Abstract
The ARF tumor suppressor participates in a p53-dependent apoptotic pathway that is stimulated in response to some oncogenic stimuli. The E2F1 transcription factor is a critical downstream target of the Rb tumor suppressor and, when active, can promote proliferation as well as apoptosis. The finding that E2F1 transcriptionally regulates the ARF gene has led to the suggestion that ARF contributes to E2F1-induced apoptosis. Counter to this hypothesis, this study demonstrates not only that ARF is unnecessary for E2F1 to induce apoptosis but also that inactivation of ARF actually enhances the ability of E2F1 to promote apoptosis. Inactivation of ARF also cooperates with E2F1 activity to promote entry into the S phase of the cell cycle. This relationship between ARF and E2F1 is demonstrated in transgenic epidermis in vivo and in mouse embryo fibroblast cultures in vitro. In contrast, the ability of Myc to induce apoptosis is diminished in the absence of ARF. E2F1 induces the accumulation of p53 in the absence of ARF, and this is associated with the phosphorylation of p53 on several residues. These findings demonstrate that ARF is a negative regulator of E2F1 activity and is not required for E2F1-induced apoptosis.
ARF (p19ARF in mice, p14ARF in humans) is one of two tumor suppressors encoded at the INK4a locus, a chromosomal region frequently deleted in human tumors (21, 23, 36, 46). Mice specifically lacking ARF, but retaining functional p16INK4a, are predisposed to developing tumors, the most common being sarcomas and lymphomas (22, 23). ARF functions as a major modifier of various p53-dependent signaling pathways through its ability to inhibit mdm2 (8, 25, 37, 66). mdm2 antagonizes p53 by blocking its transcriptional activity, promoting its translocation to the cytoplasm, and targeting it for degradation by the proteasome (45, 53). ARF may inhibit mdm2 by sequestering mdm2 in the nucleolus and promoting mdm2 degradation (44, 56, 62, 65). The mdm2 gene is a transcriptional target of p53 and thus participates in a negative-feedback loop that normally maintains p53 at low levels (3, 63). ARF activates p53 by disrupting the p53-mdm2 feedback loop (45, 53).
ARF mediates the activation of p53 in response to oncogenic signals such those emanating from Ras, E1A, and Myc (8, 37, 66). Mouse embryonic fibroblasts (MEFs) null for ARF do not undergo replicative senescence and can be transformed by Ras alone in the absence of an immortalizing oncogene such as E1A or Myc (23, 52). In primary keratinocytes, ARF is required for the p53-dependent cell cycle arrest program induced by Ras (29). Moreover, the activation of p53 and the promotion of apoptosis by Myc and E1A are impaired in the absence of ARF (8, 66). Transgenic mice expressing Myc under the control of the immunoglobulin enhancer develop B-cell lymphoma at an accelerated rate when ARF is hemizygous or nullizygous (10, 51). This suggests that the ability to activate p53 in response to oncogenic signals underlies the tumor suppressor function of ARF.
The mechanism by which at least some oncogenic signals stimulate ARF is thought to be through the activation of E2F-dependent transcription. An E2F DNA-binding site is located in the ARF promoter region, and overexpression of E2F1 transcriptionally activates the ARF gene (3a, 7). In cultured human fibroblasts, transcriptional activation of ARF by E2F1 leads to the induction of a senescence-like state (9). In other cell types, overexpression of E2F1, like inactivation of Rb, leads to both deregulated proliferation and p53-dependent apoptosis (20, 64). It has been postulated that the transcriptional activation of ARF by E2F1 is one mechanism by which E2F1 promotes p53-dependent apoptosis (40, 53).
In addition to aberrant oncogenic signaling, p53 is activated in response to a variety of other stress stimuli including DNA damage, hypoxia, and ribonucleotide depletion (13). These different stresses appear to activate p53 through unique pathways. For example, ionizing radiation and drugs that induce double-strand breaks activate p53 through a pathway that involves the ataxia-telangiectasia mutated (ATM) kinase. ATM phosphorylates p53 directly on serine 15 (2, 36). In addition, ATM phosphorylates and activates the Chk2 kinase, which in turn phosphorylates p53 on serine 20 (5, 15, 54). UV radiation also activates p53 by inducing phosphorylation of p53 on serines 15 and 20 through a pathway that involves the ATM-related kinase, ATR, and perhaps the Chk1 kinase (31, 57). These N-terminal phosphorylation events inhibit the interaction between mdm2 and p53, which allows p53 to accumulate and activate transcription (55). It has been demonstrated that ARF can modify the response to some of these other stress signals that activate p53 (25).
Our laboratory has generated transgenic mouse lines expressing E2F1 or Myc under the control of a keratin 5 (K5) promoter (41, 48). The K5 promoter is active in the basal cell layer of the epidermis and several other epithelial tissues (47). Both K5 E2F1 and K5 Myc transgenic mice exhibit hyperplasia, hyperproliferation, and aberrant apoptosis in their epidermis at levels that correlate with the levels of transgene expression (41, 48). Apoptosis induced by E2F1 or Myc in transgenic epidermis is dependent on functional p53 (42, 48). K5 E2F1 and K5 Myc transgenic mice are both predisposed to spontaneous tumor development, and this is accelerated in a p53 heterozygous or nullizygous background (41, 43, 48). Consistent with ARF being a transcriptional target of E2F1, K5 E2F1 transgenic mice overexpress ARF in their epidermis (60).
To determine the role of ARF in E2F1-induced apoptosis, we introduced the K5 E2F1 transgene into an ARF null background. Surprisingly, inactivation of ARF did not impair the ability of E2F1 to promote apoptosis in this model system but actually enhanced this activity. In contrast, Myc-induced apoptosis was impaired in transgenic epidermis lacking ARF. Inactivation of ARF also cooperated with E2F1 to stimulate proliferation in transgenic epidermis. ARF functioned as an inhibitor of E2F1-induced apoptosis and proliferation in a cell-autonomous and non-cell-type-specific manner since similar results were obtained using primary MEF cultures in vitro. Finally, E2F1 is demonstrated to induce the accumulation of p53 in the absence of ARF, and this is associated with the phosphorylation of p53 on multiple residues. These findings demonstrate that ARF differentially regulates apoptosis induced by E2F1 and Myc and suggests that one pathway used by E2F1 to promote apoptosis involves the phosphorylation of p53.
MATERIALS AND METHODS
Mice and MEF cultures.
The generation of K5 E2F1 and K5 Myc transgenic mice has been described previously (41, 48). Briefly, the transgenes contain the bovine K5 promoter (47), the rabbit β-globin intron 2, and the simian virus 40 polyadenylation signal, and either the human E2F1 cDNA or a murine genomic fragment containing Myc exons 2 and 3. K5 E2F1 (lines E2F1.0 and E2F1.1) and K5 Myc (line MM5) transgenic mice in the SENCAR inbred strain SSIN background were crossed to ARF−/− mice (23) to generate K5 E2F1 or K5 Myc transgenic mice heterozygous for ARF. F1 transgenic mice heterozygous for ARF were then crossed to ARF heterozygous mice to generate K5 E2F1 or K5 Myc transgenic and nontransgenic siblings that were either wild type, heterozygous, or nullizygous for ARF. After 6 weeks of age, sibling mice were sacrificed and skin sections were collected for analysis. ARF heterozygous mice were mated together to generate sibling-matched embryos for primary MEF cultures that were ARF+/+ or ARF−/−. MEFs were harvested from 14-day-old embryos and cultured to passage 3 to 5 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) before being used in infection experiments.
BrdU incorporation.
Mice were injected intraperitoneally with 170 μl of 20 mM bromodeoxyuridine (BrdU) and allowed to incorporate it for 20 min before being sacrificed. Skin samples were fixed in formalin, paraffin embedded, and sectioned. Immunohistochemistry was performed on skin sections using an antibody specific for BrdU (Becton Dickinson) as previously described (41). For determining the percent BrdU incorporation, interfollicular basal layer keratinocytes were examined and the numbers of unstained and stained cells were determined.
TUNEL assay.
Terminal deoxynucleotidyl transferase-mediated deoxyuridine 5"-triphosphate-biotin nick end labeling (TUNEL) assays were performed with formalin-fixed, paraffin-embedded skin sections and an ApopTag in situ apoptosis detection kit (Oncor, Gaithersburg, Md.) as specified by the manufacturer. TUNEL-positive epidermal keratinocytes were visualized by peroxidase-diaminobenzidine staining, and the average number of positive cells per 10 mm of interfollicular epidermis was determined.
Activated caspase-3 immunohistochemistry.
Skin sections were fixed in formalin, paraffin embedded, and sectioned. After deparaffinization and hydration, sections were incubated with primary caspase-3 antibody (1:2,000) (R&D Systems) for 30 min. The slides were then washed and incubated with secondary anti-rabbit antibody and enzyme conjugate from the Histostatin-plus kit (Zymed) for 10 min each. Caspase-3-positive cells were visualized using the kit's diaminobenzidine mixture, rinsed, and counterstained with hemotoxylin for 12 to 15 s. Apoptosis was scored as the average number of activated caspase-3-positive cells per 10 mm of interfollicular epidermis.
Viral infections.
Recombinant adenoviruses encoding human E2F1, human p53, and green florescent protein (GFP) (AdE2F1, Adp53, and AdGFP) were a kind gift of Timothy Kowalik (University of Massachusetts Medical School) and have been described previously (27). MEFs were plated at 200,000 cells/100-mm plate and cultured overnight in DMEM with 10% FBS. Cultures were then incubated in DMEM with 0.1% FBS 24 h prior to infection. On the day of infection, the cells were washed, counted, and cultured in 1 ml of serum-free medium containing AdE2F1, AdGFP, and/or Adp53 at the multiplicity of infection (MOI) indicated in each experiment. After 1 h of infection, 9 ml of DMEM with 0.1% FBS was added, and the cells were incubated for an additional 24 h or longer if indicated. To measure apoptosis, cells were alcohol fixed and stained with 4",6-diamidino-2-phenylindole (DAPI) and subjected to microscopic examination. Cells with condensed, abnormally shaped, and/or fragmented nuclei were scored as apoptotic. To measure S-phase entry, cells were incubated with 10 μM BrdU for 1 h, fixed in ethanol, and immunohistochemically stained for BrdU incorporation. The percentage of cells that were BrdU positive was determined by examining cells microscopically.
Western blot analysis.
Epidermal whole-cell lysates were collected from dorsal skin by separating the epidermis from dermis and scraping the epidermis onto a glass plate on ice. Epidermal tissue and cell lysates were collected in lysis buffer (50 mM Tris-HCl, 1% IGEPAL, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg each of aprotinin, leupeptin, and pepstatin per ml, 1 mM Na3 VO4, 1 mM NaF) and freeze-thawed, and the supernatant was collected by high-speed centrifugation. Protein samples were quantified using the BCA protein assay kit (Pierce Endogen; no. 23225). The lysates were electrophoresed through a sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane by wet transfer. Immunoblotting was performed with rabbit antisera specific for E2F1 (C-20; Santa Cruz Biotechnology) and β-tubulin (H-2350; Santa Cruz Biotechnology) and monoclonal antibodies specific for Ras (Transduction Laboratories), p53, or phosphorylated forms of p53 (Oncogene Science Ab-3 and Cell Signaling Technology no. 9282, 9284, 9287S, and 9281).
RESULTS
Inactivation of ARF enhances apoptosis and proliferation in K5 E2F1 transgenic epidermis.
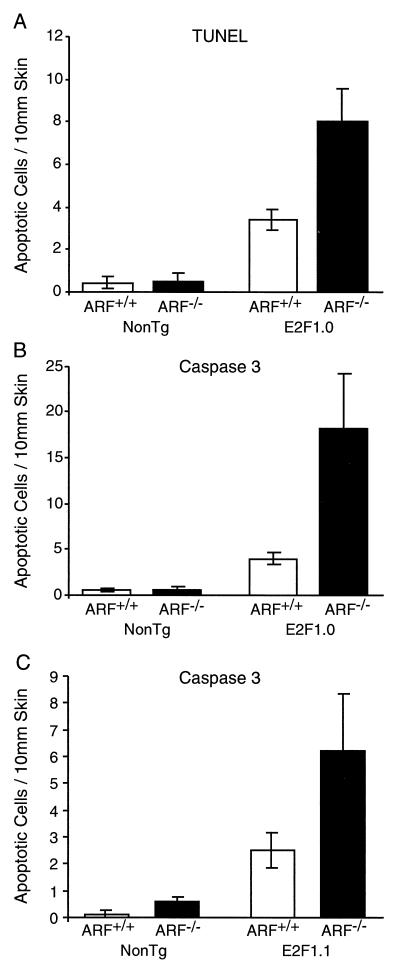
To directly examine the role of ARF in regulating the apoptotic activity of E2F1 in vivo, transgenic mice expressing E2F1 under the control of the K5 promoter (41) were crossed with mice lacking a functional ARF allele (23). K5 E2F1 transgenic mice heterozygous for ARF were then crossed to ARF heterozygous mice to generate sibling K5 E2F1 transgenic and nontransgenic mice that were either wild type, heterozygous, or nullizygous for a functional ARF gene. K5 E2F1 transgenic mice have previously been shown to have increased proliferation and aberrant, p53-dependent apoptosis in their epidermis (41, 42). Consistent with ARF being a transcriptional target of E2F1 (3a, 7), K5 E2F1 transgenic mice overexpress ARF in their epidermis (60). At 6 weeks of age the mice were sacrificed and skin samples were taken for analysis. Unexpectedly, the absence of ARF did not impair E2F1-induced apoptosis in transgenic epidermis as measured by in situ TUNEL analysis (Fig. 1A). Instead, apoptosis was increased more than twofold in K5 E2F1 (line E2F1.0) transgenic epidermis null for ARF compared to line E2F1.0 transgenic epidermis wild type for ARF. To confirm these results, skin sections from the same group of mice were analyzed for the presence of the activated form of caspase-3 by immunohistochemical staining (Fig. 1B and 2). Again, apoptosis as measured by caspase-3 activation was increased (approximately fourfold) in line E2F1.0 transgenic epidermis null for ARF compared to line E2F1.0 transgenic epidermis wild type for ARF. Similar results were obtained when an independent K5 E2F1 transgenic line expressing a lower level of E2F1 (line E2F1.1) was crossed into an ARF null background (Fig. 1C).
FIG. 1.
Inactivation of ARF enhances apoptosis in K5 E2F1 transgenic epidermis. (A) The TUNEL assay was performed on skin sections from nontransgenic (NonTg) and K5 E2F1 transgenic (line E2F1.0) mice that were either wild type for ARF (ARF+/+) or null for ARF (ARF−/−). The average number of TUNEL-positive cells per 10 mm of skin is presented for samples taken from at least four independent mice in each group. (B) Skin sections from the same mice as above were immunohistochemically stained for the activated form of caspase-3. The average number of caspase-3-positive cells per 10 mm of linear skin is presented. (C) Caspase-3 immunohistochemistry was performed on skin section from nontransgenic (NonTg) and K5 E2F1 transgenic (line E2F1.1) mice that were either wild type for ARF (ARF+/+) or null for ARF (ARF−/−). The average number of caspase-3-positive cells per 10 mm of linear skin is presented from samples taken from at least four independent mice in each group. Bars represent standard error.
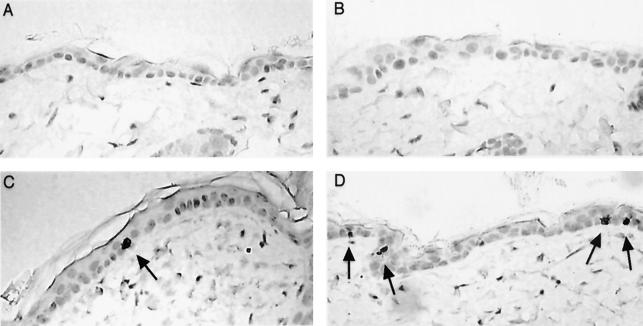
FIG. 2.
Immunohistochemical staining of activated caspase-3 in skin samples. Skin samples were taken from mice of the following genotypes: nontransgenic, ARF+/+ (A), nontransgenic, ARF−/− (B), line E2F1.0, ARF+/+ (C), and line E2F1.0, ARF−/− (D). Sections were immunohistochemically stained for the activated form of caspase-3 and photomicrographed.
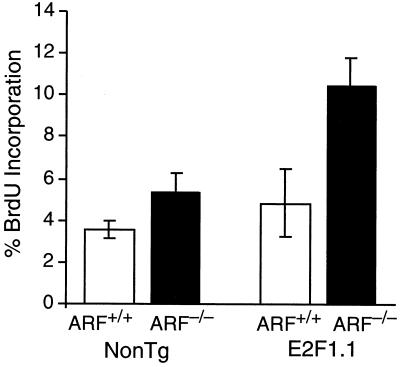
To examine the effect of ARF status on proliferation induced by E2F1, K5 E2F1 transgenic mice (line E2F1.1) were injected with BrdU 20 min prior to sacrifice. Skin samples were sectioned and immunohistochemically stained for BrdU incorporation. Consistent with previous results (41), line E2F1.1 mice had a barely detectable increase over wild-type mice in the percentage of BrdU-positive basal-layer keratinocytes in the interfollicular epidermis (Fig. 3). Interestingly, mice null for ARF also appeared to have a slight hyperproliferation of interfollicular epidermal keratinocytes. The slight increases in the average percentage of BrdU-positive keratinocytes in the epidermis of line E2F1.1 or ARF-null mice were not statistically significant based on the number of skin samples examined. In contrast, there was a statistically significant increase (over twofold) in the percentage of interfollicular basal-layer keratinocytes in S phase in line E2F1.1 mice that were also null for ARF compared to the proliferative index in wild-type, line E2F1.1, or ARF−/− epidermis (Fig. 3). This demonstrates that mild overexpression of E2F1 cooperates with inactivation of ARF to stimulate proliferation in mouse epidermis.
FIG. 3.
Overexpression of E2F1 and inactivation of ARF cooperate to stimulate proliferation in mouse epidermis. Line E2F1.1 transgenic mice (E2F1.1) and nontransgenic (NonTg) sibling controls that were either wild type for ARF (ARF+/+) or null for ARF (ARF−/−) were injected with BrdU and allowed to incorporate the BrdU for 20 min before being sacrificed. Skin sections were immunohistochemically stained for BrdU, and the average percentage of cells in the interfollicular epidermis positive for BrdU from at least four independent mice in each group was calculated. The E2F1.1, ARF−/− group was significantly different from the other groups by one-way analysis of variance with a Tukey Honestly Significantly Different Post-Hoc test with a significance level of 0.05.
Inactivation of ARF enhances E2F1-induced apoptosis and S-phase entry in MEFs.
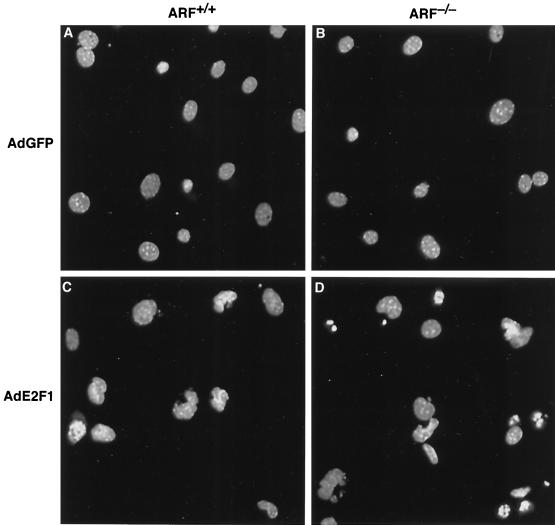
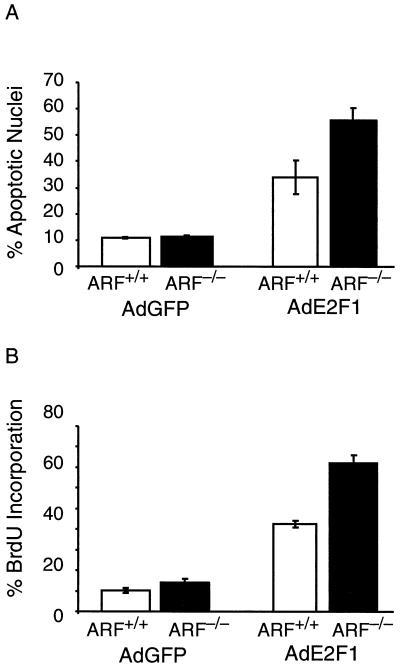
To determine the role of ARF in modulating E2F1 activity in another cell type, primary MEF cultures were generated from 14-day-old ARF-null embryos and wild-type sibling controls. These primary MEF cultures were cultured to passage 3 to 5 in 10% serum and then shifted to medium containing 0.1% serum. At 24 h after serum starvation, MEF cultures were infected with AdE2F1 or AdGFP at an MOI of 100. At 24 h following infection, the cells were fixed and stained with DAPI. They were then examined microscopically, and apoptotic cells were identified morphologically as cells with condensed, abnormally shaped, and fragmented nuclei (Fig. 4). At 24 h postinfection, wild-type MEF cultures infected with AdE2F1 had approximately a threefold increase in the percentage of apoptotic cells over the percentage of apoptotic cells in AdGFP or mock-infected wild-type MEF cultures (Fig. 5A). In ARF-null MEF cultures infected with AdE2F1, the increase in the percentage of apoptotic cells over background levels was greater than fivefold at 24 h postinfection. Thus, ARF is also not required for E2F1 to induce apoptosis in MEF cultures, and, again, ARF appears to negatively regulate E2F1-induced apoptosis in this cell type.
FIG. 4.
Morphological changes in apoptotic MEFs infected with AdE2F1. Early-passage MEF cultures isolated from ARF+/+ (A and C) and ARF−/− (B and D) sibling embryos were infected with AdGFP (A and B) or AdE2F1 (C and D) at an MOI of 100. At 24 h postinfection, the cells were stained with DAPI and examined microscopically. The nuclei of apoptotic cells appear condensed, abnormally shaped, and/or fragmented.
FIG. 5.
Inactivation of ARF enhances apoptosis and proliferation induced by E2F1 in MEF cultures. (A) Early-passage primary MEF cultures wild type or null for ARF were infected with AdGFP or AdE2F1 at an MOI of 100. At 24 h postinfection, the cells were ethanol fixed and stained with DAPI. Nuclei that appeared condensed, fragmented, or abnormally shaped were scored as apoptotic. The average percentage of cells that were apoptotic from four independent experiments is presented. (B) Infection of MEF cultures was performed as for panel A except that an MOI of 50 was used. The cells were fixed 24 h postinfection following a 1-hour incubation in 10 mM BrdU. The cells were immunohistochemically stained using an antibody specific for BrdU and microscopically examined. The average percentage of cells incorporating BrdU in three separate experiments is presented.
To determine if inactivation of ARF would also enhance E2F1-induced S-phase entry in primary embryonic fibroblasts, serum-deprived wild-type and ARF-null MEF cultures were infected with AdE2F1 or AdGFP at an MOI of 50. At 24 h postinfection, cells were allowed to incorporate BrdU and fixed. Cells were immunohistochemically stained for BrdU and examined microscopically. In ARF-null MEF cultures infected with AdE2F1, an average of over 70% of the cells were induced to enter S phase, compared to approximately 40% of the cells in S phase in wild-type MEF cultures infected with AdE2F1 (Fig. 5B). Together, these findings demonstrate that the function of ARF as a negative regulator of E2F1-induced apoptosis and proliferation is cell autonomous and not cell type specific.
Inactivation of ARF suppresses apoptosis in K5 Myc transgenic mice.
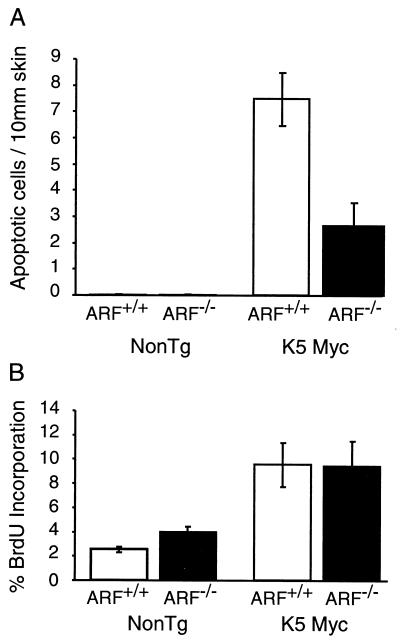
Zindy et al. previously demonstrated that p53-dependent apoptosis induced by Myc in primary MEF cultures is in part mediated through ARF (66). To determine the role of ARF in Myc-induced apoptosis in epidermal tissue in vivo, transgenic mice expressing Myc under the control of the K5 promoter were crossed to ARF-null mice. Previous studies demonstrated that K5 Myc transgenic mice have aberrant apoptosis in their epidermis and that this apoptosis is almost completely suppressed in a p53-null background (48). Epidermis from K5 Myc transgenic mice lacking ARF was found to have a threefold decrease in the number of apoptotic keratinocytes compared to epidermis from K5 Myc transgenic mice with functional ARF (Fig. 6A). Thus, consistent with previous in vitro studies, apoptosis in Myc transgenic mice is partially dependent on ARF. These findings demonstrate that ARF differentially regulates apoptosis mediated by Myc and E2F1.
FIG. 6.
Inactivation of ARF reduces apoptosis in K5 Myc transgenic epidermis. (A) The TUNEL assay was performed on skin sections from young adult nontransgenic (NonTg) and K5 Myc transgenic mice (line MM5) that were either wild type for ARF (ARF+/+) or null for ARF (ARF−/−). The average number of TUNEL-positive cells per 10 mm of skin is presented for samples taken from at least four independent mice in each group. (B) K5 Myc transgenic and nontransgenic (NonTg) control mice, wild type for ARF (ARF+/+) and null for ARF (ARF−/−), were injected with BrdU 20 min prior to sacrifice. Skin samples were immunohistochemically stained for BrdU incorporation, and the percentage of interfollicular basal layer keratinocytes staining positive was determined. The average percentage of cells that were BrdU positive from at least four independent mice in each group is presented.
The absence of ARF had no effect on the ability of Myc to stimulate proliferation in transgenic epidermis (Fig. 6B). This is in contrast to the negative effect of ARF on proliferation stimulated by E2F1 in transgenic epidermis. Thus, while ARF appears to be a general negative regulator of E2F1 activity, ARF specifically modulates the apoptotic activity but not the proliferative activity of Myc.
E2F1 induces the accumulation and phosphorylation of p53 in the absence of ARF.
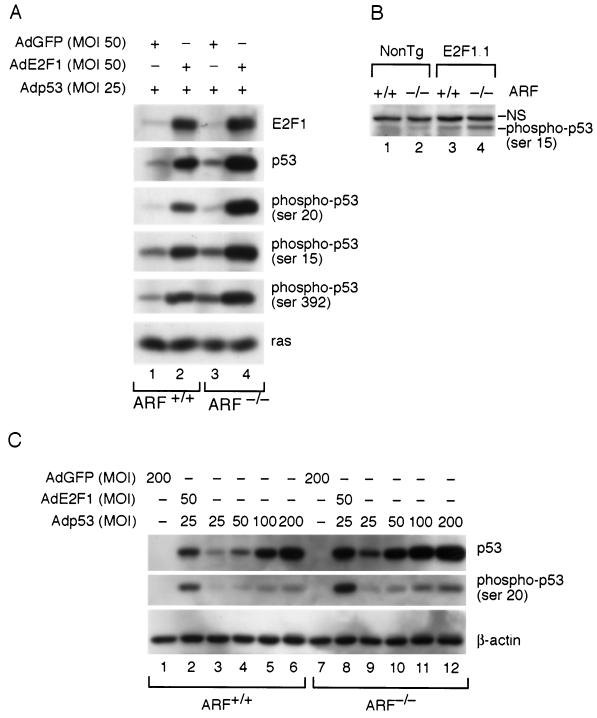
Previous studies have demonstrated that E2F1-induced apoptosis in K5 E2F1 transgenic epidermis and in MEF cultures is largely dependent on functional p53 (27, 42, 64). Consistent with p53 activation by E2F1, Western blot analysis revealed that p53 protein levels were upregulated in MEF cultures infected with AdE2F1 (Fig. 7A). For these experiments, wild-type and ARF-null MEFs were coinfected with a low MOI of a recombinant adenovirus expressing human p53. This was necessary because several of the p53 antibodies used in these experiments do not efficiently recognize mouse p53. The increase in p53 protein levels in response to E2F1 overexpression occurred in both ARF+/+ and ARF−/− backgrounds. In both cases, p53 levels were induced between 2.5- and 3-fold when E2F1 was ectopically expressed. Therefore, ARF is not required for E2F1 to induce the accumulation of p53.
FIG. 7.
E2F1 induces the phosphorylation of p53. (A) Primary MEF cultures wild type for ARF (lanes 1 and 2) or null for ARF (lanes 3 and 4) were coinfected with Adp53 (MOI of 25) and either AdGFP (lanes 1 and 3; MOI, 50) or AdE2F1 (lanes 2 and 4; MOI, 50). At 24 h postinfection, lysates were made and Western blot analysis was performed using antibodies specific for E2F1, p53, phosphoserine 15 p53, phosphoserine 20 p53, phosphoserine 392 p53, or ras where indicated. (B) Western blot analysis was performed on epidermal lysates (100 μg) from mice that were nontransgenic (lanes 1 and 2) or K5 E2F1 transgenic (lanes 3 and 4) and either wild type (lanes 1 and 3) or null (lanes 2 and 4) for ARF by using an antibody specific for the phosphoserine 15 form of p53. NS, nonspecific band. (C) Primary MEFs, either wild type for ARF (lanes 1 to 6) or null for ARF (lanes 7 to 12), were infected with the following recombinant adenoviruses: lanes 1 and 7, AdGFP (MOI, 200); lanes 2 and 8, AdE2F1 (MOI, 50) plus Adp53 (MOI, 25); lanes 3 and 9, Adp53 (MOI, 25); lanes 4 and 10, Adp53 (MOI, 50); lanes 5 and 11, Adp53 (MOI, 100); lanes 6 and 12, Adp53 (MOI, 200). At 24 h following infection, cell lysates were made and Western blot analysis was performed using antibodies specific for p53 (top panel), the serine 20-phosphorylated form of p53 (middle panel), or β-actin (bottom panel).
It should be noted that infection with the same MOI of AdE2F1 consistantly resulted in modestly higher levels (1.3- to 1.7-fold) of E2F1 protein accumulation in ARF-null MEFs than in wild-type MEFs (Fig. 7A). This phenomenon occurred when multiple batches of sibling-matched MEF cultures were used. This finding is consistent with findings of others, which demonstrated that the expression of ARF inhibits E2F1 protein accumulation (11, 34). It is also possible, however, that this is an artifact of the recombinant adenovirus system. For instance, adenoviruses may infect ARF−/− cells more efficiently than they infect ARF+/+ cells. The basal levels of p53, in the absence of exogenous E2F1, was also higher in ARF-null MEFs than in wild-type MEFs following infection with the same MOI of Adp53 (Fig. 7C). Again, this may be an artifact of the experimental system or the result of a redundant compensatory mechanism that regulates p53 protein levels in the absence of ARF.
Several forms of stress, including DNA damage, induce the accumulation and activity of p53 by stimulating p53 phosphorylation. To determine if the phosphorylation of p53 might be involved in p53 activation by E2F1, phosphospecific antibodies were used to examine the status of p53 in E2F1-overexpressing MEF cultures. Increased levels of several phosphorylated forms of p53 were observed in MEF cultures overexpressing E2F1, including forms phosphorylated on serines 15, 20, and 392 (Fig. 7A, lower panals). Infection with AdE2F1, but not the AdGFP control, increased the amount of phosphorylated forms of p53 in both ARF+/+ and ARF−/− MEF cultures. Increased levels of the serine 15-phosphorylated form of p53 could also be detected in lysates from the epidermis of K5 E2F1 transgenic mice, and this was again independent of ARF status (Fig. 7B).
The increased levels of phosphorylated forms of p53 in E2F1-overexpressing cells were not simply a consequence of more p53 protein being phosphorylated at basal levels. Infection of ARF+/+ or ARF−/− MEFs with Adp53 alone at an MOI of 100 resulted in comparable or higher levels of total p53 protein compared with coinfection with AdE2F1 and Adp53 at an MOI of 25 (Fig. 7C, compare lanes 2 and 5 and lanes 8 and 11). However, the amount of p53 phosphorylated at serine 20 was twofold smaller in the cells infected with the high MOI of Adp53 than in cells coinfected with AdE2F1 and the lower MOI of Adp53. Even infection with Adp53 at a MOI of 200 resulted in lower levels of phoshorylated p53 than did coinfection with AdE2F1 and Adp53 at a MOI of 25 even though the total amount of p53 was larger in the cells infected with the high MOI of Adp53. Thus, ectopic expression of E2F1 leads to a greater percentage of p53 being in the phosphorylated form. These findings are consistent with the idea that E2F1 stimulates the phosphorylation of p53 and that this results in the accumulation of p53 in an ARF-independent manner.
DISCUSSION
Role of ARF in regulating apoptosis induced by E2F1 and Myc.
Loss of Rb function, as occurs in many cancers, results in deregulated cellular proliferation and an increased propensity of cells to undergo apoptosis. Depending on the context, the induction of apoptosis in response to Rb inactivation can be either p53 dependent or independent (1, 33, 35). Experimental data demonstrate that E2F1 contributes to both the increased proliferation and the p53-dependent apoptosis that occurs as a consequence of Rb inactivation (32, 38, 58). A caveat of the studies presented here is that E2F1 is overexpressed rather than deregulated by loss of Rb function. However, we have previously demonstrated that there is a relatively modest increase in E2F1 DNA-binding activity in K5 E2F1 transgenic keratinocytes, which corresponds to a level of E2F1 DNA-binding activity observed in many tumor cell lines (42). Moreover, Rb binds and regulates a large number of cellular factors, including other members of the E2F family. An advantage of using overexpression to mimic deregulation is that one can examine the role of a single Rb target, such as E2F1, in regulating cellular proliferation and apoptosis.
Like Rb inactivation, overexpression of E2F1 induces apoptosis by both p53-dependent and -independent pathways. For example, overexpression of E2F1 efficiently induces apoptosis in several human tumor cell lines lacking functional p53 (12, 16, 17, 19, 39). In addition, endogenous E2F1 contributes to the p53-independent apoptosis that occurs in response to T-cell receptor activation (30). Transcriptional activation of the p53-related factor p73 by E2F1 has been implicated in both of these cases of p53-independent apoptosis induced by E2F1 (19, 30). In both of the experimental systems used in this study (primary MEF cultures and transgenic epidermis), the ability of E2F1 to induce apoptosis is largely dependent on functional p53 (26, 42, 64). It has been widely suggested that ARF participates in the p53-dependent apoptosis induced by E2F1. In sharp contrast to this suggestion, the data presented here demonstrate that apoptosis induced by E2F1 in these p53-dependent systems does not require ARF. In fact, ARF appears to be a negative regulator of the apoptotic and proliferative activities of E2F1.
The observation that ARF appears to inhibit E2F1 activity might help explain recent studies demonstrating that ARF has cell cycle-regulatory functions that are independent of mdm2 and p53. Carnero et al. showed that ARF can induce growth arrest in the presence of a dominant negative mutant of p53 (4). ARF could also induce growth arrest in p53-defecient fibroblasts (4). Weber et al. demonstrated that triple-knockout mice lacking ARF, mdm2, and p53 developed tumors at greater frequency than did double-knockout mice lacking only p53 and mdm2 (61). Moreover, exogenous ARF expression inhibited proliferation in MEFs from triple-knockout mice, demonstrating that ARF can regulate cell cycle progression in the absence of both p53 and mdm2 (61). It is quite possible that the additional cell cycle-regulatory properties of ARF demonstrated in those studies is related to the finding shown here that ARF suppresses E2F1 activity.
It is possible that the increase in the apoptotic and proliferative activities of E2F1 as a consequence of ARF inactivation results from an indirect effect on other cell cycle regulators. However, lack of ARF does not enhance proliferation and reduces apoptosis in response to Myc. This suggests that ARF may specifically repress E2F1 activity. Two recent reports suggest that ARF physically interacts with E2F1, as well as with E2F2 and E2F3 (11, 34). Binding of ARF to E2F1 correlated with increased E2F1 protein turnover in a proteasome-dependent manner and an inhibition of E2F1-dependent transcription. Moreover, it was suggested that ARF sequesters E2F1 in nucleoli as an additional means of negatively regulating E2F1 (34).
Analysis of K5 Myc transgenic mice demonstrates that in contrast to apoptosis induced by E2F1, ARF does play a significant role in promoting apoptosis in response to Myc in mouse epidermal tissue. This finding is consistent with previous reports demonstrating that Myc induction of p53-dependent apoptosis in MEFs is partially dependent on ARF (66). Thus, Myc and E2F1 appear to utilize different pathways to activate p53 and induce apoptosis. The fact that ARF can either positively or negatively modulate apoptosis in response to different oncogenic signals demonstrates that the role of ARF in regulating p53-dependent activities is more complex than current models predict.
ARF has also been implicated in the ability of E1A to activate p53 and induce apoptosis (8). E1A is a viral oncoprotein that binds to and inactivates the Rb tumor suppressor protein. E1A induces the activity of E2F1 by dissociating E2F1, as well as other E2F family members, from Rb-containing inhibitory complexes. Although E1A activates E2F1, E1A and E2F1 also appear to induce apoptosis through different mechanisms since E1A-mediated apoptosis is partially dependent on ARF (8) while E2F1-mediated apoptosis is inhibited by ARF. The fact that E1A and E2F1 are not equivalent in this respect may not be surprising, given that E1A, as well as Rb, has many other activities that are independent of E2F1. It is also quite possible that under some conditions, ARF does participate in p53-dependent apoptosis induced by E2F1.
Induction of p53 by E2F1.
We have found that p53 accumulates in response to deregulated E2F1 activity in the absence of ARF. This is consistent with a study by others which also found that overexpression of E2F1 increased p53 protein levels in cells lacking ARF (66). The mechanism by which E2F1 induces p53 in the absence of ARF is probably related to our finding that E2F1 stimulates the phosphorylation of p53 on several residues. Several forms of stress, including DNA damage, activate p53 through the stimulation of p53 phosphorylation. Phosphorylation of p53 on N-terminal residues, particularly serine 20, inhibits the ability of mdm2 to bind and promote the degradation of p53 (6, 55, 59). Phosphorylation of p53 on N-terminal residues may also promote the acetylation of p53, which stimulates the DNA-binding activity of p53 (14, 28, 49). In addition, phosphorylation of p53 at serine 392 enhances the DNA-binding activity of p53, perhaps through promoting p53 tetramerization (18, 24, 50).
It is formally possible that E2F1 induces the accumulation of p53 by a mechanism that does not involve ARF or phosphorylation and that the increased levels of phosphorylated forms of p53 are the result of more p53 being phosphorylated at basal levels. This does not appear to be the case, however. When p53 levels were increased by infecting cells with a high MOI of Adp53, this p53 protein was phosphorylated at lower levels than was p53 from cells coinfected with E2F1 and a low MOI of p53. Therefore, the increase in phosphorylated forms of p53 observed in E2F1-overexpressing cells appears to be the cause, rather than a consequence, of p53 protein accumulation. This suggests that E2F1 induces an activity that leads to the phosphorylation of p53.
The finding that p53 is phosphorylated in response to E2F1 overexpression provides a possible mechanism by which E2F1 could induce p53-dependent apoptosis in the absence of ARF. Recent studies by Kowalik and coworkers have demonstrated a close association between the ability of E2F1 to stimulate p53 phosphorylation and the ability of E2F1 to induce apoptosis. E2F2, which stimulates ARF expression but does not stimulate p53 phosphorylation, does not induce apoptosis in MEF cultures (H. A. Rogoff et al., submitted for publication). Moreover, E2F1 DNA-binding and transcriptional activation mutants that do not induce apoptosis in MEF cultures also do not induce p53 phosphorylation. Future studies will be aimed at identifying the mechanism by which E2F1 stimulates p53 phosphorylation and the kinase(s) responsible for this event.
Acknowledgments
We are especially grateful to Tim Kowalik for helpful discussions, reagents, and training. We thank Jen Smith for outstanding technical assistance, Dale Weiss and coworkers for animal care, Judy Ing and Chris Yone for artwork, and Shawnda Sanders for preparation of the manuscript.
This work was supported by grants from the National Institute of Health (CA79648 to D.G.J., CA42157 to C.J.C., NIEHS center grant ES007784, and CA16672). The work was also supported by Tobacco Settlement Funds as appropriated by the Texas State Legislature. J.L.R. is supported in part by an American Legion Auxiliary Fellowship.
J.L.R. and J.T.P. contributed equally to this work.
REFERENCES
- 1.Almasan, A., Y. Yin, R. E. Kelly, E. Y. Lee, A. Bradley, W. Li, J. R. Bertino, and G. M. Wahl. 1995. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc. Natl. Acad. Sci. USA 92:5436-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 3.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 3b.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 4.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. [DOI] [PubMed] [Google Scholar]
- 5.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 6.Chehab, N. H., A. Malikzay, E. S. Stavridi, and T. D. Halazonetis. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 96:13777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Stanchina, E., M. E. McCurrach, F. Zindy, S.-Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimri, G. P., K. Itahana, M. Acosta, and J. Campisi. 2000. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol. Cell. Biol. 20:273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eymin, B., L. Karayan, P. Seite, C. Brambilla, E. Brambilla, C. J. Larsen, and S. Gazzeri. 2001. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 10:1033-1041. [DOI] [PubMed] [Google Scholar]
- 12.Fueyo, J., C. Gomez-Manzano, W. K. A. Yung, T. J. Liu, R. Alemany, T. J. McDonnell, X. Shi, J. S. Rao, V. A. Levin, and A. P. Kyritsis. 1998. Overexpression of E2F-1 in glioma triggers apoptosis and suppresses tumor growth in vitro and in vivo. Nat. Med. 4:685-690. [DOI] [PubMed] [Google Scholar]
- 13.Giaccia, A. J., and M. B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 14.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 15.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh, J.-K., S. Fredersdorf, T. Kouzarides, K. Martin, and X. Lu. 1997. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 11:1840-1852. [DOI] [PubMed] [Google Scholar]
- 17.Hunt, K. K., J. Deng, T.-J. Liu, M. Wilson-Heiner, S. G. Swisher, G. Clayman, and M.-C. Hung. 1997. Adenovirus-mediated overexpression of the transcription factor E2F-1 induces apoptosis in human breast and ovarian carcinoma cell lines and does not require p53. Cancer Res. 57:4722-4726. [PubMed] [Google Scholar]
- 18.Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 19.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin, Jr. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, D. G., J. K. Schwarz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 21.Kamb, A., N. A. Gruis, J. Weaver-Feldhaus, Q. Liu, K. Harshman, S. V. Tavtigian, E. Stockert, R. S. Day III, B. E. Johnson, and M. H. Skolnick. 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436-440. [DOI] [PubMed] [Google Scholar]
- 22.Kamijo, T., S. Bodner, E. van de Kamp, D. H. Randle, and C. J. Sherr. 1999. Tumor spectrum in ARF-deficient mice. Cancer Res. 59:2217-2222. [PubMed] [Google Scholar]
- 23.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor, M., and G. Lozano. 1998. Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc. Natl. Acad. Sci. USA 95:2834-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, S. H., J. Moritsugu, and G. M. Wahl. 2000. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc. Natl. Acad. Sci. USA 97:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalik, T. F., J. DeGregori, G. Leone, L. Jakoi, and J. R. Nevins. 1998. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth & Differ. 9:113-118. [PubMed]
- 27.Kowalik, T. F., J. DeGregori, J. K. Schwarz, and J. R. Nevins. 1995. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J. Virol. 69:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 29.Lin, A. W., and S. W. Lowe. 2001. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc. Natl. Acad. Sci. USA 98:5025-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lissy, N. A., P. K. Davis, M. Irwin, W. G. Kaelin, and S. F. Dowdy. 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407:642-645. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Y., and E. Zacksenhaus. 2000. E2F1 mediates ectopic proliferation and stage-specific p53-dependent apoptosis but not aberrant differentiation in the ocular lens of Rb deficient fetuses. Oncogene 19:6065-6073. [DOI] [PubMed] [Google Scholar]
- 33.Macleod, K. F., Y. Hu, and T. Jacks. 1996. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 15:6178-6188. [PMC free article] [PubMed] [Google Scholar]
- 34.Martelli, F., T. Hamilton, D. P. Silver, N. E. Sharpless, N. Bardeesy, M. Rokas, R. A. DePinho, D. M. Livingston, and S. R. Grossman. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgenbesser, S. D., B. O. Williams, T. Jacks, and R. A. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72-74. [DOI] [PubMed] [Google Scholar]
- 36.Nobori, T., K. Miura, D. J. Wu, A. Lois, K. Takabayashi, and D. A. Carson. 1994. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368:753-756. [DOI] [PubMed] [Google Scholar]
- 37.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125-126. [DOI] [PubMed] [Google Scholar]
- 38.Pan, H., C. Yin, N. J. Dyson, E. Harlow, L. Yamasaki, and T. V. Dyke. 1998. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol. Cell. Biol. 2:283-292. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, A. C., S. Bates, K. M. Ryan, K. Helin, and K. H. Vousden. 1997. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 11:1853-1863. [DOI] [PubMed] [Google Scholar]
- 40.Phillips, A. C., and K. H. Vousden. 2001. E2F-1 induced apoptosis. Apoptosis 6:173-182. [DOI] [PubMed] [Google Scholar]
- 41.Pierce, A. M., S. M. Fischer, C. J. Conti, and D. G. Johnson. 1998. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumor development. Oncogene 16:1267-1276. [DOI] [PubMed] [Google Scholar]
- 42.Pierce, A. M., I. B. Gimenez-Conti, R. Schneider-Broussard, L. A. Martinez, C. J. Conti, and D. G. Johnson. 1998. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc. Natl. Acad. Sci. USA 95:8858-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce, A. M., R. Schneider-Broussard, I. B. Gimenez-Conti, J. L. Russell, C. J. Conti, and D. J. Johnson. 1999. E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Mol. Cell. Biol. 19:6408-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H.-W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2s inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 45.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 46.Quelle, D. E., F. Zindy, R. A. Ashman, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez, A., A. Bravo, J. L. Jorcano, and M. Vida. 1994. Sequences 5" of the bovine keratin 5 gene direct tissue-and-cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation. 58:53-64. [DOI] [PubMed] [Google Scholar]
- 48.Rounbehler, R. J., R. Schneider-Broussard, C. J. Conti, and D. G. Johnson. 2001. Myc lacks E2F1's ability to suppress skin carcinogenesis. Oncogene 20:5341-5349. [DOI] [PubMed] [Google Scholar]
- 49.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakaguchi, K., H. Sakamoto, M. S. Lewis, C. W. Anderson, J. W. Erickson, E. Appella, and D. Xie. 1997. Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry 36:10117-10124. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt, C. A., M. E. McCurrach, E. de Stanchina, R. R. Wallace-Brodeur, and S. W. Lowe. 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27-37. [DOI] [PubMed] [Google Scholar]
- 53.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689-3695. [PubMed] [Google Scholar]
- 54.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 55.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 56.Tao, W., and A. J. Levine. 1999. P19(ARF) stabilizes p53 by blocking nucleocytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai, K. Y., Y. Hu, K. F. Macleod, D. Crowley, L. Yamasaki, and T. Jacks. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2:293-304. [DOI] [PubMed] [Google Scholar]
- 59.Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, D., J. L. Russell, and D. G. Johnson. 2000. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Mol. Cell. Biol. 20:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. 2000. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 14:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 63.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 64.Wu, X., and A. J. Levine. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA 91:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 66.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]