Abstract
Hepatitis B virus X (HBX) is essential for the productive infection of hepatitis B virus (HBV) in vivo and has a pleiotropic effect on host cells. We have previously demonstrated that the proteasome complex is a cellular target of HBX, that HBX alters the proteolytic activity of proteasome in vitro, and that inhibition of proteasome leads to enhanced viral replication, suggesting that HBX and proteasome interaction plays a crucial role in the life cycle and pathogenesis of HBV. In the present study, we tested the effect of HBX on the proteasome activities in vivo in a transgenic mouse model in which HBX expression is developmentally regulated by the mouse major urinary promoter in the liver. In addition, microarray analysis was performed to examine the effect of HBX expression on the global gene expression profile of the liver. The results showed that the peptidase activities of the proteasome were reduced in the HBX transgenic mouse liver, whereas the activity of another cellular protease was elevated, suggesting a compensatory mechanism in protein degradation. In the microarray analysis, diverse genes were altered in the HBX mouse livers and the number of genes with significant changes increased progressively with age. Functional clustering showed that a number of genes involved in transcription and cell growth were significantly affected in the HBX mice, possibly accounting for the observed pleiotropic effect of HBX. In particular, insulin-like growth factor-binding protein 1 was down-regulated in the HBX mouse liver. The down-regulation was similarly observed during acute woodchuck hepatitis virus infection. Other changes including up-regulation of proteolysis-related genes may also contribute to the profound alterations of liver functions in HBV infection.
Hepatitis B virus (HBV) is a member of the hepadnaviridae family that includes the hepatitis viruses of the woodchuck, ground squirrel, tree squirrel, Peking duck, and heron. HBV has a fourth open reading frame, termed the hepatitis B virus X (HBX) gene. The HBX gene is well conserved among the mammalian hepadnaviruses and codes for a 16.5-kDa protein. The protein can activate the transcription of a variety of viral and cellular genes (3, 9). Since HBX does not bind to DNA directly, its activity is thought to be mediated via protein-protein interactions. HBX has been shown to enhance transcription through AP-1 and AP-2 (5, 27) and to activate various signal transduction pathways (10, 11). Several studies have also identified possible cellular targets of HBX, including members of the CREB/ATF family (23), the TATA-binding protein (25), RNA polymerase subunit RPB5 (8), the UV-damaged DNA-binding protein (28), and the mitochondrial protein (32). HBX has also been shown to interact with p53 and inhibit its function (30, 31). Furthermore, X protein is necessary for the establishment of productive infection in vivo (7, 39). Recent results also demonstrated that signaling through calcium may mediate a function of HBX in viral replication (6).
We have previously demonstrated that the proteasome complex is a cellular target of HBX (16, 17) and that this interaction is functionally important in the pleiotropic effect of HBX (37, 38). Two subunits of the 26S proteasome complex, PSMA7 and PSMC1, were identified as putative targets of HBX by using a yeast two-hybrid system. This interaction is specific using an in vitro binding assay, comigration in sucrose gradient, and coimmunoprecipitation (16, 37). Functional assay showed that HBX could lead to the inhibition of peptidase and proteinase activities of the proteasome complex in tissue culture (16). The transactivating activity of HBX was specifically inhibited by proteasome-specific inhibitors, such as MG132 and lactacystin, in a dose-dependent manner, whereas calpain protease inhibitor ALLM has no effect. Because proteasome plays a crucial role in diverse cellular functions ranging from cell differentiation, cell cycle control, signal transduction, stress response, transcriptional activation, DNA repair, apoptosis, and antigen presentation, the interaction between HBX and the proteasome complex may represent an important pathway for the biological functions of HBX.
In the woodchuck model, we demonstrated that X-deficient mutants of woodchuck hepatitis virus (WHV) are not completely defective, behaving like attenuated viruses (38). Our experiments also suggested that the function of HBX in HBV replication may be medicated through a proteasome-dependent pathway (36). In cells infected with either the recombinant adenovirus-HBV or baculovirus-WHV, the replication of the wild-type virus was not affected, while the replication of X-minus virus of either HBV or WHV was enhanced and restored to the wild-type level by proteasome inhibitors.
To further understand the role of HBX in the viral life cycle and the pathogenesis of HBV, we studied the effects of HBX on proteasome activities in vivo and the global gene expression profiles by microarray analysis in a transgenic mouse model expressing the HBX gene. In this model, the HBX expression is developmentally regulated by the mouse major urinary protein (MUP) promoter in the liver (15). We here demonstrate that the proteasome activities were inhibited in the HBX mice and an unrelated cellular protease complex, the Tricon protease, was activated at the same time, suggesting a compensatory mechanism of the cellular proteolysis pathways. Microarray analysis revealed that that a broad spectrum of genes was altered in the HBX mouse livers. Functional clustering showed that a number of transcription factors, cell growth regulators, proteolysis components, and detoxification enzymes were significantly affected in HBX mice.
MATERIALS AND METHODS
Animals.
The MUP-HBX transgenic mouse line was provided by Francis Chisari (The Scripps Research Institute, La Jolla, Calif.) (15) and was bred and maintained at the National Institutes of Health (NIH) animal facility. Expression of HBX was controlled by the liver-specific MUP promoter, which is developmentally regulated. Syngeneic FVB/n mice were used as wild-type controls. Animals were sacrificed at various times during development as illustrated in the figures. Livers were harvested and snap-frozen for further analysis. A different HBX transgenic mouse line, whose HBX expression is regulated by the endogenous HBV promoter/enhancer, was obtained from James Ou (University of Southern California, Los Angeles, Calif.) (18). Woodchucks were infected with WHV as described previously (38). In this previous study, naïve adult woodchucks were infected with infectious WHV inoculum and monitored for infection with various serologic markers. Serial liver biopsies were obtained prior to and at various times following WHV infection. Tissues were stored frozen for subsequent RNA isolation.
Reagents and proteasome inhibitors.
Proteasome peptidase substrates Sue-Leu-Leu-Val-Tyr-amino-4-methylcoumarin (AMC) (LLVY), Boc-Leu-Arg-Arg-AMC (LRR), and Z-Leu-Leu-Glu-βNa (LLE) were obtained from BACHEM Bioscience Inc. (Torrance, CA). Proteasome-specific inhibitors MG132 and lactacystin were obtained from BIOMOL Research Laboratoroies (Plymouth Meeting, PA). Tricorn protease peptide substrate H-Ala-Ala-Phe-AMC (AAF) was purchased from BACHEM Bioscience Inc. (Torrance, CA).
RNA preparations and Northern blot analysis.
Total RNA from the liver of the transgenic mouse was prepared by the guanidium isothiocyanate-acid-phenol method (ULTRASPEC RNA Isolation Kit; BIOTECX Laboratories, Houston, TX), partially purified by two ethanol precipitation, and stored precipitated at −80°C until use. For Northern blot analysis, total RNA was analyzed by 1% formaldehyde agarose gel electrophoresis and hybridized with a 32P-labeled HBV-specific probe.
Analysis of proteasome and Tricorn protease activities.
The proteasome complexes from livers of both transgenic mice and wild-type controls were extracted and purified as previously described (16). The peptidase activities of the proteasome complex were measured using three kinds of fluorogenic peptide substrates as described previously (16). In brief, 3 μg of the proteasome preparation was incubated with 0.1 mM LLVY, LRR, or LLE in a 50-μl total volume of reaction buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP) at 37°C for 20 min. The reactions were stopped by adding 1 ml of 1% sodium dodecyl sulfate. The resulting fluorescence was measured with a spectrofluorometer (Packard Instruments, Downers Grove, IL). The Tricorn protease activity was determined using synthesized AAF as a substrate (13). For activity gel analysis, partially purified proteasome was electrophoresed on a 4% native acrylamide gel in 90 mM Tris-borate-5 mM MgCl2-1 mM ATP-0.5 mM dithiothreitol-0.1 mM EDTA. After electrophoresis, the gel was incubated with 0.2 mM LLVY at 37°C for 15 min and visualized with a UV transilluminator. The bands representing 26S and 20S proteasomes were photographed and quantitated by using ImageQuant software (Molecular Dynamics).
cDNA microarray hybridization and data processing.
The indirect labeling method was used for labeling the cDNA with fluorescence dye Cy3 or Cy5 in the experiment. Briefly, 25 μg of total RNA was reverse transcribed to cDNA, which was then used as the template for coupling with fluorescence dyes. RNA from six wild-type mice was pooled as one sample and labeled with Cy3, whereas RNA from HBX mice was labeled individually with Cy5. Then, Cy5-labeled sample was mixed with age-matched Cy3-labeled pooled control sample and purified for cDNA microarray hybridization. Five or six HBX transgenic mice from each time point were used in the experiment. As an internal control for the microarray analysis, RNA from control mice at each time point was labeled with Cy3 or Cy5 and then combined as a probe for hybridization.
cDNA microarrays were purchased from the National Cancer Institute microarray facility (Frederick, MD). It contains 9,800 cDNA clones. The chips were hybridized with the above-labeled probes at 60°C overnight and washed in graded SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffers. Then, the chips were scanned with a GenePix 4000 microarray scanner (Axon Instruments, Union City, CA). Each spot was converted to a numerical ratio of Cy5 to Cy3 based on the intensity of the fluorescence. Using Genepix 4.0 software, a TIFF file (image of the hybridized cDNA spots on the chip) and a GPR file (Genepix analysis of the relative intensity of each spot) were created and uploaded to our database maintained by the Center for Information Technology at NIH for further analysis.
Data analysis and statistics.
In our experiment, we have observed that variations of the ratios of Cy5/Cy3 intensity in the self-hybridized internal controls, which represent the intrinsic errors of the hybridization, could influence the final results of analysis if not corrected. This raised the question that using a cutoff ratio without a normalization against the internal control could potentially eliminate the genes of interest and include those of little value (19, 20). To address this issue, we adopted a computational system that would allow us to dissect the changes in a statistical manner, namely, by comparing the average ratios of each spot from a number of samples with that of its corresponding control. The BRB ArrayTools (version 3.01) developed by G. Simon and A. Lam (Biometric Research Branch, National Cancer Institute, NIH) was used for the analysis. The class comparison tool used the F test to examine the number of genes that were differentially expressed in the livers of HBX transgenic and wild-type mice. The analysis was done on all the genes on the chips that survived the basic screening. Average ratios of Cy5 over Cy3 intensity from five or six mice at each time point were compared with those of the paired internal controls. To further reduce the variability, only genes within ratios between 0.8 and 1.25 in internal control chips and with a P of <0.001 between the HBX transgenic and wild-type mice were deemed significant and used for further analysis. Genes that passed the above test in the HBX mice were then grouped together based on their functional characteristics and plotted with the hierarchical clustering tool embedded in the software.
Quantitative real-time PCR analysis.
To validate the results of microarray analysis, a few genes that showed significant changes, including insulin-like growth factor-binding protein 1 (IGFBP-1), interferon consensus binding protein 1, and serine protease inhibitor 1, were selected for further real-time reverse transcription (RT)-PCR TaqMan analysis using the ABI Prism 7700 Sequence Detector System (PE Applied Biosystems, Foster City, CA). Briefly, cDNA was generated using 200 ng total RNA in a 20-μl linear RT reaction with the First-Strand cDNA Synthesis System for Quantitative RT-PCR Kit (Marligen Biosciences, Ljamsville, MD) as templates. TaqMan probes and primers for the genes of interest were purchased from PE Applied Biosystems, and the manufacturer's manual was followed. Standard curves for each individual target amplicon were constructed using a common mouse cDNA. All PCR assays were performed in duplicate, and results are represented by the mean values for five or six mice. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene against which all samples were normalized. All three genes analyzed were consistent with the changes of microarray results.
For TaqMan real-time PCR analysis of IGFBP-1 in the woodchuck liver, IGFBP-1 primers and probe were custom designed and synthesized as described above. Since the sequence of the woodchuck IGFBP-1 gene is not available from the data bank, primer and probe sequences were selected from the conserved regions of both human and mouse IGFBP-1 cDNA and examined with API Primer Express software for validity in TaqMan PCR. RT-PCR of woodchuck liver RNA showed a correctly amplified woodchuck IGFBP-1 cDNA fragment. The respective of sequences were as follows: forward primer, 5′-TACCTGCCAAACTGCAACAAG; reverse primer, 5′-GGTAGACGCACCAGCAGAGT; probe, 5′-TTATCACAGCAGACAGTGTGA.
The expression of insulin-like growth factor 1 (IGF-1) was quantified by real-time PCR in both the mouse and woodchuck livers as described above. Mouse IGF-1 primers and probe purchased from PE Applied Biosystems worked well in both the mouse and woodchuck samples.
RESULTS AND DISCUSSION
Developmental expression of HBX in the transgenic mice.
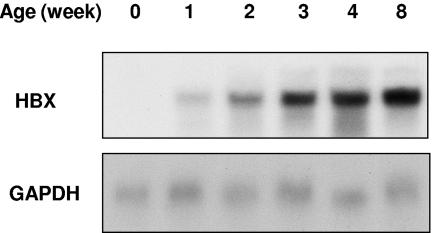
To determine the gene expression pattern of HBX in the transgenic mice, total RNA was extracted from livers of the mice at various ages (weeks 0, 1, 2, 3, 4, and 8) and analyzed by Northern blotting. As shown in Fig. 1, the expression of HBX in the transgenic mice was detected at week 1 of age and reached a maximal level after week 4 in the livers of the transgenic mice.
FIG. 1.
HBX expression in the livers of transgenic mice. Total RNAs were isolated from livers of HBX transgenic mice at various ages as indicated and subjected to Northern blot analysis (10 μg total RNA per lane). The blot was then stripped and reprobed with GAPDH as an internal loading control.
Inhibition of proteasome activities in the livers of transgenic mice.
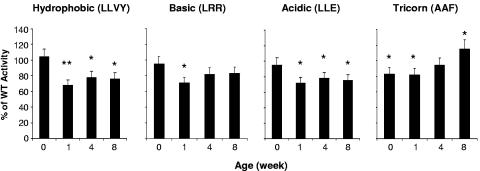
To analyze the effects of HBX on proteasome activities in vivo, livers were harvested from both the transgenic and wild-type mice at various ages. The activities of the proteasome and the Tricorn protease were determined. For peptidase activity assays, the proteasome activities of the livers from the transgenic mice were about 70% of those of the wild-type mice from the age of week 1, while there was no difference at week 0 (Fig. 2). To test the specificity of the effect, the activity of the Tricorn protease that is distinct from the proteasome activities was also assayed. The Tricorn protease is a large protease complex that is resistant to proteasome inhibitors and can functionally replace proteasome activity in adapted cells (13). As shown in Fig. 2, the Tricorn protease activity of the liver from the transgenic mouse was about 80% of that of the wild-type controls at early ages (weeks 0 and 1). Then it started to increase at the age of 4 weeks to about 95% and 8 weeks to 115% of the wild-type activity, suggesting an adaptive response of the mice to the effect of HBX on proteasome activities in vivo.
FIG. 2.
Inhibition of hepatic proteasome activities in HBX transgenic mice. For determination of the peptidase activities of the hepatic proteasomes, three peptidase substrates (LLVY, LRR, and LLE) were used to determine the proteasome activities from the livers of both transgenic mice and the wild-type controls (WT). A total of 3 μg of purified proteasome complexes was used for each reaction. Data shown are the mean values of triplicate determinations based on the percentage of the activity in the transgenic animal over that of age-matched wild-type controls. Standard errors of the mean (SEM) are shown as error bars. To determine the Tricorn protease activities, 3 μg of purified cell lysate from the livers of both transgenic mice and wild-type controls was assayed with the AAF substrate. The results are the means of three independent determinations with the SEM. Asterisks indicate differences with statistical significance (*, P < 0.05; **, P < 0.01) between the HBX and WT mice by using Student's t test.
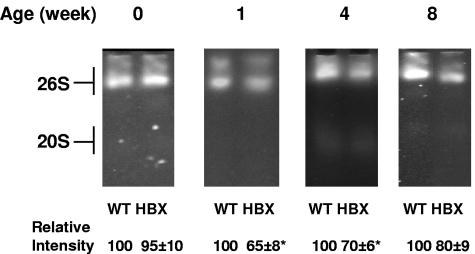
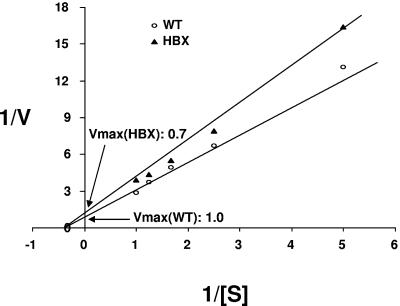
To further demonstrate the effects of HBX on proteasome activities, the liver samples were analyzed by activity gel assay. Similar to the results of the peptidase activity assay, no significant difference at week 0 was observed. From week 1 on, the proteasome activities of both 20S and 26S proteasome complexes of the livers from the transgenic mice were inhibited significantly (Fig. 3). The maximal inhibition occurred at week 1 (up to 40%), which coincides with expression of the HBX transgene. The less inhibition noted at a later time point (week 8) could represent a compensatory mechanism in response to the effect of HBX on the proteasome, despite the apparent higher level of HBX later. As discussed below, such a mechanism could occur in the form of increased proteasome subunit synthesis. Enzymatic study of the partially purified proteasomes showed that the Vmax of the protease activities from the transgenic mice at the age of 1 week was about 70% of that of the wild-type controls (Fig. 4). The Vmaxs of all three peptidase proteases were inhibited. Therefore, HBX affects the proteasome activities through inhibition of the Vmax of the proteasome activities.
FIG. 3.
Analysis of proteasome activity by activity gel assay. For the activity gel assay, 10 μg of a partially purified proteasome complex from the livers of both transgenic mice and wild-type controls (WT) was separated on a 4.0% native polyacrylamide electrophoresis gel. After electrophoresis, the gels were incubated with LLVY for 15 min at 37°C. Representative gels are shown at the top, and the means of the relative intensities of three independent experiments are shown at the bottom with standard errors of the mean. Asterisks indicate differences with statistical significance (*, P < 0.05) between the HBX and WT mice by using Student's t test.
FIG. 4.
Enzymatic analysis of the proteasome complex. To analyze the kinetics of the protease activities of the proteasome complex, the activities of the proteasome complex from the livers of both transgenic mice and wild-type controls (WT) were tested at various doses of peptidase substrates. Data shown are representative of those from the livers of mice at the age of week 1. Three peptidase substrates (LLVY, LRR, and LLE) were analyzed, and only LLVY was shown. Results are representative of three independent experiments.
Global gene expression in livers of HBX transgenic mice.
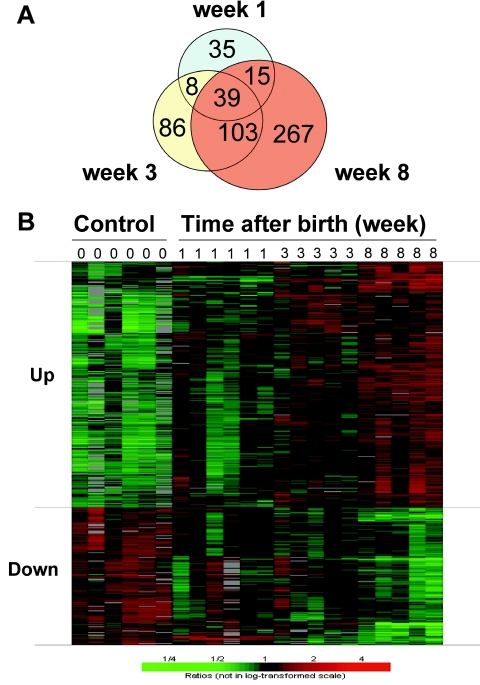
To better define the mechanism underlying the biological function of HBX in vivo, microarray analysis was performed on the livers of HBX transgenic mice. Because the HBX transgene is developmentally regulated, the gene expression patterns could be assessed at different time points to correlate with the expression of the HBX transgene in an attempt to discern the diverse functional effects of HBX. The number of genes differentially expressed in the HBX transgenic mice increased gradually with age after birth, from 97 (64 up, 33 down) at week 1, to 236 (166 up, 70 down) at week 3, and to 424 (272 up, 152 down) at week 8. The ratio of up-regulated genes versus down-regulated genes was approximately 2 to 1 at all of the time points. This progressive change was consistent with the time course of the expression of the HBX transgene (Fig. 1). In addition, the majority of the changes at later time points overlapped with those of the earlier time points (Fig. 5A) and exhibited a clear trend of progressive increase in the numbers of up- or down-regulated genes from weeks 1 to 8 (Fig. 5B), suggesting that the changes are due to a cumulative effect of HBX expression on the liver. This progressive trend also supports a temporal association among HBX expression, inhibition of proteasome activities, and the global gene expression changes that are probably a secondary effect of HBX.
FIG. 5.
Global gene expression in the livers of HBX transgenic mice. (A) The number of genes that changed significantly (P < 0.001 compared with control) in HBX transgenic mice increased with age after birth. There were 97 genes at week 1, which increased to 236 at week 3 and 424 at week 8 after birth. The majority of these genes overlapped with earlier time points. (B) Hierarchical clustering of the 424 significantly changed genes selected from the 8-week-old mice. Genes were divided into two subgroups as up- or down-regulation. The figure shows a clear trend of gradual change in the direction of up- or down-regulated genes during the course of development. The time after birth is indicated as week 1, 3, and 8, with 0 as the control. Each row represents a single gene, and each column represents one sample. Green, down-regulation; red, up-regulation. The scale is plotted from 1/4 to 4.
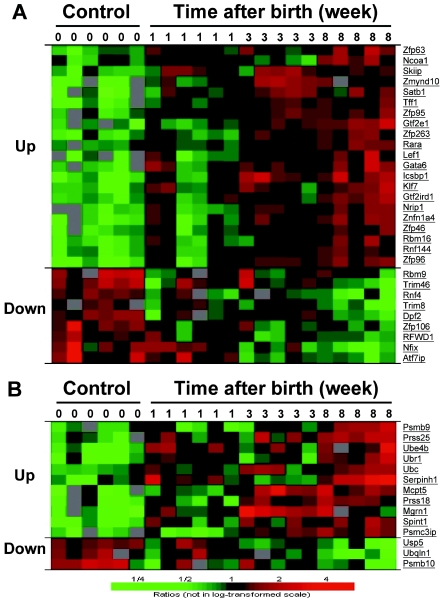
HBX has been known for its complexity in biological activities (2, 24). The extensive gene alteration in HBX mice seemed to substantiate the notion. To further define the implication of these changes in the biological function of HBX, we grouped these genes based on their major biological functions that are relevant to HBX activities for further analysis. Since HBX can affect transcriptional activation and cell growth, we first examined transcriptional factors and cell growth regulators. As indicated in Fig. 6A, the expression of a number of these factors was altered in HBX mice, with two-thirds of them up-regulated. Many of theses genes were general transcriptional factors such as genes encoding zinc finger proteins. The up-regulation of a large number of these factors may account for the broad changes of the gene expression by HBX, which in turn can lead to profound alterations of liver functions.
FIG. 6.
Hierarchical clustering of gene expression in HBX transgenic mouse livers. (A) Changes of transcriptional factors in HBX transgenic mouse livers. A total of 30 genes that were significantly altered at 8 weeks after birth belong to this group. (B) Genes related to protein degradation. A total of 14 genes that were significantly altered fall into this group, with 11 genes up-regulated and 3 genes down-regulated. The genes are presented in the same way as in Fig. 5.
The second group we identified with significant alteration was the genes related to proteolysis (Fig. 6B). The majority of genes in the cluster showed an increased expression in the HBX mice. Several genes responsible for ubiquitination were significantly up-regulated, possibly as a result of the transactivation and proteasome inhibition activities of HBX. Also up-regulated were genes encoding nonproteasome proteases. This was consistent with the compensatory increase of Tricorn protease activity in response to the inhibition of the proteasome activity observed in the HBX transgenic livers. Several proteasome subunits (Psmc3ip, Psmb9) were up-regulated by HBX. On the other hand, the proteasome unit (Psmb10) and ubiquilin, a gene that has a role in facilitating the degradation of ubiquitinated proteins through proteasome, were down-regulated.
In SAGE-based analysis of the transcriptional profiles on normal primary human hepatocytes expressing HBX protein by transient transfection, Wu et al. observed that the up-regulated genes fell into three categories, including genes encoding ribosomal protein, transcription factors with the zinc finger motif, multiple proteasome subunits, ubiquitin, and ubiquitin-associated proteins (33). Some of these changes showed a pattern similar to that found in our transgenic mouse model expressing the HBX protein, although the genes with significant changes are different. This difference is probably a result of disparate experimental systems (33, 34). Our model is probably more physiologically relevant because it is an in vivo model. The changes in these gene groups relevant to HBX functions provide a valuable clue to understanding the complicated functions of this elusive viral protein.
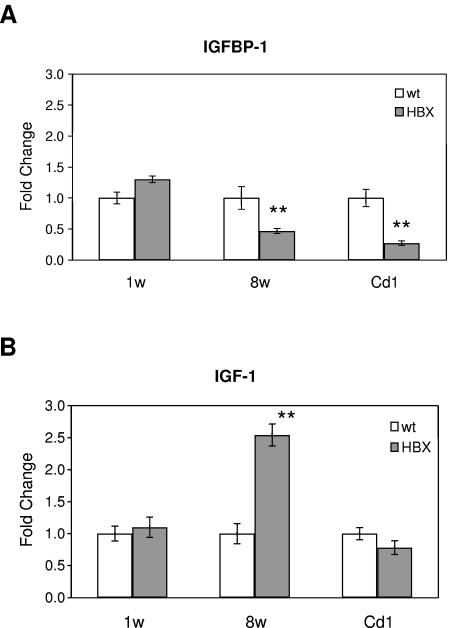
HBX protein has been implicated in the hepatocarcinogenesis of both the HBX transgenic mouse model and human livers with chronic hepatitis B virus infection, but the mechanism has not been fully elucidated. In our microarray analysis, we identified a significant down-regulation of IGFBP-1 in the livers of HBX transgenic mice. The change was further validated by quantitative TaqMan real-time PCR analysis in two HBX transgenic mouse models (Fig. 7A). IGFBP-1 is produced mainly in the liver and regulates the biological functions of the IGF (1). It has been demonstrated that IGF plays an important role in liver cancer development by enhancing cell growth and tumor transformation (1). This evidence indicates that IGFBP-1 may function as a negative modulator by neutralizing the effect of IGF in tumorigenesis. It was found that the overexpression of IGFBP-1 in transgenic mice inhibits hepatic neoplasia (22). Reduced expression of IGFBP-1 was also detected in various human cancers such as hepatocelluar carcinoma and prostate cancer (14, 21, 29). Therefore, the down-regulation of IGFBP-1 in HBX transgenic mouse livers may contribute to the carcinogenic potential of HBX.
FIG. 7.
Quantitative real-time PCR analysis of IGFBP-1 (A) and IGF-1 (B) expression in HBX transgenic livers. Gene expression levels of IGFBP-1 and IGF-1 were quantified by TaqMan RT-PCR. Liver samples from 1- and 8-week-old FVB/n mice (background for the MUP-HBX transgenic mice) and 8-week-old Cd1 mice (background for the second line of HBX transgenic mice) were analyzed as controls. HBX transgenic mice samples were compared with the corresponding control mice with the same background. Five or six mice were analyzed in each group, and the values are averaged from the individual mice. The data (means ± standard deviations) are displayed as fold changes of transgenic over control mice. Asterisks indicate differences with statistical significance (**, P < 0.01) between the HBX and wild-type (wt) mice by using Student's t test.
The expression of IGF and IGFBP has been shown to be regulated in a complex and interdependent manner; dysregulation of one is often reflected by corresponding changes in the expression of the other (26). Therefore, we examined the expression of IGF-1 in the HBX transgenic mouse liver. The IGF-1 expression was indeed up-regulated by quantitative PCR in the MUP-HBX mouse liver at week 8 compared to that at week 1 (Fig. 7B). However in the Cd1 HBX mice, the hepatic expression of IGF-1 was not significantly different from that of the control mouse liver. This lack of increase in IGF-1 expression probably reflects the difference in the mouse strain and the HBX expression profile between the two HBX mouse models.
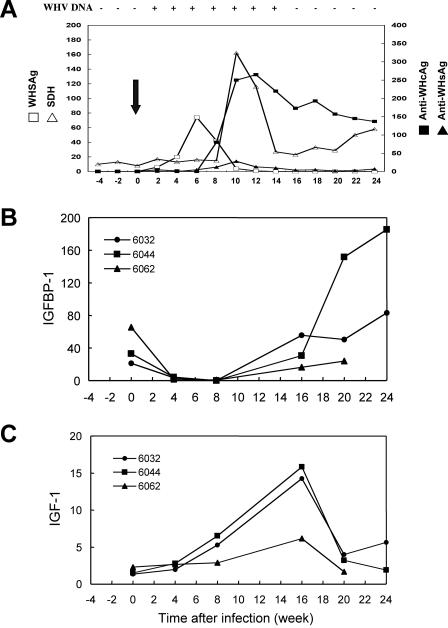
To further study the biological relevance of IGFBP-1 down-regulation by HBX in HBV infection, we examined the expression of IGFBP-1 during the acute phase of WHV infection in a woodchuck model. As shown in Fig. 8, the level of IGFBP-1 expression decreased significantly following WHV infection and was maintained at a low level until week 16, when WHV DNA became undetectable, and then rebounded to a higher level above the basal level. Similar to the MUP-HBX transgenic mice, the hepatic expression of IGF-1 also demonstrated an increase during the acute infection (Fig. 8C). However, this increase appeared to occur later than the decrease of IGFBP-1, suggesting a secondary change in the response to reduced IGFBP-1 expression. All three animals tested exhibited a similar trend. The correlation of IGFBP-1 down-regulation with WHV infection further supports the effect of HBX on IGFBP-1 expression in the context of viral infection and suggests an important role of IGFBP-1 in the pathogenesis of HBV infection.
FIG. 8.
Expression of IGFBP-1 and IGF-1 in woodchuck liver infected with WHV. Naïve adult woodchucks were infected with WHV as described previously (38). Blood and liver biopsy samples were collected at various time points before and after the virus inoculation. A representative course of WHV infection with serologic markers and liver function tests is shown (arrow indicates time of infection) (A). Quantitative real-time PCR analysis of the expression for IGFBP-1 (B) and IGF-1 (C) was performed using total RNA isolated from the liver biopsy samples of three separates woodchucks (numbered as 6032, 6044, and 6062) at various time points during acute WHV infection as indicated in the figure. The values of IGFBP-1 and IGF-1 expression were normalized to the level of GAPDH.
In line with the oncogenic potential of HBX, we have also noted the down-regulation of a number of mitochondria ribosomal genes and genes involved in the detoxification process, including cytochrome P-450, glutathione transferase, UDP-glucuronosyltransferase, and peroxiredoxin. The changes of these genes may lead to a reduced capability of detoxification by the liver and, possibly, contribute to the hepatocarcinogenic effect of HBV infection. Relevant to the function of this gene group, it is worth noting that the expression of selenium-binding protein 1 (Selenbp1) in the HBX mice was significantly reduced. Involvement of mammalian selenium-containing proteins in the detoxification of the liver has been well documented, such as in the case of glutathione transferase (4). HBX has also been shown to down-regulate the expression of selenoprotein P in a hepatoma cell line in vitro, possibly explaining the induction of tumor necrosis factor alpha expression by HBX (35). Although the function of Selenbp1 is not entirely clear, it is plausible that it is involved the biological function of selenoproteins either as a regulator or a coeffector (12). Therefore, the down-regulation of Selenbp1 by HBX may play a role in the pathogenesis of HBV infection and warrants further study.
Our study provided evidence for the inhibitory effect of HBX on the proteasome activities in vivo, which may be one of the mechanisms that lead to the profound changes in the global gene expression patterns of hepatocytes. These perturbations occur in a manner that is consistent with the pleiotropic functions of HBX and render a potential molecular mechanism for the biological role of HBX in HBV infection and hepatocellular carcinoma development.
Acknowledgments
We thank Frank Chisari and James Ou for providing the HBX transgenic mice, Xin Wang for assistance with microarray analysis, and the members of the Liver Diseases Branch of NIDDK, NIH, for helpful discussions.
This work was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
REFERENCES
- 1.Alexia, C., G. Fallot, M. Lasfer, G. Schweizer-Groyer, and A. Groyer. 2004. An evaluation of the role of insulin-like growth factors (IGF) and of type-I IGF receptor signalling in hepatocarcinogenesis and in the resistance of hepatocarcinoma cells against drug-induced apoptosis. Biochem. Pharmacol. 68:1003-1015. [DOI] [PubMed] [Google Scholar]
- 2.Andrisani, O. M., and S. Barnabas. 1999. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int. J. Oncol. 15:373-379. [DOI] [PubMed] [Google Scholar]
- 3.Aufiero, B., and R. Schneider. 1990. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 9:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behne, D., and A. Kyriakopoulos. 2001. Mammalian selenium-containing proteins. Annu. Rev. Nutr. 21:453-473. [DOI] [PubMed] [Google Scholar]
- 5.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H.-S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong, J., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5. s subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colgrove, R., G. Simon, and D. Ganem. 1989. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J. Virol. 63:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross, J. C., P. Wen, and W. Rutter. 1993. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc. Natl. Acad. Sci. USA 90:8078-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doria, M., N. Klein, R. Lucito, and R. J. Schneider. 1995. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 14:4747-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giometti, C. S., X. Liang, S. L. Tollaksen, D. B. Wall, D. M. Lubman, V. Subbarao, and M. S. Rao. 2000. Mouse liver selenium-binding protein decreased in abundance by peroxisome proliferators. Electrophoresis 21:2162-2169. [DOI] [PubMed] [Google Scholar]
- 13.Glas, R., M. Bogyo, J. S. McMaster, M. Gaczynska, and H. L. Ploegh. 1998. A proteolytic system that compensates for loss of proteasome function. Nature 392:618-622. [DOI] [PubMed] [Google Scholar]
- 14.Gong, Y., L. Cui, and G. Y. Minuk. 2000. The expression of insulin-like growth factor binding proteins in human hepatocellular carcinoma. Mol. Cell. Biochem. 207:101-104. [DOI] [PubMed] [Google Scholar]
- 15.Guidotti, L. G., B. Matzke, C. Pasquinelli, J. M. Shoenberger, C. E. Rogler, and F. V. Chisari. 1996. The hepatitis B virus (HBV) precore protein inhibits HBV replication in transgenic mice. J. Virol. 70:7056-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J., J. Kwong, E. C. Sun, and T. J. Liang. 1996. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, C. M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317-320. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J. W., and X. W. Wang. 2003. Gene expression profiling of preneoplastic liver disease and liver cancer: a new era for improved early detection and treatment of these deadly diseases? Carcinogenesis 24:363-369. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. W., Q. Ye, M. Forgues, Y. Chen, A. Budhu, J. Sime, L. J. Hofseth, R. Kaul, and X. W. Wang. 2004. Cancer associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology 39:518-527. [DOI] [PubMed] [Google Scholar]
- 21.Kondoh, N., T. Wakatsuki, A. Ryo, A. Hada, T. Aihara, S. Horiuchi, N. Goseki, O. Matsubara, K. Takenaka, M. Shichita, K. Tanaka, M. Shuda, and M. Yamamoto. 1999. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 59:4990-4996. [PubMed] [Google Scholar]
- 22.Lu, S., and M. C. Archer. 2003. Insulin-like growth factor binding protein-1 over-expression in transgenic mice inhibits hepatic preneoplasia. Mol. Carcinog. 36:142-146. [DOI] [PubMed] [Google Scholar]
- 23.Maguire, H. F., J. P. Hoeffler, and A. Siddiqui. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, S. 2001. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 25.Qadri, I., H. F. Maguire, and A. Siddiqui. 1995. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc. Natl. Acad. Sci. USA 92:1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider, M. R., L. H., Wu, M., Hoeflich, A., and E. Wolf. 2000. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. FASEB J. 14:629-640. [DOI] [PubMed] [Google Scholar]
- 27.Seto, E., P. J. Mitchell, and T. S. Yen. 1990. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature 344:72-74. [DOI] [PubMed] [Google Scholar]
- 28.Sitterlin, D., T. H. Lee, S. Prigent, P. Tiollais, J. S. Butel, and C. Transy. 1997. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 71:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennant, M. K., R. L. Vessella, C. C. Sprenger, R. A. Sikes, V. Hwa, L. D. Baker, and S. R. Plymate. 2003. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1/mac 25) is reduced in human prostate cancer and is inversely related to tumor volume and proliferation index in Lucap 23.12 xenografts. Prostate 56:115-122. [DOI] [PubMed] [Google Scholar]
- 30.Truant, R., J. Antunovic, J. Greenblatt, C. Prives, and J. A. Cromlish. 1995. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 69:1851-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X. W., K. Forrester, H. Yeh, M. A. Feitelson, J. R. Gu, and C. C. Harris. 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 91:2230-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waris, G., K.-W. Huh, and A. Siddiqui. 2001. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-κB via oxidative stress. Mol. Cell. Biol. 21:7721-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, C. G., M. Forgues, S. Siddique, J. Farnsworth, K. Valerie, and X. W. Wang. 2002. SAGE transcript profiles of normal primary human hepatocytes expressing oncogenic hepatitis B virus X protein. FASEB J. 16:1665-1667. [DOI] [PubMed] [Google Scholar]
- 34.Wu, C. G., D. M. Salvay, M. Forgues, K. Valerie, J. Farnsworth, R. S. Markin, and X. W. Wang. 2001. Distinctive gene expression profiles associated with hepatitis B virus x protein. Oncogene 20:3674-3682. [DOI] [PubMed] [Google Scholar]
- 35.Yi, Y. S., S. G. Park, S. M. Byeon, Y. G. Kwon, and G. Jung. 2003. Hepatitis B virus X protein induces TNF-alpha expression via down-regulation of selenoprotein P in human hepatoma cell line, HepG2. Biochim. Biophys. Acta 1638:249-256. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Z., U. Protzer, Z. Hu, J. Jacob, and T. J. Liang. 2004. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J. Virol. 78:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Z., N. Torii, Z. Hu, J. Jacob, and T. J. Liang. 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 108:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]