Abstract
Herpes simplex virus mutants lacking the γ134.5 gene are not destructive to normal tissues but are potent cytolytic agents in human tumor cells in which the activation of double-stranded RNA-dependent protein kinase (PKR) is suppressed. Thus, replication of a Δγ134.5 mutant (R3616) in 12 genetically defined cancer cell lines correlates with suppression of PKR but not with the genotype of RAS. Extensive analyses of two cell lines transduced with either dominant negative MEK (dnMEK) or constitutively active MEK (caMEK) indicated that in R3616 mutant-infected cells dnMEK enabled PKR activation and decreased virus yields, whereas caMEK suppressed PKR and enabled better viral replication and cell destruction in transduced cells in vitro or in mouse xenografts. The results indicate that activated MEK mediates the suppression of PKR and that the status of MEK predicts the ability of Δγ134.5 mutant viruses to replicate in and destroy tumor cells.
The studies described in this report stem from attempts to use avirulent mutants of herpes simplex virus type 1 (HSV-1) to destroy cancer cells in situ in inoperable human tumors. The basis for the selection of these mutants for cytoreductive therapy of cancer is the ability of genetically engineered mutants to destroy tumor cells without accompanying destruction of normal cells. Of the many mutants tested, the most promising to date are mutants based on the deletion of the γ134.5 gene. The product of this gene, the infected cell protein (ICP) 34.5, is a multifunctional protein whose most preeminent function is to block a major host response to infection. In brief, after the onset of viral DNA synthesis, infected cells accumulate large amounts of complementary viral RNA transcripts (29, 31). The consequence of this accumulation is the activation of double-stranded RNA-dependent protein kinase (PKR). In infected cells, activated PKR phosphorylates the α-subunit of the eukaryotic translation initiation factor 2 (eIF-2α), resulting in total shutoff of protein synthesis (30). Most viruses have evolved mechanisms to block this pathway of host defense (20). In the case of HSV-1, ICP34.5 acts as a phosphatase accessory factor to recruit protein phosphatase 1α to dephosphorylate eIF-2α (25). As a consequence, protein synthesis continues unimpeded in cells in which the PKR pathway is impaired (10, 33). Mutants derived from Δγ134.5 viruses are highly attenuated in animal model systems, and phase I clinical studies have demonstrated that Δγ134.5 mutants can be administered safely at escalating doses in patients with malignancy (35, 42). However, a major impediment to the widespread use of these mutants for cancer therapy is the observation that in animal model systems human tumor cells differ widely with respect to their ability to support the replication of Δγ134.5 mutants (2, 6, 12, 15, 38). The objective of the studies reported herein was to define the tumor genotype that confers susceptibility to Δγ134.5 mutant viruses. Relevant to this report are the following:
(i) PKR appears to play a key role in conferring resistance to Δγ134.5 mutants. The importance of PKR to a cell's innate antiviral response to viral infection is underscored by the observation that Δγ134.5 mutants replicate to near-wild-type levels in murine embryonal fibroblast (MEF) cells derived from mice lacking PKR. Moreover, Δγ134.5 mutants are virulent in PKR−/− mice, but not in wild-type mice (33). In addition, exogenous alpha interferon effectively suppresses Δγ134.5 mutant replication in PKR+/+ MEFs but has no effect in PKR−/− MEFs, while wild-type HSV-1 was reported to be resistant to the antiviral effects of interferon in these cells (9, 10). Therefore, replication of mutants lacking γ134.5 is largely dependent on the ability of cells to activate PKR-dependent pathways of host cell defense.
(ii) PKR also exerts potent growth-suppressive effects and apoptotic cell death induced by multiple stimuli (16, 48). Alternatively, inhibition of PKR function by overexpression of catalytically inactive mutants of PKR and eIF-2α transforms NIH 3T3 cells (4, 5, 36) as well as primary human cells when coexpressed with large T antigen and human telomerase reverse transcriptase, in a manner similar to the necessary mitogenic signal transmitted by activated RAS (23, 39).
(iii) Transformation of NIH 3T3 cells by oncogenic RAS activators significantly increases the permissiveness of this mouse fibroblast cell line to wild-type HSV-1, resulting in decreased phosphorylation of PKR and eIF-2α compared with infection of untransformed NIH 3T3 cells (19). In addition, growth factor withdrawal also induces PKR activation, eIF-2α phosphorylation, and apoptosis in several growth factor-dependent hematopoetic cell lines (28). Growth factor withdrawal also downregulates the activity of mitogen-activated protein kinase (MAPK) kinase (MEK), a critical downstream RAS effector kinase, while overexpression of constitutively active MEK (caMEK) mutants protects growth factor-dependent cell lines from multiple apoptotic stimuli, including growth factor withdrawal (32, 46, 47).
MEK is a key regulatory kinase activated by MAPK kinase kinases (A-RAF, B-RAF, and C-RAF) that functions to promote cell survival (3, 47, 49). Accordingly, MEK and its only known substrate, MAPKs (ERK1 and ERK 2), are constitutively activated in a large percentage of tumors as a consequence of dysregulated growth factor secretion (41), tyrosine kinase receptor activation (26), activating mutations in RAS isoforms, and somatic activating missense mutations of B-RAF (17). It is noteworthy in this regard that pharmacologic inhibition of MEK is currently under intense investigation for targeted therapy of tumors (44).
In light of the evidence that MEK plays a key role in blocking the pernicious effects of activated PKR, we sought to determine whether differential PKR activation correlated with the ability of tumor cells to support the replication of Δγ134.5 mutant viruses and, as well, the role of MEK kinase in regulating this process. We report that PKR activation is suppressed in a subset of cancer cells, thereby rendering them susceptible to viral replication and cytolysis by Δγ134.5 mutant virus R3616. Using pharmacologic inhibitors of MEK and catalytically active and inactive mutants of MEK, we show that constitutive MEK activity suppresses the viral activation of PKR. Finally, we propose that the status of MEK predicts the ability of tumor cells to support the replication and ultimate destruction of human tumors by Δγ134.5 mutant viruses.
MATERIALS AND METHODS
Molecular constructs.
caMEK- and dominant negative MEK-1 (dnMEK)-containing plasmids, designated pNC84 and pNC92, were provided by J. Charron (Quebec, Canada). Their construction is detailed elsewhere (34). Briefly, mutant cDNAs of human MEK-1 containing mutations on serine residues at positions 218 and 222 were changed to negatively charged aspartate residues. These mutations mimic the effect of phosphorylation at positions 218 and 222, resulting in constitutive activation of MEK-1 (MAPK kinase) function (27). In contrast, alanine substitutions at the same residues functionally blocks phosphorylation by upstream MAPK kinase kinases, resulting in downregulation of endogenous MAPK activity (34). The mutant MEK-1 cDNAs contain an in-frame FLAG epitope at the N terminus under the transcriptional control of a cytomegalovirus (CMV) promoter in the pCMV-Tag2b mammalian expression vector (QIAGEN Inc., Valencia, CA). Orientation and the cDNA insert sequence were confirmed by DNA sequencing.
Cell culture.
PC-3 and DU145 cells (human prostate cancer), Panc-1, BxPc3, and MiaPaCa2 cells (human pancreatic cancer), MCF7 and MDA-MB-231 cells (human breast cancer), DLD-1 and WiDr cells (colorectal cancer), Hep3B cells (human hepatoma), and Vero cells (green monkey kidney) were originally obtained from the American Type Culture Collection (Manassas, VA). The Huh7 hepatoma cell line was originally obtained from J. R. Wands (Harvard Medical School, Boston, MA). The HT1080 (human fibrosarcoma) cell line containing one wild-type and one oncogenic (Q61K) N-ras allele (1, 40) was also obtained from the American Type Culture Collection (Manassas, VA). HT1080 cells having lost the activated mutant N-ras allele were obtained from E. J. Stanbridge (Irvine, CA) and have been described previously and published as MCH603 (40). The above cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO/Invitrogen Corporation, Grand Island, NY)-10% fetal calf serum (Intergen, Purchase, NY)-1% penicillin-streptomycin at 37°C and 7% CO2.
Viruses.
HSV-1(F) is the prototype wild-type HSV-1 strain used in our laboratories (18). The derivation and properties of the recombinant virus R3616, which lacks both copies of the γ134.5 gene (11), and recombinant R2636, carrying the luciferase gene driven by the glycoprotein C (gC) promoter (gC-luc) in place of the γ134.5 gene, were reported elsewhere (37).
Construction of stable cell lines.
Plasmids, designated pNC84 and pNC92, containing the respective N-terminal FLAG-tagged (Asp218, Asp222 MEK-1) or (Ala218 and Ala222 MEK-1) cDNAs under the transcriptional control of a CMV promoter and the neomycin resistance gene were used to select for G418-resistant, FLAG-MEK-expressing clonal transfectants. Plasmids pNC84 and pNC92 were transfected into replicate cultures of HT1080 or MiaPaCa2 cells on 60-mm dishes using Superfect reagent (QIAGEN Inc., Valencia, CA). Briefly, 5 μg of plasmid DNA was diluted in 300 μl of serum- and antibiotic-free DMEM, complexed with Superfect reagent (20 μl) for 10 min at room temperature, and added to cells at 37°C for 6 h, after which medium was removed and replaced with DMEM containing 10% calf serum. After 24 h of incubation, the cells were harvested and suspended in 5 ml of DMEM containing 10% fetal calf serum, and 1 ml of this cellular suspension was grown on separate 100-mm dishes in a total volume of 10 ml of DMEM containing 10% calf serum supplemented with antibiotics and 800 μg/ml of G418 (Geneticin; Gibco BRL). Medium containing G418 was replaced every 4 days until approximately 2 weeks, when cell colonies were visible and could be selected for clonal expansion using sterile cloning cylinders as described previously (22). The level of FLAG-MEK expression was assessed by immunoblotting 20 μg of equilibrated lysates from isolated clones using a monoclonal antibody to the FLAG epitope (Sigma-Aldrich Co., St. Louis, MO). Clonal transfectants derived from the HT1080 parent cell line designated HT-caMEK and HT-dnMEK refer to clones HT 84-4 and HT 92-6, while clonal transfectants from the MiaPaCa2 parent cell line designated Mia-caMEK and Mia-dnMEK refer to clones Mia 84-9 and Mia 92-10.
Viral infection.
Cells were seeded onto 60-mm dishes at 1 × 106 cells per dish. The next day, cells were generally exposed to the viruses (1 or 10 PFU per cell) for 2 h at 37°C and then removed and replaced with medium containing 1% calf serum. The infection continued at 37°C for the length of time indicated for each experiment. Cells were either labeled for de novo protein synthesis, harvested for immunoblotting, or collected for assaying viral recovery on Vero cell monolayers as previously described (11).
[35S]methionine labeling.
For metabolic labeling experiments, at 11 h after infection cells were washed once in warm medium 199V containing 1% calf serum lacking methionine (Sigma Chemical Co., St. Lois, Mo.) and incubated for an additional hour in 199V methionine-free medium, after which cells were overlaid with medium 199V lacking methionine but supplemented with 100 μCi of [35S]methionine (specific activity, >1,000 Ci/mmol; Amersham Pharmacia Biotech) per ml and incubated for an additional 2 h. The cells were then harvested at 14 h after infection, solubilized in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 100 μM sodium orthovanadate, 1 μg leupeptin per ml, and 1 mM phenylmethylsulfonyl fluoride) and sonicated for 10 seconds, and insoluble material was clarified by centrifugation. Total protein from the supernatant was quantified by the Bradford method, and 20 μg of equilibrated protein was subjected to electrophoresis in denaturing 12% (vol/vol) polyacrylamide gels, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Bedford, MA), and subjected to autoradiography.
Immunoblotting.
Experiments to analyze the accumulation of viral proteins and phosphorylation of ERK, PKR, and eIF-2α were performed on whole-cell lysates harvested on ice at either 12 or 14 h after infection with lysis buffer, sonicated for 10 seconds, and clarified by centrifugation. Total protein from the supernatant was quantified by the Bradford method (Bio-Rad Laboratories, Hercules, CA), and 20 μg of equilibrated protein was subjected to electrophoresis in 12% or 7.5% (vol/vol) denaturing polyacrylamide gels, transferred to PVDF membranes (Millipore Corporation, Bedford, MA), blocked, and reacted with primary antibody followed by appropriate secondary antibody.
Antibodies.
Polyclonal antibodies to the total and phosphorylated forms of PKR (Thr446), eIF-2α (Ser51), and ERK (Thr202/Tyr204) were purchased from Cell Signaling Technology (Beverly, MA). Monoclonal antibody to the FLAG epitope (clone M2) was purchased from Sigma-Aldrich (St. Louis, MO). Polyclonal antibody to ICP27 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibody to glycoprotein C was purchased from Fitzgerald Industries International, Inc. (Concord, MA). Antibodies to US11 and UL42 were described elsewhere (43, 45). Secondary antibodies (Cell Signaling Technology, Beverly, MA) were conjugated to horseradish peroxidase. Protein bands were visualized using SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Rockford, IL).
Inhibitor studies.
For experiments employing the MEK inhibitor PD98059, HT1080 cells were starved overnight in serum-free medium and then exposed to 40 μM PD98059 (EMD Biosciences, San Diego, CA) or dimethyl sulfoxide (DMSO; 1:1,000 dilution) 6 h prior to and during infection. At 12 h after infection, whole-cell lysates were created as described above for immunoblotting.
In vitro viral recovery.
Cells were exposed to viruses (1 PFU per cell) for 2 h in serum-free medium at 37°C, after which the supernatant was aspirated and cells were overlaid with 2 ml of DMEM containing 1% calf serum and incubated at 37°C. At 36 h after infection, 2 ml of sterile skim milk was added to triplicate samples and plates were frozen at −80°C. Frozen cell suspensions were thawed and sonicated three times for 15 seconds each, and titers were determined on Vero cells.
HT-caMEK and HT-dnMEK xenograft regression studies.
HT-dnMEK and HT-caMEK tumor xenografts were established into the right flank of 5- to 6-week-old female, athymic nude mice (Fredrickson Cancer Research Institute, Bethesda, MD) by injection of 107 cells in 100 μl of warm phosphate-buffered saline. After 1 week, tumor xenografts grew to approximately 250 mm3 and were randomized to seven animals per treatment group. Mice were injected with 5 × 107 PFU of R3616 with a Hamilton syringe intratumorally. Tumor xenografts were measured biweekly with calipers, and tumor volume was calculated with the formula (l × w × h)/2, which is derived from the formula for an ellipsoid (d3)/g (24).
Bioluminescence imaging.
HT-dnMEK and HT-caMEK tumor xenografts were established in the left and right hindlimbs, respectively, of athymic nude mice by injection of 1 × 107 cells in 100 μl of warm phosphate-buffered saline. All animal studies were performed in accordance with the University of Chicago Animal Care and Use Committee standards. Once tumors grew to an average volume of 350 mm3, 9 × 108 PFU of virus R2636 in a total volume of 100 μl was injected intraperitoneally (i.p.) using a 30-gauge needle. At 5 days after i.p. injection, imaging of firefly luciferase in mice was performed on a charge-coupled device camera (Roper Scientific Photometrics, Tucson, AZ). Animals were injected i.p. with 15 mg/kg of body weight with d-luciferin (Biotium, Hayward, CA). After 5 min, animals were anesthetized with i.p. injection of ketamine (75 mg/kg) and xylazine (5 mg/kg) for imaging, which was performed 10 min after the injection of d-luciferin.
Quantification of bioluminescence imaging data.
The relative intensities of transmitted light from animals infected with virus R2636 are represented as pseudocolor images, with intensity ranging from low (blue) to high (red). Gray-scale images were superimposed on the pseudocolor images using MetaMorph image analysis software (Fryer Company, Huntley, Ill.). Data for total photon flux were calculated using an area-under-the-curve analysis (MetaMorph).
RESULTS
The replication of R3616 (Δγ134.5) mutant virus in human tumor cell lines is cell line dependent and correlates with constitutive activation of MEK.
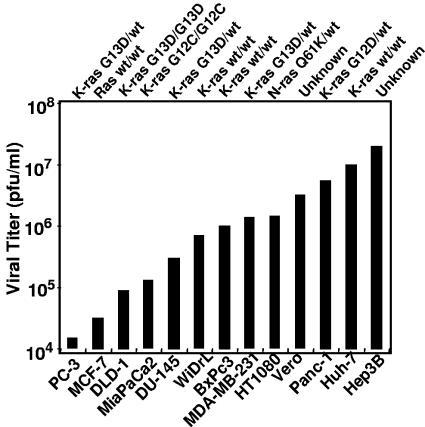
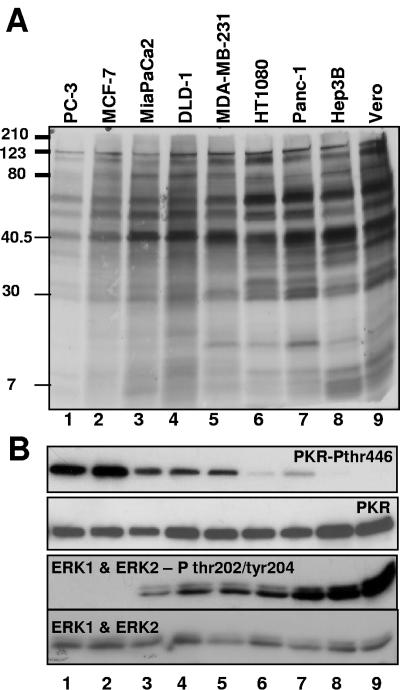
Replicate cultures of 13 cell lines derived from human tumors were exposed to R3616 (1 PFU/cell). The cells were harvested at 36 h after infection, and viral yields were measured by plaque assays on Vero cell monolayers. As shown in Fig. 1, the yields of R3616 mutant virus were highly variable, ranging from 1 × 104 to 3 × 107 PFU/ml. To determine whether the variability in virus yields was reflected in the accumulation of viral proteins, cultures of human tumor cell lines were exposed to R3616 (10 PFU/cell). Vero cells were included as an example of a nonmalignant cell type that supports replication of γ134.5-deficient viruses. At 11 h after infection, the cells were rinsed, starved of methionine for 1 hour, and then incubated in methionine-free medium supplemented with 100 μCi of [35S]methionine per ml for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in denaturing polyacrylamide gels, transferred to a PVDF membrane, and exposed to autoradiography film. As shown in Fig. 2A, the accumulation of viral proteins was reduced in cell lines that restricted viral replication compared to cell lines where viral yields were abundant.
FIG. 1.
R3616 viral replication is variable in cancer cell lines from diverse tumor types. Cells were exposed to 1 PFU/cell of R3616 in serum-free medium for 2 h, after which medium containing virus was removed and fresh medium containing 1% calf serum was added. At 36 h after infection, R3616 viral recovery was determined by standard plaque assay.
FIG. 2.
Differential protein synthesis and activation of PKR in R3616-infected cancer cell lines inversely correlates with constitutive MEK activation in uninfected cancer cell lines. (A) Cell lines were infected with 10 PFU/cell of R3616. At 11 h after infection, the cells were rinsed, starved of methionine for 1 h, and then incubated in methionine-free medium supplemented with 100 μCi of [35S]methionine per ml for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in denaturing polyacrylamide gels, transferred to a PVDF membrane, and exposed to autoradiography film. (B) Cells were infected with 10 PFU/cell of R3616, and whole-cell lysates harvested at 12 h after infection were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with an antibody that recognizes the autophosphorylated form of PKR on threonine 446. In the lower panel, after overnight serum starvation uninfected total whole-cell lysates were resolved by SDS-PAGE and immunoblotted with an antibody against the total and phosphorylated forms of ERK on threonine 202 and tyrosine 204.
To correlate the differences in the accumulation of viral proteins with the activation of PKR, replicate cultures of cell lines shown in Fig. 2A were exposed to R3616 (10 PFU/cell) for 14 h. Lysates were harvested, and 20 μg of equilibrated whole-cell lysate was electrophoretically separated in denaturing polyacrylamide gels, transferred to PVDF membranes, and reacted with antibody specific for the phosphorylated form of PKR on Thr446. As shown in the upper panels of Fig. 2B, PKR phosphorylation was elevated in the cell lines which yielded reduced viral protein accumulation (e.g., PC-3 and MCF-7) and lowest in cell lines that exhibited increased levels of viral protein accumulation (e.g., HT1080, Panc-1, Hep-3B, and Vero), while total PKR levels were similar.
The presence of known activating mutations within the commonly mutated oncogenic (K-, H-, and N-Ras) isoforms, however, did not directly correlate with the observed differences in viral recovery from the representative cell lines tested in Fig. 1. Therefore, we examined the constitutive activity of the downstream effectors of Ras, MEK and its substrate, ERK, which when inhibited results in the loss of the inhibitory functions of Ras on PKR (19). To determine endogenous constitutive MEK activity, uninfected cells were plated to confluence, serum starved for 12 h, and then immunoblotted for the phosphorylated and total forms of the MEK substrates, p42 and p44 MAPK (ERK 2 and ERK1, respectively) (Fig. 2B, lower panels). Cell lines that demonstrated increased protein synthesis and suppressed PKR activation following infection with mutant R3616 demonstrated elevated baseline levels of ERK phosphorylation. In contrast, in cancer cell lines that demonstrated PKR activation, inhibited protein synthesis, and decreased viral recovery following infection with R3616 demonstrated decreased or undetectable levels of ERK phosphorylation.
N-Ras mutation enables efficient replication of R3616 mutant virus in human fibrosarcoma cells.
To test the hypothesis that Ras/MEK/MAPK signaling suppresses PKR function, we measured the replication of R3616 mutant virus in two human fibrosarcoma cell lines that differ only by the expression of an oncogenic mutant allele of N-ras. HT1080 cells contain an endogenous activating mutant allele of N-ras, whereas the MCH603 cell line, a variant of HT1080 cells in which the mutant allele has been deleted, contains only wild-type N-ras (40). Activated MEK is a prerequisite for the Ras-dependent aggressive tumorigenic phenotype of HT1080 cells, and the two cell lines differ dramatically in the constitutive levels of MEK activation, as well as other downstream members of the Ras signaling pathway (21). The viral yields at 36 h after infection with HSV-1(F) and R3616 (1 PFU/cell) are shown in Fig. 3. The salient feature of these results is twofold. First, the yield of HSV-1(F) from the MCH603 cell line was approximately 10-fold lower than that obtained from HT1080 cells (3.1 × 107 compared to 3.5 × 106). Second, the yield of R3616 mutant virus in HT1080 cells was similar to that of wild-type virus (1.8 × 107 versus 3.1 × 107), indicating that the γ134.5 function was not necessary during the course of infection in this cell line. In contrast, the yield of R3616 mutant virus was approximately 10-fold lower than that of wild-type virus in MCH603 cells (4.8 × 105 compared to 3.5 × 106). Therefore, the presence of an activating N-ras mutation enhanced the replication of both wild-type and mutant virus, and the effect was greater on the virus lacking the γ134.5 gene.
FIG. 3.
Deletion of mutant N-ras in human fibrosarcoma cells restricts viral replication. Replicate cultures of HT1080 and MCH603 cells were infected with 1 PFU of R3616 or HSV-1(F) virus per cell in serum-free medium for 2 h, after which medium containing virus was removed and replaced with fresh medium containing 1% calf serum. At 36 h after infection, viral recovery was determined by standard plaque assay.
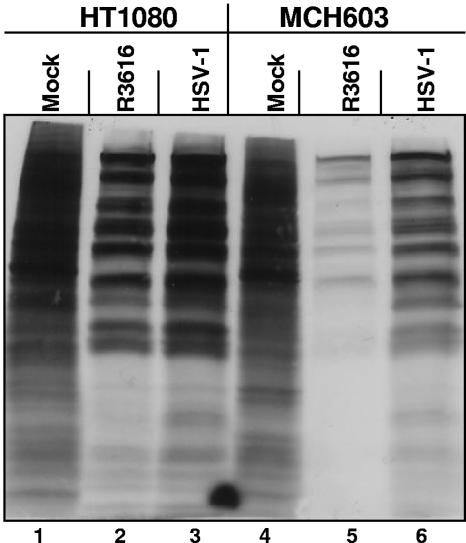
To determine whether virus yields correlate with overall levels of the accumulation of viral proteins, replicate cultures of HT1080 or MCH603 cells were mock infected or exposed to viruses R3616 or HSV-1(F) (10 PFU/cell). At 11 h after infection, the cells were rinsed, starved of methionine for 1 hour, and then supplemented with 100 μCi/ml of [35S]methionine for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in denaturing polyacrylamide gels, transferred to PVDF membranes, and subjected to autoradiography. The results shown in Fig. 4 are congruent with viral yields obtained from the two cell lines. Specifically, the abundance of labeled proteins in MCH603 cells infected with wild-type virus was significantly greater than that observed in the same cells infected with R3616 mutant virus but lower than the amounts of proteins accumulating in HT1080 cells infected with either mutant or wild-type virus.
FIG. 4.
Diminished [35S]methionine metabolic labeling in virus-infected human fibrosarcoma cells deleted for mutant N-ras. Replicate cultures of HT1080 and MCH603 cells were infected with 10 PFU of R3616 or HSV-1(F) virus per cell. At 11 h after infection, the cells were rinsed, starved of methionine for 1 h, and then incubated in methionine-free medium supplemented with 100 μCi of [35S]methionine per ml for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in denaturing polyacrylamide gels, transferred to a PVDF membrane, and exposed to autoradiography film.
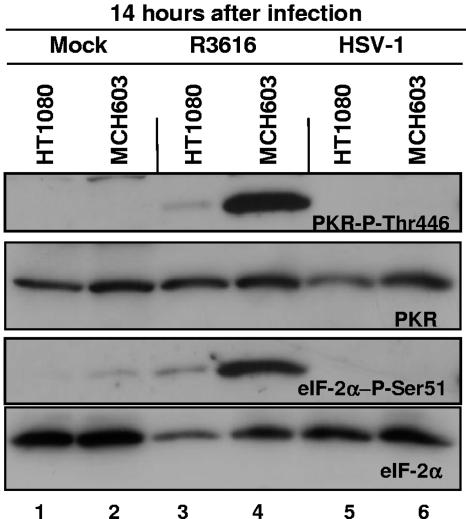
Lastly, the question of whether virus yields and levels of viral proteins accumulating in infected cells correlated with activation of PKR and phosphorylation of eIF-2α was addressed. Electrophoretically separated proteins of lysates from cells infected with R3616 and HSV-1(F) (10 PFU/cell) were harvested at 14 h after infection and probed with antibodies to PKR and the phosphorylated forms of PKR on Thr446 and eIF2α on Ser51. Αs shown in Fig. 5, both PKR and eIF-2α were phosphorylated in MCH603 cells infected with R3616 mutant virus. In contrast, only trace amounts of phosphorylated PKR and eIF-2α were detected in infected HT1080 cells.
FIG. 5.
Increased PKR and eIF-2α phosphorylation in human fibrosarcoma cells deleted for mutant N-ras during R3616 infection. Replicate cultures of HT1080 and MCH603 cells were exposed to 10 PFU of R3616 or HSV-1(F) virus per cell. Cells were harvested at 14 h after infection and processed as described in Materials and Methods. The electrophoretically separated proteins were immunoblotted with antibodies for the phosphorylated form of PKR on threonine 446 and the phosphorylated form of eIF-2α on serine 51, as well as for total PKR and eIF-2α.
Inhibition of MEK by PD98059 results in increased levels of PKR phosphorylation, decreased viral protein accumulation, and diminished replication of mutant virus R3616.
To determine if MEK mediates the observed mutant Ras-dependent suppression of PKR activation and resultant accumulation of viral proteins in HT1080 cells infected with mutant virus R3616, we compared the relative expression of a representative α (ICP27), β (UL42), and γ2 (glycoprotein C) viral protein in cells treated with a specific chemical inhibitor of MEK-1 (PD98059). Replicate cultures of HT1080 cells were serum starved overnight prior to exposure to equal volumes of DMSO or PD98059 (40 μM) for 6 h prior to infection with R3616 mutant virus (10 PFU/cell). DMSO or drug treatment was then continued until the cells were harvested at 12 h after infection, lysed, subjected to electrophoresis in denaturing polyacrylamide gels, transferred to PVDF membranes, and reacted with antibody to ICP27, UL42, or gC. As shown in Fig. 6, treatment with PD98059 had a slight effect on the accumulation of ICP27 and UL42 proteins but caused a very dramatic decrease in the amounts of gC that accumulated in HT1080 cells infected with R3616. To test whether the decrease in the accumulation of gC correlated with activation of PKR, the electrophoretically separated lysates were also probed with antibody to the autophosphorylated form of PKR on Thr446. The presence of PD98059 prior to and during infection with R3616 increased the amount of activated PKR in HT1080 cells (Fig. 6).
FIG. 6.
Inhibition of MEK by the addition of PD98059 increases PKR autophosphorylation and suppresses the accumulation of a γ2 viral protein (gC) in HT1080 cells infected with R3616. Replicate cultures of serum-starved HT1080 cells were infected with 10 PFU of R3616 viruses per cell in the presence or absence of 40 μM PD98059 as described in Materials and Methods. Cells were harvested at 12 h after infection, and the electrophoretically separated proteins were immunoblotted with antibodies for immediate-early [α (ICP27)], early [β(UL42)], and late [γ(gC)] viral proteins. The same lysates were immunoblotted for the total and phosphorylated forms of ERK1 and ERK2 on threonine 202/tyrosine 204 and PKR on threonine 446.
These results are consistent with the earlier report that in wild-type virus-infected cells PKR activation is concurrent with the onset of viral DNA synthesis and enhanced transcription of late genes. In R3616 mutant virus-infected cells, the phosphorylation of eIF-2α by PKR causes a significant reduction of viral proteins whose accumulation is dependent on viral DNA synthesis (14). In contrast, viral proteins whose synthesis is not dependent on the onset of viral DNA synthesis (e.g., ICP27 and UL42 protein) were minimally affected by the activation of PKR.
Finally, to determine if MEK inhibition affects viral replication, DMSO or PD98059 (40 μM) was added to replicate cultures of HT1080 cells 6 h prior to and during infection with R3616 (1 PFU/cell). The cells were harvested at 36 h after infection, and viral yields were measured by plaque assays on Vero cell monolayers. In the presence of PD98059 the yield of R3616 mutant virus was approximately 15-fold lower than in the presence of DMSO (4.14 × 106 compared to 1.67 × 105 PFU/ml).
Viral activation of PKR by mutant R3616 is suppressed in tumor cell lines that overexpress constitutively active MEK, while expression of dominant negative MEK increases PKR activation and restricts R3616 viral replication.
To study the potential relationship between MEK kinase activity and PKR activation in R3616-infected cancer cells, we created cell lines that stably express either caMEK or, conversely, dnMEK from two tumor cell lines that differ dramatically in the magnitude of endogenous MEK activity and the ability to support R3616 viral replication. MEK is constitutively active in the HT1080 human fibrosarcoma cell line. This cell line, as shown in Fig. 1 to 3, is also highly permissive to R3616 viral replication and demonstrates suppressed viral activation of PKR. In contrast, the MiaPaCa2 cell line, which was derived from a patient with poorly differentiated malignant pancreatic adenocarcinoma, contains oncogenic K-ras mutations in both alleles but demonstrates nearly undetectable levels of constitutively active MEK (50). The MiaPaCa2 cell line severely restricts R3616 viral replication and demonstrates robust PKR activation during R3616 viral infection.
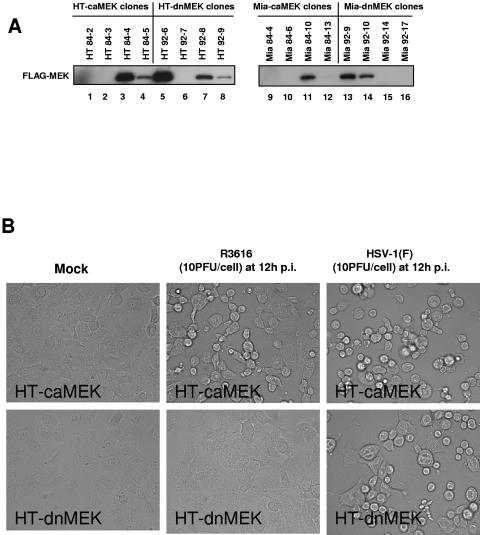
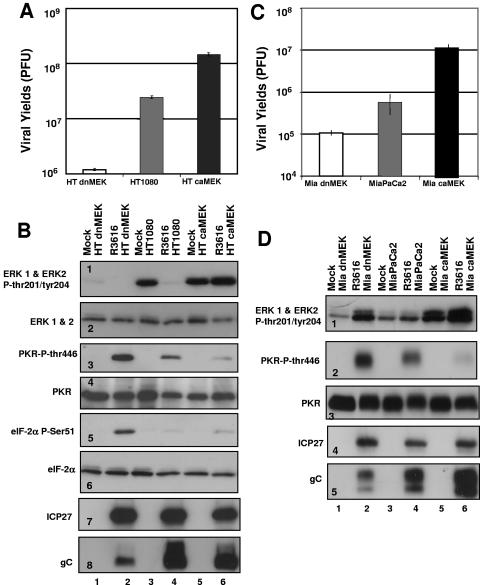
The relative levels of FLAG-tagged MEK expression are shown Fig. 7A. Clones derived from the HT1080 parent cell line, designated HT 84-4 (lane 3) and HT 92-6 (lane 5), expressing equivalent amounts of FLAG-tagged caMEK and dnMEK, respectively, were chosen for further analysis and are referred to as HT-caMEK and HT-dnMEK. Clones derived from the MiaPaCa2 cell line, designated Mia 84-10 (lane 11) and Mia 92-9 (lane 13), express equivalent levels of FLAG-tagged caMEK and dnMEK, respectively, and are referred to as Mia-caMEK and Mia-dnMEK. As shown in Fig. 7B, when the mutant MEK-expressing HT1080 stable cell lines were infected with mutant R3616 (10 PFU/cell), there were appreciable differences in cytopathic effects (CPE). HT-caMEK cells exhibited CPE at 12 h after infection, while dnMEK-expressing cells did not. Both cell lines, however, exhibited CPE upon infection with HSV-1(F) (10 PFU/cell). Next, we compared viral recovery from the stable transfectants generated from HT1080 and MiaPaCa2 cells after exposure of the cells to 1 PFU of R3616 virus per cell. There was a greater-than-200-fold increase in viral titer in R3616-infected caMEK cells compared with dnMEK cells (1.18 × 106 compared to 1.46 × 108 PFU/ml for the HT1080 transfectants and 1.05 × 105 compared to 1.10 × 107 PFU/ml for the MiaPaCa2 transfectants) (Fig. 8A and C).
FIG. 7.
FLAG-MEK expression levels in HT1080 and MiaPaCa2 stable transfectants. Differences in cytopathic effects in virus infected caMEK and dnMEK stable cell lines are shown. (A) Equivalent amounts of whole-cell lysates from caMEK and dnMEK HT1080 and MiaPaCa2 stable transfectants were immunoblotted for FLAG-tagged caMEK and dnMEK using a monoclonal antibody to the FLAG-epitope. (B) Replicate cultures of HT-caMEK and HT-dnMEK cells were infected with 10 PFU of mock, R3616, or HSV-1(F) virus per cell. Photos were taken at 12 h after infection.
FIG. 8.
Effects of dnMEK and caMEK overexpression on R3616 viral recovery and PKR function during R3616 infection. (A) Replicate cultures of HT-dnMEK, HT1080, and HT-caMEK cells were exposed to 1 PFU of R3616 virus per cell in serum-free medium for 2 h, after which medium containing virus was removed and fresh medium containing 1% calf serum was added. At 36 h after infection, R3616 viral recovery was determined by standard plaque assay. (C) R3616 viral recovery from replicate cultures of Mia-dnMEK, MiaPaCa2, and Mia-caMEK at 36 h after infection. (B) To determine the influence of mutant MEK expression on PKR activation, replicate cultures of HT-dnMEK, HT1080, and HT-caMEK cells were exposed to 10 PFU of R3616 virus per cell. Cells were harvested at 12 h after infection and processed as described in Materials and Methods. Electrophoretically separated proteins were immunoblotted with antibodies for total and the phosphorylated forms of ERK1 and ERK2 on threonine 202 and tyrosine204, PKR on threonine 446, and eIF-2α on serine 51. The same lysates were immunoblotted with antibodies for immediate-early [α(ICP27)] and late [γ(gC)] viral proteins. (D) Immunoblotting was performed on replicate lysates of Mia-dnMEK, MiaPaCa2, and Mia-caMEK cells.
Lastly, three series of experiments were done to determine whether the enhancement of replication of the R3616 mutant virus in caMEK cells correlated with the increase in the accumulation of viral proteins and inhibition of PKR activation. In the first experiment, dnMEK- and caMEK-expressing cell lines and their respective parent cell lines were exposed to 10 PFU per cell of mutant virus R3616 (Fig. 8). The cells were harvested 12 h after infection, solubilized, subjected to electrophoresis in denaturing polyacrylamide gels, and reacted with antibodies to PKR, eIF-2α, and the phosphorylated forms of PKR on Thr446 and eIF-2α on Ser51, respectively. Baseline differential MEK activities in uninfected dnMEK- and caMEK-expressing cells and the parental cell lines were established by immunoblotting whole-cell lysates with antibody to ERK1/ERK2 and the phosphorylated form of ERK1/ERK2 on Thr202 and Try204 (Fig. 8B, panel 1, and D, panel 1). As shown in Fig. 8B, panel 3, and D, panel 2, levels of phosphorylated PKR and eIF-2α were higher in dnMEK-expressing lines infected with the R3616 mutant virus than the parental cell line or the caMEK-expressing cell lines. Conversely, activated PKR was nearly undetectable in caMEK-expressing cells infected with the R3616 mutant virus.
In the second series of experiments, electrophoretically separated lysates of caMEK- or dnMEK-expressing cells lines that had been infected with the R3616 mutant virus and processed as described above were reacted with antibody to α (ICP27) and γ2 (glycoprotein C) proteins. As shown in Fig. 8B, panel 7, and D, panel 4, the accumulation of ICP27 was similar in both the stably transfected mutant cell lines and the parental cell lines, suggesting that the expression of MEK-1 mutants did not significantly affect the accumulation of ICP27, a protein expressed prior to the onset of viral DNA synthesis. However, consistent with the result shown in Fig. 6 with chemical inhibition of MEK, the accumulation of gC was markedly decreased in dnMEK-expressing cell lines at 12 h after infection compared with the parent or caMEK-expressing stable cells (Fig. 8B, panel 8, and D, panel 5).
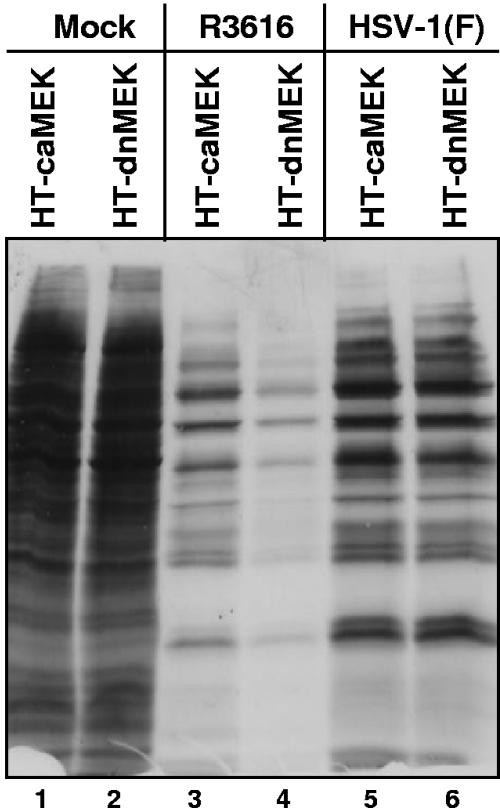
Lastly, caMEK- or dnMEK-overexpressing HT1080 cell lines were exposed to 10 PFU of virus HSV-1(F) or mutant R3616. At 11 h after infection, the cells were rinsed, starved of methionine for 1 hour and then supplemented with 100 μCi/ml of [35S]methionine for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in denaturing polyacrylamide gels, transferred to PVDF membranes, and exposed to autoradiography film. As shown in Fig. 9, the accumulation of labeled proteins was similar in HT-caMEK (lane 5) or HT-dnMEK (lane 6) cells during infection with HSV-1(F). In contrast, the accumulation of labeled proteins in HT-dnMEK cells (lane 4) was diminished compared with HT-caMEK cells (lane 3) infected with the R3616 mutant virus.
FIG. 9.
Diminished [35S]methionine metabolic labeling in R3616-infected human fibrosarcoma cells expressing dnMEK. Replicate cultures of HT-caMEK and HT-dnMEKcells were infected with 10 PFU of R3616 or HSV-1(F) virus per cell. At 11 h after infection, mock- and virus-infected cells were rinsed, starved of methionine for 1 hour, and then incubated in methionine-free medium supplemented with 100 μCi of [35S]methionine per ml for two additional hours. At 14 h after infection, 20 μg of equilibrated protein lysates was electrophoretically separated in a denaturing polyacrylamide gel, transferred to a PVDF membrane, and exposed to autoradiography film.
Intratumoral inoculation of R3616 mutant virus results in tumor regression in tumors expressing caMEK but not in tumors expressing dnMEK.
To determine if differential replication correlated with a reduction of tumor size, we measured tumor volumes of untreated and R3616-treated HT-caMEK and HT-dnMEK tumor xenografts. HT-dnMEK and HT-caMEK tumor xenografts were grown to an average volume of 250 mm3 and injected with a single dose of 5 × 107 PFU of R3616 or buffer on day zero. At 31 days after infection with the R3616 mutant virus, only one of seven animals had a palpable HT-caMEK tumor (100 mm3), in comparison to untreated HT-caMEK tumors, which averaged 4,300 ± 730 mm3 (mean ± standard error of the mean). In contrast, all (seven of seven) of the HT-dnMEK tumors were palpable, with an average tumor volume of 830 ± 210 mm3, and untreated HT-dnMEK average tumor volumes averaged 4,000 ± 660 mm3.
Systemic administration of a recombinant virus R2636 expressing the gC-Luc construct targets tumor tissue overexpressing constitutively active MEK.
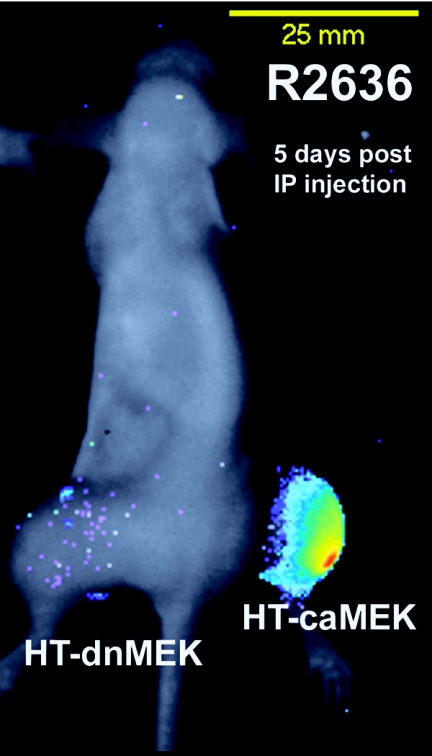
To test the hypothesis that differential MEK activity confers tumor-selective viral replication upon systemic delivery of a γ134.5-deficient virus, bilateral hindlimb tumor xenografts were grown by injecting the left and right hindlimbs of athymic nude mice with 5 × 106 cells of the HT-dnMEK and HT-caMEK cell lines, respectively. In order to image viral replication in vivo, we utilized R2636, a γ134.5-deficient virus that expresses the firefly luciferase gene under the transcriptional control of the HSV-1 gC promoter, a representative γ promoter (37). In tissue that restricts viral replication, the accumulation of the firefly luciferase gene product expressed with the kinetics of a γ gene such as gC would be decreased over successive replicative cycles by PKR-mediated shutoff of protein synthesis. However, Δγ134.5 mutant virus-infected caMEK-xenografted tumor cells, which support viral replication and gC expression, would be expected to support R2636 replication and express gC-luciferase enzyme activity. At 5 days after i.p. delivery of R2636, bioluminescence localized to the right hindlimb, which corresponded to the caMEK-xenografted tumor (3,692 photons/mm2/s), while the dnMEK tumor demonstrated 95-fold less photon expression (39 photons/mm2/s). Also, there was no detectable bioluminescence outside of the caMEK-expressing tumors by 5 days post-i.p. injection (Fig. 10).
FIG. 10.
Bioluminescence of systemically delivered R2636 in mice growing bilateral dnMEK- and caMEK-expressing tumor xenografts. HT-dnMEK and HT-caMEK tumors were established in the left and right hindlimbs of athymic nude mice. Once tumors reached an average volume of 350 mm3, animals were given a single intraperitoneal injection of 9 × 108 PFU of R2636 virus. Bioluminescence imaging was performed 5 days after intraperitoneal injection.
DISCUSSION
Activation of PKR constitutes a major host defense to infection by viruses. The key consequence of activation of PKR is phosphorylation of eIF-2α and the ensuing shutoff of protein synthesis. Viruses have evolved numerous mechanisms to block either the activation of PKR or the consequences of the phosphorylation of eIF-2α. The importance of this pathway of resistance to infection is reflected in the observation that some viruses have evolved multiple mechanisms to thwart either activation or the consequence of activation of PKR. In the case of HSV-1, two genes suppress the effects of PKR. One viral protein, US11, if expressed early, blocks the activation of PKR (7). The more important viral protein that has evolved to subvert the consequences of activation of PKR is ICP34.5 (11). Thus, ICP34.5 recruits protein phosphatase 1α and redirects it to dephosphorylate eIF-2α, and infection proceeds unimpeded (8, 13). Since Δγ34.5 mutants fail to replicate efficiently and are readily inhibited by the interferon pathway, they tend to be avirulent in normal tissues but destructive in some, but not in all, tumor tissues. An investigation into the differential susceptibility of various human tumor cell lines led to the conclusion that susceptibility to the Δγ134.5 mutants hinges on the endogenous kinase activity of the MEK protein. Since the Δγ134.5 mutants are at present the most promising therapeutic viruses for treatment of intractable malignancies, it was of interest to examine the requirements for the replication of Δγ134.5 mutants in some detail. We report the following:
(i) Examination of 12 cell lines derived from a variety of human tumors indicates that there is no correlation between viral yields and the RAS genotype of these cells. Previously, Farassati el al. demonstrated that transformation of NIH 3T3 cells with oncogenic RAS activators enhanced the susceptibility of this nonpermissive mouse fibroblast cell line to wild-type HSV-1 (19). However, the transformed phenotype resulting from overexpression of RAS oncogenes in mouse cells and the resultant increase in HSV-1 permissiveness likely do not accurately reflect the tumorigenic phenotype of human tumor cell lines derived from diverse tissue types. Our data indicate that while the presence of a naturally occurring oncogenic RAS mutation did not correlate with the ability of a Δγ134.5 mutant to replicate in human cancer cell lines, viral replication and the synthesis of viral proteins did correlate with activation of MEK, as measured by the amounts of ERK1 and ERK2 phosphorylated at Thr202/Tyr204 and inversely with the phosphorylation of Thr446 of PKR.
(ii) A more detailed examination focused on HT1080 cells, which contain an oncogenic mutant allele of RAS, and MCH603 cells, a variant derived from HT1080 in which the oncogenic RAS allele but not the wild-type allele was deleted. The three key findings in Fig. 3 to 5 show that in HT1080 cells the Δγ134.5 virus mutant R3616 produced higher viral yields and more viral proteins than its sibling cell line, MCH603. Moreover, while the levels of total PKR were similar in both cell lines, the amounts of activated PKR and phosphorylated eIF-2α were significantly higher in MCH603 cells infected with R3616 mutant than in the infected HT1080 cells. Thus, activation of PKR was suppressed in HT1080 cells but not in MCH603 cells. Since we had excluded the genotype of RAS as the basis for suppression of PKR, our attention focused on the status of the downstream RAS effector kinase, MEK. Evidence supporting a role for MEK is suggested by the results shown in Fig. 6. Thus, suppression of phosphorylation of ERK1 and ERK2 by the MEK inhibitor PD98059 resulted in activation of PKR and reduction in the yields of late proteins as exemplified by gC. These results are consistent with the findings of Farassati et al., in which the addition of the MEK inhibitor PD98059 to Ras-transformed NIH 3T3 cells partially decreased wild-type HSV-1 viral yields and viral protein synthesis (19).
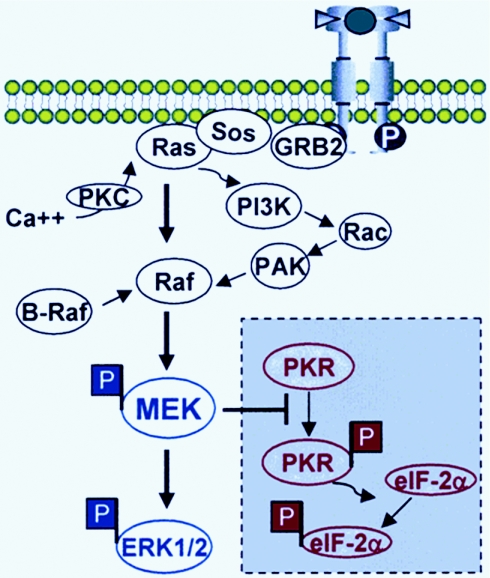
(iii) The role of MEK in the suppression of PKR became apparent from the studies illustrated in Fig. 8, 9, and 10. The hypothesis tested in these experiments was that activation of MEK correlates with suppression of activation of PKR and enhanced replication of the Δγ134.5 mutant R3616 in human tumor cell lines in vitro and in tumor xenografts in nude mice. To this end the permissive HT1080 cell line was transduced with dominant negative or constitutively active MEK constructs. The expectation was that the dnMEK construct would endow the HT1080 cells with the phenotype of MCH603 cells. The results were in part consistent with this model. Thus, dnMEK reduced virus yields and production of R3616 mutant virus. However, two findings were unexpected. Foremost, constitutively active MEK enhanced the replication of the R3616 mutant beyond the levels observed in the untransduced parent HT1080 cells. Second, ERK1 and ERK2 were phosphorylated in cells transduced with constitutively active MEK and then infected, but not in the parent HT1080 cell line infected with R3616 virus. To determine whether these results were unique to HT1080 cells, we performed identical studies using a relatively nonpermissive cell line, MiaPaCa2, derived from a patient with malignant human pancreatic adenocarcinoma. As illustrated in Fig. 1, these cells were somewhat more restrictive than HT1080 cells with respect to the replication of the R3616 mutant. The results of the studies on this cell line parallel the results obtained with HT1080 cells. These results led us to two conclusions. First, as illustrated in Fig. 11, while MEK is downstream of the pathway of activation of RAS, it is also activated by alternative pathways of activation of Raf. Hence, activation of MEK could be both dependent and independent of activation of RAS. Second, the studies on HT1080, HT-dnMEK, and HT-ca MEK cells clearly indicate that MEK directly or indirectly suppresses the activation of PKR. PKR activation is not suppressed in HT-dnMEK cells, while it is fully suppressed in HT-caMEK cells. One plausible explanation for the permissive phenotype of the parental HT1080 cells is that activation of MEK is transient and that is all that is required to block at least in part the activation of PKR. Another explanation is that suppression of PKR involves the formation of complexes that require posttranslationally modified MEK rather than purely enzymatically active protein and that these would be in greater abundance in HT-caMEK cells than in HT1080 cells.
FIG. 11.
Model for the interaction between activated MEK and the suppression of PKR function during viral infection of tumor cells by Δγ134.5 mutant viruses. Earlier studies demonstrated that activation of the ERK kinase (MEK)/ERK pathway by either oncogenic activating mutations of RAS isoforms, point mutations within B-RAF alleles, or receptor tyrosine kinase activation/overexpression is involved in transformation and tumor progression. In addition, Ras-independent activation of Raf/MEK/ERK signaling is cell and tumor type specific. This report shows that activated MEK suppresses PKR autophosphorylation and effectively blocks PKR-mediated eIF-2α phosphorylation. Tumor cells with activated MEK/ERK signaling therefore are exquisitely permissive to Δγ134.5 mutant viral replication and oncolysis.
The significance of the studies described in this report stems from two considerations. Foremost, they indicate that MEK plays a key role in the regulation of activation of PKR. The mechanism by which MEK affects the status of PKR remains to be determined. Second, they indicate that the success of tumor therapies based on the application of cytolytic viruses lacking the Δγ134.5 gene hinge on the status of MEK and on the potential for manipulating the activity of MEK in the tumor bed inoculated with these viruses.
Acknowledgments
We thank Eric J. Stanbridge (Irvine, Calif.) for providing the MCH603 cell line, David Smith (Fryer) for help with animal imaging and analysis, Samuel Hellman for helpful discussion, and Edwardine Lebay and Marija Pejovic for technical help.
This work was supported by NIH grant to R.R.W. (CA7193307-07).
We declare no conflicts of interest.
REFERENCES
- 1.Anderson, M. J., G. Casey, C. L. Fasching, and E. J. Stanbridge. 1994. Evidence that wild-type TP53, and not genes on either chromosome 1 or 11, controls the tumorigenic phenotype of the human fibrosarcoma HT1080. Genes Chromosomes Cancer 9:266-281. [DOI] [PubMed] [Google Scholar]
- 2.Andreansky, S., L. Soroceanu, E. R. Flotte, J. Chou, J. M. Markert, G. Y. Gillespie, B. Roizman, and R. J. Whitley. 1997. Evaluation of genetically engineered herpes simplex viruses as oncolytic agents for human malignant brain tumors. Cancer Res. 57:1502-1509. [PubMed] [Google Scholar]
- 3.Ballif, B. A., and J. Blenis. 2001. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 12:397-408. [PubMed] [Google Scholar]
- 4.Barber, G. N., R. Jagus, E. F. Meurs, A. G. Hovanessian, and M. G. Katze. 1995. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J. Biol. Chem. 270:17423-17428. [DOI] [PubMed] [Google Scholar]
- 5.Barber, G. N., M. Wambach, S. Thompson, R. Jagus, and M. G. Katze. 1995. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol. Cell. Biol. 15:3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, J. J., K. A. Delman, B. M. Burt, A. Mariotti, S. Malhotra, J. Zager, H. Petrowsky, S. Mastorides, H. Federoff, and Y. Fong. 2002. Comparison of safety, delivery, and efficacy of two oncolytic herpes viruses (G207 and NV1020) for peritoneal cancer. Cancer Gene Ther. 9:935-945. [DOI] [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, R., G. Y. Gillespie, L. Soroceanu, S. Andreansky, S. Chatterjee, J. Chou, B. Roizman, and R. J. Whitley. 1995. Comparison of genetically engineered herpes simplex viruses for the treatment of brain tumors in a scid mouse model of human malignant glioma. Proc. Natl. Acad. Sci. USA 92:1411-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, A. V., and B. Roizman. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J. Virol. 78:4185-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, G., M. E. Brett, and B. He. 2001. Val193 and Phe195 of the gamma 1 34.5 protein of herpes simplex virus 1 are required for viral resistance to interferon-alpha/beta. Virology 290:115-120. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 12.Chou, J., A. P. Poon, J. Johnson, and B. Roizman. 1994. Differential response of human cells to deletions and stop codons in the γ134.5 gene of herpes simplex virus. J. Virol. 68:8304-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou, J., and B. Roizman. 1994. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc. Natl. Acad. Sci. USA 91:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, S. M., S. J. Advani, J. D. Bradley, Y. Kataoka, K. Vashistha, S. Y. Yan, J. M. Markert, G. Y. Gillespie, R. J. Whitley, B. Roizman, and R. R. Weichselbaum. 2002. The use of a genetically engineered herpes simplex virus (R7020) with ionizing radiation for experimental hepatoma. Gene Ther. 9:75-80. [DOI] [PubMed] [Google Scholar]
- 16.Clemens, M. J. 2004. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene 23:3180-3188. [DOI] [PubMed] [Google Scholar]
- 17.Davies, H., G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton, and P. A. Futreal. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949-954. [DOI] [PubMed] [Google Scholar]
- 18.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 19.Farassati, F., A. D. Yang, and P. W. Lee. 2001. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat. Cell Biol. 3:745-750. [DOI] [PubMed] [Google Scholar]
- 20.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29-46. [DOI] [PubMed] [Google Scholar]
- 21.Gupta, S., R. Plattner, C. J. Der, and E. J. Stanbridge. 2000. Dissection of Ras-dependent signaling pathways controlling aggressive tumor growth of human fibrosarcoma cells: evidence for a potential novel pathway. Mol. Cell. Biol. 20:9294-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, S., and E. J. Stanbridge. 2001. Paired human fibrosarcoma cell lines that possess or lack endogenous mutant N-ras alleles as experimental model for Ras signaling pathways. Methods Enzymol. 333:290-306. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 24.Hallahan, D. E., H. J. Mauceri, L. P. Seung, E. J. Dunphy, J. D. Wayne, N. N. Hanna, A. Toledano, S. Hellman, D. W. Kufe, and R. R. Weichselbaum. 1995. Spatial and temporal control of gene therapy using ionizing radiation. Nat. Med. 1:786-791. [DOI] [PubMed] [Google Scholar]
- 25.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino, R., Y. Chatani, T. Yamori, T. Tsuruo, H. Oka, O. Yoshida, Y. Shimada, S. Ari-i, H. Wada, J. Fujimoto, and M. Kohno. 1999. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene 18:813-822. [DOI] [PubMed] [Google Scholar]
- 27.Huang, W., and R. L. Erikson. 1994. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc. Natl. Acad. Sci. USA 91:8960-8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, T., R. Jagus, and W. S. May. 1994. Interleukin 3 stimulates protein synthesis by regulating double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 91:7455-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacquemont, B., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J. Virol. 15:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katze, M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75-78. [DOI] [PubMed] [Google Scholar]
- 31.Kozak, M., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. IX. Evidence for accumulation of abundant symmetric transcripts in nuclei. J. Virol. 15:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Gall, M., J. C. Chambard, J. P. Breittmayer, D. Grall, J. Pouyssegur, and E. Van Obberghen-Schilling. 2000. The p42/p44 MAP kinase pathway prevents apoptosis induced by anchorage and serum removal. Mol. Biol. Cell 11:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansour, S. J., J. M. Candia, J. E. Matsuura, M. C. Manning, and N. G. Ahn. 1996. Interdependent domains controlling the enzymatic activity of mitogen-activated protein kinase kinase 1. Biochemistry 35:15529-15536. [DOI] [PubMed] [Google Scholar]
- 35.Markert, J. M., M. D. Medlock, S. D. Rabkin, G. Y. Gillespie, T. Todo, W. D. Hunter, C. A. Palmer, F. Feigenbaum, C. Tornatore, F. Tufaro, and R. L. Martuza. 2000. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 7:867-874. [DOI] [PubMed] [Google Scholar]
- 36.Meurs, E. F., J. Galabru, G. N. Barber, M. G. Katze, and A. G. Hovanessian. 1993. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 90:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mezhir, J., S. J. Advani, K. D. Smith, T. E. Darga, A. P. Poon, H. Schmidt, M. C. Posner, B. Roizman, and R. R. Weichselbaum. Ionizing radiation activates late herpes simplex virus 1 promoters via the p38 pathway in tumors treated with oncolytic viruses. Cancer Res. 65:9479-9484. [DOI] [PubMed]
- 38.Nakamura, H., H. Kasuya, J. T. Mullen, S. S. Yoon, T. M. Pawlik, S. Chandrasekhar, J. M. Donahue, E. A. Chiocca, R. Y. Chung, and K. K. Tanabe. 2002. Regulation of herpes simplex virus γ134.5 expression and oncolysis of diffuse liver metastases by Myb34.5. J. Clin. Investig. 109:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins, D. J., and G. N. Barber. 2004. Defects in translational regulation mediated by the alpha subunit of eukaryotic initiation factor 2 inhibit antiviral activity and facilitate the malignant transformation of human fibroblasts. Mol. Cell. Biol. 24:2025-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plattner, R., M. J. Anderson, K. Y. Sato, C. L. Fasching, C. J. Der, and E. J. Stanbridge. 1996. Loss of oncogenic ras expression does not correlate with loss of tumorigenicity in human cells. Proc. Natl. Acad. Sci. USA 93:6665-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouyssegur, J., V. Volmat, and P. Lenormand. 2002. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64:755-763. [DOI] [PubMed] [Google Scholar]
- 42.Rampling, R., G. Cruickshank, V. Papanastassiou, J. Nicoll, D. Hadley, D. Brennan, R. Petty, A. MacLean, J. Harland, E. McKie, R. Mabbs, and M. Brown. 2000. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 7:859-866. [DOI] [PubMed] [Google Scholar]
- 43.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sebolt-Leopold, J. S., and R. Herrera. 2004. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4:937-947. [DOI] [PubMed] [Google Scholar]
- 45.Sheaffer, A. K., W. W. Hurlburt, J. T. Stevens, M. Bifano, R. K. Hamatake, R. J. Colonno, and D. J. Tenney. 1995. Characterization of monoclonal antibodies recognizing amino- and carboxy-terminal epitopes of the herpes simplex virus UL42 protein. Virus Res. 38:305-314. [DOI] [PubMed] [Google Scholar]
- 46.Shimamura, A., B. A. Ballif, S. A. Richards, and J. Blenis. 2000. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr. Biol. 10:127-135. [DOI] [PubMed] [Google Scholar]
- 47.von Gise, A., P. Lorenz, C. Wellbrock, B. Hemmings, F. Berberich-Siebelt, U. R. Rapp, and J. Troppmair. 2001. Apoptosis suppression by Raf-1 and MEK1 requires MEK- and phosphatidylinositol 3-kinase-dependent signals. Mol. Cell. Biol. 21:2324-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed] [Google Scholar]
- 49.Xia, Z., M. Dickens, J. Raingeaud, R. J. Davis, and M. E. Greenberg. 1995. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270:1326-1331. [DOI] [PubMed] [Google Scholar]
- 50.Yip-Schneider, M. T., A. Lin, D. Barnard, C. J. Sweeney, and M. S. Marshall. 1999. Lack of elevated MAP kinase (Erk) activity in pancreatic carcinomas despite oncogenic K-ras expression. Int. J. Oncol. 15:271-279. [DOI] [PubMed] [Google Scholar]