Abstract
In Saccharomyces cerevisiae, pheromone response requires Ste5 scaffold protein, which ensures efficient G-protein-dependent recruitment of mitogen-activated protein kinase (MAPK) cascade components Ste11 (MAPK kinase kinase), Ste7 (MAPK kinase), and Fus3 (MAPK) to the plasma membrane for activation by Ste20 protein kinase. Ste20, which phosphorylates Ste11 to initiate signaling, is activated by binding to Cdc42 GTPase (membrane anchored via its C-terminal geranylgeranylation). Less clear is how activated and membrane-localized Ste20 contacts Ste11 to trigger invasive growth signaling, which also requires Ste7 and the MAPK Kss1, but not Ste5. Ste50 protein associates constitutively via an N-terminal sterile-alpha motif domain with Ste11, and this interaction is required for optimal invasive growth and hyperosmotic stress (high-osmolarity glycerol [HOG]) signaling but has a lesser role in pheromone response. We show that a conserved C-terminal, so-called “Ras association” (RA) domain in Ste50 is also essential for invasive growth and HOG signaling in vivo. In vitro the Ste50 RA domain is not able to associate with Ras2, but it does associate with Cdc42 and binds to a different face than does Ste20. RA domain function can be replaced by the nine C-terminal, plasma membrane-targeting residues (KKSKKCAIL) of Cdc42, and membrane-targeted Ste50 also suppresses the signaling deficiency of cdc42 alleles specifically defective in invasive growth. Thus, Ste50 serves as an adaptor to tether Ste11 to the plasma membrane and can do so via association with Cdc42, thereby permitting the encounter of Ste11 with activated Ste20.
In eukaryotes, cell surface receptors elicit responses that affect diverse cellular processes via mitogen-activated protein kinase (MAPK) cascades (37, 69). A MAPK cascade comprises three tiers of conserved protein kinases that act sequentially: a MAPK kinase kinase (MAPKKK) phosphorylates and activates a MAPK kinase (MAPKK), which, in turn, phosphorylates and activates a MAPK. However, this chain can be extended, because among MAPK targets are MAPK-activated protein kinases (76) and among MAPKKK activators are upstream protein kinases, such as a p21-activated protein kinase (9, 16).
Budding yeast (Saccharomyces cerevisiae) uses MAPK modules to adapt rapidly to environmental changes, such as nutrient limitation (filamentous growth signaling) (84), hyperosmotic stress (high-osmolarity glycerol [HOG] signaling) (91), cell wall damage (Pkc1-dependent signaling) (20), presence of mating-inducing peptides (pheromone signaling) (90), and conditions that evoke meiosis in diploid cells (sporulation) (6, 89). Nutrient deprivation triggers a developmental transition from yeast-form (round) cells to cells capable of invasive or pseudohyphal growth (23, 63). Diploids starved for nitrogen elongate, alter their budding pattern, and increase their adhesiveness, forming prominent chains of pseudohyphae that, on a plate, project from a colony and penetrate the agar surface. Similarly, haploids starved for carbon (as when grown for long periods of time on rich medium) elongate, change budding pattern, become more cohesive, and invade agar. These dimorphic switches are controlled by inputs from 5′-AMP-activated protein kinase (Snf1) (42) and a specific 3′,5′-cyclic-AMP-dependent protein kinase isoform (Tpk2) (19), which act in concert with a MAPK cascade (47).
The MAPK module required for filamentous growth, i.e., Ste11 (MAPKKK), Ste7 (MAPKK), and Kss1 (MAPK), responds to signals from two small GTP-binding proteins, Ras2 and Cdc42, and Cdc42 appears to act downstream of Ras2 (57). One function of Cdc42 is to activate the p21-activated protein kinase, Ste20 (45). Ste20 then activates Ste11 (18), which, in turn, activates Ste7 and Kss1 (14, 50). Kss1, along with two corepressors, Dig1/Rst1 and Dig2/Rst2 (3, 13, 83), directly represses the Ste12 transcription factor (2). Ste12 is targeted to the promoters of the genes necessary for filamentous growth in concert with a second DNA-binding transcription factor, Tec1 (52). Activation of Kss1 alleviates its own and Dig-mediated repression and, in conjunction with Snf1 and Tpk2 action on other transcriptional activators and repressors, induces expression of genes, such as FLO11/MUC1, required for robust invasive growth (48, 78). The level of Ste12 is also modulated by yet another protein kinase, Srb10/Ssn3 (ortholog of human Cdk8), thereby influencing filamentous growth (59).
Ste20, Ste11, and Ste7 also have essential functions (and Kss1 plays a supporting role) in pheromone response (90). Moreover, Ste20 and Ste11 have critical functions in one branch of the HOG pathway (91). This branch requires an apparent osmosensor (Sho1), a polytopic membrane protein that associates with a transmembrane mucin-like protein (Msb2) (15). Sho1 binds a unique MAPKK, Pbs2, thereby tethering Pbs2 at the plasma membrane, where it can become activated by Ste11, which also reportedly associates with Sho1 (94). The second branch converges on Pbs2 via a different osmosensor mechanism that resembles prokaryotic two-component signaling systems and activates two other MAPKKKs (Ssk2 and Ssk22). Once Pbs2 is activated by either route, it phosphorylates and activates a unique MAPK, Hog1. Defects that prevent any output (e.g., pbs2Δ or hog1Δ) confer severe osmosensitivity, whereas the role of each branch is revealed only in a background in which the other branch has been inactivated (61).
Given that Ste20 and Ste11 act in three different MAPK pathways, an important question is how activation of these protein kinases is coupled to the initial events that trigger each pathway. Ste20 is activated by GTP-bound Cdc42, which is firmly anchored in the plasma membrane via important features of its C terminus: a CAAX box that directs attachment of a geranylgeranyl (C20) chain in thioether linkage to the sulfur atom of the Cys (followed by proteolytic removal of the AAX residues), masking of the negative charge of the carboxyl group of the resulting prenylated Cys by methylesterification, and four conserved basic residues (situated immediately proximal to the CAAX box) that interact with the head groups of acidic phospholipids (36, 74). In the pheromone response pathway, a dedicated scaffold protein, Ste5 (21), ensures that Ste11 is delivered to activated Ste20 at the plasma membrane because Ste5 binds Ste11, Ste7, and the appropriate MAPK (Fus3), as well as associates with the farnesylated plasma membrane-bound Gβγ released upon ligand occupancy of pheromone receptors (90). In marked contrast, how Ste11 encounters Ste20 in the filamentous growth signaling pathway has been unclear. Genetic analysis hinted that the STE50 gene product (72) might play a role.
A complete null mutation (ste50Δ) severely compromises both filamentous growth signaling (35, 71) and the Ste20-dependent branch of the HOG pathway (60, 67) but has only a modest effect on pheromone response (92, 93). Ste50 is small (346 residues). It contains an N-terminal sterile-alpha motif (SAM) domain (residues 32 to 101) (64) followed by a spacer and a putative C-terminal Ras association (RA) domain (residues 235 to 327) (65). Structures for the SAM domain have been determined at atomic resolution (7, 27, 44). The SAM domain of Ste50 binds heterotypically to a SAM domain in the N-terminal regulatory region of Ste11 (27, 64), and these two proteins are associated under all conditions tested (67, 71) via this SAM-SAM interaction (35, 92). The RA domain is dispensable for Ste50-Ste11 association (67, 92), yet removal of the RA domain has been shown to compromise the Ste20-dependent branch of the HOG pathway (67). The role, if any, of the RA domain in filamentous growth signaling has not been explored, nor has any biochemical function for the RA domain yet been ascribed. The studies described here address both of these issues.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
The yeast strains used were constructed in the Σ1278b background (Table 1). Gene disruptions were confirmed by PCR using gene-specific primers (38), and absence of the corresponding DNA or gene product was further verified by Southern analysis or immunoblotting, respectively. Plasmids were introduced using the lithium acetate transformation method (11). Yeasts were cultivated in standard rich (YP) and defined minimal (SC) media (11) containing either 2% glucose (dextrose), 2% raffinose/0.4% sucrose, or 2% galactose as the carbon source and supplemented with appropriate nutrients to maintain selection for plasmids. Synthetic low ammonia/dextrose (SLAD) plates (3, 49) were made with agar washed with distilled deionized water before autoclaving and supplemented with nutrients as required. Plasmids were constructed and site-directed mutants were generated using standard molecular biology techniques (see the supplemental material for further details).
TABLE 1.
S. cerevisiae strains
| Strain | Salient property | Genotype | Source or reference |
|---|---|---|---|
| JCY100 | Wild type | MATaleu2Δ::hisG his3Δ::hisG trp1Δ::hisG ura3-52 | 14 |
| JCY102 | Wild-type diploid | MATa/MATα (JCY100 × JCY101) | 3 |
| YDV105a | ste50Δ | JCY100 ste50Δ::HIS3 | This work |
| YDV200b | ste20Δ | JCY100 ste20Δ::LEU2 | This work |
| YDV205c | ste50Δ ste20Δ | YDV200 ste50Δ::HIS3 | This work |
| YDV500d | ste11Δ | JCY100 ste11Δ::ura3 | This work |
| YDV505e | ste50Δ ste11Δ | YDV500 ste50Δ::HIS3 | This work |
| FP67 | ssk2Δ ssk22Δ ste50Δ | MATα ura3 leu2Δ trip1Δ his3 ssk2::LEU2 ssk22::LEU2 ste50Δ::HIS3 | 67 |
| YTIMK92 | CDC42(I46M) | MATα cdc42Δ::HIS3 ura3-52 his3Δ::hisG leu2Δ::hisG::LEU2::CDC42(I46M) | 56 |
| YTIMK94 | CDC42(S71P) | MATα cdc42Δ::HIS3 ura3-52 his3Δ::hisG leu2Δ::hisG::LEU2::CDC42(S71P) | 56 |
| KKY691 | CDC42(E100A) | MATα cdc42-119:LEU2 his3Δ::hisG leu2Δ0 ura3Δ0 trp1Δ0::hisG | 40 |
Derived from JCY100 by transformation with a ste50Δ::HIS3 PCR product generated using primers DV3 and DV4.
Derived from JCY100 by transformation with a 4.6-kb XhoI fragment excised from plasmid pYES-ste20Δ::LEU2.
Derived from YDV200 by transformation with a 2.2-kb ste50Δ::HIS3 PCR product generated using primers ste50-3 and ste50-4.
Derived from JCY100 by transformation with plasmid pRS306-ste11Δ::URA3 (gift of B. Errede, University of North Carolina, Chapel Hill) linearized with SnaBI; ura3− derivatives were selected on 5-fluoro-orotic acid plates (8).
Derived from YDV500 by transformation with a 2.2-kb ste50Δ::HIS3 PCR product generated using primers ste50-3 and ste50-4.
Ste50 purification, preparation of anti-Ste50 antibodies, and immunoblotting.
A culture (2 liters) of Escherichia coli strain BL21(DE)(pLysS) (Stratagene) carrying plasmid pDT1 expressing a glutathione S-transferase (GST)-Ste50 fusion protein was grown at 30°C to an A600 of ∼0.8 and then induced by addition of 300 μM IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration). After 3 h, cells were chilled on ice, harvested by centrifugation at 4°C, and washed and resuspended in 40 ml ice-cold buffer A (140 mM NaCl, 2.7 mM KCl, 1 mM EDTA, 0.1% Triton X-100, 15% glycerol, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]). After one freeze-thaw cycle, 5 mM dithiothreitol (DTT) and 1 mM phenylmethylsulfonyl fluoride (PMSF) were added, and cell lysis was completed by sonication on ice. To ensure complete solubilization of the GST-Ste50 chimera, 2% Triton X-100 and 1.5% Na-N-laurylsarcosine (final concentrations) were added. GST-Ste50 was captured by passing the extract twice over a bed (5 ml) of glutathione-Sepharose (Amersham-Pharmacia) in a small column equilibrated with buffer A. After the column was washed with 100 ml buffer A, 50 ml buffer B (150 mM NaCl, 50 mM Tris-HCl [pH 7.5]), and 50 ml buffer B containing 2.5 mM CaCl2, the residual buffer was drained, the bottom of the column was clamped, and then 5 ml buffer B containing 2.5 mM CaCl2 and 200 U of highly purified thrombin (22) (a gift of J. W. Fenton II, New York State Department of Public Health, Wadsworth Center, Albany) was run into the column until liquid just started to appear at the outflow. After incubation of the column for 12 to 16 h at room temperature (29), the cleavage product (Ste50) was eluted with buffer B and concentrated at 4°C to 3 mg/ml by ultrafiltration in a Centricon device (Amicon, Inc.).
Purified Ste50 (1 mg) was used as the immunogen in an emulsion with Ribi's adjuvant (Sigma Chemical) and injected subcutaneously into each of two adult female New Zealand White rabbits. Each rabbit received booster injections (500 μg purified Ste50) every 4 weeks. After the third inoculation, serum from a bleed (5 ml) of each rabbit was tested for the presence of specific anti-Ste50 antibodies by immunoblotting against extracts prepared from exponentially growing cultures of STE50+ (JCY100), ste50Δ (YDV105), and STE50-overexpressing [JCY100(pDT2)] cells. Final bleeds were drawn 2 weeks after the fourth booster injection. The immunoglobulin G fraction of the resulting serum with the lowest nonspecific background, as judged by immunoblotting, was purified and concentrated by ammonium sulfate precipitation (31).
For immunoblots, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to an Immobilon-P membrane (Millipore) by using a semidry transfer cell (Bio-Rad). Membranes were incubated overnight at room temperature with the appropriate antibody, followed by incubation with a 1:5,000 dilution of the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature, and then visualized by using a commercial chemiluminescence detection system (NEN Life Science Products). The concentrated rabbit polyclonal anti-Ste50 antibodies were used at a 1:5,000 dilution, anti-v-H-Ras antibody (Oncogene Research Products) (which recognizes yeast Ras2) was used at 1:160, affinity-purified anti-Cdc42 antibodies (a gift of D. I. Johnson, University of Vermont, Burlington) was used at 1:1,000, and a monoclonal antihemagglutinin (anti-HA) antibody (Covance) was used at 1:1,000.
Bioassays.
Haploid invasive growth was assessed qualitatively using the standard agar penetration assay at 30°C (75). Overnight cultures (3 ml) were diluted with water to 2 × 107 cells per ml (A660 of ∼1), and portions (typically 50 μl) were spread in wedge-shaped sectors on an SCD plate lacking appropriate nutrients to maintain selection for plasmids. After incubation for 24 to 48 h, the cells were replica plated onto a YPD plate and incubated for at least 3 days. The plate was photographed before and after being washed under a gentle stream of distilled water. Diploid pseudohyphal growth was assessed using the standard colony morphology assay (25, 26). To evaluate filamentous growth signaling more quantitatively, expression of an FRETy1-lacZ reporter gene was measured (14). To assess osmosensitivity, overnight cultures were diluted to 2 × 108 cells per ml, portions (2.5 μl) of appropriate dilutions (1/5 and 1/25) of the cell suspension were spotted onto YPD plates lacking or containing 1.5 M sorbitol, and the cells were grown for 2 days at 30°C (67).
In vitro protein binding assays.
To prepare purified Ras2-(His)6, a culture (500 ml) of E. coli strain BL21(DE3)-RIL (Stratagene) carrying plasmid pDT21 was grown at 30°C to A600 of ∼0.8 and then induced by addition of 0.4 mM IPTG (final concentration). After 3 h, cells were chilled on ice, harvested by centrifugation at 4°C, washed and resuspended in 14 ml of buffer C (300 mM NaCl, 10% glycerol, 50 mM NaH2PO4 [pH 6.5]), and subjected to one freeze-thaw cycle. A protease inhibitor tablet (without EDTA; Boehringer-Mannheim) and lysozyme (20 mg) were added, the cell suspension was incubated for 45 min on ice, and then lysis was completed by sonication on ice. The resulting extract was mixed with a 50:50 slurry (4 ml) of washed Ni2+-saturated nitrilotriacetate (Ni-NTA) beads (QIAGEN) and incubated on a roller drum at 4°C for 2 h. The beads were then decanted into a small column (1.8-cm diameter) at 4°C, and after settling, the column was drained and then the flowthrough was reapplied to the column once more. After washing with 10 ml buffer C and 20 ml buffer C containing 20 mM imidazole, Ras2-(His)6 was eluted with 30 ml buffer C containing 250 mM imidazole and 2-ml fractions were collected. To exchange the buffer, pooled peak fractions (as judged by Coomassie blue staining of samples subjected to SDS-PAGE) were applied to a column (10 cm by 2.5 cm in diameter) of Sephadex G-25 (fine) preequilibrated with 6.25 mM EDTA-50 mM Tris-HCl (pH 7.5) and eluted with the same buffer. Fractions (1 ml) were collected, the peak fractions were pooled, and the resulting purified Ras2-(His)6 (≥80% of total protein) was used for all subsequent studies. Prior to loading Ras2-(His)6 with nucleotide, a protease inhibitor tablet (without EDTA) was added, the solution was adjusted to 2 mM 2-mercaptoethanol (final concentration), and a final concentration of 8 mM GTPγS or 8 mM GDPβS was added (from 50 mM stock solutions at pH 8.0). After incubation with gentle agitation at room temperature for 1 h, 10 mM MgCl2 (final concentration) was added and the samples were chilled on ice. Samples (200 μl; 0.5 mg/ml) were quick-frozen in liquid N2 and stored at −80°C prior to use.
To prepare purified (His)6-Cdc42, a culture (1 liter) of BL21(DE3)-RIL carrying plasmid pDT53 was grown at 27°C to an A600 of ∼0.8 and induced by addition of 50 μM IPTG (final concentration). After 12 h, cells were chilled, harvested by centrifugation at 4°C, washed and resuspended in buffer D (300 mM NaCl, 10% glycerol, 1 mM 2-mercaptoethanol, 1 mM PMSF, 1 μg/ml leupeptin, 1 mM benzamidine, 1 μM pepstatin A, 1.5 μg/ml aprotinin, 50 mM NaH2PO4 [pH 7.3]), and subjected to one cycle of freeze-thawing, and then lysis was completed by passage through a French pressure cell at 20,000 lb/in2. After addition of 2 mM imidazole (final concentration), the extract was mixed with a 50:50 slurry (5 ml) of washed Ni-NTA beads and incubated on a roller drum for 1 h at 4°C. The beads were then decanted into a small column (2.5-cm diameter) at 4°C, and after settling, the column was drained and then the flowthrough was reapplied to the column once more. After washing with 50 ml buffer D and 25 ml buffer D containing 5 mM imidazole, (His)6-Cdc42 was eluted with 25 ml buffer D containing 250 mM imidazole and 2-ml fractions were collected. To exchange the buffer, pooled peak fractions were applied to a column (11 cm by 2.5 cm in diameter) of Sephadex G-25 (fine) preequilibrated with 10% glycerol-0.5 mM DTT-50 mM Tris-HCl (pH 7.5) and eluted with the same buffer. Fractions (1.5 ml) were collected, the peak fractions were pooled, and the resulting purified (His)6-Cdc42 (≥30% of total protein) was used for all subsequent studies. To load (His)6-Cdc42 with nucleotide, a protease inhibitor tablet (without EDTA) was added, and then a final concentration of 1 mM GTPγS or 1 mM GDPβS was added, followed by 5 mM EDTA (final concentration). After incubation on a roller drum for 1 h, 10 mM MgCl2 (final concentration) was added and the samples were chilled on ice for 30 min. Samples (200 μl; 1.8 mg/ml) were snap-frozen and stored at −80°C prior to use. In the same manner as just described, (His)6-Cdc42(I46M) (expressed from plasmid pDT167) was purified (4 mg/ml; ≥80% of total protein) from E. coli (except that cells were lysed by addition of 1 mg/ml lysozyme before freeze-thawing and sonication and no imidazole was added during initial passage of the lysate over the Ni-NTA column), loaded with nucleotide, and stored.
To measure binding of Ras2-(His)6 to bead-bound proteins, cultures (20 ml) of BL21(DE3)-RIL expressing GST, GST-Ste50-RA, GST-Ste50-RA(I267A L268A), GST-Ste50-RA(K254A), or GST-RalGDS-RA from plasmid pGEX4T-1, pDT32, pDT44, pDT45, or pGEX2T-RalGDS98 (33), respectively, were grown at 30°C to an A600 of ∼0.8 and induced by addition of 0.3 mM IPTG (final concentration). After 3 h, cells were chilled, harvested by centrifugation at 4°C, washed and resuspended in buffer A containing 1 mM DTT and 1 mM PMSF, and lysed by one freeze-thaw cycle followed by sonication. The resulting extracts were mixed with a 50:50 slurry (150 μl) of glutathione-Sepharose beads, incubated on a roller drum for 1.5 h at 4°C, and washed three times (1 ml each) with buffer A and one time with 1 ml buffer E (100 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM PMSF, 2 mM 2-mercaptoethanol, 50 mM Tris-HCl [pH 7.5]). The washed beads were resuspended in 200 μl buffer E, dispensed in two equal portions into plastic microcentrifuge tubes (Eppendorf), and adjusted to 0.2 mg/ml bovine serum albumin (final concentration). One tube received the Ras2 · GTPγS preparation (50 μl); the other received the Ras2 · GDPβS preparation (50 μl). Both tubes were then adjusted with buffer E to a final volume of 300 μl and incubated on a roller drum at 4°C for 1.5 h. The beads were washed three times (1 ml each) with buffer E, and bead-associated proteins were solubilized in 40 μl 1× SDS-PAGE sample buffer, resolved by SDS-PAGE, and analyzed by Coomassie blue staining and immunoblotting with appropriate antibodies.
To measure binding of (His)6-Cdc42 to bead-bound proteins, cultures of BL21(DE3)-RIL expressing GST, GST-RalGDS-RA, GST-Ste50-RA, GST-Ste50-RA(I267A L268A), GST-Ste50-RA(K254A), GST-Ste20-CRIB, and GST-Gic2-CRIB from plasmids pGEX4T-1, pGEX2T-RalGDS98, pDT32, pDT44, pDT45, pDT46, and pMP2003 (gift of M. Peter, ETH, Zürich, Switzerland), respectively, were grown, harvested, washed, and lysed as described in the preceding paragraph. The corresponding GST fusion proteins were purified by adsorption to glutathione-Sepharose beads as described above, except that buffer F (100 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 mM DTT, 50 mM Tris-HCl [pH 7.5]) was used instead of buffer E for the fourth (final) wash. The washed beads were resuspended in 200 μl buffer F, and the protein content was measured using the dye binding method of Bradford (10). If necessary, empty beads were added to adjust the concentration of GST fusion protein to ∼1.3 mg/ml (final volume, 300 μl). Equal portions (110 μl) of each bead suspension were dispensed into microcentrifuge tubes and adjusted to 0.1 mg/ml bovine serum albumin (final concentration). One tube received the Cdc42 · GTPγS preparation (∼2 μg); the other, received the Cdc42 · GDPβS preparation (∼2 μg). Both tubes were adjusted with buffer F to a final volume of 400 μl and incubated on a roller drum at 4°C for 2.5 h. After washing four times (1 ml each) with buffer F, bead-bound proteins were analyzed as described above. Binding of GTPγS- and GDPβS-bound Cdc42(I46M) to GST, GST-RalGDS-RA, GST-Ste50-RA, and GST-Ste20-CRIB was carried out in an identical manner.
Radioactive Ste11 was prepared by in vitro translation using a TNT coupled reticulocyte lysate system (Promega) with pGEM-4Z-Ste11 as a template, SP6 polymerase, and a mixture of [35S]methionine and [35S]cysteine (New England Nuclear or Amersham). To measure binding of 35S-Ste11 to bead-bound proteins, cultures (100 ml) of BL21(DE3)-RIL expressing GST, GST-Ste50, GST-Ste50(I267A L268A), GST-Ste50(K254A), GST-Ste50(E261A D262A), and GST-Ste50(R274A N276A) from plasmids pGEX-4T-1, pDT1, pDT17, pDT18, pDT19, and pDT20, respectively, were grown, harvested, and lysed as described above. The corresponding GST fusions were purified using glutathione-Sepharose beads as described above, except that the fourth wash was omitted. To beads resuspended in buffer A (160 μl) was added 35S-Ste11 (40 μl) containing (final concentrations) bovine serum albumin (1 mM), PMSF (1 mM), and leupeptin (1 μg/ml). After incubation on a roller drum for 1.5 h at 4°C, beads were washed four times with 1 ml buffer A, and bead-bound proteins were resolved by SDS-PAGE and visualized by autoradiography.
Localization.
Cell fractionation was used to analyze biochemically the subcellular distribution of Ste50 and its derivatives. Strain YDV105 carrying appropriate plasmids was grown in selective medium (8 ml) at 30°C in a gyratory water bath to mid-exponential phase (A600 of ∼0.8). Preparation of cell extracts and fractionation by differential centrifugation were carried out as described previously (17). After being washed with distilled water, cells were suspended in 100 μl buffer G (150 mM NaCl, 0.3 M sorbitol, 2 μg/ml leupeptin, 1 μM pepstatin A, 1 mM benzamidine, 2 μg/ml aprotinin, 1 mM PMSF, 50 mM Tris-HCl [pH 8]) and lysed by vigorous vortex mixing with glass beads. The resulting extracts were diluted with additional (50 μl) buffer G, and the beads and cell debris were removed by centrifugation at 500 × g for 4 min at 4°C. The clarified extract was then subjected to centrifugation at 10,000 × g for 10 min at 4°C, and the resulting pellet was resuspended in a volume of buffer equivalent to the volume of the supernatant fraction. The 10,000 × g supernatant fraction was subjected to centrifugation at 100,000 × g, and the resulting pellet was resuspended in a volume equivalent to that of the supernatant fraction. Equal volumes of the pellet and supernatant fractions (corresponding, in the supernatant fraction from the 10,000 × g centrifugation, to ∼50 μg total protein) were resolved on a 12% SDS-polyacrylamide gel and analyzed by immunoblotting.
Fluorescence microscopy was used to examine subcellular localization of green fluorescent protein (GFP)-tagged Ste50 derivatives visually. Strain YDV105 or YDV505 carrying a plasmid expressing a GFP-tagged protein was grown in a gyratory water bath at 30°C in selective medium (5 ml) to mid-exponential phase (A600 of ∼0.7). To visualize nuclear and mitochondrial DNAs, 4′,6-diamidino-2-phenylindole (DAPI) was added (1 μg/ml, final concentration) during the last hour of growth. After being washed with phosphate-buffered saline, cells were examined using an inverted fluorescence microscope (Nikon Eclipse TE300) equipped with a 100×/1.3 Plan Fluor oil objective and appropriate band pass filters to detect either DAPI or GFP fluorescence. Images were captured using a cooled charge-coupled device camera (Hamamatsu ORCA) and recorded digitally (ImageProPlus software; Phase3 Imaging Systems, Inc.).
RESULTS
Ste50 functions downstream of Ras2, Cdc42, and Ste20.
In filamentous growth signaling, Ste50 functions downstream of Ras2 but upstream of Ste11 (71). To verify these findings and to determine where Ste50 functions with respect to Cdc42 and Ste20, we examined whether the absence of Ste50 blocks pathway stimulation (assessed by FRETy1-lacZ reporter gene expression) evoked by constitutively active genes: Ras2(G19V) (14), Cdc42(G12V) (95), ΔN-Ste20 (70), or Ste11(T596I) (STE11-4 mutation) (82). These activating mutations elevated reporter expression in wild-type cells, as expected; however, only the STE11-4 allele was able to do so in ste50Δ cells (Fig. 1). Thus, Ste50 functions downstream of Ras2, Cdc42, and Ste20 but upstream of (or at the same level as) Ste11, in agreement with the fact that Ste11 and Ste50 form a complex (27).
FIG. 1.
Ste50 functions downstream of Ras2, Cdc42, and Ste20. Strain JCY100 (STE50+) or YDV105 (ste50Δ) carrying both a low-copy (CEN) plasmid expressing from the GAL1 promoter the constitutively active variant of the indicated protein and a plasmid (YEpT-FTyZ) containing the FRETY1-lacZ reporter gene were induced for 3 h on galactose (2%) medium, and then β-galactosidase activity present in extracts of the cells was measured as described in Materials and Methods. Values represent the averages of measurements, made in duplicate, on extracts prepared from three independent transformants. Error bars indicate the standard deviation.
The Ste50 RA domain is essential for filamentous growth.
Ste50 and Ste11 associate constitutively via their respective N-terminal SAM domains, and this interaction is important for filamentous growth signaling (35, 67, 71, 92). In contrast, function of the C-terminal RA domain of Ste50 has not been explored. Strikingly, the RA-domain portion of Ste50 is even more highly conserved among yeast species than the SAM domain (Fig. 2A). By comparison, between different classes of signaling proteins, RA domains are much more variable (Fig. 2B). In some of these other proteins (Fig. 2B), the RA domain has been shown to interact with Ras (or Rap) (32, 58, 62, 79, 88), whereas in others (e.g., Myr5), the RA domain does not (39).
FIG.2.
The RA domain of Ste50. (A) An alignment of the primary structure of S. cerevisiae Ste50 with the homologous proteins from seven other fungi, as indicated. As a means to emphasize the most highly conserved portions of the sequence, those residues that are identical in at least seven out of the eight proteins are indicated as a white letter on a black background. Conservative substitutions are indicated as a black letter on a gray background. (B) An alignment of eight RA domains (65). Numbering refers to residue positions in the Ste50 sequence. Residues in Ste50 mutated to Ala are marked with a star.
First, to test its importance, the Ste50 RA domain was altered by site-directed mutagenesis (Fig. 2B) or removed altogether [this truncation, Ste50Δ(219-346), was designated Ste50-ΔRA]. The point mutations introduced (K254A, E261A D262A, I267A L268A, and R274A N276A) were based, in part, on alignment of the Ste50 RA domain with the RA domain of mammalian RalGDS (Fig. 2B) and, in part, on modeling the Ste50 RA domain onto the crystal structures of two Ras · RalGDS RA-domain complexes (32, 88). Equivalent site-directed mutations in RalGDS alter its interaction with Ras (32, 88). As a negative control, we mutated a residue (Ser240) in Ste50 that, based on the position of the apparently equivalent residue in the Ras · RalGDS RA-domain complexes, should be within the interface but should not contribute a strong contact.
To determine the effect of these RA-domain alterations on function, normal Ste50 and each of the mutants was expressed from a low-copy-number (CEN) vector under control of the native STE50 promoter in ste50Δ cells. As judged by the standard bioassay, the absence of Ste50 prevented agar invasion and this defect was fully complemented by either wild-type Ste50 or Ste50(S240A), to a level indistinguishable from that displayed by wild-type (STE50+) cells (Fig. 3A). By contrast, each of the other four point mutants showed a defect, from severe to moderate, in agar invasion. The most defective mutant was Ste50(I267A L268A), followed by Ste50(K254A), whereas the defect of Ste50(R274A N276A) was intermediate and that of Ste50(E261A D262A) was mild (Fig. 3A). Reassuringly, the mutants displayed the same order of severity in the reporter gene assay (Fig. 3D). Immunoblotting with rabbit polyclonal anti-Ste50 antibodies indicated that the mutants were expressed at a somewhat lower level than wild-type Ste50 (although some of these apparent differences may be attributable to destruction or alteration of antigenic sites in the mutant polypeptides) (Fig. 3A, right panel). Nevertheless, there was no correlation between the level of expression and function. For example, Ste50(K254A), which is clearly defective for function, was expressed at a higher level than Ste50(S240A), which was fully functional. Most convincingly, even the most defective mutants [Ste50(I267A L268A), Ste50(K254A), and Ste50-ΔRA] were still unable to rescue the invasion defect of ste50Δ cells even when expressed from a multicopy (2μm DNA) plasmid (Fig. 3B) at a level much higher than that of endogenous Ste50 (Fig. 3B, right panel). Thus, malfunction of the mutant proteins was not due to lower expression.
FIG. 3.
Site-directed mutations in or removal of the Ste50 RA domain prevents invasive growth. (A) Left panel, strain JCY100 (wild type [WT]) or YDV105 (ste50Δ) was transformed with either empty low-copy-number (CEN) vector, YCplac111 (24), or YCpL-STE50 (pDT4), YCpL-STE50(S240A) (pDT7), YCpL-STE50(I267A L268A) (pDT8), YCpL-STE50(K254A) (pDT11), YCpL-STE50(E261A D262A) (pDT12), or YCpL-STE50(R274A N276A) (pDT13). Second panel, total growth (after 3 days incubation on YPD medium at 30°C). Third panel, invasive growth (after washing). Right panel, an immunoblot showing expression levels (detected with polyclonal anti-Ste50 serum). (B and C) Left panels, strain JCY100 (WT) or YDV105 (ste50Δ), as indicated, was transformed with either empty high-copy-number (2μm DNA) vector, YEplac181 (24), or YEpL-STE50 (pDT24), YEpL-STE50(K254A) (pDT88), YEpL-STE50(I267A L268A) (pDT87), or YEpL-STE50-ΔRA (pDT90), which express the indicated genes from the STE50 promoter on the same multicopy vector. Second panels, total growth (after 3 days incubation on YPD medium at 30°C). Third panels, invasive growth (after washing). Right panel (B only), an immunoblot showing protein expression levels (detected with polyclonal anti-Ste50 serum). (D) Strain JCY100 (WT) or YDV105 (ste50Δ), each harboring plasmid YEpT-FTyZ containing the FRETY1-lacZ reporter gene, was transformed with the indicated plasmids described for panel A or with YCpL-STE50-ΔRA (pDT70) and grown to an A600 of ∼0.8, and then β-galactosidase activity present in extracts of the cells was measured as described in Materials and Methods. Values represent the averages of measurements, made in duplicate, on extracts prepared from two (or, in some cases, three) independent transformants. Error bars indicate the standard deviation. (E) Purified GST, GST-Ste50, GST-Ste50(I267A L268A), GST-Ste50(K254A), GST-Ste50(E261A D262A), and GST-Ste50(R274A N276A) were immobilized on glutathione-agarose beads, as indicated, and assayed for their ability to bind radioactive Ste11. Input Ste11 and that retained on the beads were detected by autoradiography, as described in Materials and Methods.
Moreover, in vitro, radioactive Ste11 bound to all four of the invasion-defective RA-domain point mutants equally well and with an affinity comparable to that of wild-type Ste50 (Fig. 3E), consistent with prior observations that complete deletion of the Ste50 RA domain does not affect Ste50 association with Ste11 (67, 92). Furthermore, if the RA-domain mutants can associate with Ste11 in vivo but RA-domain alteration prevents some other function of Ste50 in invasive growth signaling, then overexpression of the RA-domain mutants should exhibit a dominant-negative effect in normal cells. Consistent with this prediction, when overexpressed in a wild-type strain, each of the three RA-domain mutants tested exerted a readily detectable inhibition of invasive growth, whereas overexpression of wild-type Ste50 had no detectably deleterious effect (Fig. 3C).
As a complementary and independent means to demonstrate that the RA domain has a distinct target in filamentous growth signaling, the RA domain (residues 219 to 346) was highly expressed in a wild-type MATa/MATα diploid in which pseudohyphal growth was stimulated by Ras2(G19V). Prominent flares of pseudohyphae were observed in colonies overexpressing Ras2(G19V) alone, as expected (Fig. 4). In contrast, in cells co-overexpressing Ras2(G19V) and the RA domain, pseudohyphal protrusions were greatly suppressed at every colony size examined, suggesting that the isolated RA domain is indeed able to titrate out its binding partner, thereby preventing efficient signaling. Neither of two RA-domain point mutants (K254A and I267A L1268A) overexpressed in the same fashion was able to suppress pseudohyphal filament formation (Fig. 4), further confirming that the RA domain is responsible for exerting this inhibition. The same dominant-negative effect of the isolated RA domain was also observed in haploid cells. Overexpression of the Ste50 RA domain completely blocked stimulation of reporter gene expression induced by Ras2(G19V), Cdc42(G12V), and ΔN-Ste20 but was not able to fully squelch stimulation of reporter gene expression induced by activated Ste11(T596I) (Table 2), in agreement with our epistasis analysis (Fig. 1). We concluded that interaction of the C-terminal RA domain of Ste50 with one or more targets is essential for filamentous growth signaling. Moreover, given that deletion or certain point mutations of the RA domain cause a phenotype as severe as that of a complete ste50Δ null mutation, the RA domain of Ste50 has a function in signal propagation just as important as the role that its N-terminal SAM domain plays in mediating its interaction with Ste11.
FIG. 4.
RA-domain-containing fragment of Ste50 suppresses Ras2(G19V)-stimulated pseudohyphal growth. Strain JCY102 (MATa/MATα) (3) was transformed with YCpU-RAS2(G19V) and either empty vector pAD4M (54), pAD4M-HA3-STE50-RA (pDT63), pAD4M-HA3-STE50-RA(K254A) (pDT64), or pAD4M-HA3-STE50-RA(I267A L268A) (pDT65); grown on low-nitrogen (SLAD) plates for 2.5 days at 30°C; and examined for the presence of pseudohyphal filaments. Representative colonies of three different sizes were photographed. Equivalent results were obtained with RA-domain constructs lacking the (HA)3 tag (data not shown).
TABLE 2.
Ste50 RA-domain overexpression blocks filamentous growth signalinga
| Filamentous growth stimulated by: | β-Galactosidase activity (Miller units, mean ± SD)b
|
% Inhibitionb | |
|---|---|---|---|
| Empty vector (A) | Vector + Ste50-RA (B) | ||
| Empty vector | 3307 ± 468 | 3158 ± 312 | 5 ± 17 |
| Ras2(G19V) | 5635 ± 1130 | 3229 ± 588 | 97 ± 29 |
| Cdc42(G12V) | 4872 ± 426 | 3164 ± 482 | 100 ± 37 |
| ΔN-Ste20 | 5304 ± 845 | 2793 ± 669 | 118 ± 38 |
| Ste11(T596I) | 5033 ± 482 | 3934 ± 92 | 55 ± 26 |
Strain JCY100 (Table 1) was transformed with plasmids that express from the GAL1 promoter constitutively active variants of either Ras2 (pJW83), Cdc42 (pDV1998), Ste20 (pSR1993), or Ste11 (pDT126), as indicated, and either an empty vector (pAD4M) or the same vector expressing the RA domain of Ste50 from the strong constitutive ADH1 promoter (pDT37). Cells were then grown to mid-exponential phase, induced as described in the legend to Fig. 1, and harvested, and the amount of β-galactosidase activity present was assayed as described in Materials and Methods.
Percent inhibition represents the reduction of the induced activity observed in the presence of the RA domain, i.e., 100 − 100 × [(B − background [i.e., empty vector, B])/(A − background [i.e., empty vector, A])]. Values given represent the averages of two to four independent trials each performed in duplicate or triplicate, and the error in A and B (standard deviation of the mean) was propagated in the final estimate of percent inhibition by using standard formulae (63a).
The Ste50 RA domain interacts with Cdc42 and not with Ras2.
Ras2 functions upstream of Cdc42, Ste20, and Ste11-Ste50 (57). However, the mechanism of signaling downstream of Ras2 is not known. Our results were consistent with the possibility that the RA domain might, as its name implies, mediate association of Ste50 with Ras2, and Ras2 might, thereby, influence localization, activity, or stability of Ste11. To test this idea, the ability of purified Ras2-(His)6, loaded with either GTPγS or GDPβS (Fig. 5A, input), to bind to GST alone, to GST fused to the RA domain of a known Ras-binding protein (mammalian RalGDS), or to GST fused to the Ste50 RA domain (both wild type and mutant) immobilized on glutathione beads (Fig. 5A, lower panel) was assessed. As expected, GST was unable to bind either Ras2 · GTPγS or Ras2 · GDPβS, whereas the GST-RalGDS-RA domain construct bound Ras2 · GTPγS strongly and did not bind Ras2 · GDPβS detectably (Fig. 5A, upper panels), demonstrating that the Ras2 preparation was functional and behaved with the expected specificity. Neither Ras2 · GTPγS nor Ras2 · GDPβS was able to associate with either wild-type or mutant GST-Ste50-RA. This in vitro result was consistent with our inability to detect an interaction in vivo between Ste50 and nonfarnesylated yeast Ras2 or nonfarnesylated and/or activated (GTPase-defective) variants of mammalian H-ras by using the two-hybrid method under conditions where c-Raf displayed a readily detectable interaction with both the yeast and mammalian Ras variants (L. Bardwell and J. Thorner, unpublished results) and was also in agreement with the inability of another group to detect interaction between Ste50 and mammalian Ras(G12V) by using the same in vivo method (67).
FIG. 5.
The Ste50 RA domain interacts with Cdc42 but not with Ras2. (A) Purified GST, GST-Ste50-RA, GST-Ste50-RA(I267A L268A), GST-Ste50-RA(K254A), or GST-RalGDS-RA was immobilized on glutathione-agarose beads at equivalent levels, as judged by staining with Coomassie blue (bottom panel), and assayed for the ability to retain purified yeast Ras2-(His)6 preloaded with either GDPβS (upper panel) or GTPγS (middle panel). Input Ras2 and that retained on the beads were detected by immunoblotting with an anti-Ras monoclonal antibody, as described in Materials and Methods. (B) Purified GST, GST-RalGDS-RA, GST-Ste50-RA, GST-Ste50-RA(K254A), GST-Ste50-RA(I267A L268A), and GST-Ste20-CRIB were immobilized on glutathione-agarose beads at equivalent levels, as judged by staining with Coomassie blue (bottom panel), and assayed for the ability to retain purified yeast (His)6-Cdc42 preloaded with either GDPβS (upper panel) or GTPγS (middle panel). Input Cdc42 and that retained on the beads were detected by immunoblotting with polyclonal anti-Cdc42 antibodies, as described in Materials and Methods. (C) Same as panel B, with the indicated GST fusion proteins (bottom panel), except purified mutant (His)6-Cdc42(I46M) preloaded with either GDPβs (upper panel) or GTPγs (middle panel) was used in the binding assay instead of wild-type Cdc42.
Given the essential role of Cdc42 in filamentous growth signaling, our results were also consistent with the possibility that this small GTPase, rather than Ras, might associate with the Ste50 RA domain. For example, overexpression of the RA domain completely blocked stimulation of reporter gene expression induced by Cdc42(G12V) in haploid cells (Table 2). Moreover, given that sequence conservation within the RA-domain family is quite low (Fig. 2B), it seemed possible that some RA domains might interact with small GTPases other than Ras. To test this possibility, the ability of purified (His)6-Cdc42, loaded with either GTPγS or GDPβS (Fig. 5B, input), to bind to GST and the other GST fusions described above and, as a positive control, GST fused to a known Cdc42-binding domain, the CRIB motif of Ste20 (45) (Fig. 5B, lower panel), was assessed. GST was unable to bind either Cdc42 · GTPγS or Cdc42 · GDPβS. In contrast to its avid and specific binding of GTP-bound Ras2, GST-RalGDS-RA displayed little or no binding to Cdc42. As expected, GST-Ste20(CRIB) bound almost exclusively to Cdc42 · GTPγS (Fig. 5B, upper panel). These controls demonstrated that the Cdc42 preparation was functional and behaved with the proper specificity. Strikingly, GST-Ste50-RA was able to bind Cdc42, albeit less strongly than the Ste20 CRIB motif (Fig. 5B, upper panel). Moreover, unlike the CRIB motif, the Ste50 RA domain interacted equivalently with either GTP- or GDP-bound Cdc42. Thus, Ste50 joins a growing list of proteins that are able to interact with Cdc42 in a nucleotide-independent manner (28). In support of the physiological relevance of this interaction, GST-Ste50-RA(I267A L268A), the point mutant which showed the most severe defects in the agar invasion and filamentous signaling assays, reduced binding of either Cdc42 · GTPγS or Cdc42 · GDPβS to background levels, whereas GST-Ste50-RA(K254A), a much weaker allele as judged by both criteria, displayed a reproducible, but only modest, reduction in binding.
A KCAAX box can replace the Ste50 RA domain.
The observed interaction between the RA domain of Ste50 and Cdc42 was intriguing. Since Cdc42 is firmly anchored at the plasma membrane, the observed moderate affinity and nucleotide-insensitive binding of the RA domain to Cdc42 might provide a mechanism for Ste11-Ste50 complexes to associate transiently with the plasma membrane, thereby permitting Ste11 activation whenever activated Ste20 is also present. The hypothesis that Cdc42-dependent membrane delivery of the Ste50-Ste11 complex is a physiological role of the RA domain of Ste50 led to several readily testable predictions. First, this idea predicts that attachment of a membrane-targeting element should rescue the signaling defect of Ste50 mutants lacking a functional RA domain. For this purpose, the C-terminal nine residues (KKSKKCAIL) of Cdc42 were added in frame to the C termini of the Ste50 RA-domain point mutants and to Ste50-ΔRA, as well as to normal Ste50 as a control (Fig. 6A). This nine-residue segment of Cdc42 (abbreviated KCAAX box) is the minimal element responsible for prenylation and plasma membrane-specific targeting of Cdc42 (17, 36, 74).
FIG.6.
A C-terminal KCAAX box rescues the defects of Ste50 RA-domain mutants. (A) Schematic diagram of the Ste50 constructs used. KCAAX corresponds to the nine C-terminal residues of Cdc42 (KKSKKCAIL). Site-directed mutations are indicated by stars. Positions of the SAM and RA domains are indicated. (B) Left panel, strain YDV105 (ste50Δ) was transformed with either empty low-copy-number (CEN) vector, YCplac111 (24), or YCpL-STE50 (pDT4), YCpL-STE50(I267A L268A) (pDT8), YCpL-STE50(I267A L268A)-KCAAX (pDT92), YCpL-STE50-ΔRA (pDT70), or YCpL-STE50-ΔRA-KCAAX (pDT93), as indicated. Middle panel, total growth (after 3 days incubation on YPD medium at 30°C). Right panel, invasive growth (after washing). (C) Same as in panel B, except strain YDV105 was transformed with high-copy-number (2μm DNA) vectors, YEplac181 (24), YEpL-STE50 (pDT24), YEpL-STE50(I267A L268A) (pDT87), YEpL-STE50(I267A L268A)-KCAAX (pDT109), YEpL-STE50-ΔRA (pDT90), or YEpL-STE50-ΔRA-KCAAX (pDT110), as indicated. (D) Left panel, strains YDV105 (ste50Δ), YDV505 (ste50Δ ste11Δ), and YDV205 (ste50Δ ste20Δ) were transformed either with empty vector, YEplac112 (24), or with YEpT-STE50-ΔRA-KCAAX (pDT175). Middle panel, total growth (after 3 days incubation on YPD medium at 30°C). Right panel, invasive growth (after washing). (E) Strain YDV105 (ste50Δ), harboring plasmid YEpT-FTyZ containing the FRETY1-lacZ reporter gene, was transformed with the indicated plasmids described for panel B and grown to an A600 of ∼0.8, and then β-galactosidase activity present in extracts of the cells was measured as described in Materials and Methods. Values represent the averages of measurements, made in duplicate, on extracts prepared from two (or, in some cases, three) independent transformants. Error bars indicate the standard deviation. (F) Strain FP67 (ste50Δ ssk2Δ ssk22Δ) was transformed with either a high-copy-number (2μm DNA) vector, YEplac195 (24), or YEpU-STE50 (pDT151), YEpU-STE50(I267A L268A) (pDT149), YEpU-STE50(I267A L268A)-KCAAX (pDT147), YEpU-STE50-ΔRA (pDT150), or YEpU-STE50-ΔRA-KCAAX (pDT148), as indicated. After overnight growth at 30°C, cultures were adjusted to the same A600 and serial dilutions were spotted on YPD plates lacking (left) or containing (right) 1.5 M sorbitol. After growth at 30°C for 2 days, the plates were photographed.
Ste50-KCAAX behaved identically to native Ste50 in both the agar invasion and pseudohyphal growth assays (data not shown). Unlike the corresponding RA-domain mutants which do not support invasive growth in ste50Δ cells, the KCAAX derivatives of the exact same mutants were able to restore agar invasion nearly as well as wild-type Ste50 (Fig. 6B), even when expressed under control of the STE50 promoter on a low-copy-number (CEN) plasmid and even though under these conditions the KCAAX derivatives were expressed at a lower level than Ste50 itself, as judged by immunoblotting (data not shown). When expressed from the STE50 promoter on a multicopy (2μm DNA) plasmid, the RA-domain mutants were still defective (as observed before [Fig. 3B]), whereas the KCAAX derivatives of the same RA-domain mutants fully complemented the agar invasion defect of the ste50Δ strain (Fig. 6C). Likewise, as judged by the reporter gene assay, expression of Ste50-ΔRA-KCAAX was able to rescue the filamentous growth signaling defect of ste50Δ cells just as well as wild-type Ste50 (Fig. 6E). Thus, membrane tethering via a KCAAX box functionally substitutes for the absence of RA-domain function in supporting invasive growth, consistent with the idea that the role of the observed RA-domain-Cdc42 interaction is delivery of the Ste11-Ste50 complex to the membrane.
If the observed complementation occurs via the known elements of the invasive growth signaling pathway and is not caused by some other, unexpected mechanism that bypasses the normal pathway, then rescue of the agar invasion defect of ste50Δ cells conferred by the KCAAX derivatives should require the function of both Ste11 and Ste20. Indeed, we found that both Ste20 and Ste11 function were required for either Ste50-ΔRA-KCAAX (Fig. 6D) or Ste50-RA(I267A L268A)-KCAAX (data not shown) to promote invasion. In this regard, it is noteworthy that absence of Ste20 causes a stronger defect in agar invasion than absence of Ste11, as has also been observed by others (47, 56, 75), suggesting that Ste20 activates both a major Ste11-dependent route and a minor Ste11-independent mechanism that stimulates invasive growth (this Ste11-independent route may involve another, as-yet-unidentified, MAPKKK).
It has been reported that the RA domain of Ste50 has an important role in the Ste11-dependent branch of HOG pathway signaling (67, 92). Therefore, we tested whether the KCAAX box constructs could rescue the osmosensitivity of triple mutant cells that lack both the Ste50-dependent branch (ste50Δ) and the other branch (ssk2Δ ssk22Δ) of the HOG pathway. Indeed, unlike that of Ste50(I267A L268A) or Ste50-ΔRA, expression of either Ste50(I267A L268A)-KCAAX or Ste50-ΔRA-KCAAX in the ssk2Δ ssk22Δ ste50Δ strain fully restored normal osmoresistance (Fig. 6F).
A KCAAX box enhances membrane targeting.
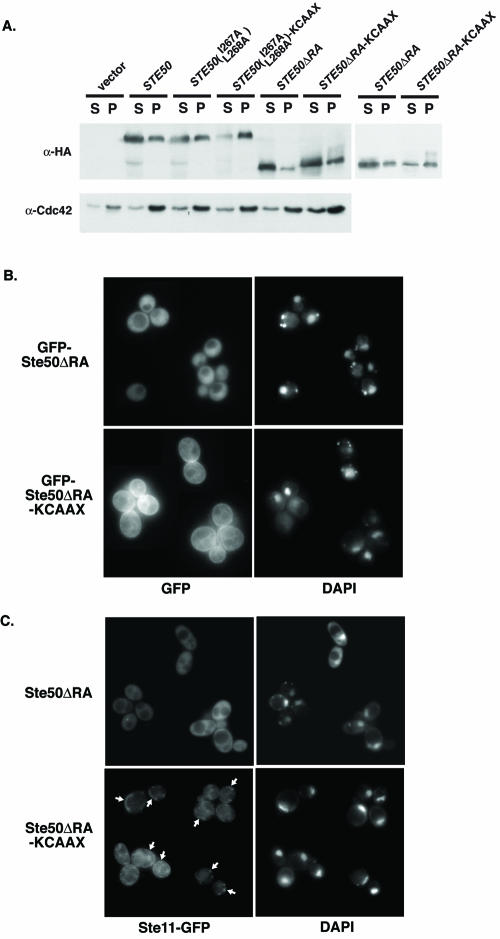
To confirm that appending a KCAAX box increases the amount of Ste50 associated with the plasma membrane, we first used subcellular fractionation. Extracts of cells expressing Ste50, Ste50 mutants, or the KCAAX derivatives of the same mutants were separated by differential centrifugation at 10,000 × g into a membrane-containing particulate fraction and a soluble fraction. As expected, a known plasma membrane-associated KCAAX-containing protein (Cdc42) partitioned more in the particulate fraction than in the soluble fraction. Likewise, for Ste50 and its derivatives, the expected trends were generally observed. For example, in the representative experiment shown, the presence of a KCAAX box significantly shifted more of any given Ste50-derived protein into the particulate fraction (Fig. 7A). Conversely, the absence of the RA domain shifted more Ste50 into the soluble fraction (Fig. 7A, compare intact Ste50 to Ste50-ΔRA). The same distribution was observed when the Ste50-related species remaining in the 10,000 × g supernatant fraction were subjected to subsequent sedimentation at 100,000 × g and thereby separated into secondary particulate and soluble fractions (data not shown).
FIG. 7.
The KCAAX box enhances membrane recruitment. (A) Strain YDV105 (ste50Δ) was transformed with either high-copy-number (2μm DNA) vector, YEplac181, or YEpL-HA3-STE50 (pDT36), YEpL-HA3-STE50(I267A L268A) (pDT112), YEpL-HA3-STE50(I267A L268A)-KCAAX (pDT113), YEpL-HA3-STE50-ΔRA (pDT114), or YEpL-HA3-STE50-ΔRA-KCAAX (pDT115), as indicated; grown to an A600 of ∼0.8, lysed; and fractionated at 10,000 × g into the supernatant (S) and pellet (P) fractions as described in Materials and Methods. Proteins in these fractions were resolved by SDS-PAGE and analyzed by immunoblotting with either a mouse anti-HA antibody or rabbit polyclonal anti-Cdc42 serum, as indicated. Right panel, a separate analysis of Ste50-ΔRA and Ste50-ΔRA-KCAAX performed with independent transformants. (B) Strain YDV105 (ste50Δ) was transformed with plasmids expressing GFP-STE50-ΔRA (pDT100) or GFP-STE50-ΔRA-KCAAX (pDT134), as indicated, and grown to mid-exponential phase at 30°C. DAPI (1-μg/ml final concentration) was added as a vital stain to reveal the position of the nucleus, and growth was continued for at least 1 h more. Cells then were washed with phosphate-buffered saline and examined under a fluorescence microscope, as described in Materials and Methods. (C) Strain YDV505 (ste50Δ ste11Δ) was transformed with both pFP176 expressing Ste11-GFP from the STE11 promoter on a CEN plasmid and either pDT90 expressing Ste50-ΔRA from the STE50 promoter on a 2μm DNA plasmid or pDT110 expressing Ste50-ΔRA-KCAAX in the same fashion, as indicated. Cells were treated and examined under a fluorescence microscope as for panel B. Arrows highlight puncta, islands, and swatches of plasma membrane-associated Ste11-GFP fluorescence.
As a second, independent means to show that a KCAAX box enhanced plasma membrane targeting, Ste50-ΔRA, Ste50-ΔRA-KCAAX, Ste50(I267A L268A), and Ste50(I267A L268A)-KCAAX were each fused to the C terminus of GFP (81), expressed from the strong constitutive TPI1 promoter to ensure ready detection, and viewed directly by standard epifluorescence microscopy. Like GFP-Ste50 [and unlike either GFP-Ste50-ΔRA or GFP-Ste50(I267A L268A)], both GFP-Ste50-ΔRA-KCAAX and GFP-Ste50(I267A L268A)-KCAAX fully complemented the filamentous growth defect of ste50Δ cells (data not shown). GFP-Ste50-ΔRA showed diffuse cytoplasmic staining that was excluded from the vacuole but not from the nucleus (Fig. 7B), and GFP-Ste50 and GFP-Ste50(I267A L268A) showed the same distribution (data not shown). Access to the nuclear compartment was not surprising given the small size of these chimeras. By contrast, GFP-Ste50-ΔRA-KCAAX displayed prominent rim staining at the cell periphery, which is diagnostic of plasma membrane association, as well as decorated internal membranes (Fig. 7B). This fluorescence pattern is very similar to that reported for GFP-Cdc42 (74). An essentially identical staining pattern at the cell periphery and on internal membranes, albeit somewhat less bright, was observed for GFP-Ste50(I267A L268A)-KCAAX (data not shown).
If Ste50 associates tightly with Ste11 via heterotypic SAM-SAM interaction, then plasma membrane tethering of Ste50 should bring more Ste11 to the plasma membrane. To test this prediction, the effect of copiously expressing Ste50-ΔRA or Ste50-ΔRA-KCAAX in cells coexpressing a limiting amount of Ste11-GFP was examined (Fig. 7C). In cells coexpressing Ste11-GFP and Ste50-ΔRA, fluorescence was confined to the cytosol and none was detected at the cell periphery. In contrast, in cells coexpressing Ste11-GFP and Ste50-ΔRA-KCAAX, we reproducibly observed distinct puncta, patches, and occasionally prominent swatches of Ste11-GFP fluorescence at the cell periphery (Fig. 7C). These observations further support the conclusion that attachment of a KCAAX box restores function to Ste50 mutants lacking an RA domain because it promotes membrane association of Ste11-Ste50 complexes.
The Ste50 RA domain associates with a surface of Cdc42 distinct from its switch I and II loops.
Proteins that interact exclusively with GTP · Cdc42 must make contact with the switch I and switch II loops of Cdc42 (36) (Fig. 8B), which are forced into the conformation productive for effector binding only when GTP is bound (77). In this regard, we found that the CRIB domains of Ste20 (Fig. 5B) and Gic2 (data not shown) interact almost exclusively with GTP-bound Cdc42. By contrast, the RA domain of Ste50 interacted with both GDP- and GTP-bound Cdc42 (Fig. 5B). Hence, a second prediction of this observed association is that the Ste50 RA domain should interact with residues in Cdc42 whose conformation is not affected by nucleotide binding, namely, residues on a face of Cdc42 distinct from the switch I and II loops. This supposition further suggested that cdc42 alleles that are competent to interact with Ste20 but not competent to interact with Ste50 might exist. Such a Cdc42 mutant should be able to support pheromone signaling because it can still recruit and activate Ste20 at the plasma membrane, which can then encounter Ste11 bound to the Ste5 scaffold protein tethered at the plasma membrane by its interaction with Gβγ. However, such an Ste50 binding-defective Cdc42 mutant should not be able to support filamentous growth signaling, because there would be no way to convey Ste50-Ste11 complexes to the activated Ste20 at the plasma membrane.
FIG. 8.
Membrane tethering of Ste50 rescues the invasive growth defect of cdc42 alleles specifically defective for RA-domain association. (A) Left panel, either strain YDV105 (ste50Δ) (right-hand third) or strain YTIMK92 [cdc42(I46M)], strain KKY691 [cdc42(E100A)], or strain YTIMK94 [cdc42(S71P)] (remaining two-thirds), as indicated, was transformed with a high-copy-number (2μm DNA) vector, YEplac195 (24), or YEpU-STE50(I267A L268A)-KCAAX (pDT147), YEpU-STE50-ΔRA-KCAAX (pDT148), YEpU-STE50(I267A L268A) (pDT149), or YEpU-STE50-ΔRA (pDT150), as shown. Middle panel, total growth (after 6 days incubation on YPD medium at 30°C). Right panel, invasive growth (after washing). (B) Ribbon diagram of a three-dimensional model of the structure of S. cerevisiae Cdc42, generated with Swiss-Model (30), based on alignment of the amino acid sequence of yeast Cdc42 with human Cdc42 (80% identity) and the crystal structures of Cdc42Hs (Protein Data Bank entries 2NGR, 1GRN, 1AN0, and 1A4R), drawn with MOLSCRIPT (41). Switch I and II elements are shown in yellow. Side chains of Ile46, Ser71, and Glu100 are indicated as ball-and-stick representations in turquoise. The N- and C-terminal ends of the Cdc42 polypeptide are also indicated.
Two cdc42 mutations, Cdc42(I46M) (56) and Cdc42(E100A) (40; K. Kozminski, personal communication), have been described that are defective in invasive growth signaling, as judged by reporter gene assay or other criteria, but are not compromised for other Cdc42 functions. As we expected, both mutations lie outside the switch I and switch II loops (Fig. 8B). As judged by the two-hybrid method, Cdc42(I46M) interacts with Ste20 as well as wild-type Cdc42, whereas other alleles defective for both pheromone and invasive growth signaling, e.g., Cdc42(S71P) located in the switch II loop (Fig. 8B), show a greatly reduced affinity for Ste20 by the same method (56). Correspondingly, we found that purified Cdc42(I46M) was able to associate in vitro with the CRIB domain of Ste20 just as well as normal Cdc42 and did so in a strictly GTP-dependent manner (Fig. 5C). Conversely, and in agreement with our second prediction, this cdc42 allele, unlike wild-type Cdc42, was unable to bind detectably to the RA domain of Ste50 (Fig. 5C).
A third prediction of the hypothesis that the RA-domain-Cdc42 interaction is important for delivering Ste50-Ste11 complexes to the plasma membrane is that the defect of invasive growth-deficient cdc42 alleles should be bypassed by KCAAX constructs that tether Ste50 to the plasma membrane in a Cdc42-independent manner. To test this idea, cells expressing each of three cdc42 mutants as the sole source of Cdc42 were transformed with plasmids expressing either Ste50(I267A L268A), Ste50(I267A L268A)-KCAAX, Ste50-ΔRA, or Ste50-ΔRA-KCAAX. Although all three cdc42 mutations maintain cell viability, none of them supports invasive growth, even though endogenous Ste50 is present (Fig. 8A). None of them supports invasive growth when either Ste50(I267A L268A) or Ste50-ΔRA is overexpressed in the same cells. Strikingly, however, when either Ste50(I267A L268A)-KCAAX or Ste50-ΔRA-KCAAX was overexpressed in the cells expressing Cdc42(I46M) and Cdc42(E100A), the ability to invade agar was substantially restored (Fig. 8A). These in vivo results, and the fact that Cdc42(I46M) does not bind to the Ste50 RA domain in vitro [Cdc42(E100A) was not tested], are consistent with the idea that the major defect of the cdc42(I46M) and cdc42(E100A) alleles in filamentous growth signaling is an inability to associate with the RA domain of Ste50 and, thus, lack of recruitment of Ste11-Ste50 complexes to the membrane. Consistent with other evidence that Cdc42(S71P) is defective in interacting with Ste20 (56) and with the essential role that Ste20 plays in this signaling pathway (Fig. 6D), the agar invasion defect of cells expressing Cdc42(S71P) was not rescued detectably by either Ste50(I267A L268A)-KCAAX or Ste50-ΔRA-KCAAX (Fig. 8A).
DISCUSSION
Our findings reveal a novel mechanism in MAPK signaling. Our results indicate that the RA domain of Ste50 is responsible for recruitment of Ste11-Ste50 complexes to the plasma membrane. This membrane tethering is necessary for signaling in both the filamentous growth and HOG signaling pathways and seems to be achieved by binding of the RA domain to either the GDP- or GTP-bound form of the small Rho-type GTPase Cdc42 (Fig. 9). The most telling evidence in support of this model is as follows. First, the isolated RA domain binds nucleotide-loaded Cdc42 in vitro but does not bind nucleotide-loaded Ras2. Second, the isolated RA domain, when overexpressed, exerts a dominant-negative effect on invasive growth signaling in both haploids and diploids and can completely squelch pathway stimulation by activated Cdc42(G12V). Third, the signaling defect of Ste50 mutated in or totally lacking its RA domain, which cannot interact with Cdc42, can be fully rescued by artificial membrane tethering of the same proteins. Fourth, certain cdc42 alleles are specifically unable to support invasive growth signaling, and one such mutant tested cannot bind to the Ste50 RA domain of Ste50, whereas it still binds to the Ste20 CRIB domain. Fifth, the defect of these invasive growth-specific cdc42 alleles is suppressed by membrane tethering of Ste50 that bypasses the need for the RA-domain-Cdc42 interaction.
FIG. 9.
Schematic depiction of the proposed function of the Cdc42-Ste50-RA domain and Ste11-Ste50-SAM domain interactions in filamentous growth and HOG pathway signaling. See text for further explanation.
For pheromone response, proximity of activated Ste20 and Ste11, bound to the Ste5 scaffold protein (21), is ensured by the fact that the C terminus of Ste20 (46) and the RING-H2 domain of Ste5 (34) both bind to free Gβγ (released when pheromone receptors are occupied by agonist). Activated Cdc42 is generated in the same locale, in part because the Far1 scaffold protein, which binds the GEF (Cdc24) for Cdc42, also associates with free Gβγ (12). GTP-bound Cdc42 binds to the Ste20 CRIB domain, activating Ste20 catalytic activity and reinforcing its membrane anchoring (45). In the filamentous growth and HOG pathways, activated Ste20 presumably also resides at the plasma membrane because it is bound via its CRIB domain to GTP · Cdc42. However, Ste5 is completely dispensable for both of these signaling pathways (47, 60). Hence, another mechanism is required for encounter between Ste11 and active membrane-bound Ste20. Ste50 appears to fulfill this role. The RA-domain-Cdc42 interaction is guanine nucleotide-insensitive and seems close to the limit of detection for GST pull-down assays (88a), suggesting modest affinity (Kd of ≤1 μM). This constitutive, but relatively weak, interaction may allow Ste11-Ste50 complexes to come on and off Cdc42 at the plasma membrane, in agreement with the partitioning of Ste50 seen in our subcellular fractionation (Fig. 7A). The transient nature of such Ste11-Ste50-Cdc42 ternary complexes in vivo is further supported by the fact that GFP-Ste50, which is fully functional in our hands, displays rather diffuse cytoplasmic fluorescence and does not show distinct staining at the cell periphery, even in haploids undergoing invasive growth (data not shown). It has been reported that a different construct, Ste50-GFP, also displays diffuse cytoplasmic fluorescence but converts to a punctuate pattern when cells are exposed to high-osmolarity conditions (67). Perhaps Ste50 localizes more stably to the membrane during HOG pathway signaling, but whether the punctate pattern seen represents authentic membrane association was not explored (67). Our data clearly show that the observed interactions of Ste11 with Pbs2 (66) and with Sho1 (94) are not sufficient, in the absence of the RA domain of Ste50, to allow for efficient Ste11 activation in the HOG pathway (Fig. 6F). Hence, Ste50-mediated membrane delivery of Ste11 is critical for both invasive growth and HOG signaling.
Transient association and dissociation of Ste50-Ste11 complexes with Cdc42 ensures that downstream signaling can occur whenever a sufficient population of activated Ste20 is present (Fig. 9). For example, such continuous generation and release of activated Ste11-Ste50 complexes from the membrane would permit encounter with Ste7 in the cytosol. Such a route seems advantageous for promoting pseudohyphal growth in diploids, where Ste7 is not bound to Ste5 (because STE5 is not expressed in MATa/MATα cells [51]). Even in haploids, at least some Ste7 is always present in the cytoplasm, because Ste5 undergoes continuous nucleocytoplasmic shuttling (43, 53). Once activated (87), Ste7 must enter the nucleus, because the MAPK required for filamentous growth (Kss1) resides there (50) bound to the Ste12 transcription factor in ternary complexes with the Dig repressors (2, 3). Ste7 has a high-affinity docking site by which it binds Kss1 (1). In response to hyperosmotic stress, activated Ste11 need not dissociate from the membrane to gain access to its target, because Pbs2 is tethered to the SH3 domain in the cytoplasmic tail of the Sho1 osmosensor, an integral membrane protein (66, 68, 73). Pbs2 has a high-affinity docking site by which it binds Hog1 (66).
Our experiments show that addition of a completely different membrane anchor, i.e., the KCAAX box of Cdc42, targets Ste50 to the plasma membrane and, most importantly, compensates completely for loss of the Ste50 RA domain, providing independent confirmation that membrane localization is a critical aspect of the function of Ste50 and its RA domain during both filamentous growth and HOG pathway signaling. Likewise, the fact that addition of the KCAAX box to Ste50 RA-domain mutants suppresses two different mutations (I46M and E100A) in Cdc42 that are specifically defective in filamentous growth signaling provides independent confirmation of two aspects of our model. First, these cdc42 alleles are defective in filamentous growth signaling because they cannot interact properly with the RA domain of Ste50, and this specific inference was directly confirmed for Cdc42(I46M) (Fig. 5C). Second, association with Cdc42 to deliver the Ste50-Ste11 complex to the membrane must therefore be at least one of the critical functions of the Ste50 RA domain. Based on the crystal structure of GTP-bound Cdc42 (Fig. 8B), I46 is located on a side of Cdc42 opposite from its switch I and II loops, and E100 is located on a helix that projects away from the switch I and II segments, consistent with the idea that Ste50 interacts with Cdc42 via a mode that differs from that of those effectors that bind preferentially to the GTP-bound state of Cdc42. Finally, the fact that high-level expression of Ste50-ΔRA-KCAAX in the presence of a limiting amount of Ste11-GFP greatly increases the amount of fluorescence present at the plasma membrane confirms by an independent means that the SAM domain of Ste50 is sufficient to form a stable complex with Ste11 and further supports the conclusion that recruitment of Ste50 to the plasma membrane mediated by interaction of its RA domain with Cdc42 will also deliver Ste11 to this location.
Our results suggest that other RA domains also may interact with members of the superfamily of small GTPases other than Ras (or even with other proteins), given that sequence conservation between RA-domain family members is quite low (65). Detailed structural information at atomic resolution will be necessary to discern what features of the Ste50 RA domain confer selectivity for binding Cdc42 and/or mitigate against binding to Ras. Based on four RA-domain · Ras (or Rap1A) complexes that have been characterized both biochemically and structurally, association is largely electrostatic (80): in the main, basic side chains in the RA domain interact with acidic residues in the G protein. For these RA domains, mutagenesis has shown that the positively charged residues are important for interaction with Ras (32, 58, 62, 79, 88). Hence, it is tempting to speculate that RA domains that are unable to bind Ras, like those in Ste50 (as shown here) and mammalian Myr5 (39), are unable to do so because they lack Arg or Lys residues at certain critical positions (corresponding to S240 and/or N276 in Ste50) (Fig. 2B). It remains to be seen what other classes of RA domains interact, like that in Ste50, with Cdc42 per se. Whether the RA domain of the apparent Ste50 ortholog in Schizosaccharomyces pombe, Ste4 (Fig. 2B), associates with Cdc42 has not been tested. S. pombe Ste4 does form tight complexes with the fission yeast Ste11 homolog (Byr2), just as Ste50 does with Ste11. Interestingly, unlike S. cerevisiae Ste11, Byr2 itself contains an RA domain (4, 85) that is able to interact with GTP-bound Ras (5, 55, 85). Thus, it is possible that, in S. pombe, Ste4-Byr2 complexes may be under the dual control of both Cdc42 and Ras.
As we have shown here, in two different MAPK signaling pathways (invasive growth and hyperosmotic stress response), Cdc42 functions like a general adaptor protein that recruits a critical complex of signaling proteins (Ste50-Ste11) to the plasma membrane. Thus, our findings demonstrate that, in addition to acting in a switch-like manner that depends on the nature of the guanine nucleotide bound, Cdc42 can also perform a physiologically important function in a guanine nucleotide-insensitive manner. Similarly, another small Rho-type GTPase, Rac, has been shown to interact with a mammalian scaffolding protein, OSM, in a nucleotide-insensitive manner (86).
Supplementary Material
Acknowledgments
We thank L. Bardwell, B. Cairns, R. W. Davis, D. Drubin, B. Errede, T. Handel, C. Inouye, D. I. Johnson, K. Kozminski, G. S. Martin, H.-U. Mösch, S. O'Rourke, M. Peter, F. Posas, J. Rine, H. Saito, J. Shimoni, D. Voora, and M. Whiteway for generous gifts of reagents and/or useful advice.
This work was supported by postdoctoral fellowship 99-20069Y from the American Heart Association, NIH-NRSA postdoctoral fellowship GM20022, and NCI postdoctoral traineeship CA09041 (to D.M.T.); by a University of California President's Undergraduate Research Fellowship (to J.E.B.); and by NIH research grant GM21841 and resources provided by the Berkeley campus Cancer Research Laboratory (to J.T.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, L., J. G. Cook, D. M. Voora, A. R. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95:15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, M. M., H. Tu, L. Van Aelst, and M. Wigler. 1996. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol. Cell. Biol. 16:5597-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauman, P., Q. C. Cheng, and C. F. Albright. 1998. The Byr2 kinase translocates to the plasma membrane in a Ras1-dependent manner. Biochem. Biophys. Res. Commun. 244:468-474. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharjya, S., P. Xu, R. Gingras, R. Shaykhutdinov, C. Wu, M. Whiteway, and F. Ni. 2004. Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex. J. Mol. Biol. 344:1071-1087. [DOI] [PubMed] [Google Scholar]
- 8.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 9.Bokoch, G. M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743-781. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 13.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 14.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 15.Cullen, P. J., W. J. Sabbagh, E. Graham, I. M. M., E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. J. Sprague. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 17.Davis, C. R., T. J. Richman, S. B. Deliduka, J. O. Blaisdell, C. C. Collins, and D. I. Johnson. 1998. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J. Biol. Chem. 273:849-858. [DOI] [PubMed] [Google Scholar]
- 18.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 20.Duran, A., and C. Nombela. 2004. Fungal cell wall biogenesis: building a dynamic interface with the environment. Microbiology 150:3099-3103. [DOI] [PubMed] [Google Scholar]
- 21.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 114:3967-3978. [DOI] [PubMed] [Google Scholar]
- 22.Fenton, J. W., II, M. J. Fasco, and A. B. Stackrow. 1977. Human thrombins. Production, evaluation, and properties of alpha-thrombin. J. Biol. Chem. 252:3587-3598. [PubMed] [Google Scholar]
- 23.Gagiano, M., F. F. Bauer, and I. S. Pretorius. 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2:433-470. [DOI] [PubMed] [Google Scholar]
- 24.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 27.Grimshaw, S. J., H. R. Mott, K. M. Stott, N. P. R., K. A. Evetts, L. J. Hopkins, D. Nietlispach, and D. Owen. 2004. Structure of the sterile alpha motif (SAM) domain of the Saccharomyces cerevisiae mitogen-activated protein kinase pathway-modulating protein STE50 and analysis of its interaction with the STE11 SAM. J. Biol. Chem. 279:2192-2201. (Erratum, 279:9672.) [DOI] [PubMed] [Google Scholar]
- 28.Grohmanova, K., D. Schlaepfer, D. Hess, P. Gutierrez, M. Beck, and R. Kroschewski. 2004. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J. Biol. Chem. 279:48495-48504. [DOI] [PubMed] [Google Scholar]
- 29.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 30.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 31.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Huang, L., F. Hofer, G. S. Martin, and S. H. Kim. 1998. Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol. 5:422-426. [DOI] [PubMed] [Google Scholar]
- 33.Huang, L., X. Weng, F. Hofer, G. S. Martin, and S. H. Kim. 1997. Three-dimensional structure of the Ras-interacting domain of RalGDS. Nat. Struct. Biol. 4:609-615. [DOI] [PubMed] [Google Scholar]
- 34.Inouye, C., N. Dhillon, and J. Thorner. 1997. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278:103-106. [DOI] [PubMed] [Google Scholar]
- 35.Jansen, G., F. Buhring, C. P. Hollenberg, and M. Ramezani Rad. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:102-117. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 38.Jones, J. S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 39.Kalhammer, G., M. Bahler, F. Schmitz, J. Jockel, and C. Block. 1997. Ras-binding domains: predicting function versus folding. FEBS Lett. 414:599-602. [DOI] [PubMed] [Google Scholar]
- 40.Kozminski, K. G., A. J. Chen, A. A. Rodal, and D. G. Drubin. 2000. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell 11:339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 42.Kuchin, S., V. K. Vyas, and M. Carlson. 2003. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 31:175-177. [DOI] [PubMed] [Google Scholar]
- 43.Künzler, M., J. Trueheart, C. Sette, E. Hurt, and J. Thorner. 2001. Mutations in the YRB1 gene encoding yeast ran-binding-protein-1 that impair nucleocytoplasmic transport and suppress yeast mating defects. Genetics 157:1089-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwan, J. J., N. Warner, T. Pawson, and L. W. Donaldson. 2004. The solution structure of the S. cerevisiae Ste11 MAPKKK SAM domain and its partnership with Ste50. J. Mol. Biol. 342:681-693. [DOI] [PubMed] [Google Scholar]
- 45.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22:2939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 47.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 48.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma, D., J. G. Cook, and J. Thorner. 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol. Biol. Cell 6:889-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacKay, V. L. 1983. Cloning of yeast STE genes in 2 microns vectors. Methods Enzymol. 101:325-343. [DOI] [PubMed] [Google Scholar]
- 52.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 53.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98:501-512. [DOI] [PubMed] [Google Scholar]
- 54.Martin, G. A., D. Viskochil, G. Bollag, P. C. McCabe, W. J. Crosier, H. Haubruck, L. Conroy, R. Clark, P. O'Connell, R. M. Cawthon, et al. 1990. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63:843-849. [DOI] [PubMed] [Google Scholar]
- 55.Masuda, T., K. Kariya, M. Shinkai, T. Okada, and T. Kataoka. 1995. Protein-kinase Byr2 is a target of Ras1 in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 270:1979-1982. [DOI] [PubMed] [Google Scholar]
- 56.Mösch, H. U., T. Kohler, and G. H. Braus. 2001. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol. Cell. Biol. 21:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mösch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nassar, N., G. Horn, C. Herrmann, A. Scherer, F. McCormick, and A. Wittinghofer. 1995. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375:554-560. [DOI] [PubMed] [Google Scholar]
- 59.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187-190. [DOI] [PubMed] [Google Scholar]
- 60.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Rourke, S. M., I. Herskowitz, and E. K. O'Shea. 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18:405-412. [DOI] [PubMed] [Google Scholar]
- 62.Pacold, M. E., S. Suire, O. Perisic, S. Lara-Gonzalez, C. T. Davis, E. H. Walker, P. T. Hawkins, L. Stephens, J. F. Eccleston, and R. L. Williams. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103:931-943. [DOI] [PubMed] [Google Scholar]
- 63.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 63a.Parratt, L. G. 1961. Probability and experimental errors in science. John Wiley & Sons, Inc., New York, N.Y.
- 64.Ponting, C. P. 1995. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4:1928-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponting, C. P., and D. R. Benjamin. 1996. A novel family of Ras-binding domains. Trends Biochem. Sci. 21:422-425. [DOI] [PubMed] [Google Scholar]
- 66.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 67.Posas, F., E. A. Witten, and H. Saito. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18:5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raman, M., and M. H. Cobb. 2003. MAP kinase modules: many roads home. Curr. Biol. 13:R886-R888. [DOI] [PubMed] [Google Scholar]
- 70.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramezani Rad, M., G. Jansen, F. Buhring, and C. P. Hollenberg. 1998. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet. 259:29-38. [DOI] [PubMed] [Google Scholar]
- 72.Ramezani-Rad, M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr. Genet. 43:161-170. [DOI] [PubMed] [Google Scholar]
- 73.Reiser, V., S. M. Salah, and G. Ammerer. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620-627. [DOI] [PubMed] [Google Scholar]
- 74.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 76.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rudolph, M. G., A. Wittinghofer, and I. R. Vetter. 1999. Nucleotide binding to the G12V-mutant of Cdc42 investigated by X-ray diffraction and fluorescence spectroscopy: two different nucleotide states in one crystal. Protein Sci. 8:778-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheffzek, K., P. Grunewald, S. Wohlgemuth, W. Kabsch, H. Tu, M. Wigler, A. Wittinghofer, and C. Herrmann. 2001. The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast. Structure 9:1043-1050. [DOI] [PubMed] [Google Scholar]
- 80.Sheinerman, F. B., and B. Honig. 2002. On the role of electrostatic interactions in the design of protein-protein interfaces. J. Mol. Biol. 318:161-177. [DOI] [PubMed] [Google Scholar]
- 81.Shulewitz, M. J., C. J. Inouye, and J. Thorner. 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stevenson, B. J., N. Rhodes, B. Errede, and G. F. Sprague, Jr. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 6:1293-1304. [DOI] [PubMed] [Google Scholar]
- 83.Tedford, K. S., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 84.Truckses, D. M., L. S. Garrenton, and J. Thorner. 2004. Jekyll and Hyde in the microbial world. Science 306:1509-1511. [DOI] [PubMed] [Google Scholar]
- 85.Tu, H., M. Barr, D. L. Dong, and M. Wigler. 1997. Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol. 17:5876-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uhlik, M. T., A. N. Abell, N. L. Johnson, W. Sun, B. D. Cuevas, K. E. Lobel-Rice, E. A. Horne, M. L. Dell'Acqua, and G. L. Johnson. 2003. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 5:1104-1110. [DOI] [PubMed] [Google Scholar]
- 87.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3:1051-1059. [DOI] [PubMed] [Google Scholar]
- 88.Vetter, I. R., T. Linnemann, S. Wohlgemuth, M. Geyer, H. R. Kalbitzer, C. Herrmann, and A. Wittinghofer. 1999. Structural and biochemical analysis of Ras-effector signaling via RalGDS. FEBS Lett. 451:175-180. [DOI] [PubMed] [Google Scholar]
- 88a.Vikis, H. G., and K. L. Guan. 2004. Glutathione-S-transferase-fusion based assays for studying protein-protein interactions. Methods Mol. Biol. 261:175-186. [DOI] [PubMed] [Google Scholar]
- 89.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, Y., and H. G. Dohlman. 2004. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science 306:1508-1509. [DOI] [PubMed] [Google Scholar]
- 91.Westfall, P. J., D. R. Ballon, and J. Thorner. 2004. When the stress of your environment makes you go HOG wild. Science 306:1511-1512. [DOI] [PubMed] [Google Scholar]
- 92.Wu, C., E. Leberer, D. Y. Thomas, and M. Whiteway. 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell 10:2425-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu, G., G. Jansen, D. Y. Thomas, C. P. Hollenberg, and M. Ramezani Rad. 1996. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 20:773-783. [DOI] [PubMed] [Google Scholar]
- 94.Zarrinpar, A., R. P. Bhattacharyya, M. P. Nittler, and W. A. Lim. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 14:825-832. [DOI] [PubMed] [Google Scholar]
- 95.Ziman, M., J. M. O'Brien, L. A. Ouellette, W. R. Church, and D. I. Johnson. 1991. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.