Abstract
Cyclin-dependent kinase 9 (Cdk9) of fission yeast is an essential ortholog of metazoan positive transcription elongation factor b (P-TEFb), which is proposed to coordinate capping and elongation of RNA polymerase II (Pol II) transcripts. Here we show that Cdk9 is activated to phosphorylate Pol II and the elongation factor Spt5 by Csk1, one of two fission yeast CDK-activating kinases (CAKs). Activation depends on Cdk9 T-loop residue Thr-212. The other CAK—Mcs6, the kinase component of transcription factor IIH (TFIIH)—cannot activate Cdk9. Consistent with the specificities of the two CAKs in vitro, the kinase activity of Cdk9 is reduced ∼10-fold by csk1 deletion, and Cdk9 complexes from csk1Δ but not csk1+ cells can be activated by Csk1 in vitro. A cdk9T212A mutant is viable but phenocopies conditional growth defects of csk1Δ strains, indicating a role for Csk1-dependent activation of Cdk9 in vivo. A cdk9T212A mcs6S165A strain, in which neither Cdk9 nor Mcs6 can be activated by CAK, has a synthetic growth defect, implying functional overlap between the two CDKs, which have distinct but overlapping substrate specificities. Cdk9 forms complexes in vivo with the essential cyclin Pch1 and with Pcm1, the mRNA cap methyltransferase. The carboxyl-terminal region of Cdk9, through which it interacts with another capping enzyme, the RNA triphosphatase Pct1, is essential. Together, the data support a proposed model whereby Cdk9/Pch1—the third essential CDK-cyclin complex described in fission yeast—helps to target the capping apparatus to the transcriptional elongation complex.
Cyclin-dependent kinases (CDKs) first emerged as controllers of cell division but have also been implicated in processes not strictly coupled to the cell cycle, most notably transcription by RNA polymerase II (Pol II) (4). In metazoans, Cdk9 and cyclin T constitute positive transcription elongation factor b (P-TEFb), which phosphorylates both the carboxyl-terminal domain (CTD) of Rpb1, the largest subunit of Pol II, and Spt5, a subunit of the elongation factor DRB sensitivity-inducing factor, to overcome kinetic blocks to elongation (21). The requirement for Cdk9 in facilitating elongation is probably a general one (66) and is posited to be a quality control mechanism to ensure that nascent transcripts are not elongated unless and until mRNA-capping enzymes and other processing machinery can be recruited (9, 10, 49, 52).
In the budding yeast Saccharomyces cerevisiae, the Bur1/Bur2 CDK-cyclin pair and a heterotrimeric CDK, CTDK-I, show roughly equal homology between their catalytic subunits (Bur1 and Ctk1, respectively) and metazoan Cdk9 (46). The BUR1 gene is essential (56), whereas neither BUR2 nor any of the genes encoding CTDK-I subunits is required for viability (68, 77). Cdk9 of the fission yeast Schizosaccharomyces pombe was identified in a two-hybrid interaction screen with Pct1, the RNA triphosphatase component of the mRNA-capping apparatus (52). Cdk9 can form an active kinase complex with the essential fission yeast cyclin Pch1, and the two proteins expressed together in S. cerevisiae complemented a bur1 deletion (19, 52), but the physiologic cyclin partner of Cdk9 in S. pombe remained to be identified.
CDKs depend to various degrees on phosphorylation within the activation segment, or T loop, of the catalytic subunit by a CDK-activating kinase (CAK). The dedicated cell cycle CDKs, exemplified by Cdk1, absolutely require T-loop phosphorylation (22, 37), whereas CDKs involved in transcription need this modification for full catalytic activity and/or for stability but not for their essential functions (29, 31, 78). The CAKs fall into two classes. In metazoans, the major CAK is a heterotrimeric complex of Cdk7, cyclin H, and the RING finger protein Mat1, which is also part of general transcription factor IIH (TFIIH), which phosphorylates the Pol II CTD (23). In contrast, the sole CAK in budding yeast is Cak1, a monomeric enzyme related only distantly to the CDK family (27). Cak1 activates both Cdk1 and the Cdk7 ortholog Kin28 (17, 29), which unlike its metazoan counterpart is a dedicated TFIIH-associated CTD kinase that does not activate CDKs (12, 73).
S. pombe has one CAK from each class: the Mcs6/Mcs2/Pmh1 complex, which is homologous to Cdk7/cyclin H/Mat1, and Csk1, a single-subunit enzyme most closely related to Cak1 (2, 5, 14, 24, 33, 34, 67). Both enzymes can activate Cdk1 (34, 63), a redundancy that probably explains why csk1+ is dispensable for viability (43) and why mutations in genes encoding Mcs6 complex subunits do not impair CDK activation or impede entry into mitosis unless combined with other mutations, such as csk1Δ (5, 14, 24, 33, 34, 43, 63). The Mcs6 complex is required for viability, however, suggesting it has another essential target, which is likely to be Pol II. The growth defects caused by csk1 deletion (3, 25, 63) might reflect specialized requirements for Csk1-mediated activation of Cdk1 and/or activity of Csk1 towards proteins that the Mcs6 complex does not phosphorylate.
Activation of metazoan P-TEFb by a CAK has not been demonstrated, but mutating Thr-186 in the human Cdk9 T loop abolished activity and, paradoxically, binding to a ribonucleoprotein inhibitor (7). In budding yeast, a temperature-sensitive bur1 mutation was suppressed by overexpression of CAK1, and phosphorylation by Cak1 stimulated the kinase activity of Bur1 in vitro, dependent on the Thr-240 residue of the Bur1 T loop. That stimulation is apparently important in vivo; a bur1T240A allele only partially complemented bur1Δ (78). More recently, Cak1 was shown to activate Ctk1, and a mutation in ctk1 preventing T-loop phosphorylation caused a defect in the entry into stationary phase (50). Changing Thr-212 within the T loop of fission yeast Cdk9 to alanine abolished heterologous complementation of bur1Δ, whereas a mutation of the same residue to glutamic acid rendered it cold and temperature sensitive (52, 53).
The regulation of Cdk9 by upstream kinases (CAKs) in fission yeast has not been investigated. Moreover, its role(s) in regulating gene expression and possibly coordinating mRNA-processing events with transcription remains to be elucidated. Here we show specificity within the CAK-CDK network of S. pombe; Csk1, but not the Mcs6 complex, activates Cdk9/Pch1 complexes in vitro and in vivo. The nonphosphorylatable T-loop mutant cdk9T212A grows poorly on minimal media and is cold sensitive, essentially phenocopying csk1Δ. We observe a synthetic interaction between cdk9T212A and the analogous mcs6S165A T-loop mutation, suggesting that the essential Cdk9 and Mcs6 complexes, which have partially overlapping substrate specificities in vitro, have redundant as well as unique functions in controlling gene expression in vivo. Finally, we provide support for the idea that Cdk9 couples transcription to mRNA capping (9, 10, 49, 52) by demonstrating (i) that the carboxyl-terminal, Pct1-interacting region of Cdk9 (52) is required for viability and (ii) that Cdk9 stably associates in vivo with the guanine-N7 methyltransferase component of the fission yeast mRNA-capping apparatus in ∼500-kDa complexes that are released from larger complexes by RNase digestion.
MATERIALS AND METHODS
Expression and purification of recombinant proteins.
Mutant Cdk9/Pch1 complexes were expressed with recombinant baculoviruses (see the supplemental material) and purified as described previously (52). Recombinant Csk1 was produced by infecting Sf9 insect cells with appropriate viruses according to standard methods (60). After being harvested by centrifugation, cells were resuspended in hypotonic lysis buffer (50 mM Tris-HCl [pH 7.8], 20 mM NaCl, 1 μg/ml leupeptin, 2 μg/ml aprotinin, 0.5 mM phenylmethylsulfonyl fluoride) and disrupted with a Dounce homogenizer; 5 M NaCl and 1 M imidazole were then added to final concentrations of 300 mM and 10 mM, respectively. Insoluble material was removed by centrifugation for 1 h at 100,000 × g. The soluble extract was applied to a Ni2+-nitrilotriacetic acid-agarose column equilibrated in buffer A (50 mM Tris-HCl [pH 7.8], 300 mM NaCl, 10 mM imidazole, and 10% glycerol), the column was washed with buffer A, and bound proteins were eluted with buffer B (50 mM Tris-HCl [pH 7.8], 300 mM NaCl, 200 mM imidazole, and 10% glycerol). The eluate was concentrated threefold in a Centricon YM-10 instrument and applied to a Superdex 200 gel filtration column equilibrated in buffer C (25 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, and 10% glycerol). The fractions containing Csk1 were pooled, and the protein concentration was determined by the Bradford method with bovine serum albumin as the standard. Mcs6/Mcs2 complexes were purified by a similar protocol.
Kinase assays.
The Spt5 and Rpb1 kinase activities of unphosphorylated mutant Cdk9/Pch1 complexes were measured as described previously for wild-type Cdk9/Pch1 (52). To examine the direct phosphorylation of wild-type and mutant Cdk9/Pch1 complexes by Csk1, reaction mixtures (30 μl) containing 10 μM HEPES (pH 7.4), 1 mM DTT, 10 mM MgCl2, 50 μM ATP including 1 to 5 mCi [γ-32P]ATP, ∼2 μg of wild-type or mutant Cdk9/Pch1 complex, and Csk1 were incubated for 15 min at 22°C. To measure activation of wild-type and mutant Cdk9/Pch1 by Csk1, activation mixtures (30 μl) containing 10 mM HEPES (pH 7.4), 1 mM DTT, 10 mM MgCl2, 1 mM ATP, ∼150 ng wild-type or mutant Cdk9/Pch1, and Csk1 were incubated for 15 min at 22°C. One-tenth of the activation mixture was withdrawn and added to a labeling mixture (27 μl) containing 10 mM HEPES (pH 7.4), 1 mM DTT, 10 mM MgCl2, 50 μM ATP including 5 μCi [γ-32P]ATP, and 4 μg of either glutathione S-transferase (GST)-Spt5(801-990) or GST-Rpb1 CTD, which was incubated for 10 min at 22°C. Phosphorylation reactions were stopped by adding sodium dodecyl sulfate (SDS) to a 1% final concentration. The products were analyzed by electrophoresis in a 10% polyacrylamide gel. Phosphorylated proteins were detected by autoradiography of dried gels and quantified by scanning with a FUJIX BAS2500 phosphorimager.
The specific activities of unphosphorylated mutant Cdk9/Pch1 complexes towards Spt5 and Rpb1 were measured as described previously for wild-type Cdk9/Pch1 (52). Reaction mixtures containing 50 mM Tris-acetate (pH 6.0), 1 mM DTT, 2.5 mM MnCl2 or 10 mM MgCl2, 50 μM [γ-32P]ATP, 4 μg of GST-Spt5(801-990) or GST-Rpb1 CTD, and wild-type or mutant Cdk9/Pch1 complexes as specified were incubated for 1 h at 22°C. The extent of substrate phosphorylation was quantified by scanning the dried gel with a FUJIX phosphorimager and plotted as a function of input protein. The apparent specific activity of each mutant Cdk9/Pch1 complex was determined from the average slope of two or three independent titration curves in the linear range of enzyme dependence and expressed as percent values relative to those of wild-type Cdk9/Pch1. To calculate specific activities of mutant Cdk9/Pch1 complexes, the amounts of mutant Cdk9 polypeptides were normalized to that of wild-type Cdk9. The stained gels were scanned with a FUJI FLA-5000 fluorescent image analyzer, and the densities of Cdk9 polypeptides were quantified in Image Gauge 4.0. The relative amounts of D184N and T212E were determined from the Coomassie blue-stained gel shown in Fig. 1A, whereas those of Cdk9ΔC and T212A were determined after TEV protease treatment (see Fig. S1 in the supplemental material). Because Cdk9ΔC comigrates with undigested His-Pch1, the ∼43-kDa band was excised and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) to estimate the relative amount of Cdk9(1-385). The estimated total amount of Cdk9T212A included both the full-length polypeptide and the ∼43-kDa proteolytic fragment.
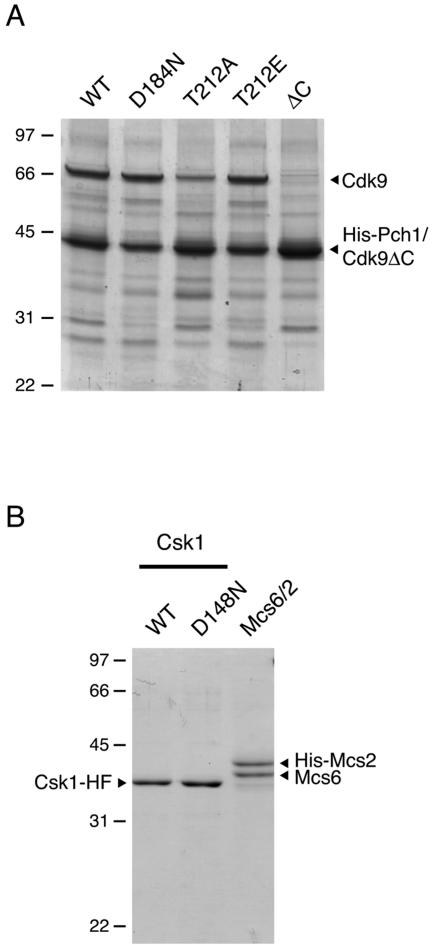
FIG. 1.
Purification of Cdk9/Pch1 and CAKs. (A) Wild-type (WT) and D184N, T212A, T212E, and Cdk9(1-385) (ΔC) mutant versions of Cdk9 were coexpressed with His-Pch1 in insect cells and purified by metal-affinity chromatography. Aliquots (4 μg) of the purified proteins were electrophoresed in a 12% polyacrylamide gel containing 0.1% SDS. Polypeptides were visualized by Coomassie blue staining. The positions and sizes (in kilodaltons) of marker proteins are indicated at the left. The polypeptides corresponding to Cdk9 and His-tagged Pch1 are denoted by arrowheads at the right. (Note that Cdk9ΔC comigrates with His-Pch1.) (B) Csk1-His-Flag, the wild type or the D148N mutant, or Mcs6/His-Mcs2 complex, as indicated at the top, were purified by metal-affinity and gel-exclusion chromatographies. Aliquots of each preparation (2 μg total protein) were analyzed by SDS-polyacrylamide gel electrophoresis and staining with Coomassie blue. The band corresponding to Csk1-His-Flag (Csk1-HF) is indicated at the left, and the mobilities of Mcs6 and His-Mcs2 are indicated at the right.
General yeast methods.
Fission yeast cell culturing, transformation, sporulation, tetrad dissection, and extract preparation were performed according to standard methods (44). Cells were grown in yeast extract medium with supplements (YES) or in Edinburgh minimal medium (EMM). To determine population doubling times, cells were grown in YES at 30°C from a starting density of ∼5 × 105 cells/ml, and growth rate constants were determined from exponential curves obtained by determining best fit to the experimental data. Doubling times were calculated from the equation n = noekt, where n is the cell number, no is the initial cell number, k is the growth rate constant, and t is time. For analysis of Cdk9-Myc complexes by coimmunoprecipitation and gel filtration, we prepared extracts in a modified lysis buffer consisting of 25 mM HEPES (pH 7.4), 150 mM NaCl, 50 mM NaF, 60 mM β-glycerophosphate, 2 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1.3 mM benzamidine, 0.1 mM Na3VO4, 4 mg/ml leupeptin, and one protease inhibitor cocktail tablet (Roche) per 50 ml buffer. Extracts were centrifuged for 1 h at ∼100,000 × gav and applied to a Superdex 200 10/30 HR gel filtration column (Pharmacia) as previously described (31).
Disruption, mutagenesis, and tagging of cdk9+.
To disrupt the cdk9+ locus, DNA fragments corresponding to regions 5′ and 3′ of the coding region were amplified by PCR from the S. pombe genomic DNA template. The flanking regions were subcloned on either side of a kanMX marker in plasmid pFA6akanMX6. A linear fragment was used to transform a diploid strain maintained by ade− complementation on EMM minus adenine. Transformants were selected on YES plus 200 mg/ml G418 and tested for correct gene targeting by PCR. The cdk9+/cdk9::kanMX heterozygous diploids were induced to sporulate on maltose extract (ME) medium.
To introduce a carboxyl-terminal Myc epitope tag by homologous integration at the chromosomal cdk9+ locus, we generated a linear fragment with ∼80 bp of the cdk9+ 3′-terminal coding region fused in frame with DNA encoding 13 copies of the Myc epitope in tandem with the kanMX marker followed by another ∼80 bp of genomic sequence downstream of the cdk9+ gene as described previously (1). The epitope-tagging cassette was amplified from plasmid pFA6a-13myc-kanMX6 (gift of Jian-Qiu Wu, Yale University). The transformation with the PCR product and the selection of correct integrants on YES plus 100 mg/ml G418 were carried out as described previously (1).
To introduce the T212A mutation, we transformed a haploid strain with an AccI restriction fragment containing the mutant sequence amplified by PCR from the mutant cDNA (52) in tandem with the kanMX marker and flanking sequence amplified from the 3′ untranslated region. We selected G418-resistant colonies and screened for correct integration. The presence of the mutation was initially detected by digestion with NgoMIV (for which a recognition site was introduced during the mutagenesis) of a PCR fragment amplified from genomic DNA and confirmed by sequencing.
Immunological methods.
Cdk9-13Myc was detected by probing immunoblots with monoclonal antibody (MAb) 9E10 (Covance). Total Rpb1 CTD was detected with MAb 8WG16 (Covance), and phospho-isoforms were detected with MAbs H5 and H14 (Covance). We performed immunoprecipitation with MAb 9E10 bound to protein G-agarose (Amersham Pharmacia Biotech). Immunoprecipitates were washed three times with 10 mM HEPES (pH 7.4), 150 mM NaCl, and 0.1% Triton X-100 and twice with 50 mM Tris-acetate (pH 6.0), 2.5 mM MnCl2, and 1 mM DTT and tested for Spt5 kinase activity or subjected to immunoblotting.
Identification of Cdk9-associated proteins by mass spectrometry.
Proteins excised from gels were digested with trypsin, the mixtures fractionated on a Poros 50 R2 RP microtip, and resulting peptide pools analyzed by MALDI-reflectron TOF MS using a Bruker UltraFlex TOF/TOF instrument (Bruker Daltonics, Bremen, Germany) as described previously (16, 75). Selected experimental masses (m/z) were taken to search the S. pombe segment of a nonredundant protein database (NR; 6,786 entries; National Center for Biotechnology Information, Bethesda, MD) by use of the PeptideSearch (Matthias Mann, Southern Denmark University, Odense, Denmark) algorithm with a mass accuracy restriction of better than 40 ppm and a maximum of one missed cleavage site allowed per peptide. Mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples by use of an UltraFlex instrument in “lift” mode. Fragment ion spectra were taken to search NR using the MASCOT MS/MS ion search program (Matrix Science Ltd., London, United Kingdom). Any identification thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
RESULTS
Cdk9 is activated by Csk1, but not by Mcs6/Mcs2, in vitro.
We generated wild-type and mutant Cdk9/Pch1 complexes in insect Sf9 cells infected with recombinant baculoviruses (Fig. 1A) and measured their specific activities towards either the CTD of Rpb1 (the largest subunit of Pol II) or a carboxyl-terminal fragment of Spt5 that contains the nonapeptide repeats phosphorylated by Cdk9 (53) (Table 1). A fraction of Cdk9T212A was proteolyzed in the insect cells; we showed by differential susceptibility to TEV protease (which cleaves the His-Pch1 fusion protein) and mass spectrometry that the ∼43-kDa polypeptide in this preparation was a mixture of His-Pch1 and a proteolytic fragment of Cdk9 containing the intact amino-terminal kinase domain (Fig. 1A; see Fig. S1 in the supplemental material; also data not shown). Nonetheless, this mutant, which failed to rescue bur1Δ (52), had Mg2+-dependent activity comparable to that of the wild-type kinase towards both substrates. In addition, the Cdk9T212E/Pch1 complex, which complemented the bur1Δ mutation partially at 30°C and not at 37°C or 18°C, was more active than the wild-type kinase in vitro.
TABLE 1.
Effects of point mutations and C-terminal truncation of Cdk9 on the specific activity of Cdk9/Pch1 complex
| Mutation | % Specific activity ofa:
|
|||
|---|---|---|---|---|
| Spt5 (aa 801-990) with:
|
Rpb1 CTD with:
|
|||
| MgCl2 | MnCl2 | MgCl2 | MnCl2 | |
| T212A | 87 | 240 | 75 | 120 |
| T212E | 310 | 300 | 200 | 320 |
| D184N | <0.5 | <0.5 | <0.1 | <0.1 |
| Cdk9ΔC | 24 | 73 | 57 | 106 |
The apparent specific activity of each mutant Cdk9/Pch1 complex was determined from the average slope of two or three independent protein titration curves in the linear range of enzyme dependence and expressed as percent values relative to those of wild-type Cdk9/Pch1. The specific activities of wild-type Cdk9/Pch1 complex (defined as 100%) were as follows. Towards GST-Spt5 (aa 801 to 990), they were 0.8 pmol phosphoproduct per ng kinase complex with Mn2+ as the cofactor and 0.26 pmol/ng with Mg2+. Towards GST-Rpb1 CTD, they were 0.22 pmol/ng with Mn2+ and 0.16 pmol/ng with Mg2+. See Materials and Methods for a full description of measurements of specific activity.
The relative activities in vitro of the different Cdk9 variants (activity of T212E > wild type ≈ T212A in the presence of Mg2+) suggested that Thr-212 was a site of activating phosphorylation and moreover that it was phosphorylated inefficiently, if at all, by endogenous insect cell kinases. We were unable to detect phosphorylation of Thr-212 by mass spectrometry in wild-type Cdk9/Pch1 complexes purified from Sf9 cells (our unpublished observations). We therefore tested whether Cdk9/Pch1 was a substrate for either of the two fission yeast CAKs, which were also produced with recombinant baculoviruses (Fig. 1B). Preincubating Cdk9/Pch1 with Csk1, but not with Mcs6/Mcs2, resulted in stimulation of kinase activity towards Spt5 (Fig. 2A). In control reactions, Csk1 did not detectably phosphorylate either Spt5 (lane 7) or Rpb1 (data not shown), whereas Mcs6/Mcs2 was active towards Rpb1 (lanes 13 and 14) but not towards Spt5 (lane 12). The stimulation of Cdk9 activity was ∼10-fold towards either substrate (Fig. 2B; see Fig. S3A in the supplemental material) and was abolished by the inactivating D148N mutation in Csk1 (72).
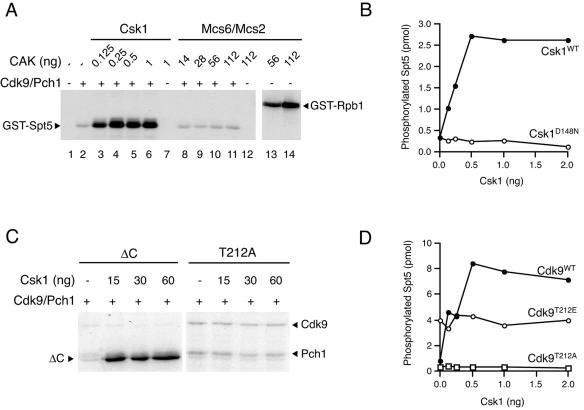
FIG. 2.
Activation of Cdk9/Pch1 by Csk1 in vitro. (A) Wild-type Cdk9/Pch1 complexes (150 ng per reaction) were preincubated with indicated amounts of purified Csk1 (lanes 3 to 6) or Mcs6/Mcs2 (lanes 8 to 11) plus cold ATP and tested for activity towards GST-Spt5(801-990). Activity of untreated Cdk9/Pch1 is shown in lane 2, and reaction mixtures lacking Cdk9/Pch1 but containing either Csk1 or Mcs6/Mcs2 were analyzed in lanes 7 and 12, respectively. The Mcs6/Mcs2 complex is active towards the Rpb1 CTD (lanes 13 and 14). The mobilities of radiolabeled GST-Spt5 and -Rpb1 polypeptides are indicated by arrowheads at the left and at the right, respectively. (B) Activity of wild-type Cdk9/Pch1 towards GST-Spt5(801-990) after preincubation with increasing amounts of wild-type or kinase-dead (D148N) Csk1 plotted as a function of the input Csk1 protein. (C) Csk1 phosphorylates Cdk9 in a manner dependent on Thr-212 in the T loop. The Cdk9/Pch1 complex (2 μg), Cdk9ΔC, or Cdk9T212A, as indicated above each panel, was incubated alone (first lane in each panel) or with increasing amounts (indicated at top) of wild-type Csk1 in the presence of [γ-32P]ATP. The electrophoretic mobility of the Cdk9ΔC polypeptide is indicated by the arrowhead at left, whereas the mobilities of radiolabeled full-length Cdk9 and Pch1 polypeptides are indicated by arrowheads at right. (D) Activation of Cdk9 by Csk1 is Thr-212 dependent. Activities of wild-type Cdk9, Cdk9T212A, and Cdk9T212E complexes towards GST-Spt5(801-990) after preincubation with increasing amounts of wild-type Csk1 are plotted as a function of the input Csk1 protein.
Cdk9 phosphorylation and activation by Csk1 depend on Thr-212 of the T loop.
We next sought to confirm that Csk1 phosphorylated Cdk9 directly. The complex of wild-type Cdk9 with Pch1 was capable of autophosphorylation on both the catalytic and cyclin subunits (see Fig. S2 in the supplemental material) as previously reported (53). The autophosphorylation was suppressed in a Cdk9ΔC mutant that retained the kinase domain but lacked a 206-amino-acid carboxyl-terminal extension required for interaction with Pct1 (52), allowing us to detect phosphorylation of the catalytic subunit by Csk1 (Fig. 2C). In contrast, Csk1 did not phosphorylate Cdk9T212A above the background signal due to autophosphorylation (Fig. 2C). Based on these results (and data shown in Fig. S2 in the supplemental material), we conclude that Csk1 phosphorylates Cdk9 directly, dependent on Thr-212 of the Cdk9 T loop.
The wild-type and T212A mutant forms of Cdk9 had similar basal activities towards either Spt5-derived (Fig. 2D and Table 1) or Rpb1-derived (Table 1; see Fig. S3B in the supplemental material) substrates. In contrast, Cdk9T212E was approximately fourfold more active than the wild-type enzyme in the absence of CAK. Csk1 caused an ∼10-fold activation of the wild type, but not of either T212 mutant enzyme, towards both Spt5 (Fig. 2D) and Rpb1 (see Fig. S3B in the supplemental material). Csk1 similarly enhanced the activity of Cdk9ΔC, but not that of Cdk9D184N (data not shown). Thus, both phosphorylation and enzymatic activation depend on the threonine residue at the position within the activation loop conserved in other CAK-dependent CDKs, indicating that Csk1 is a CAK for Cdk9.
Csk1 is a Cdk9-activating kinase in vivo.
To determine whether the activation of Cdk9 depends on Csk1 in vivo, we tagged Cdk9 at its carboxyl terminus with 13 copies of the Myc epitope. The resulting cdk9-13Myc strain was indistinguishable from the wild type with respect to growth and cell morphology (data not shown). Cdk9-Myc could be detected as an ∼110-kDa polypeptide in immunoblots of whole-cell extracts (Fig. 3A), and anti-Myc immunoprecipitates from the tagged strain exhibited kinase activity towards a GST-Spt5 fusion protein (Fig. 3B). We generated cdk9-13Myc csk1Δ strains and measured Cdk9 protein and activity levels in cells lacking Csk1. Cdk9 abundance was consistently increased by two- to fourfold in extracts from strains deleted of csk1+ (Fig. 3A). In contrast, the kinase activity recovered in Cdk9 complexes from a csk1Δ strain was reduced by >10-fold relative to a csk1+ strain, even though more Cdk9 protein was recovered per μg of total input protein from the csk1Δ extract (Fig. 3B).
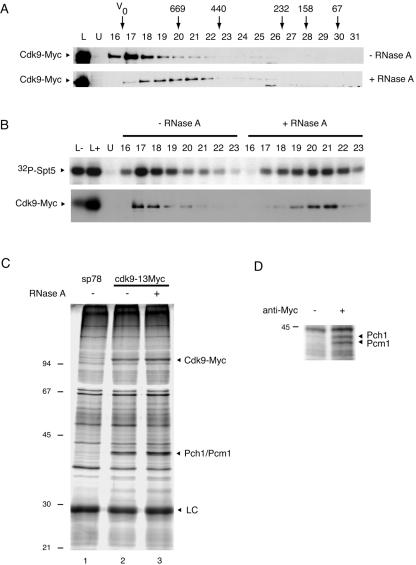
FIG. 3.
Cdk9 is activated by Csk1 in vivo. (A) Expression of Cdk9 increases upon deletion of csk1+ as measured by immunoblotting increasing amounts (indicated above each lane) of S. pombe whole-cell extracts from strains of the indicated genotypes. Both csk1+ and csk1Δ strains carry the cdk9-13Myc allele, whereas sp78 (first lane) is an untagged control strain. The mobility of Cdk9-13Myc is indicated by the arrowhead at the left. (B) The specific activity of Cdk9 is decreased in csk1Δ strains relative to csk1+ strains. Cdk9 was immunoprecipitated from increasing amounts of extracts of cells of the two different genetic backgrounds (indicated at the top) and assayed for kinase activity towards GST-Spt5(801-990) and recovery of Cdk9-Myc protein by immunoblotting with anti-Myc antibody. (C) Cdk9 complexes from the csk1+ and csk1Δ strains (50 μg total extract protein) were isolated by immunoprecipitation, preincubated in the presence of Mg-ATP with no protein added or with indicated amounts of Csk1, and tested for activity towards GST-Spt5(801-990) and recovery of Cdk9-Myc protein by immunoblotting with anti-Myc antibody. The relative intensities of phosphorimager signals are indicated below each lane, with the signal obtained in the csk1Δ sample in the absence of Csk1 treatment defined as 1.0. The first two lanes of the top panel contain GST-Spt5(801-990) phosphorylated by purified, baculovirus-derived Cdk9/Pch1 without (first lane) or with (second lane) activation by Csk1. (In panels B and C, the mobilities of GST-Spt5 and Cdk9-13Myc are indicated by arrowheads at the left.)
To confirm that decreased Cdk9-associated kinase activity was due to lack of T-loop phosphorylation, we attempted to activate Cdk9 recovered from csk1Δ or csk1+ cells with CAK in vitro. Treatment of Cdk9 complexes from a cdk9-13Myc csk1Δ strain with recombinant Csk1 restored Spt5 kinase levels to ∼70% of that in untreated complexes from a cdk9-13Myc csk1+ strain (Fig. 3C), indicating that cells lacking Csk1 had assembled Cdk9/cyclin complexes requiring only T-loop phosphorylation for full activity. Inefficient phosphorylation of Cdk9 immobilized in immune complexes or the presence of a population of Cdk9 molecules incapable of being activated could explain why we could not restore activity fully to wild-type levels. In contrast, complexes from csk1+ cells were refractory to activation by CAK in vitro, suggesting they contained little or no unphosphorylated, cyclin-bound Cdk9. The data indicate that Cdk9 is activated in vivo by Csk1 but not by Mcs6.
Cdk9 is essential for viability.
To ascertain the requirement, if any, for Cdk9 function in S. pombe, we disrupted one copy of cdk9+ in a diploid strain with a kanMX drug resistance marker and induced the resulting cdk9+/cdk9Δ heterozygote to sporulate. Tetrad analysis revealed a 2:2 segregation of viability, and all viable progeny were G418 sensitive, indicating that cdk9+ is essential (data not shown). Similar results were recently reported in a deletion analysis of S. pombe genes encoding known and suspected kinases (3). Cdk9 is thus the third essential CDK described in fission yeast, the other two being Cdk1 (also known as Cdc2) and Mcs6.
Importance of Cdk9 T-loop phosphorylation in vivo.
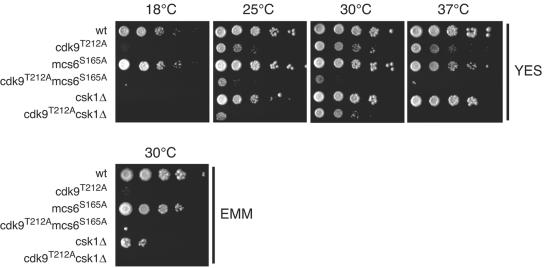
To test whether Cdk9 requires T-loop phosphorylation to perform its essential function, we replaced the wild-type cdk9+ gene with the mutant cdk9T212A allele. Although we were able to recover viable haploid cdk9T212A mutants, indicating that T-loop phosphorylation is not essential, the mutants grew slowly (the doubling time at 30°C in rich medium was ∼3.8 h, while that for a wild-type strain was ∼2.8 h; see Materials and Methods). The growth defect was exacerbated at 18°C or at 37°C (Fig. 4), suggesting further impairment of and/or greater dependence on Cdk9 activity at both extremes of temperature. The temperature sensitivity was leaky and caused by the failure to accelerate cell division at elevated temperature (data not shown). Neither a csk1Δ nor an mcs6S165A strain, in which the site phosphorylated by Csk1 within the Mcs6 T loop is mutated (24, 34), was similarly temperature sensitive (Fig. 4) or slow growing in rich medium (doubling times for csk1Δ and mcs6S165A were ∼2.7 and ∼3.1 h, respectively). Like cdk9T212A, however, csk1Δ strains were cold sensitive, as was also recently reported (3), but an mcs6S165A strain was not (Fig. 4). A strain lacking csk1+ was also retarded in growth on minimal medium compared to a wild-type strain (3) (Fig. 4); at 30°C, the optimal temperature for growth on rich medium, cdk9T212A exhibited a similar defect on minimal medium. In contrast, mcs6S165A had no discernible effect on growth in minimal medium (Fig. 4). There may be subtle or cryptic (see below) effects on cell growth due to the reduction in TFIIH-associated kinase activity produced by the mcs6S165A mutation (25, 63). Under the conditions tested here, however, the failure to activate Cdk9 appears to be the predominant biochemical disruption in csk1Δ strains.
FIG. 4.
The cdk9T212A mutant phenocopies csk1 deletion. The cold sensitivity and the poor growth on minimal medium of the Cdk9 T-loop mutant are shown. Tenfold serial dilutions of the cdk9+ (wt), cdk9T212A, mcs6S165A, cdk9T212A mcs6S165A, csk1Δ, and cdk9T212A csk1Δ strains were grown as indicated on YES at 18°C, 25°C, 30°C, or 37°C for 14, 8, 4, or 6 days, respectively, or on minimal medium (EMM) at 30°C for 8 days.
A synthetic interaction between cdk9 and mcs6 T-loop mutants.
At the permissive temperature of 30°C, the growth of the cdk9T212A csk1Δ double mutant was similar to that of the cdk9T212A single-mutant parent (Fig. 4). At 37°C, however, we observed a synthetic interaction, indicating that the full activity of other Csk1 targets, such as Mcs6 and Cdk1, becomes more important at high temperatures. Combining cdk9T212A and mcs6S165A exacerbated the growth defect under all conditions; in dissected tetrads, double-mutant progeny gave rise to pinpoint colonies (data not shown) and exhibited severe growth defects at all temperatures (Fig. 4). The doubling time of this strain was ∼5.4 h at 30°C in rich medium, approximately twice that of a wild-type strain. The synthetic phenotype produced by combining mutations in mcs6 and cdk9 implies partial overlap in function between the two essential CDKs.
Cdk9/Pch1 and Mcs6/Mcs2 have distinct but overlapping substrate preferences.
In metazoans, TFIIH-associated Cdk7 is specific for Ser-5 within the heptad repeat of the Pol II CTD (59, 61, 71). Cdk9 has been reported to prefer Ser-2, but under certain conditions, such as binding to human immunodeficiency virus Tat protein, it can switch its specificity to phosphorylate Ser-5 (80). The situation in vivo is likely to be even more complex; for example, both kinases are influenced by the length of, position within, and prior phosphorylation of the CTD array (26, 55, 58). To investigate the site specificity of fission yeast Cdk9 and Mcs6, we phosphorylated the CTD in vitro with either enzyme and measured the immunoreactivity of the reaction products with the phosphoisoform-specific monoclonal antibodies H5 and H14. Incubation of the GST-Rpb1 fusion with Csk1-activated Cdk9/Pch1 complexes, but not with active Mcs6/Mcs2 complexes, gave rise to a strong signal in an immunoblot probed with the H5 antibody (Fig. 5A). Although reactivity with H5 cannot be taken as evidence of phosphorylation exclusively at Ser-2 (26), this result nevertheless suggests preferences for different sites within the GST-Rpb1 fusion protein by Cdk9 and Mcs6 in vitro. Both enzymes, however, phosphorylated Ser-5 (Fig. 5B), as can be concluded from an increase in immunoreactivity with antibody H14 (26). Thus, Mcs6 and Cdk9 have distinct but partially overlapping substrate specificities within the Pol II CTD.
FIG. 5.
The Cdk9 and Mcs6 complexes have distinct but overlapping substrate specificities. As indicated above each lane, aliquots of reaction mixtures containing (i) Cdk9/Pch1 complex (29 ng) either mock treated or activated with Csk1 during a preincubation or (ii) active Mcs6/Mcs2 complex (11 ng) were incubated with GST-Rpb1 fusion protein (3 μg) under standard radioactive kinase assay conditions (top) or with 1 mM cold ATP (middle and bottom). Reaction products were electrophoresed in 10% SDS-polyacrylamide gels and detected either by autoradiography (32P-CTD) (A and B, top) or by immunoblotting with phospho-specific antibodies H5 (A, middle) or H14 (B, middle) or with antibody 8WG16 (A and B, bottom), which recognizes unphosphorylated CTD.
Cdk9 associates with the mRNA cap methyltransferase and RNA in vivo.
To test whether the carboxyl-terminal segment of Cdk9, which mediates interaction with Pct1 in a yeast two-hybrid assay (52), is required for an essential function of Cdk9 in vivo, we transformed a cdk9+/cdk9::kanMX heterozygous diploid with plasmids encoding either wild-type Cdk9 or Cdk9ΔC and induced sporulation. By random spore analysis, we recovered G418-resistant transformants at the expected frequency (∼50% of all transformants) with the plasmid encoding wild-type Cdk9 but did not recover any with the plasmid encoding Cdk9ΔC (Table 2). Thus, the carboxyl terminus of Cdk9, which is dispensable for kinase activity (Table 1) and for regulation by CAK (Fig. 2C), is required for cell viability, perhaps as a protein-protein interaction domain.
TABLE 2.
Plasmid-based complementationa of cdk9Δ
| Plasmid | No. of progeny colonies
|
|
|---|---|---|
| G418 resistant | G418 sensitive | |
| pREP3x | 0 | 40 |
| pREP3x-cdk9+ | 35 | 45 |
| pREP3x-cdk9ΔC | 0 | 40 |
The heterozygous, leu− cdk9+/cdk9::kanMX diploid was transformed with the indicated plasmids carrying a budding yeast LEU1 marker conferring leucine prototrophy and containing no insert (pREP3x), a cDNA encoding full-length Cdk9 (pREP3x-cdk9+), or a cDNA encoding the first 385 amino acids of Cdk9 (pREP3x-cdk9ΔC). Transformed diploids were selected for growth on medium lacking leucine and then induced to sporulate in ME medium. Random spore analysis was performed on the progeny according to standard methods (44); totals of 40 colonies (for pREP3x and pREP3x-cdk9ΔC) or 80 colonies (for pREP3x-cdk9+) were screened for resistance to G418 in the medium, with the results indicated in the second column.
To identify the cyclin partner of Cdk9 and to investigate possible interactions with the mRNA-capping machinery in vivo, we analyzed the polypeptide composition of Cdk9-containing complexes in extracts of fission yeast cells expressing Myc-tagged Cdk9 under the control of its own promoter. All detectable Cdk9 protein (Fig. 6A) and associated Spt5 kinase activity (Fig. 6B) appeared to be in a large complex, which migrated just after the excluded volume during Superdex 200 gel filtration chromatography. We detected polypeptides present in anti-Myc immunoprecipitates in the cdk9-13Myc strain, but not in the untagged wild-type strain, by silver staining of SDS-polyacrylamide gels (Fig. 6C). In addition to the ∼110-kDa Cdk9-Myc fusion protein (confirmed by mass spectrometry), we consistently observed a doublet at ∼43 kDa. The entire region containing this cluster was excised and digested with trypsin, and the released peptides were subjected to mass spectrometry, which revealed two major components: the mRNA cap methyltransferase Pcm1 (62) (16 peptides identified, which covered 32.9% of the amino acid sequence of Pcm1) and the cyclin Pch1 (19) (7 peptides, 32.8% sequence coverage). We repeated the immunoprecipitation on a larger scale and ran longer gels to increase separation; under these conditions, Pch1 and Pcm1 were resolved into two discrete bands (identities confirmed by mass spectrometry), which appeared to be in near 1:1 stoichiometry (Fig. 6D). These results verified that Pch1 is a bona fide cyclin partner of Cdk9 and showed for the first time in any organism a physiologic association between Cdk9 and a component of the mRNA-capping machinery.
FIG. 6.
Cdk9 associates with the cyclin Pch1, the mRNA-capping apparatus, and RNA in vivo. (A) Whole-cell extracts of the cdk9-13Myc strain were incubated for 15 min at 30°C without (top) or with (bottom) 100 μg/ml RNase A and fractionated by Superdex 200 gel exclusion chromatography. Cdk9-Myc was detected by immunoblotting with anti-Myc MAb 9E10. Size markers used to calibrate the column are indicated at top in kDa; V0, excluded volume. Lane L contains column input; lane U contains whole-cell extract from the untagged, wild-type strain. (B) The kinase activity associated with Cdk9-Myc shifts with the protein upon RNase digestion. Selected Cdk9-containing fractions (indicated above each lane) from the chromatography shown in panel A were immunoprecipitated with 9E10 and tested for kinase activity towards GST-Spt5(801-990) (32P-Spt5) by autoradiography (top) and recovery of Cdk9-Myc protein by immunoblotting (bottom). (C) Polypeptides immunoprecipitated with MAb 9E10 covalently coupled to protein G-agarose from extracts (5 mg total protein) of an untagged control strain (sp78) or the cdk9-13Myc strain without (−) or with (+) RNase A treatment were separated in SDS-polyacrylamide (10%) gels and visualized by silver staining. Identities of fission yeast proteins identified by mass spectrometry in excised gel slices corresponding to ∼110 kDa (Cdk9-Myc) and ∼43 kDa (Pcm1 and Pch1) are indicated at the right. LC, immunoglobulin light chain. (D) An anti-Myc immunoprecipitate or a mock precipitate (from which the antibody was omitted) from 25 mg total protein extracted from the cdk9-13Myc strain was analyzed in a longer 10% polyacrylamide denaturing gel; polypeptides were visualized by silver staining and identified by mass spectrometry. The cyclin Pch1 was identified as a major component of the slower-migrating band, and the methyltransferase Pcm1 was the major component of the faster-migrating band (which also contained Pch1 as a minor component).
One molecule each of Cdk9-Myc, Pch1, and Pcm1 would yield an aggregate mass of ∼200 kDa, making the chromatographic behavior of the endogenous Cdk9 complex difficult to explain. Metazoan P-TEFb associates with 7SK RNA in an inactive complex (47, 76). We therefore treated extracts from the cdk9-13Myc strain with RNase A prior to gel filtration. Both Cdk9-Myc protein (Fig. 6A) and the associated kinase activity (Fig. 6B) shifted to a smaller apparent size (∼500 kDa) with this treatment, indicating that the larger Cdk9-containing complex is a ribonucleoprotein. There was no apparent change in the kinase activity recovered by immunoprecipitation of the fractions after RNase digestion. Although that might indicate that the RNA component of the S. pombe Cdk9-containing ribonucleoprotein is not an inhibitor analogous to the metazoan 7SK RNA-HEXIM1 complex (7, 42, 47, 76, 79), we cannot yet rule out the possibility that the RNA and/or associated inhibitory proteins dissociate from the Cdk9 complex during immunoprecipitation and subsequent washing. Likewise, the polypeptide composition of the Cdk9-containing complexes after immunoprecipitation was not altered by prior RNase digestion (Fig. 6C, compare lanes 2 and 3), suggesting that the interactions of Cdk9 with Pch1 and Pcm1 do not depend on RNA.
DISCUSSION
Function of S. pombe Cdk9: a capping connection.
In higher eukaryotes, promoter-proximal pausing relieved by Cdk9-dependent phosphorylation is implicated in the regulation of heat shock gene transcription in Drosophila melanogaster (39) and in the mechanism by which human immunodeficiency virus coopts the Pol II transcription apparatus (57). A more general role is likely for Cdk9 in enforcing dependency of elongation by Pol II on the recruitment of mRNA-processing enzymes (8, 10, 49, 66). An association of fission yeast Cdk9 with the capping machinery was suggested by its interaction with Pct1 when both proteins were expressed in budding yeast or incubated together in vitro (52). Here we have shown that the truncated Cdk9ΔC, which cannot interact with Pct1 (52), is active as a kinase (Table 1) and can be phosphorylated and activated by Csk1 (Fig. 2C and data not shown) but fails to rescue a cdk9Δ strain when expressed from a plasmid (Table 2). Moreover, Cdk9 forms stable complexes in vivo with the cap methyltransferase Pcm1 (Fig. 6C and D). Taken together, the data suggest that a physical connection exists between the capping apparatus and the Cdk9 complex and support a role for Cdk9 in a quality control—an elongation checkpoint—on mRNA synthesis (52).
Whereas the other two capping enzymes, Pct1 and the guanylyltransferase Pce1, interact independently with the Rpb1 CTD (51, 69) and with Spt5 (54), Pcm1 did not interact in a two-hybrid assay with Rpb1, Spt5, or any other component of the capping machinery (Y. Pei and S. Shuman, unpublished observations). In S. cerevisiae, the cap methyltransferase Abd1 can bind directly to the phosphorylated CTD of Pol II in vitro (11, 41). Direct association between Pcm1 and the Cdk9/Pch1 complex in fission yeast could provide an alternative mechanism to target cap methylation to the nascent transcript. In budding yeast, the guanylyltransferase Ceg1 dissociates early in elongation, but Abd1 tracks with Pol II throughout the coding regions of transcribed genes (30, 64) and directly influences the function and CTD phosphorylation patterns of elongating Pol II independently of its enzymatic activity (65). Stable interaction of Pcm1 and the S. pombe P-TEFb ortholog Cdk9 could provide a means to retain the methyltransferase in the Pol II elongation complex, where it might perform analogous functions.
Fission yeast Cdk9 interacted with Pct1 when the two proteins were coexpressed as fusion proteins in S. cerevisiae (52), but we did not find definitive evidence for the presence of Pct1 or Pce1 in Cdk9 complexes immunoprecipitated from the cdk9-13Myc strain of S. pombe. Therefore, if Cdk9, Pch1, and Pcm1 are in a complex with 1:1:1 stoichiometry, we can account at present for only ∼40% of its apparent size after RNase digestion (Fig. 6A). Two recent reports suggested that mammalian P-TEFb-HEXIM complexes were capable of multimerization (15, 35). Further experiments will be needed to characterize and quantify the apparently stoichiometric Cdk9-Pcm1 interaction we have detected (e.g., under different growth conditions or in different genetic backgrounds), to answer the question of whether Cdk9 complexes multimerize in fission yeast, and to detect possibly substoichiometric amounts of other capping enzymes.
Regulation of S. pombe Cdk9 by the upstream CAK Csk1.
Cdk9 in fission yeast partners with the cyclin Pch1 (Fig. 6C) and requires phosphorylation by Csk1 for full enzymatic and biological activity; this requirement could be readily demonstrated because Csk1 is not essential for activation of the cell cycle CDK. In contrast, a demonstration of the Cak1 dependence of Bur1 and Ctk1 in vivo depended on the presence of a CAK bypass allele of CDC 28, which encodes the cell cycle CDK that is the sole essential target of Cak1 in budding yeast (13, 50, 78). A direct comparison of the csk1Δ and cdk9T212A phenotypes suggests that Cdk9 is a critical target of Csk1 in vivo.
The cdk9T212A mutant phenocopies the cold sensitivity and poor growth on minimal media of csk1Δ but grew more slowly in rich media and was also temperature sensitive (Fig. 4 and data not shown). There are several possible explanations for the increased severity of cdk9T212A relative to csk1Δ (and that of cdk9T212A mcs6S165A relative to cdk9T212A csk1Δ) that are not mutually exclusive: (i) effects on stability and/or activity of both Cdk9 and Mcs6 caused by alanine substitution in the T loop independent of phosphorylation; (ii) compensation for the decreased activity of Cdk9 in csk1Δ cells by overexpression of the protein (Fig. 3A), which may not occur in the cdk9T212A or cdk9T212A mcs6S165A mutants; and (iii) low levels of phosphorylation by another kinase in the absence of Csk1. The susceptibility of Cdk9T212A to proteolysis in insect cells (Fig. 1A see Fig. S1 in the supplemental material) is consistent with the first explanation and raises the possibility that the cdk9T212A phenotype is exacerbated by loss of the carboxyl-terminal segment, potentially uncoupling kinase activity from interaction with capping enzymes.
Apparent redundancy in a CDK network: interaction between cdk9 and mcs6.
The synthetic interaction between mcs6 and cdk9 suggests a degree of functional overlap between two components of the CAK-CDK network. Although the simplest model to explain that overlap is one involving redundancy of action on their common substrate, the CTD of Rpb1, the overlap could also stem from the two kinases working on different substrates (e.g., Cdk9 and Mcs6 phosphorylating Spt5 and the Pol II CTD, respectively), which might themselves have overlapping functions in transcription. Measurements of steady-state CTD phosphorylation in the different strains did not help to resolve this question: when growing exponentially, the single mcs6 and cdk9 T-loop mutants and the double mutant all had detectable Ser-5 phosphorylation of bulk Pol II (data not shown). Severe slow-growth phenotypes were previously reported in kin28 bur1 and kin28 ctk1 double-mutant strains of budding yeast (38). In Drosophila, both pharmacologic inhibition of P-TEFb and inactivation of a temperature-sensitive Cdk7 were necessary to abolish the Ser-5 phosphorylation of Pol II engaged in transcribing heat shock genes (48). Thus, studies of P-TEFb and TFIIH orthologs in widely divergent eukaryotes suggest that when one kinase is compromised, the other can partially compensate.
Deletion of csk1+ reduces Mcs6 activity approximately threefold in vivo (24, 43). Phosphorylation of the analogous site in human or Drosophila Cdk7 confers thermal stability and an ∼20-fold stimulation of enzymatic activity (31). The mcs6S165A mutation is virtually silent, however, unless combined with cdk9T212A (Fig. 4). Although the two CDKs perform unique, essential functions and probably phosphorylate Pol II in temporally and spatially distinct patterns, the synthetic interaction between mcs6 and cdk9 implies an inherent flexibility, perhaps to allow combinatorial control of gene expression, which appears as redundancy when probed genetically. An analogy can be made to apparently redundant mechanisms of cell cycle control: just as cells can withstand loss of one or more cyclins and still maintain orderly cell cycle progression (45), impairment of one CDK in the transcriptional machinery might be tolerated as long as another is fully functional.
CAK-CDK network wiring: evolutionary implications.
There has been fundamental conservation in the molecular mechanisms of eukaryotic cell cycle control and transcriptional regulation. In both spheres, the roles of effector CDKs have remained largely unchanged, although expansion of the CDK family in metazoans has been accompanied by specialization to perform subsets of functions carried out by a single CDK in fungi (45). In contrast, the organization of the CAK-CDK network has diverged (Fig. 7).
FIG. 7.
An expanded map of the CAK-CDK network in yeasts and metazoans. The upstream, general CAK (white) is Cak1 in budding yeast, Csk1 in fission yeast, and apparently absent (as indicated by the question mark) in metazoans. The TFIIH-associated CDK (light gray) is Kin28 in budding yeast, Mcs6 in fission yeast, and Cdk7 in metazoans. In the last two cases, it is also a CAK capable of activating the cell cycle CDKs (dark gray), which in S. pombe renders the upstream CAK nonessential. Cdk9 orthologs (black) of budding yeast (Bur1 and Ctk1) and fission yeast (Cdk9) are activated by Cak1 and Csk1, respectively, but the Cdk9-activating kinase remains unidentified in metazoans. See the text for further details.
In budding yeast, the substrates of Cak1 include all CDKs known to depend on T-loop phosphorylation for full activity. The metazoan cell division machinery likewise depends on a single CAK; Cdk7 is required for entry into mitosis in flies and worms (32, 74), and its depletion eliminates most or all CAK activity in cell extracts (18, 32, 40). No transcriptional CDK, however, is known to be activated by Cdk7. Cdk7 itself is incapable of autophosphorylation within the T loop (20). The CDK associated with the Pol II mediator (Cdk8 in metazoans, Srb10 in budding yeast) bypasses the need for CAK altogether with an aspartic acid substitution for the phosphoacceptor residue (36, 70). Cdk9 is a priori a candidate for CAK-dependent activation but might not be phosphorylated by Cdk7 (6, 7, 28), apparently leaving vacant a niche for an upstream kinase.
Fission yeast contains orthologs of both the Cdk7 complex and Cak1. Of the three S. pombe CDKs known to be activated by T-loop phosphorylation, only Cdk1 is a substrate of the Mcs6 complex, but all three—Cdk1, Mcs6, and Cdk9—are substrates of Csk1 (references 24 and 34 and this report). Csk1 is thus a general CAK, analogous to Cak1. Csk1 is dispensable for viability because the only essential function of CAK, activation of Cdk1 (13), is also provided by the Mcs6 complex (34, 63), whereas the other CDKs can perform their essential functions without T-loop phosphorylation (references 25, 29, 63, and 78 and this report). Fission yeast strains lacking csk1+, however, have profound growth defects that are mimicked by mutation of the Cdk9 T loop, indicating that the full CAK-dependent activity of Cdk9 is required in the face of nutrient limitation or suboptimal environmental conditions.
Supplementary Material
Acknowledgments
We are grateful to Matthew Gamble, Hilary Gerber, and Stéphane Larochelle for critical review of the manuscript; to Karen Lee and Julia Saiz for providing strains, reagents, and advice; and to Hediye Erdjument-Bromage and Paul Tempst for mass spectrometric identification of proteins. S.S. is an American Cancer Society Research Professor.
This work was supported by NIH grant GM52470 to S.S. and by American Cancer Society grant RSG-99-043-044-CCG and NIH grant GM56985 to R.P.F.
Footnotes
Supplemental material for this article may be found at http://mcb.asm/org/.
REFERENCES
- 1.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 2.Bamps, S., T. Westerling, A. Pihlak, L. Tafforeau, J. Vandenhaute, T. P. Mäkelä, and D. Hermand. 2004. Mcs2 and a novel CAK subunit Pmh1 associate with Skp1 in fission yeast. Biochem. Biophys. Res. Commun. 325:1424-1432. [DOI] [PubMed] [Google Scholar]
- 3.Bimbó, A., Y. Jia, S. L. Poh, R. K. Karuturi, N. den Elzen, X. Peng, L. Zheng, M. O'Connell, E. T. Liu, M. K. Balasubramanian, and J. Liu. 2005. Systematic deletion analysis of fission yeast protein kinases. Eukaryot. Cell 4:799-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregman, D. B., R. G. Pestell, and V. J. Kidd. 2000. Cell cycle regulation and RNA polymerase II. Front. Biosci. 5:D244-D257. [DOI] [PubMed] [Google Scholar]
- 5.Buck, V., P. Russell, and J. B. A. Millar. 1995. Identification of a cdk-activating kinase in fission yeast. EMBO J. 14:6173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D., and Q. Zhou. 1999. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol. Cell. Biol. 19:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, Y. L., H. Cao, J. M. Jacque, M. Stevenson, and T. M. Rana. 2004. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/cyclinT1). J. Virol. 78:2517-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, Y. L., E. Coronel, C. K. Ho, S. Shuman, and T. M. Rana. 2001. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 276:12959-12966. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, Y. L., C. K. Ho, N. Saha, B. Schwer, S. Shuman, and T. M. Rana. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10:585-597. [DOI] [PubMed] [Google Scholar]
- 11.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damagnez, V., T. P. Mäkelä, and G. Cottarel. 1995. Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J. 14:6164-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulac, C., A. A. Michels, A. Fraldi, F. Bonnet, V. T. Nguyen, G. Napolitano, L. Lania, and O. Bensaude. 2005. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J. Biol. Chem. 280:30619-30629. [DOI] [PubMed] [Google Scholar]
- 16.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. McNulty, S. A. Carr, and P. Tempst. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826:167-181. [DOI] [PubMed] [Google Scholar]
- 17.Espinoza, F. H. E., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi, and D. O. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fesquet, D., N. Morin, M. Dorée, and A. Devault. 1997. Is cdk7/cyclin H/MAT1 the genuine cdk activating kinase in cycling xenopus egg extracts? Oncogene 15:1303-1307. [DOI] [PubMed] [Google Scholar]
- 19.Furnari, B., P. Russell, and J. Leatherwood. 1997. pch1+, a second essential C-type cyclin gene in Schizosaccharomyces pombe. J. Biol. Chem. 272:12100-12106. [DOI] [PubMed] [Google Scholar]
- 20.Garrett, S., W. A. Barton, R. Knights, P. Jin, D. O. Morgan, and R. P. Fisher. 2001. Reciprocal activation by cyclin-dependent kinases 2 and 7 is directed by substrate specificity determinants outside the T loop. Mol. Cell. Biol. 21:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garriga, J., and X. Grana. 2004. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337:15-23. [DOI] [PubMed] [Google Scholar]
- 22.Gould, K. L., S. Moreno, D. J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper, J. W., and S. J. Elledge. 1998. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 12:285-289. [DOI] [PubMed] [Google Scholar]
- 24.Hermand, D., A. Pihlak, T. Westerling, V. Damagnez, J. Vandenhaute, G. Cottarel, and T. P. Mäkelä. 1998. Fission yeast Csk1 is a CAK-activating kinase (CAKAK). EMBO J. 17:7230-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermand, D., T. Westerling, A. Pihlak, J.-Y. Thuret, T. Vallenius, M. Tiainen, J. Vandenhaute, G. Cottarel, C. Mann, and T. P. Mäkelä. 2001. Specificity of Cdk activation in vivo by the two Caks Mcs6 and Csk1 in fission yeast. EMBO J. 20:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279:24957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaldis, P. 1999. The cdk-activating kinase (CAK): from yeast to mammals. Cell. Mol. Life Sci. 55:284-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 29.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the kin28p subunit of yeast TFIIH by cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larochelle, S., J. Chen, R. Knights, J. Pandur, P. Morcillo, H. Erdjument-Bromage, P. Tempst, B. Suter, and R. P. Fisher. 2001. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 20:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larochelle, S., J. Pandur, R. P. Fisher, H. K. Salz, and B. Suter. 1998. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 12:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, K. M., I. Miklos, H. Du, S. Watt, Z. Szilagyi, J. E. Saiz, R. Madabhushi, C. J. Penkett, M. Sipiczki, J. Bähler, and R. P. Fisher. 2005. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol. Biol. Cell 16:2734-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, K. M., J. E. Saiz, W. A. Barton, and R. P. Fisher. 1999. Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs). Curr. Biol. 9:441-444. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., J. P. Price, S. A. Byers, D. Cheng, J. Peng, and D. H. Price. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280:28819-28826. [DOI] [PubMed] [Google Scholar]
- 36.Liao, S.-M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 37.Lim, H. H., C. J. Loy, S. Zaman, and U. Surana. 1996. Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for start in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4573-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuoka, M., J. Kato, R. P. Fisher, D. O. Morgan, and C. J. Sherr. 1994. Activation of cyclin-dependent kinase-4 (CDK4) by mouse MO15-associated kinase. Mol. Cell. Biol. 14:7265-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molz, L., and D. Beach. 1993. Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J. 12:1723-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 45.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 46.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 48.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13:55-65. [DOI] [PubMed] [Google Scholar]
- 49.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 50.Ostapenko, D., and M. J. Solomon. 2005. Phosphorylation by Cak1 regulates the C-terminal domain kinase Ctk1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:3906-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pei, Y., S. Hausmann, C. K. Ho, B. Schwer, and S. Shuman. 2001. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J. Biol. Chem. 276:28075-28082. [DOI] [PubMed] [Google Scholar]
- 52.Pei, Y., B. Schwer, and S. Shuman. 2003. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 278:7180-7188. [DOI] [PubMed] [Google Scholar]
- 53.Pei, Y., and S. Shuman. 2003. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J. Biol. Chem. 278:43346-43356. [DOI] [PubMed] [Google Scholar]
- 54.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639-19648. [DOI] [PubMed] [Google Scholar]
- 55.Pinhero, R., P. Liaw, K. Bertens, and K. Yankulov. 2004. Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur. J. Biochem. 271:1004-1014. [DOI] [PubMed] [Google Scholar]
- 56.Prelich, G., and F. Winston. 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 59.Rickert, P., J. L. Corden, and E. Lees. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093-1102. [DOI] [PubMed] [Google Scholar]
- 60.Rosenblatt, J., H. De Bondt, J. Jancarik, D. O. Morgan, and S.-H. Kim. 1993. Purification and crystallization of human cyclin-dependent kinase 2. J. Mol. Biol. 230:1317-1319. [DOI] [PubMed] [Google Scholar]
- 61.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermuelen, J.-P. Tassan, L. Schaeffer, E. A. Nigg, J. H. J. Hoeijmakers, and J.-M. Egly. 1994. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 79:1093-1101. [DOI] [PubMed] [Google Scholar]
- 62.Saha, N., B. Schwer, and S. Shuman. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274:16553-16562. [DOI] [PubMed] [Google Scholar]
- 63.Saiz, J. E., and R. P. Fisher. 2002. A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr. Biol. 12:1100-1105. [DOI] [PubMed] [Google Scholar]
- 64.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder, S. C., D. A. Zorio, B. Schwer, S. Shuman, and D. Bentley. 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13:377-387. [DOI] [PubMed] [Google Scholar]
- 66.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spåhr, H., O. Khorosjutina, V. Baraznenok, T. Linder, C. O. Samuelsen, D. Hermand, T. P. Mäkelä, S. Holmberg, and C. M. Gustafsson. 2003. Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J. Biol. Chem. 278:51301-51306. [DOI] [PubMed] [Google Scholar]
- 68.Sterner, D. E., J. M. Lee, S. E. Hardin, and A. L. Greenleaf. 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takagi, T., E. J. Cho, R. T. Janoo, V. Polodny, Y. Takase, M. C. Keogh, S. A. Woo, L. D. Fresco-Cohen, C. S. Hoffman, and S. Buratowski. 2002. Divergent subunit interactions among fungal mRNA 5′-capping machineries. Eukaryot. Cell 1:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tassan, J.-P., M. Jaquenoud, P. Léopold, S. J. Schultz, and E. A. Nigg. 1995. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc. Natl. Acad. Sci. USA 92:8871-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 273:6769-6775. [DOI] [PubMed] [Google Scholar]
- 72.Tsakraklides, V., and M. J. Solomon. 2002. Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J. Biol. Chem. 277:33482-33489. [DOI] [PubMed] [Google Scholar]
- 73.Valay, J.-G., M. Simon, M.-F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249:535-544. [DOI] [PubMed] [Google Scholar]
- 74.Wallenfang, M. R., and G. Seydoux. 2002. cdk-7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99:5527-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winkler, G. S., A. Kristjuhan, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99:3517-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 77.Yao, S., A. Neiman, and G. Prelich. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao, S., and G. Prelich. 2002. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol. Cell. Biol. 22:6750-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 80.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Biol. Cell 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.