Abstract
We previously demonstrated that the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) potently activates the cellular c-Jun N-terminal kinase (JNK) pathway by sequentially engaging an unknown adaptor, TRAF6, TAB1/TAK1, and JNKKs. We now show that BS69, a MYND domain-containing cellular protein, is the missing adaptor that bridges LMP1 and TRAF6, as the MYND domain and a separate region of BS69 bind to the carboxyl termini of LMP1 and TRAF6, respectively. While LMP1 promotes the interaction between BS69 and TRAF6, the complex formation between LMP1 and TRAF6 is BS69 dependent. A fraction of LMP1 and BS69 is constitutively colocalized in the membrane lipid rafts. Importantly, knockdown of BS69 by small interfering RNAs specifically inhibits JNK activation by LMP1 but not tumor necrosis factor alpha. Although overexpression of either BS69 or a mutant LMP1 without the cytoplasmic carboxyl tail is not sufficient to activate JNK, interestingly, when BS69 is covalently linked to the mutant LMP1, the chimeric protein restores the ability to activate JNK. This indicates that the recruitment and aggregation of BS69 is a prerequisite for JNK activation by LMP1.
Epstein-Barr virus (EBV) is a transforming DNA virus (45). The main cell types infected by EBV are human B lymphocytes and nasopharyngeal epithelial cells (15, 40, 48). Although up to 95% of world population is EBV positive, most of them will be healthy carriers for the rest of their lives, due to the effective surveillance of their immune systems (41, 48). However, in certain immune system-compromised individuals, the presence of EBV is thought to contribute to several malignancies including Burkitt's lymphoma, Hodgkin's disease, and nasopharyngeal carcinoma (NPC) (41, 45, 48). Although NPC is relatively rare in most part of the world (e.g., <1 per 100,000 Caucasians in Western countries), it is quite prevalent in southern China, including Hong Kong where the incidence rate is >20 per 100,000 (34).
EBV readily transforms quiescent human B cells in vitro, resulting in formation of the immortalized lymphoblastoid cell lines (45). Nine latent viral antigens including six nuclear antigens (EBNA1 to -6) and three membrane proteins (latent membrane protein 1 [LMP1], LMP2A, and LMP2B) are expressed in lymphoblastoid cell lines (45). Among them, LMP1 is most extensively studied and is well established to be an oncogenic protein. LMP1 is a 386-amino-acid (386-aa) viral protein with six transmembrane domains and both its amino and carboxyl tails facing the cytoplasm (Fig. 1A). When overexpressed in fibroblasts and epithelial cells, LMP1 could transform these cells (3, 16, 50). When specifically introduced into epidermis and lymphocytes in transgenic mice, the mice displayed epithelial hyperplasia and an increased incidence for lymphoma, respectively (31, 52). In addition, a recombinant EBV with a truncated LMP1 fails to transform resting human B cells in vitro (26, 27). Thus, to understand the molecular mechanisms underlying the EBV-associated pathogenesis, it is crucial for us to first understand the impact of LMP1 in host cells.
FIG. 1.
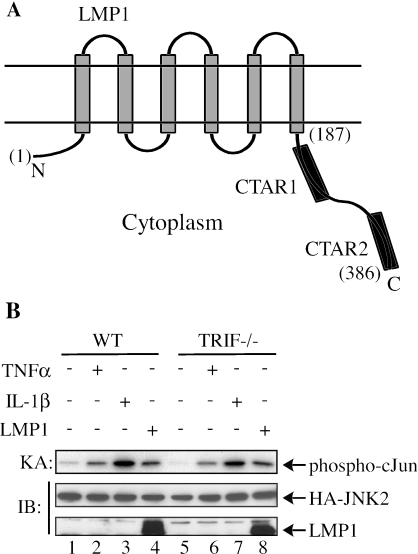
TRIF is not essential in the LMP1-mediated JNK pathway. (A) Schematic representation of LMP1. N and C indicate the amino and carboxyl termini, respectively. The two horizontal parallel lines represent the lipid bilayer of the plasma membrane. The numbers in parenthesis indicate the positions of amino acids. (B) The wild-type (WT) and TRIF−/− MEFs were separately cotransfected with HA-JNK2, with either an empty vector or LMP1. Before harvest, cells were either left untreated or treated with TNF-α or IL-1β (20 ng/ml for 10 min). HA-JNK2 was subjected to immune complex kinase assays (KA). IB, immunoblot.
Two subregions in the 200-aa (i.e., aa 187 to 386) cytoplasmic carboxyl tail of LMP1, namely, the C-terminal activating region 1 (CTAR1) and CTAR2 (Fig. 1A), play important roles in LMP1-mediated cell transformation and signaling (15, 40, 48). CTAR1 contains a typical tumor necrosis factor (TNF) receptor (TNFR)-associated factor (TRAF)-binding motif; this motif is required for CTAR1 binding to TRAF1, -2, -3, and -5, which are members of an important family of proteins involved in cytokine signaling (7, 15, 40, 48). CTAR1 is capable of activating the phosphatidylinositide-3 kinase/Akt-mediated pathway and, to a lesser extent, the NF-κB pathway (11, 20, 39). In a few selected cell types where TRAF1 is expressed, CTAR1 is also capable of moderately activating the c-Jun N-terminal kinase (JNK) pathway (13). In contrast, CTAR2 is known to be responsible for the majority of the JNK and NF-κB activity induced by LMP1 (15, 40, 48). CTAR2 was found to interact with TNFR-associated death domain protein (TRADD) and receptor-interacting protein, two key proteins indispensable for the TNF-α-mediated NF-κB and JNK pathways (18, 22, 24). In addition, overexpression of the “dominant-negative” TRADD or TRAF2 was found to inhibit the LMP1-induced JNK and NF-κB pathways (12, 23, 25, 29). Thus, LMP1 was previously thought to functionally mimic members of the TNFR superfamily in signaling (15, 40, 48). However, several recent reports argued against a role for TRAF2 and TRADD in the LMP1-mediated JNK and NF-κB pathways (29, 37, 49, 54). Using cells derived from different knockout mice and the small interference RNA (siRNA) technique, we recently showed that the LMP1-mediated JNK pathway is distinct from that utilized by members of the TNFR superfamily as LMP1 does not require TRADD, TRAF2, and receptor-interacting protein to activate JNK (49). Instead, LMP1 selectively engages TRAF6, TAB1/TAK1, and JNKK1/2 to activate JNK (49). Although members of the interleukin-1 receptor (IL-1R)/Toll-like receptor superfamily also selectively utilize TRAF6/TAB1/TAK1 to activate JNK (1), LMP1 differs from them in that LMP1 does not require myeloid differentiation factor 88, IL-1 receptor-associated kinase 1 (IRAK1), and IRAK4 to engage TRAF6 (49). As LMP1 does not seem to directly interact with TRAF6 (49), it remains unclear how LMP1 transmits signal to TRAF6.
BS69, a multidomain-containing (i.e., PHD, Bromo, PWWP, and MYND) cellular protein (Fig. 2A), was originally identified as an adenoviral early region 1A (E1A)-interacting protein and shown to inhibit the E1A-mediated transcription (17, 38). The carboxyl MYND domain of BS69 is predicted to adopt a two-zinc-finger-like structure and shown to interact with several target molecules containing the PXLXP motif (2). BRAM1, an alternatively spliced variant of BS69 (Fig. 2A), retains the full MYND domain and is shown to interact with the BMP receptor 1A (32).
FIG. 2.
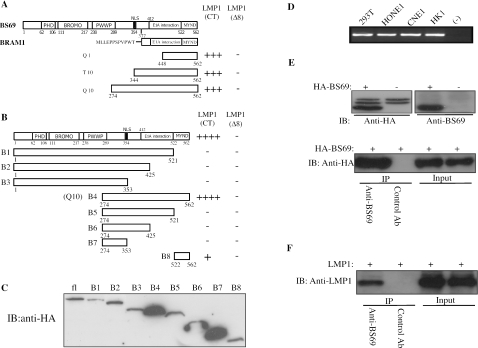
BS69 interacts with LMP1 in yeast and mammalian cells. (A and B) Schematic representation of BS69, BRAM1, and various truncated fragments of BS69. The numbers underneath each bar indicate the positions of amino acids in BS69. NLS, nuclear localization sequence; CT, carboxyl terminus. The interaction of full-length BS69 and its truncated derivatives with either LMP1(CT) or LMP1(Δ8) was analyzed by yeast two-hybrid assays, and the results are summarized next to each construct on the right. Q1, T10, and Q10 were three BS69 fragments isolated from our yeast two-hybrid screen. ++++ and +++ represent the appearance of yeast colonies in 3 and 5 days, respectively, after being streaked on QDO plates. ++ and + indicate the appearance of yeast colonies in 3 and 5 days, respectively, on TDO plates (these clones did not grow on QDO plates). −, no growth on either QDO or TDO plates. (C) The expression levels of different BS69 fragments in yeast were detected by immunoblotting. fl, full-length. (D) Total RNA was extracted from 293T, HONE1, CNE1, and HK1 cells; an equal amount of RNA was subjected to RT-PCR analysis. (−), negative control without the reverse transcriptase. (E) 293T cells were either mock transfected or transfected with HA-BS69. (Top) WCEs were subjected to direct immunoblot analysis. (Bottom) WCEs were immunoprecipitated separately with either the anti-BS69 antibody or a control antibody (Ab), followed by immunoblotting with an anti-HA antibody. (F) 293T cells were transfected with LMP1. Cells were cross-linked with dithiobis(succinimidylpropionate) for 5 min before harvest. The lysates were immunoprecipitated with either the anti-BS69 antibody or a control antibody, followed by immunoblotting with the anti-LMP1 antibody.
We show here that BS69, but not BRAM1, serves as a specific adaptor directly linking LMP1 and TRAF6. A fraction of LMP1 and BS69 constitutively colocalize in membrane lipid rafts. Furthermore, BS69 is specifically required for LMP1-mediated JNK activation.
MATERIALS AND METHODS
Cell lines, DNA constructs, and reagents.
293T, HeLa, and TRIF−/− (TRIF is TIR domain-containing adaptor protein inducing beta interferon) mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100-U/ml penicillin, and 100-μg/ml streptomycin in a 37°C incubator with 5% CO2. LMP1, HA-BS69, and HA-BRAM1 were described previously (6, 38). Both the full-length and the truncated BS69 cDNA fragments were inserted in frame into pGADT7 (BD Biosciences) to generate BS69-AD fusion constructs. Yeast bait constructs were constructed by inserting the PCR fragments encoding either the entire carboxyl terminus of LMP1(CT) (aa 187 to 386) or its truncation mutant without the carboxyl terminal 8 aa (aa 187 to 378) [i.e., LMP1(Δ8)] into pGBKT7 (BD Biosciences), respectively. Gal4-DNA-binding domain (DBD)-TRAF6 (Gal4-DBD-TRAF6) and Gal4-DBD-TRAF6(C) were generated by inserting the PCR fragments encoding either the full-length or TRAF(C) domain (aa 351 to 522) of TRAF6 into pGBKT7, respectively. Xpress-tagged TRAF6 (xp-TRAF6) was constructed by inserting the cDNA fragments into pcDNA3.1c. The vector-based BS69 small hairpin RNAs (shRNAs) were constructed by inserting the following two double-stranded oligonucleotides into the pSuper vector between BglII and HindIII sites (5): (i) human BS69 (N terminus, bp 196 to 214) forward, 5′ GATCCCCTGCCATTTGCCTGGAGAGGTTCAAGAGACCTCTCCAGGCAAATGGCATTTTTGGAAA; (ii) human BS69 (C terminus, bp 1437 to 1455) forward, 5′ GATCCCCCATGCAGGGTGAGATGGACTTCAAGAGAGTCCATCTCACCCTGCATGTTTTTGGAAA (both sense and antisense BS69 sequences are underlined). The LMP1 (1-186)-BS69 chimera was generated using fusion PCR by linking LMP1 (1-186) with the full-length HA-BS69, and then the PCR product was digested with EcoRI and BamHI and cloned into the PCMV5 vector. All constructs generated above were verified by DNA sequencing. TNF-α and IL-1β were purchased from R&D Systems.
Transfection and cell lysis.
Cells were transfected with various plasmids using either Lipofectamine Plus reagents (for 293T and HeLa cells) or Lipofectamine 2000 (Invitrogen) (for MEF cells) according to the manufacturer's instruction. Twenty-four hours after transfection, the cells were lysed in lysis buffer (50 mM HEPES [pH 7.6], 10% glycerol, 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 100 mM NaF, 20 mM p-nitrophenyl phosphate, 20 mM β-glycerol phosphate, 2 mM dithiothreitol, 50 μM sodium vanadate, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstatin), followed by removal of insoluble debris with a bench-top centrifuge to obtain whole-cell extracts (WCEs).
Antibodies.
Mouse monoclonal antibodies to HA (Santa Cruz), JNK (BD Biosciences), β-tubulin (Sigma), and TRAF6 (Santa Cruz); rabbit polyclonal antibodies to Xpress, caveolin-1 (Santa Cruz), phospho-p38, phospho-JNK, and total p38 (Cell Signaling); and the goat polyclonal antibody to TRADD (Santa Cruz) were used in this study. Monoclonal anti-LMP1 was described previously (6). A rabbit polyclonal antibody to BS69 was raised with an amino-terminal region (aa 1 to 265) of BS69 as an antigen.
Coimmunoprecipitation assays.
293T cells were cotransfected with various plasmids. Thirty-six hours after transfection, the cells were cross-linked with 20-μg/ml of dithiobis(succinimidylpropionate) (Pierce) for 10 min, followed by lysis in RIPA buffer (25 mM HEPES [pH 7.4], 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin). Protein A-Sepharose beads were incubated with 400 μg of extracts and 2 μg of appropriate antibodies for 2 h at 4°C. After being washed extensively with RIPA buffer, we eluted the bound proteins by boiling them and subjected them to SDS-polyacrylamide gel electrophoresis and immunoblotting.
Immune complex protein kinase assays.
For the JNK kinase assays, we followed the protocols as described previously (53).
Yeast two-hybrid screening.
We first transformed the yeast strain AH109 expressing Gal4-DBD-LMP1(CT) (aa 187 to 386) fusion protein with a mouse 17-day-embryo MATCHMAKER cDNA library (no. 638846; BD Biosciences) following the manufacturer's protocol. Half of the cells were plated on triple synthetic dropout plates (TDO) lacking leucine, tryptophan, and histidine and supplemented with 5 mM 3-amino-1,2,4-triazole. The other half was plated on quadruple synthetic dropout plates (QDO) lacking leucine, tryptophan, histidine, and adenine. A total of 180 colonies were obtained from primary screening. Yeast fish plasmid DNA were then purified, transformed into Escherichia coli DH5α, and recovered on LB-agar plates containing 50 μg/ml ampicillin. Distinctive plasmids (judged by insert size and restriction enzyme digestion pattern) were separately retransformed into AH109 containing either pGBKT7 (empty bait vector), pGBKT7-p53 (negative control), or pGBKT7-LMP1(CT) to confirm their specific interaction with LMP1(CT). Thirty-two LMP1(CT)-specific clones were subjected to DNA sequencing to reveal their identities.
Isolation of lipid rafts by sucrose gradient centrifugation.
Lipid rafts were isolated by sucrose gradient centrifugation as described by Yasui et al. with some modifications (56). Briefly, 2 × 107 cells were lysed on ice for 30 min in 0.5 ml of TENT buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 20 mM p-nitrophenyl phosphate, 20 mM β-glycerol phosphate, 2 mM dithiothreitol, 50 μM sodium vanadate, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin). Cell lysates were then mixed with 0.5 ml of 90% ice-cold sucrose in TENT. One milliliter of the mixture was placed at the bottom of a centrifuge tube and overlaid with 1 ml (each) of 30% and 5% sucrose in TENT. After centrifugation in a Hitachi preparative ultracentrifuge (Himac CP80MX) with a Sorvall TST 60.4 rotor at 170,000 × g at 4°C for 18 h, 0.3-ml fractions were aspirated from the top of the gradient and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting.
Semiquantitative RT-PCR.
Total RNA was extracted from cells using TRI REAGENT (Molecular Research Center, Inc., Cincinnati, Ohio). Reverse transcription-PCR (RT-PCR) was performed as previously described by using AmpliTaq Gold (Applied Biosystems) (44). The PCR program started with an initial denaturation at 95°C for 10 min, followed by 40 cycles (each consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s) of amplification, and ended with a final extension at 72°C for 10 min. The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as a control with only 25 cycles of amplification. The sequences of the primers used are as follows: BS69 (forward), 5′ GTCTCGAGTCCACGGTATG; BS69 (reverse), 5′ AACACCTCTCCAGGCAAATG; GAPDH (forward), 5′ ATCTCTGCCCCCTCTGCTGA; GAPDH (reverse), 5′ GGATGACCTTGCCCACAGCC.
RESULTS
TRIF is not involved in the LMP1-mediated JNK pathway.
To look for the missing adaptor that may bridge TRAF6 and LMP1, we first focused on a few known TRAF6-interacting proteins including IRAK1, IRAK4, and TRIF (1). As we had already shown that neither IRAK1 nor IRAK4 were involved in the LMP1-mediated JNK pathway (49), we then turned to TRIF, a TRAF6-interacting adaptor molecule mainly involved in toll-like receptor 3/4 signaling (46, 55). The wild-type and TRIF−/− MEFs were separately cotransfected with HA-JNK together with either an empty vector or LMP1. As expected, both the TNF-α- and IL-1β-mediated JNK activation was not affected in TRIF−/− cells (Fig. 1B) (55). Similarly, LMP1 also activated JNK in cells with or without TRIF (Fig. 1B, lanes 4 and 8). This suggests that TRIF is unlikely the adaptor to bridge LMP1 and TRAF6.
BS69 interacts with LMP1 in both yeast and mammalian cells.
Next, we performed a yeast two-hybrid screening using the cytoplasmic carboxyl tail of LMP1 (i.e., aa 187 to 386) as bait. Among the 32 positive clones we identified, 12 of them encoded three different but overlapping carboxyl fragments of BS69, a multidomain-containing cellular protein (Fig. 2A) (38). Interestingly, none of the three BS69 fragments interacted with a control LMP1 bait missing the carboxyl-terminal eight amino acids [i.e., LMP1(Δ8)] by yeast two-hybrid assays, suggesting that the carboxyl terminal eight amino acids are necessary for LMP1 binding to BS69.
To map the minimal region on BS69 that binds to LMP1, different truncated BS69 fragments were generated and tested in the yeast two-hybrid assays. Although two longer BS69 fragments with intact carboxyl termini (i.e., full-length and B4) interacted well with LMP1, a short BS69 fragment consisting of only the carboxyl-terminal MYND domain (i.e., B8) also weakly interacted with LMP1 (Fig. 2B). In contrast, all other BS69 fragments without the carboxyl terminal MYND domain (i.e., B1 to B3 and B5 to B7) completely failed to interact with LMP1 (Fig. 2B). As a control, we showed that all BS69 constructs were expressed in yeast cells (Fig. 2C). As several cellular proteins contain the MYND domain, to test whether the MYND of BS69 specifically interacts with LMP1, we also tested the interaction of LMP1 with BLU, an MYND-containing tumor suppressor implicated in several cancers including NPC (44). No such interaction was detected by yeast two-hybrid assays (our unpublished data). Thus, our data above indicate that the MYND domain of BS69 is necessary and sufficient to specifically interact with LMP1.
BS69 was previously shown to be expressed in a few tissues previously examined (17). To find out whether BS69 is expressed in 293T cells and several NPC-derived cell lines (i.e., HONE1, CNE1, and HK1), total RNA was extracted from these cell lines, and RT-PCR was performed. As shown in Fig. 2D, BS69 mRNA was expressed in 293T, HONE1, CNE1, and HK1 cells.
Next, we tested whether LMP1 interacts with the endogenous BS69 in mammalian cells. To facilitate our study, we first raised a polyclonal BS69 antibody recognizing a region at its amino terminus. Although the antibody could not detect the endogenous BS69 in 293T whole-cell extracts in Western blotting, it did recognize the transfected BS69 and could effectively immunoprecipitate it (Fig. 2E). We then transfected LMP1 into 293T cells and prepared the whole-cell extracts. We subjected the whole-cell extracts to immunoprecipitation with either the anti-BS69 antibody or a control antibody. We showed that LMP1 was only coprecipitated by the anti-BS69 antibody but not by the control antibody (Fig. 2F).
A fraction of LMP1 and BS69 constitutively colocalizes in membrane lipid rafts.
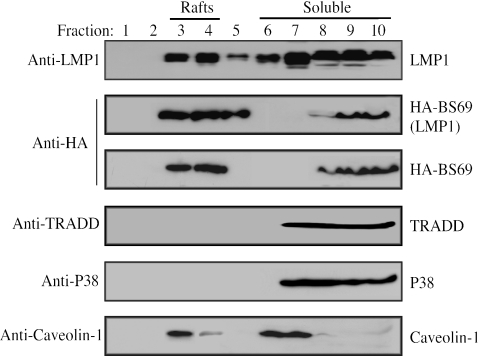
It has been well established that a fraction of LMP1 resides in membrane lipid rafts (9, 10, 19, 28). We then examined whether BS69 is localized to lipid rafts or not with or without a cotransfected LMP1. As shown in Fig. 3, we confirmed that a fraction of LMP1 was indeed present in the lipid raft fractions, as indicated by the presence of caveolin-1, a known resident of lipid rafts (35). Interestingly, with or without LMP1, a fraction of BS69 was constitutively present in lipid raft fractions (Fig. 3, panels 2 and 3). As a negative control, we showed that neither TRADD nor p38 mitogen-activated protein kinase was present in the lipid raft fractions, in agreement with previous reports (19).
FIG. 3.
A fraction of BS69 is localized in membrane lipid rafts. 293T cells were separately transfected with LMP1, HA-BS69, or both. Cells were extracted in TENT buffer, and the lysates were subjected to ultracentrifugation in a sucrose gradient. Samples from each fraction were subjected to immunoblot analysis with various primary antibodies as indicated.
BS69 interacts with TRAF6 in yeast and mammalian cells.
We next checked whether BS69 interacts with TRAF6. We first tested whether different BS69 truncation clones (i.e., those used in the experiments shown in Fig. 2B) interacted with either the full-length TRAF6 or its carboxyl-terminal TRAF domain [i.e., TRAF6(C), aa 351 to 522] in yeast two-hybrid assays. As shown in Fig. 4A, all BS69 fragments containing amino acids 274 to 353 bound to TRAF6, with the full-length BS69 and B4 showing the strongest interaction (as manifested by the shorter time it took for yeast cells to grow in quadruple dropout plates). In contrast, those BS69 fragments (i.e., Q1, T10, and B8) missing the region (aa 274 to 353) failed to interact with TRAF6. Furthermore, we found that the full-length BS69 and B4 interacted with TRAF6(C) (Fig. 4A), suggesting that the carboxyl TRAF domain of TRAF6 interacts with BS69. The fact that those BS69 fragments missing the carboxyl terminus interacted only with the full-length TRAF6 but not TRAF6(C) suggested that both the amino terminus of TRAF6 and the carboxyl terminus of BS69 facilitate the interaction between TRAF6(C) and the region in BS69 spanning amino acids 274 to 353.
FIG. 4.
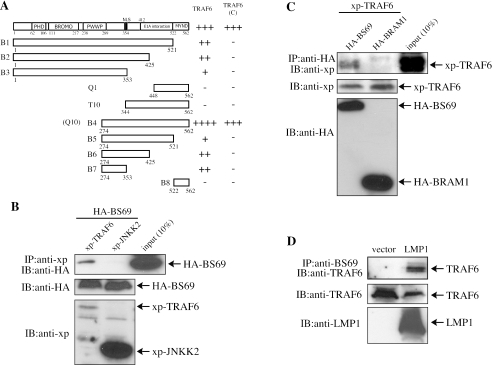
BS69 interacts with TRAF6 in both yeast and mammalian cells. (A) The schematic representation of the full-length and truncated BS69 and a summary of their interaction with either the full-length or the TRAF(C) domain of TRAF6 in yeast. The definitions for the plus and minus signs are the same as that in the legend to Fig. 2. (B to D) 293T cells were cotransfected with various plasmids as indicated. WCEs were immunoprecipitated with either the anti-xp (B), anti-HA (C), or anti-BS69 (D) antibodies, and the coprecipitated proteins were detected by immunoblotting.
To further confirm whether BS69 interacts with TRAF6 in mammalian cells, we cotransfected 293T cells with HA-BS69 together with either an xp-TRAF6 or JNKK2 (control). HA-BS69 was specifically coimmunoprecipitated with TRAF6 but not JNKK2 (Fig. 4B). Based on our yeast two-hybrid results, BRAM1, the splice variant of BS69, was not expected to interact with TRAF6, due to its lack of the region spanning amino acids 274 to 353 (Fig. 2A). To test whether this was the case in mammalian cells, we cotransfected 293T cells with xp-TRAF6 together with either BS69 or BRAM1. Indeed, xp-TRAF6 was specifically coprecipitated by BS69 but not by BRAM1 (Fig. 4C). We next tested whether the endogenous BS69 and TRAF6 interacted with each other. Interestingly, we found that the endogenous BS69 and TRAF6 did not significantly interact with each other in cells without LMP1. However, in the presence of LMP1, BS69 indeed formed a stable complex with TRAF6 (Fig. 4D).
BS69 is required for LMP1 to recruit TRAF6.
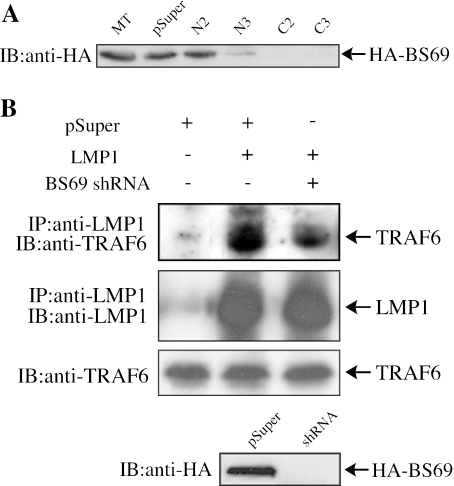
We previously showed that LMP1 can form a complex with the endogenous TRAF6 in mammalian cells (49). We next tested whether BS69 was essential for the formation of such a complex. We resorted to siRNA to knock down the endogenous BS69 by constructing two vector-based shRNAs, which target separate regions at the amino and carboxyl termini of BS69, respectively (5). Two shRNA clones targeting either end of BS69 were chosen with N3, C2, and C3 (the prefix N and C denoting the amino- and carboxyl-terminal shRNAs, respectively) containing the correct targeting sequences. In contrast, clone N2 contains 2-bp mutations due to errors generated during cloning. When different control and BS69-specific shRNAs were transfected together with HA-BS69 into 293T cells, N3, C2, and C3 shRNA efficiently reduced expression of HA-BS69 (Fig. 5A). In contrast, the empty vector and N2 had no obvious effect (Fig. 5A). When we examined BS69 mRNA by RT-PCR, we also found that N3, C2, and C3 but not N2 led to a significant decrease in BS69 mRNA levels (our unpublished data). To evaluate the role of BS69 in complex formation between LMP1 and TRAF6, we transfected LMP1 into 293T cells with or without a BS69-specific shRNA (C3). Although the endogenous TRAF6 was specifically coprecipitated by LMP1 in the absence of BS69 siRNA, less TRAF6 was coprecipitated by LMP1 in the presence of the BS69-specific shRNA (Fig. 5B). Our data suggest that the complex formation between LMP1 and TRAF6 is BS69 dependent.
FIG. 5.
BS69 is required for the complex formation between LMP1 and TRAF6. (A) The empty shRNA vector (pSuper) or different BS69 shRNA constructs were transfected to 293T cells twice, followed by transfection of an expression vector encoding HA-BS69. WCEs were subjected to immunoblotting with the anti-HA antibody. MT, mock transfected; N2, an ineffective BS69 shRNA clone with 2-bp mutations in its sequence; N3, the correct BS69 shRNA construct targeting a region in the amino terminus of BS69; C2 and C3, two correct clones of BS69 shRNA targeting a region in the carboxyl terminus of BS69. (B) 293T cells were first transfected twice with either pSuper or BS69 shRNA (C3), followed by transfection of an empty vector or LMP1. WCEs were first immunoprecipitated with the anti-LMP1 antibody; both the immunoprecipitated LMP1 and the coprecipitated TRAF6 were sequentially detected by immunoblotting.
BS69 is specifically required for LMP1-mediated JNK activation.
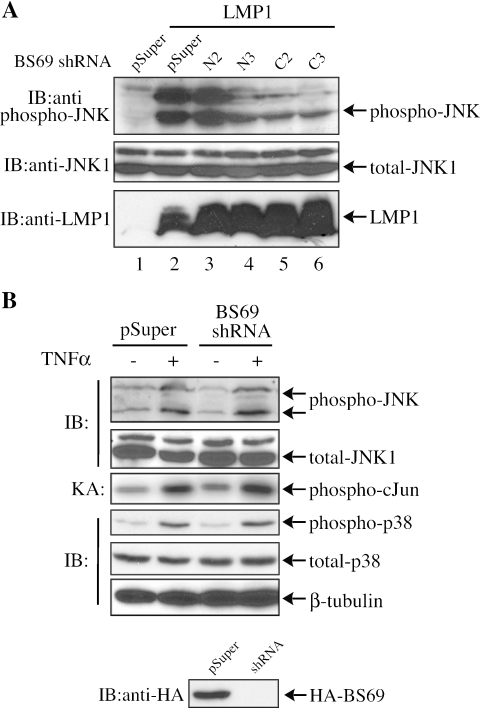
Since BS69 scaffolds both LMP1 and TRAF6, we next asked whether it plays any functional role in the LMP1-mediated JNK pathway. We cotransfected 293T cells with different control and BS69-specific shRNAs with or without LMP1. In the presence of either an empty vector (i.e., pSuper) or N2 (the noneffective mutated BS69 siRNA), the endogenous JNK was potently phosphorylated (i.e., activated), as measured by a specific antibody recognizing the dually phosphorylated JNK (Fig. 6A, lanes 2 and 3). However, in the presence of either N3, C2, or C3 (i.e., the effective BS69 shRNAs), the extent of JNK phosphorylation was significantly decreased (Fig. 6A, lanes 4 to 6). Similar results were also achieved in a direct JNK kinase assay (our unpublished data).
FIG. 6.
BS69 is specifically involved in the LMP1-meidated JNK pathway. (A) 293T cells were transfected with different shRNAs twice as indicated, followed by transfection of LMP1. WCEs were subjected to immunoblotting with both the anti-phospho-JNK and anti-total JNK antibodies. (B) 293T cells were transfected twice with pSuper or BS69 siRNA. Twenty-four hours after the last transfection, cells were treated with 20-ng/ml TNF-α for 10 min. WCEs were subjected to immunoblotting with anti-phospho-p38, anti-total-p38, anti-phospho-JNK, and anti-total JNK antibodies. Anti-β-tubulin was used here as a loading control. Endogenous JNK1 was also immunoprecipitated from WCEs and subjected to kinase assays (KA).
We then asked whether BS69 plays any role in the TNF-α-mediated mitogen-activated protein kinase pathways. 293T cells were transfected with either an empty vector (pSuper) or a BS69-specific shRNA (C3) with or without TNF-α treatment. As shown in Fig. 6B, neither the TNF-α-mediated JNK activation (panels 1 and 3) nor p38 activation (panel 4) was affected by the BS69-specific shRNA. Our data suggest that BS69 specifically functions in the LMP1-mediated JNK pathway.
Recruitment and oligomerization of BS69 are required for LMP1-induced JNK activation.
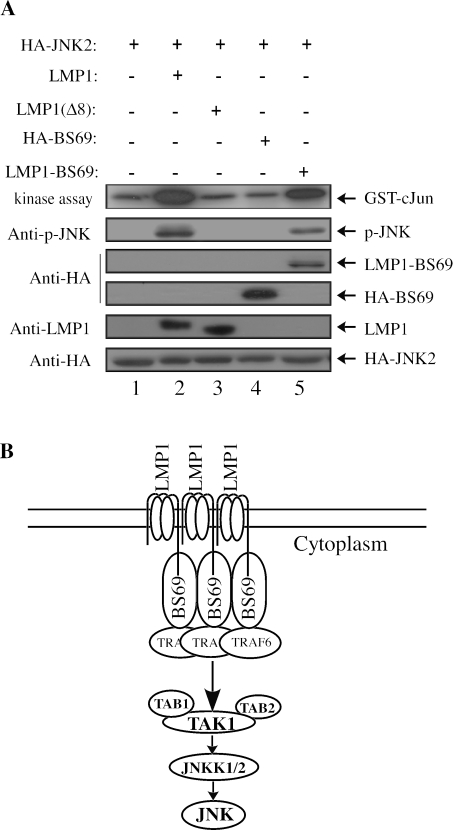
Previous studies by several groups including our own clearly showed that the CTAR2 domain is mainly responsible for LMP1-induced JNK activation (14, 30, 49). Our data above showed that a main role of CTAR2 in the LMP1-mediated JNK pathway is to recruit BS69. To test whether BS69 can physically and functionally replace CTAR2 in inducing JNK activation, we started with a mutant LMP1 without the entire cytoplasmic carboxyl tail [i.e., LMP1 (1-186)], which by itself is completely defective in JNK activation (14, 29). We then fused BS69 in frame to this mutant LMP1 (1-186). To test whether the chimeric protein could restore JNK activation, we transfected 293T cells with HA-JNK2, together with either LMP1, LMP1(Δ8), BS69, or the chimeric LMP1 (1-186)-BS69, as indicated in Fig. 7A. As expected, LMP1 potently activated JNK, whereas LMP1(Δ8) failed to do so (Fig. 7A, compare lanes 2 and 3). Overexpression of BS69 alone was not sufficient to activate JNK either (Fig. 7A, lane 4). Interestingly, the expression of the chimeric LMP1 (1-186)-BS69 in cells restored JNK activation (Fig. 7A, lane 5). This result further confirmed that BS69 participates in the LMP1-mediated JNK pathway and suggested that recruitment and oligomerization of BS69 are prerequisites for JNK activation by LMP1.
FIG. 7.
Fusion of BS69 to an LMP1 mutant without the cytoplasmic carboxyl terminus restores JNK activation. (A) 293T cells were cotransfected with HA-JNK, together with other expression vectors as indicated. HA-JNK was immunoprecipitated from cell lysates and subjected to in vitro kinase assays. Separately, a part of the cell lysates was also subjected to immunoblotting with various primary antibodies as indicated. (B) The key signal transducers in the LMP1-mediated JNK pathway and the flow of the signal are depicted in the cartoon. Although LMP1, BS69, and TRAF6 are each drawn as a trimer, we do not know for sure whether this is the case (except for TRAF6). We simply imply here that they all need to oligomerize to activate the JNK pathway.
DISCUSSION
BS69 but not BRAM1 bridges LMP1 and TRAF6 for JNK activation.
Although BS69 was identified a decade ago, its biological role remains largely unknown at present. So far, BS69 has been shown to interact with a few viral proteins (e.g., E1A and EBNA2) and cellular protein (e.g., c-Myb, Ets2, and EMSY) and may function as a transcriptional corepressor (2, 17, 21, 33, 38, 51). We now provide evidence showing that BS69 also serves an important role as an adaptor in viral protein-mediated intracellular signal transduction pathways. It is likely that LMP1 may contribute to EBV-mediated pathogenesis by interfering with the normal cellular function of BS69. Although BS69 already contains several known protein-protein interaction domains (Fig. 2A) (38), in this study, we identified another novel domain of BS69 (i.e., aa 274 to 353) involved in binding TRAF6 (Fig. 4). We also showed that the MYND domain of BS69 is indispensable for its interaction with the carboxyl terminus of LMP1, which is in agreement with a recent finding in which BRAM1, an alternatively spliced form of BS69, was also shown to interact with LMP1 (8). This is not unexpected, since both BS69 and BRAM1 contain the intact MYND domain. Although BRAM1 interacts with LMP1, we show that it does not interact with TRAF6 (Fig. 4). In HEK293 cells, BRAM1 mRNA was barely detected, as judged by RT-PCR with a forward primer annealing to the 5′ end of the gene encoding 12 aa unique to BRAM1 (32), whereas BS69 mRNA could be readily detected (our unpublished data). Thus, BRAM1 may function as a regulatory molecule to fine tune the BS69-mediated cellular signaling pathways in a cell type-dependent manner. We think that it is BS69 (but not BRAM1) that normally participates in the LMP1-initiated signaling pathways. This is further supported by the fact that N3, a BS69-specific shRNA targeting a region present only in BS69 but not in BRAM1, still effectively inhibits LMP1-induced JNK activation (Fig. 6B). Interestingly, although PXLXP and PXEXX (aromatic-acidic residue) have been defined as the consensus sequences in binding proteins for the MYND domain of BS69 and the TRAF(C) domain of TRAF6, respectively (2, 57), no such motifs are found in LMP1 and BS69. This suggests that the modes of BS69's interaction with both LMP1 and TRAF6 are unique in nature and remain to be structurally elucidated.
As LMP1 also requires TRAF6 to activate NF-κB (37), we initially hypothesized that BS69 is also involved in the LMP1-mediated NF-κB pathway. However, in experiments where the BS69-specific shRNA efficiently reduced the LMP1-mediated JNK activation, we failed to observe a consistent decrease in the LMP1-medited NF-κB activation (our unpublished data). This could be due to technical reasons, as NF-κB activation by LMP1 is difficult to detect by either the IκB kinase assays or the NF-κB-dependent luciferase reporter assays of several cell lines (e.g., HeLa or MEFs), except in 293T cells (our unpublished data). Differential expression of a key factor in different cell types may account for this. Alternatively, a scaffolding molecule other than BS69 is specifically involved in the LMP1-mediated NF-κB pathway. The exact role of BS69 in the LMP1-mediated NF-κB pathway remains to be further clarified.
Oligomerization of BS69 is a prerequisite for JNK activation by LMP1.
Protein oligomerization is an important yet recurring theme in various ligand-induced signal transduction pathways (36, 43, 47). Many receptors including the epidermal growth factor receptor, G-protein-coupled receptors, and TNF receptor form oligomers on cell membrane upon ligand binding, which initiates various downstream signal transduction pathways. Similarly, many intracellular signal transducers are capable of transmitting signals only after oligomerization. For example, members of the TRAF family proteins are known to function by oligomerization (4, 42). BS69, the adaptor in the LMP1-mediated JNK pathway, also seems to function by oligomerization, as expression of BS69 alone fails to activate JNK. When BS69 is covalently linked to the transmembrane domain of LMP1, which is known to promote LMP1 aggregation on cell membranes, the fusion protein significantly activates the JNK pathway (Fig. 7). This result tells us two things. First, oligomerization of BS69 is required to activate JNK. Considering the fact that BS69 recruits TRAF6 to activate JNK and that TRAF6 is known to function as oligomers (4), it is not difficult for us to understand why oligomerization of BS69 is needed. Second, the main function of the CTAR2 domain in the LMP1-mediated JNK pathway is to recruit BS69. Thus, a clear picture of the LMP1-mediated JNK pathway is emerging in which autonomous aggregation of LMP1 on host cell membrane recruits BS69 and facilitates its oligomerization. The oligomerized BS69 in turn recruits and promotes aggregation of TRAF6, eventually leading to TAK1 and JNK activation (Fig. 7B).
In the future, it would be conceivably beneficial to screen for small molecules that disrupt the interaction between LMP1 and BS69. These molecules could be therapeutically useful in interfering with the LMP1-mediated JNK pathway and in inhibiting EBV-mediated pathogenesis.
Acknowledgments
We thank R. Bernards for BS69 and BRAM1 expression vectors, G. Natoli for the TRAF6 expression vector, T. Roberts for the suggestion of the LMP1-BS69 chimera experiment, and Wanda Shen and Carol Wong for the production of BS69 antibodies.
This work was supported by grants from Hong Kong Research Grant Council (HKUST6129/04 M and HKUST3/03C to Z.W.) and by the Areas of Excellence scheme established under the University Grants Committee of the Hong Kong Special Administrative Region, China (project no. AoE/B-15/01).
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signaling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Ansieau, S., and A. Leutz. 2002. The conserved Mynd domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 277:4906-4910. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 4.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Chan, B. C., K. F. To, J. C. Pang, Y. F. Chung, K. W. Lo, J. H. Tong, D. W. Huang, P. L. Lim, and Y. L. Chui. 2002. Generation of monoclonal antibodies against Hong Kong nasopharyngeal carcinoma-associated Epstein-Barr virus latent membrane protein 1 (LMP1). Int. J. Cancer 102:492-498. [DOI] [PubMed] [Google Scholar]
- 7.Chung, J. Y., Y. C. Park, H. Ye, and H. Wu. 2002. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115:679-688. [DOI] [PubMed] [Google Scholar]
- 8.Chung, P. J., Y. S. Chang, C. L. Liang, and C. L. Meng. 2002. Negative regulation of Epstein-Barr virus latent membrane protein 1-mediated functions by the bone morphogenetic protein receptor IA-binding protein, BRAM1. J. Biol. Chem. 277:39850-39857. [DOI] [PubMed] [Google Scholar]
- 9.Clausse, B., K. Fizazi, V. Walczak, C. Tetaud, J. Wiels, T. Tursz, and P. Busson. 1997. High concentration of the EBV latent membrane protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virology 228:285-293. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, W. F., III, T. R. Geiger, and J. M. Martin. 2003. Transmembrane domains 1 and 2 of the latent membrane protein 1 of Epstein-Barr virus contain a lipid raft targeting signal and play a critical role in cytostasis. J. Virol. 77:3749-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos, A. G., S. M. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 73:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eliopoulos, A. G., E. R. Waites, S. M. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 15.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435-444. [DOI] [PubMed] [Google Scholar]
- 16.Fahraeus, R., L. Rymo, J. S. Rhim, and G. Klein. 1990. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature 345:447-449. [DOI] [PubMed] [Google Scholar]
- 17.Hateboer, G., A. Gennissen, Y. F. Ramos, R. M. Kerkhoven, V. Sonntag-Buck, H. G. Stunnenberg, and R. Bernards. 1995. BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. EMBO J. 14:3159-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi, M., K. M. Izumi, and E. Kieff. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. USA 98:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 21.Hughes-Davies, L., D. Huntsman, M. Ruas, F. Fuks, J. Bye, S. F. Chin, J. Milner, L. A. Brown, F. Hsu, B. Gilks, T. Nielsen, M. Schulzer, S. Chia, J. Ragaz, A. Cahn, L. Linger, H. Ozdag, E. Cattaneo, E. S. Jordanova, E. Schuuring, D. S. Yu, A. Venkitaraman, B. Ponder, A. Doherty, S. Aparicio, D. Bentley, C. Theillet, C. P. Ponting, C. Caldas, and T. Kouzarides. 2003. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 115:523-535. [DOI] [PubMed] [Google Scholar]
- 22.Izumi, K. M., E. D. Cahir McFarland, A. T. Ting, E. A. Riley, B. Seed, and E. D. Kieff. 1999. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol. Cell. Biol. 19:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaye, K. M., O. Devergne, J. N. Harada, K. M. Izumi, R. Yalamanchili, E. Kieff, and G. Mosialos. 1996. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc. Natl. Acad. Sci. USA 93:11085-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye, K. M., K. M. Izumi, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J. Virol. 69:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaykas, A., K. Worringer, and B. Sugden. 2001. CD40 and LMP-1 both signal from lipid rafts but LMP-1 assembles a distinct, more efficient signaling complex. EMBO J. 20:2641-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieser, A., C. Kaiser, and W. Hammerschmidt. 1999. LMP1 signal transduction differs substantially from TNF receptor 1 signaling in the molecular functions of TRADD and TRAF2. EMBO J. 18:2511-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 16:6478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulwichit, W., R. H. Edwards, E. M. Davenport, J. F. Baskar, V. Godfrey, and N. Raab-Traub. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. USA 95:11963-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurozumi, K., M. Nishita, K. Yamaguchi, T. Fujita, N. Ueno, and H. Shibuya. 1998. BRAM1, a BMP receptor-associated molecule involved in BMP signaling. Genes Cells 3:257-264. [DOI] [PubMed] [Google Scholar]
- 33.Ladendorff, N. E., S. Wu, and J. S. Lipsick. 2001. BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-Myb. Oncogene 20:125-132. [DOI] [PubMed] [Google Scholar]
- 34.Lee, A. W., W. Foo, O. Mang, W. M. Sze, R. Chappell, W. H. Lau, and W. M. Ko. 2003. Changing epidemiology of nasopharyngeal carcinoma in Hong Kong over a 20-year period (1980-99): an encouraging reduction in both incidence and mortality. Int. J. Cancer 103:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Legler, D. F., O. Micheau, M. A. Doucey, J. Tschopp, and C. Bron. 2003. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFalpha-mediated NF-κB activation. Immunity 18:655-664. [DOI] [PubMed] [Google Scholar]
- 36.Lemmon, M. A., and J. Schlessinger. 1994. Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci. 19:459-463. [DOI] [PubMed] [Google Scholar]
- 37.Luftig, M., E. Prinarakis, T. Yasui, T. Tsichritzis, E. Cahir-McFarland, J. Inoue, H. Nakano, T. W. Mak, W. C. Yeh, X. Li, S. Akira, N. Suzuki, S. Suzuki, G. Mosialos, and E. Kieff. 2003. Epstein-Barr virus latent membrane protein 1 activation of NF-κB through IRAK1 and TRAF6. Proc. Natl. Acad. Sci. USA 100:15595-15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masselink, H., and R. Bernards. 2000. The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene 19:1538-1546. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosialos, G. 2001. Cytokine signaling and Epstein-Barr virus-mediated cell transformation. Cytokine Growth Factor Rev. 12:259-270. [DOI] [PubMed] [Google Scholar]
- 41.Moss, D. J., C. Schmidt, S. Elliott, A. Suhrbier, S. Burrows, and R. Khanna. 1996. Strategies involved in developing an effective vaccine for EBV-associated diseases. Adv. Cancer Res. 69:213-245. [DOI] [PubMed] [Google Scholar]
- 42.Park, Y. C., V. Burkitt, A. R. Villa, L. Tong, and H. Wu. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398:533-538. [DOI] [PubMed] [Google Scholar]
- 43.Pawson, T., and P. Nash. 2000. Protein-protein interactions define specificity in signal transduction. Genes Dev. 14:1027-1047. [PubMed] [Google Scholar]
- 44.Qiu, G. H., L. K. Tan, K. S. Loh, C. Y. Lim, G. Srivastava, S. T. Tsai, S. W. Tsao, and Q. Tao. 2004. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene 23:4793-4806. [DOI] [PubMed] [Google Scholar]
- 45.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 46.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304-4310. [DOI] [PubMed] [Google Scholar]
- 47.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 48.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 49.Wan, J., L. Sun, J. W. Mendoza, Y. L. Chui, D. P. Huang, Z. J. Chen, N. Suzuki, S. Suzuki, W. C. Yeh, S. Akira, K. Matsumoto, Z. G. Liu, and Z. Wu. 2004. Elucidation of the c-Jun N-terminal kinase pathway mediated by Estein-Barr virus-encoded latent membrane protein 1. Mol. Cell. Biol. 24:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 51.Wei, G., A. E. Schaffner, K. M. Baker, K. C. Mansky, and M. C. Ostrowski. 2003. Ets-2 interacts with co-repressor BS69 to repress target gene expression. Anticancer Res. 23:2173-2178. [PubMed] [Google Scholar]
- 52.Wilson, J. B., W. Weinberg, R. Johnson, S. Yuspa, and A. J. Levine. 1990. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 61:1315-1327. [DOI] [PubMed] [Google Scholar]
- 53.Wu, Z., J. Wu, E. Jacinto, and M. Karin. 1997. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol. Cell. Biol. 17:7407-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie, P., B. S. Hostager, and G. A. Bishop. 2004. Requirement for TRAF3 in signaling by LMP1 but not CD40 in B lymphocytes. J. Exp. Med. 199:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 56.Yasui, T., M. Luftig, V. Soni, and E. Kieff. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc. Natl. Acad. Sci. USA 101:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye, H., J. R. Arron, B. Lamothe, M. Cirilli, T. Kobayashi, N. K. Shevde, D. Segal, O. K. Dzivenu, M. Vologodskaia, M. Yim, K. Du, S. Singh, J. W. Pike, B. G. Darnay, Y. Choi, and H. Wu. 2002. Distinct molecular mechanism for initiating TRAF6 signaling. Nature 418:443-447. [DOI] [PubMed] [Google Scholar]