Abstract
The FoxO forkhead transcription factors FoxO4 (AFX), FoxO3a (FKHR.L1), and FoxO1a (FKHR) represent important physiological targets of phosphatidylinositol-3 kinase (PI3K)/protein kinase B (PKB) signaling. Overexpression or conditional activation of FoxO factors is able to antagonize many responses to constitutive PI3K/PKB activation including its effect on cellular proliferation. It was previously shown that the FoxO-induced cell cycle arrest is partially mediated by enhanced transcription and protein expression of the cyclin-dependent kinase inhibitor p27kip1 (R. H. Medema, G. J. Kops, J. L. Bos, and B. M. Burgering, Nature 404:782-787, 2000). Here we have identified a p27kip1-independent mechanism that plays an important role in the antiproliferative effect of FoxO factors. Forced expression or conditional activation of FoxO factors leads to reduced protein expression of the D-type cyclins D1 and D2 and is associated with an impaired capacity of CDK4 to phosphorylate and inactivate the S-phase repressor pRb. Downregulation of D-type cyclins involves a transcriptional repression mechanism and does not require p27kip1 function. Ectopic expression of cyclin D1 can partially overcome FoxO factor-induced cell cycle arrest, demonstrating that downregulation of D-type cyclins represents a physiologically relevant mechanism of FoxO-induced cell cycle inhibition.
The FoxO subfamily of forkhead transcription factors comprises three functionally related proteins, AFX, FKHR, and FKHRL1, which have recently been renamed FoxO4 (AFX), FoxO1a (FKHR), and FoxO3a (FKHRL1), respectively (21). They represent mammalian orthologues of a Caenorhabditis elegans transcription factor, DAF-16, which in the worm regulates longevity and responses to environmental stress (14, 18). Genetic screens established that DAF-16 function is suppressed by a pathway involving the C. elegans homologue of the insulin receptor DAF-2, a phosphatidylinositol-3 kinase (PI3K)-like protein, AGE-1, and the protein kinase B (PKB) serine/threonine kinases AKT1 and AKT2 (10, 28, 37, 38).
In mammals the PI3K/PKB pathway is stimulated by a variety of growth factors, by cytokines, and by cell-matrix interactions, and it controls many biological functions including cell proliferation, survival, and insulin responses (27). Importantly, constitutive activation of this pathway facilitates tumor formation both by supporting S-phase entry and by conferring resistance to apoptotic signals which normally restrict uncontrolled cell growth (7, 47).
In agreement with the findings for C. elegans, recent studies have provided evidence that at least some of the physiological consequences of PI3K/PKB activation in mammals are mediated by negative regulation of FoxO factors (24). PKB was shown to phosphorylate all three FoxO factors directly on two or three critical residues (T24, S253, and S316 in FoxO1a) (4, 15, 25, 45), which results in nuclear exclusion and inhibition of FoxO factor-mediated gene expression (1, 3, 44). PKB and FoxO factors thus may fulfill antagonistic functions in vivo. Consistent with a proposed antiapoptotic and proliferative effect of the PI3K/PKB pathway, forced expression or activation of FoxO factors triggers apoptotic responses or a cell cycle arrest, depending on the cell system studied (24).
We and others have previously reported that FoxO factor-induced withdrawal from the cell cycle occurs in G1 phase and results from increased transcription of the cyclin-dependent kinase inhibitor p27kip1 (33, 36). Although deficiency for p27kip1 greatly reduced the antiproliferative potential of FoxO factors, we nevertheless noticed that p27kip1-deficient mouse embryonic fibroblasts (MEFs) still showed reduced DNA synthesis rates upon FoxO4 expression (33), indicating that DAF-16-like forkheads may also control the functions of other important regulators of G1-S transition.
Progression from G1 to S phase is most importantly regulated by the combined actions of the G1 cyclin/CDK complexes cyclin D/CDK4/6 and cyclin E/CDK2 (11, 42). In early G1 the kinase activities of CDK4/6 and CDK2 are low, primarily due to absence of their cyclin partners, which are barely expressed at this stage and are required for activation. In the presence of mitogens, levels of D-type cyclins gradually rise in mid-to-late G1 (11). As a result, active cyclin D/CDK4/6 complexes which initiate phosphorylation of pRb, an important transcriptional repressor of G1-S progression, are formed, thereby relieving its inhibitory effect on E2F-mediated gene expression, which is crucial for initiation of DNA replication and entry into S phase (16, 51). An early consequence of E2F-activated gene transcription is an increase of cyclin E levels resulting in the formation of active cyclin E/CDK2 complexes, which augment and maintain pRb hyperphosphorylation. At this stage, cells become irreversibly committed to entering S phase and no longer depend on further growth factor stimulation (11, 42).
In an attempt to elucidate whether FoxO factors can utilize alternative (p27kip1-dependent and -independent) mechanisms to inhibit cellular proliferation, we analyzed the effect of FoxO factor introduction on the activity of crucial regulators of the G1-S transition more extensively. We show that retrovirally mediated expression of FoxO factors results in a prominent inhibition of CDK4-dependent phosphorylation of the S-phase repressor protein pRb. This occurs independently of functional p27kip1 and correlates with decreased protein levels of the D-type cyclins D1 and D2. The reduction in protein expression is preceded by a decrease in cyclin D mRNA levels, and promoter studies utilizing human cyclin D1 and cyclin D2 promoter constructs revealed that activation of a conditionally active FoxO3a mutant results in efficient repression of basal cyclin D1 and D2 promoter activities. Thus, modulation of cyclin D levels by FoxO factors occurs through transcriptional regulation. We furthermore demonstrate that ectopic expression of cyclin D1 partially protects cells from FoxO factor-induced cell cycle arrest, emphasizing the functional relevance of our findings.
MATERIALS AND METHODS
Plasmids.
All retroviral constructs used in this study have a pBabe.puro backbone and have been described before (26, 33). Expression vectors for cyclin D1, HA-CDK4, HA-CDK4KD, CD20, and p21 are derivatives from pCMV and have also been described earlier (31, 49). Myc-tagged pRb was a gift from R. Bernards. DsRed2-C1 is commercially available from Clontech Laboratories Inc. For transient expression of wild-type and mutant FoxO factors, the following plasmids were used: pMT-2-HA-FoxO4, pMT-2-HA-FoxO4.DB, pMT-2-HA-FoxO4.A3, and pcDNA-FoxO3a.A3 (33). Cyclin D1 promoter constructs −1748D1 and −962D1 were a kind gift of F. McCormick and have been described previously (48). The various cyclin D2 promoter constructs (−1624D2, D2Δ-892, and D2Δ-444) were a kind gift of P.G. Milner and have been described previously (2). The 6×DBE-luciferase vector (12) was kindly provided by T. Furuyama.
Cells and cell culture.
Primary MEFs from p27kip1−/− and wild-type embryos were a gift from B. Scheyen. Primary MEFs from pRb−/−/p107−/−/p130−/− triple-knockout embryos (TKO) were a kind gift of Floris Foijer. Immortalized wild-type and p27kip1-deficient MEFs (39) were kindly provided by D. Peeper. The FoxO3a.A3-ER-expressing DL23 colon carcinoma cell line and the DLD-1 parental cell line have been described recently (26). NIH 3T3 cells as well as Phoenix cells were from common stocks from The Netherlands Cancer Institute.
For generation of the cyclin D1-expressing D1-7 MEF cell clone and the corresponding polyclonal vector cell line, immortalized wild-type MEFs were grown in six-well plates and cotransfected with pCMV.cyclin D1 or pCMV empty vector and limiting amounts of pBabe.hygro. Following a selection procedure of 14 days in Dulbecco's modified Eagle's medium containing 150 μg of hygromycin B (Calbiochem)/ml, single clones were picked and expanded. Exogenous cyclin D1 expression levels were analyzed at a low concentration of serum (32 h at 0.03% serum). Under these conditions, endogenous cyclin D1 expression is suppressed (33) while exogenous cyclin D1 expressed under the control of the constitutive cytomegalovirus (CMV) promoter is not affected (data not shown).
All primary cells and cell lines were cultured in Dulbecco's modified Eagle's medium (Life Technologies) containing standard supplements. For activation of FoxO3a.A3-ER in the DL23 cell line, 4-hydroxy tamoxifen (4-OHT) (Sigma) was directly added to fresh medium from a stock solution with a concentration of 100 μM to a final concentration of 100 nM.
Antibodies, immunoprecipitations, and immunoblots.
Anti-cyclin E (sc-481), anti-CDK4 (sc-270), anti-phospho S780 pRb (sc-12901), and anti-cyclin D2 (sc-452) were purchased from Santa Cruz. Anti-pRb (14001A) was from PharMingen, and anti-p27kip1 (K25020) was from Transduction Laboratories. Monoclonal anti-cyclin D1 antibody (5D4) was obtained from Immunotech. For detection of hemagglutinin (HA)-tagged FoxO-factors, a biotinylated anti-HA mouse monoclonal antibody (BMG-3F10) from Roche was used. Myc-tagged pRb was immunoprecipitated and detected with a monoclonal antibody against the Myc-epitope (9E10; Upstate). Immunoprecipitations and Western blotting were performed as described previously (32).
Northern blots and RT-PCR.
Total RNA for Northern blots and reverse transcription (RT)-PCRs was isolated by a standard guanidine isothiocyanate extraction method. For Northern detection of cyclin D1 mRNA, 20 μg of each RNA sample was separated by gel electrophoresis on formaldehyde-agarose gels and after blotting to nylon membranes hybridized with a radiolabeled cDNA probe prepared from a 1.2-kb HindIII fragment of a human cyclin D1 cDNA insert in pRC/CMV (20).
For the detection of cyclin D1, cyclin D2, and actin by RT-PCR, 10 μg of DNase I-treated RNA was reverse transcribed by using 20 U of avian myeloblastosis virus reverse transcriptase (Roche) according to the manufacturer's instructions. Products were purified by column purification (QiaQuick-PCR purification kit; Qiagen) and levels of cyclin D1, cyclin D2, and actin cDNA were detected by semiquantitative PCR (56°C, 25 to 35 cycles) and subsequent electrophoresis on 2% agarose gels. The following primers were used: cyclin D1, 5′-CGAGGAGCTGCTGCAAATGG-3′ (forward) and 5′-GGTATCAAAATGCTCCGGAGAGG-3′ (reverse); cyclin D2, 5′-AGCAGCGGGAGAAGCTGTCTCTGATCC-3′ (forward) and 5′-ATGGACGCGTCTCTC-TCTTTCGGCC-3′ (reverse); actin, 5′-GACATGGAGAAAATCTGGCA-3′ (forward), and AATGTCACGCACGATTTCCC-3′ (reverse). In all cases, PCRs were shown to be in a linear range by performing parallel control PCRs with increasing template cDNA concentrations.
Transfections.
For transfections of immortalized MEFs, cells were grown in six-well dishes and transfected with FuGENE 6 (Roche) according to the manufacturer's instructions. Total amounts of 2.5 μg of DNA and 5 μl of FuGENE 6 were used per well. In cotransfection experiments a minimum of 0.25 μg of each plasmid was used. DL23 cells were also transfected with FuGENE 6. In this case, cells were seeded in 6-cm-diameter dishes and transfected with a mixture of DNA and FuGENE 6 in the ratio of 2 μg to 4 μl. Phoenix cells and NIH 3T3 cells were grown in 10-cm-diameter plates and transfected by standard procedures by the calcium phosphate method.
Retroviral infections of MEFs.
Recombinant retroviruses were produced by transfection of the relevant retroviral constructs into the Phoenix virus packaging cell line. Conditioned medium containing infective retrovirus was collected in two harvests after 36 and 42 h and was either used fresh or snap-frozen on solid CO2 and stored at −80°C for up to 1 month. For infection, exponentially growing MEF cultures were in two consecutive rounds exposed to a 1:1 dilution of filtered virus supernatant and fresh medium in presence of 6 μg of polybrene/ml. Routinely, infection efficiencies of about 90% were obtained as determined by puromycin selection.
Cell cycle distribution.
For analysis of cell cycle distribution, NIH 3T3 cells were transfected with the appropriate expression plasmids in combination with CMV.CD20. Thirty-two hours after transfection, cells were treated for 16 h with 250 ng of nocodazole/ml to trap cycling cells in G2-M. DNA profiles of CD20-positive cells were obtained by combined CD20 and DNA staining followed by fluorescence-activated cell sorter (FACS) analysis (31). To examine the effect of FoxO factor expression on DNA synthesis, retrovirally infected MEFs were pulsed with 1 μM BrdU for 30 min and BrdU positivity was analyzed by flow cytometry as previously described (32).
Colony formation assays.
Primary or immortalized MEFs were transfected with the relevant expression plasmids in combination with DsRed2-C1 or alternatively infected with an appropriate retrovirus. In the case of the transfections, cells were trypsinized 42 h posttransfection and cells of each combination were equally reseeded at multiple dilutions. The remnants were used to determine transfection rates quantifying DsRed positivity by flow cytometry and to prepare total lysates. From the reseeded cells the old medium was removed after 18 h and replaced by selection medium containing 2 μg of puromycin/ml. Cultures were maintained in selection medium until the end of the observation period about 10 to 14 days after transfection. Cells were then fixed in 100% methanol (30 min at room temperature) and stained for 1 h with 0.1% crystal violet (Sigma). The stained colonies were counted, and values were corrected for the measured transfection efficiencies. In the infection experiments, cells were counted the day after infection by using an automatic cell counter (Casy 1; Schärfe System) and were equally reseeded into selection medium at densities ranging from 2 × 105 to 5 × 104 cells/10-cm-diameter dish. Colony formation assays were then performed as detailed above.
Cyclin E/CDK2 kinase assays.
Cyclin E/CDK2 kinase assays were performed as previously described (32).
Promoter studies.
Promoter studies were performed by following a dual-luciferase reporter protocol provided with a luciferase detection kit from Promega. In short, DL23 cells were seeded in 6-cm-diameter plates and transfected in duplicate with 2 μg of the relevant promoter-firefly luciferase reporter constructs in combination with 2.5 ng of a thymidine kinase promoter-Renilla luciferase reporter (pRL-TK; Promega). Eighteen hours after transfection, cells were stimulated with 4-OHT as described above, and they were lysed 24 h thereafter. Transfection-corrected promoter activities were obtained by dual measurement of firefly and Renilla luciferase activities in a luminometer.
RESULTS
FoxO factor expression inhibits proliferation in the absence of functional p27kip1.
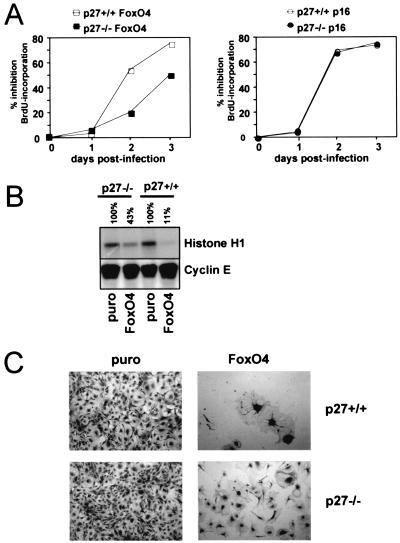
We previously demonstrated that a variety of cell types respond to forced expression of FoxO transcription factors with a rapid block of cell cycle progression at the G1-S transition (33). This effect was attributed mainly to enhanced p27kip1 expression; however, to some extent DNA synthesis rates were also found to be reduced in p27kip1-deficient cells upon FoxO4 introduction (33). This suggested the existence of an alternative, p27kip1-independent pathway that contributes to the antiproliferative potential of FoxO factors. To confirm this hypothesis, we first aimed to study the consequence of FoxO4 expression on DNA replication of p27kip1-deficient and wild-type MEFs, respectively. For this, MEFs of both genotypes were alternatively infected with either a FoxO4-containing retrovirus or a control retrovirus, and DNA synthesis rates were compared by quantitative analysis of BrdU incorporation. Consistent with earlier observations (33), both p27kip1+/+ and p27kip1-deficient cells responded to FoxO4 expression with impaired BrdU incorporation. Compared to that obtained with wild-type cells, the reduction obtained with p27kip1−/− MEFs was less pronounced but became more apparent at later time points after infection (Fig. 1A, left panel). The weaker response in p27kip1-deficient MEFs was not due to a generally decreased sensitivity to G1 inhibitors, since retroviral expression of the CDK4-specific inhibitor p16INK4A resulted in a comparable repression of BrdU incorporation in wild-type and p27kip1−/− cells (Fig. 1A, right panel). When immune complex kinase assays for cyclin E-associated kinase were performed, cyclin E/CDK2 complex activity was also found to be inhibited in both cell subtypes. In agreement with the BrdU incorporation experiments, inhibition occurred less efficiently in p27kip1−/− cells (Fig. 1B). Thus, although in a physiological context p27kip1 induction amplifies the antiproliferative potential of FoxO-factors, their capacity to block G1-S transition does not rely on p27kip1 function alone.
FIG. 1.
FoxO factor-induced cell cycle arrest does not depend on p27kip1 expression alone. MEFs from wild-type (p27+/+) or p27kip1-deficient (p27−/−) embryos were infected with an HA-FoxO4-containing retrovirus (FoxO4), a p16INK4a-containing retrovirus, or the parental retrovirus (puro) as indicated. (A) Time course of FoxO4-mediated (left panel) or p16-induced (right panel) DNA synthesis inhibition as determined by quantification of BrdU incorporation. Shown is the percent inhibition of BrdU incorporation compared to pBabe.puro-infected cells as a function of time. BrdU positivity of pBabe.puro-infected cells was around 50% at each time point, with no significant differences seen between the two genotypes (not shown). (B) Immunocomplex kinase assay for cyclin E-associated kinase using histone H1 as a substrate for immunoprecipitated cyclin E/CDK2 complexes. Lysates were taken 48 h postinfection. The percentage above each lane indicates the relative kinase activity compared to the control samples. Equal loading of immunoprecipitated cyclin E/CDK2 complexes was confirmed by Western blotting for cyclin E. (C) Light microscopic image showing morphology and densities of cells from the different infections after 10 days. A representative microscopic field is depicted. Each experiment was reproduced at least twice. Similar results were obtained with a retrovirus encoding a constitutively active FoxO3a mutant, HA-FoxO.A3 (data not shown).
For a better evaluation of the long-term consequences of FoxO expression on cellular proliferation, colony formation assays were performed. Again, wild-type or p27kip1-deficient MEFs were alternatively infected with a FoxO4-containing retrovirus or with an empty retroviral construct. Twenty-four hours postinfection, equal cell numbers of each combination were reseeded into selective medium, and the growth behaviors of the differently infected cells were monitored over a period of 10 days. While both vector-infected p27kip1-deficient and wild-type MEFs grew dense towards the end of the observation period, FoxO4-expressing cells in either case exhibited strikingly lower densities (Fig. 1C). Consistent with weaker suppression of DNA synthesis, p27kip1−/− MEFs appeared slightly less responsive to FoxO4 expression than did their wild-type counterparts. Identical results were obtained when experiments were performed with a retrovirus encoding a constitutive active FoxO3a mutant (data not shown). These data demonstrate that p27kip1-deficient cells, like their wild-type counterparts, undergo cell cycle arrest upon sustained FoxO factor expression, albeit with slower kinetics.
FoxO expression results in a reduction of cyclin D/CDK4 activity.
We next wanted to address the question of which mechanism underlies p27kip1-independent S-phase inhibition by FoxO forkheads. The initial trigger for progression from G1 to S phase in response to growth factor stimulation is increased activity of cyclin D/CDK4 complexes due to upregulation of D-type cyclins (11, 42). Consequently, FoxO factor-mediated S-phase inhibition could result from negative regulation of cyclin D/CDK4 activity. To test this concept, we analyzed CDK4 activation upon expression of FoxO4 in NIH 3T3 cells, wild-type MEFs, and p27kip1−/− MEFs. We took advantage of phosphospecific antibodies raised against residue S780 of the CDK4 target pRb. Phosphorylation of this residue was reported to occur in a CDK4-specific manner in vivo and effectively prevented the S-phase limiting interaction of pRb with E2F1 in vitro (22). Besides providing information on the activation status of cyclin D/CDK4 complexes, investigation of S780 phosphorylation therefore additionally allows a rough estimation of whether the antiproliferative capacity of FoxO factors might result from stabilization of pRb function.
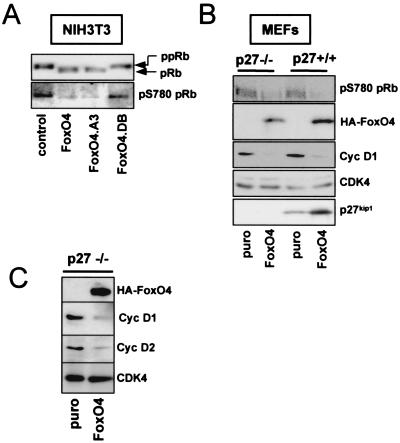
Figure 2A shows the results of transient transfection experiments performed with NIH 3T3 cells. Expression of either full-length FoxO4 or a constitutive active FoxO4 mutant, FoxO4.A3, which lacks the three inhibitory phosphorylation sites for PKB, caused a dramatic reduction in S780 phosphorylation of coexpressed pRb. In contrast, no effect was seen on pRb-S780 phosphorylation when FoxO4.DB, a mutant of FoxO4 lacking the transactivation domain, was transfected. This mutant still binds DNA but is unable to activate FoxO4-mediated gene expression (33), suggesting that inhibition of CDK4 activity depends on the transactivation capacity of FoxO4.
FIG. 2.
FoxO factor expression results in reduced protein levels of cyclins D1 and D2 and inhibits activity of cyclin D/CDK4 complexes in vivo. (A) NIH 3T3 cells were transiently transfected with an expression vector for Myc-tagged pRb in combination with either an empty control plasmid, a vector encoding full-length HA-FoxO4, or the indicated FoxO4 mutants, respectively. Forty-eight hours after transfection, exogenous Myc-pRb was immunoprecipitated from total lysates by using a monoclonal anti-Myc antibody, and CDK4-mediated phosphorylation of immunoprecipitated Myc-pRb was analyzed by Western blotting with a phosphospecific antibody against S780 (22). Similar loading of immunoprecipitated Myc-pRb was confirmed by staining the same blot with antiserum against total pRb. Phosphorylated pRb (ppRb) (upper band) migrates more slowly than unphosphorylated pRb (pRb) (lower band). (B) MEFs from wild-type (p27+/+) or p27kip1-deficient (p27−/−) embryos were infected with an HA-FoxO4-containing retrovirus (FoxO4) or a control retrovirus (puro). Forty-eight hours later, cells were harvested and total lysates were analyzed by Western blotting for the presence of HA-FoxO4, cyclin D1 (Cyc D1), or p27kip1. The cyclin D1 blot was additionally immunostained for CDK4 as a loading control. The activity of cyclin D/CDK4 complexes was judged by immunoblotting for phospho-S780pRb (pS780pRb). (C) Immortalized p27kip1−/− MEFs were infected as described for panel B and analyzed for protein expression of cyclin D1 (Cyc D1), cyclin D2 (Cyc D2), and HA-FoxO4 by Western blotting. CDK4 served as a loading control.
Similar to what was observed with NIH 3T3 cells, retrovirally mediated FoxO4 expression decreased pRb-S780 phosphorylation in primary MEFs (Fig. 2B, upper panel). This occurred irrespective of p27kip1 gene function, since a comparable decrease of pRb S780 phosphorylation was observed in wild-type and p27kip1-deficient cells. These data indicate that inhibition of cyclin D/CDK4 function contributes to the FoxO-induced cell cycle arrest in a p27kip1-independent fashion.
Activation of FoxO factors leads to downregulation of D-type cyclins.
An important determinant controlling the activity of CDKs within the cell cycle is availability of their cyclin interaction partners (42). Consequently, we considered modulation of cyclin D protein levels as a putative mechanism of FoxO factor-mediated CDK4 inhibition. For evaluation of this concept, protein lysate of vector-infected and FoxO4 retrovirus-infected MEFs was analyzed for differences in D-type cyclin expression. Of the three mammalian D-type cyclins, MEFs expressed cyclin D1 and cyclin D2, while no cyclin D3 could be detected (data not shown). Hence, we restricted our study to cyclin D1 and cyclin D2. Figure 2B illustrates that FoxO4 expression resulted in a pronounced decrease of cyclin D1 protein in the absence and presence of functional p27kip1. Likewise, expression of FoxO4 reduced protein levels of cyclin D2, irrespective of p27kip1 function (Fig. 2C). These data indicate that FoxO factors inhibit CDK4 function at the level of cyclin D.
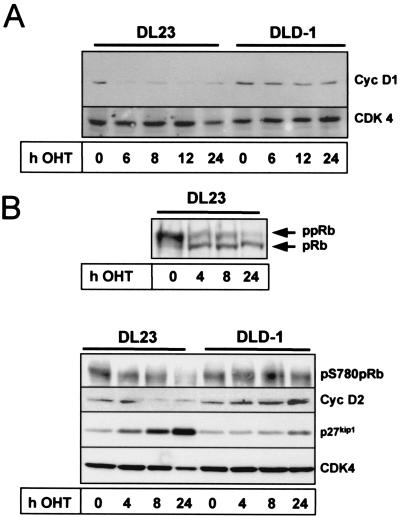
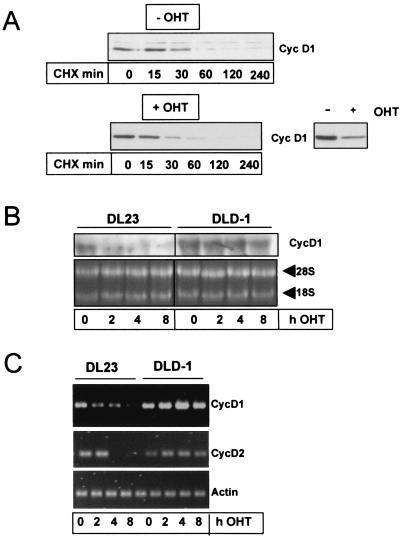
We next addressed whether inhibition of D-type cyclins is a general phenomenon accompanying forkhead-mediated cell cycle arrest or whether this is a cell type-specific characteristic of fibroblasts. An earlier study reported generation of a human colon carcinoma cell line (DL23) stably expressing a nonphosphorylatable, constitutively active mutant of FoxO3a (FoxO3a.A3) fused to a modified form of the estrogen receptor hormone binding domain (FoxO3a.A3-ER) (26). This FoxO3a.A3-ER fusion protein is rendered active by addition of the estrogen receptor agonist 4-OHT to the medium (9), and 4-OHT treatment leads to rapid p27kip1 induction and cell cycle arrest in these cells (26) (Fig. 3B). The DL23 cell line therefore appeared ideal to study the consequence of FoxO3a activation on cyclin D expression in an independent cell system. Figure 3A shows that incubation of DL23 cells with 4-OHT diminished cyclin D1 protein levels as early as 6 h after stimulation. This effect was specific for FoxO3a activation since 4-OHT treatment did not alter the levels of cyclin D1 protein in the parental DLD-1 vector cell line (Fig. 3A). Similar to what was observed with cyclin D1, 4-OHT treatment reduced cyclin D2 protein levels in the FoxO3a.A3-ER-expressing DL23 cell line but did not affect cyclin D2 expression in the corresponding DLD-1 vector cell line (Fig. 3B, lower panel). Again, no cyclin D3 protein was detectable in either case (data not shown), indicating that by downregulation of cyclin D1 and cyclin D2, FoxO3a reduced all functionally relevant D-type cyclins in these cells. Analysis of total pRb phosphorylation revealed that cyclin D reduction in the DL23 cell line correlated with concurrent accumulation of hypophosphorylated pRb (Fig. 3B, upper panel), suggesting that pRb activation and hence cell cycle arrest had commenced. Western blot experiments using the phosphospecific antibody for S780 of the pRb protein validated that FoxO3a-mediated inhibition of pRb phosphorylation also affected CDK4-specific phosphorylation sites of the pRb protein (Fig. 3B, lower panel). This strongly suggests that attenuation of D-type cyclin expression also contributes to the antiproliferative response to FoxO forkhead transcription factors in nonfibroblast cells.
FIG. 3.
Conditional activation of FoxO3a results in downregulation of cyclin D1 and D2 proteins and inhibition of CDK4 activity in human colon carcinoma cells. DL23 and DLD-1 cells were left untreated (0 h) or treated with 100 nM 4-OHT for the indicated time periods. Total lysates were prepared and subsequently analyzed by Western blotting for the presence of cyclin D1 (Cyc D1) (A), or cyclin D2 (Cyc D2) and p27kip1 (B, lower panel). Additionally, total phosphorylation of pRb (ppRb) (B, upper panel) or of phospho-pRb S780 (pS780pRb) (B, lower panel) was determined by using an antibody against total pRb or a phosphospecific antiserum against S780 of the pRb protein, respectively. Equal loading for panels A and B was confirmed by Western blotting for CDK4.
FoxO-induced cyclin D downregulation is not dependent on cellular quiescence.
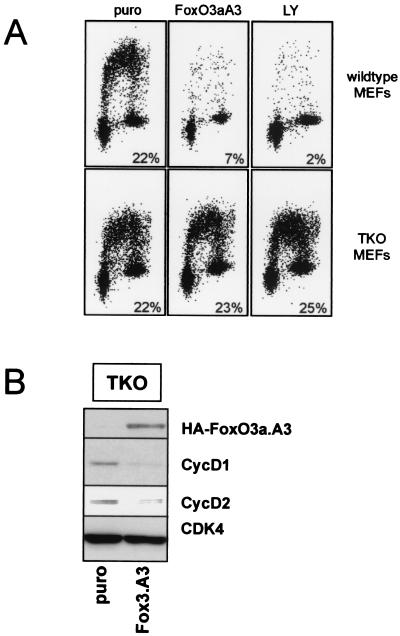
Recently, we found that cells enter a G0 state upon activation of FoxO3a (26). Typically, cyclin D expression is low or absent in cells that have entered a quiescent state (42). Thus, it could well be that the downregulation of cyclins D1 and D2 that is observed in response to FoxO activation occurs as an indirect consequence of a FoxO-driven entry into G0. To rule out this possibility, we utilized cells lacking pRb and the related pocket proteins p107 and p130 (TKO cells). It was recently shown that these TKO cells are not arrested in response to FoxO activation and do not enter quiescence (26). Indeed, infection of these cells with a FoxO3a-encoding retrovirus failed to block BrdU-incorporation, while the same retrovirus efficiently reduced BrdU incorporation in wild-type MEFs (Fig. 4A). Similar differences were found when these cells were treated with LY294002, leading to activation of endogenous FoxO factors (Fig. 4A). Nevertheless, examination of cyclin D1 and D2 protein levels showed that expression of both was reduced in response to active FoxO3a (Fig. 4B). This demonstrates that downregulation of cyclins D1 and D2 occurs independently of FoxO3a-induced quiescence and excludes the possibility that the effect on cyclin D is secondary to an arrest in G0.
FIG. 4.
FoxO-induced downregulation of cyclin D does not depend on induction of quiescence. MEFs deficient for pRb/p107/p130 pocket proteins (TKO) were infected with a FoxO3a.A3-containing retrovirus (FoxO3A3) or a control retrovirus (puro) (A and B) or treated with 10 μM LY294002 (LY) (A). (A) FACS analysis of BrdU incorporation 48 h after infection with the indicated retrovirus or 24 h after LY294002 treatment. (B) Expression levels of cyclin D1, cyclin D2, and HA-FoxO3a.A3 proteins in lysates prepared 48 h postinfection as determined by Western blotting. An immunoblot for CDK4 is shown as a control for equal loading.
FoxO-dependent repression of D-type cyclin expression involves transcriptional regulation.
FoxO factor-mediated downregulation of D-type cyclins could be due to increased protein degradation. PKB is known to stabilize cyclin D1 protein via direct phosphorylation of GSK3, leading to inhibition of GSK3 activity (8). It is possible that expression of a nonphosphorylatable FoxO3a mutant could interfere with PKB-mediated GSK3 phosphorylation and, as such, influence cyclin D levels. However, addition of 4-OHT to DL23 cells did not result in inhibition of PKB-mediated GSK3 phosphorylation (data not shown), ruling out the possibility that the FoxO3a mutant exerts a dominant negative effect on PKB activity. To further demonstrate that FoxO3a does not affect protein stability of cyclins D1 and D2, we analyzed protein turnover before and after FoxO3a activation. Protein degradation was monitored by blocking de novo protein synthesis by the addition of cycloheximide to the cells. While overall cyclin D1 protein levels were significantly reduced 6 h after FoxO3a activation (Fig. 5A), we found that the half-life of cyclin D1 was not affected under these conditions (∼30 min with or without 4-OHT) (Fig. 5A). Similar results were obtained for cyclin D2 (not shown). These data indicate that protein stability of cyclins D1 and D2 is not affected by FoxO3a and suggest that regulation occurs at the level of transcription. To confirm this, we studied mRNA levels for cyclins D1 and D2 at different time points after induction with 4-OHT. As is depicted in Fig. 5B, cyclin D1 mRNA levels dropped very rapidly in response to FoxO3a activation. As early as 2 h after FoxO3a activation, we could already observe a >50% drop in the mRNA levels on Northern blots. Similar results were obtained by RT-PCR for cyclin D1 (Fig. 5C). In addition, RT-PCR analysis of cyclin D2 mRNA levels showed that cyclin D2 transcription is also inhibited, albeit with somewhat slower kinetics (Fig. 5C). No effect of 4-OHT on cyclin D1 and cyclin D2 mRNA expression in the control DLD-1 cell line was seen (Fig. 5B and C). Taken together, these data demonstrate that FoxO3a regulates cyclin D1/D2 expression at the level of transcription.
FIG. 5.
Activation of FoxO3a represses cyclin D transcription but does not affect protein stability of cyclin D1. (A) DL23 cells were either pretreated with 100 nM 4-OHT (+OHT, lower panel) or left untreated (−OHT, upper panel). After 6 h, cells were incubated with 10 μg of cycloheximide/ml (CHX) to prevent de novo protein synthesis. At the indicated time points after CHX addition, cells were lysed and the effect of 4-OHT-mediated FoxO3a activation on the stability of cyclin D1 protein was assessed by Western blotting. The inset demonstrates the effectiveness of 4-OHT-mediated FoxO3a activation in this experiment, as 4-OHT treatment alone resulted in reduced cyclin D1 protein levels compared to those of uninduced cells (lower right panel). To compare protein degradation in the presence or absence of 4-OHT, a longer exposure of the blot from the 4-OHT-stimulated cells is shown. (B and C) DL23 cells or DLD-1 cells were stimulated with 100 nM 4-OHT for the indicated times, and total RNA was isolated. The samples were then analyzed for mRNA levels of cyclin D1, cyclin D2, and actin (control) by semiquantitative RT-PCR (C) or for cyclin D1 mRNA by Northern blotting (B). As a loading control for panel B, an ethidium bromide staining of the 28S and 18S ribosomal RNAs is shown. RT-PCRs were shown to be in a linear range by performing parallel control PCRs with increasing template cDNA concentrations (not shown).
Transcriptional repression of D-type cyclins appears not to involve direct binding of FoxO factors to cyclin D1 or D2 promoters.
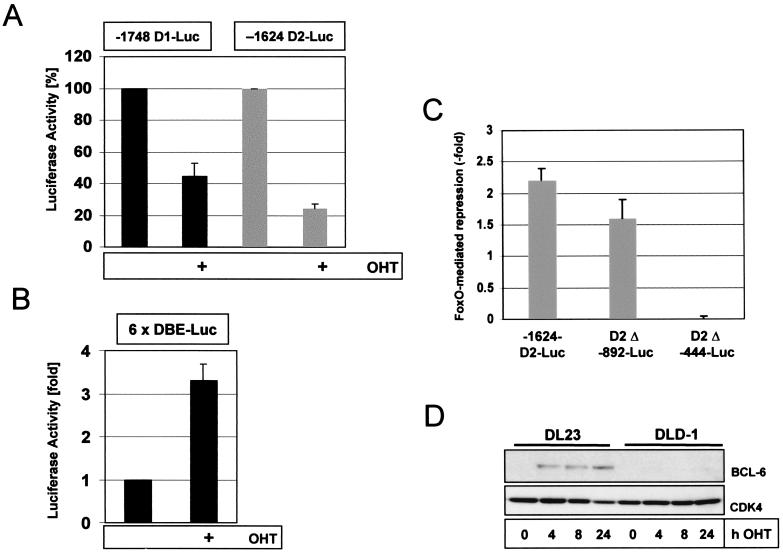
In order to study the mechanism of transcriptional repression in more detail, we performed promoter analysis to narrow down the FoxO-responsive region involved in transcriptional repression. To this end, we performed promoter-reporter gene assays utilizing luciferase reporter constructs under the control of either human cyclin D1 or cyclin D2 promoter sequences. These plasmids were transfected into the 4-OHT-responsive DL23 cell line, and the consequence of 4-OHT treatment on promoter transactivation was analyzed. As illustrated in Fig. 6A, incubation of these cells with 4-OHT resulted in a strong inhibition of promoter activities of the two full-length cyclin D constructs (−1748 D1-Luc and −1624 D2-luc, respectively). Parallel experiments using a FoxO-factor-responsive luciferase promoter construct bearing six tandem repeats of DAF-16 binding sites (6×DBE) (12) confirmed enhanced FoxO3a activity resulting from addition of 4-OHT to the medium (Fig. 6B). Similarly to DL23 cells, expression of a constitutively active FoxO3a.A3 mutant resulted in strongly decreased basal activity of the full-length Cyc D2-luc promoter construct in p27kip1−/− MEFs (data not shown), suggesting that repression of cyclin D-promoter transactivation does not depend on p27kip1 induction.
FIG. 6.
Conditional activation of FoxO3a decreases basal cyclin D1 and cyclin D2 promoter activities in human colon carcinoma cells. (A) Repression of cyclin D1 (−1748D1-luc) and cyclin D2 (−1624D2-luc) promoter activities by 4-OHT-induced FoxO3a activation (24 h). Luciferase values obtained with the untreated samples were set to 100%. (B) Confirmation of FoxO3a activation by 4-OHT using a forkhead-responsive luciferase plasmid containing six tandem repeats of DAF-16 binding elements (6×DBE-Luc) (12). (C) Effect of progressive deletion of cyclin D2-promoter sequences on FoxO3a.A3-mediated repression of cyclin D2-promoter-driven luciferase reporter activity in DLD1 colon carcinoma cells. Cells were transiently transfected with the indicated cyclin D2-luciferase reporter constructs (D2-luc) in combination with a FoxO3a.A3 expression vector or a control plasmid, and luciferase activity was assessed 48 h later. Depicted is the relative fold repression of luciferase activity by FoxO3a.A3 compared to control plasmid cotransfection. Graphs shown in panels A to C represent the average results from two independent experiments performed in duplicate, and error bars represent the standard error. (D) DL23 or DLD1 cells were stimulated with 100 nM 4-OHT and lysed at the indicated time points. Lysates were then analyzed for Bcl-6 expression by Western blotting. Levels of cyclin D2 and p27 in the lysates were examined in parallel, confirming the effectiveness of 4-OHT-mediated FoxO3a activation (Fig. 3B).
Progressive deletion of the cyclin D2 promoter fragment used to drive luciferase expression revealed that the FoxO-responsive element lies somewhere between −892 and −444 bp upstream of the transcription start site (Fig. 6C). Since this region does not contain any obvious FoxO binding sites, transcriptional regulation of cyclins D1 and D2 by FoxO apparently either takes place through FoxO interaction with other transcription factors present at these promoter elements or is more indirectly mediated through FoxO-dependent induction of a transcriptional repressor protein. One likely candidate for repression is bcl-6, a potent transcriptional repressor, recently shown to be induced by FoxO4a (46). Interestingly, bcl-6 has been described as repressing expression of cyclin D2 (41). Therefore, we analyzed expression of bcl-6 after activation of FoxO3a in DL23 cells. Expression of bcl-6 was rapidly induced (Fig. 6D), consistent with the data obtained with other cell systems (46). However, the bcl-6 binding element in the cyclin D2 promoter (−1,200 bp) lies well outside the region required for FoxO-mediated repression (Fig. 6C). Therefore, although our data cannot rule out an involvement of bcl-6 in FoxO-induced repression of cyclin D2, it seems not to be the primarily responsible factor for transcriptional downregulation. Also, the rapid kinetics of downregulation of cyclin D mRNA rather imply a more direct mode of repression.
Restoration of cyclin D/CDK4 activity overcomes a FoxO-induced cell cycle arrest.
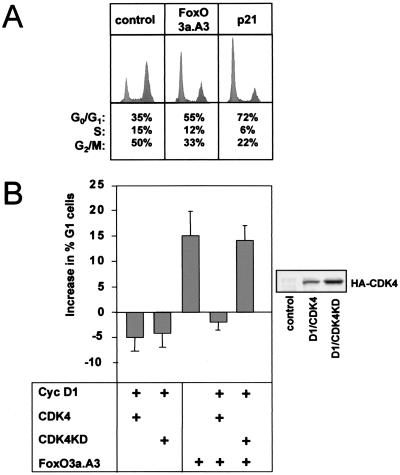
Our data presented so far strongly suggest that reduction of D-type cyclin expression and subsequent inhibition of CDK4 function importantly contribute to FoxO-induced cell cycle inhibition. We thus examined whether ectopic expression of cyclin D1 and CDK4 proteins could overcome FoxO-induced G1 arrest. For this, NIH 3T3 cells were transiently transfected with empty vector FoxO3a.A3, and the consequence of cyclin D1 cotransfection in combination with wild-type CDK4 was determined by DNA profile analysis. Consistent with previously published results (33), expression of FoxO3a.A3 resulted in a G1 arrest represented by a 15 to 20% increase in the proportion of cells in G1 (Fig. 7A). Coexpression of cyclin D1/CDK4 almost completely reversed the G1 arrest induced by FoxO3a.A3 (Fig. 7B). Since cyclin D1/CDK4 complexes can efficiently bind p27kip1 (17), this reversion might be a mere consequence of titration of p27kip1. To rule out this possibility, a kinase dead mutant of CDK4 (CDK4KD) (49) was cotransfected. Interestingly, coexpression of CDK4KD in combination with cyclin D1 was not able to restore the proliferation capacity of FoxO.A3-transfected cells (Fig. 7B), demonstrating that the cyclin D1/CDK4-mediated rescue of FoxO-induced G1 arrest is not a simple titration effect but requires CDK4 activity. Similar effects were seen when cyclin D2 was used in these experiments instead of cyclin D1 or when FoxO4a was used instead of FoxO3a (not shown).
FIG. 7.
Coexpression of functional CDK4 and cyclin D1 proteins rescues FoxO-induced G1 arrest in NIH 3T3 cells. NIH 3T3 fibroblasts were transfected with empty vector (control) or the indicated expression plasmids and cotransfected with limiting amounts of a vector encoding the surface marker CD20. Thirty-two hours later, cells were treated for 16 h with nocodazole to trap cells in G2-M and stained with propidium iodide. DNA profiles of CD20-positive cells were recorded by FACS analysis. (A) Induction of G1 arrest by FoxO3a.A3 expression. As a positive control for G1 arrest, cells were transfected with an expression plasmid for the CDK inhibitor p21Cip1/Waf1. (B) Effect of cyclin D1 (Cyc D1)/wild-type CDK4 (CDK4) or Cyc D1/kinase dead CDK4 (CDK4KD) cotransfection on FoxO3a.A3-induced G1 arrest. The bars indicate percent increases in G1 cells + standard deviation compared to the ratio of G1 cells after transfection with an empty control vector. Data are obtained from three independent experiments. Expression of exogenous HA-CDK4 and HA-CDK4KD was confirmed by Western blotting using a monoclonal anti-HA antibody (right).
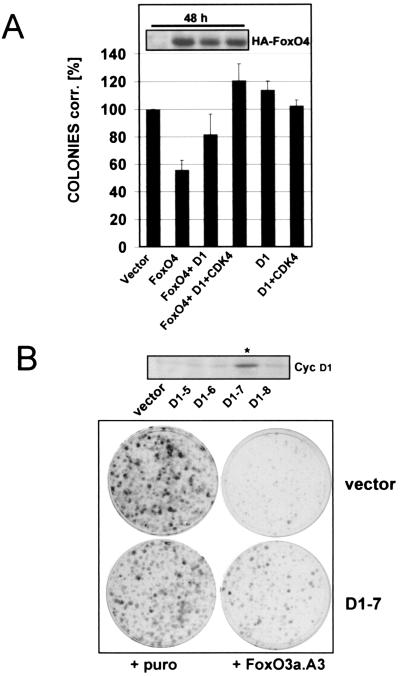
The experiments described above provide evidence that overexpression of cyclin D1/CDK4 can overcome short-term cell cycle effects of FoxO factors but do not allow a judgment as to whether restoration of CDK4 activity protects FoxO-expressing cells from cell cycle arrest in the long run. To prove the functional relevance of our findings, we therefore analyzed whether sustained expression of cyclin D1 alone or in combination with CDK4 could overcome the permanent cell cycle effects of FoxO factors. For this purpose, colony formation assays with transfected immortalized wild-type MEFs were performed as detailed in Materials and Methods. Figure 8A illustrates that in a time period of 10 to 14 days expression of FoxO4 reduced colony formation of positively transfected cells to about 50% of the vector control. Cotransfection of cyclin D1 alone could partially reverse the antiproliferative effect of FoxO4, while coexpression of CDK4 and cyclin D1 fully restored the growth capacity of FoxO4-transfected cells (Fig. 8A). Similar results were obtained when the experiments were performed with an expression vector encoding constitutively active FoxO3a.A3 (not shown). Additional colony formation experiments in which wild-type MEFs stably expressing cyclin D1 (6D1-7) were infected with a FoxO3a.A3-containing retrovirus confirmed that cyclin D1 expression could partially rescue FoxO3a.A3-induced cell cycle arrest (Fig. 8B). Compared to the parental vector cell line, FoxO3a.A3 expression inhibited colony formation less efficiently in the cyclin D1-expressing 6D1-7 clone, while infection of both the vector cell line and 6D1-7 cells with an empty retroviral construct gave rise to comparable colony numbers.
FIG. 8.
Exogenous cyclin D1 expression partially overcomes FoxO factor-induced cell cycle arrest. (A) Immortalized wild-type MEFs were transfected with pBabe.puro control vector (vector) or pBabe.puro-HA-FoxO4 (FoxO4) alone or in combination with cyclin D1 (D1) or cyclin D1/HA-CDK4 (D1/CDK4), respectively. Each combination was additionally cotransfected with limiting amounts of the fluorescent marker DsRed, and transfection rates were determined after 48 h by quantification of DsRed-positive cells by flow cytometry. Cells from each transfection were reseeded at equal dilutions into puromycin-containing selection medium, and colony formation assays were performed as detailed in Materials and Methods. Obtained colonies were counted, and numbers were corrected for transfection efficiency. Average values from two independent experiments are shown as percentages of corrected colonies + standard error. As a reference, values obtained with pBabe.puro empty vector were arbitrarily set to 100%. Western blot analysis of total lysates confirmed similar expression levels of ectopic HA-FoxO4 at the time point of reseeding (48 h after transfection [inset]). Similar results were obtained when experiments were alternatively performed with a different FoxO4 expression plasmid or when a retroviral construct expressing FoxO3a.A3 was used (data not shown). (B) Monoclonal immortalized wild-type MEFs stably expressing cyclin D1 (D1-7) or the corresponding polyclonal vector cell line (vector) were infected with empty pBabe.puro retrovirus or pBabe.puro encoding HA-FoxO3a.A3. The following day, cells from each infection were reseeded at equal numbers into selection medium and colony formation assays were performed as described in Materials and Methods. Crystal violet-stained colonies after 10 days of growth in selection medium are shown. The upper panel displays a Western blot for cyclin D1 demonstrating cyclin D1 overexpression of the selected D1-clone (indicated with an asterisk) compared to cyclin D expression of the negative clones and the utilized vector cell line.
These data demonstrate that sustained cyclin D1 expression is capable of conferring partial protection from FoxO-induced cell cycle arrest, suggesting that downregulation of D-type cyclins represents a physiologically relevant mechanism of FoxO-dependent cell cycle inhibition.
DISCUSSION
The mechanism by which winged helix/forkhead transcription factors of the FoxO subfamily induce cell cycle arrest is not fully understood.
In the past we and others have shown that forced expression of FoxO factors results in transcriptional activation of the CDK inhibitor p27kip1 (33, 36), which correlated with reduced BrdU incorporation and low cyclin E/CDK2 kinase activity, indicative of a cell cycle arrest at the G1-S transition (33). Although p27kip1−/− MEFs showed greatly reduced responsiveness to FoxO expression, we did not observe a complete protection from FoxO factor-induced S-phase inhibition (33), indicating that a p27kip1-independent pathway contributes to the FoxO-induced cell cycle arrest.
Here we show that this pathway involves downregulation of D-type cyclin expression and subsequent inhibition of CDK4-mediated phosphorylation of the S-phase inhibitory protein pRb.
Comparing short-term and long-term consequences of FoxO factor expression in p27kip1-deficient versus wild-type MEFs, we found that p27kip1 mutant cells were initially less responsive to FoxO factor-induced S-phase inhibition than were their wild-type counterparts (Fig. 1A and B) but ultimately became very efficiently arrested (Fig. 1C). From this we conclude that p27kip1 induction is not required for FoxO-induced cell cycle arrest but significantly accelerates the rate at which FoxO factors exert their antiproliferative effect. Such a notion is fully compatible with a previously postulated role of p27kip1 induction in this process (33, 36) but underscores that a second p27kip1-independent mechanism is crucially involved in FoxO factor-induced cell cycle arrest.
Our data indicate that this mechanism involves inhibition of CDK4 activity. Both in the absence and in the presence of functional p27kip1, FoxO factor expression interfered with phosphorylation of pRb on residue S780 (Fig. 2A and B and Fig. 3B), a specific consensus site for CDK4-mediated phosphorylation that is not targeted by CDK2 complexes (22). Interestingly, phosphorylation at this residue was reported to abolish interaction of pRb and E2F1 in vivo (22). Dephosphorylation of this site is thus predicted to promote interaction between pRb and E2F1 and perhaps other E2F family members. As a result, FoxO factor expression is expected to cause a shift in the distribution of the E2F protein pool from transcriptionally active, free E2F towards transcriptionally inactive pRb/E2F complexes, which ultimately would shut off S-phase-specific gene expression (16). Accordingly, the antiproliferative capacity of FoxO factors most likely depends on stabilization of pRb function. This invites the conclusion that FoxO factors should not be able to trigger cell cycle arrest in cells lacking pRb function. Experiments from our previous study challenge this notion, since FoxO4 expression was able to induce a cell cycle arrest in pRb−/− MEFs (33). However, the pRb family of pocket proteins includes two additional members, p130 and p107, which are able to repress E2F-dependent gene expression (16). Our data with triple-knockout fibroblasts, lacking all functional pocket proteins, demonstrate that p107 and p130 indeed act redundantly with pRb in a FoxO-induced cell cycle arrest. Consistent with this notion is the finding that these TKO cells do not enter quiescence upon serum starvation, while pRb-deficient fibroblasts do (6). In this respect it is important to note that the D-type cyclins are not the sole cell cycle regulatory targets of FoxO factors (33, 36), while such a mechanism would have been expected to be fully Rb dependent (23, 29, 31).
FoxO factor-dependent decrease of CDK4 activity was accompanied by downregulation of cyclin D1 and D2 protein levels (Fig. 2B and C and Fig. 3A and B). Again this occurred irrespective of p27kip1 function, demonstrating that this is not a secondary effect resulting from p27kip1 induction (Fig. 2A and B). Taking into account that CDK4 activity essentially depends on the presence of its cyclin D partners (11, 34, 42), we conclude that FoxO factor-induced repression of CDK4 activity results from downregulation of D-type cyclin levels.
This downregulation occurs via transcriptional regulation, as we were able to demonstrate that protein stability was not affected while mRNA levels as well as cyclin D1/2 promoter activities were reduced. At present, we do not know whether transcriptional repression occurs directly or indirectly via FoxO-mediated transcriptional activation of a repressor. Expression levels of bcl-6, a known transcriptional repressor for cyclin D2 (41) (but described as inducing expression of cyclin D1 [43]), were rapidly induced after 4-OHT treatment, consistent with recent data from others (46). However, the bcl-6 binding element identified in the cyclin D2 promoter (41) lies outside the region involved in FoxO-mediated transcriptional repression as determined by this study (Fig. 6C).
During the revision of our manuscript others have reported the identification of cyclin D1 and cyclin D2 as transcriptional targets of FoxO1 (40) by microarray analysis. Their data are consistent with our findings, in that transcriptional repression does not seem to involve direct binding of FoxO to promoter elements in the cyclin D promoter. However, chromatin immunoprecipitation assays suggest that binding occurs indirectly. In our reporter assays the −962CD1 promoter fragment is still efficiently suppressed (data not shown), while they have performed ChIP assays with primers spanning the region between bp −1148 and −840. This suggests that a DNA binding element present between bp −962 and −840 could be responsible for indirect recruitment of FoxO factors. Interestingly, within this 122-bp fragment lies a 12-bp regulatory element (bp −930 to −911) (19) that appears to be conserved in the cyclin D2 promoter (bp −587 to −575) (2). Importantly, this regulatory element falls within the region of the cyclin D2 promoter (bp −892 to −444) that we mapped as the FoxO-responsive region. In addition, a binding site for caudal-related homeodomain transcription factors (cdx) is present in the FoxO-responsive region of the cyclin D2 promoter, and cdx-1 has been reported to repress expression of cyclins D1 and D2 (30). The underlying mechanism for this repression has not been addressed but could very well involve a functional interaction with FoxO factors. Clearly, further mutagenesis and comparison with the FoxO-responsive elements in the cyclin D1 and D2 promoters will be required in order to elucidate the exact mechanism of transcriptional repression. Nevertheless, the present data seem to be most compatible with a model where a transcription factor bound to the cyclin D promoter recruits FoxO, which in turn represses transcription. In this respect it is of interest that a recent report showed that FoxO factors can interact with a variety of nuclear receptors and function as transcriptional corepressor (52).
In rescue experiments, we observed that overexpression of cyclin D1 could partially overcome the antiproliferative effect of FoxO-factor expression in immortalized wild-type MEFs (Fig. 8). This clearly demonstrates that downregulation of D-type cyclins indeed is a physiologically relevant mechanism by which FoxO factors induce a cell cycle arrest. Since cyclin D1 and cyclin D2 fulfill very similar functions during progression through G1 (11), it is unlikely that the remaining antiproliferative response to FoxO factors results from a unique function of cyclin D2 that cannot be complemented by cyclin D1 expression. Rather, we would propose that p27kip1 induction accounts for the residual responsiveness to FoxO factor-mediated cell cycle arrest. Since we failed to perform colony formation assays in p27kip1−/− MEFs because of increased cell death upon cyclin D1 overexpression in these cells (data not shown), we currently cannot decide whether p27kip1 induction is responsible for the residual cell cycle inhibition or if another yet-unknown mechanism is involved. However, it is unlikely that the observed rescue by cyclin D1 results from mere outtitration of p27kip1 protein, since coexpression of cyclin D1 with CDK4KD was not able to suppress the inhibitory effect of FoxO.A3 mutants on S-phase entry in NIH 3T3 cells (Fig. 7B).
FoxO factors are negatively regulated by the PI3K/PKB pathway (24). Interestingly, PKB exerts its proliferative effect both by induction of cyclin D1 and by downregulation of p27kip1 expression (27). In either case a transcriptional regulation mechanism is involved (13, 33). While it was previously shown that PKB-mediated repression of p27kip1 transcription results from inactivation of FoxO factors (33), the data presented here furthermore suggest a role of FoxO factors in the regulation of cyclin D1 transcription by PKB. Besides stimulating cyclin D1 transcription, PKB was additionally shown to increase cyclin D1 levels via enhanced translation of cyclin D1 transcripts (35) and modulation of cyclin D1 protein turnover by inhibiting GSK-3-mediated cyclin D1 phosphorylation (8). Hence, PKB can regulate cyclin D expression at multiple levels. Such a multiple-control mechanism would guarantee the best possible effectiveness and is predicted to trigger a sharp increase of D-type cyclin levels upon activation of PKB, which is essential to pass the G1 checkpoint and to permit progression to S phase.
In summary our data strongly support the notion that forced FoxO factor expression essentially antagonizes the proliferative potential of the PI3K/PKB pathway. This pathway plays an important role in tumorigenesis (47), as demonstrated by the large variety of tumor-associated mutational events that affect this pathway (50). Clear examples of this are the inactivation of the tumor suppressor PTEN, a negative regulator of PI3K signaling (5, 7), and the frequent amplification and constitutive activation of PKB (47) in a large fraction of human tumors. Our data indicate that deregulation of this pathway will not only affect the expression of p27kip1 but also modulate expression of cyclins D1 and D2 and is therefore expected to have a major impact on the control of proliferation. In this respect, our work may importantly contribute to the development of novel therapeutic strategies for human malignancies that result from constitutive activation of the PI3K/PKB pathway.
Acknowledgments
This work was supported by a grant from the Dutch Scientific Organization (NWO-90128139).
We thank F. Foijer, B. Scheyen, F. McCormick, M. Kitagawa, and P. Milner for providing useful reagents and W. R. Sellers for sharing unpublished data.
REFERENCES
- 1.Biggs, W. H., III, J. Meisenhelder, T. Hunter, W. K. Cavenee, and K. C. Arden. 1999. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. USA 96:7421-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks, A. R., D. Shiffman, C. S. Chan, E. E. Brooks, and P. G. Milner. 1996. Functional analysis of the human cyclin D2 and cyclin D3 promoters. J. Biol. Chem. 271:9090-9099. [DOI] [PubMed] [Google Scholar]
- 3.Brownawell, A. M., G. J. Kops, I. G. Macara, and B. M. Burgering. 2001. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 21:3534-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 5.Cantley, L. C., and B. G. Neel. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 96:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Cristofano, A., and P. P. Pandolfi. 2000. The multiple roles of PTEN in tumor suppression. Cell 100:387-390. [DOI] [PubMed] [Google Scholar]
- 8.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijkers, P. F., R. H. Medema, C. Pals, L. Banerji, N. S. Thomas, E. W. Lam, B. M. Burgering, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell. Biol. 20:9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman, J. B., B. Albinder, T. Shroyer, and C. Kenyon. 1995. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141:1399-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekholm, S. V., and S. I. Reed. 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12:676-684. [DOI] [PubMed] [Google Scholar]
- 12.Furuyama, T., T. Nakazawa, I. Nakano, and N. Mori. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gille, H., and J. Downward. 2040. 1999. Multiple ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem. 274:22033-22040. [DOI] [PubMed] [Google Scholar]
- 14.Guarente, L., and C. Kenyon. 2000. Genetic pathways that regulate ageing in model organisms. Nature 408:255-262. [DOI] [PubMed] [Google Scholar]
- 15.Guo, S., G. Rena, S. Cichy, X. He, P. Cohen, and T. Unterman. 1999. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 274:17184-17192. [DOI] [PubMed] [Google Scholar]
- 16.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. W., S. J. Elledge, K. Keyomarsi, B. Dynlacht, L. H. Tsai, P. Zhang, S. Dobrowolski, C. Bai, L. Connell-Crowley, E. Swindell, et al. 1995. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 6:387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hekimi, S., J. Burgess, F. Bussiere, Y. Meng, and C. Benard. 2001. Genetics of lifespan in C. elegans: molecular diversity, physiological complexity, mechanistic simplicity. Trends Genet. 17:712-718. [DOI] [PubMed] [Google Scholar]
- 19.Herber, B., M. Truss, M. Beato, and R. Muller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 9:2105-2107. [PubMed] [Google Scholar]
- 20.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 21.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 22.Kitagawa, M., H. Higashi, H. K. Jung, I. Suzuki-Takahashi, M. Ikeda, K. Tamai, J. Kato, K. Segawa, E. Yoshida, S. Nishimura, and Y. Taya. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15:7060-7069. [PMC free article] [PubMed] [Google Scholar]
- 23.Koh, J., G. H. Enders, B. D. Dynlacht, and E. Harlow. 1995. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 375:506-510. [DOI] [PubMed] [Google Scholar]
- 24.Kops, G. J., and B. M. Burgering. 2000. Forkhead transcription factors are targets of signalling by the proto-oncogene PKB (C-AKT). J Anat. 197(Part 4):571-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 26.Kops, G. J., R. H. Medema, J. Glassford, M. A. Essers, P. F. Dijkers, P. J. Coffer, E. W. Lam, and B. M. Burgering. 2002. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 22:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawlor, M. A., and D. R. Alessi. 2001. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 28.Lin, K., J. B. Dorman, A. Rodan, and C. Kenyon. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319-1322. [DOI] [PubMed] [Google Scholar]
- 29.Lukas, J., D. Parry, L. Aagaard, D. J. Mann, J. Bartkova, M. Strauss, G. Peters, and J. Bartek. 1995. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503-506. [DOI] [PubMed] [Google Scholar]
- 30.Lynch, J., E. R. Suh, D. G. Silberg, S. Rulyak, N. Blanchard, and P. G. Traber. 2000. The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by down-regulation of D-type cyclins. J. Biol. Chem. 275:4499-4506. [DOI] [PubMed] [Google Scholar]
- 31.Medema, R. H., R. E. Herrera, F. Lam, and R. A. Weinberg. 1995. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl. Acad. Sci. USA 92:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medema, R. H., R. Klompmaker, V. A. Smits, and G. Rijksen. 1998. p21waf1 can block cells at two points in the cell cycle, but does not interfere with processive DNA-replication or stress-activated kinases. Oncogene 16:431-441. [DOI] [PubMed] [Google Scholar]
- 33.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 34.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 35.Muise-Helmericks, R. C., H. L. Grimes, A. Bellacosa, S. E. Malstrom, P. N. Tsichlis, and N. Rosen. 1998. Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J. Biol. Chem. 273:29864-29872. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994-999. [DOI] [PubMed] [Google Scholar]
- 38.Paradis, S., and G. Ruvkun. 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12:2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peeper, D. S., T. M. Upton, M. H. Ladha, E. Neuman, J. Zalvide, R. Bernards, J. A. DeCaprio, and M. E. Ewen. 1997. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386:177-181. [DOI] [PubMed] [Google Scholar]
- 40.Ramaswamy, S., N. Nakamura, I. Sansal, L. Bergeron, and W. R. Sellers. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2:81-91. [DOI] [PubMed] [Google Scholar]
- 41.Shaffer, A. L., X. Yu, Y. He, J. Boldrick, E. P. Chan, and L. M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199-212. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 43.Shvarts, A., T. R. Brummelkamp, F. Scheeren, E. Koh, G. Q. Daley, H. Spits, and R. Bernards. 2002. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 16:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takaishi, H., H. Konishi, H. Matsuzaki, Y. Ono, Y. Shirai, N. Saito, T. Kitamura, W. Ogawa, M. Kasuga, U. Kikkawa, and Y. Nishizuka. 1999. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc. Natl. Acad. Sci. USA 96:11836-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, E. D., G. Nunez, F. G. Barr, and K. L. Guan. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274:16741-16746. [DOI] [PubMed] [Google Scholar]
- 46.Tang, T. T., D. Dowbenko, A. Jackson, L. Toney, D. A. Lewin, A. L. Dent, and L. A. Lasky. 2002. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J. Biol. Chem. 277:14255-14265. [DOI] [PubMed] [Google Scholar]
- 47.Testa, J. R., and A. Bellacosa. 2001. AKT plays a central role in tumorigenesis. Proc. Natl. Acad. Sci. USA 98:10983-10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 49.van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050-2054. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez, F., and W. R. Sellers. 2000. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim. Biophys. Acta 1470:M21-M35. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 82:323-330. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, H. H., R. E. Herrera, E. Coronado-Heinsohn, M. C. Yang, J. H. Ludes-Meyers, K. J. Seybold-Tilson, Z. Nawaz, D. Yee, F. G. Barr, S. G. Diab, P. H. Brown, S. A. Fuqua, and C. K. Osborne. 2001. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 276:27907-27912. [DOI] [PubMed] [Google Scholar]