Abstract
Insulin-like growth factor binding protein 1 (IGFBP-1) is a hypoxia-inducible gene that plays an important role in regulating embryonic growth and development under hypoxic stress. The molecular mechanisms underlying hypoxia-induced IGFBP-1 gene expression in the embryonic tissues are not well understood. Here we report that the hypoxia-inducible factor 1 (HIF-1) pathway is established in early embryogenesis and mediates hypoxia-induced IGFBP-1 expression. Hypoxia increased the HIF-1 activity, and HIF-1α overexpression or CoCl2 treatment resulted in elevated IGFBP-1 expression in zebra fish embryos. Although the zebra fish IGFBP-1 promoter contains 13 consensus hypoxia response elements (HREs), deletion and mutational analysis revealed that only the HRE positioned at −1090/−1086 is required for the hypoxia and HIF-1 induction. Further experiments revealed that there is an HIF-1 ancillary sequence (HAS) adjacent only to the functional HRE. Mutation of this HAS greatly reduced the responsiveness of the IGFBP-1 promoter to hypoxia and HIF-1. The HAS does not directly bind to HIF-1 or affect the binding of the HRE to HIF-1. The HAS is bound to a nuclear protein(s), and this HAS binding activity is reduced by hypoxia. These results suggest that HIF-1 mediates hypoxia-induced IGFBP-1 gene expression in early development by selectively interacting with the −1090/−1086 HRE and its adjacent HAS.
Hypoxia triggers the transcription of regulatory genes that promote O2 delivery and anaerobic metabolism, suppress major energy-requiring processes, and inhibit growth and development in animals ranging from invertebrates to mammals (14, 15, 40). Hypoxia also influences several human pathological processes, such as tumorigenesis and intrauterine growth restriction (IUGR). The majority of these transcriptional responses to hypoxia are mediated by the hypoxia-inducible factor 1 (HIF-1) complex. HIF-1 is a heterodimeric complex composed of HIF-1α and HIF-1β (44, 45). HIF-1β, also known as aryl hydrocarbon receptor nuclear translocator, is constitutively expressed and insensitive to O2 availability. When oxygen levels are high, HIF-1α is bound to the von Hippel-Lindau tumor suppressor (pVHL) and targeted for ubiquitination and proteosomal degradation (30, 34). Hypoxic conditions inhibit this degradation, which allows HIF-1α to accumulate in the cell (16, 18, 27). HIF-1α is then translocated to the nucleus, dimerizes with HIF-1β, binds to DNA, and activates target gene expression (3). Hypoxia response elements (HREs) are cis-regulatory DNA sequences that specifically bind to HIF-1 and are required for transcriptional induction upon hypoxia exposure.
Insulin-like growth factor binding protein 1 (IGFBP-1) is a hypoxia-inducible gene. Earlier studies have shown that circulating levels of IGFBP-1 are elevated in IUGR fetuses (7, 12, 42, 43). In vitro studies using cultured human cells and in vivo studies using rodent and fish models suggest that IGFBP-1 gene expression is elevated under hypoxic conditions (13, 29, 31, 35, 41). Since IGFBP-1 binds IGFs and inhibits IGF action on cell growth in vitro (4, 9) and because IGFBP-1 overexpression reduced birth weights in mice (5, 11, 36), it was postulated that elevated IGFBP-1 plays a major role in hypoxia-induced IUGR by binding IGFs and inhibiting their growth-promoting activities (41). Using the transparent and free-living zebra fish embryo as a model system, we have recently shown that (i) hypoxia strongly up-regulates IGFBP-1 expression and delays growth and developmental rate in zebra fish embryos; (ii) IGFBP-1 knockdown partially abrogates these hypoxic effects, whereas IGFBP-1 overexpression decreases growth and developmental rates under normoxia; and (iii) reintroduction of IGFBP-1 to the knocked down embryos restores the hypoxic effects (19). These findings provide strong evidence arguing that up-regulation of IGFBP-1 by hypoxia plays a key role in coordinating embryonic growth rate and developmental timing in response to environmental oxygen availability. Although there is in vitro evidence that overexpression of HIF-1α in cultured human hepatoma (HepG2) cells increases human IGFBP-1 promoter activity (41), how hypoxia triggers IGFBP-1 gene expression in vivo is not clear, and the cis-regulatory elements responsible for hypoxia-induced IGFBP-1 transcription in vivo are not well defined.
The objectives of this study are (i) to determine when the HIF-1 pathway becomes operational in early development and whether it plays a role in mediating hypoxia-induced IGFBP-1 gene expression in zebra fish embryos and (ii) to identify the key cis-regulatory element(s) responsible for hypoxia-induced IGFBP-1 transcription in vivo. Our results indicate that the HIF-1 pathway is established and operational in the earliest stages of vertebrate development and that it plays a major role in mediating hypoxia-induced IGFBP-1 gene expression in zebra fish embryos in vivo. Intriguingly, although the zebra fish IGFBP-1 promoter contains 13 consensus HREs, only one of these HREs is required for the hypoxia/HIF-1 regulation of IGFBP-1 gene expression in vivo. We show that a HIF-1 ancillary sequence (HAS) adjacent to the functional HRE is critical for its hypoxia/HIF-1 responsiveness.
MATERIALS AND METHODS
Materials.
All chemicals and reagents were purchased from Fisher Scientific (Pittsburgh, PA) unless noted otherwise. DNA polymerase was purchased from Promega (Madison, WI). Restriction endonucleases were purchased from New England BioLabs (Beverly, MA). Superscript II reverse transcriptase (RT) and oligonucleotide primers were purchased from Invitrogen Life Technologies, Inc. (Carlsbad, CA).
Experimental animals and procedures.
Wild-type zebra fish (Danio rerio) were maintained on a 14-h light-10-h dark cycle at 28°C and fed twice daily. Embryos were obtained by natural crossing. Fertilized eggs were raised in embryo medium at 28.5°C and staged according to the methods of Kimmel et al. (21). To inhibit embryo pigmentation, embryo medium was supplemented with 0.003% (wt/vol) 2-phenylthiourea (21). Hypoxia treatment was conducted as described previously (19). Cobalt chloride (CoCl2; Sigma, St. Louis, MO) was dissolved in embryo medium at concentrations ranging from 1 × 10−6 M to 1 × 10−2 M. Embryos at 12 h postfertilization (hpf) were transferred to and kept in these CoCl2 solutions for 24 h. The samples were collected and immediately frozen in liquid nitrogen and stored at −80°C. The experiments were conducted in accordance with procedures approved by the Animal Care and Use Committee, University of Michigan.
Molecular cloning.
Genomic DNA was isolated from adult zebra fish (AB-line) and used as a template for genomic PCR to amplify the promoter region of the zebra fish IGFBP-1 gene. A primer pair (5′-TAA GGT ACC ATT AAT GTA GAT GTG AAG CAT TTT CCT ATA CGT TTG GAC-3′ and 5′-AAG AAG CTT ACC GGT CCC AAA GCA CGG CTC AGA ATA AAT AGA TAA-3′) was designed based on Ensembl zebra fish genome data resources (http://www.ensembl.org/Danio_rerio/). The amplified PCR product was subcloned into pCR2.1-TOPO vector (Invitrogen) and sequenced. Sequences were analyzed using TESS (http://www.cbil.upenn.edu/tess) (37). To clone the full open reading frame of zebra fish HIF-1α cDNA, a primer set (5′-TT GCC GCC ACC ATG G AT ACT GGA GTT GTC ACT G-3′ and 5′-TCA GTT GAC TTG GTC CAG AG-3′) was designed based on the available sequence information (GenBank/EMBL/DDBJ accession number AY326951). The N terminus of zebra fish HIF-1α was fused with a hemagglutinin (HA) tag by PCR-based amplification using a primer set (5′-ACT AAG CTT GCC GCC ACC ATG TAC CCA TAC GAT GTT CCA GAT TAC GCT GAT ACT GGA GTT GTC ACT GA-3′ and 5′-GAT GCT GTG ACC GGT CAG AT-3′). The amplified PCR fragment was subcloned into the pcDNA 3.1 vector (Invitrogen) and sequenced.
Plasmid construction.
The isolated IGFBP-1 promoter region from −2025 to +89 (relative to the cap site) was digested by KpnI and HindIII and subcloned into a promoterless luciferase vector (pGL-basic2; Promega) in the sense orientation to generate p2025Luc. The p2025Luc construct was digested with PmeI, NruI, XhoI, or BstXI, end filled with Klenow polymerase, and ligated to generate p1430Luc, p1225Luc, p1128Luc, and p282Luc, respectively. To create p148Luc and p50Luc, PCR was carried out using genome DNA as a template with the following primer sets: p248Luc, 5′-ACT GCT AGC AAA GAA CAA GAA GAC TGG ATC CCC GGC A-3′ and 5′-AAG AAG CTT ACC GGT CCC AAA GCA CGG CTC AGA ATA AAT AGA TAA-3′; p50Luc, 5′-GAG GCT AGC AGC GGG ACC CAG TGT GCG TAT AAA TAC-3′ and 5′-AAG AAG CTT ACC GGT CCC AAA GCA CGG CTC AGA ATA AAT AGA TAA-3′. The amplified PCR products were digested by NheI and HindIII and ligated into pGL-basic2.
To analyze the functional role of the two HREs positioned at −1090/−1086 and −1070/−1066, two sets of mutants were constructed by site-directed mutagenesis using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). In the first set of mutants, each HRE (A/G CGTG) was mutated (underlined) to A/G AAAG using specific primers (5′-GCG CTC TGT GGC ACC GAAAGC ACT CGC CCT GTG G-3′ and 5′-CAC TCG CCC TGT GGT AAAAGA CCT CAC TCA GGT C-3′) to create p1128mut1Luc and p1128mut2Luc, respectively. Complementary sequences of the shown primers were used as forward primers. p1128mut3Luc was also constructed by mutating both HREs using HREmut1 in the p1128mut2Luc background. In the second set of mutants, the HRE sequence positioned at −1090/−1086 (ACGTG) was mutated to GCGTG using HREmut4 (5′-CAC TCG CCC TGT GGT GCG TGA CCT CAC TCA GGT C-3′) in the p1128mut1Luc background, creating p1128mut4Luc. In a similar manner, p1128mut5Luc was constructed by mutating the HRE positioned at −1070/−1066 (GCGTG) to ACGTG in the p1128mut2Luc background using HREmut5 (5′-GCG CTC TGT GGC ACC ACG TGC ACT CGC CCT GTG G-3′). To investigate whether the distance of an HRE from a TATA box is important, p1113Luc and p1143Luc were constructed using 5′-GTA ATG AGG TGA GAG ACA CAA CGC TTG CGT GCA CTC GCC CTG TGG TAC GT-3′ for p1113Luc and 5′-GTG CAC TCG CCC TGT GGT CAC TCG CCC TGT GGT ACG TGA CCT CAC TCA GG-3′ for p1143Luc. To test the possible involvement of a putative cyclic AMP response element (CRE), p1128mut6Luc was generated using CREmut1 (5′-CCC TGT GGT ACG TGA CAAAAC TCA GGT CAG GAC G-3′). To examine the possible requirement of an HAS in the hypoxic induction, the HAS (CAGGT) located at −1099/−1103 was mutated to TTTTT using HASmut1 (5′-GTA CGT GAC CTC ACT TTTTTC AGG ACG GCA CCC C-3′) to create p1128mut7Luc. Similarly, p1128mut8Luc and p1128mut9Luc were constructed by mutating the HAS of p1128mut1Luc and p1128mut2Luc using the same primer. In order to test whether HAS is sufficient, a HAS was introduced 8 bp upstream of the nonfunctional HRE at −1070/−1066 by using HASmut2 (5′-GTG CAC TCG CCCAGGT GTA AAA GAC CTC ACT TTTT TCA GGA CGG CA-3′) to create p1128mut10Luc and p1128mut11Luc.
Cell culture and DNA transfection.
ZFL cells, a cell line derived from adult zebra fish liver, were obtained from the American Type Culture Collection (Manassas, VA) and grown in a mixture of 50% Leibovitz's L-15 medium, 35% Dulbecco's modified Eagle's medium with 4.5 g/liter glucose, and 15% Ham's F-12 supplemented with 15 mM HEPES, 0.15 mg/ml of sodium bicarbonate, 0.01 mg/ml insulin, 50 ng/ml epidermal growth factor, penicillin (100 U/ml), streptomycin (100 μg/ml), and 5% fetal bovine serum at 28.0°C in 20% O2 or 5% CO2. Human hepatoma cells (HepG2), obtained from the American Type Culture Collection, were grown in minimal essential medium and Dulbecco's modified Eagle's medium supplemented with penicillin, streptomycin, and 10% fetal bovine serum at 37.0°C in 20% O2 and 5% CO2.
Twenty-four hours after plating into six-well plates, cells were transfected with 2 μg reporter construct using Fugene 6 (Roche, Indianapolis, IN). For cotransfection experiments, 1 μg reporter plasmid was cotransfected with 1 μg zebra fish HIF-1α or the empty pcDNA 3.1 vector. To control for transfection efficiency, 100 ng Renilla luciferase reporter plasmid (pRL-SV40) was cotransfected. Twenty-four hours after transfection, cells were subjected to hypoxia (1% O2) or normaloxygen (20% O2) in a humidified modular incubation chamber (Billups-Rothenberg Inc., Del Mar, CA). Luciferase activity was measured using the dual-luciferase reporter assay system (Promega) following the manufacturer's instruction. A human enolase promoter (p2.1) was used as a positive control, and p2.4, which has a mutated HRE, was used as a negative control for the HIF-1 response (38).
In vivo promoter analysis.
To analyze IGFBP-1 promoter activity in vivo, two DNA fragments covering IGFBP-1 promoter region (−2025 to + 89 and −1128 to +89) were subcloned into a promoterless pEGFP-N1 vector to create p2025:GFP (pIGFBP-1:GFP) and p1128:GFP, respectively. In addition, p1128mut2:GFP was constructed by mutating the HRE at position −1090/−1086 as described above. After linearization, each of these plasmids (100 pg) was microinjected into zebra fish embryos at the one- to two-cell stage as described previously (19). Green fluorescent protein (GFP)-positive embryos were selected at 24 hpf and transferred to normoxic or hypoxic water. After 24 h, visible GFP-positive cells were counted under a fluorescence microscope. GFP protein levels were measured by Western blotting using a GFP antibody (Torrey Pines Biolabs, Houston, TX) following a published method (19). As a control, Western blotting was performed using an antitubulin antibody (Sigma).
Whole-mount in situ hybridization, Northern blotting, and RT-PCR.
Whole-mount in situ hybridization, Northern blotting, and RT-PCR were carried out as reported previously (28). For RT-PCR analysis, total RNA was extracted from wild-type zebra fish embryos at various stages using Tri reagent (Molecular Research Center, Inc., Cincinnati, OH). After DNase treatment (Invitrogen), the RNA (2.5 μg) was reverse transcribed with random hexamer primers and Superscript II reverse transcriptase (Invitrogen). Sequences of primer sets were as follows: HIF-1α, 5′-CTA CAA TGA TGT CAT GCT GCC-3′ and 5′-ACA CAG AGT GAG TGG CAG AA-3′; HIF-1β, 5′-ATG GCA GAC CAA AGA ATG GA-3′ and 5′-GAA GAG GAA ACC ATC AGC AG-3′; IGFBP-1, 5′-CTT CTG AAC TTC TTC TGG GTG G-3′ and 5′-CTT CTG AAC TTC TTC TGG GTG G-3′; ornithine decarboxylase (ODC), 5′-TCA ATC CCA TCT CTT CCA TTC G-3′ and 5′-TCC GTT TTG CTG GCA CAG TC-3′. PCR cycles and amounts of templates were optimized for each primer set in pilot experiments. PCR cycles were as follows: 94°C for 3 min, 30 cycles (HIF-1α, HIF-1β, and ODC) or 35 cycles (IGFBP-1) of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min. PCR products were analyzed by electrophoresis followed by ethidium bromide staining. Cloned zebra fish HIF-1α, HIF-1β, and ODC DNA fragments were used as positive controls, and a no-template control was the negative control.
Overexpression of HIF-1α in developing zebra fish embryos.
Capped zebra fish HIF-1α mRNA was synthesized with the mMESSAGE mMACHINE kit (Ambion Inc., Austin, TX). The capped mRNA was microinjected at 1 μg/μl. Injected embryos were sampled 12 h later. Total RNA was extracted and reverse transcribed, and the IGFBP-1 mRNA levels were measured by RT-PCR as described above.
Electrophoretic mobility shift assay (EMSA).
The nuclear extract from cultured HepG2 cells was prepared following the procedure of Semenza and Wang (39), except that the dialysis step was omitted. To prepare nuclear extract from zebra fish embryos, 100 deyolked embryos raised in normoxic or hypoxic water were homogenized in 750 μl low-salt buffer containing 10 mM Tris-HCl (pH 7.8), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), and protein inhibitors (0.4 mM phenylmethylsulfonyl fluoride and 2 μg/ml each of pepstatin, aprotinin and leupeptin). The lysates were then centrifuged at 1,000 × g for 5 min at 4°C. The pellets were reconstituted in high-salt buffer containing 20 mM Tris-HCl (pH 7.9), 420 mM KCl, 1.5 mM MgCl2, 20% glycerol, 0.5 mM DTT, and protein inhibitors and incubated on a rotator at 4°C for 2 h. The samples were frozen quickly in liquid nitrogen and stored at −80°C until use. The protein contents of the nuclear extracts were quantified using the BCA protein assay kit (Pierce, Rockford, IL).
For EMSA, oligonucleotides derived from the zebra fish promoter (WT1, 5′-CAC TCG CCC TGT GGT ACG TGA CCT CAC TCA GGT C-3′; WT2, 5′-CGC CCT GTG GTA CGT GAC CTC ACT CAG GTC AGG ACG G-3′; HAS, 5′-ACC TCA CTC AGG TCA GGA CGG C-3′) were end labeled using T4 polynucleotide kinase (Promega) and [32P]ATP (ICN) and purified using a G-25 column (Amersham Bioscience, Piscataway, NJ). The radiolabeled double-stranded probe (5 fmol) was incubated with the nuclear extracts prepared from zebra fish embryo (5 μg) or HepG2 cells (1 μg) in binding buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 4% glycerol, 0.5 mM DTT, and 0.5 μg poly(dI-dC)-poly(dI-dC) in a final volume of 20 μl. For competition, 50 or 500 fmol unlabeled double-stranded probe or various competing oligonucleotides (sequences are shown below in Fig. 5A and 8A and C) was added to the binding reaction mixture. To prove the binding activity was indeed due to HIF-1, a human HIF-1α antibody (Lab Vision Co., Fremont, CA) or an HA antibody (Covance Research Products Inc., Berkeley, CA) was added into the binding reaction mixture prior to the probe. An equal amount of mouse immunoglobulin G (IgG) was used as a control. After 20 min of incubation at room temperature, the DNA-protein complexes were separated by electrophoresis on a 4% polyacrylamide gel electrophoresis gel in 1× Tris-borate-EDTA buffer. The gels were dried and exposed to X-ray film.
FIG. 5.
Both ACGTG and GCGTG are functional HREs. (A) Sequences of the wild-type (WT1) and mutant oligonucleotides used in the EMSA. The HRE is indicated in bold, and altered sequences are underlined. (B) Both forms of HRE (ACGTG and GCGTG) bind HIF-1. 32P-labeled double-stranded WT1 oligonucleotide was incubated with nuclear extracts prepared from zebra fish embryos or HepG2 cells. For competition, unlabeled WT1 or MutA1 or MutB1 was added in a 10- or 100-fold molar excess, respectively. The HIF-1-DNA complexes were separated by electrophoresis and visualized by autoradiography. (C) The two distinct HREs and their surrounding sequences in the wild-type p1128Luc and two mutants. The HRE sequences are indicated in bold, and mutated sequences are underlined. (D) HepG2 cells were cotransfected with the zebra fish HIF-1α-expressing construct or empty pcDNA3.1 together with individual constructs indicated in panel C. Twenty-four hours after transfection, cells were kept in 20% O2 or switched to 1% O2 (hypoxia) for 24 h. Luciferase activity was measured, and transfection efficiency was normalized by Renilla luciferase activity. Values are means ± standard errors (n = 4 to 6). *, P < 0.01 between the HIF-1α-transfected and the empty vector-transfected group; †, P < 0.01 between the normoxia and hypoxia groups.
FIG. 8.
The adjacent HAS does not affect HIF-1-HRE binding but is bound to a nuclear protein under normoxic conditions. (A) Sequences of the oligonucleotide probe (WT2) and various competitors used. The HRE and HAS are indicated in bold, and mutated sequences are underlined. (B) The HAS does not affect HIF-1-HRE binding. 32P-labeled double-stranded WT2 was incubated with nuclear extracts prepared from HepG2 cells kept in 20% or 1% O2. For competition, unlabeled WT2 or MutA2 or MutB2 was added in a 10- or 100-fold molar excess, respectively. In the last two lanes, human HIF-1α antibody or mouse IgG (negative control [NC]) was added prior to the probe. (C) Sequences of the HAS probe and various competitors used. The HRE and HAS are indicated in bold, and mutated sequences are underlined. (D)32P-labeled double-stranded HAS was incubated with nuclear extracts prepared from HepG2 cells kept in 20% O2. For competition, unlabeled HAS, mutated HAS (MuHAS), or HRE was added in a 10- or 100-fold molar excess, respectively. The protein-DNA complexes were separated by electrophoresis and visualized by autoradiography.
Statistics.
Values are means ± standard errors. Differences among groups were analyzed with Student's t test or by one- or two-way analysis of variance followed by Fisher's protected least significant difference test using StatView software (SAS Institute, Cary, NC). Statistical significance was accepted at a P level of <0.05.
RESULTS
The HIF-1 pathway is functional and mediates hypoxia-induced IGFBP-1 gene expression in zebra fish embryos.
To determine whether and when the HIF-1 pathway is operational in early development, we analyzed HIF-1α and -1β mRNA expression during zebra fish embryogenesis. Although zebra fish HIF-1α and -1β cDNA sequences have been deposited in GenBank (accession numbers AY326951 and NM131577), there is no published information on their spatial or temporal expression patterns, regulation, or biological actions. RT-PCR analysis indicated that both HIF-1α and HIF-1β mRNAs were expressed abundantly throughout embryogenesis (Fig. 1A). Whole-mount in situ hybridization analysis revealed a ubiquitous distribution pattern of HIF-1α mRNA in zebra fish embryos at all stages examined under normoxic conditions, although higher levels of HIF-1α mRNA were observed in the anterior portion of the embryos at 24 hpf (Fig. 1B).
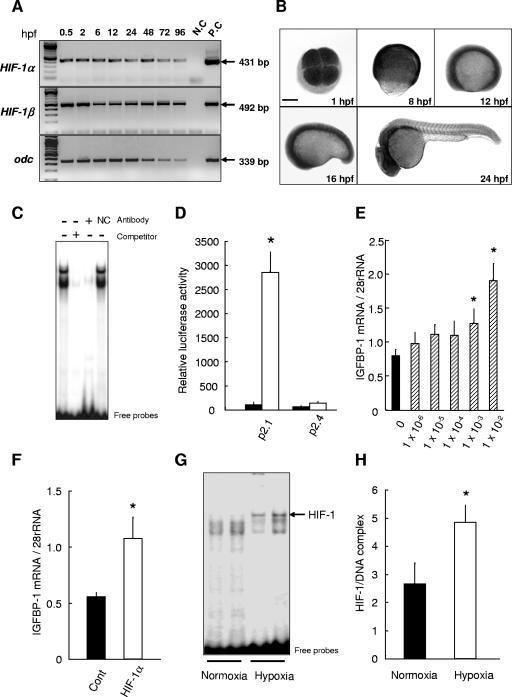
FIG. 1.
The HIF-1 pathway is operational in early embryogenesis and mediates hypoxia-induced IGFBP-1 transcription. (A) RT-PCR analysis of HIF-1α and HIF-1β mRNA expression in zebra fish embryos of the indicated stages. N.C, negative control; P.C, positive control. (B) Whole-mount in situ hybridization analysis of zebra fish HIF-1α mRNA in embryos of the indicated stages. Bar, 200 μm. (C) Zebra fish HIF-1α specifically binds to a probe containing the functional HRE in the zebra fish IGFBP-1 promoter. EMSA was conducted using nuclear extracts prepared from HepG2 cells transfected with a zebra fish HA:HIF-1α expression plasmid. Addition of an excess amount (100-fold) of unlabeled probe or HA antibody, but not mouse IgG (NC), abolished the binding. (D) Zebra fish HIF-1α activates human enolase promoter activity in an HRE-dependent manner. HepG2 cells were transfected with a zebra fish HIF-1α expression plasmid (open bars) or the empty vector (filled bars) together with a human enolase promoter reporter construct (p2.1) or a human enolase promoter reporter construct with the HRE mutated (p2.4). Forty-eight hours after transfection, luciferase activity was measured as described in Materials and Methods. Transfection efficiency was normalized by measuring Renilla luciferase activity. Values are means ± standard errors (SE) (n = 3 to 4). *, P < 0.01. (E) CoCl2, a HIF-1 inducer, increases IGFBP-1 mRNA expression in zebra fish embryos in vivo. At 24 hpf, embryos were treated with CoCl2 for 24 h at the indicated concentrations. At the end of the treatment, total RNA was extracted and subjected to Northern blotting analysis. IGFBP-1 mRNA levels were normalized by 28S rRNA levels. Values are means ± SE (n = 3). *, P < 0.05. (F) Injection of zebra fish HIF-1α mRNA into zebra fish embryos increases IGFBP-1 gene expression in vivo. Zebra fish HIF-1α mRNA (1 ng/μl) was microinjected into zebra fish embryos at the one- to two-cell stage. Twelve hours later, embryos were collected for RNA extraction. IGFBP-1 mRNA levels were measured by semiquantitative RT-PCR. Values are means ± SE (n = 5). *, P < 0.05. (G) Hypoxia activates the HIF-1 pathway in zebra fish embryos in vivo. EMSA was performed using nuclear extracts prepared from zebra fish embryos kept in normoxic or hypoxic water. (H) Densitometric analysis of the results in panel G. Values are means ± SE (n = 6). *, P < 0.05.
To test the ability of zebra fish HIF-1α to bind to an HRE, nuclear extracts isolated from HepG2 cells transfected with a zebra fish HA:HIF-1α expression plasmid were subjected to EMSA using an oligonucleotide probe containing a functional HRE (WT1). As shown in Fig. 1C, strong DNA binding activity specifically bound to the probe was detected. Addition of an HA antibody, but not mouse IgG, eliminated this binding, suggesting that the HA-tagged zebra fish HIF-1α binds to the probe. We next tested the transactivation activity of zebra fish HIF-1α using a well-established HIF-1 reporter construct, the human enolase promoter p2.1 (38). Coexpression of zebra fish HIF-1α resulted in a 27.5-fold increase in the human enolase promoter activity (Fig. 1D). When the HRE in the human enolase promoter construct was mutated (p2.4), this induction was abolished (Fig. 1D), suggesting that zebra fish HIF-1α is a functionally conserved protein that can activate gene transcription in an HRE-dependent manner.
To determine whether zebra fish HIF-1 can stimulate IGFBP-1gene expression in vivo, zebra fish embryos were treated with various concentrations of CoCl2, a chemical HIF-1 inducer. The effect of CoCl2 treatment on the levels of IGFBP-1 mRNA expression was determined by Northern blot analysis. As shown in Fig. 1E, CoCl2 increased IGFBP-1 mRNA levels in a dose-dependent manner. Next, zebra fish HIF-1α mRNA was synthesized and delivered into zebra fish embryos by microinjection. This resulted in a significant increase in the levels of endogenous IGFBP-1 mRNA (Fig. 1F). Injection of GFP mRNA did not have such an effect (data not shown). To show that the HIF-1 pathway is operational in early development, EMSA experiments were performed using nuclear extracts prepared from zebra fish embryos kept in normoxic or hypoxic water. As shown in Fig. 1G and H, hypoxia treatment caused a significant increase in levels of the HIF-1-DNA complex in zebra fish embryos. Taken together, these results suggest that the HIF-1 pathway is fully operative in early embryogenesis and that it activates IGFBP-1 gene expression in zebra fish embryos under hypoxic stress.
The zebra fish IGFBP-1 promoter contains the hypoxia responsive cis-regulatory element(s).
To determine the molecular mechanism(s) underlying hypoxia-induced zebra fish IGFBP-1 gene expression, we isolated a 2.1-kb DNA of the zebra fish IGFBP-1 gene promoter region (GenBank/EMBL/accession number AB181657). This DNA fragment was subcloned into a promoterless GFP reporter plasmid. Injection of the linearized pIGFBP-1:GFP plasmid into zebra fish embryos resulted in mosaic GFP expression at low levels (Fig. 2A). Hypoxia treatment markedly increased the number of cells with visible GFP signals in these embryos (Fig. 2A; see also Fig. 4, below). As an independent control, a GFP construct driven by the cytomegalovirus (CMV) promoter was injected into zebra fish embryos. While mosaic GFP expression was detected, no marked difference was found in the number of GFP-expressing cells or GFP protein levels between normoxia and hypoxia in these control animals (data not shown). Western blot analysis was also performed to measure the total GFP protein levels. Hypoxia markedly increased the GFP protein levels in the embryos injected with the pIGFBP-1:GFP plasmid (Fig. 2B; see also Fig. 4, below). In contrast, hypoxia treatment resulted in a decrease in the levels of tubulin protein (Fig. 2B) and several other cellular proteins (data not shown), indicating a global decrease in protein synthesis under hypoxia. These results suggest that the zebra fish IGFBP-1 5′-untranslated region contains the promoter activity and the hypoxia responsive cis-regulatory element(s).
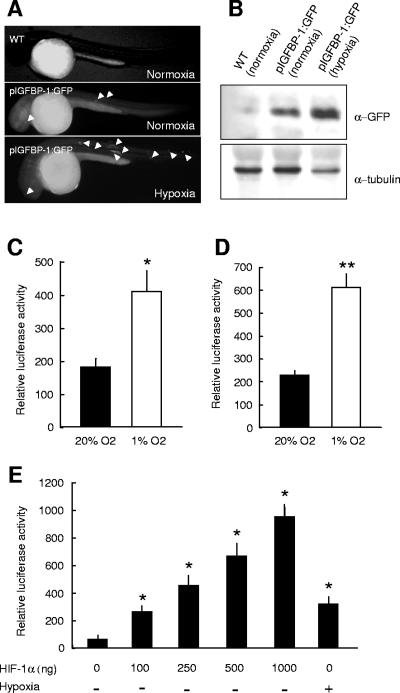
FIG. 2.
In vivo and in vitro analysis of hypoxia responsiveness of the zebra fish IGFBP-1 promoter. (A) In vivo analysis. Embryos at the one- to two-cell stage were microinjected with linearized pIGFBP-1:GFP DNA, a GFP reporter construct driven by the zebra fish IGFBP-1 promoter region (p2025). The injected embryos were raised for 12 hpf and transferred to normoxic or hypoxic water. Twenty-four hours later, GFP-expressing cells were observed under a fluorescence microscope. The upper panel is a wild-type control embryo kept in normoxic water. The middle and lower panels show a representative pIGFBP-1:GFP-injected embryo kept in normoxic and hypoxic water, respectively. Note the increase in visible GFP-expressing cells (indicated by arrowheads) in response to hypoxia. (B) pIGFBP-1:GFP-injected 12-hpf embryos were subjected to a 24-h hypoxia treatment. Pooled embryos were subjected to Western blotting analysis using a GFP antibody. The expression of tubulin protein was also measured using a tubulin antibody. (C and D) In vitro analysis of the hypoxia responsiveness of the cloned zebra fish IGFBP-1 promoter in cultured ZFL (C) and HepG2 (D) cells. Cells were transfected with the p2025Luc construct. Twenty-four hours after transfection, they were switched to hypoxia (1% O2) or kept in normoxia (20% O2). After another 24 h, the cells were lysed and the luciferase activity was measured as described in Materials and Methods. Transfection efficiency was normalized by measuring Renilla luciferase activity. Values are means ± standard errors (SE) (n = 3). *, P < 0.05; **, P < 0.01. (E) Overexpression of zebra fish HIF-1α increases IGFBP-1 promoter activity in cultured HepG2 cells. pCMV-HIF-1α at the concentration indicated was transfected into HepG2 cells with a zebra fish IGFBP-1 promoter construct (p1128Luc). The difference in pCMV-HIF-1α DNA was compensated using empty vector DNA. At 48 h after transfection, luciferase activity was measured. Transfection efficiency was normalized by Renilla luciferase activity. Values are means ± SE (n = 3). *, P < 0.05.
FIG. 4.
The HRE positioned at −1090/−1086 is required for hypoxia-induced IGFBP-1 gene expression in vivo. (A) In vivo analysis of the involvement of the −1090/−1086 HRE in hypoxia-induced IGFBP-1 gene transcription. Embryos at the one- to two-cell stage were microinjected with p1128:GFP or p1128mut2:GFP, two GFP reporter gene constructs driven by wild-type p1128 or p1128mut2 shown in Fig. 3. The injected embryos were raised for 24 hpf and transferred to normoxic (normoxia) or hypoxic (hypoxia) water. Twenty-four hours later, visible GFP-expressing cells were counted under a fluorescence microscope. (B) Quantification of results shown in panel A. Filled bars represent the normoxia group, and open bars represent the hypoxia groups. Values are means ± standard errors (SE) (n = 69 to 94). *, P < 0.01. (C) Western blotting analysis of p1128:GFP- or p1128mut2:GFP-injected embryos. (D) Densitometric analysis result of the results shown in panel C. The expression levels of GFP relative to tubulin are shown. Filled bars represent the normoxia groups, and open bars represent the hypoxia groups. Values are means ± SE (n = 5). *, P < 0.05.
To further study the hypoxia-induced IGFBP-1 promoter activity in vitro, the IGFBP-1 promoter region was subcloned into a promoterless luciferase vector and transfected into ZFL cells and human HepG2 cells. As shown in Fig. 2C, hypoxia caused a significant increase (2.3-fold; P < 0.05) in reporter gene activity in ZFL cells. A 2.8-fold, highly significant increase (P < 0.001) was seen in HepG2 cells (Fig. 2D). A similar hypoxic induction was reported for the human IGFBP-1 promoter in HepG2 cells (41). Time course experiments indicated that 4 h was needed to obtain a significant increase in zebra fish IGFBP-1 promoter activity, and the maximal response was seen after 24 h of hypoxia treatment (data not shown). Since the hypoxic responsiveness of the IGFBP-1 gene is a conserved mechanism and because HepG2 cells can be cultured without insulin (a known regulator of mammalian IGFBP-1 genes) and are easily transfectable, HeG2 cells were used in subsequent studies. As shown in Fig. 2E, cotransfection of a zebra fish HIF-1α expression plasmid in HepG2 cells resulted in dose-dependent increases in zebra fish IGFBP-1 promoter activity, further demonstrating the utility of HeG2 cells in studying the zebra fish IGFBP-1 promoter. At the concentration of 100 ng DNA, zebra fish HIF-1α caused an increase comparable to the hypoxia-induced increase. At higher doses, zebra fish HIF-1α caused even greater increases (Fig. 2E).
The zebra fish IGFBP-1 promoter contains multiple consensus HREs, but only one HRE is used in its response to hypoxia and HIF-1.
Although hypoxia has been shown to affect the IGFBP-1 gene expression of a wide variety of fish genes (13, 29), a functional HRE has not been characterized in any fish gene to date (32). In mammals, the minimal HRE is (A/G)CGTG (3). We searched the zebra fish IGFBP-1 promoter region and identified 13 canonical mammalian HREs, including 7 in the forward orientation and 6 in reverse orientation. These HREs are located at −1502/−1498, −1461/−1457, −1436/−1432, −1435/−1431, −1412/−1408, −1368/−1364, −1222/−1218, −1209/−1205, −1090/−1086, −1070/−1066, −215/−211, −206/−202, and −57/−53, relative to the cap site (Fig. 3A). To determine whether these and/or other cis elements are responsible for hypoxia and HIF-1-induced IGFBP-1 gene expression, a series of deletion constructs were generated. These reporter constructs, i.e., p2025Luc, p1430Luc, p1225Luc, p1128Luc, p282Luc, and p148Luc, contain 13, 9, 7, 5, 3, and 1 HRE, respectively. p50Luc has no HRE site. As shown in Fig. 3B, deletion of the sequence between −2025 and −1430 caused a modest but statistically significant increase (P < 0.01) in basal promoter activity, suggesting the presence of a negative regulatory element(s) in this region. This deletion, however, had little effect on the hypoxia responsiveness. Further deletion of the sequence −1430 to −1225 and from −1225 to −1128 significantly decreased the basal activity (P < 0.05). Again, little effect was seen on the hypoxia-induced increase. Because of the reduced basal activity, the hypoxia induction appeared even greater. Further deletion of sequence from −1128 to −282 did not cause a further decrease in the basal promoter activity, but it abolished the hypoxia responsiveness. p148Luc was similar to p282Luc. These results suggest the sequence between −1128 and −282 contains the cis-regulatory element(s) for the hypoxia responsiveness.
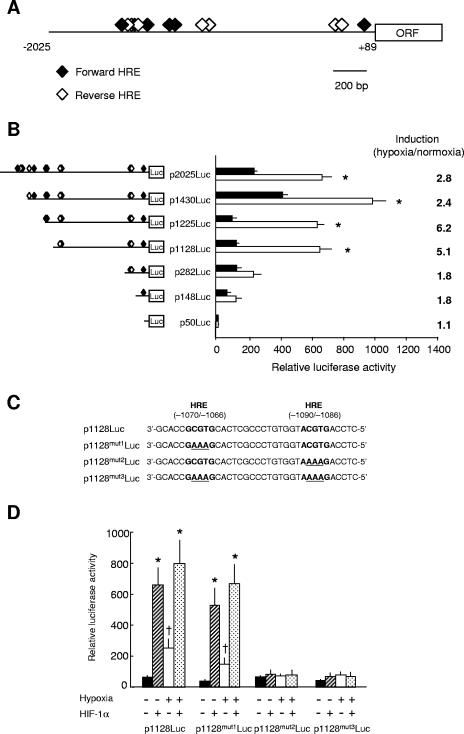
FIG. 3.
The zebra fish IGFBP-1 promoter contains 13 HREs, but only the −1090/−1086 HRE is required for the hypoxia or the HIF-1 response. (A) Location and direction of the 13 canonical HRE sites identified in the zebra fish IGFBP-1 promoter region. The HREs in forward orientation are indicated as filled diamonds, and reverse HREs are shown as open diamonds. (B) Deletion analysis of the zebra fish IGFBP-1 promoter. HepG2 cells were transfected with a series of luciferase reporter constructs containing the indicated portion of the IGFBP-1 promoter region. Twenty-four hours after transfection, cells were kept in 20% O2 (filled bars) or transferred to 1% O2 (open bars) for 24 h, and luciferase activity was measured. Transfection efficiency was normalized by measuring Renilla luciferase activity. Values are means ± standard errors (SE) (n = 3). *, P < 0.05. (C) The two HREs and their surrounding sequences in wild-type p1128Luc and various mutants. Because the two HREs are in reverse orientation, the sequences are presented the 3′-to-5′ direction. The HREs are indicated in bold, and mutated sequences are underlined. (D) Mutational analysis of the two HREs. HepG2 cells were cotransfected with the constructs indicated in panel C and the zebra fish HIF-1α-expressing construct or the empty pcDNA3.1. Twenty-four hours after transfection, cells were kept in 20% O2 or switched to 1% O2 (hypoxia) for 24 h, and luciferase activity was measured. Transfection efficiency was normalized by Renilla luciferase activity. Values are means ± SE (n = 3 to 4). *, P < 0.01 between the HIF-1α- and vector-transfected groups; †, P < 0.01 between the normoxic and hypoxic groups.
There are two HREs, both in the reverse orientation, between −1128 and −282. They are positioned at −1090/−1086 and −1070/−1066 (Fig. 3C). To test whether one or both of them are involved in the hypoxia response, either one or both of the HREs were mutated (Fig. 3C). As shown in Fig. 3D, hypoxia treatment caused a fourfold increase (P < 0.01) in the activity of p1128Luc. Overexpression of zebra fish HIF-1α resulted in a 10.5-fold increase (P < 0.01) in the activity of p1128Luc. Hypoxia did not cause a further increase in the HIF-1α-transfected group, suggesting that HIF-1α overexpression is sufficient to account for hypoxia-induced IGFBP-1 promoter activity. Mutation of the HRE at −1070/−1066 (p1128mut1Luc) did not affect the responsiveness to hypoxia. Likewise, this mutant showed an increase in response to HIF-1α coexpression similar to wild-type p1128Luc. Mutation of the HRE at −1090/−1086 (p1128mut2Luc), however, completely abolished the hypoxia- and HIF-1α-induced increase. The double mutant p1128mut3Luc acted like p1128mut2Luc (Fig. 3D), indicating that only the HRE at −1090/−1086 is required for the hypoxic induction.
To investigate whether the HRE at position −1090/−1086 is required for hypoxia-induced IGFBP-1 expression in vivo, an enhanced GFP reporter construct driven by the minimal zebra fish promoter region (p1128:GFP) was generated. Injection of p1128:GFP plasmid into zebra fish embryos resulted in mosaic GFP expression at low levels under normoxic conditions (Fig. 4A). Hypoxia treatment caused a highly significant (P < 0.001), 4.2-fold increase in the number of visible GFP-positive cells (Fig. 4B). Western immunoblot analysis showed that p1128:GFP-injected embryos also had a significant increase in the total GFP protein levels in response to hypoxia (Fig. 4C and D). Mutation of the −1090/−1086 HRE did not alter the basic expression levels of GFP, but it completely abolished the hypoxia response. These data suggest that the HRE at −1090/−1086 is required for the hypoxia- and HIF-1-induced zebra fish IGFBP-1 expression in developing zebra fish embryos in vivo.
The selective use of the −1090/−1086 HRE cannot be attributed to its sequence or location.
The two HREs in p1128Luc are only 15 bp apart. Yet, only the HRE at −1090/−1086 is required for the response to hypoxia and HIF-1. Since the HRE at −1070/−1066 is GCGTG and the HRE at −1090/−1086 is ACGTG, and because a functional HRE has not been reported in any teleost species (32), we wondered whether the single nucleotide difference could account for the observed functional difference. To determine whether both forms of HRE can bind HIF-1, EMSA experiments were performed using the oligonucleotides shown in Fig. 5A. The labeled probe (WT1) containing the functional HRE bound to nuclear protein or protein complexes in the nuclear extracts prepared from hypoxia-treated zebra fish embryo (Fig. 5B). This binding was specific to the HRE, because addition of excess amounts of unlabeled WT1 displaced the binding, while Mut A1, the same oligonucleotide with the HRE mutated, did not. MuB1, which has a single nucleotide mutation in the HRE from ACGTG toGCGTG, was equally effective as WT1 in replacing the binding, suggesting that zebra fish HIF-1 can bind to both forms of the HRE.
To determine whether the single nucleotide difference causes any functional difference, the nucleotide A at position −1090 was changed into G in the p1128mut1 background, resulting in p1128mut4Luc (Fig. 5C). If the sequence ACGTG is critical, then changing it to GCGTG should abolish the hypoxia and HIF-1 responsiveness. Conversely, the nucleotide G at position −1066 was changed into A in the p1128mut2 background, resulting in the creation of p1128mut5Luc (Fig. 5C). If the sequence ACGTG is critical, then this mutation should restore the hypoxia and HIF-1 responsiveness. When the activities of these mutants were examined, however, p1128mut4Luc had similar responses to hypoxia and HIF-1α expression as wild-type p1128Luc (Fig. 5D). p1128mut5Luc, like p1128mut2, did not show any increase in response to hypoxia/HIF-1α overexpression (Fig. 5D), suggesting that both HREs (ACGTG and GCGTG) are functional in a fish gene when positioned at the right location (e.g., −1090/−1086).
These data led us to hypothesize alternatively that the precise location (or distance from the TATA box) of an HRE may be critical. To test this idea, two constructs, p1113Luc and p1143Luc, were engineered. In p1113Luc, the −1090/−1086 HRE is changed into −1075/−1071 by deleting 15 nucleotides (nt) (Fig. 6A). In p1143Luc, an additional 15 nt is added between the two HREs, thus moving the functional HRE to position −1105/−1101 without altering the location of the −1070/−1066 HRE. When the activities of these two mutants were tested, they acted like the wild-type p1128Luc (Fig. 6B). These data suggest that moving the functional HRE by 15 nt in either direction does not affect the responsiveness of the IGFBP-1 promoter to hypoxia or HIF-1α.
FIG. 6.
The selective use of the −1090/−1086 HRE cannot be attributed to its location. (A) Diagram showing the locations of the two HREs in p1128Luc (wild type), p1113Luc, and p1143Luc; the latter two were generated by deleting or adding 15 bp. The functional HRE is represented by an open diamond, and the nonfunctional HRE is indicated by a filled diamond. (B) Moving the functional HRE by 15 nt in either direction has no effect on the hypoxia or HIF-1 responsiveness. HepG2 cells were cotransfected with individual reporter constructs indicated in panel A and a zebra fish HIF-1α expression plasmid or the empty vector. Values are means ± standard errors (n = 3). *, P < 0.05 between the HIF-1α- and the empty vector-transfected groups; †, P < 0.05 between the normoxic and hypoxic groups.
A HAS adjacent to the functional HRE is critical for hypoxic induction.
To determine whether the sequence(s) surrounding the −1090/−1086 HRE is critical, we analyzed its neighboring sequence for potential DNA binding sites. A putative CRE contiguous to the functional HRE was identified (Fig. 7A). CRE-binding protein (CREB) and HIF-1 are known to use a common coactivator, CREB binding protein/p300, to initiate the transcription of target genes (2, 6, 20). It was also reported that an increase in cyclic AMP levels potentiates the hypoxic induction of the mouse lactate dehydrogenase A (LDHA) promoter and that mutation of the CRE reduces the hypoxia responsiveness (8). We therefore investigated the possible involvement of the putative CRE. As shown in Fig. 7B, mutation of the CRE in the zebra fish IGFBP-1 promoter (p1128mut6Luc) did not change its responses to hypoxia or HIF-1α overexpression, suggesting that this CRE is not involved in hypoxic induction in IGFBP-1 promoter activity.
FIG. 7.
A HAS adjacent to the functional HRE, but not a CRE, is critical for the hypoxia and HIF-1 responsiveness of the IGFBP-1 promoter. (A) The two HREs and the adjacent CRE sequences in the wild-type p1128Luc and p1128Lucmut6. The HREs are underlined, and CRE sequence is in bold letters. The mutated sequences are in bold and underlined. (B) Mutation of the CRE does not alter the hypoxia or HIF-1 responsiveness of the IGFBP-1 promoter. HepG2 cells were cotransfected with individual reporter constructs indicated in panel A and a zebra fish HIF-1α expression plasmid or the empty vector. Values are means ± standard errors (SE) (n = 4). *, P < 0.05 between the HIF-1α- and the empty vector-transfected groups; †, P < 0.05 between the normoxic and hypoxic groups. (C) The two HREs and the HAS in the wild-type p1128Luc and four mutants. The HREs and HAS are indicated in bold, and mutated sequences are underlined. (D) Mutational analysis of the HAS. HepG2 cells were cotransfected with individual reporter constructs indicated in panel C and a zebra fish HIF-1α expression plasmid or the empty vector. Values are means ± SE (n = 3 to 6). *, P < 0.05 between the HIF-1α- and the empty vector-transfected groups; †, P < 0.05 between the normoxic and hypoxic groups.
In addition to the CRE, a putative HAS is positioned adjacent to the functional HRE (Fig. 7C). HAS was initially identified in the vascular endothelial growth factor (VEGF) promoter and was shown to be involved in the HIF-1-induced VEGF expression (22, 23, 25). In that case, the HAS may form an imperfect inverted repeat with an HRE with a spacer of 8 nt (23). In the zebra fish IGFBP-1 promoter region, a putative HAS is positioned at −1099/−1103, precisely 8 nt upstream of the functional HRE. No HAS-like sequence was found in the proximity of the other 12 HREs in the zebra fish IGFBP-1 promoter. We asked whether the putative HAS is functional and plays any role in the selective use of the functional HRE. p1128mut7Luc and p1128mut8Luc, two mutants generated by altering the HAS in wild-type p1128Luc and p1128mut1Luc backgrounds (Fig. 7C), were used to address this question. Mutation of the HAS in either the wild-type p1128Luc or p1128mut1Luc background greatly reduced, but did not abrogate, the hypoxic response to hypoxia and/or HIF-1α overexpression (Fig. 7D). When the activity of p1128mut9, an HRE and HAS double mutant, was tested, it did not respond to either hypoxia or HIF-1α overexpression (Fig. 7D). These results suggest that the HAS is important for the magnitude of hypoxia responsiveness of the zebra fish IGFBP-1 promoter. To determine whether the HAS core sequence is sufficient to confer the function of an HRE, we generated another mutant construct (p1128mut10Luc) in which the −1090/−1086 HRE and its adjacent HAS were mutated and the minimal HAS (CAGGT) was introduced 8 nt upstream of the −1070/−1066, nonfunctional HRE (Fig. 7C). If the HAS core sequence were solely responsible for the function of the −1090/−1085 HRE, p1128mut10Luc would restore the hypoxia/HIF-1-induced promoter activity. As shown in Fig. 7D, this mutant did not show improved hypoxia/HIF-1 responsiveness. Since the HRE at −1070/−1066 is GCGTG and the HRE at −1090/-1086 is ACGTG, we next changed the nucleotide G at position −1066 into A in the p1128mut10Luc background, resulting in the creation of p1128mut11Luc. p1128mut11Luc also failed to show improved hypoxia/HIF-1 induction (Fig. 7D), suggesting that the −1103/−1099 HAS is required, but not sufficient, for the functional involvement of the −1090/−1086 HRE in hypoxia/HIF-1 regulation.
Since mutating the HAS greatly reduces the magnitude of hypoxia responsiveness of the IGFBP-1 promoter, we wondered whether the HAS can directly interact with the HIF-1 complex or whether it influences the HIF-1 and HRE binding. To test this idea, an EMSA was performed using an oligonucleotide probe that contains the functional HRE, HAS, and their surrounding sequence (WT2) (Fig. 8A). As shown in Fig. 8B, two major bands were detected in the nuclear extracts isolated from hypoxia-treated HepG2 cells, but not from those kept under normoxic conditions. These binding activities were displaced by adding excess amounts of unlabeled probe (WT2), but not by Mut A2, the same oligonucleotide with a mutated HRE (Fig. 8A and B). Furthermore, addition of a human HIF-1α antibody prior to the probe eliminated these hypoxia-induced bands, while mouse IgG had no such effect (Fig. 8B), indicating that these hypoxia-induced HRE binding activities indeed contained HIF-1α. When MutB2, an oligonucleotide with a mutated HAS, was added in excess, it was as effective as WT2 in displacing the HIF-1/DNA complexes (Fig. 8A and B). MutC2, in which both the HRE and HAS were mutated, did not alter the HIF-1 and HRE binding. These results suggest that having an adjacent HAS does not significantly influence the binding of an HRE to HIF-1. During the course of these experiments, a strong DNA binding activity was consistently observed in the nuclear extract isolated from normoxic cells (Fig. 8B). This activity was specific because it was replaced by adding an excess amount of unlabeled WT2 (data not shown). This activity was predominantly detected under normoxic conditions (Fig. 8B). To determine whether there is a nuclear protein specifically bound to the HAS site under normoxia, HAS, an oligonucleotide probe only containing the HAS, and its surrounding sequence were used for further EMSA experiments (Fig. 8C). This probe was found to interact with the same protein complex (Fig. 8D). Addition of excess amounts of unlabeled HAS displaced this binding activity, whereas Mut HAS, which carries a 4-bp mutation in the HAS, was much less efficient in competing for binding (Fig. 8C and D). Importantly, an oligonucleotide containing the functional HRE was unable to compete for binding (Fig. 8C and D). These results indicate the presence of a nuclear protein that can specifically interact with the HAS under normoxic conditions.
DISCUSSION
In this study, we have demonstrated that the HIF-1 pathway is fully functional in the early stages of development and it mediates the hypoxia-induced IGFBP-1 gene expression in zebra fish embryos in vivo. Detailed in vivo and in vitro analyses of the zebra fish IGFBP-1 promoter revealed two key cis-regulatory elements required for hypoxia/HIF-1-regulated IGFBP-1 gene transcription. We showed that the zebra fish IGFBP-1 promoter contains 13 canonical HREs, but only the HRE at position −1090/−1086 is required for the hypoxia/HIF-1 regulation. Disruption of this HRE, but not the other 12 HREs, abolishes the hypoxia/HIF-1-induced transcriptional response of the IGFBP-1 promoter. Furthermore, we provide evidence that a HAS adjacent to the −1090/−1086 HRE is critical for the hypoxia response. The HAS does not directly interact with HIF-1 or affect the binding of its neighboring HRE to HIF-1. Rather, the HAS is bound to a hypoxia-regulated nuclear protein.
The ability of the IGFBP-1 gene to respond to hypoxia has been documented in a variety of vertebrate species, ranging from fish to humans (13, 29, 31, 35, 41). A previous in vitro study has shown that coexpression of a luciferase reporter construct driven by the human IGFBP-1 promoter and a constitutively active form of human HIF-1α in HepG2 cells results in a fourfold induction in reporter activity (41). In this study, we have extended this in vitro observation and we have shown that the HIF-1pathway mediates hypoxia-induced zebra fish IGFBP-1 gene expression in developing zebra fish embryos in vivo. Several lines of experimental evidence support this conclusion. First, the HIF-1 pathway is operational in zebra fish embryos. The major components of the HIF-1 pathway are abundantly expressed in most, if not all, embryonic tissues, and hypoxia increases the levels of functional HIF-1. These results suggest that the regulation of HIF-1α in response to environmental oxygen levels is established very early in zebra fish embryogenesis. Knockout studies in the mouse model have shown that HIF-1 is essential for normal mammalian fetal development, in addition to its key role in oxygen homeostasis in adult stages (1, 17, 26). Second, the zebra fish HIF-1α is a functionally conserved protein and can bind to an HRE and activate HRE-dependent gene transcription when tested in vitro. Third, cotransfection of the zebra fish IGFBP-1 promoter construct with a zebra fish HIF-1α in HepG2 cells leads to a 10-fold increase in the IGFBP-1 promoter activity. More importantly, overexpression of zebra fish HIF-1α in developing zebra fish significantly increases endogenous IGFBP-1 mRNA levels. Likewise, CoCl2, a chemical HIF-1α inducer, causes a concentration-dependent increase in IGFBP-1 mRNA levels inzebra fish embryos.
Tazuke et al. (41) reported that an HRE in the first intron of the human IGFBP-1 gene is required for the hypoxia response in cultured HepG2 cells. In the case of the zebra fish IGFBP-1 gene, however, there is no consensus HRE in the first intron. Instead, we have identified 13 canonical HREs in the promoter region. This region possesses basal promoter activity and contains the cis-regulatory element(s) responsible for the hypoxia responsiveness when tested in vitro and in vivo. Although the zebra fish IGFBP-1 promoter contains 13 canonical HREs, disruption of the HRE positioned at −1090/−1086, but not the other 12 HREs, abolishes the hypoxia/HIF-1-induced transcriptional response of the IGFBP-1 promoter in vitro. The functional importance of the −1090/−1086 HRE in hypoxia-induced IGFBP-1 expression was further confirmed by in vivo promoter analysis. While hypoxia induced a fourfold increase in the reporter activity in vivo, mutation of the −1090/−1086 HRE completely abolished the hypoxia response. To our knowledge, this is the first in vivo study demonstrating the critical role of an HRE in hypoxia-induced gene expression in a developing vertebrate embryo.
The finding that only 1 of the 13 canonical HREs is used for the hypoxic induction of IGFBP-1 gene transcription is both interesting and puzzling. We were particularly intrigued by the fact that there is a canonical HRE located at −1070/−1066, 15 nt away from the functional HRE. We considered three possible explanations underlying this high degree of selectivity: (i) the HRE at −1070/−1066 has a single nucleotide difference (GCGTG versus ACGTG), and this may account for the observed functional difference; (ii) the precise location (or distance to the TATA box) of an HRE is critical; and (iii) the neighboring sequence surrounding an HRE is critical for its function. The first hypothesis was rejected because EMSA and functional analyses revealed that HIF-1 can bind to both forms of HRE and GCGTG can act as a functional HRE when introduced at the right location (e.g., −1090/−1086). Thus, both GCGTG and ACGTG can be functional HREs in a fish gene. The second possibility was also rejected, because p1113Luc and p1143Luc, two reporter constructs in which the location of the functional HRE is moved by 15 nt in either direction, are fully responsive to hypoxia/HIF-1. Our conclusion that the location of an HRE is not critical for its function is also in line with the finding that the functional HRE in the human IGFBP-1 gene is located in the first intron (41). To test the third possibility, we examined a consensus CRE and HAS located in the proximity of the functional HRE. Although mutation of an adjacent CRE in the mouse LDHA promoter was previously reported to reduce its hypoxia responsiveness (8), mutation of the CRE in the zebra fish IGFBP-1 promoter has no effect on its hypoxia responsiveness. When the HAS is altered, however, the magnitude of hypoxia responsiveness of the zebra fish IGFBP-1 promoter is greatly reduced. Mutation of both the functional HRE and HAS abolishes responsiveness to either hypoxia or HIF-1α overexpression, suggesting that cooperative interactions between the functional HRE and its adjacent HAS may be critical to confer hypoxia responsiveness. The importance of an adjacent HAS in determining a functional HRE is also supported by our sequence analysis results. Among the 13 HREs found in the zebra fish IGFBP-1 promoter region, a HAS motif was present only in the proximity of the functional HRE. In addition, a HAS motif was found next to the functional HRE located in the first intron of the human IGFBP-1 gene (Table 1). Likewise, a consensus HAS is found adjacent to an HRE in the mouse and rat IGFBP-1 genes, although they are separated by 9 or 10 nt (Table 1) rather than the 8 nt found in the zebra fish IGFBP-1 promoter. Whether a HAS is involved in HRE function in a mammalian IGFBP-1 gene needs to be investigated in the future. While the adjacent HAS is clearly required for the hypoxia- and HIF-1-induced zebra fish IGFBP-1 promoter activity, introducing the minimal HAS (CAGGT) 8 nt upstream of the −1070/−1066 HRE was insufficient to restore the hypoxia and HIF-1 regulation, suggesting that another sequence(s) or factor(s) may be also involved in the selective use of an HRE.
TABLE 1.
Presence of a HAS site adjacent to the functional HRE in known IGFBP-1 promoter regions
| Gene | Promoter regiona |
|---|---|
| Human IGFBP-1 | 5′-GCACGGTCTTGGCAGGACGTGCTC-3′ |
| Mouse IGFBP-1 | 3′-AGGACGTGCATGCAGCCCAGATGG-5′ |
| Rat IGFBP-1 | 3′-CACACGTGCTTTCTAGGCACGTCA-5′ |
| Zebra fish IGFBP-1 | 3′-GGTACGTGACCTCACTCAGGTCAG-5′ |
| Human VEGF | 5′-CATACGTGGGCTCCAACAGGTCCT-3′ |
The HRE is shown in bold, and the HAS is underlined.
It remains an open question how the HAS acts to modulate HRE function. Our EMSA results indicate that the HAS does not directly interact with the HIF-1 protein complex, because no HIF-1/DNA complexes were detected by EMSA when the HAS probe was used. Likewise, in the EMSA experiments using the WT2 probe (containing both the HRE and HAS), MutB2, an oligonucleotide with a mutated HAS, was as effective as the wild-type probe in displacing the HIF-1/DNA complexes, suggesting that an adjacent HAS does not significantly influence the binding of HIF-1 to the HRE. Interestingly, there is a nuclear protein or protein complex which binds specifically to the HAS region. This nuclear binding activity or factor does not interact with the HRE. A previous study has shown that a HAS identified in the human VEGF promoter also binds to a constitutive, non-HIF-1 protein in A172 and other cells (23). In this study, we found that this HAS binding activity was reduced or inhibited by hypoxia in HepG2 cells. Since the HAS binding activity is predominantly seen under normoxic conditions, we suspect that it may facilitate the selective binding of HIF-1 to its neighboring HRE, perhaps by providing a platform or docking site for the HIF-1 complex. At present, the molecular identity of this HAS binding factor(s) is unknown. HIF-1 has been reported to interact with ATF-1/CREB-1 in the mouse LDHA gene (8), with AP-1 in the VEGF gene and the tyrosine hydroxylase gene (10, 24), and with the orphan receptor hepatic nuclear factor 4 in the erythropoietin gene (33). These transcription factors are unlikely to be the HAS binding factor, because they are either constitutively bound to an HRE or are induced by hypoxia while the HAS binding activity is inhibited or reduced under hypoxia. In addition, mutation of the CRE in the zebra fish IGFBP-1 gene has no effect on the hypoxic induction. Clearly, more studies are needed to identify and characterize this HAS binding factor and determine whether and how it modulates HRE function. Future studies aiming to elucidate the role of the HAS and its binding protein(s) in regulating IGFBP-1 gene expression should deepen our understanding of hypoxia-regulated gene expression in early development.
Acknowledgments
We thank H. Hirata, S. Takahashi, and H. Kamei for discussions and help, K. Inoki, T. Iwashita, and K. Sakaki for providing reagents, and E. J. Clowney and A. J. Maguire for critical reading of the manuscript.
This work was supported in part by NSF grant IBN 0110864 to C.D. S.K. was supported by a fellowship from the Japan Society for Promotion of Science (11824) and by NSF grant IBN 0110864.
REFERENCES
- 1.Adelman, D. M., E. Maltepe, and M. C. Simon. 1999. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes Dev. 13:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany, Z., L. E. Huang, R. Eckner, S. Bhattacharya, C. Jiang, M. A. Goldberg, H. F. Bunn, and D. M. Livingston. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. USA 93:12969-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruick, R. K. 2003. Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 17:2614-2623. [DOI] [PubMed] [Google Scholar]
- 4.Clemmons, D. R. 2001. Use of mutagenesis to probe IGF-binding protein structure/function relationships. Endocr. Rev. 22:800-817. [DOI] [PubMed] [Google Scholar]
- 5.Crossey, P. A., C. C. Pillai, and J. P. Miell. 2002. Altered placental development and intrauterine growth restriction in IGF binding protein-1 transgenic mice. J. Clin. Investig. 110:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dames, S. A., M. Martinez-Yamout, R. N. De Guzman, H. J. Dyson, and P. E. Wright. 2002. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA 99:5271-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fant, M., C. Salafia, R. C. Baxter, J. Schwander, C. Vogel, J. Pezzullo, and F. Moya. 1993. Circulating levels of IGFs and IGF binding proteins in human cord serum: relationships to intrauterine growth. Regul. Pept. 48:29-39. [DOI] [PubMed] [Google Scholar]
- 8.Firth, J. D., B. L. Ebert, and P. J. Ratcliffe. 1995. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J. Biol. Chem. 270:21021-21027. [DOI] [PubMed] [Google Scholar]
- 9.Firth, S. M., and R. C. Baxter. 2002. Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23:824-854. [DOI] [PubMed] [Google Scholar]
- 10.Galson, D. L., T. Tsuchiya, D. S. Tendler, L. E. Huang, Y. Ren, T. Ogura, and H. F. Bunn. 1995. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol. Cell. Biol. 15:2135-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gay, E., D. Seurin, S. Babajko, S. Doublier, M. Cazillis, and M. Binoux. 1997. Liver-specific expression of human insulin-like growth factor binding protein-1 in transgenic mice: repercussions on reproduction, ante- and perinatal mortality and postnatal growth. Endocrinology 138:2937-2947. [DOI] [PubMed] [Google Scholar]
- 12.Giudice, L. C., F. de Zegher, S. E. Gargosky, B. A. Dsupin, L. de las Fuentes, R. A. Crystal, R. L. Hintz, and R. G. Rosenfeld. 1995. Insulin-like growth factors and their binding proteins in the term and preterm human fetus and neonate with normal and extremes of intrauterine growth. J. Clin. Endocrinol. Metab. 80:1548-1555. [DOI] [PubMed] [Google Scholar]
- 13.Gracey, A. Y., J. V. Troll, and G. N. Somero. 2001. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 98:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillemin, K., and M. A. Krasnow. 1997. The hypoxic response: huffing and HIFing. Cell 89:9-12. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S. T., K. C. Vo, D. J. Lyell, G. H. Faessen, S. Tulac, R. Tibshirani, A. J. Giaccia, and L. C. Giudice. 2004. Developmental response to hypoxia. FASEB J. 18:1348-1365. [DOI] [PubMed] [Google Scholar]
- 16.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 17.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 19.Kajimura, S., K. Aida, and C. Duan. 2005. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc. Natl. Acad. Sci. USA 102:1240-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann, and T. F. Schilling. 1995. Stages of embryonic development of the zebra fish. Dev. Dyn. 203:253-310. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, H., A. Weisz, Y. Kurashima, K. Hashimoto, T. Ogura, F. D'Acquisto, R. Addeo, M. Makuuchi, and H. Esumi. 2000. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood 95:189-197. [PubMed] [Google Scholar]
- 23.Kimura, H., A. Weisz, T. Ogura, Y. Hitomi, Y. Kurashima, K. Hashimoto, F. D'Acquisto, M. Makuuchi, and H. Esumi. 2001. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276:2292-2298. [DOI] [PubMed] [Google Scholar]
- 24.Kvietikova, I., R. H. Wenger, H. H. Marti, and M. Gassmann. 1995. The transcription factors ATF-1 and CREB-1 bind constitutively to the hypoxia-inducible factor-1 (HIF-1) DNA recognition site. Nucleic Acids Res. 23:4542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Y., S. R. Cox, T. Morita, and S. Kourembanas. 1995. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ. Res. 77:638-643. [DOI] [PubMed] [Google Scholar]
- 26.Maltepe, E., J. V. Schmidt, D. Baunoch, C. A. Bradfield, and M. C. Simon. 1997. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature 386:403-407. [DOI] [PubMed] [Google Scholar]
- 27.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maures, T., S. J. Chan, B. Xu, H. Sun, J. Ding, and C. Duan. 2002. Structural, biochemical, and expression analysis of two distinct insulin-like growth factor I receptors and their ligands in zebra fish. Endocrinology 143:1858-1871. [DOI] [PubMed] [Google Scholar]
- 29.Maures, T. J., and C. Duan. 2002. Structure, developmental expression, and physiological regulation of zebra fish IGF binding protein-1. Endocrinology 143:2722-2731. [DOI] [PubMed] [Google Scholar]
- 30.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 31.McLellan, K. C., S. B. Hooper, A. D. Bocking, P. J. Delhanty, I. D. Phillips, D. J. Hill, and V. K. Han. 1992. Prolonged hypoxia induced by the reduction of maternal uterine blood flow alters insulin-like growth factor-binding protein-1 (IGFBP-1) and IGFBP-2 gene expression in the ovine fetus. Endocrinology 131:1619-1628. [DOI] [PubMed] [Google Scholar]
- 32.Nikinmaa, M., and B. B. Rees. 2005. Oxygen-dependent gene expression in fishes. Am. J. Physiol. Regul Integr Comp. Physiol. 288:R1079-R1090. [DOI] [PubMed] [Google Scholar]
- 33.Norris, M. L., and D. E. Millhorn. 1995. Hypoxia-induced protein binding to O2-responsive sequences on the tyrosine hydroxylase gene. J. Biol. Chem. 270:23774-23779. [DOI] [PubMed] [Google Scholar]
- 34.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 35.Popovici, R. M., M. Lu, S. Bhatia, G. H. Faessen, A. J. Giaccia, and L. C. Giudice. 2001. Hypoxia regulates insulin-like growth factor-binding protein 1 in human fetal hepatocytes in primary culture: suggestive molecular mechanisms for in utero fetal growth restriction caused by uteroplacental insufficiency. J. Clin. Endocrinol. Metab. 86:2653-2659. [DOI] [PubMed] [Google Scholar]
- 36.Rajkumar, K., D. Barron, M. S. Lewitt, and L. J. Murphy. 1995. Growth retardation and hyperglycemia in insulin-like growth factor binding protein-1 transgenic mice. Endocrinology 136:4029-4034. [DOI] [PubMed] [Google Scholar]
- 37.Schug, J., and G. C. Overton. 1997. TESS: transcription element search software on the WWW. Technical report CBIL-TR-1997-1001-v0.0. Computational Biology and Informatics Laboratory, University of Pennsylvania, Philadelphia.
- 38.Semenza, G. L., B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, and A. Giallongo. 1996. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271:32529-32537. [DOI] [PubMed] [Google Scholar]
- 39.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seta, K. A., and D. E. Millhorn. 2004. Functional genomics approach to hypoxia signaling. J. Appl. Physiol. 96:765-773. [DOI] [PubMed] [Google Scholar]
- 41.Tazuke, S. I., N. M. Mazure, J. Sugawara, G. Carland, G. H. Faessen, L. F. Suen, J. C. Irwin, D. R. Powell, A. J. Giaccia, and L. C. Giudice. 1998. Hypoxia stimulates insulin-like growth factor binding protein 1 (IGFBP-1) gene expression in HepG2 cells: a possible model for IGFBP-1 expression in fetal hypoxia. Proc. Natl. Acad. Sci. USA 95:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unterman, T., R. Lascon, M. B. Gotway, D. Oehler, A. Gounis, R. A. Simmons, and E. S. Ogata. 1990. Circulating levels of insulin-like growth factor binding protein-1 (IGFBP-1) and hepatic mRNA are increased in the small for gestational age (SGA) fetal rat. Endocrinology 127:2035-2037. [DOI] [PubMed] [Google Scholar]
- 43.Unterman, T. G., R. A. Simmons, R. P. Glick, and E. S. Ogata. 1993. Circulating levels of insulin, insulin-like growth factor-I (IGF-I), IGF-II, and IGF-binding proteins in the small for gestational age fetal rat. Endocrinology 132:327-336. [DOI] [PubMed] [Google Scholar]
- 44.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]