kinase (HSV-TK) cassette (35) for negative selection. From the CDP/Cux locus, a 696-bp HindIII (intron 19) and XhoI (exon 20; 12 bp into the homeodomain) fragment was used as 5" homology and a 6-kb BamHI fragment was used as 3" homology.
Homologous recombination into ES cells.
CJ7 ES cells were electroporated with 50 μg of linearized targeting vector followed by a week of selection with G418 (200 μg/ml) and ganciclovir (2 μM). G418/ganciclovir doubly resistant clones were screened by Southern blot analysis of BamHI-digested genomic DNA and probed with a radiolabeled fragment located 5" to the targeted sequence. One correctly targeted heterozygous ES clone had a normal diploid karyotype and was subsequently used to inject blastocysts.
Generation of chimeric mice carrying the mutant allele.
C57BL/6J blastocysts were injected with the ES clone and transferred to the uterus of day 2.5 pseudopregnant female mice. Chimeras identified by agouti coat color were mated with C57BL/6J females to generate F1 progeny. The genotypes of F1 mice were analyzed by Southern blotting with BamHI-digested genomic DNA from tail biopsies, as described above. The genotypes of F1 mice were analyzed by Southern blotting with BamHI-digested genomic DNA from tail biopsies, as described above. Multiplex PCR analysis was also performed to determine mice genotypes, using CDP/Cux intron 19 forward primer HD-F (5"-CAGGGTTTTAT TTGGGGGC TTT TT-3"), which produced a 596-bp product with exon 20 reverse primer HD-R1 (5"-AAGTTCCTCGATGGTTT TT-3") from the wild-type allele or a 1.06-kb product with neo reverse primer HD-R2 (5"-GCATCGCCTTCTATCCGCCTTCTTG-3") from the mutant allele. PCRs were performed by adding genomic DNA to a mixture containing 10 mM Tris-HCl, 150 mM KCl, 2.5 mM MgCl2, a 0.25 mM concentration of each deoxynucleoside triphosphate (dNTP), 25 pmol of each primer, and 5 U of each Taq polymerase (Promega Corp., Madison, Wis.). Following a 2-min denaturation at 95°C, PCRs were amplified for 30 cycles (94°C, 30 s; 62°C, 30 s; and 72°C, 1 min). F1 mice were crossed to generate F2 litters, and the genomic PCR analysis described above was performed to detect the wild-type (0.6 kb) or mutant allele (1.0 kb).
Preparation of mouse embryonic fibroblasts (MEFs).
Embryos were harvested from female mice that were at day 12.5 of gestation. A part of the embryo was removed for genotyping. The remainder of the embryo was passed through an 18-gauge needle with 1 ml of trypsin to break up the tissue and was incubated for 5 to 10 min at 37°C in a CO2 incubator. Primary fibroblasts were then plated with 20 ml of complete Dulbecco's minimal essential medium (10% fetal bovine serum).
Growth curve of MEFs.
Embryonic fibroblasts derived from the same litter were plated at 4 × 106 cells per plate. On the indicated days, MEFs were trypsinized and the number of cells per plate was determined by sampling each plate four times.
Preparation of protein extracts.
Lungs and brains were harvested from adult mice, rapidly frozen, and reduced to powder in liquid nitrogen by a Bessman tissue pulverizer (Fisher Scientific, Pittsburgh, Pa.). The tissue powder was transferred to a 50-ml conical tube and thawed on ice with 30 ml of ice-cold buffer R (10 mM KCl, 10 mM HEPES [pH 7.5], 0.5% Triton, 300 mM sucrose, and 3 mM MgCl2). The suspension was then passed through a Sweeney filter (Millipore, Bedford, Mass.) to separate released cells from the extracellular matrix. Cells were pelleted at 1,500 rpm using a low-speed, swing-out centrifuge (IEC-4B). To prepare whole-cell extract, the cell pellet was resuspended in 200 μl of radioimmunoprecipitation assay buffer (1× phosphate-buffered saline [PBS], 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]), and centrifuged at 15,000 × g for 20 min at 4°C. The supernatant was rapidly frozen in liquid N2 in 25-μl aliquots and was stored at −70°C as whole-cell lysate. To prepare nuclear extract, the cell pellet was resuspended in 1 ml of buffer A (10 mM HEPES [pH 7.5], 10 mM KCl), transferred to a 1.5-ml tube, and centrifuged at 3,000 × g for 1 min. The nuclear pellets were extracted with 300 to 600 μl of buffer C (400 mM KCl, 25 mM HEPES [pH 7.5], and 25% glycerol) for 30 min on ice and rapidly frozen in 50-μl aliquots. All buffers used were complemented with a protease inhibitor cocktail (Boehringer Mannheim, Mannheim, Germany).
Western blot analysis.
The concentration of cell extracts was determined by using the Coomassie Protein Assay Reagent (Pierce Chemical Co., Rockford, Ill.) according to the manufacturer's instructions. Total protein (20 μg) was mixed with loading buffer (6.25 mM Tris HCl, pH 6.8, 10% glycerol, 2% SDS, 2% β-mercaptoethanol, and bromophenol blue), boiled for 5 min, loaded onto a 5% gel, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to polyvinylidene diflouride membranes (Immobilon-P; Millipore Corp.) and were Western blotted at room temperature. Membranes were blocked for 1 h in PBS-T buffer (PBS containing 0.1% Tween 20) and 5% fat-free dry milk, incubated with primary antibodies at 1:1,000 (guinea pig polyclonal antibody against full-length CDP or Santa Cruz goat polyclonal antibody against CDP C terminus) for 1 h in PBS-T buffer containing 1% milk, and lastly incubated with secondary antibodies at 1:5,000 dilution in PBS-T buffer containing 1 or 5% milk for 45 min. Three 15-min washes were performed with PBS-T after each incubation.
RNA analysis.
Total cellular RNA was isolated from proliferating MEFs with the TRIzol Reagent (Life Technologies, Rockville, Md.) according to the manufacturer's instructions. To perform reverse transcription-PCR (RT-PCR), a mixture of 2 μg of total cellular RNA, 2.5 μM oligo(dT), 1 mM dNTP, and 10 U of Moloney murine leukemia virus reverse transcriptase (Promega Corp.) was incubated for 1 h at 37°C followed by 15 min of heat inactivation at 95°C. The resulting complementary DNA (cDNA) mixture (2 μl) was used in PCRs containing 1 mM dNTP, 1 mM MgCl2, 10 U of Taq polymerase (Promega Corp.), and primers (20 μM) which spanned the C terminus of CDP. Following a 5-min denaturation at 95°C, PCRs were amplified for 30 cycles (94°C, 30 s; 60°C, 30 s; and 72°C, 30 s) and terminated with a final elongation step at 72°C for 10 min. Primers for murine CDP/Cux included primer 22F (5"-TCTCCGACCTCCTTGCCCG-3"); primer 23F (5"-GCCCCCAGCCACAACACCA-3"); primer 23R (5"-TGGTGTTGTGGCTGGGGGC-3"); primer 24F (5"-GGAGAGGACGCCGCTACC-3"); primer 24R (5"-GGCTTCCAGCTTGAATCTCC-3"); primer NeoR1 (5"-CCATCAGAAGCTGACTC-3"); and primer NeoR2 (5"-GAAGAACGAGATCAGCAGCC-3"). RT-PCR products were visualized in a 1% agarose/ethidium bromide gel and transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech, Arlington Heights, Ill.) for Southern blot analysis. Blots were hybridized with a random-primed (Prime-It kit; Stratagene, La Jolla, Calif.), 32P-labeled cDNA probe for Cux (1.7-kb EcoRI/DraII fragment spanning Cut repeat 3 to C terminus) at 65°C overnight. The blot was washed and subjected to autoradiography.
For Northern blot analysis, total RNA (20 μg) was electrophoresed in a 1% agarose gel and transferred to Hybond-N+. Blots were hybridized with random-primed (Prime-It kit; Stratagene), 32P-labeled cDNA probes for histone H4 (1-kb fragment including the entire coding region) at 65°C overnight. After washing, the blots were subjected to autoradiography.
S1 nuclease assays were performed according to the protocol of the manufacturer (Ambion, Austin, Tex.) with minor modifications. Total RNA was extracted from proliferating MEFs. Oligonucleotides complementary to the mRNA cap sites of the mouse genes encoding histones H1 (5" GCG AGA AAA CTA AAA GAA ATC TAA CGC GAA GAG CAA ATT TTG CGA TGC TCC 3") and H4.1 (also referred to as FO108) were used as probes (38). A cytoplasmic β-actin probe served as an internal control. The probes were [γ-32P]ATP labeled by T4 polynucleotide kinase, and approximately 100 fmol of each was used per reaction. The probes were first denatured at 94°C and were then hybridized to RNA (10 μg) overnight at 16°C. The samples were subsequently digested for 40 min at 37°C by S1 nuclease (Ambion). The reaction was stopped by addition of stop solution, and digested fragments were purified by ethanol precipitation. The pellets were dissolved in loading buffer and separated in a 6% denaturing polyacrylamide gel (SequaGel, Hessle Hull, England) alongside undigested probes. Gels were exposed and analyzed by phosphorimaging (PhosphorImager Storm 840; Molecular Dynamics, Inc., Sunnyvale, Calif.).
EMSA.
The electrophoretic mobility shift assay (EMSA) was performed as described previously (41). The histone H4/Site II probe and gp91-phox probe were EcoRI-HindIII inserts from plasmids pFP202 and pFp-Puc, respectively. The Sp1 probe is the consensus binding sequence [5"-ATTCGATCGGGGCGGGGCGAGC-3"]. Protein/DNA binding reactions with H4/Site II and gp91-phox probes were performed by combining 32P-labeled probe DNA (10 nmol), nonspecific competitor DNA [1 μg of poly(G/C) and 100 ng of poly(I/C)], and nuclear extract (1 to 10 μg). Protein/DNA binding reactions with the Sp1 probe were performed with 1 μg of poly(G/C) as nonspecific competitor DNA. Oligonucleotide competition assays were performed in the presence of a 100-fold-molar excess (1 pmol) of the unlabeled TM-3 oligonucleotide (self-competition with wild-type H4/Site II) (3), the gp91-phox oligonucleotide (28), or the Sp1 oligonucleotide. Nonspecific competition was performed with the SUB-11 oligonucleotide containing a mutated H4/Site II (3). Immunoreactivity EMSAs were performed by preincubating antibodies (0.5 to 2 μg) with proteins on ice for 15 min prior to the addition of probe DNA. Antibody against cyclin E (Santa Cruz Biotechnology, Santa Cruz, Calif.) was used as a nonspecific antibody control. Separation of protein/DNA complexes was performed in a 4% (80:1) polyacrylamide gel.
FACS analysis.
Proliferating MEFs were harvested for fluorescence-activated cell sorter (FACS) analysis with trypsin-EDTA (Life Technologies), washed three times with PBS, and stained with a propidium iodide solution (2 mM MgCl2, 20 μg of propidium iodide/ml, and 50 μg of RNase/ml). Cell cycle phase estimates were obtained with a FACS Scan Cytometer equipped with pulse-processing electronics (Becton Dickinson, San Diego, Calif.). Per sample, 15,000 doublet discrimination events were collected and analyzed using MODFIT software (Verity House Software, Topsham, Maine). The data were subsequently analyzed by the Tukey HSD statistical test.
Immunofluorescence microscopy.
Cell extraction of proliferating MEFs was performed as described previously (5). Briefly, cells were rinsed twice with ice-cold PBS and fixed in 3.7% formaldehyde in PBS for 10 min on ice. After rinsing twice with PBS, cells were permeabilized in 0.1% Triton X-100 in PBS and rinsed twice with PBSA (0.5% bovine serum albumin in PBS) followed by antibody staining. Cells were incubated with a dilution (1:100) of immunoglobulin G-purified guinea pig antibody against full-length CDP and rabbit anti-Sp1 antibody for 16 h at 4°C. Coverslips were rinsed 4 times with PBSA before incubation with a dilution (1:200) of fluorescein isothiocyanate donkey anti-guinea pig antibody and Alexa 594 goat anti-rabbit antibody for 1 h at 37°C. Cells were rinsed four times with PBSA and then stained with 4",6"-diamidino-2-phenylindole (DAPI) (mixture: 0.1% DAPI and 0.1% Triton X-100 in PBSA) for 1 min. Coverslips were washed once with 0.1% Triton in PBSA and twice with PBS. Immunostaining of whole-cell preparations was recorded using an epifluorescence microscope attached to a charge-coupled device camera, and the digital images were analyzed with the Metamorph software programs. HeLa cells were transfected with pcDNA-Cuxfl (10 μg) or pcDNA-CuxΔC (10 μg). After 24 h of transfection with Superfect (QIAGEN Inc., Valencia, Calif.), cells were processed for immunofluorescence microscopy. Cell extraction of transfected HeLa cells was performed as described above. Cells were incubated with a dilution (1:500) of rabbit antibody against the Xpress epitope (Invitrogen) for 1 h at 37°C, followed by incubation with a dilution (1:200) of donkey anti-rabbit antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) for 1 h at 37°C.
Confocal immunofluorescence microscopy.
Cell extraction of proliferating MEFs was performed as described above. Cells were incubated with a dilution (1:50) of guinea pig anti-CDP antibody and a dilution (1:500) of polyclonal rabbit anti-CASP antibody. Cells were subsequently incubated with a dilution (1:200) of fluorescein isothiocyanate donkey anti-guinea pig and a dilution (1:400) of Alexa red 594 goat anti-rabbit antibody (Molecular Probes, Eugene, Oreg.). Samples were examined using the Leica True Confocal Scanning Spectrophotometer.
Histology.
Tissues were fixed in Bouin's fixative or in 10% buffered neutral formalin, processed, and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin, examined, and photographed.
Serum testosterone levels.
Blood was collected by cardiac puncture immediately after the animal was sacrificed by cervical dislocation. Blood samples were kept on ice for 10 min and subsequently subjected to centrifugation at 4°C for 10 min at 6,000 × g. The supernatant was aliquoted and stored at −70°C until assayed. Serum samples were measured using a Coat-A-Count Total Testosterone kit as directed by the manufacturer (Diagnostic Products Corp., Los Angeles, Calif.). Controls were littermates or age-matched males.
Scanning electron microscopy.
Multiple dorsal hairs were manually removed from a homozygous mutant and a control mouse, both 5-month-old females. These hairs, representing the four major hair types (zigzag, guard, auchene, and awl), were prepared for scanning electron microscopy as previously described (32, 48) and were screened for abnormalities.
RESULTS
Loss of CDP/Cux C terminus results in high neonatal lethality and severe growth retardation.
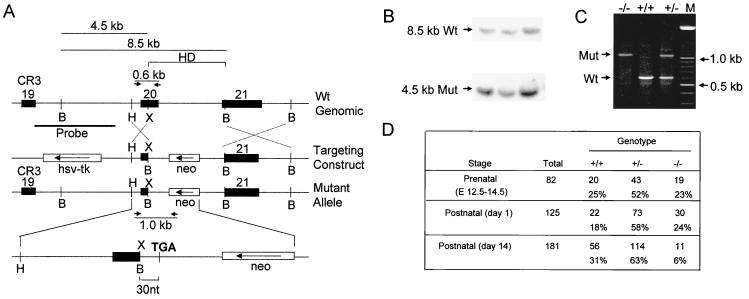
We generated a targeting construct designed to functionally inactivate CDP/Cux by forcing premature translational termination in exon 20, which encodes the beginning of the homeodomain, a DNA binding region of CDP/Cux (ΔC; Fig. 1A). The genotypes of heterozygous mice generated from ES clones with the correctly targeted allele were determined by Southern blot analysis (Fig. 1B). Heterozygous mice were crossed, and PCR analysis demonstrates that the mutation is transmitted to heterozygous and homozygous offspring at the expected frequency (Fig. 1C and D).
FIG. 1.
Targeted mutation of mouse CDP/Cux gene by homologous recombination. (A) Top, a schematic diagram of the wild-type (Wt) CDP/Cux allele with the positions of exons 19 to 21 indicated. Arrows denote the positions of the primers used in PCR-based genotyping. Middle, targeting construct with the neo gene in an antisense orientation. Bottom, predicted mutant allele resulting from homologous recombination between the wild-type allele and the targeting construct. Ten amino acids encoded by 30 nucleotides (nt) of the polylinker sequence in the targeting construct are added to the open reading frame of the mutant Cux transcript before reaching the stop codon (TGA). A BamHI site introduced in the construct gives a diagnostic fragment of 4.5 kb. (B) Southern blot analysis of three heterozygous embryonic stem cell clones displaying both the 8.5- and the 4.5-kb BamHI fragments corresponding to the wild-type (Wt) and mutant (Mut) allele, respectively. (C) Genotyping of pups from the F2 generation with primers in intron 19 (HD-F), exon 20 (HD-R1), and the neomycin cassette of the targeting vector (HD-R2). The HD-R1 sequence is in the region of exon 20 that is deleted from the mutant allele. All genotypes are represented among the progeny. (D) Genotype distribution of progeny from heterozygous matings. Data for postnatal day 1 litters are based on those litters in which the genotypes of all born pups were determined. E, embryonic day; HD, homeodomain; CR3, Cut repeat 3; B, BamHI; H, HindIII; X, XhoI; and M, molecular weight marker lane.
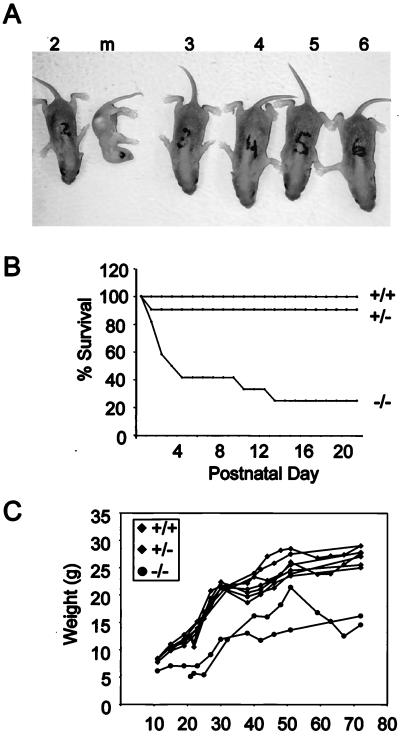
Mice heterozygous for the CDP/Cux ΔC mutation are normal in appearance and are fertile. Homozygous mutant (ΔC−/−) pups appear indistinguishable from their littermates at birth. The CDP/Cux ΔC−/− pups nursed normally, and there was milk evident in their stomachs during the first 2 or 3 days after birth, but they failed to thrive. By the end of the 1st week after birth, many ΔC−/− pups appear considerably smaller than their littermates and most die at this stage (Fig. 2A). More than 70% of ΔC−/− pups failed to survive to weaning age (Fig. 2B). Homozygous mutant mice have increased susceptibility to bacterial infections and suffer from purulent rhinitis characterized by mucosal and submucosal purulent infiltrates within the nasal turbinates (data not shown). In addition, histopathological examination of bone marrow and sternebrae reveals a relative hyperplasia of myeloid cell types in ΔC−/− mice (27a). The homozygous mutant mice that do survive to adulthood have a normal life span but are severely growth retarded and weigh 30 to 50% less than their normal littermates (Fig. 2C). Thus, the CDP/Cux ΔC mutation has an effect on pup viability, general growth, and susceptibility to bacterial infections.
FIG. 2.
High postnatal lethality and stunted growth in CDP/Cux ΔC homozygous mutant mice. (A) A litter with a moribund ΔC−/− pup (m) was photographed within the 1st week after birth. Wild-type (no. 3 and 6) and heterozygous (no. 2, 4, and 5) pups are depicted. (B) Of 60 mice screened, more than 70% of homozygous mutant mice die by postnatal day 10. Wild-type and nullizygous mice, n = 13; heterozygous mice, n = 34. (C) ΔC−/− mice exhibit stunted growth and weigh significantly less than their littermates. Graph represents the growth curve of seven males from two litters.
Expression of the mutant CDP/Cux allele.
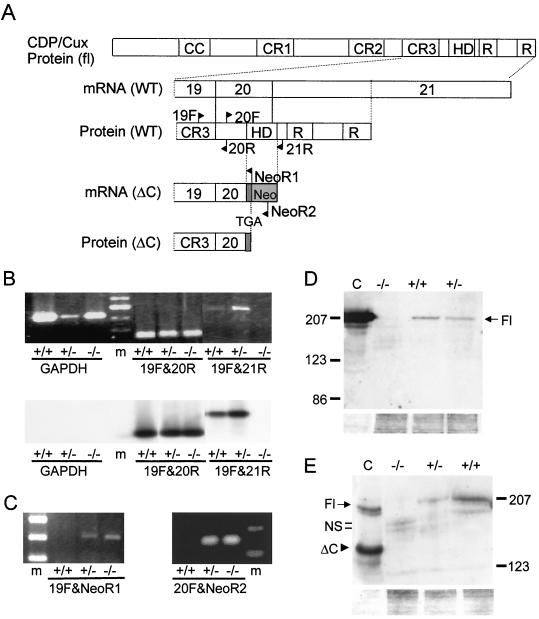
To assess the effect of the mutation on CDP/Cux expression, CDP/Cux transcripts were analyzed by using RT-PCR with primers spanning exons 19 to 21 (Fig. 3A). Total RNA was isolated from proliferating MEFs derived from homozygous mutant (ΔC−/−), heterozygous (ΔC+/−), and wild-type (ΔC+/+) embryos. We detected the expected RT-PCR product (258 bp) with primers 19F and 20R in exons 19 and 20 in all RNA samples, indicating that CDP/Cux transcripts were made in all three types of MEFs. A 441-bp product was detected using primer pair 19F and 21R spanning exons 19 to 21 in ΔC+/+ and ΔC+/− samples. However, this product was absent in ΔC−/− samples, establishing that insertional mutagenesis created the designed truncation in the 3" end of the mutant CDP/Cux transcripts (Fig. 3B, top panel). Southern blot analysis showed that a probe spanning exons 19 to 21 hybridized with the RT-PCR products from CDP/Cux primers but not with products from GAPDH primers (Fig. 3B, bottom panel). Furthermore, two chimeric RT-PCR products (307 bp with primers 19F and NeoR1 and 377 bp with primers 20F and NeoR2) were detected with primers spanning exon 19 and the neomycin cassette in heterozygous and homozygous mutant samples but not in wild-type samples as expected (Fig. 3C). Taken together, our RT-PCR data indicate that the targeted allele expresses a chimeric truncated transcript in which the homeodomain encoding sequences were replaced with sequences from the neo cassette.
FIG. 3.
Full-length CDP/Cux mRNA and protein are not expressed in ΔC−/− mice. (A) A schematic diagram of wild-type (WT) and mutant Cux mRNA and protein. Arrows indicate the positions of the primers used in RT-PCR analysis. Dark gray rectangle represents 30 nucleotides that are added to the mutant transcript from the polylinker sequence of the targeting construct. fl, full length; CC, coiled-coil domain; CR, cut repeat; R, repressor domains; TGA, stop codon; HD, homeodomain. (B and C) Total lung RNA (2 μg) was used in RT-PCR assays with various primers spanning exons 19 to 21 of the Cux gene. (B) Bottom, Southern blot of RT-PCR products using a radiolabeled probe spanning exons 20 to 21. (D) Top, Western blot of lung extracts (30 μg) with a polyclonal antibody against CDP C terminus. Bottom, Coomassie stain of the SDS-PAGE gel after transfer to the polyvinylidene difluoride membrane used in the above Western blot. Control lane (lane C), whole-cell extract of HeLa cells transfected with pcDNA-Cux. (E) Top, Western blot with a polyclonal antibody against full-length CDP. Bottom, Coomassie stain of the SDS-PAGE gel. Control lane (lane C), whole-cell extract of HeLa cells transfected with pcDNA-Cux(fl) and pcDNA-Cux(ΔC). Fl, full-length Cux; NS, nonspecific bands; and ΔC, C-terminal truncated Cux.
To characterize the expression from the mutant CDP/Cux transcript, we analyzed whole-cell protein extracts derived from lungs of wild-type, heterozygous, and homozygous mutant mice using antibodies against the C terminus of CDP (Fig. 3D) or the full-length protein (Fig. 3E). A major band at ∼200 kDa, which comigrates with the overexpressed, full-length Cux protein in the control lanes, is detected with both antibodies in extracts from wild-type and heterozygous but not homozygous mutant mice. Coomassie staining of the SDS-PAGE gels shows that comparable amounts of protein were blotted (bottom panels of Fig. 3D and E). These data indicate that the C terminus of CDP/Cux is absent in ΔC−/− mice and that a full-length protein is not produced in these mice. In the extracts derived from ΔC−/− mice, we note that the antibody against full-length CDP/Cut detects diffuse and faint bands at ∼160 to 170 kDa (Fig. 3E). These bands do not comigrate with the overexpressed truncated protein, which migrates at approximately the same molecular mass (∼123 kDa) predicted for the endogenous ΔC Cux protein that is expected to be produced in mice. Similar bands are observed when an antibody is used against a C-terminal epitope that is not present in the ΔC protein (Fig. 3E). Thus, we conclude that these faint bands are nonspecific. Although the truncated protein is produced at a level below detection by our Western blot analysis, immunofluorescence microscopy data indicate that it is expressed at low levels in MEFs and in other cell types (27a). The low expression of the ΔC protein in the MEFs could be due to production of an antisense transcript originating from the neo cassette that is transcribed in the reverse orientation. Irrespective of the ΔC protein levels expressed in the homozygous mutant mice, further molecular analysis reveals a significant loss of CDP/Cux nuclear functions in these mice.
The CDP/Cux-containing HiNF-D complex is absent in ΔC−/− mice.
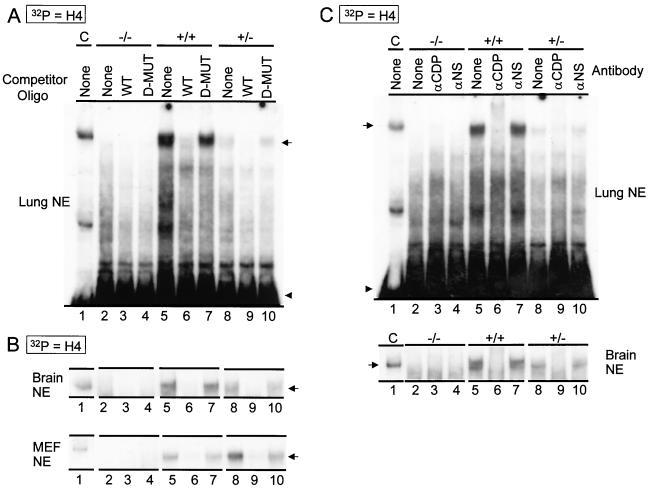
To investigate whether the ΔC protein retains the ability to form protein/DNA complexes, we performed EMSAs with nuclear extracts from MEFs, as well as from the lung and brain of wild-type, heterozygous, and homozygous mutant mice (Fig. 4). Wild-type CDP protein has been shown previously to interact with Site II of the cell cycle-regulated histone H4 gene as a multicomponent protein/DNA complex (HiNF-D) containing cyclin A, CDC2/CDK1, and pRb/p105 (39, 43). EMSAs show a low-mobility complex (HiNF-D) in wild-type and heterozygous extracts but not in homozygous mutant extracts (Fig. 4A), suggesting that the ΔC protein has lost the ability to form protein/DNA complexes. The identity of the HiNF-D complex was established by competition with wild-type and mutant oligonucleotides (Fig. 4A) and by immunoreactivity in EMSAs with a CDP/Cut antibody and a nonspecific antibody as a control (Fig. 4C).
FIG. 4.
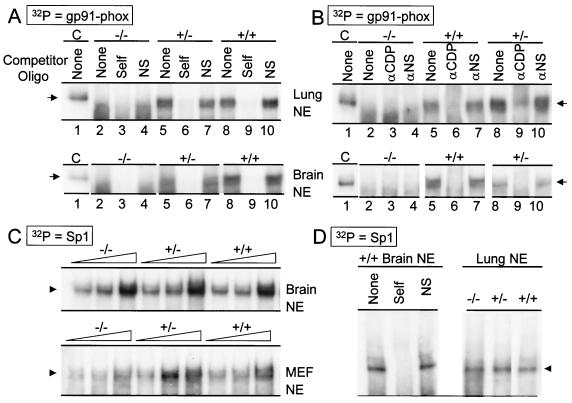
Absence of DNA binding activity of HiNF-D complex in homozygous mutant nuclear extracts. EMSA was performed with 32P-labeled double-stranded oligonucleotides spanning the Site II/Cell Cycle Element of histone H4.1 promoter and nuclear extract (NE) (3 μg) prepared from adult lung, adult brain, and proliferating embryonic fibroblasts (as indicated). (A and B) Competition with unlabeled wild-type (WT) oligonucleotides and mutant (D-MUT) oligonucleotides (SUB-11) which have no significant HiNF-D binding activity. (C) Immunomobility shift assay with antibody against full-length CDP and a nonspecific antibody as control. Arrows indicate the HiNF-D complex. Arrowheads indicate unbound labeled probe. Control lane (lane C), HeLa nuclear extract was used as a positive control for the presence of HiNF-D complex (39).
We also tested the ability of the ΔC protein to bind the myeloid-specific gp91-phox promoter, because CDP/Cux has previously been shown to bind this promoter in complex with the pRb-related p107. Our data show that the probe forms a CDP/Cux-containing complex with protein from ΔC+/+ and ΔC+/− but not ΔC−/− mice (Fig. 5A and B). We performed EMSAs with an Sp1 probe using nuclear extract from MEFs, as well as lung and brain tissues from mice of all genotypes. The results show a similar Sp1 binding activity in all protein preparations (Fig. 5C and D), suggesting that the quality of the nuclear extract is comparable for all genotypes. We conclude that deletion of the CDP/Cux C terminus results in a molecular phenotype reflected by the absence of CDP/Cux-containing protein/DNA complexes.
FIG. 5.
ΔC mutant CDP/Cux does not retain DNA binding activity. (A and B) EMSA with lung and brain nuclear extracts (NE) (3 μg) was performed with 32P-labeled double-stranded oligonucleotides containing gp91-phox promoter sequences. Control lane (lane C), HeLa nuclear extract was used as a positive control for the presence of HiNF-D complex (39). (A) Competition with unlabeled wild-type and nonspecific (SUB-11) oligonucleotides. (B) Immunomobility shift assay with antibody against full-length CDP and a nonspecific antibody (2 μg) as control. Arrows indicate CDP-containing complexes. (C and D) EMSA was performed with 32P-labeled oligonucleotides containing the Sp1 consensus binding site. (C) Increasing amounts (1, 2, and 4 μg) of adult lung and embryonic fibroblast extracts were used. Arrowheads indicate Sp1-containing complexes. (D) Left panel, competition with unlabeled wild-type Sp1 and nonspecific competitor (SUB-11) oligonucleotides. Right panel, EMSA with lung extracts (3 μg).
Levels of CDP/Cux ΔC protein in the nucleus are significantly reduced in ΔC−/− cells.
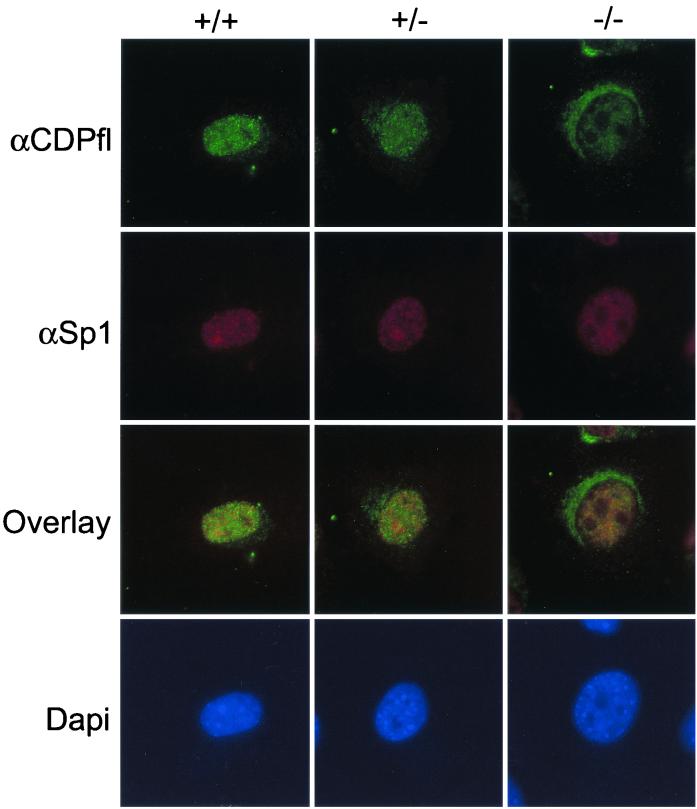
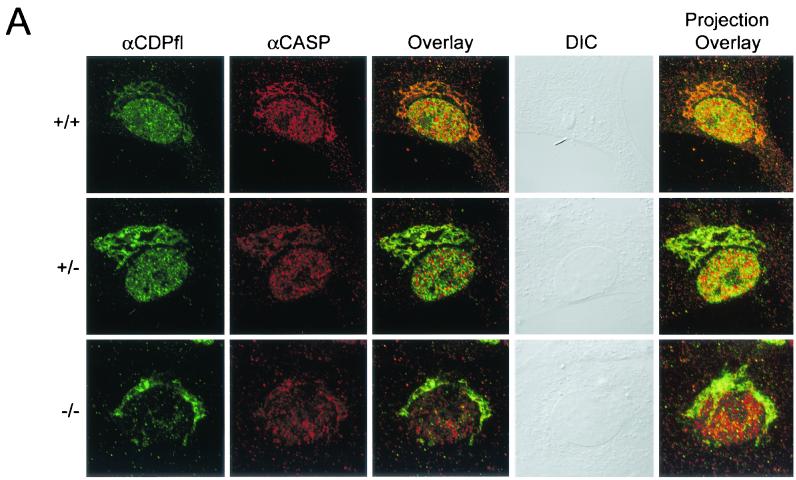
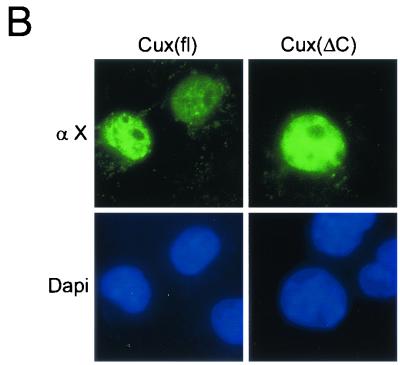
To address the possibility that the absence of CDP-containing protein/DNA complexes is due to their exclusion from the nucleus, we performed immunofluorescence microscopy with a polyclonal antibody against full-length CDP/Cux. We observed strong nuclear immunofluorescence for CDP/Cux in wild-type and heterozygous MEFs, but the nuclear signal is significantly reduced in homozygous mutant MEFs (Fig. 6). No specific signal was detected in MEFs when the primary antibody was omitted (data not shown). We note that when we increased the stringency of primary antibody incubation, nuclear staining was less apparent (data not shown). To ensure that overall protein antigenicity was preserved during cellular extraction, MEFs were also incubated with antibody against Sp1. Strong Sp1 nuclear staining was observed in ΔC−/− MEFs, as well as ΔC+/+ and ΔC+/− MEFs (Fig. 6). To verify that the ΔC protein is localized inside the nucleus, rather than on its surface, we performed confocal microscopy with antibody against full-length CDP. A nuclear signal is detected in ΔC−/− MEFs, albeit at a greatly reduced intensity compared with the wild-type signal. Thus, we conclude that the ΔC protein produced in mutant mice retains the ability to localize to the nucleus (Fig. 7A). A faint and diffuse cytoplasmic signal is present in ΔC+/+ MEFs, but a pronounced reticular pattern is observed in ΔC−/− MEFs with antibody against full-length CDP (Fig. 6 and 7). Consistent with the reticular staining pattern, the N terminus coding sequences remaining in the mutant allele encode an extensive coiled-coil domain which has the potential to form filamentous structures. This coiled-coil domain is also present in an alternatively spliced variant called CASP. It is likely that the polyclonal antibody against full-length CDP can recognize CASP as well. To determine which protein is responsible for the perinuclear signal, we performed confocal microscopy with a CASP-specific antibody. A perinuclear signal was detected by the CASP-specific antibody and colocalized with the cytoplasmic staining observed with the antibody against full-length CDP (Fig. 7A). In addition, we transfected HeLa cells with an epitope (Xpress)-tagged recombinant Cux protein analogous to the truncated protein produced in the ΔC mutant mice (Fig. 7B). The expressed truncated protein was localized to the nucleus, as determined by immunofluorescence microscopy with antibody against the Xpress epitope. Taken together, our findings establish that deletion of the 3" end of the CDP/Cux gene results in a significant loss of nuclear functions of the gene product.
FIG. 6.
Reduced levels of ΔC mutant CDP/Cux protein are detected in the nucleus. Cells grown on gelatin-coated coverslips were fixed, permeabilized, and incubated for 16 h at 4°C with an antibody against full-length CDP or an antibody against Sp1 at a 1:100 dilution. DAPI staining of chromatin was included to visualize nuclei.
FIG. 7.
Significantly reduced levels of ΔC protein are observed in the nucleus of homozygous mutant MEFs. (A) Cultured embryonic fibroblasts were subjected to confocal microscopy. Cells grown on gelatin-coated coverslips were fixed, permeabilized, and incubated with an antibody against full-length (fl) CDP or an antibody against CASP. DIC, differential interference contrast. (B) HeLa cells expressing epitope-tagged (Xpress) full-length and C-terminally truncated (ΔC) Cux proteins were analyzed by digital immunofluorescence microscopy using an anti-Xpress antibody (α X).
Embryonic fibroblasts homozygous for the CDP/Cux ΔC mutation exhibit normal cell growth.
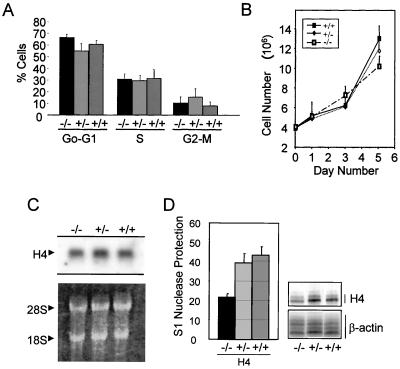
CDP/Cux has been shown to regulate the promoters of genes involved in cell growth regulation, including histone H4, c-myc, and p21 (7, 11, 42, 43). We examined whether the functional loss of CDP in the nucleus affects cell growth control by using FACS analysis to assess the DNA content of the cells and the relative number of cells present in specific cell cycle stages. The results did not reveal a significant difference in growth characteristics between nonsynchronized proliferating nullizygous and wild-type MEFs (P > 0.05) (Fig. 8A). Furthermore, we determined the growth rates of ΔC−/−, ΔC+/−, and ΔC+/+ MEFs and found them to be comparable (Fig. 8B). We also analyzed total RNA for histone H4 expression, which is tightly coupled to DNA replication and is a specific marker for cells in the S phase. As probe, we used a DNA segment encompassing the entire coding sequence, which cross-hybridizes with mRNA transcribed from the 20 to 30 known histone H4 gene copies in mammals. Consistent with the FACS results, Northern blot analysis reveals overall histone H4 mRNA levels from wild-type, heterozygous, and homozygous mutant MEFs to be comparable (Fig. 8C), indicating that a similar proportion of the cells in each population is in S phase.
FIG. 8.
ΔC homozygous mutant MEFs exhibit normal growth characteristics and reduced histone H4.1 and H1 mRNA levels. (A) Cell cycle distribution of proliferating homozygous mutant MEFs and wild-type MEFs was comparable. Cell cycle estimates were obtained by FACS analysis for proliferating MEFs stained with a propidium iodide solution. (B) ΔC−/− MEFs exhibit normal growth rates. MEFs were plated in triplicate at 4 × 106 cells per plate on day 0, and the number of cells per plate was determined on days 1, 3, and 5. (C) Total histone H4 mRNA is expressed at comparable levels in proliferating wild-type and ΔC−/− MEFs. Top, Northern blot of total RNA (20 μg) was performed using a probe spanning the entire coding region of the human histone H4 gene. Bottom, rRNA (28S and 18S indicated) was visualized by using ethidium bromide. (D) S1 nuclease protection assays show a slight decrease in histone H4.1 mRNA expression in homozygous mutant MEFs. Labeled oligonucleotides complementary to the cap site of H4.1 were hybridized to total RNA (10 μg) from proliferating MEFs and were digested with the S1 nuclease. A β-actin probe served as an internal control. Left, the histogram represents a composite of the ratios of the histone levels in five experiments. Right, autoradiograph of a representative S1 nuclease protection experiment.
We have previously focused on one specific histone H4 gene in human and mouse (referred to as FO108 or H4.1) to understand the regulatory role of CDP/Cux in cell cycle-controlled transcription (38, 43). We performed S1 nuclease protection assays to selectively detect expression of the mouse H4.1 gene. Similar to its human counterpart, the murine H4.1 gene is cell cycle regulated and interacts with CDP/Cux (38). S1 nuclease protection analysis was performed with the RNA described above using a 50-bp probe encompassing the divergent 5" flanking region of H4.1. The results suggest that expression of the H4.1 gene is moderately reduced in ΔC−/− MEFs compared to ΔC+/+ MEFs (Fig. 8D). In parallel we also performed S1 analysis to monitor the expression of a histone H1 gene which is also known to bind CDP/Cux (38). Similar to the results for the H4.1 gene, we observed a moderate decrease in H1 expression in ΔC−/− MEFs (data not shown). The moderate reduction in the expression of H4.1 and H1 genes is consistent with the role of CDP/Cux in regulating histone gene transcription but also suggests that there are compensatory mechanisms. It has been well established that histone gene expression is regulated by both transcriptional and posttranscriptional mechanisms and that mRNA stabilization may offset transcriptional changes (29).
The CDP/Cux ΔC−/− mice reduced fertility in nullizygous males.
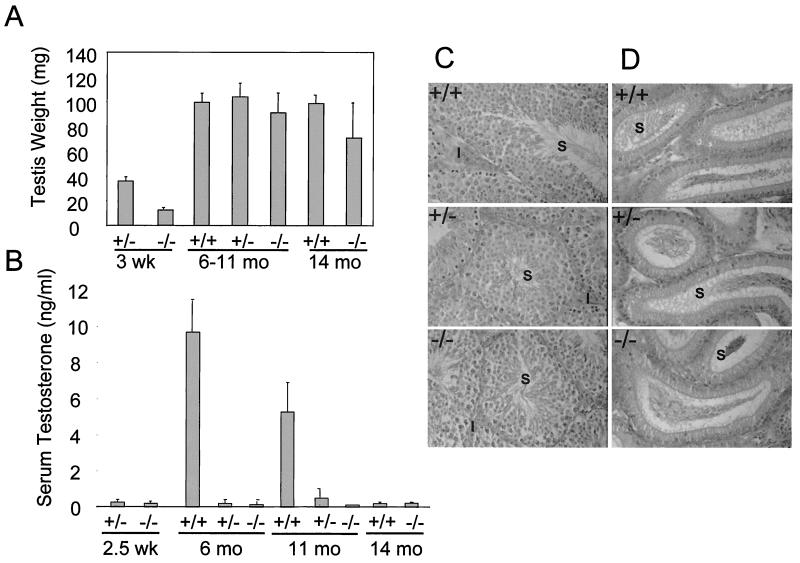
One of the most striking features of the CDP/Cux ΔC mouse is that reproductive fitness is greatly compromised in the homozygous mutant males. In general, ΔC−/− males rarely produce offspring, even when mated with wild-type females. For example, only one of 25 ΔC−/− males sired two litters of pups. To assess whether ΔC−/− mice have defects in testicular development or function, we performed gross anatomical and histological studies. The weights of the testes dissected from ΔC+/+, ΔC+/−, and ΔC−/− adult mice were comparable (Fig. 9A). The testes and epididymis appear normal by gross examination and histological studies with hematoxylin-and-eosin sections. Abundant maturing germ cells were observed in the seminiferous tubules and epididymis of wild-type, heterozygous, and homozygous mutant male littermates at 7 months (Fig. 9C and D). Serum testosterone levels in ΔC+/− and ΔC−/− males were significantly lower than those in ΔC+/+ male littermates (Fig. 9B). Heterozygous males are fertile and yet display the same reduced levels of testosterone as the homozygous mutant males. These results indicate that serum testosterone levels alone cannot account for the reduced fertility of the homozygous mutant males.
FIG. 9.
ΔC mutant males with normal testicular morphology have reduced serum testosterone levels. (A) Testes were weighed from mice at the ages of 3 weeks (four mice), 10 to 11 months (nine mice), and 14 months (four mice). (B) Total serum testosterone levels from mice at age 2 to 3 weeks (six mice), 6 months (three mice), 10 to 11 months (11 mice), and 14 months (four mice) were measured by radioimmunoassay. Micrograph (magnification, ×25) of hematoxylin-and-eosin-stained sections of testis (C) and epididymis (D) from male littermates at 7 months. S, sperm; I, interstitial cells.
The development of the GI tract and other internal organs is unperturbed in ΔC−/− mice.
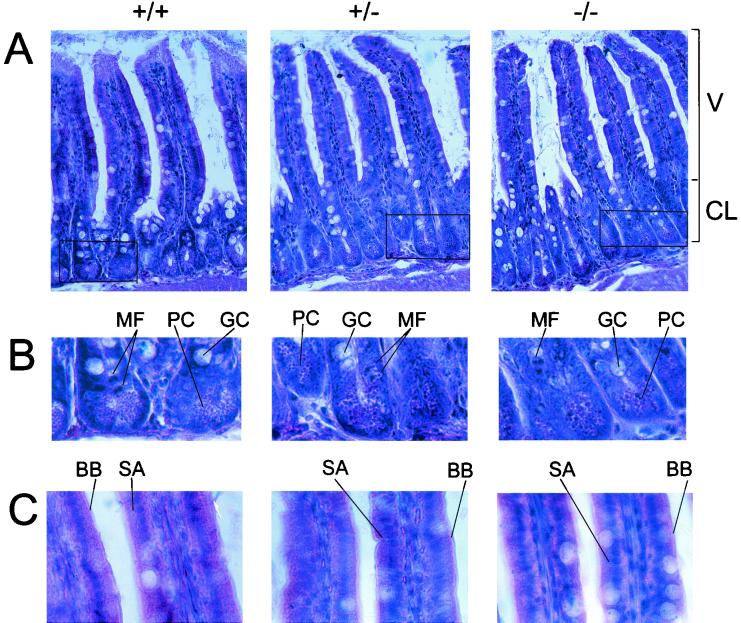
CDP/Cux RNA is expressed in the brain and all major internal organs except in the adult liver (37). In addition, many homeobox-containing genes have been shown to play essential roles in the developing mouse brain (44). However, hematoxylin-and-eosin-stained sections revealed no histological abnormalities in the major internal organs or the brains of ΔC−/− mice (data not shown). Although observed to feed normally, ΔC−/− mice are severely growth retarded. This runted phenotype may be a result of defective nutrient absorption. Therefore we performed a thorough histological examination of the GI tract in these mice. One potential cause of malabsorption is structural defects in the small intestine, where the majority of nutrient absorption occurs. Careful examination of histological sections stained with hematoxylin and eosin revealed no structural abnormalities in the small intestine of mutant mice (Fig. 10). The height of the villi (Fig. 10A) in ΔC−/− mice appears normal. In addition, a comparable number of narrow stem cells (Fig. 10B), undergoing mitotic activity to reconstitute the cell population of the crypt and villus, are observed in the crypts of Lieberkuhn. Also present are the major cell types with protective and absorptive functions in the small intestine, namely, the mucus-secreting goblet cells (Fig. 10B) and the surface absorptive cells (Fig. 10B). The structure of the brush border (Fig. 10C), where many of the enzymatic activities of digestion occur, appears intact in the ΔC−/− mutant mice. No abnormalities were observed upon examination of the other organs of the gastrointestinal (GI) tract, such as the stomach and the colon (data not shown). Another potential cause of malabsorption is bacterial overgrowth in the GI tract, which would be expected to cause inflammatory changes in the wall of the gut. We did not observe inflammation in the GI tract of mutant mice, and the numbers of Peyer's patches were comparable in all the mice examined (data not shown). Thus, we conclude that the ΔC mutation does not disturb the development of these organs.
FIG. 10.
Sections of the small intestine from ΔC−/− mice have normal histology. Photomicrographs of hematoxylin-and-eosin sections (7 μm) of the small intestine. (A) Villi (V) and crypts of Lieberkuhn (CL). (B) Enlarged view of the boxed area in panel A depicting stem cell mitotic figures (MF), Paneth cells (PC), and goblet cells (GC). (C) High-power micrograph of the villi. SA, surface absorptive cells; BB, a columnar type of epithelium with a brush border.
CDP/Cux ΔC mutation results in abnormal dermis and hair loss.
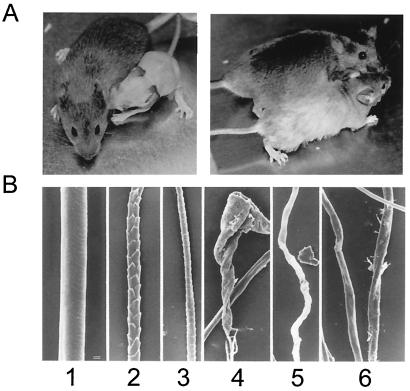
Homozygous mutant pups begin to shed hair the 2nd to 3rd week after birth. By 1 month after birth, the mice are completely bald except for very thin hair covering parts of the ventral region and head (Fig. 11A). Regrowth of the coat hair occurs gradually over a period of several months and is more evident in female than in male ΔC−/− mice. The regrown coat hair on ΔC−/− mice has a distinctive light gray color and appears longer than hair of wild-type animals. In contrast to wild-type controls in which there was a thick mat of normal hair, the mutant skin was covered by hairs, but they were scant and distorted, appearing to be very slender and broken off (data not shown). The wild-type controls had normal awl and guard hairs, which were straight and of uniform diameter along their length with distinct cuticular scales (Fig. 11B, panels 1 to 3). Regardless of where they are found on the body, the hair fibers from ΔC−/− mice are essentially uniform in size, with a variety of deformities, including kinky, twisted, and flattened characteristics (Fig. 11B). The overriding pattern on these mutant mice is that the cuticle is formed or partially formed on some fibers but is missing on others (data not shown), suggesting that the inner root sheath may not be normal. Furthermore, no vibrissae are evident in the muzzle skin of nullizygous mice, only lance-shaped broken ends of hair fibers and irregularly wavy hairs that are short and similar to the coat hair (data not shown). As the homozygous mutant mice aged, progressive alopecia was observed with follicular dystrophy which was scattered among anagen follicles, resulting in dilated follicles containing twisted, broken, and disrupted fibers at all levels (data not shown). Occasionally, dystrophic fibers pushed out of growing follicles, twisted below the level of the sebaceous gland, and entered the dermis and hypodermis, resulting in an inflammatory response in and around the fiber (data not shown). In summary, our findings suggest that hair fiber types produced in ΔC−/− mice are all of one type with loss of other types or that all hairs produced are so deformed that they cannot be separated into the various anatomical fiber types. Thus, the genetic mutation that we introduced into the CDP/Cux gene locus apparently causes abnormal formation of the coat and vibrissa hair fibers.
FIG. 11.
Hair loss in homozygous mutant CDP/Cux-ΔHD mice. (A) ΔC−/− mice with extensive hair loss compared to its wild-type sibling at 1 month (left) and partial regrowth of abnormal hair at 4 months (right). (B) Scanning electron micrographs of plucked hair from 5-month-old mice. The wild-type mouse had normal awl (panel 1) and guard hairs (panels 2 and 3). In contrast, its homozygous mutant littermate had severely deformed hair fibers that could not be typed (panels 4 to 6). Deformed hairs were corkscrew-like in appearance with no evidence of cuticular scales. Bar = 10 μm.
DISCUSSION
To investigate the in vivo role of CDP/Cux, we characterized mice in which the homeodomain and C terminus of the protein were genetically deleted (ΔC mice). The nuclear signal originating from the truncated Cux protein is dramatically decreased in homozygous mutant mice, suggesting a significant reduction of its nuclear activity. Consistent with the proposed role of CDP/Cux in the regulation of histone genes (13, 43), altered expression of specific histone H4 and H1 genes was observed in embryonic fibroblasts homozygous for the ΔC mutation. The genetic mutation in the CDP/Cux gene resulted in decreased immune function, hyperplasia of myeloid cell lines, abnormal formation of the hair fibers, and reduced fertility. These data suggest that CDP/Cux is required for normal dermal tissue development, reproduction and the ability to resist microbial infections. A previous study characterizing genetically targeted mice that lack only Cut repeat 1 (due to exon skipping) described a mild phenotype consisting of curly vibrissae, wavy hair, and high pup loss due to impaired lactation in homozygous mutant mothers (34). The mice characterized in the present study which lack the entire C terminus of CDP/Cux have a pronounced and more severe phenotype.
Heterozygous mice are fertile and appear indistinguishable from wild-type animals. This finding suggests that haploinsufficiency of the wild-type allele has no phenotypic effect and that the mutant allele does not act in a dominant negative manner. Homozygous mutant (ΔC−/−) mice are viable at birth and are born at the expected frequency but have a high level of neonatal lethality. Although the cause of the lethality remains unclear, it does not appear to be a result of impaired feeding because milk is clearly visible in the abdominal cavity of moribund pups. The small proportion of surviving ΔC−/− mice has a normal life span, but these animals are severely growth retarded. The cause of this failure to thrive is presently unknown. However, growth retardation is not due to general defects in cell proliferation because the cell growth characteristics of the homozygous mutant embryonic fibroblasts appear to be normal. Furthermore, there do not appear to be anatomical or histological abnormalities in the GI tract of the ΔC−/− mice that may lead to defective absorption of nutrients.
Aberrant coat hair, hair loss, and a high level of male infertility were the most obvious tissue-specific defects in ΔC−/− mice. Interestingly, homozygous deletion of the mouse ovo1 gene, which encodes a zinc finger transcription factor, has a similar phenotype, including growth retardation, aberrant coat hair, and a reduced ability to reproduce (9). In addition, ΔC−/− mice have similarities to the lanceolate hair (lah) mutant mice that are runted and alopecic and lacking vibrissae. The lah mutant mice also have follicular dystrophy and lance-shaped, broken ends of hair fibers in the muzzle skin (31). Several homeobox-containing genes have been shown to be differentially expressed in the dermis and epidermis of fetal and adult skin (10, 30). For example, when the homeodomain protein Msx-2 is overexpressed under the control of a keratin promoter in transgenic mice, the animals have a thickened epidermis, shorter coat hair, and a reduced matrix region (45). Thus, our data contribute to the list of regulatory factors involved in general growth control and dermal tissue development.
We find a considerable reduction in the fertility of ΔC−/− mice, and many factors may potentially contribute to this reduced fertility. However, it is that unlikely serum testosterone is directly related to the reduced fertility of ΔC−/− males, because fertile heterozygous males display the same low testosterone levels as observed in the homozygous males. Adult homozygous mutant males are smaller than wild-type and heterozygous females; this difference in size may affect the mating interactions and thus lower the mating frequency. Furthermore, ΔC−/− mice have purulent rhinitis, which could disrupt their sense of smell. Interestingly, the ΔC−/− mice have many phenotypes in common with the p73-deficient mice, such as runted appearance, high rates of mortality, severe rhinitis, and reduced fertility. As has been shown for the p73-deficient mice, it is possible that the lack of interest of ΔC−/− male mice in sexually mature females is due to defects in the sensory pathways, such as the absence of expression of pheromone receptors V1R and V2R (49). A previous study suggested that a truncated CDP/Cux protein uniquely expressed in the testis may be involved in testis development and the regulation of gene expression during spermatogenesis (37). However, our histological sections of the adult testis and epididymis of nullizygous mice reveal maturing sperm cells in both organs. Hence, we suggest that perturbation of the sensory pathways may contribute to the reduced fertility in the ΔC−/− male mice.
In addition to its proposed role as a transcriptional repressor, CDP/Cux may also function as an activator of histone genes in somatic cells. The maximal expression of cell cycle-regulated histone genes closely precedes DNA replication, a time at which CDP/Cux DNA binding activity is the highest (7). CDP/Cux has been shown to be a component of the promoter complex HiNF-D, which may play a role in the transcriptional upregulation of several histone genes during the G1-to-S transition of the cell cycle (3, 40, 43). In this study, we demonstrate that the HiNF-D complex is absent in homozygous mutant MEFs which exhibit a moderate reduction of H4 and H1 gene expression. The results are consistent with the postulated role of CDP/Cux as a transcriptional regulator of histone genes.
The expression of DHFR and other genes required for the G1-to-S transition is regulated by E2F transcription factors (19). E2F activity is controlled by its interaction with the retinoblastoma protein family (pRb), cyclins, and cyclin-dependent kinases (12, 47). Immuno-EMSA studies have suggested that CDP/Cux may interact with E2F-interacting proteins, including cyclin A, pRb, and CDC2/CDK2 in the histone promoter complex HiNF-D (39, 43). Of note, upregulation of histone reporter constructs does not require the binding of E2F proteins to histone promoter sequences, suggesting an E2F-independent mechanism for gene regulation at the G1-to-S transition point. E2F4 accounts for most of the endogenous pRb-, p107-, and p130-associated E2F activity, and its repression of E2F-responsive genes suggests a critical role in cell cycle exit or in the maintenance of a quiescent state (12, 24). Remarkably, loss of E2F4 had no effect on cell cycle arrest or proliferation. We note that the phenotype of E2F4 knockout mice has many similarities to that of the CDP/Cux ΔC−/− mice (15). In addition to high pup loss resulting from chronic rhinitis and otitis, E2F4 knockout mice also have severe growth retardation and reduced fertility.
Many lines of evidence support the involvement of transcription factors, such as E2F4, p107, p130, and CDP/Cux, in the regulation of cell growth and differentiation. However, mice with genetic lesions in these genes display limited tissue-specific defects, indicating that genetic compensation and/or tissue-specific factors may be rate limiting in the penetrance of cell growth-related phenotypes.
Acknowledgments
We thank C. Cardasis, C. Lengner, and A. Fischer for help with histology. We also thank S. Zaidi, R. Xie, S. Gupta, A. Javed, and F. Adamidou for valuable discussion.
This work was supported by NIH grant GM32010.
REFERENCES
- 1.Andres, V., B. Nadal-Ginard, and V. Mahdavi. 1992. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development 116:321-334. [DOI] [PubMed] [Google Scholar]
- 2.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2 dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 177:453-464. [DOI] [PubMed] [Google Scholar]
- 3.Aziz, F., A. J. van Wijnen, P. S. Vaughan, S. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of IRF-2 (HiNF-M) CDP/cut (HiNF-D) and H4TF-2 (HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol. Biol. Reprod. 25:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Barberis, A., G. Superti-Furga, and M. Busslinger. 1987. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell 50:347-359. [DOI] [PubMed] [Google Scholar]
- 5.Choi, J.-Y., A. J. van Wijnen, F. Aslam, J. D. Leszyk, J. L. Stein, G. S. Stein, J. B. Lian, and S. Penman. 1998. Developmental association of the β-galactoside-binding protein galectin-1 with the nuclear matrix of rat calvarial osteoblasts. J. Cell Sci. 111:3035-3043. [DOI] [PubMed] [Google Scholar]
- 6.Coqueret, O., G. Berube, and A. Nepveu. 1996. DNA binding by cut homeodomain proteins is down-modulated by protein kinase C. J. Biol. Chem. 271:24862-24868. [DOI] [PubMed] [Google Scholar]
- 7.Coqueret, O., G. Berube, and A. Nepveu. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 17:4680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coqueret, O., N. Martin, G. Berube, M. Rabbat, D. W. Litchfield, and A. Nepveu. 1998. DNA binding by cut homeodomain proteins is down-modulated by casein kinase II. J. Biol. Chem. 273:2561-2566. [DOI] [PubMed] [Google Scholar]
- 9.Dai, X., C. Schonbaum, L. Degenstein, W. Bai, A. Mahowald, and E. Fuchs. 1998. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12:3452-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Detmer, K., H. J. Lawrence, and C. Largman. 1993. Expression of class I homeobox genes in fetal and adult murine skin. J. Investig. Dermatol. 101:517-522. [DOI] [PubMed] [Google Scholar]
- 11.Dufort, D., and A. Nepveu. 1994. The human Cut homeodomain protein represses transcription from the c-myc promoter. Mol. Cell. Biol. 14:4251-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 13.El-Hodiri, H. M., and M. Perry. 1995. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol. Cell. Biol. 15:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holthuis, J., T. A. Owen, A. J. van Wijnen, K. L. Wright, A. Ramsey-Ewing, M. B. Kennedy, R. Carter, S. C. Cosenza, K. J. Soprano, J. B. Lian, et al. 1990. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science 247:1454-1457. [DOI] [PubMed] [Google Scholar]
- 15.Humbert, P. O., C. Rogers, S. Ganiatsas, R. L. Landsberg, J. M. Trimarchi, S. Dandapani, C. Brugnara, S. Erdman, M. Schrenzel, R. T. Bronson, and J. A. Lees. 2000. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell 6:281-291. [DOI] [PubMed] [Google Scholar]
- 16.Jack, J., D. Dorsett, Y. Delotto, and S. Liu. 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113:735-747. [DOI] [PubMed] [Google Scholar]
- 17.Jack, J. W. 1985. Molecular organization of the cut locus of Drosophila melanogaster. Cell 42:869-876. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, T. K., and B. H. Judd. 1979. Analysis of the cut locus of Drosophila melanogaster. Genetics 92:485-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krek, W., M. E. Ewen, S. Shirodkar, Z. Arany, W. G. Kaelin, Jr., and D. M. Livingston. 1994. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78:161-172. [DOI] [PubMed] [Google Scholar]
- 20.Last, T. J., M. Birnbaum, A. J. van Wijnen, G. S. Stein, and J. L. Stein. 1998. Repressor elements in the coding region of the human histone H4 gene interact with the transcription factor CDP/cut. Gene 221:267-277. [DOI] [PubMed] [Google Scholar]
- 21.Li, S., B. Aufiero, R. L. Schiltz, and M. J. Walsh. 2000. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc. Natl. Acad. Sci. USA 97:7166-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lievens, P. M., J. J. Donady, C. Tufarelli, and E. J. Neufeld. 1995. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J. Biol. Chem. 270:12745-12750. [DOI] [PubMed] [Google Scholar]
- 23.Mailly, F., G. Bérubé, R. Harada, P.-L. Mao, S. Phillips, and A. Nepveu. 1996. The human Cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol. Cell. Biol. 16:5346-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moberg, K., M. A. Starz, and J. A. Lees. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Neufeld, E. J., D. G. Skalnik, P. M. Lievens, and S. H. Orkin. 1992. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1:50-55. [DOI] [PubMed] [Google Scholar]
- 27.Owen, T. A., J. Holthuis, E. Markose, A. J. van Wijnen, S. A. Wolfe, S. R. Grimes, J. B. Lian, and G. S. Stein. 1990. Modifications of protein-DNA interactions in the proximal promoter of a cell-growth-regulated histone gene during onset and progression of osteoblast differentiation. Proc. Natl. Acad. Sci. USA 87:5129-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in ccAAT displacement protein mutant mice. Blood 98:3658-3667. [DOI] [PubMed] [Google Scholar]
- 28.Skalnik, D. G., E. C. Strauss, and S. H. Orkin. 1991. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 266:16736-16744. [PubMed] [Google Scholar]
- 29.Stein, G. S., J. L. Stein, A. J. van Wijnen, and J. B. Lian. 1996. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 20:41-49. [DOI] [PubMed] [Google Scholar]
- 30.Stelnicki, E. J., L. G. Komuves, D. Holmes, W. Clavin, M. R. Harrison, N. S. Adzick, and C. Largman. 1997. The human homeobox genes MSX-1, MSX-2, and MOX-1 are differentially expressed in the dermis and epidermis in fetal and adult skin. Differentiation 62:33-41. [DOI] [PubMed] [Google Scholar]
- 31.Sundberg, J. P., D. Boggess, C. Bascom, B. J. Limberg, L. D. Shultz, B. A. Sundberg, L. E. King, Jr., and X. Montagutelli. 2000. Lanceolate hair-J (lahJ): a mouse model for human hair disorders. Exp. Dermatol. 9:206-218. [DOI] [PubMed] [Google Scholar]
- 32.Sundberg, J. P., A. A. Erickson, D. R. Roop, and R. L. Binder. 1994. Ornithine decarboxylase expression in cutaneous papillomas in SENCAR mice is associated with altered expression of keratins 1 and 10. Cancer Res. 54:1344-1351. [PubMed] [Google Scholar]
- 33.Superti-Furga, G., A. Barberis, G. Schaffner, and M. Busslinger. 1988. The −117 mutation in Greek HPFH affects the binding of three nuclear factors to the CCAAT region of the gamma-globin gene. EMBO J. 7:3099-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tufarelli, C., Y. Fujiwara, D. C. Zappulla, and E. J. Neufeld. 1998. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 200:69-81. [DOI] [PubMed] [Google Scholar]
- 35.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 36.Valarche, I., J. P. Tissier-Seta, M. R. Hirsch, S. Martinez, C. Goridis, and J. F. Brunet. 1993. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development 119:881-896. [DOI] [PubMed] [Google Scholar]
- 37.Vanden Heuvel, G. B., S. E. Quaggin, and P. Igarashi. 1996. A unique variant of a homeobox gene related to Drosophila cut is expressed in mouse testis. Biol. Reprod. 55:731-739. [DOI] [PubMed] [Google Scholar]
- 38.van der Meijden, C. M. J., P. S. Vaughan, A. Staal, W. Albig, D. Doenecke, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 1998. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta 1442:82-100. [DOI] [PubMed] [Google Scholar]
- 39.van Wijnen, A. J., F. Aziz, X. Grana, A. De Luca, R. K. Desai, K. Jaarsveld, T. J. Last, K. Soprano, A. Giordano, J. B. Lian, et al. 1994. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc. Natl. Acad. Sci. USA 91:12882-12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Wijnen, A. J., C. Cooper, P. Odgren, F. Aziz, A. De Luca, R. A. Shakoori, A. Giordano, P. J. Quesenberry, J. B. Lian, G. S. Stein, and J. L. Stein. 1997. Cell cycle-dependent modifications in activities of pRb-related tumor suppressors and proliferation-specific CDP/cut homeodomain factors in murine hematopoietic progenitor cells. J. Cell. Biochem. 66:512-523. [DOI] [PubMed] [Google Scholar]
- 41.van Wijnen, A. J., J. B. Lian, J. L. Stein, and G. S. Stein. 1991. Protein/DNA interactions involving ATF/AP1-, CCAAT-, and HiNF-D-related factors in the human H3-ST519 histone promoter: cross-competition with transcription regulatory sites in cell cycle controlled H4 and H1 histone genes. J. Cell. Biochem. 47:337-351. [DOI] [PubMed] [Google Scholar]
- 42.van Wijnen, A. J., T. A. Owen, J. Holthuis, J. B. Lian, J. L. Stein, and G. S. Stein. 1991. Coordination of protein-DNA interactions in the promoters of human H4, H3, and H1 histone genes during the cell cycle, tumorigenesis, and development. J. Cell. Physiol. 148:174-189. [DOI] [PubMed] [Google Scholar]
- 43.van Wijnen, A. J., M. F. van Gurp, M. C. de Ridder, C. Tufarelli, T. J. Last, M. Birnbaum, P. S. Vaughan, A. Giordano, W. Krek, E. J. Neufeld, J. L. Stein, and G. S. Stein. 1996. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA 93:11516-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollmer, J. Y., and R. G. Clerc. 1998. Homeobox genes in the developing mouse brain. J. Neurochem. 71:1-19. [DOI] [PubMed] [Google Scholar]
- 45.Wang, W. P., R. B. Widelitz, T. X. Jiang, and C. M. Chuong. 1999. Msx-2 and the regulation of organ size: epidermal thickness and hair length. J. Investig. Dermatol. Symp. Proc. 4:278-281. [DOI] [PubMed] [Google Scholar]
- 46.Wright, K. L., R. T. Dell'Orco, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1992. Multiple mechanisms regulate the proliferation-specific histone gene transcription factor HiNF-D in normal human diploid fibroblasts. Biochemistry 31:2812-2818. [DOI] [PubMed] [Google Scholar]
- 47.Xu, M., K. A. Sheppard, C. Y. Peng, A. S. Yee, and H. Piwnica-Worms. 1994. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol. Cell. Biol. 14:8420-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanaka, R., C. Barlow, J. Lekstrom-Himes, L. H. Castilla, P. P. Liu, M. Eckhaus, T. Decker, A. Wynshaw-Boris, and K. G. Xanthopoulos. 1997. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc. Natl. Acad. Sci. USA 94:13187-13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 50.Yoon, S. O., and D. M. Chikaraishi. 1994. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J. Biol. Chem. 269:18453-18462. [PubMed] [Google Scholar]